Summary
During mammalian embryogenesis, de novo hematopoiesis occurs transiently in multiple anatomical sites including yolk sac, dorsal aorta, and heart tube. A long unanswered question is whether these local transient hematopoietic mechanisms are essential for embryonic growth. Here, we show that endocardial hematopoiesis is critical for cardiac valve remodeling as a source of tissue macrophages. Colony formation assay from explanted heart tubes and genetic lineage tracing with the endocardial specific Nfatc1-Cre mouse revealed that hemogenic endocardium is a de novo source of tissue macrophages in the endocardial cushion, the primordium of the cardiac valves. Surface marker characterization, gene expression profiling, and ex vivo phagocytosis assay revealed that the endocardially-derived cardiac tissue macrophages play a phagocytic/antigen presenting role. Indeed, genetic ablation of endocardially-derived macrophages caused severe valve malformation. Together, these data suggest that transient hemogenic activity in the endocardium is indispensable for the valvular tissue remodeling in the heart.
eTOC Blurb
Shigeta et al. found that a subset of macrophages in the heart valves originates locally from the fetal endocardium. This local type of macrophages is more phagocytic than other macrophages and critical for the valve remodeling during cardiogenesis.
Graphical Abstract

Introduction
During mammalian embryogenesis, hematopoiesis occurs transiently in multiple anatomical sites (Dzierzak and Bigas, 2018; Orkin and Zon, 2008; Palis, 2016). A long unanswered question is whether each of these transient hematopoiesis is essential for the embryonic growth. Hemogenic endocardial cells arise in the developing heart tube and contribute to transient hematopoiesis including primitive erythroid and transient definitive progenitors (Nakano et al., 2013; Yzaguirre and Speck, 2016; Zamir et al., 2017). This endocardial hematopoiesis is dependent on a homeobox transcription factor, Nkx2–5 (also known as tinman in Drosophila), suggesting that they are the mammalian homologue of tinman-dependent hematopoiesis in Drosophila (Han and Olson, 2005; Mandal et al., 2004). However, while tinman+ hematopoiesis predominantly generates phagocytes and plays a dominant role in Drosophila hematopoiesis, genetic lineage tracing in mouse suggests that Nkx2–5-derived hematopoietic cells are found only in 4% and 12% of circulating erythroid and myeloid, respectively, in a narrow time-window (Nakano et al., 2013). Given its relatively minor contribution to the circulating blood, it is unclear whether hemogenic endocardium is indispensable for the embryonic growth in mammals.
Endocardial cells are heterogeneous in their origin and function (Baldwin, 1996; Harris and Black, 2010; Milgrom-Hoffman et al., 2011; Moretti et al., 2006; Nakano et al., 2013; Stanley et al., 2002). The hemogenic activity of endocardial cells is found mainly in the cushion endocardium at E9.5–11.5 in mouse (Nakano et al., 2013; Yzaguirre and Speck, 2016). Interestingly, this spatiotemporally coincides with the initiation of valve formation. At this stage, the cushion endocardial cells start the epithelial-mesenchymal transition (EMT) to generate mesenchymal cells. The accumulation of mesenchymal cells in the cushion results in thick and cellularized primitive valves, which subsequently undergo remodeling throughout embryonic and neonatal stages and eventually form thin acellular mature valves. Apoptotic cells are identified in the valve primordia during mid-late gestational stages (Aikawa et al., 2006; Icardo, 1990; Pexieder, 1975; Wang et al., 2017), although it is not well understood how these apoptotic cells are eliminated. The spatiotemporal coincidence of the initiation of valve formation and hemogenic activity of the endocardium led us to hypothesize that the hemogenic endocardium is a source for the phagocytes required for the valve remodeling.
In this report, we demonstrate that endocardium is a de novo source of a subset of cardiac tissue macrophages during embryogenesis. Analyses of surface markers, genome-wide expression profiling and phagocytosis assay suggest that endocardially-derived cardiac tissue macrophages (EcTMs) are more phagocytic than macrophages of non-endocardial origin. Genetic ablation of EcTMs by endocardial specific deletion of Csf1r/CD115/MCSF-R resulted in severe valve anomaly with excess mesenchymal cells in both aortic and mitral valves, suggesting that EcTMs are indispensable for the valvular remodeling. This result suggests that local transient hematopoiesis is critical for the local tissue remodeling.
Result
The heart tube is a de novo source of macrophages
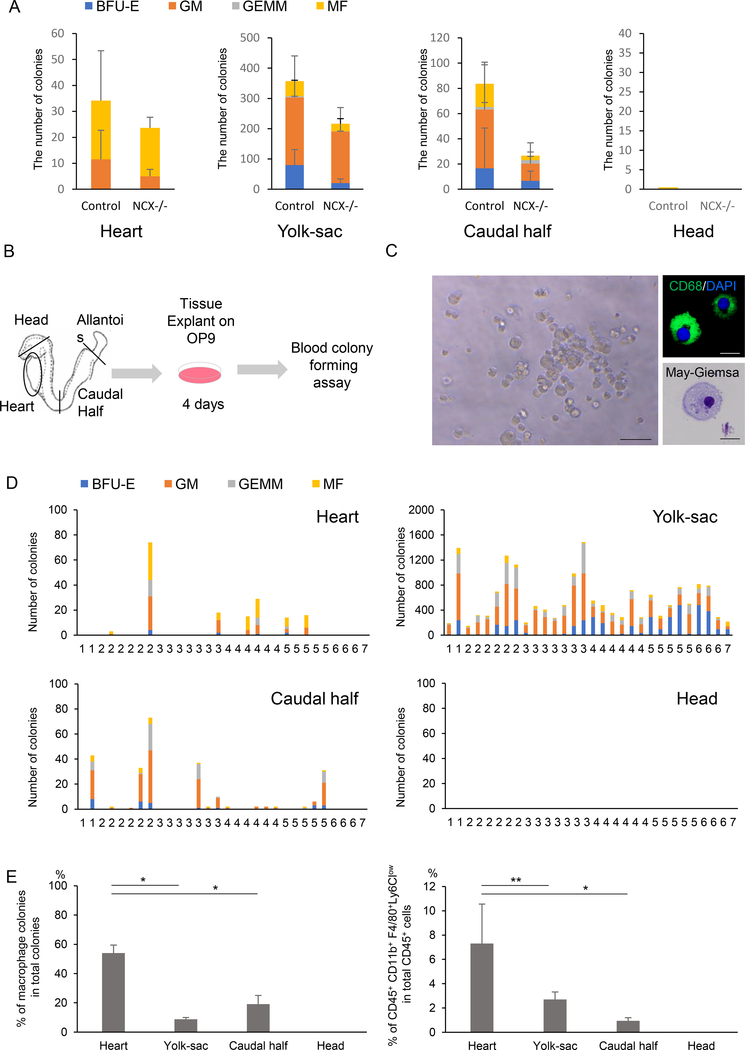
Defining the origin of hematopoietic cells is complicated by their mobile nature. Once effective heartbeat is initiated at around 8 somite stages (~E8.5), blood cells may migrate to the heart. To examine whether the heart tube generates macrophages in situ, two ex vivo colony formation assays were performed. First, the heart explants from Ncx1-mutant embryos were examined for their colony-forming activity. Ncx1 is a sodium-calcium exchanger specifically expressed in the heart. Ncx1-mutant embryos die at around E10.5 due to the lack of systemic circulation (Koushik et al., 2001), which makes them a useful model for examining local hematopoietic emergence. Consistent with a previous report (Nakano et al., 2013), hematopoietic colonies were grown from Ncx1-mutant hearts (Fig. 1A). Interestingly, significant number of macrophage colonies was generated even in the absence of circulation (Fig. 1A). Second, the colony-forming activity of the heart tube was examined using the heart explants at pre-circulation stages (1–7 somite stages or ~E8.0), when the heart tube is not contaminated by hematopoietic cells from circulation (Fig. 1B). The heart tubes were pre-cultured on an OP9 feeder layer, which supports erythro-myeloid differentiation, followed by methylcellulose culture supplemented with cytokines/growth factors (Kodama et al., 1994). Consistent with the results from Ncx1-mutants (Fig. 1A), macrophage colonies with typical morphology were observed from the heart tubes and other established hemogenic tissues (Fig. 1C, D). They were manually picked up, prepared by Cytospin and validated by CD68 and May-Giemsa stainings (Fig. 1C). Interestingly, the heart explant generated more macrophage colonies (Fig. 1E, left) and macrophages (Fig. 1E, right) than other tissues. The results from these two complementary studies suggest that the heart is a de novo source of macrophages.
