Abstract
The development of efficacious anticancer therapeutics is difficult due to the heterogeneity of the cellular response to chemotherapy. Anticancer peptides (ACPs) are promising drug candidates that have been shown to be active against a range of cancer cells. However, few ACP studies focus on tumour single-cell heterogeneities. In order to address this need, we developed a microfluidic device and an imaging procedure that enable the capture, monitoring, and analysis of several hundred single cells for the study of drug response. MCF-7 human breast adenocarcinoma cells were captured in hydrodynamic traps and isolated in individual microchambers of less than 100 pL volume. With pneumatic valves, different sets of microchambers were actuated to expose the cells to various drugs. Here, the effect of three membranolytic ACPs – melittin, aurein 1.2 and aurein 2.2 – was investigated by monitoring the efflux of calcein from single MCF-7 cells. The loss of membrane integrity was observed with two different strategies that allow either focusing on one cell for mechanistic studies or parallel analysis of hundreds of individual cells. In general, the device is applicable to the analysis of the effect of various drugs on a large number of different cell types. The platform will enable us in the future to determine the origin of heterogeneous responses on pharmacological substances like ACPs within cell populations by combining it with other on-chip analytical methods.
Introduction
Cell heterogeneity is biologically motivated as it enables a cell population to adapt to external influences.1 However, it is also considered to drive the emergence of drug resistance and several diseases, including cancer.2,3 Drug resistance in heterogeneous tumour cell populations represents a challenge in tumour therapy, since all genetically and phenotypically different subpopulations have to be killed by therapy. Even small surviving subpopulations may cause repopulation and refractory tumors.4–6
Membranolytic anticancer peptides (ACPs) have recently been proposed as promising therapeutics due to their ability to target cells by non-enzymatic membranolysis impeding the emergence of resistant cells.7,8 A widely studied subclass of ACPs are amphipathic, short helical peptides (10 to 30 amino acids), characterized by an overall positive charge, which makes them prone to interactions with anionic cell membranes.9 The overexpression of anionic molecules such as glycoproteins and glycolipids on the surface of cancer cells and the exposure of the negatively charged phospholipids phosphatidylserine and phosphatidylethanolamine on the outer membrane leaflet have been proposed to be the selection mechanisms for the targeted action of ACPs towards cancer cells.8,10 Melittin, the principal component of bee venom, is an extensively studied model ACP consisting of 26 amino acids.11 Melittin was shown to exhibit high membranolytic activity, but low selectivity towards cancer cells.12,13 Aurein 1.2 and aurein 2.2 are well-characterized antimicrobial peptides present in the skin secretions of Australian frogs, with additional anticancer activity against a broad range of different cancer types.14
Up to now, the characterization of ACPs has focused on the analysis of cells in bulk. These measurements are conducted with several thousands of cells and can only provide information about average cell behaviour.15,16 Information gained by studies of individual cells can reveal formerly unknown effects, such as the resistance of small portions of a cell population to certain drugs. However, only a few approaches investigate the time-resolved effect of ACPs at the single-cell level.17,18
The main reason for the absence of single-cell tests is the lack of available simple assay platforms. Flow cytometry is still the most widely used method to investigate heterogeneity in cell populations. Commercial systems are remarkably fast, have ultra-high throughput and enable multiplexing and sorting, but the technique is not suitable for the analysis of time-dependent processes.19,20 Hence, long-term investigations or repeated measurements of individual cells after treatment with chemical or pharmacological substances by flow cytometry are not feasible.
In contrast, microfluidic devices for the capture and analysis of single cells allow performance of time-resolved experiments and are used in a wide range of applications. Mechanical, magnetic, acoustic, di-electrophoretic, or antibody-based capture of single cells has already been presented.21 Passive size-based capture of cells was initially presented by Di Carlo et al. and benefits from the fact that no external devices or any laborious coating procedures are necessary.22 The isolation of cells in miniature compartments can be realized by droplet microfluidics or by the integration of pneumatic valves into microfluidic devices.23 These devices must have the additional capacity to exchange the aqueous environment of single or certain sets of cells on demand to precisely start or stop experimental procedures, such as washing steps or drug exposure. In the field of droplet microfluidics, most single-cell studies focus on cell isolation or barcoding of cells followed by end-point analysis.24,25 Recently, some new approaches to monitor cells in droplets for extended time periods have been presented, but fluid exchange in droplets remains difficult.26–28 In contrast, integrated pneumatic valves as initially developed by Quake and co-workers allow for rapid and repeated fluid exchange29 and opened the way for highly complex systems, e.g. for integrated genetic analysis.30,31 New approaches have also been introduced for the contactless capture of single cells including exchange of fluids.32,33 However, these approaches are not suitable for highly parallel analysis and there is still a need for flexible systems that allow straightforward monitoring of a large number of isolated cells.34
To study the effect of ACPs at the single-cell level with high time resolution, we developed a microfluidic chip with a footprint of 20 × 28 mm2 for the simultaneous analysis of up to 612 individual cells. To simplify the use of the platform, we employed mechanical cell capture using hydrodynamic traps and isolation of cells using pneumatic doughnut-shaped valves. In contrast to other platforms including our own previous work,35 one large channel houses all chambers, reducing the shear stress on the cells and avoiding problems like clogging and hereby induced alterations in the fluid flow – a problem often experienced with multiple parallel channels. The pneumatic valves enabled us to expose the cells to the ACPs melittin, aurein 1.2, and aurein 2.2 with precise timing as well as to monitor their membranolytic effect on single MCF-7 breast cancer cells applying fluorescence microscopy and automated image analysis (Fig. 1).
