As part of the Biomedical Data Translator program, the National Center for Advancing Translational Sciences (NCATS) has assembled 11 teams comprising nearly 200 team members drawn from 28 institutions. Roughly 1 year into the feasibility phase, we describe the program's conception, the rapid coalescence of teams and team members, the novel mechanisms of interaction and communication, and the emergent collaborative culture and community that we believe are driving the early success of the program.
Overview and structure of the translator program
Clinical and translational research has reached an inflection point in the availability of clinical, environmental, socio‐environmental, and both mechanistic and phenomenological biomolecular data and the potential impact of such data on improving public health. Despite this ostensible availability, many such data remain siloed, unlinked, disorganized, and obscured by discipline‐specific differences in terminology and representation—a condition that has been termed the “Chasm of Semantic Despair.”1
To address these challenges, the Biomedical Data Translator Consortium (Table S1) has undertaken a feasibility assessment directed at the creation of a prototype “Translator” system capable of integrating existing biomedical data sets as “Knowledge Sources” and “translating” those data into insights to accelerate translational research, generate new hypotheses, and drive innovations in clinical care and drug discovery. A little over 1 year into the feasibility assessment, the program has made significant progress in the research and development of the prototype system. We believe that this success reflects, to a large extent, the ability of NCATS to rapidly establish a committed and united team within a program that involves 28 institutions and nearly 200 team members who are distributed geographically and who, frankly, have competing interests. Herein, we reflect on what we believe are the key enabling factors for this collaborative culture and community to develop.
Shared vision
A seemingly innocuous question frames the shared scientific vision of the Translator program: What is disease? At first glance, the answer seems straightforward; however, further reflection reveals that the answer is not so simple (cf., refs.2, 3). Indeed, this is the question that Dr. Christopher Austin, Director of NCATS, posed to team members at the kickoff meeting of the Translator program. The general argument is that, although biomedical research has unprecedentedly advanced, profound challenges remain, including limited translation of basic science and clinical observations into tangible improvements to human health; a failure‐prone and expensive research‐and‐development process for new clinical interventions; rising drug‐development costs; and low adoption rates for interventions that are demonstrably beneficial.4 The Translator program aims to address these challenges, in part, by promoting data‐driven clinical regrouping of patients to refine our conceptualization of disease by mechanistically grouping patients according to shared molecular and cellular biomarkers and who is likely to respond to specific interventions.
Translator team members have embraced the vision of the Translator program, and this adoption has fostered a collective communitarian spirit and desire to tackle important, challenging issues. This communitarian spirit was fostered by NCATS since the inception of the Translator program and continues to serve as a bedrock of program success.
Nimble funding mechanism and management approach
The Translator program is funded via an atypical funding mechanism: an Other Transaction Award (OTA).5 The OTA is not a contract, cooperative agreement, or grant. Rather, the OTA offers much greater flexibility and allows NCATS to engage traditional partners (i.e., academic institutions) in novel ways, engage nontraditional research partners (e.g., any adult applicant, regardless of professional affiliation or educational background), and negotiate terms and conditions that focus team efforts, spur innovation, facilitate collaborative problem solving, and support rapid changes in direction in response to project needs.
As an example of this nimbleness, the Translator program was initiated with five external teams and one internal NCATS team. After about a year, NCATS saw a need to accelerate progress in specific new directions. Thus, they rapidly brought in five new teams, enabled by the OTA, that were selected based on a truly innovative process: a series of challenge questions that applicants were required to “solve” over a 2 week period before being invited to submit a full application. The intent was to level the playing field between existing awardees and prospective new applicants and to vet the technical skills and commitment of prospective teams prior to soliciting full applications. Accepted applicants then competed by virtual interviews, during which each team demonstrated proof‐of‐concept software developed during the application process (i.e., at risk, prior to funding). Although this experience was described variously at the time by applicants as “crazy” and “novel,” the approach undeniably accelerated progress on the Translator program.
The OTA also requires regular review and approval of milestones and associated deliverables before release of each new round of funding. Notably, NCATS supports changes in direction if they are justified and in line with the vision of the program. This management approach has effectively propelled interdisciplinary team science. Indeed, the program has evolved at lightning speed relative to more traditional sources of federal funding (Figure 1).
Figure 1.
