Abstract
Eukaryotic genomes are organized into chromatin, divided into structurally and functionally distinct euchromatin and heterochromatin compartments. The high level of compaction and the abundance of repeated sequences in heterochromatin pose multiple challenges for the maintenance of genome stability. Cells have evolved sophisticated and highly controlled mechanisms to overcome these constraints. Here, we summarize recent findings on how the heterochromatic state influences DNA damage formation, signaling and repair. By focusing on distinct heterochromatin domains in different eukaryotic species, we highlight heterochromatin contribution to the compartmentalization of DNA damage repair in the cell nucleus and to repair pathway choice. We also describe the diverse chromatin alterations associated with the DNA damage response in heterochromatin domains and present our current understanding of their regulatory mechanisms. Finally, we discuss the biological significance and the evolutionary conservation of these processes.
Keywords: chromatin reorganization, DNA damage repair, heterochromatin, nuclear domains
Introduction
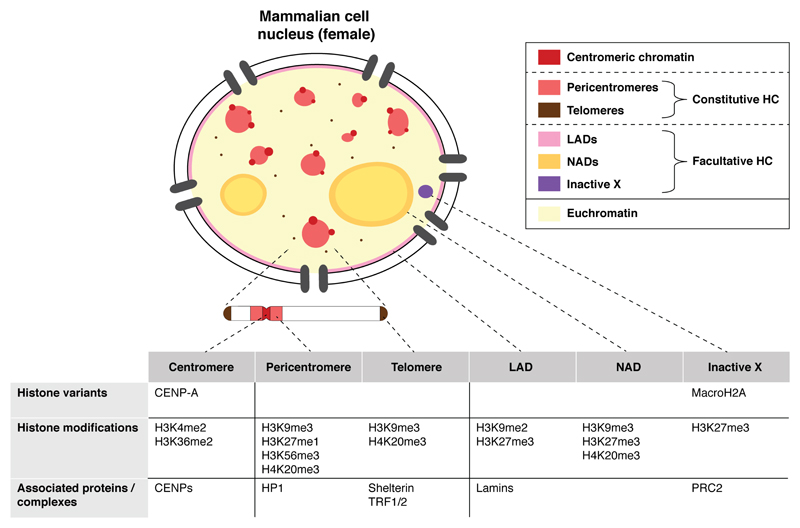
In eukaryotic cell nuclei, the genetic information is packaged in the form of chromatin (Kornberg 1977) where DNA wraps around histone proteins to form nucleosomes (Luger et al. 1997) and higher-order structures (Bonev and Cavalli 2016). The different levels of chromatin organization are central to cell function as they constitute key vectors of epigenetic information, which dictates cell identity (Allis and Jenuwein 2016). Among higher-order chromatin structures, heterochromatin domains are critical chromatin compartments with a major influence on chromosome segregation and stability (Allshire and Madhani 2017). Originally defined as chromosomal regions that remain compact throughout the cell cycle (Heitz 1928), heterochromatin domains are generally gene-poor, mostly transcriptionally silent and are characterized by specific sets of histone modifications and associated proteins. Recent advances in super-resolution microscopy have provided a refined three-dimensional picture of chromatin in vivo at nanoscale resolution, revealing that heterochromatin domains are formed by larger, denser and less mobile nucleosome clutches compared to euchromatin (Ricci et al. 2015; Nozaki et al. 2017; Ou et al. 2017). Beyond these general features, heterochromatin actually exists in various forms that are structurally and functionally distinct: while constitutive heterochromatin remains condensed and mostly transcriptionally silent throughout development and cell divisions (Saksouk et al. 2015), facultative heterochromatin corresponds to regions of the genome where gene silencing is dynamically regulated (Trojer and Reinberg 2007). A typical example of facultative heterochromatin is the inactive X chromosome in female mammals (Gendrel and Heard 2014), but it also includes genomic regions that interact with specific nuclear structures, such as the lamina-associated domains (LADs) located at the nuclear periphery (van Steensel and Belmont 2017) and nucleolus-associated domains (NADs; Matheson and Kaufman 2016). Constitutive heterochromatin is found at subtelomeric regions (Schoeftner and Blasco 2009) and at pericentromeres (Saksouk et al. 2015), which surround repetitive centromeric DNA (McKinley and Cheeseman 2016). Each of these heterochromatin domains is defined epigenetically by specific histone post-translational modifications, histone variants and associated proteins (Fig. 1), in addition to DNA methylation, which contributes to transcriptional silencing.
Figure 1. Main heterochromatin domains and their distinctive features in mammalian cells.
Constitutive and facultative heterochromatin domains are depicted and their characteristic histone variants, modifications and associated proteins are listed. Although it is not heterochromatin per se, we also consider centromeric chromatin, which is rich in repetitive sequences and surrounded by constitutive heterochromatin domains. CENP: centromere protein, HC: heterochromatin, HP1: heterochromatin protein 1, LAD: lamina-associated domain, NAD: nucleolus-associated domain, PRC2: polycomb repressive complex 2, TRF1/2: telomeric repeat binding factor 1/2.
In recent years, a growing number of studies focused on understanding how heterochromatin domains are established during development and then perpetuated through replication and cell division. Another major challenge for heterochromatin maintenance is the response to DNA damage, which poses a constant threat to both genome and epigenome stability (Dabin et al. 2016). Furthermore, with the exception of LADs, heterochromatin is highly enriched for repetitive sequences, including tandem satellite sequences and transposable elements (Padeken et al. 2015), which compromises faithful DNA replication and repair, with a risk of aberrant homologous recombination between ectopic repeats leading to chromosome rearrangements and aneuploidy (Peng and Karpen 2008). Silencing of transposable elements through heterochromatinization is also critical for genome stability (Padeken et al. 2015). The issue of genome and epigenome maintenance is thus particularly prominent in heterochromatin.