Figure 1. Endocardium is a de novo source of cardiac tissue macrophages.
A. Number of hematopoietic colonies from controls and Ncx1-mutant embryos that lack heartbeat, showing the hematopoietic activity in the heart tube in the absence of circulation. Data represent mean ± SE. n=7 for each tissue.
B. Schematic representation of the colony-forming assay from ex vivo organ explant at pre-circulation stages. The yolk sac, heart tube, caudal half (including future AGM region) and head were dissected at somite stages 1–7, when the effective circulation is not established yet.
C. Representative macrophage colony grown from the embryonic heart explanted at E8.0, pre-cultured on OP9, and propagated in methylcellulose medium supplemented with cytokines. Right panels are representative CD68 immunostaining and May-Giemsa staining. Scale bar = 20 μm
D. Hematopoietic colonies retrieved from various tissues at various somite stages. Each column represents colonies from one tissue.
E. Quantification of the macrophage colony counts (left) and the percentage of CD45+ CD11b+ F4/80+ Ly6Clowmacrophages per CD45 positive cells by flow cytometry (right) grown from E8.0 tissue explants. Data represent mean ± SE. n=7. *P < 0.0001 and **P < 0.0005 by unpaired t-test.
BFU-E, burst-forming unit-erythroid; GM, granulocyte-macrophage; GEMM, granulocyte-erythroid-macrophage-megakaryocyte; MF, macrophage.
Endocardial cells contribute to the macrophages in vivo
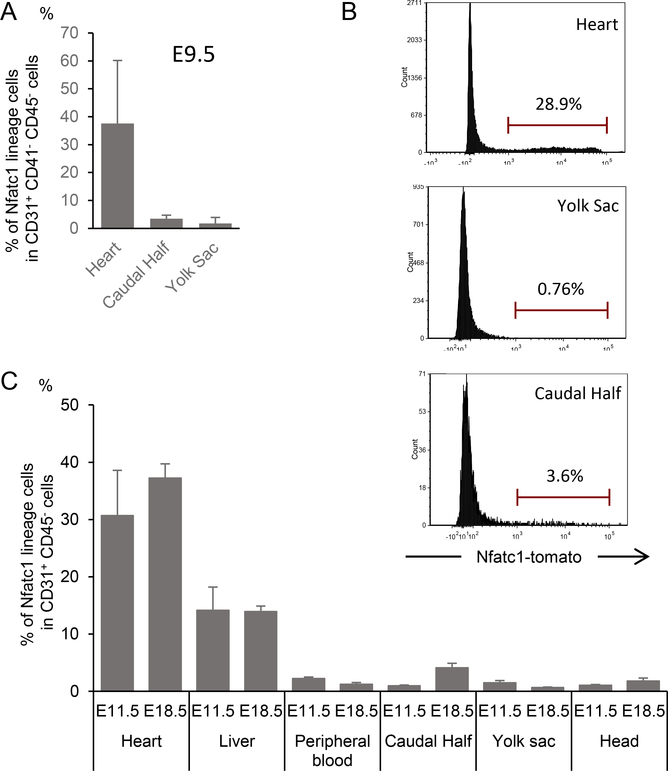
Next, the in vivo fate of endocardial cells was traced using Nfatc1 as a marker. Nfatc1 is expressed specifically in the endocardium from E9.5 to E13.5 and Nfatc1-Cre knockin line labels the endocardium in highly specific and sensitive manner (Wu et al., 2012). To further confirm the specificity, the Nfatc1lineage contribution was quantified by flow cytometry using Nfatc1Cre/+;R26Tomato reporter/+ embryos at E9.5. As CD31+ CD41+ hematopoietic progenitors are found in circulation at this stages, pure endocardial cells were gated as CD31+ CD41− CD45− in this experiment. As shown in Fig. 2A, B, Nfatc1-derived Tomato+ cells were found in the purified endocardial/endothelial cells in the heart but only marginally in two other established de novo site for hematopoiesis, the yolk sac and caudal half (containing aorta-gonad-mesonephros (AGM) region) at E9.5and E18.5 (Fig. 2A, B). Consistent with previous observation, The progenies of Nfatc1-positive endocardial cells were found in the endothelium of the fetal liver, which is not a de novo hematopoietic source (Fig. 2C) (Zhang et al., 2016). Therefore, Nfatc1-Cre knockin line is useful to trace the hematopoietic fate of the endocardial cells.
Figure 2. Nfatc1cre/+ line specifically labels endocardial cells.
A The percentage of Nfatc1-derived cells within total endothelial cells in embryonic tissues during embryogenesis measured by flow cytometry for Tomato, CD31, CD41 and CD45 in Nfatc1cre/+; Rosa26Tomato reporter/+ embryos. Tomato-positive cells are identified in the CD31+ CD41− CD45− pure endocardial cells in heart tissue at E9.5. CD41+ fraction was excluded as CD31 is expressed in a subset of CD41+ circulating progenitors at this stage. Data represent mean ± SE.
B. Representative histogram of Nfatc1-lineage-labeled cells within CD31+ CD45− endothelial (endocardial) cells in the heart, yolk-sac, and caudal half from Nfatc1cre/+; Rosa25Tomato reporter/+ at E18.5.
C. The percentage of Nfatc1-derived cells within total endothelial cells in embryonic tissues during embryogenesis measured by flow cytometry for Tomato, CD31, and CD45 in Nfatc1cre/+; Rosa26Tomato reporter/+ embryos. Tomato-positive cells are identified in the CD31+ CD45− pure endocardial cells in heart tissue. Liver endocardial cells, known to be partially derived from endocardial cells, are also Tomato-positive. Data represent mean ± SE.