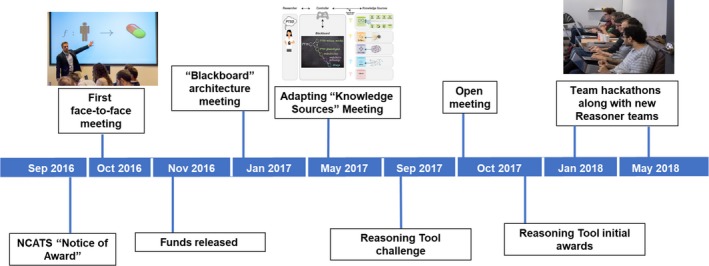
Timeline for the Translator program as of the May 2018 hackathon.
The speed at which the program has progressed, the high expectations of team members, and the unusual, bottom‐up management approach have encouraged self‐selection and self‐organization of teams, who simultaneously express enthusiasm, confusion, and exhaustion. In addition, although a shared vision and mission drive the work of Translator team members, a “fear‐of‐missing‐out” ethic and a creative tension stimulate team members to continue working intensely and collaboratively.
Collaborative culture and community
In order to realize the shared vision, the Translator program requires a team‐science approach, with highly coordinated teams and diverse backgrounds and skill sets. The National Institutes of Health, the National Research Council, and other scientific bodies are increasingly emphasizing the importance of team science in large‐scale research projects, in terms of fostering innovation and accelerating productivity.6, 7 Successful teams are associated with a variety of characteristics, including a shared vision, effective leadership, trust, open communication and debate, resolution of language barriers and conflict, and general enthusiasm for the scientific challenge driving the research.8 Although we did not formally assess team science in the context of the Translator program, our collective years of scientific experience support our assertion that the Translator program shares the key characteristics of successful team‐science initiatives and serves as an exemplar of effective team science. We describe below several key challenges and solutions that enabled a collaborative interdisciplinary culture and community to rapidly form within the program.
Challenges
NCATS and Translator team members realized early in the project that communication can be challenging among team members who do not necessarily share the same vocabularies and languages, both literally and with respect to subtleties and nuances across biomedical fields. Moreover, the integration of diverse personalities and varying interpersonal sensitivities presents formidable challenges, especially as most teams did not know each other at the start of the program yet were required to coordinate efforts and work quickly and collectively toward the shared vision. The steep learning curve and rapid speed at which the program has progressed present challenges in attrition and the need to maintain engaged team members throughout the term of the project. Logistical challenges also abound, including coordination and scheduling among team members who reside in multiple time zones and have different preferred forms of communication. A related challenge is the identification of the ideal number of working groups, as well as the size and composition of those groups. In part due to the speed with which the program has progressed, team members also have found it challenging to coordinate milestones and deliverables across teams and align the goals of the Translator program with the goals of their own non‐Translator research projects. Finally, the program does not have a top‐down structure, which represents both a strength and a challenge by necessitating self‐organization among team members who are often more comfortable and familiar with hierarchically structured programs.
Solutions
An initial “success” of the Translator program was the adoption of colors (red, orange, green, blue, purple, and grey) to name the six initial Translator teams at the outset of the project. This strategy was conceptualized by NCATS, but highly welcomed by Translator team members, as it equated teams and simplified discourse, particularly among teams comprising multiple institutions and principal investigators. A similar approach was taken for the five teams who later joined the program (alpha, gamma, infrared, ultraviolet, and X‐ray).
Trust also has been critical to the early success of the Translator program. Trust has been fostered, in part, by the fact that NCATS Translator staff themselves represent one of the six initial Translator teams. Moreover, while NCATS had expectations of certain teams and team members working together more closely than others, teams and team members were given the freedom to explore new collaborations within the confines of the larger Translator program and to allow collaborations to form organically. This freedom instilled enthusiasm among Translator team members and fostered trust and collaboration. In addition, NCATS has encouraged open and frank discussions and constructive criticism among all team members, including NCATS, and this encouragement has promoted trust and accelerated research and development of the prototype Translator system.