Here, we review recent advances in our understanding of DNA damage formation, signaling and repair in heterochromatin domains, and describe heterochromatin reorganization associated with the DNA damage response. We focus mainly on the response to DNA double-strand breaks (DSBs) and UV photoproducts in diverse eukaryotic cell systems, including yeast, Drosophila and mammalian cells. We highlight that even though they share common features, not all heterochromatin domains are treated equal following a genotoxic stress challenge.
DNA damage formation in heterochromatin domains
Chromatin organization in the cell nucleus has a significant impact on the DNA damage response, from damage formation to repair. Indeed, chromatin loops were recently identified as a source of topoisomerase 2-mediated DNA breaks in mammalian cells, putting forward chromatin organization as a major driver of genome fragility (Canela et al. 2017). Heterochromatin organization in particular markedly impacts genome stability, as illustrated by higher mutation rates in human cancer cells, both in constitutive (Schuster-Böckler and Lehner 2012) and facultative heterochromatin (Jäger et al. 2013). Furthermore, mutation patterns strongly associate with nuclear organization, with heterochromatin at the nuclear periphery, LADs in particular, displaying higher mutation frequencies in various cancer types (Smith et al. 2017). These studies suggest that DNA damage formation and/or repair is influenced by higher-order chromatin organization in the cell nucleus. Over the last few years, several studies have addressed how tridimensional chromatin organization and compaction affect the susceptibility of DNA to damage. In vitro manipulation of chromatin compaction by adjusting magnesium concentration on permeabilized human nuclei and on mitotic chromosomes revealed that the levels of DSBs induced by ionizing radiation in compact chromatin were 5 to 50-fold lower than in decondensed chromatin, implying that chromatin compaction protects genomic DNA from radiation damage (Takata et al. 2013). The question of DSB generation in different chromatin domains was then tackled in vivo both in mouse and human cells. For this, several genome-wide approaches were developed for mapping DSBs across the genome at single-nucleotide resolution, including BLESS (Crosetto et al. 2013), END-seq (Canela et al. 2016) and DSBCapture (Lensing et al. 2016), which established the higher susceptibility of transcriptionally active euchromatin to endogenous DSB formation. In contrast, breaks induced by aphidicolin were enriched in centromeric and pericentromeric chromatin, most likely reflecting the higher sensitivity of DNA repeats to replication stress. Mechanistic insights into how heterochromatin may hinder endogenous break induction are still lacking. The low levels of transcription in heterochromatin may preserve this chromatin compartment from transcription-induced genome instability {Gaillard:2016fw}. In terms of molecular players, a recent study in Drosophila put forward linker histone H1 as preventing the accumulation of R-loop-induced DNA damage in heterochromatin (Bayona-Feliu et al. 2017). Further work is still needed to fully dissect the mechanisms that control DSB distribution between euchromatin and heterochromatin domains.
While the genome-wide distribution of DSBs is established, contrasting reports continue to emerge regarding the formation of UV-induced DNA lesions in mammalian genomes. Single-nucleotide resolution mapping of cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6-4) pyrimidone photoproducts (6-4PPs) by HS-Damage-seq in UV-irradiated human fibroblasts (Hu et al. 2017) showed that the distribution of both types of UV lesions was essentially uniform throughout the genome. In contrast, a concomitant study using a similar genome-wide mapping approach in human fibroblasts showed that lamina-associated heterochromatin at the nuclear periphery was more vulnerable to UV damage than euchromatin (García-Nieto et al. 2017). Furthermore, immunofluorescence-based detection of UV damage revealed that 6-4PP were excluded from pericentromeric heterochromatin in mouse fibroblasts (Han et al. 2016), suggesting that the highly condensed heterochromatin environment may interfere with the formation of some UV lesions. Thus, it is not yet entirely clear whether the UV mutation signature observed in human cancer cells (Smith et al. 2017) results from higher damage formation or from slower repair in heterochromatin. Therefore, the role of nuclear organization and chromatin compaction on DNA damage formation remains an important field of study with broad implications for our understanding of genome stability and mutational landscapes.
Impact of heterochromatin on DNA damage signaling
One of the earliest consequences of DNA damage infliction is the recruitment of DNA damage signaling kinases, which initiates a complex cascade of events leading to cell cycle checkpoint activation. Among the many targets of these kinases, the histone variant H2A.X gets rapidly phosphorylated in large chromatin domains surrounding DSBs, giving rise to γH2A.X foci (Rogakou et al. 1998), which serve as a platform for recruiting downstream checkpoint and repair factors (Smeenk and van Attikum 2013). While this is a general response to DNA damage, several studies in yeast and mammalian cells originally showed that silenced chromatin domains were refractory to H2A.X phosphorylation (Cowell et al. 2007; Kim et al. 2007) and hampered DNA damage checkpoint signaling (Brunton et al. 2011). However, closer examination of the DDR in a time-resolved fashion later showed that H2A.X (H2A.v in Drosophila) was phosphorylated within pericentromeric heterochromatin domains in mouse and Drosophila cells, while subsequent steps of damage signaling occurred outside heterochromatin domains after a relocation of the breaks to the periphery of the domains (Chiolo et al. 2011; Jakob et al. 2011; Tsouroula et al. 2016; Janssen et al. 2016) or even to the nuclear periphery in Drosophila cells (Ryu et al. 2015; Ryu et al. 2016). Noteworthy, such relocation specifically affects DSBs repaired by recombination, as discussed in the following sections. In plant cells, the situation is more complex with the existence of a heterochromatin-specific histone variant H2A.W.7, which is phosphorylated in response to damage, while H2A.X phosphorylation takes place primarily in euchromatin (Lorković et al. 2017).