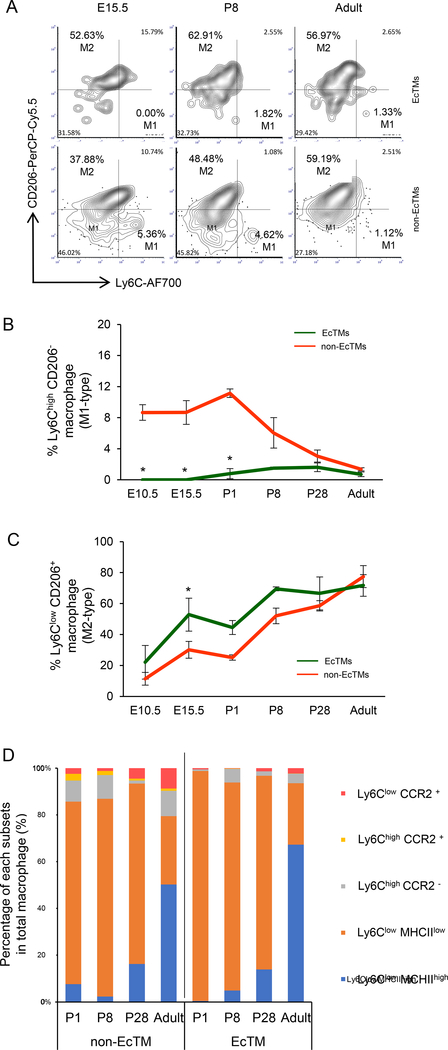
Immunostaining of Nfatc1Cre/+; R26YFP reporter/+ hearts revealed that YFP-labeled CD45+ cells were enriched in the endocardial cushion. These cells showed mesenchymal morphology (Fig. 3A, B) and coexpressed pan-macrophage markers (CD68, F4/80, Csf1r) and an M2 macrophage marker (CD206), suggesting their macrophage identity (Fig. 3C-I, Fig. S1A). These Nfatc1-lineage-labeled macrophages were identified from E10.5 and often found immediately under endocardial layer in the heart tube (Fig. S1A). CD68, Csf1r, and CD206 were expressed from E10.5 and F4/80 (a marker for mature macrophages) was found after E13.5 (Fig. S1B-E). Semi-quantitative cell counting suggests that, at E13.5, 58% of F4/80+ macrophages in the cushion were YFP-positive (Fig. S1F). At the postnatal stages, YFP+ macrophages were enriched in the aortic and mitral valves and atrial myocardia (Fig. S1G-J). Although the hemogenic activity of the endocardium is transient (Nakano et al., 2013), lineage-labeled macrophages were identified in the postnatal valve mesenchyme derived from cushion mesenchyme (Fig. 3J-Y). Flow cytometry analyses revealed that endocardially-derived cardiac tissue macrophages (defined by CD45+ CD11b+ F4/80+ Nfatc1-lineage+ in the Nfatc1Cre/+; R26Tomato reporter/+ heart; hereafter EcTMs) contributed to 2.6–17.4% of CD45+ CD11b+ F4/80+ macrophages in the whole heart during embryonic stages (Fig. 4A, bar graph). While the number of EcTMs did not change significantly during postnatal stages, non-Nfatc1-derived macrophages (Nfatc1-lineage− fraction of CD45+ CD11b+ F4/80+ cells; hereafter non-EcTMs) decreased after postnatal day 8, resulting in a gradual increase in the proportion of EcTMs to 37.9% by adult stages (Fig. 4A, black line).
Figure 3. Endocardial cells contribute to cardiac tissue macrophages.
A, B. Immunofluorescent staining for CD45 (red), cTnT (white) and YFP (green) of Nfatc1cre/+; Rosa26YFP reporter/+ cushion mesenchyme of AV canal (A) and outflow tract (B) at E13.5. YFP-labeled CD45+ myeloid cells with mesenchymal morphology are enriched at the border of cushion mesenchyme and cTnT+ myocardium. Scale bar = 10 μm
C-Y. Immunofluorescent staining for macrophage markers (CD68, F4/80 and CD206; red) of the cushion and valvular mesenchyme of Nfatc1cre/+; Rosa26YFP reporter/+ mouse at E10.5 (C-E), E15.5 (F-I), P1 (J-M), P8 (N-Q), P28 (R-U) and adult (V-Y) stages. Nfatc1-derived macrophages are found throughout embryonic and postnatal stages. Scale bar = 10 μm. See also FigureS1
Figure 4. Spatiotemporal distribution of EcTMs.
A. The number of EcTMs and non-EcTMs per 100,000 live cells and their percentages at each developmental stage. Data represent mean ± SE. n=3, each.
B. Percent contribution of Nfatc1-derived cells to macrophages in the heart, liver, yolk sac (YS), caudal half (CH), head and circulating blood of Nfatc1cre/+; Rosa26Tomato/+ embryos. Nfatc1-derived cells were identified at E9.5 in the heart but not in other tissues. n = 2–4 for each data point.
C-F. Percent contribution of Nfatc1-derived cells to (C) monocytes (CD45+ CD11bhigh F4/80low Ly6Chigh Csf1r+), (D) granulocytes (CD45+ CD11bhigh F4/80− Ly6G+ Csf1r−), (E) erythro-myeloid progenitors (EMPs; CD45+ CD41+ c-kit+), and (F) hematopoietic stem cells (HSCs; Lin− c-kit+ Sca1+ CD150+ CD48−) in the heart, liver, yolk sac (YS), caudal half (AGM), head and circulating blood of Nfatc1cre/+; Rosa26Tomato/+ embryos. n = 2 and 4 for E15.5 and E18.5, respectively. See also FigureS2A
By flow cytometry, Nfatc1-derived macrophages (EcTMs) were identified at E9.5 in the heart and at E10.5 in circulation (Fig. 4B). However, in any organs examined, Nfatc1-derived cells were hardly identified in monocytes (CD45+ CD11bhigh F4/80low Ly6Chigh Csf1r+), granulocytes (CD45+ CD11bhigh F4/80− Ly6G+ Csf1r−), erythro-myeloid progenitors (EMPs; CD45+ CD41+ c-kit+), or hematopoietic stem cells (HSCs; Lineage− c-kit+ Sca1+ CD150+ CD48−) (Fig. 4C-F, S2A), suggesting that endocardially-derived macrophages are not generated through monocytes or multipotent stem/progenitors. Thus, (1) macrophage colonies arise from the heart tube ex vivo even in the absence of circulation (Fig. 1), (2) a fraction of cardiac tissue macrophages are labeled by Nfatc1-Cre knockin line that is highly specific and sensitive to the endocardium (Fig. 2), and (3) Nfatc1-labeled macrophages appear in the heart tube prior to other organs (Fig. 4B). These data support that Nfatc1-labeled macrophages originate from the endocardium.
To further examine whether EcTMs are derived from multipotent stem/progenitors, Runx1 was conditionally deleted in endocardial specific manner. Runx1 is a key regulator of definitive hematopoiesis. Runx1 mutant embryos die at around E12.5 due to the absence of entire definitive hematopoietic cells (North et al., 1999; North et al., 2002). Nfatc1cre/+; Runx1 fl/fl mutants survived until adult stages with no detectable phenotype. Analysis of Nfatc1cre/+; Runx1 fl/fl mutant hearts revealed no significant decrease in the number of macrophages in the mutant hearts compared to the control littermates at E10.5 and 11.5 (Fig. S2B). Thus, EcTMs are not dependent on Runx1, suggesting that they are not definitive in nature.
Endocardially-derived macrophages are more phagocytic than macrophages of other origins in the heart
Macrophages are activated in response to the local tissue environment and differentiate into specific subsets including pro-inflammatory M1 macrophages (Ly6Chigh CD206−) and alternatively-activated M2 macrophages (Ly6Clow CD206+) that play phagocytic and pro-tissue reparatory roles (Pinto et al., 2012). Surface marker characterization revealed that M1 macrophages were not present in EcTMs during embryonic stages whereas 8.6–11.2% of non-EcTMs were M1 (Fig. 5A, B, P < 0.05). On the other hand, the percentage of M2 macrophages was significantly higher in EcTMs compared to non-EcTMs at E15.5 (Fig. 5C). These data suggest that EcTMs are more M2 type during embryonic stages. In both EcTMs and non-EcTMs, the percentage of M2 macrophages progressively increased during embryonic and postnatal development (Fig. 5C). The surface characteristics of EcTMs and non-EcTMs gradually became less distinct towards adult stages (Fig. 5B, C, Fig. S2C). Expression of MHCII (a marker for antigen-presenting macrophages) and CCR2 (a marker for monocyte-derived macrophages) was examined to further characterize the postnatal maturation of EcTMs. In both EcTMs and non-EcTMs, phagocytic-type macrophages (Ly6Clow MHCIIlow)(Epelman et al., 2014) were dominant from P1 to 28, followed by adoption of an antigen presenting role (Ly6Clow MHCIIhigh) at adult stages (Fig. 5D). However, EcTMs and non-EcTMs were clearly distinct in that CCR2+ macrophages (monocyte-derived) were rarely found among EcTMs. Although possibly oversimplified, surface marker categorization supports that EcTMs play more phagocytic role than macrophages of other origins during heart development.