NCATS also has provided leadership and direction in a manner that is team‐oriented and not top‐down. NCATS has managed to balance its roles as both program leader and integrated team member by providing guidance and direction, while also acting as a partner and equal stakeholder focused on pursuing a shared goal. Specific examples of NCATS engagement include: designating an NCATS team member as liaison for each Translator team; hosting regular weekly teleconference meetings with each Translator team; arranging regular working‐group meetings; providing regular communications; posting regular “homework” assignments; and convening five all‐hands meetings over the first 16 months of the feasibility assessment. These meetings have proven crucial to set priorities as a group, report progress, and develop a shared action plan. Moreover, the meetings have not been rigidly structured and have been reorganized on an ad hoc basis, thus functioning more as “unconferences.”9
Frequent communication via multiple communication platforms also has been essential for the early success of the program. For instance, the program has used WebEx, Skype, Zoom, Email, Slack, Gitter, and other communication channels to foster communication and accommodate team‐member preferences and time zones. To illustrate these communication efficiencies, we report Slack activity by user and subject for the first year of the program across 30 Translator‐specific Slack channels, including 10 with substantial activity in 2018 (Figure 2).
Figure 2.
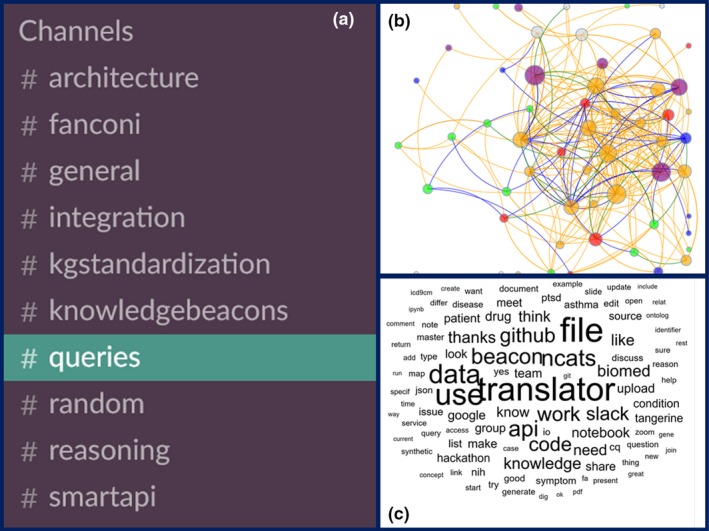
Slack communications among the five initial Translator teams. Data were captured for 30+ Slack channels, including 10 channels (A) with substantial ongoing activity as of Spring 2018. The communication network (B) depicts individual contributors, with node colors representing Translator team colors and edge colors representing different types of communication. Node size is proportional to the sum of the number of links and the number of broadcast messages associated with each node. For interactive exploration, please see the visualization. The word cloud (C) shows the 100 most frequently used words across Translator team communication messages on Slack over the same time period. The larger the word, the more frequently it was used.
A final factor that has fostered a collaborative interdisciplinary culture and sense of community has been the focus on use cases that span the disease‐frequency spectrum: Fanconi anemia, a rare disease, and asthma, a common disease. By strategically restricting the primary scientific focus of the feasibility assessment, Translator team members were able to maintain a singular focus on a common shared goal through targeted inquiries designed to advance understanding of the pathophysiology of disease and guide the design of the prototype Translator system. Moreover, the restricted focus on two use cases allowed team members to more rapidly overcome disciplinary language barriers and leverage the rich array of expertise, tools, and perspectives provided by team members from different disciplines, thereby gaining insights and identifying solutions to scientific and technical challenges that otherwise might not have been possible during the feasibility phase of the program. For example, team members worked collaboratively to identify creative ways of revealing clinical data in a manner that is regulatory compliant, openly accessible, and aligned with the technical constraints of the prototype Translator system. Likewise, team members with expertise in clinical practice, clinical data, ontologies, and analytics worked together to successfully tackle challenging use‐case questions that individual team members were unable to solve on their own. A final example of the benefit of collaborative interdisciplinary teams is the unique insights and shifts in perception that have been fostered by the integrated work of team members with expertise in different data sources (e.g., clinical data, environmental exposures data, and chemical data).
Moving forward
We assert that the Translator program owes its success to a combination of a shared vision, a nimble funding mechanism and management approach, and a culture and sense of community that are conducive to close interdisciplinary collaboration on a project that progresses rapidly and often changes direction. We believe that the processes used to enable the Translator program may serve as instructive guides to other multi‐institutional and multi‐disciplinary efforts, thereby enabling other teams to solve challenges that would be difficult to address using traditional funding mechanisms and a top‐down organizational structure.