Furthermore, dynamic chromatin compaction appears to play an important regulatory role in DNA damage signaling. Indeed, tethering heterochromatin factors to a LacO array in the absence of DNA damage in human cells induces local chromatin condensation and is sufficient to activate early steps in DNA damage signaling but not downstream effectors (Burgess et al. 2014). While the exact molecular mechanism by which chromatin condensation initiates early damage signaling is unknown, it might involve the repressive histone mark H3K9me3 and its ability to stimulate the acetyltransferase activity of Tip60, which then contributes to the activation of the DNA damage signaling kinase ataxia-telangiectasia mutated (ATM) (Sun et al. 2005; Sun et al. 2009). However, these assumptions are based on studies performed in euchromatin domains, and further studies are needed to clarify the role of heterochromatin compaction in damage signaling.
Altogether, these studies demonstrate that heterochromatin is permissive for DNA damage signaling and that heterochromatin features including histone marks and chromatin compaction exert a positive role in response to DNA damage by contributing to checkpoint activation.
Impact of heterochromatin on DNA repair efficiency
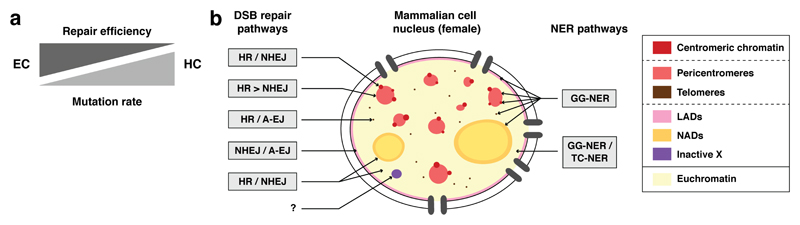
In the highly compartmentalized eukaryotic nucleus, both the chromatin state and the nuclear position of DNA lesions have a significant impact on repair pathway choice and repair efficiency (Kalousi and Soutoglou 2016). In this regard, compact heterochromatin domains may be seen as a barrier to repair factor recruitment, underlying higher mutation rates (Fig. 2a). Indeed, it was observed that excision of CPDs, the main UV photoproducts repaired by the nucleotide excision repair (NER) pathway (Marteijn et al. 2014), was significantly slower in H3K9me3-containing chromatin in human cells (Han et al. 2016). Recently, a high-throughput sequencing method, known as XR-seq, was used to analyze oligonucleotide fragments excised during NER in UV-irradiated fibroblasts, further establishing the slower repair associated with heterochromatin regions (Adar et al. 2016). Furthermore, transcription-coupled NER does not operate in poorly transcribed heterochromatin domains. These differences in NER efficiency underly cancer-associated mutagenesis, with an increased mutation density in heterochromatin regions and a reduced mutation rate in euchromatin that is abrogated by loss-of-function of NER factors (Polak et al. 2014; Zheng et al. 2014). Similarly, a lower efficiency of mismatch repair (Jiricny 2013) contributes to higher mutation rates in heterochromatin (Supek and Lehner 2015). Although early steps of DNA break repair proceed efficiently in pericentromeric heterochromatin (Chiolo et al. 2011; Jakob et al. 2011) slower DSB repair has been observed at chromocenters in mouse cells, where, about 25% of radiation-induced DSBs are repaired with slow kinetics and they predominantly localize at the vicinity of pericentromeric heterochromatin domains (Goodarzi et al. 2008). In contrast, sequence-specific DSBs induced by the I-SceI endonuclease in Drosophila are repaired with similar kinetics in euchromatin and pericentromeric heterochromatin (Janssen et al. 2016). This may reflect differences between species or between DNA ends, radiation-induced breaks requiring more processing than endonuclease-induced breaks. In the future, DSB genome-wide mapping techniques (Crosetto et al. 2013; Canela et al. 2016; Lensing et al. 2016) will be instrumental for analyzing DSB repair efficiency and pathway choice in distinct chromatin compartments.
Figure 2. DNA damage repair in heterochromatin domains.
a Balance between repair efficiency and mutation rates in euchromatin (EC) and heterochromatin (HC). b Compartmentalization of DNA double-strand break (DSB) repair and nucleotide excision repair (NER) pathways in the mammalian cell nucleus. A-EJ: alternative end-joining, GG-NER: global genome NER, HR: homologous recombination, LAD: lamina-associated domain, NAD: nucleolus-associated domain, NHEJ: non-homologous end-joining, TC-NER: transcription-coupled NER.