Figure 5. Surface marker characteristics of EcTMs.
A. Representative flow cytometric profiles of Ly6c and CD206 expression in EcTMs (top) and nonEcTMs (bottom).
B. The percentage of M1 macrophages (Ly6Chigh CD206−) in EcTMs and non-EcTMs. M1-type cells are absent in EcTMs during embryonic stages. On the other hand, M1-type subset is present in non-EcTMs during embryonic stages but gradually decreases after birth. See also FigureS2C
C. The percentage of M2 macrophages (Ly6Clow CD206+) in EcTMs and non-EcTMs. The percentage of M2 macrophages progressively increases in both EcTMs and non-EcTMs. EcTMs show a higher percentage of M2-type cells during embryonic stages, but non-EcTMs gradually catch up after birth. Overall, the surface marker characteristics are significantly different during embryonic stages but become less distinct at adult stages. Data represent mean ± SE. n=3, each. *P < 0.05 by paired t-test. See also FigureS2C
D. Characterization of tissue macrophage subsets after birth. Ly6Clow MHCIIlow with phagocytosis potential is the majority in both EcTMs and non-EcTMs up to P28 when valvular remodeling completes. MHCIIhigh antigen-presenting macrophages become dominant at adult stages. Only few percentages of EcTMs are CCR2+, suggesting that they are not derived from circulating monocytes. n=3, each.
To further examine the role of EcTMs, we performed RNA-Seq on these cells at postnatal day 28, when the total number of cardiac tissue macrophages is at peak. 40 genes were differentially expressed with a Padj < 0.05 between the two groups: 36 were higher and 4 genes were lower in EcTMs compared to non-EcTMs (Fig. 6A). Gene Ontology analysis revealed that EcTMs were enriched for genes related to antigen presentation, lysosomes, and phagosomes (H2-Ab1, H2-Aa, H2-Eb1, CD74, Laptm5; Fig. 6B, C). To examine their phagocytic function, EcTMs and non-EcTMs were sorted from Nfatc1Cre/+; Rosa26Tomato reporter/+ hearts at P8 and incubated with FITC-labeled latex beads. Consistent with the gene expression data, EcTMs showed 44.4% higher phagocytic activity than non-EcTMs (Fig. 6D, P < 0.05). Moreover, electron microscopy analysis revealed large phagocytic cells with phagosomes engulfing apoptotic cell debris in the cushion mesenchyme at E13.5 (de Lange et al., 2004), although filopodia were less developed compared to typical tissue macrophages in adult (Fig. 6E-J). Together, these data suggest that EcTMs predominantly play a phagocytic role in the endocardial cushion and valvular mesenchyme.
Figure 6. Phagocytic role of EcTMs.
A. Volcano plots of RNA-seq from EcTMs and non-EcTMs. 36 and 4 genes are expressed at significantly higher and lower level in EcTMs, respectively. n=4, each
B. Clustered heatmap of the genes enriched in EcTMs. n=4, each
C. Gene ontology analysis of the genes enriched in EcTMs.
D. Macrophage phagocytosis assay. EcTMs and non-EcTMs are sorted from P8 hearts and incubated with FITC-labeled latex beads. The fraction of phagocytic macrophages were higher in EcTMs. Data represent mean ± SE. n=5. *P < 0.05 by paired t-test.
E-J. Hematoxylin and Eosin (H-E) staining (E, H) and electron microscopy images (F, G, I, J) of the endocardial cushion at E13.5. Magnification is 4800x (F), 7200x (G), 3600x (I), and 7200x (J) magnification. Note the large cells (white arrows) with phagosomes containing apoptotic cells (black arrows) and particles (black arrowhead). Scale bar = 2.3 um
Endocardially-derived macrophages are required for the valve formation
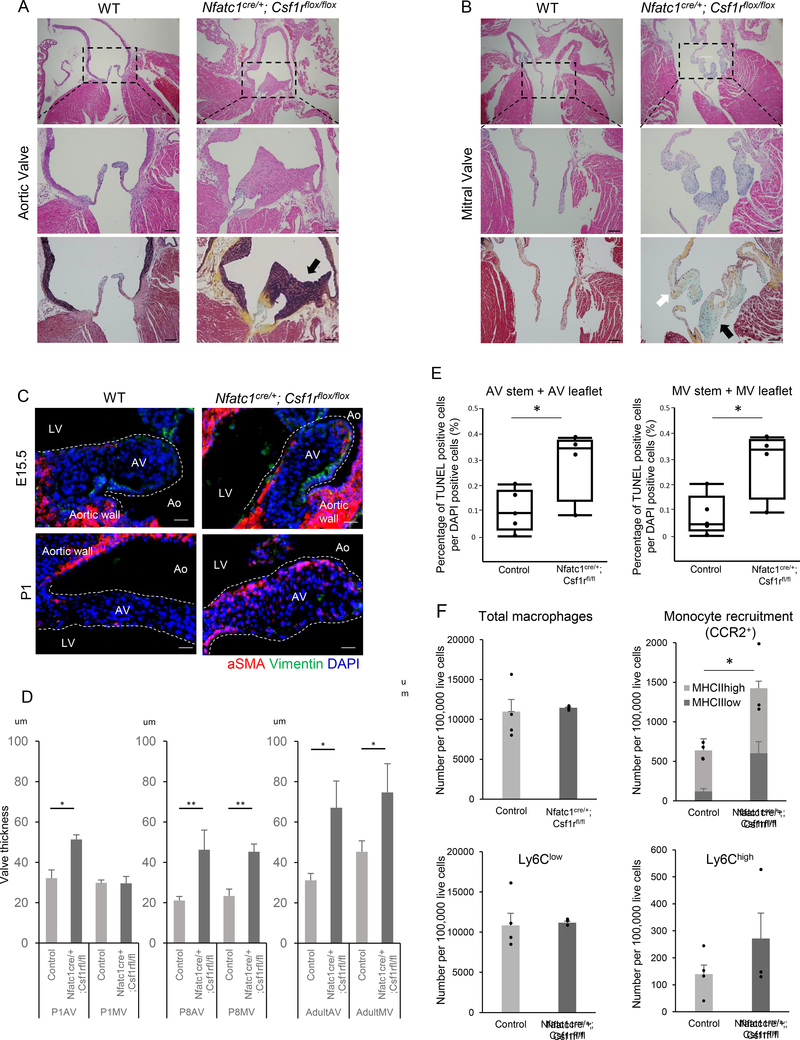
Previous reports suggest that tissue macrophages of different origins compensate for each other in diseased tissues (Bain et al., 2016; Epelman et al., 2014). To examine whether EcTMs are dispensable for the valve formation, we generated mutant mice in which EcTMs were ablated by deleting Csf1r (also known as MCSF receptor and CD115) in the Nfatc1+ endocardial lineage. Csf1 and Csf1r constitute an autocrine system essential for macrophage development, and Csf1r is specifically expressed in the macrophage lineage. Germ-line knockout mice of Csf1 and Csf1r show embryonic lethality with nearly identical phenotypes due to complete ablation of the macrophages (Chitu and Stanley, 2006; Ginhoux et al., 2010; Sauter et al., 2014). Nfatc1cre/+; Csf1rfl/fl mutant mice were underrepresented at weaning (Fig. S3A, P < 0.05,) and had enlarged hearts (Fig. S3B, C). Histological analysis of the survivors revealed a spectrum of valvular anomalies including ectopic elastic tissue with hypercellularity in aortic valves and typical myxomatous changes with the accumulation of collagen and mucin in mitral valve leaflets (Fig. 7A, B). Less severe phenotypes include thickening of the valves with an increase in both cellular and matrix components although the matrix pattern was not significantly affected until adulthood (Fig. S4A, B). Typical bicuspid aortic valve, calcification or right heart valve deformity was not identified in our analyses (Fig. S4C, D). The valvular thinckening and anomalies were obvious at neonatal stages by histological analyses and 3D imaging by light-sheet microscopy (Fig. 7D, S5A). However, expression of αSMA and Vimentin, markers for activated valve interstitial cells, were higher in the mutants at E15.5 and P1, indicating that a pathological activation of interstitial cells starts prior to the morphological deformity (Fig. 7C, D, Fig. S5B). TUNEL staining revealed that apoptotic cells were not cleared efficiently in valvular mesenchyme in Nfatc1cre/+; Csf1rfl/fl hearts at P1, suggesting an inefficient clearance of apoptotic cells (Fig. 7E). Cell proliferation assay revealed no evidence of increase in pH3+ cells in mutant aortic or mitral valves in surviving P8 mutants (Fig. S5C). Inefficient clearance of apoptotic cells likely contributed to the activation of mesenchymal cells in the valves in our mutants. No sign of cardiac contractile dysfunction was noticed in surviving adult mutants by μMRI or echo (Fig. S6A, B). Interestingly, significant prolongation of PR interval was observed on EKG (Fig. S6C). These results suggest that EcTMs are required for the formation of the valves through their phagocytic role. Notably, no significant morphological abnormality was identified in any other organs (Fig. S3E, F), indicating that these locally-derived tissue macrophages are specifically required for local tissue remodeling.
