Abstract
Purpose
Current understanding of local disease pathophysiology in AMD is limited. Analysis of the human disease-affected tissue is most informative, as gene expression, expressed quantitative trait loci, microenvironmental, and epigenetic changes can be tissue, cell type, and location specific. Development of a novel translational treatment and prevention strategies particularly for earlier forms of AMD are needed, although access to human ocular tissue analysis is challenging. We present a standardized protocol to study rapidly processed postmortem donor eyes for molecular biochemical and genomic studies.
Methods
We partnered with the Utah Lions Eye Bank to obtain donor human eyes, blood, and vitreous, within 6 hours postmortem. Phenotypic analysis was performed using spectral-domain optical coherence tomography (SD-OCT) and color fundus photography. Macular and extramacular tissues were immediately isolated, and the neural retina and retinal pigment epithelium/choroid from each specimen were separated and preserved. Ocular disease phenotype was analyzed using clinically relevant grading criteria by a group of four ophthalmologists incorporating data from SD-OCT retinal images, fundus photographs, and medical records.
Results
The use of multimodal imaging leads to greater resolution of retinal pathology, allowing greater phenotypic rigor for both interobserver phenotype and known clinical diagnoses. Further, our analysis resulted in excellent quality RNA, which demonstrated appropriate tissue segregation.
Conclusions
The Utah protocol is a standardized methodology for analysis of disease mechanisms in AMD. It uniquely allows for simultaneous rigorous phenotypic, molecular biochemical, and genomic analysis of both systemic and local tissues. This better enables the development of disease biomarkers and therapeutic interventions.
Keywords: age-related macular degeneration, donor eye phenotype, local disease mechanisms
AMD is a blinding condition of aging that can lead to permanent loss of fine, central vision.1–4 The clinical AMD phenotype, particularly of advanced disease, is heterogeneous. We now have greater appreciation for the retinal-level changes that occur in the spectrum of AMD phenotype, from subretinal drusen to retinal atrophy and subretinal neovascular changes. This is due in large part to the advancements in clinical imaging modalities allowing for visualization of the retina on the “microanatomic” level. This has led to refinement of diagnostic and prognostic classifications for AMD, most notably including spectral domain-optical coherence tomography (SD-OCT)–based prognostic scales.5,6 Although significant advances have been made in the diagnostic AMD arena, treatment of AMD has not reached a level whereby we can slow disease progression or restore vision in a meaningful way, particularly for patients with nonexudative (dry) AMD. As a result, AMD is a significant medical problem and remains a leading cause of irreversible blindness in the aging population.
Our understanding of the systemic pathophysiologic disease mechanisms for AMD have, to date, been primarily informed by genetic epidemiologic studies in human populations (for review, please see DeAngelis et al.).3,7–16 Certainly these analyses have led to identification of important environmental exposures, genetic variation, and serum biomarkers associated with AMD. Furthermore, analysis in a human population is inherently more translational given the limitations of animal models of disease.17 However currently, these discoveries have not led to broadly applicable advances in AMD prevention or treatment.18–20 In fact, even the most current clinical trials based on data from systemic analysis of AMD molecular etiology, such as the recent complement pathway trials, have not resulted in improved patient outcomes (https://www.roche.com/media/store/releases/med-cor-2017-09-08b.htm).21–23 One reason for the translational failure of our past efforts may be the inability to perform a systemic disease analysis and analysis of disease affected tissues (i.e., the retina, RPE, and choroid) within a given donor and across many donors simultaneously. This is problematic, however, given that fresh donor tissue is not accessible in patients, and biopsy is not clinically indicated in otherwise structurally intact eyes. Therefore, access to affected fresh tissues has been challenging to disease analysis.24
In addition to limited access to human ocular tissues, the ability to analyze postmortem tissues in a clinically relevant standardized manner has presented challenges. Although histologic, immunocytochemical, pathologic, and gene expression work on tissue affected by AMD (i.e., the neural retina, RPE, and/or choroid) has begun to shed light on the molecular and cellular pathophysiology underlying AMD disease, as well as the specific structural and/or cellular contributions to disease mechanisms, it has not been without its limitations. This includes lack of information on the postmortem processing time or a death to preservation time of greater than 6 hours, as well as a lack of well-characterized phenotyping of both eyes for a given donor by more than one trained clinical specialist (postmortem) using a standardized protocol.25–35 These limitations can confound downstream experimental analysis and hence translational efforts in developing effective therapies for AMD. Specifically, lengthy or indeterminate death to preservation time can change the tissue quality such that a meaningful analysis of disease mechanisms is no longer possible.36
Two grading schemes that have been developed to standardize donor eye phenotyping data for research use are the Alabama AMD grading system for donor eyes and the Minnesota grading system. The Alabama AMD grading system37 is based on the Wisconsin Age-Related Maculopathy Grading System,38 whereas the Minnesota grading system39 is based on the 9-Step Age-Related Eye Disease Study Scale; both scales were developed based in part on fundoscopy.
With the goal of building on prior work by others, as well moving the translational value of the field forward, our group collaborated with the Utah Lions Eye Bank and pioneered work to study tissues affected by AMD: the RPE and the neural retina. We believe that if performed in a standardized fashion prioritizing tissue integrity and using a multimodal retinal imaging approach to ensure rigorous phenotype, these data can be used for molecular biochemical and genomic studies. Herein, we report a novel paradigm for the collection, analysis, and interpretation of ocular tissue. Using a highly translational approach at the outset will facilitate development of novel disease biomarkers and/or therapeutic interventions especially for intermediate AMD. The goal is improved disease prevention and treatment.