Impact of heterochromatin on DNA repair pathway choice
In line with the heterogeneity of the eukaryotic nucleus, there are regional differences in DNA repair pathways between euchromatin and heterochromatin compartments. Heterochromatin being mostly transcriptionally silent, global genome NER (GG-NER) is predominant over transcription-coupled NER (TC-NER) in heterochromatin regions with a major role of the GG-NER factor DNA damage binding protein 2 (DDB2) in promoting CPD removal from H3K9me3-containing chromatin (Han et al. 2016). Heterochromatin is also a major determinant in the regulation of DSB repair outcome (Fig. 2b). Repair of genomic DSBs is achieved either by homology-based pathways, i.e. error-free homologous recombination (HR) and mutagenic single strand annealing (SSA), or by non-homologous end joining (NHEJ), with alternative end joining (A-EJ) serving as a back-up (Mladenov et al. 2016). The repetitive nature of heterochromatin increases the risk of illegitimate recombination during repair. Therefore, a tight control of recombination events is critical in these domains. In particular, the silenced chromatin state plays a key role in repressing mitotic recombination at centromeres and telomeres, as revealed in DNA methyltransferase (DNMT)-deficient mouse cells showing increased telomeric and centromeric recombination accompanied by changes in centromere and telomere repeat length (Gonzalo et al. 2006; Jaco et al. 2008). This suggests that prevention of illicit recombination in these compartments is important to maintain centromere and telomere integrity. Likewise, telomere hyper-recombination and subsequent chromosomal fusions in mouse embryonic stem cells are prevented by the telomere-associated protein Rif1, which mediates heterochromatic silencing by maintaining H3K9me3 levels at subtelomeric regions (Dan et al. 2014). The importance of the silenced chromatin state in controlling recombination has also been observed in Drosophila cells, where completion of recombinatorial repair requires a SUMO-dependent relocation of DSBs outside H3K9me2- and HP1a-containing domains (Chiolo et al. 2011; Ryu et al. 2015; Ryu et al. 2016). Similarly, in budding yeast, silent information regulators (Sir) inhibit recombinational repair in silenced chromatin domains (Sinha et al. 2009). Interestingly, this inhibition is relieved through the eviction of Sir3p by the SWI/SNF chromatin remodeler (Sinha et al. 2009), suggesting that the constraints on recombinational repair in silenced chromatin can be alleviated by the action of chromatin remodelers. Similar to mitotic recombination, meiotic recombination is also repressed in silenced chromatin, as observed in fission yeast centromeres (Ellermeier et al. 2010). Furthermore, when recombination happens in silenced chromatin, error-free repair pathways are promoted. In budding yeast for instance, subtelomeric Sir3p-repressed chromatin promotes HR by inhibiting excessive DNA-end resection (Batté et al. 2017), and in fission yeast centromeric chromatin Rad51-dependent HR is favored over SSA (Zafar et al. 2017). Heterochromatic DSBs also rely largely on HR for their repair in G2 mouse cells (Beucher et al. 2009) and in Drosophila cultured cells, where pericentromeric heterochromatin appears to be largely repaired through Rad51-dependent HR (Chiolo et al. 2011; Tsouroula et al. 2016). Yet, in fly tissues, which are mostly in G1, NHEJ predominates over HR in pericentromeric heterochromatin (Janssen et al. 2016). Several studies have provided mechanistic insights into how DSB repair could be regulated in heterochromatin based on the involvement of heterochromatin-associated factors in euchromatin repair (Lemaître and Soutoglou 2014). In particular, heterochromatin protein 1 (HP1) has been identified as a main player in the control of DNA-end resection and shown to operate through the recruitment of Breast Cancer 1 (BRCA1) (Baldeyron et al. 2011; Soria and Almouzni 2013; Lee et al. 2013). In addition to HP1, other heterochromatin-associated factors function with BRCA1 in controlling resection, including the histone H3K9 methyltransferases SET Domain Bifurcated 1 (SETDB1) and Suppressor of Variegation 3-9 Homolog (Suv39H1/2) (Alagoz et al. 2015). Another important player in the repair of heterochromatic DSBs is p53-binding protein 1 (53BP1) (Noon et al. 2010; Kakarougkas et al. 2013). In line with this, Suppressor Of Cancer Cell Invasion (SCAI) has been identified as a 53BP1- and HP1-associated factor that promotes repair of heterochromatic DSBs by facilitating ATM-dependent signaling (Hansen et al. 2016). Together, this intricate network of molecular players is critical for preventing unscheduled repair, thus suppressing mutagenic events in heterochromatin domains.
Heterochromatin domains and compartmentalization of DNA repair
Not all heterochromatin domains have the same impact on repair pathway choice, resulting in a compartmentalization of DNA repair within the eukaryotic nucleus. This has been extensively studied in response to DSBs (Fig. 2b), by tethering DSBs to defined heterochromatin compartments (Lemaître et al. 2014) or by targeted introduction of DSBs into repetitive sequences (Torres-Rosell et al. 2007; van Sluis and McStay 2015; Harding et al. 2015; Tsouroula et al. 2016; Doksani and de Lange 2016). Thus, important differences have emerged regarding how DSBs are processed in distinct silenced chromatin compartments. In mouse cells, both centromeric and pericentromeric DSBs are repaired through HR and NHEJ, but HR is restricted to S/G2 for DSBs arising in pericentromeric heterochromatin while centromeric DSBs recruit the HR factor RAD51 throughout interphase (Tsouroula et al. 2016). Future work will address the molecular bases of these differences by assessing the importance of centromere specific histone variant and histone modifications in allowing HR of centromeric DSBs in G1 cells. Furthermore, NHEJ repair occurs inside centromeric and pericentromeric chromatin domains in mouse cells as opposed to late steps of HR, which are confined to the periphery of these domains after a relocation of the breaks (Tsouroula et al. 2016). In contrast to what observed at centromeres and pericentromeres, NHEJ does not contribute to repair of telomeric DSBs, which are processed by HR and A-EJ in mouse embryonic fibroblasts (Doksani and de Lange 2016). Stricking differences are also found among heterochromatin domains interacting with nuclear structures, with LADs being repaired by error-prone NHEJ and A-EJ (Lemaître et al. 2014), whereas nucleolar DSBs are repaired within NADs by NHEJ and HR (Torres-Rosell et al. 2007; van Sluis and McStay 2015; Harding et al. 2015). The DSB repair pathways that operate in other facultative heterochromatin domains like the inactive X chromosome still remain to be characterized. Future studies will also be needed to fully understand the molecular determinants and the biological relevance of such compartmentalization of DSB repair in the eukaryotic cell nucleus for genome and epigenome stability.