Figure 7. Valve abnormality in EcTMs-ablation mice.
A, B. H-E staining [top (4x), middle (10x)] and Movat’s pentachrome staining [bottom (10x)] of aortic (A) and mitral (B) valves from the control and Nfatc1cre/+; Csf1rfl/fl adult mice. A. Aortic valve of Nfatc1cre/+; Csf1rfl/fl is thick and hypercellular with ectopic elastic tissue (black staining, black arrow). B. Mitral valve of Nfatc1cre/+; Csf1rfl/fl is thick and hypercellular with collagen (yellow, white arrow)/mucin (blue, black arrow) deposition, suggesting a myxomatous degeneration. Scale bar = 100 μm
C. Immunofluorescent staining of aortic valves for αSMA (red) and Vimentin (green) at E15.5 and P1. Note the higher αSMA expression in valve interstitial cells of Nfatc1cre/+; Csf1rfl/fl valves. See also FigureS5B
D. Quantification of the thickness of aortic (AV) and mitral (MV) valves at P1, P8, and adult mutants and controls. Data represent mean ± SE. n = 2–4 at each data point. *P < 0.05 and **P < 0.001 by unpaired t-test.
E. Quantification of TUNEL-positive cells in the aortic valve (AV) stem and leaflet (left) and mitral valve (MV) stem and leaflet (right). Data represent mean ± SE. n=5 for control and 4 for mutants. *P < 0.05 by unpaired t-test.
F. Characterization of the macrophages in wild-type control and Nfatc1cre/+; Csf1rfl/fl adult hearts. Monocytes derived macrophages (CCR2+) are significantly increased in Nfatc1cre/+; Csf1rfl/fl, while the total number of macrophages are not significantly different. Data represent mean ± SE. n=4 for control and 3 for mutant. *P < 0.05 by unpaired t-test.
Monocyte-derived macrophages compensate the total number of macrophages in the absence of endocardially-derived macrophages
To examine how the ablation of EcTMs impacts the subpopulations of macrophages in the heart, we profiled surface marker expression in Nfatc1cre/+; Csf1rfl/fl mutant hearts. While the numbers of total, Ly6Chigh and Ly6Clow macrophages were not significantly different, monocyte-derived CCR2+ macrophages were significantly increased in Nfatc1cre/+; Csf1rfl/fl hearts (638±43 vs 1424±218 per 100,000 live cells, P < 0.05, Fig. 7F). The increase in CCR2+ macrophages was mainly due to the MHCIIlow macrophages. Thus, the total number of macrophages was compensated by monocyte recruitment in the absence of EcTMs; however, the monocyte-derived macrophages failed to rescue the remodeling of valves as the phagocytic function was not fully restored. These data suggest that EcTMs play a unique role during valvular formation.
Discussion
In this report, we found that endocardial cells give rise to a subpopulation of tissue macrophages essential for the remodeling of valves (Fig. S7). Since the discovery of primitive erythroid cells in the yolk sac a century ago, transient hematopoietic activities have been identified in multiple anatomical sites at multiple developmental stages (Dieterlen-Lievre, 2007; Dzierzak and Speck, 2008; Orkin and Zon, 2008; Yoshimoto et al., 2008). As these local sites of transient hematopoiesis make only a minor contribution to circulating blood, it has long been unknown whether the local transient hematopoietic activities are all critical for embryogenesis. Our results suggest that the biological importance of local transient hemogenic activity in the endocardium lies in facilitating the valve remodeling as a source for phagocytes. In other words, the local transient hematopoiesis is critical for the local tissue remodeling. This concept might be generalized to other transient hematopoietic sites: for example, the aorta-gonad-mesonephros (AGM) region displays the strongest hemogenic activity at the site where the two dorsal aortae fuse. Aortic sac and pharyngeal arch arteries are also enriched for Nkx2–5+/CD41+ endothelial cells at E9.5–10.5 (Nakano et al., 2013; Paffett-Lugassy et al., 2013). This symmetric structure undergoes drastic remodeling within the next few days to form a single asymmetric aorta. Thus, local transient hematopoiesis in the heart tube, dorsal aortae (AGM), and aortic sac/pharyngeal arch arteries all spatiotemporally coincides with the sites of active tissue remodeling.
Identification of the de novo source of embryonic blood cells is complicated by their mobile nature. In fact, Nfatc1 lineage-labeled macrophages are found in circulation (Fig. 4B), complicating the identification of their origin. It is worth mentioning that our data do not deny the possibility of the existence of non-Nfatc1-labeled macrophages of endocardial origin or Nfatc1-labeled macrophages of non-endocardial origin. However, several pieces of evidence support that macrophages do arise from the endocardium: First, robust macrophage colony forming activity was observed from the heart explants at pre-circulation stages that are not yet contaminated by circulating blood cells (Fig. 1). Second, our Nfatc1-Cre knockin line labels the endocardium in a highly specific and sensitive manner (Fig. 2). Third, Nfatc1-labeled macrophages appear in the heart tube prior to other organs (Fig. 4B). Although Nfatc1-labeled cells are found in the liver, the expression of Nfatc1 mRNA is reportedly downregulated after endocardial cells migrate to the liver (Zhang et al., 2016). Furthermore, the phenotypes of endocardial-specific ablation of macrophages are restricted in the heart (Fig. 7, S3), which indirectly supports their heart-specific role. Thus, although there is no ‘perfect’ Cre mouse line for such studies, our conclusion is based not solely on the Cre-based genetic tracing but also on several lines of evidence. Altogether, our data suggest that a subset of macrophages originates from the endocardium.
Our study uncovers a tissue-specific role of locally-derived macrophages. This observation challenges the current view that all embryonic tissue macrophages originate from yolk sac directly or via the fetal liver (Davies et al., 2013; Epelman et al., 2014; Fantin et al., 2010; Ginhoux et al., 2010; Guilliams et al., 2013; Hashimoto et al., 2013; Hoeffel et al., 2015; Jakubzick et al., 2013; Jenkins et al., 2011; Kierdorf et al., 2015; Lux et al., 2008; Nahrendorf and Swirski, 2013; Pinto et al., 2012; Schulz et al., 2012; Yona et al., 2013). It has been speculated that the absence of a subset of macrophages can be supplemented and compensated by the expansion of others. However, valvular degeneration in Nfatc1Cre/+; Csf1rfl/fl mutants indicates that the valvulogenic role of EcTMs cannot be compensated by expansion of CCR2+ monocyte-derived macrophages, suggesting that a functional specificity exists with the EcTMs. Thus, the diverse populations of macrophages play partially redundant and partially specific roles.