Funding
Support for the preparation of this manuscript was provided by the National Center for Advancing Translational Sciences, National Institutes of Health, through the Biomedical Data Translator program (awards 1OT3TR002019, 1OT3TR002020, 1OT3TR002025, 1OT3TR002026, 1OT3TR002027, 1OT2TR002514, 1OT2TR002515, 1OT2TR002517, 1OT2TR002520, and 1OT2TR002584). Any opinions expressed in this document are those of The Biomedical Data Translator Consortium and do not necessarily reflect the views of the National Institutes of Health, individual Translator team members, or affiliated organizations and institutions.
Conflict of Interest
The authors declared no competing interests for this work.
Supporting information
Table S1. The Biomedical Data Translator Consortium: teams and team members.
Acknowledgments
Karamarie Fecho prepared the first draft of the manuscript and served as lead author. Paul A. Clemons served as senior author. Stanley C. Ahalt, Russ B. Altman, Noel Burtt, Christopher G. Chute, Paul A. Clemons, Michel Dumontier, Tricia Francis, Gustavo Glusman, Rajarshi Guha, Melissa Haendel, Maureen Hoatlin, Trey Ideker, Theo Knijnenburg, Matthew Might, David B. Peden, Alexander Tropsha, Mathias Wawer, and Chunhua Weng provided critical feedback and editorial input. NCATS staff created Figure 1; Paul A. Clemons edited the figure. Hong Yi generated the visualization displayed in Figure 2. Paul A. Clemons and Hong Yi created Figure 2. All members of The Biomedical Data Translator Consortium contributed to the vision of the manuscript and conducted the work described therein.
References
- 1. Chute, C.G. Crossing the chasm of semantic despair: integrating knowledge and data from science and clinical practice. Clinical Research Forum – IT Roundtable, Washington DC <https://c.ymcdn.com/sites/crforum.site-ym.com/resource/resmgr/docs/IT_Roundtable/CRF_IT_Roundtable_3Nov2017re.pdf> (November 2, 2017). Accessed June 21, 2018.
- 2. Scully, J.L. What is a disease? EMBO Rep. 5, 650–653 <https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1299105> (2004). Accessed June 21, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Manrai, A.K. , Patel, C.J. & Ioannidis, J.P.A. In the era of precision medicine and big data, who is normal? JAMA. Published online April 23, 2018. <https://jamanetwork.com/journals/jama/fullarticle/2679460> (2018). Accessed June 21, 2018. [DOI] [PMC free article] [PubMed]
- 4. Wagner, J.A. et al Application of a dynamic map for learning, communicating, navigating, and improving therapeutic development. Clin. Transl. Sci. 11, 166–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. National Center for Advancing Translational Science, National Institutes of Health, Department of Health and Human Services . Other Transaction Award Policy Guide <https://ncats.nih.gov/files/NCATS-Translator-OT-Policy-Guide.pdf> (April 28, 2016). Accessed June 21, 2018.
- 6. Office of Strategic Coordination – The Common Fund, National Institutes of Health . A decade of discovery. The NIH roadmap and common fund. Washington, DC: National Institutes of Health, NIH Pub. No. 14‐8013 <https://commonfund.nih.gov/sites/default/files/ADecadeofDiscoveryNIHRoadmapCF.pdf> (2014). Accessed June 21, 2018. [Google Scholar]
- 7. Committee on the Science of Team Science, National Research Council. Board on Behavioral, Cognitive, and Sensory Sciences, Division of Behavioral and Social Sciences and Education . Enhancing the effectiveness of team science. (eds. Cooke N.J. Hilton M.L.) Washington, DC: The National Academies Press; <https://www.nap.edu/catalog/19007/enhancing-the-effectiveness-of-team-science> (2015). Accessed June 21, 2018. [PubMed] [Google Scholar]
- 8. Bennett, L.M. & Gadlin, H. Collaboration and team science: from theory to practice. J. Investig. Med. 60, 768–775 <https://www.ncbi.nlm.nih.gov/pubmed/22525233> (2012). Accessed June 21, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Budd, A. et al Ten simple rules for organizing an unconference. PLoS Comput. Biol. 11, e1003905 <https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4310607/> (2015). Accessed June 21, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Slack visualization <https://xdciviz.renci.org/translator/teamscience/slackviz> Accessed June 21, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The Biomedical Data Translator Consortium: teams and team members.