Materials and Methods
Donor Eye Tissue Repository
From 2013 to the present, we established a repository of fresh human eye tissue, blood, serum plasma, and vitreous in collaboration with the Utah Lions Eye Bank at the Moran Eye Center/University of Utah. We follow Eye Bank Association of America guidelines. Following Utah Code, Title 26 Chapter 28 (Uniform Anatomical Gift Act or UAGA), our coordination department uses the Utah donor registry to obtain authorization (Section 114), but will also reach out to authorized decision makers such as families when the individual is not found on a donor registry. Law enforcement, firefighters, emergency medical services, providers, or emergency rescuers are required by law as soon as reasonably possible to notify the appropriate organ procurement organization, tissue bank, or eye bank of a pending death or a death that occurs in Utah (Section 112). Working with the Utah Lions Eye Bank, our biorepository implements a standardized system for acquisition of eyes and peripheral blood (serum and plasma) and vitreous ≤6 hours of death, as well as retrospective collection of epidemiologic and clinical data from these donors8 (a detailed appendix can be found Supplementary Information). Because there is a short death-to-preservation time, only potential eye donors that reside 1 hour away in travel distance by car from the Utah Lions Eye Bank (at the John A. Moran Eye Center in Salt Lake City, Utah) are approached. This 1-hour travel distance by car is an operational decision made by our laboratory in collaboration with the Utah Lions Eye Bank concerning the timeline associated with providing fresh globes specifically for our laboratory's program of research. Our work and that of others demonstrated that there is a significant change in gene expression and RNA quality in postmortem ocular tissue with death to preservation times greater than 6 hours.36,40–42 Moreover, recent reports demonstrate that longer postmortem intervals can skew results of methylation studies particularly in neural tissues.43
Following the Utah Lions Eye bank's mission to Restore Hope Through Vision, the recovery program will always pursue a case for transplant as the primary goal. Part of eligibility is “ruling out” a case for transplant before pursuing research. The research exclusionary process consists on finding conditions that could be harmful to the recovery staff (e.g., herpes simplex virus [HSV] and HIV) and excluding those cases. The remaining screening that is done includes the criteria for the given laboratory that is on call to accept eyes. Our laboratory will receive all donors 60 years and older that are within the death to preservation time as stated above. During the screening period, the eye bank reviews all medical records that are available. In addition, and after consent is obtained, the next of kin or authorized decision maker is asked about the donor's history prior to recovery.
This questionnaire is used to collect medical history and risk factors including, stroke, diabetes, cardiovascular disease, Alzheimer disease status, dyslipidemia, smoking, education level, known ocular disease (AMD, glaucoma, diabetic retinopathy, retinitis pigmentosa, Lucentis and/or Avastin injections, who their eye physician was) and the name of their primary care physician. Upon death of the donor, a trained recovery technician is dispatched to the recovery location, generally at a hospital. A donor coordinator then pages the trained laboratory technician in our laboratory who is on-call once the whole globes are in transit to ensure prompt accession and processing of donor eye tissue and associated biological materials.
Ocular Phenotype Analysis
Donor eye tissue, blood, serum, plasma, and vitreous are obtained from individuals, as noted above, who have given consent for postmortem tissue donation. To perform a robust molecular analysis, the ocular phenotype relative to disease (i.e., AMD, etc.) must be assessed with a high degree of precision and correlation to current clinical practice. When ocular medical records are available, they can clarify ocular phenotype; however, they are not uniformly available or inclusive of ocular history for donor eyes. Therefore, to effectively correlate molecular findings with disease state, we need a precise and reproducible system to assess postmortem donor eye phenotype.
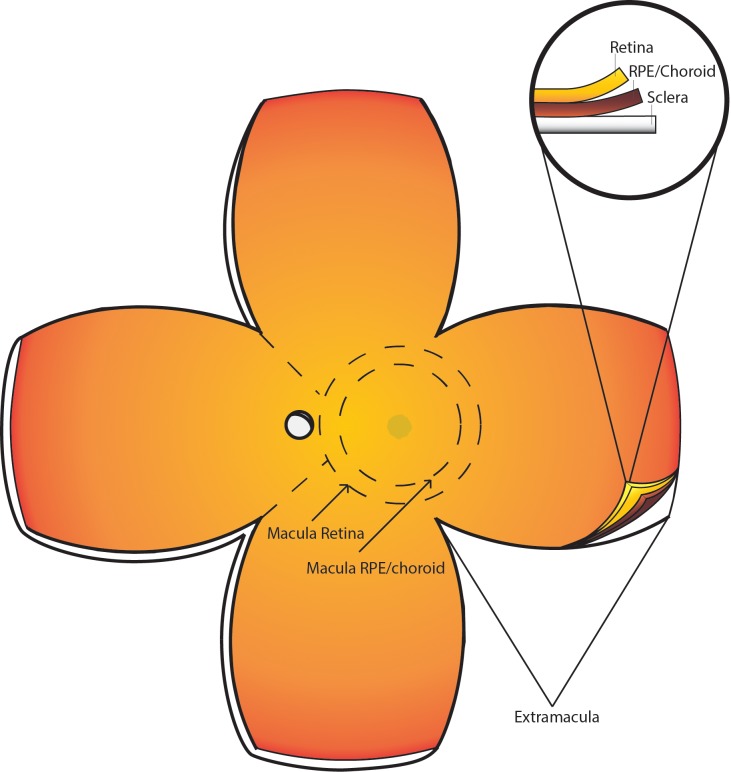
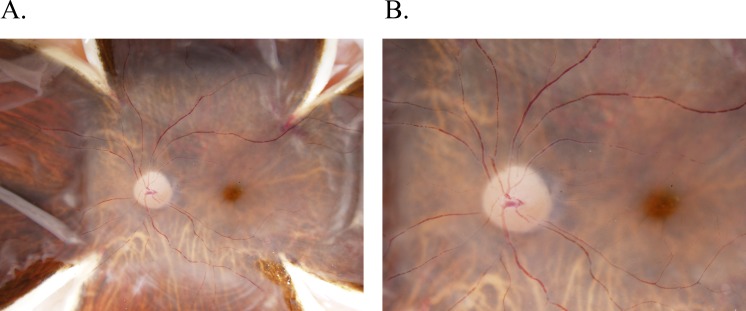
To achieve a rigorous and clinically relevant method of postmortem ocular phenotype analysis, we use imaging techniques used in the clinical setting, namely SD-OCT and color fundus photography. Images are taken in a manner consistent with the appearance of the analogous clinical images for both donor eyes. More specifically, immediately on receipt of the donor eyes, an incision is made approximately 2 mm posterior to the limbus, and Westcott scissors are used to remove the anterior ocular structures, including the cornea and lens. The remaining globe (all tissues posterior to the corneal limbus) is placed in an optical glass cuvette (cat. no. 93-G-50; Starna Cells, Atascadero, CA, USA) and submersed in saline solution (cat. no. 23-062-125; Thermo Scientific, Waltham, MA, USA). The cuvette is then placed on a platform such that the anterior aspect of the eye is facing the OCT, similar in orientation to that of a patient in a clinical setting. OCT images are then obtained using the Heidelberg Spectralis (Franklin, MA, USA). Macular images are obtained in 73 B-scans each spaced by 60 μm. The globe is then dissected into four equal lobes (Fig. 1). Incisions are made from the anterior opening, between the attachment points for each extraocular muscle, and continue toward the posterior pole of the eye until the four lobes of the eye lay flat, with macula and optic nerve in the center. The vitreous is then carefully removed from the retinal surface using both blunt and sharp dissection techniques to minimize retinal detachment. Retinal images are then obtained for each eye at 0.7× and 1.25× magnifications using an Olympus SZX16 microscope camera (Shinjuku, Tokyo, Japan) (Fig. 2).