Macrophages are heterogeneous not only in their function but also in their origin. Our data suggest that Nfatc1-derived macrophages are rather minority in the developing heart (30–40% of total macrophages at most; Fig. 4A). Recent reports suggest that there are at least two distinct origins of tissue resident macrophages; c-Myb-independent erythro-myeloid progenitors (EMPs) and c-Myb-dependent EMPs (Hoeffel and Ginhoux, 2018). Nfatc1+ endocardial cells contribute substantially to the macrophages while only marginally to monocytes or multipotent HSCs or EMPs (Fig. 4C-F). Conditional knockout study suggests that EcTM formation does not require Runx1, the key regulator for definitive hematopoiesis that is also expressed in the hemogenic endocardial cells at E10.5–12.5 (Lluri et al., 2015; Yzaguirre and Speck, 2016) (Fig. S2B). Therefore, EcTMs are unlikely generated through the monocytes or other intermediate progenitors, nor are they dependent on the definitive hematopoiesis. The phylogenetic origin of macrophages is old. In Drosophila, 95% of total blood cells are plasmatocytes that is equivalent to phagocytes/macrophages. Furthermore, these plasmatocytes are mainly generated in lymph grand adjacent to the heart tube and regulated by tinman, the Drosophila homologue of Nkx2–5 (Han and Olson, 2005; Han et al., 2006; Mandal et al., 2004). Thus, one can speculate that EcTMs in mammals are generated through a primitive mechanism conserved among species.
The observation that EcTM ablation leads to congenital valvular anomalies highlights the importance of these cells for cardiac development. Recent reports suggest the non-canonical roles of cardiac tissue macrophages including neonatal heart regeneration (Aurora et al., 2014) and electrical conduction (Hulsmans et al., 2017). Our results add another insight into the role of macrophages in the heart valve formation. The link between embryonic hematopoiesis and valve development raises a possibility that aberrant macrophage formation or function may underlie some human congenital valvular anomalies.
STAR Methods text
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Atsushi Nakano (anakano@ucla.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse lines
Nfatc1-Cre knock-in mice and Rosa26YFP reporter/+ or Rosa26Tomato reporter/+ were crossed to generate Nfatc1Cre/+; Rosa26YFP reporter/+ and Nfatc1Cre/+; Rosa26Tomato reporter/+, respectively. Nfatc1-Cre knock-in mice and floxed-Csf1r mice were crossed to generate Nfatc1Cre/+; Csf1rfl/fl mice, according to the Guide for the Care and Use of Laboratory Animals published by the US National Institute of Health (NIH Publication No. 85–23, revised 1996). Housing and experiments were performed in accordance with the Appropriate Care and Use of Laboratory Animals by the UCLA Institutional Animal Care and Use Committee. Both males and females were used in the study.
METHODS DETAILS
Preparation of frozen tissue sections and paraffin tissue sections.
In terms of frozen sections, dissected embryos and postnatal hearts were fixed in 4% paraformaldehyde for 2–3 hrs and at 4 °C overnight, respectively, followed by equilibration in 30% sucrose in PBS solution at 4 °C overnight. And then, hearts were placed in 1:1 30% sucrose/OCT (Tissue-Tek, Electron Microscopy Sciences) solution for 1–2 hr at 4 °C, in 100% OCT compound for 1hr at 4°C and embedded in 100% OCT compound in Cryomolds. The blocks were immediately frozen on dry ice with isopropanol and stored at −80 °C. The sample blocks were sectioned at 6 μm thin section with a Leica CM3050 S cryostat just before immunostaining. For paraffin section, dissected embryos and postnatal hearts were fixed in 4% paraformaldehyde for 2–3 hrs and at 4°C overnight, respectively. Fixed hearts were washed with PBS, dehydrated with ethanol, ethanol + xylene and then paraffin, and embedded. The sample blocks were sectioned at 4 μm thin section in the same way as frozen section.
Histological staining.
Paraffin sections were deparaffinized, rehydrated, pre-treated using heat mediated antigen retrieval with sodium citrate buffer (pH6) for 20mins. Both paraffin and frozen sections were placed in 10% goat serum blocking solution for 1hr at room temperature and incubated with primary antibody in 5% goat serum at 4°C overnight. Secondary fluorescent conjugated antibody reaction was completed in 2% goat serum for 1hr at room temperature. Primary antibodies used in the experiment and each ratio are listed in Key Resources Table. Pentachrome staining was performed following the manufacturer’s protocols (Mastertech, KTRMP) and Hematoxylin and Eosin staining was performed following the standard protocol.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti CD45 | BD bioscience | Cat#557659 |
| Mouse monoclonal anti CD68 | Bio rad | Cat#852700 |
| Mouse monoclonal anti F4/80 | Bio rad | Cat#MCA487A488 |
| Mouse monoclonal anti CD206 | Biolegend | Cat#141716 |
| Mouse monoclonal anti Csf1r | Invitrogen | Cat#14–1152-81 |
| Mouse monoclonal anti aSMA | Sigma | Cat#A5228–200 |
| Mouse monoclonal anti Vimentin | Biolegend | Cat#PCK-594P |
| Mouse monoclonal anti CD45 | BD biosciences | Cat#557659 |
| Mouse monoclonal anti CD11b | eBioscience | Cat#25011282 |
| Mouse monoclonal anti F4/80 | Biolegend | Cat#123133 |
| Mouse monoclonal anti CD64 | BD Biosciences | Cat#741024 |
| Mouse monoclonal anti MHCII | BD Biosciences | Cat#563415 |
| Mouse monoclonal anti CCR2 | RnD | Cat#FAB5538A |
| Mouse monoclonal anti Ly6C | BD Biosciences | Cat#561237 |
| Mouse monoclonal anti CX3CR1 | Biolegend | Cat#149007 |
| Mouse monoclonal anti CD206 | Biolegend | Cat#141716 |
| Mouse monoclonal anti CD117 | eBioscience | Cat#17–1171-83 |
| Mouse monoclonal anti CD117 | Biolegend | Cat#105826 |
| Mouse monoclonal anti Ly6G | eBioscience | Cat#15–5931-81 |
| Mouse monoclonal anti CD41 | BD Biosciences | Cat#740504 |
| Mouse monoclonal anti CD115 | BD Biosciences | Cat#743639 |
| Mouse monoclonal anti CD31 | BD Biosciences | Cat#553372 |
| Mouse monoclonal anti CD48 | Biolegend | Cat#103423 |
| Mouse monoclonal anti Ly6A/Ly6E | eBioscience | Cat#45–5981-82 |
| Mouse monoclonal anti CD150 | Biolegend | Cat#115929 |
| Critical Commercial Assays | ||
| DeadEndTM Fluorometric TUNEL System | Promega | Cat#G3250 |
| Phagocytosis Assay Kit (IgG FITC) | Cayman chemical | Cat#500290 |
| Software and Algorithms | ||
| FCS Express 4 | De Novo software | N/A |
| JMP 11.0 | SAS Institute | N/A |
| Excel 2016 | Microsoft | N/A |
| PowerPoint 2016 | Microsoft | N/A |
| Photoshop | Adobe | N/A |
| Zen | Zeiss | N/A |
TUNEL apoptotic assay.