Heterochromatin reorganization in response to DNA damage
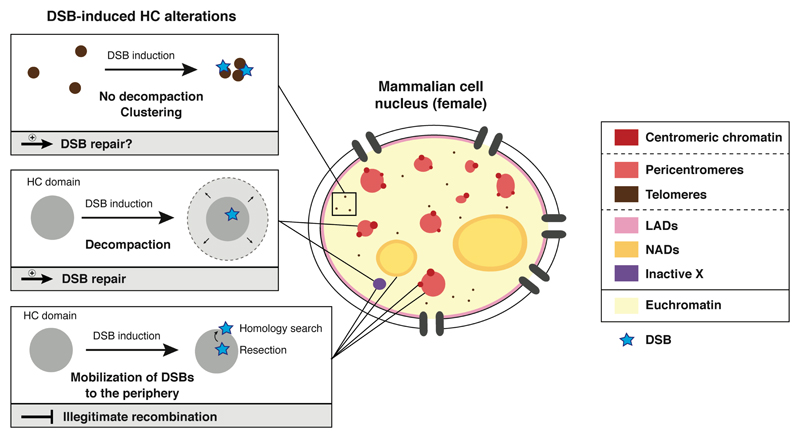
The DNA damage response is accompanied by a marked reorganization of heterochromatin (Fig. 3). In particular, decondensation of damaged heterochromatin has been observed in response to radiation- and nuclease-induced breaks, as reported for pericentromeric heterochromatin in flies (Chiolo et al. 2011) and in mouse embryonic fibroblasts (Jakob et al. 2011; Tsouroula et al. 2016), and for the inactive X chromosome in female human fibroblasts (Müller et al. 2013). This is thus a conserved response between eukaryotic species affecting both constitutive and facultative heterochromatin compartments. Future studies will address whether this is also a general response to various types of DNA lesions besides DSBs. Remarkably, the decompaction of damaged heterochromatin is not accompanied by a detectable loss of heterochromatin-specific histone marks such as H3K9me3 and H4K20me3 at the pericentromere, suggesting that heterochromatin identity may be preserved during this process (Goodarzi et al. 2011; Tsouroula et al. 2016; Natale et al. 2017). Nevertheless, more in depth studies are needed to fully characterize the local changes in histone marks upon DNA damage in constitutive heterochromatin domains. Whether facultative heterochromatin marks are maintained also remains to be determined. Notably, however, the response to DNA damage in heterochromatin is not always associated with chromatin decondensation as recently reported for uncapped telomeres (Timashev et al. 2017; Vancevska et al. 2017).
Figure 3. Heterochromatin reorganization in response to DNA damage.
Main alterations of heterochromatin domains in response to DNA double-strand breaks (DSBs, blue stars) and functional relevance. HC: heterochromatin, LAD: lamina-associated domain, NAD: nucleolus-associated domain.
Indeed, super-resolution imaging reveals that the DNA damage response elicited by removal of shelterin components occurs without substantial telomere decompaction, but is accompanied by telomere clustering. Understanding the molecular mechanisms that trigger heterochromatin decompaction in response to DNA damage may clarify the differences observed between distinct heterochromatin domains.
Among the mechanisms that may drive damaged heterochromatin decompaction, ATM-dependent phosphorylation of the heterochromatin building factor KRAB-domain associated protein 1 (KAP1) was shown to trigger euchromatin relaxation (Ziv et al. 2006) and to facilitate the repair of heterochromatic DSBs at mammalian pericentromeres (Goodarzi et al. 2008). KAP1 phosphorylation indeed results in dissociation of the chromatin remodeler Chromodomain Helicase DNA Binding Protein 3 (CHD3) (Goodarzi et al. 2011), allowing the opposing imitation switch (ISWI) remodeler to promote chromatin relaxation (Klement et al. 2014). In addition to KAP1 phosphorylation, desumoylation of KAP1 by the SUMO1/Sentrin Specific Peptidase 7 (SENP7) also regulates this pathway (Garvin et al. 2013).
Besides chromatin decompaction, another striking feature of the response to DNA damage in heterochromatin domains is the relocation of DNA lesions (Amaral et al. 2017). Indeed, the decompaction of damaged heterochromatin at pericentromeres (Chiolo et al. 2011; Janssen et al. 2016) and the inactive X (Müller et al. 2013) is accompanied with a relocation of DSBs to the periphery of heterochromatin domains and to the nuclear periphery in Drosophila. Notably, a similar relocation of DSBs has been observed at centromeric chromatin (Tsouroula et al. 2016) and nucleoli (Torres-Rosell et al. 2007; van Sluis and McStay 2015; Harding et al. 2015), DSBs being repaired by HR at the periphery of the domains. The mechanisms underlying the relocation of pericentromeric DSBs have been extensively investigated. It has been shown that DSB relocation relies at least in part on the activation of DNA damage checkpoint kinases in Drosophila and requires functional DNA end resection both in Drosophila and mouse cells (Chiolo et al. 2011; Tsouroula et al. 2016). The molecular details of how resection drives DSB mobility are still elusive. In this respect, it would be important to examine the possible contribution of chromatin remodeling factors, which promote DSB mobility in yeast (Dion and Gasser 2013). Moreover, in light of recent studies involving nuclear actin and myosin in the DNA damage response (Belin et al. 2015; Lottersberger et al. 2015; Kulashreshtha et al. 2016; Aymard et al. 2017), it will be interesting to investigate the role of cytoskeletal and motor proteins in this process. Relocation of DSBs also involves demethylation of the heterochromatin mark H3K56me3 by the Lysine Demethylase 4A (KDM4A) in Drosophila cells (Colmenares et al. 2017). Despite the strong similarities between model organisms regarding the mobility of heterochromatic DSBs, there are also mechanistic discrepancies, with pericentromeric DSBs being ultimately relocated to the nuclear periphery in Drosophila cells (Ryu et al. 2015; Ryu et al. 2016), which so far has not been observed in mouse cells (Tsouroula et al. 2016). In addition, exclusion of the RAD51 recombinase from heterochromatin domains is dependent on HP1 and Structural Maintenance of Chromosomes (SMC) 5/6 in Drosophila (Chiolo et al. 2011) and not in mouse cells (Tsouroula et al. 2016). Functionally, the dynamic relocation of DSBs resulting in their extrusion from heterochromatin domains is thought to be critical for the prevention of illegitimate recombination between heterochromatic repeats through a spatial separation between DNA end resection and homology search (Fig. 3).