Figure 1.
Butterfly dissection of the posterior donor eye. The posterior globe is cut into the depicted butterfly pattern, including sclera, RPE/choroid, and retina. Incisions are made from the anterior opening, between the attachment points for each extraocular muscle, and continue toward the posterior of the eye until the four lobes of the eye lay flat, with macula and optic nerve in the center. Macular retinal tissue is collected using an 8-mm disposable biopsy punch centered over the fovea as depicted. Additionally, a 6-mm punch is used to cut a button of RPE/choroid from within the 8-mm punch, which minimizes retina contamination to the RPE.
Figure 2.
Color fundus imaging in a normal donor eye. Following the butterfly dissection but prior to macular tissue isolation, retinal images are obtained for each eye at 0.7× (A) and 1.25× (B) magnifications using an Olympus SZX16 microscope camera illuminated with a Schott KL 1600 LED Fiber Optic Light Source Illuminator. Images are taken in the same orientation as done in a clinical setting for highest translational quality. As shown, detailed optic nerve, macula, fovea, and posterior pole vasculature can be seen.
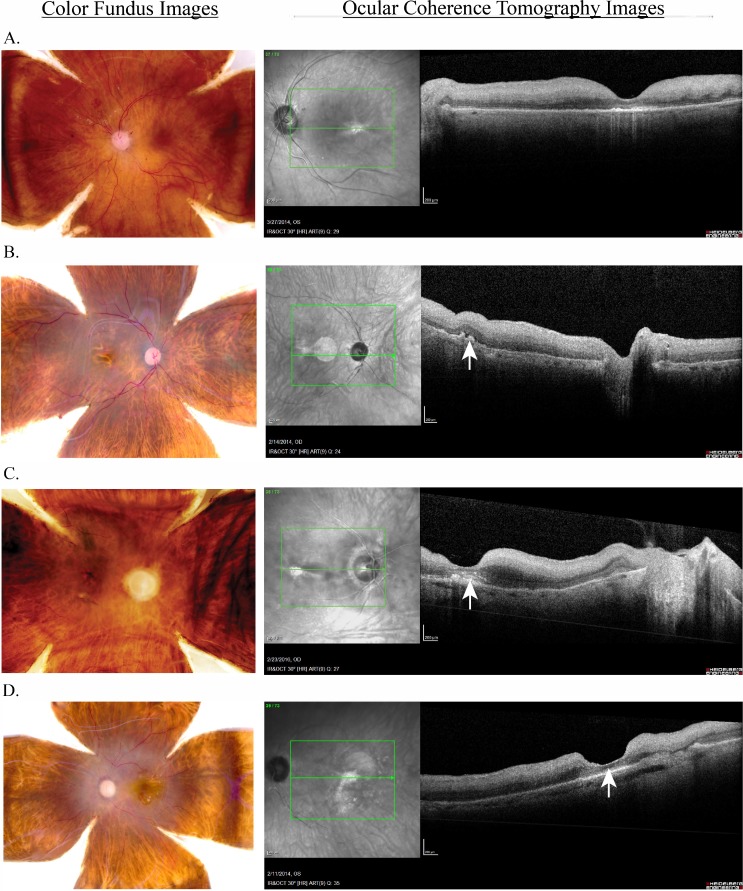
To determine the precise ocular phenotype relative to disease, analysis of each set of images is performed by a team of retinal specialists and ophthalmologists at the University of Utah School of Medicine, Moran Eye Center, and the Massachusetts Eye and Ear Infirmary Retina Service, Harvard Medical School. Even the most detailed molecular analysis of tissue incorrectly phenotyped may produce artifactual results. However, there are unique challenges that arise when attempting to phenotype postmortem ocular tissue. For example, depending on the mechanism of death, there may be a degree of retinal edema that occurs postmortem. Although this may not obscure gross morphology on color fundus photography, drusen, particularly small to intermediate drusen, may be difficult to appreciate. Therefore, a multipronged strategy accounting for the unique findings in postmortem eyes was used to develop a precise tissue characterization. This is particularly important for analysis of nonexudative AMD where not just drusen but perhaps small areas of atrophy may be best appreciated with SD-OCT imaging. Bearing this in mind, each donor eye is phenotyped by independent review of the color fundus photograph and SD-OCT imaging; discrepancies are resolved by collaboration between a minimum of three specialists to ensure a robust and rigorous phenotypic analysis. If there is disagreement between the specialists, a fourth vitreoretinal specialist will weigh in until the diagnosis is resolved. If there is not sufficient evidence to decisively conclude a diagnosis, this eye cannot be further analyzed or included for study in downstream experiments. The team also has frequent meetings with calls-ins and face-to-face meetings to resolve any concerns, when available postmortem ocular phenotypes are then correlated with available clinical data. All patients are phenotyped relative to AMD status using the clinically derived modified Age-Related Eye Disease Study severity grading scale (AREDS 1, AREDS 2, AREDS, AREDS 4a, AREDS 4b) using color fundus images, as is true for existing postmortem grading scales.44 Furthermore, SD-OCT images are used to resolve the appearance of characteristic extracellular deposits consistent with drusen in the clinical setting, fluid, atrophy, and fibrosis and differentiate artifact from pathology. Representative images in Figure 3 demonstrate the utility of using the combination of these imaging techniques for the postmortem eye as is true in clinical practice. Figure 3A demonstrates normal findings, Figure 3B shows intermediate nonexudative AMD, Figure 3C shows neovascular AMD, and Figure 3D shows geographic atrophy. Thus, our phenotyping approach uses clinically relevant systems in a multifaceted approach, appropriate for the unique factors presented by analysis of a postmortem globe. This phenotype is then compared with any known ocular diagnoses provided by the Utah Lions Eye Bank.