Apoptotic cell death on valve area was detected by a TdT-mediated dUTP nickend labeling (TUNEL) assay (DeadEnd™ Fluorometric TUNEL System, Promega) according to the protocol provided by the manufacturer. The number of TUNEL positive cells and DAPI positive cells were counted on 5 sections in each valve per one mouse. The number of TUNEL positive cells per DAPI positive cells was used for quantitation of apoptotic cell.
Preparation of single cell suspensions from heart.
Before removing P28 and adult hearts, we perfused hearts with 20ml of cold PBS. Removed hearts were rinsed with Hank’s balanced salt solution (HBSS) without calcium and magnesium (Gibco-Invitrogen), minced finely and digested with shaking for 20 min at 37°C in Tyrode buffer containing Liberase (Roche) 0.1mg/mL per 1 heart. The digested material was filtered through a 40 μM filter and suspended in HBSS supplemented with 2% FCS and 0.2% BSA at 4°C. All digested material was pelleted by centrifugation at 400g for 5min and then re-suspended in FACS buffer (PBS with 5% FCS).
Flow cytometry.
Using DAPI exclusion, only live cells were analyzed. All antibodies used for flow cytometry and each ratio are listed in Key Resources Table.. Flow cytometry was performed using LSR II (BD Biosciences). Samples were blocked with mouse Fc block (BD Biosciences) for 5 min, followed by labeling for 25 min at 4°C with each antibody.
Hematopoietic colony-forming assays.
Embryonic tissues were dissected and cultured on OP9 stromal cells for 4 days in 48-well plates in 500 μl of α-MEM (Gibco/Invitrogen) containing 20% fetal bovine serum (Hyclone), 1% penicillin/streptomycin, SCF (50 ng/ml), IL-3 (5 ng/ml), IL-6 (5 ng/ml), thrombopoietin (5 ng/ml) and Flt-3L (10 ng/ml). The tissues were then dissociated mechanically by transfer pipette and filtered to remove the stromal cells (celltreck). The filtered cells were transferred into 1.5 ml methylcellulose with SCF, IL-6, IL-3 and EPO (MethoCult 3434, Stem Cell Technologies) supplemented with thrombopoietin (5 ng/ml) to determine their colony formation potential. Colonies were scored 7–10 days later.
RNA-seq and expression analysis.
Total RNA of sorted EcTMs and non-EcTMs by FACS were extracted using the QIAGEN RNeasy plus micro kit. Libraries for RNA-Seq were prepared with Clonetech SMARTer Stranded Total RNA-Seq (Pico) Kit. The workflow consists of first-strand synthesis, template switching, adaptor ligation, cleavage of ribosomal cDNA and PCR amplification. Different adaptors were used for multiplexing samples in one lane. Sequencing was performed on an Illumina Nextseq500 a single read 75 run. Data quality check was conducted using Illumina SAV. Demultiplexing was performed with the Illumina Bcl2fastq2 v 2.17 program. Reads were aligned to the UCSC mm10 reference genome map using TopHat. DEseq2 was used to identify differentially expressed genes of EcTMs compared to non-EcTMs. 36 upregulated genes in EcTMs compared to non-EcTMs with a Padj < 0.05 were subsequently clustered and visualized using R. Gene ontology functional analyses were performed using the database for annotation, visualization, and integrated discovery (DAVID) online tool. Raw RNA-seq data of EcTMs and non-EcTMs can be downloaded from the Gene Expression Omnibus database, accession number GEO: GSE 100495.
Macrophage phagocytosis assay.
Single cell suspension including EcTMs and non EcTMs from P8 hearts of Nfatc1Cre/+; Rosa26Tomato reporter/+ were prepared as above mentioned. After the cell isolation from the individual heart, the cells were suspended at a concentration of 1 × 106 cells/ml in media consisted of DMEM and 10 % FCS and incubated with latex beads with FITC (Cayman chemical) to a dilution of 1:100 for three hours at 37 °C. After the centrifugation and removal of the supernatant, the cells were subjected to the further staining for the macrophage markers using CD45, CD11b, CD68 and F4/80. Flow cytometry analysis was performed to identify phagocytizing cells within EcTM and non EcTM populations.
Electron microscopic analysis.
Tissues were fixed in 1% glutaraldehyde, 4% paraformaldehyde in PBS and washed. After post-fixation in 1% OsO4 in PB for 1 hr, the tissues were dehydrated in a graded series of ethanol, treated with propylene oxide and embedded in Eponate 12 (Ted Pella). Approximately 60–70-nm-thick sections were cut on a Reichert–Jung Ultracut E ultramicrotome and picked up on formvar-coated copper grids. The sections were stained with uranyl acetate and Reynolds lead citrate, and examined on a JEOL 100CX electron microscope at 80 kV.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis.
All data are presented as mean ± standard error of the mean (SEM). Statistical analysis was performed on JMP software 11 and R. Paired student’s t-test was used for comparison of EcTMs and non-EcTMs (Figure 2b,c) and unpaired student’s t-test was used for comparison of control and mutant mice (Figure 4e). Spearman’s rank correlation was used for confirming the relationship between two variables (Figure S4). A P value of < 0.05 was considered statistically significant.
Supplementary Material
Highlight.
Hemogenic endocardium is a source of cardiac macrophages in cardiac valves
Endocardial macrophages are more phagocytic than macrophages from other source(s)
Genetic ablation of endocardial macrophages induces defects in valve formation
Endocardially-derived macrophages are indispensable for normal valve remodeling
Acknowledgment
Authors thank Drs. Alvaro Sagasti, Karen Lyons and Hanna K. A. Mikkola (UCLA) for the critical reading of the manuscript and Dr. Hajime Yokota for the assistance in bioinformatics analyses. Dr. Kouki Morizono (UCLA) advised macrophage phagocytosis assay. Technical supports were provided by BSCRC flow cytometry core, BSCRC sequencing core, and UCLA histology core. This work was supported by NIH (HL127427 to A.N., HL129178 and HL137241 to A.D., HL111437 and HL129727 to T.H.). Authors were supported by the fellowships from Japanese Respiratory Foundation-Pfizer (A.S.), Howard Hughs Medical Institute (V.H.), UCLA Program for Excellence in Education and Research in the Sciences (PEERS) (R.B.), Japanese Circulation Society (Y.N.) and Uehara Memorial Foundation (Y.N.).