Even though heterochromatin is markedly reorganized in response to DNA damage to control and facilitate repair, somehow surprisingly, chromatin silencing components including HP1 and H3K9me2/3 appear to accumulate at euchromatic damage sites. In particular, the heterochromatin component HP1 is required for DNA repair and is mobilized in response to DNA damage, being recruited to both UV- and laser-induced DNA lesions in a H3K9me3-independent manner in mammalian cells (Luijsterburg et al. 2009; Dinant and Luijsterburg 2009; Baldeyron et al. 2011) HP1 is loaded at DSBs together with the Suv39H1 methyltransferase, which deposits H3K9me3 resulting in local spreading of silencing marks spanning several kilobases around DSBs (Ayrapetov et al. 2014). Interestingly, deposition of silencing epigenetic marks is also favored at sites of replication stress, although the underlying mechanisms are not fully elucidated yet (Nikolov and Taddei 2015). The deposition of silencing marks at euchromatic DSBs was proposed to promote DNA damage signaling (Ayrapetov et al. 2014) and may also contribute to transcriptional silencing in response to DNA damage (Capozzo et al. 2017).
Conclusions and perspectives
DNA lesions arise in all chromatin compartments and among them compact heterochromatin domains pose major constraints to DNA damage repair. In recent years, exciting progress has been made in understanding how heterochromatin regulates DNA damage formation, signaling and repair, with the characterization of repair pathways operating in distinct heterochromatin domains. Recent studies have also identified important heterochromatin alterations that accompany the DNA damage response. However, mechanistic insights into the reorganization of damaged heterochromatin are still missing, and their functional relevance is not yet completely understood. Most importantly, whether and how the original heterochromatin state is restored after DNA damage repair is still an open question. Future studies will address this important issue and dissect the mechanisms for heterochromatin maintenance following genotoxic stress. This may also shed new light on heterochromatin instability associated with tumorigenesis and on the heterochromatin alterations that arise during cellular aging (Criscione et al. 2016).
Acknowledgements
Work in S.E.P. lab is supported by the European Research Council under Grant ERC-2013-StG-336427 “EpIn”; “Who am I?” laboratory of excellence under Grant ANR-11-LABX-0071. A.F. is funded by an international PhD fellowship from Sorbonne Paris Cité University.
Footnotes
Compliance with ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Adar S, Hu J, Lieb JD, Sancar A. Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc Natl Acad Sci USA. 2016;13:E2124–33. doi: 10.1073/pnas.1603388113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagoz M, Katsuki Y, Ogiwara H, et al. SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells. Nucleic Acids Res. 2015;43:7931–7944. doi: 10.1093/nar/gkv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Madhani HD. Ten principles of heterochromatin formation and function. Nat Rev Mol Cell Biol. 2017 doi: 10.1038/nrm.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral N, Ryu T, Li X, Chiolo I. Nuclear Dynamics of Heterochromatin Repair. Trends Genet. 2017;33:86–100. doi: 10.1016/j.tig.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F, Aguirrebengoa M, Guillou E, et al. Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat Struct Mol Biol. 2017;24:353–361. doi: 10.1038/nsmb.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, et al. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci USA. 2014;111:9169–9174. doi: 10.1073/pnas.1403565111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldeyron C, Soria G, Roche D, et al. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batté A, Brocas C, Bordelet H, et al. Recombination at subtelomeres is regulated by physical distance, double-strand break resection and chromatin status. EMBO J. 2017;36:2609–2625. doi: 10.15252/embj.201796631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayona-Feliu A, Casas-Lamesa A, Reina O, et al. Linker histone H1 prevents R-loop accumulation and genome instability in heterochromatin. Nat Commun. 2017;8:283. doi: 10.1038/s41467-017-00338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Lee T, Mullins RD. DNA damage induces nuclear actin filament assembly by Formin-2 and Spire-½ that promotes efficient DNA repair. Elife. 2015;4:e07735. doi: 10.7554/eLife.07735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, et al. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17:661–678. doi: 10.1038/nrg.2016.112. [DOI] [PubMed] [Google Scholar]
- Brunton H, Goodarzi AA, Noon AT, et al. Analysis of human syndromes with disordered chromatin reveals the impact of heterochromatin on the efficacy of ATM-dependent G2/M checkpoint arrest. Mol Cell Biol. 2011;31:4022–4035. doi: 10.1128/MCB.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RC, Burman B, Kruhlak MJ, Misteli T. Activation of DNA damage response signaling by condensed chromatin. Cell Rep. 2014;9:1703–1717. doi: 10.1016/j.celrep.2014.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Jung S, et al. Genome Organization Drives Chromosome Fragility. Cell. 2017;170:507–521.e18. doi: 10.1016/j.cell.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Sridharan S, Sciascia N, et al. DNA Breaks and End Resection Measured Genome-wide by End Sequencing. Mol Cell. 2016;63:898–911. doi: 10.1016/j.molcel.2016.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzo I, Iannelli F, Francia S, d'Adda di Fagagna F. Express or repress? The transcriptional dilemma of damaged chromatin. FEBS J. 2017;284:2133–2147. doi: 10.1111/febs.14048. [DOI] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Swenson JM, Langley SA, et al. Drosophila Histone Demethylase KDM4A Has Enzymatic and Non-enzymatic Roles in Controlling Heterochromatin Integrity. Dev Cell. 2017;42:156–169.e5. doi: 10.1016/j.devcel.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell IG, Sunter NJ, Singh PB, et al. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscione SW, Teo YV, Neretti N. The Chromatin Landscape of Cellular Senescence. Trends Genet. 