Figure 3.
Color fundus and OCT imaging of normal and diseased donor eyes provides precise visualization of AMD features. Analogous or the same eye that was used for color fundus image that is also used for the corresponding SD-OCT image as shown demonstrating (A) normal findings, (B) intermediate nonexudative AMD, (C) neovascular AMD, and (D) central geographic retinal atrophy. Color images are used to assign a classic AREDS disease classification. SD-OCT allows for clarification of drusen as seen in B or a neovascular membrane as confirmed in C and labeled with arrows. Please note the white circle present in the en face is an imaging artifact from the light source throughout.
Dissection and Isolation of Ocular Tissue
We developed a standardized dissection protocol to reliably isolate the RPE/choroid from the retina and segregate the layers into quadrants. After all imaging is complete, macular retinal tissue is collected using an 8-mm disposable biopsy punch (cat. no. 33-37; Integra, Plainsboro, NJ, USA) centered over the fovea. Additionally, a 6-mm punch is used to cut a button of RPE/choroid from within the 8-mm punch. In our experience, this smaller punch of 6 mm minimizes retina contamination to the RPE (Bowes-Rickman C, personal communication).45 Retinal tissues are separated from the underlying RPE/choroid tissues using sharp dissection under a dissecting microscope. Isolated macula retina and RPE samples are either then placed in cryotubes with RNAlater (Ambion, ThermoFisher, Waltham, MA, USA), an RNA stabilizing reagent, and stored at 4°C for 24 hours and then transferred to −80°C for long-term storage or flash frozen in liquid nitrogen and stored at −80°C for long-term storage. The peripheral retina and RPE tissues are similarly collected from each of the four individual lobes and stored separately in RNA later or flash frozen in liquid nitrogen. A total of 10 (n = 5 retina and n = 5 RPE) samples were collected from each donor eye. For a detailed protocol please see Supplementary Information, Appendix S1. Tissue samples stored in RNAlater are processed for downstream molecular analysis using a Qiagen All-Prep DNA/RNA/miRNA Universal kit (cat. no. 80224; Qiagen, Hilden, Germany). Tissue is thawed and removed from the RNAlater reagent, homogenized using the Qiagen tissue ruptor (cat. no. 9001271; Qiagen), and processed for DNA/RNA/miRNA using the manufacturer's recommended protocol. The All-Prep Universal kit (cat. no. 80224; Qiagen) can also be used to purify protein from tissue. We confirm the quality of the resulting nucleic acids as follows: RNA concentration and integrity is measured via Agilent High Sensitivity RNA ScreenTape Assay (2200 Tape Station cat. no. G2991AA; Agilent Technologies, Santa Clara, CA, USA). DNA concentration and purity is evaluated by a NanoDrop 1000 Spectrophotometer (cat. no. ND-1000; Thermo Fisher), and picogreen quantitation (Qubit 2.0 fluorometer; cat. no. Q33226; Invitrogen, ThermoFisher, Waltham, MA, USA).
Results
Of the 604 donor eyes analyzed for this study, all were Caucasian of European descent, and 60% were male. The average age for the normal donors was 75.0 years (range, 60.0 to 95.0 years), and the average age for the donors with AMD was 76.0 years (range, 60.0 to 87.0 years); 7.3% of the donors included in this study were discordant in phenotype between fellow eyes, underscoring the importance of phenotyping both eyes. This is in line with that found by Olsen et al. on phenotyping of postmortem eyes using a different approach.26 In the case of discordant phenotypes within the same donor, the more severe diseased eye was used for inclusion in the Utah Protocol study. For example, if a patient had a diagnosis of intermediate AMD consistent with AREDS 3 in one eye and normal or AREDS 1 in the contralateral eye only, the donor would have the diagnosis of intermediate AMD. Similarly, if a donor had a diagnosis of intermediate AMD/AREDS 3 in one eye and neovascular AMD/AREDS 4B in the contralateral eye, the donor would have the diagnosis of neovascular or exudative AMD, and only that eye would be used for downstream experiments.
Thirty-six percent of donors had a diagnosis of AMD; of those with AMD, 49.5% had the diagnosis of intermediate dry AMD; 15.5% had the diagnosis of geographic atrophy; 27% had the diagnosis of neovascular AMD; and 8% were mixed, having both geographic atrophy and neovascular AMD. A total of 366 eyes were normal or had no evidence of disease as described above. In addition, we had donor eyes with other diseases of aging, including diabetic retinopathy and glaucoma, which were not included in the current study (n = 110). Clinical and epidemiologic information was obtained through medical records, which included diabetes status, hypertension, hyperlipidemia, education, history of Alzheimer disease, and cardiovascular history, and was obtained for all donors included in this study. Not all donors had relevant ocular history within their medical records. For those who did, we demonstrated a high concordance rate between the postmortem diagnosis of AMD and prior ocular diagnosis (65%). Importantly, we were able to accurately detect the presence of AMD for all patients with a prior diagnosis of AMD. For some donors, we were able to detect early and intermediate AMD changes that were not noted in the medical records. Upon review, we determined this was either due to remote history with an ophthalmic provider or care provided in the absence of a dilated eye examination.
The addition of SD-OCT to fundus imaging enhanced our ability to more accurately phenotype postmortem tissue by improving the differentiation between artifact and true pathology through confirmation of the presence of structural retinal or subretinal features of AMD (characteristic extracellular deposits consistent with drusen in the clinical setting CNVM, fibrosis, and atrophy). As an example, a differential diagnosis for the color image shown in Figure 3B includes intermediate AMD versus geographic atrophy. However, addition of SD-OCT helps to resolve the lack of true retinal atrophy and presence of multiple intermediate drusen consistent with a diagnosis of intermediate AMD. Conversely, the presence of true retinal atrophy is confirmed for the image three dimensionally using SD-OCT.