Footnotes
Declration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aikawa E, Whittaker P, Farber M, Mendelson K, Padera RF, Aikawa M, and Schoen FJ (2006). Human semilunar cardiac valve remodeling by activated cells from fetus to adult: implications for postnatal adaptation, pathology, and tissue engineering. Circulation 113, 1344–1352. [DOI] [PubMed] [Google Scholar]
- Aurora AB, Porrello ER, Tan W, Mahmoud AI, Hill JA, Bassel-Duby R, Sadek HA, and Olson EN (2014). Macrophages are required for neonatal heart regeneration. J Clin Invest 124, 1382–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain CC, Hawley CA, Garner H, Scott CL, Schridde A, Steers NJ, Mack M, Joshi A, Guilliams M, Mowat AM, et al. (2016). Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat Commun 7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin HS (1996). Early embryonic vascular development. Cardiovasc Res 31 Spec No, E34–45. [PubMed] [Google Scholar]
- Chitu V, and Stanley ER (2006). Colony-stimulating factor-1 in immunity and inflammation. Curr Opin Immunol 18, 39–48. [DOI] [PubMed] [Google Scholar]
- Davies LC, Jenkins SJ, Allen JE, and Taylor PR (2013). Tissue-resident macrophages. Nat Immunol 14, 986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange FJ, Moorman AF, Anderson RH, Manner J, Soufan AT, de Gier-de Vries C, Schneider MD, Webb S, van den Hoff MJ, and Christoffels VM (2004). Lineage and morphogenetic analysis of the cardiac valves. Circ Res 95, 645–654. [DOI] [PubMed] [Google Scholar]
- Dieterlen-Lievre F (2007). Emergence of haematopoietic stem cells during development. C R Biol 330, 504–509. [DOI] [PubMed] [Google Scholar]
- Dzierzak E, and Bigas A (2018). Blood Development: Hematopoietic Stem Cell Dependence and Independence. Cell stem cell 22, 639–651. [DOI] [PubMed] [Google Scholar]
- Dzierzak E, and Speck NA (2008). Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol 9, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, Brija T, Gautier EL, Ivanov S, Satpathy AT, et al. (2014). Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 40, 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin A, Vieira JM, Gestri G, Denti L, Schwarz Q, Prykhozhij S, Peri F, Wilson SW, and Ruhrberg C (2010). Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood 116, 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, and Lambrecht BN (2013). Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med 210, 1977–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, and Olson EN (2005). Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development 132, 3525–3536. [DOI] [PubMed] [Google Scholar]
- Han Z, Yi P, Li X, and Olson EN (2006). Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development 133, 1175–1182. [DOI] [PubMed] [Google Scholar]
- Harris IS, and Black BL (2010). Development of the endocardium. Pediatr Cardiol 31, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, et al. (2013). Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, Beaudin AE, Lum J, Low I, Forsberg EC, et al. (2015). C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, and Ginhoux F (2018). Fetal monocytes and the origins of tissue-resident macrophages. Cellular immunology. [DOI] [PubMed] [Google Scholar]
- Hulsmans M, Clauss S, Xiao L, Aguirre AD, King KR, Hanley A, Hucker WJ, Wulfers EM, Seemann G, Courties G, et al. (2017). Macrophages Facilitate Electrical Conduction in the Heart. Cell 169, 510–522.e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icardo JM (1990). Development of the outflow tract. A study in hearts with situs solitus and situs inversus. Ann N Y Acad Sci 588, 26–40. [DOI] [PubMed] [Google Scholar]
- Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, Ivanov S, Duan Q, Bala S, Condon T, et al. (2013). Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity 39, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins SJ, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N, MacDonald AS, and Allen JE (2011). Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332, 1284–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdorf K, Prinz M, Geissmann F, and Gomez Perdiguero E (2015). Development and function of tissue resident macrophages in mice. Semin Immunol 27, 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama H, Nose M, Niida S, Nishikawa S, and Nishikawa S (1994). Involvement of the c-kit receptor in the adhesion of hematopoietic stem cells to stromal cells. Exp Hematol 22, 979–984. [PubMed] [Google Scholar]
- Koushik SV, Wang J, Rogers R, Moskophidis D, Lambert NA, Creazzo TL, and Conway SJ (2001). Targeted inactivation of the sodium-calcium exchanger (Ncx1) results in the lack of a heartbeat and abnormal myofibrillar organization. Faseb J 15, 1209–1211. [DOI] [PubMed] [Google Scholar]
- Lluri G, Huang V, Touma M, Liu X, Harmon AW, and Nakano A (2015). Hematopoietic progenitors are required for proper development of coronary vasculature. J Mol Cell Cardiol 86, 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux CT, Yoshimoto M, McGrath K, Conway SJ, Palis J, and Yoder MC (2008). All primitive and definitive hematopoietic progenitor cells emerging before E10 in the mouse embryo are products of the yolk sac. Blood 111, 3435–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal L, Banerjee U, and Hartenstein V (2004). Evidence for a fruit fly hemangioblast and similarities between lymph-gland hematopoiesis in fruit fly and mammal aorta-gonadal-mesonephros mesoderm. Nat Genet 36, 1019–1023. [DOI] [PubMed] [Google Scholar]
- Milgrom-Hoffman M, Harrelson Z, Ferrara N, Zelzer E, Evans SM, and Tzahor E (2011). The heart endocardium is derived from vascular endothelial progenitors. Development 138, 4777–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. (2006). Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127, 1151–1165. [DOI] [PubMed] [Google Scholar]
- Nahrendorf M, and Swirski FK (2013). Monocyte and macrophage heterogeneity in the heart. Circ Res 112, 1624–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H, Liu X, Arshi A, Nakashima Y, van Handel B, Sasidharan R, Harmon AW, Shin JH, Schwartz RJ, Conway SJ, et al. (2013). Haemogenic endocardium contributes to transient definitive haematopoiesis. Nat Commun 4, 1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, and Speck NA (1999). Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126, 2563–2575. [DOI] [PubMed] [Google Scholar]
- North TE, de Bruijn MF, Stacy T, Talebian L, Lind E, Robin C, Binder M, Dzierzak E, and Speck NA (2002). Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 16, 661–672. [DOI] [PubMed] [Google Scholar]
- Orkin SH, and Zon LI (2008). Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffett-Lugassy N, Singh R, Nevis KR, Guner-Ataman B, O’Loughlin E, Jahangiri L, Harvey RP, Burns CG, and Burns CE (2013). Heart field origin of great vessel precursors relies on nkx2.5-mediated vasculogenesis. Nat Cell Biol 15, 1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palis J (2016). Hematopoietic stem cell-independent hematopoiesis: emergence of erythroid, megakaryocyte, and myeloid potential in the mammalian embryo. FEBS letters 590, 3965–3974. [DOI] [PubMed] [Google Scholar]
- Pexieder T (1975). Cell death in the morphogenesis and teratogenesis of the heart. Adv Anat Embryol Cell Biol 51, 3–99. [DOI] [PubMed] [Google Scholar]
- Pinto AR, Paolicelli R, Salimova E, Gospocic J, Slonimsky E, Bilbao-Cortes D, Godwin JW, and Rosenthal NA (2012). An abundant tissue macrophage population in the adult murine heart with a distinct alternatively-activated macrophage profile. PLoS One 7, e36814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter KA, Pridans C, Sehgal A, Tsai YT, Bradford BM, Raza S, Moffat L, Gow DJ, Beard PM, Mabbott NA, et al. (2014). Pleiotropic effects of extended blockade of CSF1R signaling in adult mice. J Leukoc Biol 96, 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, et al. (2012). A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90. [DOI] [PubMed] [Google Scholar]
- Stanley EG, Biben C, Elefanty A, Barnett L, Koentgen F, Robb L, and Harvey RP (2002). Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3’UTR-ires-Cre allele of the homeobox gene Nkx2–5. Int J Dev Biol 46, 431–439. [PubMed] [Google Scholar]
- Wang Y, Wu B, Farrar E, Lui W, Lu P, Zhang D, Alfieri CM, Mao K, Chu M, Yang D, et al. (2017). Notch-Tnf signalling is required for development and homeostasis of arterial valves. Eur Heart J 38, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Zhang Z, Lui W, Chen X, Wang Y, Chamberlain AA, Moreno-Rodriguez RA, Markwald RR, O’Rourke BP, Sharp DJ, et al. (2012). Endocardial Cells Form the Coronary Arteries by Angiogenesis through Myocardial-Endocardial VEGF Signaling. Cell 151, 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto M, Porayette P, and Yoder MC (2008). Overcoming obstacles in the search for the site of hematopoietic stem cell emergence. Cell stem cell 3, 583–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yzaguirre AD, and Speck NA (2016). Insights into blood cell formation from hemogenic endothelium in lesser-known anatomic sites. Dev Dyn 245, 1011–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir L, Singh R, Nathan E, Patrick R, Yifa O, Yahalom-Ronen Y, Arraf AA, Schultheiss TM, Suo S, Han JJ, et al. (2017). Nkx2.5 marks angioblasts that contribute to hemogenic endothelium of the endocardium and dorsal aorta. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Pu W, Tian X, Huang X, He L, Liu Q, Li Y, Zhang L, He L, Liu K, et al. (2016). Genetic lineage tracing identifies endocardial origin of liver vasculature. Nat Genet 48, 537–543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.