2016;32:751–761. doi: 10.1016/j.tig.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Mitra A, Silva MJ, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabin J, Fortuny A, Polo SE. Epigenome Maintenance in Response to DNA Damage. Mol Cell. 2016;62:712–727. doi: 10.1016/j.molcel.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J, Liu Y, Liu N, et al. Rif1 maintains telomere length homeostasis of ESCs by mediating heterochromatin silencing. Dev Cell. 2014;29:7–19. doi: 10.1016/j.devcel.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant C, Luijsterburg MS. The emerging role of HP1 in the DNA damage response. Mol Cell Biol. 2009;29:6335–6340. doi: 10.1128/MCB.01048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V, Gasser SM. Chromatin movement in the maintenance of genome stability. Cell. 2013;152:1355–1364. doi: 10.1016/j.cell.2013.02.010. [DOI] [PubMed] [Google Scholar]
- Doksani Y, de Lange T. Telomere-Internal Double-Strand Breaks Are Repaired by Homologous Recombination and PARP1/Lig3-Dependent End-Joining. Cell Rep. 2016;17:1646–1656. doi: 10.1016/j.celrep.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C, Higuchi EC, Phadnis N, et al. RNAi and heterochromatin repress centromeric meiotic recombination. Proc Natl Acad Sci USA. 2010;107:8701–8705. doi: 10.1073/pnas.0914160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H, Aguilera A. Transcription as a Threat to Genome Integrity. Annu Rev Biochem. 2016;85:291–317. doi: 10.1146/annurev-biochem-060815-014908. [DOI] [PubMed] [Google Scholar]
- García-Nieto PE, Schwartz EK, King DA, et al. Carcinogen susceptibility is regulated by genome architecture and predicts cancer mutagenesis. EMBO J. 2017;36:2829–2843. doi: 10.15252/embj.201796717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin AJ, Densham RM, Blair-Reid SA, et al. The deSUMOylase SENP7 promotes chromatin relaxation for homologous recombination DNA repair. EMBO Rep. 2013;14:975–983. doi: 10.1038/embor.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A-V, Heard E. Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu Rev Cell Dev Biol. 2014;30:561–580. doi: 10.1146/annurev-cellbio-101512-122415. [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol. 2011;18:831–839. doi: 10.1038/nsmb.2077. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Han C, Srivastava AK, Cui T, et al. Differential DNA lesion formation and repair in heterochromatin and euchromatin. Carcinogenesis. 2016;37:129–138. doi: 10.1093/carcin/bgv247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RK, Mund A, Poulsen SL, et al. SCAI promotes DNA double-strand break repair in distinct chromosomal contexts. Nat Cell Biol. 2016;18:1357–1366. doi: 10.1038/ncb3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SM, Boiarsky JA, Greenberg RA. ATM Dependent Silencing Links Nucleolar Chromatin Reorganization to DNA Damage Recognition. Cell Rep. 2015;13:251–259. doi: 10.1016/j.celrep.2015.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz E. Das Heterochromatin der Moose. Jahrb Wiss Bot. 1928;69:762–818. [Google Scholar]
- Hu J, Adebali O, Adar S, Sancar A. Dynamic maps of UV damage formation and repair for the human genome. Proc Natl Acad Sci USA. 2017;114:6758–6763. doi: 10.1073/pnas.1706522114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaco I, Canela A, Vera E, Blasco MA. Centromere mitotic recombination in mammalian cells. J Cell Biol. 2008;181:885–892. doi: 10.1083/jcb.200803042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Conrad S, et al. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011;39:6489–6499. doi: 10.1093/nar/gkr230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, Breuer GA, Brinkman EK, et al. A single double-strand break system reveals repair dynamics and mechanisms in heterochromatin and euchromatin. Genes Dev. 2016;30:1645–1657. doi: 10.1101/gad.283028.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger N, Schlesner M, Jones DTW, et al. Hypermutation of the inactive X chromosome is a frequent event in cancer. Cell. 2013;155:567–581. doi: 10.1016/j.cell.2013.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. Postreplicative mismatch repair. Cold Spring Harbor Perspectives in Biology. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A, Ismail A, Klement K, et al. Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res. 2013;41:9719–9731. doi: 10.1093/nar/gkt729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousi A, Soutoglou E. Nuclear compartmentalization of DNA repair. Curr Opin Genet Dev. 2016;37:148–157. doi: 10.1016/j.gde.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Kim J-A, Kruhlak M, Dotiwala F, et al. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement K, Luijsterburg MS, Pinder JB, et al. Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J Cell Biol. 2014;207:717–733. doi: 10.1083/jcb.201405077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–54. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Kulashreshtha M, Mehta IS, Kumar P, Rao BJ. Chromosome territory relocation during DNA repair requires nuclear myosin 1 recruitment to chromatin mediated by ϒ-H2AX signaling. Nucleic Acids Res. 2016;44:8272–8291. doi: 10.1093/nar/gkw573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-H, Kuo C-Y, Stark JM, et al. HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response. Nucleic Acids Res. 2013;41:5784–5798. doi: 10.1093/nar/gkt231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître C, Grabarz A, Tsouroula K, et al. Nuclear position dictates DNA repair pathway choice. Genes Dev. 2014;28:2450–2463. doi: 10.1101/gad.248369.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître C, Soutoglou E. Double strand break (DSB) repair in heterochromatin and heterochromatin proteins in DSB repair. DNA Repair (Amst) 2014;19:163–168. doi: 10.1016/j.dnarep.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Lensing SV, Marsico G, Hänsel-Hertsch R, et al. DSBCapture: in situ capture and sequencing of DNA breaks. Nat Methods. 2016;13:855–857. doi: 10.1038/nmeth.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ, Park C, Goiser M, et al. Compartmentalization of DNA Damage Response between Heterochromatin and Euchromatin Is Mediated by Distinct H2A Histone Variants. Curr Biol. 2017;27:1192–1199. doi: 10.1016/j.cub.