Implementation of our rigorous tissue processing methods resulted in production of high-quality genomic material. Resulting RNA integrity numbers (RINs) for RNA extracted from neural retinal tissue averaged ≥8.0, whereas RPE/choroid averaged ≥7.4 when stored in RNAlater. RNA yield from the macular neural retina averaged 14 μg/μL, whereas RNA yield from the macular RPE/choroid averaged 2.4 μg/μL. DNA yield from macular retina stored in RNAlater averaged 15 μg/μL, and DNA yield from macular RPE/choroid stored in RNA later averaged 4 μg/μL.
We additionally demonstrated excellent tissue integrity as measured in our downstream gene expression and epigenetic experiments.7,8,46 Further, tissue expression signatures as measured by single cell nuclei sequencing obtained from tissue processed in the aforementioned manner are consistent with the specific tissue type and the individual cell type.47 This bioinformatic approach supports proper tissue segregation and fidelity of resultant genomic materials.
Discussion
Analysis of molecular AMD pathophysiology in a human population is arguably the most translational model. Systemic human tissue analysis to date, although excellent for genetic association studies, has resulted in limited success with respect to novel prevention and treatment strategies.48 This is particularly true for nonexudative AMD for which we are currently unable to significantly slow or reverse pathologic progression. Investigation of the diseased tissues is likely to yield more disease specific and translationally relevant findings. Access to these tissues in living human patients is not possible in the clinical setting.
Herein, we report a method by which diseased ocular tissue can be ascertained, collected, and processed in a standardized fashion such that (1) the tissue and molecular integrity is maintained and (2) relevance to human disease can be assessed in a rigorous fashion. In this way, analysis of diseased tissues in aggregate, as well as at the cellular level, can be performed with direct relevance to clinical disease particularly at the single nuclei gene expression level.47 Further, discoveries in the diseased tissue may better represent the fundamental pathophysiology that should be targeted therapeutically. In this way, we improve our AMD diagnosis and prognosis abilities given that systemic testing is more feasible in the patient population. Finally, analysis of the affected tissue along with epidemiologic information, blood, serum, plasma, and vitreous from the same donor may allow for greater insight into which, if any, systemically associated findings such as genetic variation and comorbid disorders are functionally relevant to the disease process. This will also allow for increased understanding of the relationship between systemic and local disease mechanisms, the degree to which AMD pathophysiology is interrelated and/or co-occurring with other diseases of aging, and the degree to which we can direct treatments on the basis of systemic analysis.
The Utah protocol is a different approach for classifying rapidly processed postmortem eyes that has some similarities and some differences from prior systems. It is a standardized protocol that adds to and in some instances improves/builds on other grading schemes (Minnesota, Alabama). First, compared with the two other repositories, we not only collect the eye tissue but also collect and process the corresponding peripheral blood, plasma, serum, and vitreous for each donor. This becomes quite important and relevant for future studies, particularly when trying to identify a relevant therapeutic biomarker. Specifically, the Utah protocol allows for understanding whether biomarkers that we identify at the genomic level (DNA, RNA, and protein) in the AMD-affected tissues can be measured systemically. This is of particular importance with respect to disease relevant extracellular RNAs, which can serve as biomarkers of disease progression.49–52 Moreover, induced pluripotent stem cells (iPSCs) can be generated from a donor's blood53–55 and then compared back with the retina and RPE cells from the donor eye, for example, by RNASeq. This is of particular importance to make appropriate cells that can be used for functional assays in the identification of disease mechanisms and therapeutics. Although the Minnesota grading scale incorporates AMD risk to advanced AMD through the nine-step AREDS severity scale with the use of color fundus photographs, the Utah grading protocol uses the four-step AREDS severity scale. This four-step progressive classification with end-stage AMD is defined as either center involving geographic atrophy (GA, 4a), or subfoveal choroidal neovascularization (CNV, 4b), or both (4c). Moreover, the Utah protocol augments color fundus photographs with SD-OCT of eyes to resolve the appearance of characteristic extracellular deposits consistent with drusen in the clinical setting, fluid, atrophy, and fibrosis, and differentiate artifact from pathology. This is similarly true of the role for SD-OCT in current clinical practice. Although the Alabama grading scale incorporates the use of histology of the fellow donor eye relative to the donor eye being phenotyped with color fundus photography and SD-OCT, this scheme accepts the idea that some measure of discordance in phenotype between the two eyes is permissible.25 Both the Minnesota grading scheme and the Utah protocol, while using different phenotyping approaches, do not incorporate histology and do not allow for any degree in discordancy or phenotypic ambiguity between the two donor eyes. This is in fact a tradeoff, keeping in mind the purpose of the biorepository. On one hand, although much can be learned from histology of the normal and diseased retina in AMD, both the Minnesota Grading Scheme and the Utah Protocol found at least 7% discordancy between the two donor eyes.56–60 This was particularly apparent when the AREDS 3 phenotype was observed in one donor eye, whereas the fellow donor eye had either the normal phenotype of AREDS0/1 or neovascular AMD, AREDS 4b. This is a rather high number for discordancy between fellow eyes when one is using the donor eyes for downstream analysis, particularly in studies of gene expression and epigenetics, if the findings are to be translatable. We believe that this is the real advance under the Utah protocol: the collection of tissue for biochemical material. Similar to that of the Alabama grading scale, a strength of our approach is the relatively short postmortem tissue processing interval. In addition, the Utah protocol uses color fundus photographs combined with SD-OCT of both eyes that are interpreted by at least three independent retinal specialists. Thus, our work increases the level of rigor, clinical applicability, and potential translational value of work with donor eyes. Further, “our proof of principle” for analysis of individual ocular tissues showing measurable and reliable differences in tissue expression in single cell types demonstrates the reliability on a molecular level such that further downstream experimental analysis is feasible and appropriate.47
Although our work has added to prior human ocular analysis protocols, we recognize some limitations. For increased comprehensive understanding of disease pathomechanisms, we will include the collection of aqueous fluid in addition to the biological materials we currently collect. Furthermore, our current study did not include donor eyes with pseudodrusen or very early AMD (AREDS 2). Although we collected and ascertained donors who we believe to have these diagnoses, further investigation is ongoing to ensure that our phenotyping methods are suitably rigorous to ensure proper classification of eyes with more mild findings. This is important, as pseudodrusen may be a clinical biomarker for the more severe forms of AMD.61–64 Moreover, depending on the presentation of the pseudodrusen, this could indicate progression to geographic atrophy or neovascular AMD. Therefore, for greatest precision, more work is required to resolve this question.
In conclusion, although the Utah protocol is a complementary approach to standardized phenotyping of donor eyes compared with those previously described, we present a novel method for analysis of ocular tissue that optimizes molecular biochemical and phenotypic integrity to a degree not currently demonstrated in the literature.26,65 We believe implementation of the Utah protocol will allow for discovery at the level of disease that will be invaluable to moving our prevention, diagnostic, and treatment paradigms forward for this disease.
Supplementary Material
Acknowledgments
The authors thank the donors and their families who have made this research possible.
Supported by the Eunice Kennedy Shriver National Institute of Child Healthy & Human Development, the Office of Research on Women's Health of the National Institutes of Health under Award K12HD085852, the Carl Marshall Reeves & Mildred Almen Reeves Foundation, Inc., The Skaggs Foundation for Research, the Macular Degeneration Foundation, Inc., National Institutes of Health Core Grant EY014800, and an Unrestricted Grant from Research to Prevent Blindness, Inc., New York, NY, to the Department of Ophthalmology & Visual Sciences, University of Utah.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosure: L.A. Owen, None; A. Shakoor, None; D.J. Morgan, None; A.A. Hejazi, None; M.W. McEntire, None; J.J. Brown, None; L.A. Farrer, None; I. Kim, None; A. Vitale, None; M.M. DeAngelis, None
References
- 1.DeAngelis MM, Lane AM, Shah CP, Ott J, Dryja TP, Miller JW. Extremely discordant sib-pair study design to determine risk factors for neovascular age-related macular degeneration. Arch Ophthalmol. 2004;122:575–580. doi: 10.1001/archopht.122.4.575. [DOI] [PubMed] [Google Scholar]
- 2.Deangelis MM, Ji F, Adams S, et al. Alleles in the HtrA serine peptidase 1 gene alter the risk of neovascular age-related macular degeneration. Ophthalmology. 2008;115:1209–1215.e7. doi: 10.1016/j.ophtha.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Owen LA, Morrison MA, Ahn J, et al. FLT1 genetic variation predisposes to neovascular AMD in ethnically diverse populations and alters systemic FLT1 expression. Invest Ophthalmol Vis Sci. 2014;55:3543–3554. doi: 10.1167/iovs.14-14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yonekawa Y, Miller JW, Kim IK. Age-related macular degeneration: advances in management and diagnosis. J Clin Med. 2015;4:343–359. doi: 10.3390/jcm4020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lei J, Balasubramanian S, Abdelfattah NS, Nittala MG, Sadda SR. Proposal of a simple optical coherence tomography-based scoring system for progression of age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2017;255:1551–1558. doi: 10.1007/s00417-017-3693-y. [DOI] [PubMed] [Google Scholar]
- 6.Garrity ST, Sarraf D, Freund KB, Sadda SR. Multimodal imaging of nonneovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2018;59:AMD48–AMD64. doi: 10.1167/iovs.18-24158. [DOI] [PubMed] [Google Scholar]
- 7.DeAngelis MM, Owen LA, Morrison MA, et al. Genetics of age-related macular degeneration (AMD) Hum Mol Genet. 2017;26:R45–R50. doi: 10.1093/hmg/ddx228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan DJ, DeAngelis MM. Differential gene expression in age-related macular degeneration. Cold Spring Harb Perspect Med. 2015;5:a017210. doi: 10.1101/cshperspect.a017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan Q, Maranville JC, Fritsche L, et al. HDL-cholesterol levels and risk of age-related macular degeneration: a multiethnic genetic study using Mendelian randomization. Int J Epidemiol. 2017;46:1891–1902. doi: 10.1093/ije/dyx189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee V, Rekhi E, Hoh Kam J, Jeffery G. Vitamin D rejuvenates aging eyes by reducing inflammation, clearing amyloid beta and improving visual function. Neurobiol Aging. 2012;33:2382–2389. doi: 10.1016/j.neurobiolaging.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Morrison MA, Silveira AC, Huynh N, et al. Systems biology-based analysis implicates a novel role for vitamin D metabolism in the pathogenesis of age-related macular degeneration. Hum Genomics. 2011;5:538–568. doi: 10.1186/1479-7364-5-6-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millen AE, Meyers KJ, Liu Z, et al. Association between vitamin D status and age-related macular degeneration by genetic risk. JAMA Ophthalmol. 2015;133:1171–1179. doi: 10.1001/jamaophthalmol.2015.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim KL, Park SP. Association between serum vitamin D deficiency and age-related macular degeneration in Koreans: clinical case-control pilot study. Medicine (Baltimore) 2018;97:e11908. doi: 10.1097/MD.0000000000011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermann MM, van Asten F, Muether PS, et al. Polymorphisms in vascular endothelial growth factor receptor 2 are associated with better response rates to ranibizumab treatment in age-related macular degeneration. Ophthalmology. 2014;121:905–910. doi: 10.1016/j.ophtha.2013.10.047. [DOI] [PubMed] [Google Scholar]
- 15.Burgess S, Davey Smith G. Mendelian randomization implicates high-density lipoprotein cholesterol-associated mechanisms in etiology of age-related macular degeneration. Ophthalmology. 2017;124:1165–1174. doi: 10.1016/j.ophtha.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan Q, Ding Y, Liu Y, et al. Genome-wide analysis of disease progression in age-related macular degeneration. Hum Mol Genet. 2018;27:929–940. doi: 10.1093/hmg/ddy002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forest DL, Johnson LV, Clegg DO. Cellular models and therapies for age-related macular degeneration. Dis Model Mech. 2015;8:421–427. doi: 10.1242/dmm.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maguire MG, Martin DF, Ying GS, et al. ; Comparison of Age-related Macular Degeneration Treatments Trials (CATT) Research Group Five-year outcomes with anti-vascular endothelial growth factor treatment of neovascular age-related macular degeneration: the comparison of age-related macular degeneration treatments trials. Ophthalmology. 