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Lottersberger F, Karssemeijer RA, Dimitrova N, de Lange T. 53BP1 and the LINC Complex Promote Microtubule-Dependent DSB Mobility and DNA Repair. Cell. 2015;163:880–893. doi: 10.1016/j.cell.2015.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, Dinant C, Lans H, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JHJ. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- Matheson TD, Kaufman PD. Grabbing the genome by the NADs. Chromosoma. 2016;125:361–371. doi: 10.1007/s00412-015-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM. The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol. 2016;17:16–29. doi: 10.1038/nrm.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov E, Magin S, Soni A, Iliakis G. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: Cell cycle and proliferation-dependent regulation. Semin Cancer Biol. 2016;37–38:51–64. doi: 10.1016/j.semcancer.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Müller I, Merk B, Voss K-O, et al. Species conserved DNA damage response at the inactive human X chromosome. Mutat Res. 2013;756:30–36. doi: 10.1016/j.mrgentox.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Natale F, Rapp A, Yu W, et al. Identification of the elementary structural units of the DNA damage response. Nat Commun. 2017;8 doi: 10.1038/ncomms15760. 15760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov I, Taddei A. Linking replication stress with heterochromatin formation. Chromosoma. 2015;125:523–533. doi: 10.1007/s00412-015-0545-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon AT, Shibata A, Rief N, Löbrich M, Stewart GS, Jeggo PA, Goodarzi AA. 553BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- Nozaki T, Imai R, Tanbo M, et al. Dynamic Organization of Chromatin Domains Revealed by Super-Resolution Live-Cell Imaging. Mol Cell. 2017;67:282–293.e7. doi: 10.1016/j.molcel.2017.06.018. [DOI] [PubMed] [Google Scholar]
- Ou HD, Phan S, Deerinck TJ, et al. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017 doi: 10.1126/science.aag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padeken J, Zeller P, Gasser SM. Repeat DNA in genome organization and stability. Curr Opin Genet Dev. 2015;31:12–19. doi: 10.1016/j.gde.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev. 2008;18:204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Lawrence MS, Haugen E, et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nat Biotechnol. 2014;32:71–75. doi: 10.1038/nbt.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci MA, Manzo C, García-Parajo MF, et al. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Ryu T, Bonner MR, Chiolo I. Cervantes and Quijote protect heterochromatin from aberrant recombination and lead the way to the nuclear periphery. Nucleus. 2016;7:485–497. doi: 10.1080/19491034.2016.1239683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu T, Spatola B, Delabaere L, et al. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol. 2015;17:1401–1411. doi: 10.1038/ncb3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksouk N, Simboeck E, Déjardin J. Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin. 2015;8:3. doi: 10.1186/1756-8935-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. A “higher order” of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J. 2009;28:2323–2336. doi: 10.1038/emboj.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Böckler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- Sinha M, Watanabe S, Johnson A, et al. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell. 2009;138:1109–1121. doi: 10.1016/j.cell.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G, van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu Rev Biochem. 2013;82:55–80. doi: 10.1146/annurev-biochem-061809-174504. [DOI] [PubMed] [Google Scholar]
- Smith KS, Liu LL, Ganesan S, et al. Nuclear topology modulates the mutational landscapes of cancer genomes. Nat Struct Mol Biol. 2017 doi: 10.1038/nsmb.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Almouzni G. Differential contribution of HP1 proteins to DNA end resection and homology-directed repair. Cell Cycle. 2013;12:422–429. doi: 10.4161/cc.23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, et al. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Xu Y, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–84. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata H, Hanafusa T, Mori T, et al. Chromatin compaction protects genomic DNA from radiation damage. PLoS ONE. 2013;8:e75622. doi: 10.1371/journal.pone.0075622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timashev LA, Babcock H, Zhuang X, de Lange T. The DDR at telomeres lacking intact shelterin does not require substantial chromatin decompaction. Genes Dev. 2017;31:578–589. doi: 10.1101/gad.294108.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Tsouroula K, Furst A, Rogier M, et al. Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Mol Cell. 2016;63:293–305. doi: 10.1016/j.molcel.2016.06.002. [DOI] [PubMed] [Google Scholar]
- van Sluis M, McStay B. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev. 2015;29:1151–1163. doi: 10.1101/gad.260703.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Belmont AS. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell. 2017;169:780–791. doi: 10.1016/j.cell.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vancevska A, Douglass KM, Pfeiffer V, et al. The telomeric DNA damage response occurs in the absence of chromatin decompaction. Genes Dev. 2017;31:567–577. doi: 10.1101/gad.294082.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafar F, Okita AK, Onaka AT, et al. Regulation of mitotic recombination between DNA repeats in centromeres. Nucleic Acids Res. 2017;45:11222–11235. doi: 10.1093/nar/gkx763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CL, Wang NJ, Chung J, et al. Transcription restores DNA repair to heterochromatin, determining regional mutation rates in cancer genomes. Cell Rep. 2014;9:1228–1234. doi: 10.1016/j.celrep.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]