2016;123:1751–1761. doi: 10.1016/j.ophtha.2016.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maguire MG, Ying G-S, Jaffe GJ, et al. Single-nucleotide polymorphisms associated with age-related macular degeneration and lesion phenotypes in the comparison of age-related macular degeneration treatments trials. JAMA Ophthalmol. 2016;134:674–681. doi: 10.1001/jamaophthalmol.2016.0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JW, Beyond VEGF. – The Weisenfeld Lecture. Invest Ophthalmol Vis Sci. 2016;57:6911–6918. doi: 10.1167/iovs.16-21201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen G, Tzekov R, Li W, Jiang F, Mao S, Tong Y. Pharmacogenetics of complement factor H Y402H polymorphism and treatment of neovascular AMD with anti-VEGF agents: a meta-analysis. Sci Rep. 2015;5:14517. doi: 10.1038/srep14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volz C, Pauly D. Antibody therapies and their challenges in the treatment of age-related macular degeneration. Eur J Pharm Biopharm. 2015;95:158–172. doi: 10.1016/j.ejpb.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 23.Yehoshua Z, de Amorim Garcia Filho CA, Nunes RP, et al. Systemic complement inhibition with eculizumab for geographic atrophy in age-related macular degeneration: the COMPLETE study. Ophthalmology. 2014;121:693–701. doi: 10.1016/j.ophtha.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamer WD, Williams AM, Pflugfelder S, Coupland SE. Accessibility to and quality of human eye tissue for research: a cross-sectional survey of ARVO members. Invest Ophthalmol Vis Sci. 2018;59:4783–4792. doi: 10.1167/iovs.18-25319. [DOI] [PubMed] [Google Scholar]
- 25.Curcio CA. Imaging maculopathy in post-mortem human eyes. Vision Res. 2005;45:3496–3503. doi: 10.1016/j.visres.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 26.Olsen TW, Liao A, Robinson HS, Palejwala NV, Sprehe N. The nine-step Minnesota grading system for eyebank eyes with age related macular degeneration: a systematic approach to study disease stages. Invest Ophthalmol Vis Sci. 2017;58:5497–5506. doi: 10.1167/iovs.17-22161. [DOI] [PubMed] [Google Scholar]
- 27.Seddon JM, McLeod DS, Bhutto IA, et al. Histopathological insights into choroidal vascular loss in clinically documented cases of age-related macular degeneration. JAMA Ophthalmol. 2016;134:1272–1280. doi: 10.1001/jamaophthalmol.2016.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ezzat M-K, Hann CR, Vuk-Pavlovic S, Pulido JS. Immune cells in the human choroid. Br J Ophthalmol. 2008;92:976–980. doi: 10.1136/bjo.2007.129742. [DOI] [PubMed] [Google Scholar]
- 29.Zanzottera EC, Messinger JD, Ach T, Smith RT, Freund KB, Curcio CA. The Project MACULA retinal pigment epithelium grading system for histology and optical coherence tomography in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2015;56:3253–3268. doi: 10.1167/iovs.15-16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim EJ, Grant GR, Bowman AS, Haider N, Gudiseva HV, Chavali VRM. Complete transcriptome profiling of normal and age-related macular degeneration eye tissues reveals dysregulation of anti-sense transcription. Sci Rep. 2018;8:3040. doi: 10.1038/s41598-018-21104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman AM, Gallo NB, Hancox LS, et al. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med. 2012;4:16. doi: 10.1186/gm315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghazi NG, Dibernardo C, Ying H, Mori K, Gehlbach PL. Optical coherence tomography of peripheral retinal lesions in enucleated human eye specimens with histologic correlation. Am J Ophthalmol. 2006;141:740–742. doi: 10.1016/j.ajo.2005.10.058. [DOI] [PubMed] [Google Scholar]
- 33.Brown NH, Koreishi AF, McCall M, Izatt JA, Rickman CB, Toth CA. Developing SDOCT to assess donor human eyes prior to tissue sectioning for research. Graefes Arch Clin Exp Ophthalmol. 2009;247:1069–1080. doi: 10.1007/s00417-009-1044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bagheri N, Bell BA, Bonilha VL, Hollyfield JG. Imaging human postmortem eyes with SLO and OCT. Adv Exp Med Biol. 2012;723:479–488. doi: 10.1007/978-1-4614-0631-0_60. [DOI] [PubMed] [Google Scholar]
- 35.Curcio CA, Zanzottera EC, Ach T, Balaratnasingam C, Freund KB. Activated retinal pigment epithelium, an optical coherence tomography biomarker for progression in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2017;58:BIO211–BIO226. doi: 10.1167/iovs.17-21872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim B-J, Sprehe N, Morganti A, Wordinger RJ, Clark AF. The effect of postmortem time on the RNA quality of human ocular tissues. Mol Vis. 2013;19:1290–1295. [PMC free article] [PubMed] [Google Scholar]
- 37.Curcio CA, Medeiros NE, Millican CL. The Alabama age-related macular degeneration grading system for donor eyes. Invest Ophthalmol Vis Sci. 1998;39:1085–1096. [PubMed] [Google Scholar]
- 38.Klein R, Davis MD, Magli YL, Segal P, Klein BE, Hubbard L. The Wisconsin age-related maculopathy grading system. Ophthalmology. 1991;98:1128–1134. doi: 10.1016/s0161-6420(91)32186-9. [DOI] [PubMed] [Google Scholar]
- 39.Davis MD, Gangnon RE, Lee L-Y, et al. The Age-Related Eye Disease Study severity scale for age-related macular degeneration: AREDS Report No. 17. Arch Ophthalmol. 2005;123:1484–1498. doi: 10.1001/archopht.123.11.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malik KJ, Chen C-D, Olsen TW. Stability of RNA from the retina and retinal pigment epithelium in a porcine model simulating human eye bank conditions. Invest Ophthalmol Vis Sci. 2003;44:2730–2735. doi: 10.1167/iovs.02-1120. [DOI] [PubMed] [Google Scholar]
- 41.Beach TG, Sue LI, Walker DG, et al. The Sun Health Research Institute brain donation program: description and experience, 1987–2007. Cell Tissue Bank. 2008;9:229–245. doi: 10.1007/s10561-008-9067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trabzuni D, Ryten M, Walker R, et al. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem. 2011;119:275–282. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sjöholm LK, Ransome Y, Ekström TJ, Karlsson O. Evaluation of post-mortem effects on global brain DNA methylation and hydroxymethylation. Basic Clin Pharmacol Toxicol. 2018;122:208–213. doi: 10.1111/bcpt.12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Age-Related Eye Disease Study Research Group. The Age-Related Eye Disease Study system for classifying age-related macular degeneration from stereoscopic color fundus photographs: the Age-Related Eye Disease Study Report Number 6. Am J Ophthalmol. 2001;132:668–681. doi: 10.1016/s0002-9394(01)01218-1. [DOI] [PubMed] [Google Scholar]
- 45.Bowes Rickman C, Ebright JN, Zavodni ZJ, et al. Defining the human macula transcriptome and candidate retinal disease genes using EyeSAGE. Invest Ophthalmol Vis Sci. 2006;47:2305–2316. doi: 10.1167/iovs.05-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pennington KL, DeAngelis MM. Epigenetic mechanisms of the aging human retina. J Exp Neurosci. 2016;9:51–79. doi: 10.4137/JEN.S25513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang Q, Dharmat R, Owen L, et al. Single-nuclei RNA-seq on human retinal tissue provides improved transcriptome profiling. bioRxiv. 2018. 468207. [DOI] [PMC free article] [PubMed]
- 48.Fritsche LG, Igl W, Bailey JNC, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen S, Jiang H, Bei Y, Xiao J, Li X. Long non-coding RNAs in cardiac remodeling. Cell Physiol Biochem. 2017;41:1830–1837. doi: 10.1159/000471913. [DOI] [PubMed] [Google Scholar]
- 50.Yang G, Yang L, Wang W, Wang J, Wang J, Xu Z. Discovery and validation of extracellular/circulating microRNAs during idiopathic pulmonary fibrosis disease progression. Gene. 2015;562:138–144. doi: 10.1016/j.gene.2015.02.065. [DOI] [PubMed] [Google Scholar]
- 51.Olivares AM, Jelcick AS, Reinecke J, et al. Multimodal regulation orchestrates normal and complex disease states in the retina. Sci Rep. 2017;7:690. doi: 10.1038/s41598-017-00788-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jun S, Datta S, Wang L, Pegany R, Cano M, Handa JT. The impact of lipids, lipid oxidation, and inflammation on AMD, and the potential role of miRNAs on lipid metabolism in the RPE. Exp Eye Res. 2018] doi: 10.1016/j.exer.2018.09.023. [published online ahead of print October 5, [DOI] [PMC free article] [PubMed]
- 53.Miyagishima KJ, Wan Q, Corneo B, et al. In pursuit of authenticity: induced pluripotent stem cell-derived retinal pigment epithelium for clinical applications. Stem Cells Transl Med. 2016;5:1562–1574. doi: 10.5966/sctm.2016-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saini JS, Corneo B, Miller JD, et al. Nicotinamide ameliorates disease phenotypes in a human iPSC model of age-related macular degeneration. Cell Stem Cell. 2017;20:635–647. doi: 10.1016/j.stem.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Micklisch S, Lin Y, Jacob S, et al. Age-related macular degeneration associated polymorphism rs10490924 in ARMS2 results in deficiency of a complement activator. J Neuroinflammation. 2017;14:4. doi: 10.1186/s12974-016-0776-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rossberger S, Ach T, Best G, Cremer C, Heintzmann R, Dithmar S. High-resolution imaging of autofluorescent particles within drusen using structured illumination microscopy. Br J Ophthalmol. 2013;97:518–523. doi: 10.1136/bjophthalmol-2012-302350. [DOI] [PubMed] [Google Scholar]
- 57.Trieschmann M, van Kuijk FJGM, Alexander R, et al. Macular pigment in the human retina: histological evaluation of localization and distribution. Eye Lond Engl. 2008;22:132–137. doi: 10.1038/sj.eye.6702780. [DOI] [PubMed] [Google Scholar]
- 58.Li M, Dolz-Marco R, Messinger JD, et al. Clinicopathologic correlation of anti-vascular endothelial growth factor-treated type 3 neovascularization in age-related macular degeneration. Ophthalmology. 2018;125:276–287. doi: 10.1016/j.ophtha.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 59.Litts KM, Messinger JD, Freund KB, Zhang Y, Curcio CA. Inner segment remodeling and mitochondrial translocation in cone photoreceptors in age-related macular degeneration with outer retinal tubulation. Invest Ophthalmol Vis Sci. 2015;56:2243–2253. doi: 10.1167/iovs.14-15838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Litts KM, Ach T, Hammack KM, et al. Quantitative analysis of outer retinal tubulation in age-related macular degeneration from spectral-domain optical coherence tomography and histology. Invest Ophthalmol Vis Sci. 2016;57:2647–2656. doi: 10.1167/iovs.16-19262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finger RP, Chong E, McGuinness MB, et al. Reticular pseudodrusen and their association with age-related macular degeneration: The Melbourne Collaborative Cohort Study. Ophthalmology. 2016;123:599–608. doi: 10.1016/j.ophtha.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 62.Sivaprasad S, Bird A, Nitiahpapand R, et al. Perspectives on reticular pseudodrusen in age-related macular degeneration. Surv Ophthalmol. 2016;61:521–537. doi: 10.1016/j.survophthal.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 63.Domalpally A, Clemons TE, Danis RP, et al. ; Writing Committee for the OPERA Study Peripheral retinal changes associated with age-related macular degeneration in the Age-Related Eye Disease Study 2: Report Number 12. Ophthalmology. 2017;124:479–487. doi: 10.1016/j.ophtha.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 64.Kaszubski PA, Ben Ami T, Saade C, et al. Changes in reticular pseudodrusen area in eyes that progressed from early to late age-related macular degeneration. Int Ophthalmol. 2018;38:503–511. doi: 10.1007/s10792-017-0485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zanzottera EC, Messinger JD, Ach T, Smith RT, Curcio CA. Subducted and melanotic cells in advanced age-related macular degeneration are derived from retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2015;56:3269–3278. doi: 10.1167/iovs.15-16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.