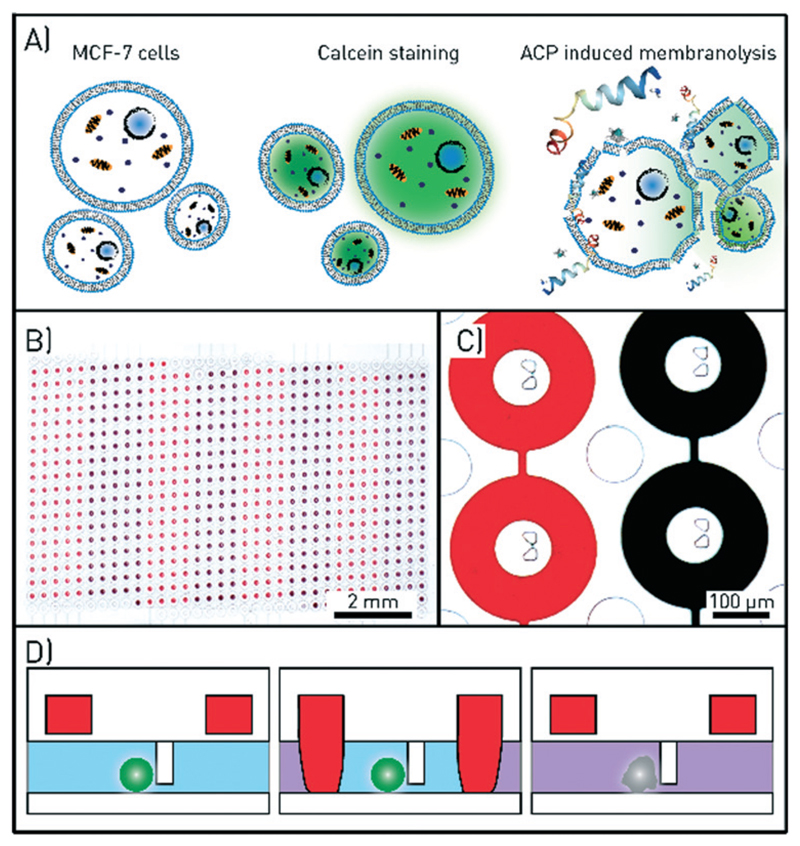
Fig. 1.
Assay and microfluidic chip design for single-cell drug testing. A) Schematics of the assay. Anticancer peptides (ACPs) are administered to MCF-7 cells loaded with calcein, which leaks out of the cells upon ACP-induced membranolysis. B) A microscopy image of the microfluidic channel with 612 chambers with central cell traps. The round valves that define the chambers when in the closed state are filled with red and black coloured solutions. Eight valve control lines are used to open and close the red and black subsets of the valves. C) Microscopy image of four microchambers and cell traps. The columns in between the chambers prevent the large channel and the chambers from collapsing. D) Schematic side view of one chamber. The actuation of the ring-valve (red) is used to isolate cell, ensure flow-free conditions during imaging, and precisely time the drug exposure.
Materials and methods
Fabrication of the microfluidic chip
We used a multilayer microfluidic chip made of poly(dimethylsiloxane), PDMS, that incorporates a fluid and a control layer for the analysis of ACPs at the single-cell level. Separate manufacturing of the two layers and bonding of the PDMS chip to a glass substrate formed the final microfluidic device with 612 microchambers (34 rows, 18 columns) defined by round valves that incorporate mechanical cell traps of 8 μm spacing in the centre of each microchamber.
First, we fabricated two master moulds by means of optical photolithography. To account for shrinkage, the structures on the mask for the control layer were enlarged by 1.6% (both masks are appended to the ESI†). For fabrication, SU-8 2015 photoresist (Microchem, Woburn, MA, USA) was spin-coated onto two 4″ silicon wafers (SI-Mat, Kaufering, Germany) to achieve a final height of 20 μm. The resist was exposed to UV light (150 mJ cm−2 at 365 nm) with an MA-6 mask aligner (Süss Microtec, Garching, Germany) through the corresponding foil mask (Selba S.A., Switzerland). After development (mr-Dev 600, Micro Resist Technology GmbH, Berlin, Germany) and hardbaking, the master forms were silanized with 1H,1H,2H,2H-perfluorodecyltrichlorosilane (ABCR, Karlsruhe, Germany) and PTFE-coated by spin-coating 0.1% poly[4,5-difluoro-2,2-bis(trifluoromethyl)-1,3-dioxole-cotetrafluoroethylene] solution in FC-40 fluorinated oil (both from Sigma-Aldrich, St. Louis, MO, USA) onto the wafer to simplify the release of PDMS from the master mould.
During chip fabrication, the PDMS oligomer and curing agent (Sylgard 184 silicone elastomer kit, Dow Corning, Midland, MI, USA) were mixed at a ratio of 10 : 1. The mixture was degassed for 15 min, poured onto the control layer master, and cured at 80 °C for 2 h. For the fluid layer, 2 mL of the PDMS mixture were spin-coated at 2250 rpm for 60 s onto the fluid master and baked at 80 °C for 1 h. This generates a 30 μm thick PDMS layer that covers the 20 μm high microfluidic channels with a 10 μm thick PDMS membrane. After the control layer was cured and peeled off the wafer, it was cut to size and 1 mm diameter-sized connection holes to the pressure control unit were punched with a biopsy puncher (Integra Miltex, York, PA, USA). Next, 1 mL pure PDMS curing agent was spincoated onto a blank 4″ silicon wafer for 30 s at 6000 rpm. The control layer was stamped onto this coated wafer and then manually aligned on top of the fluid master bearing the spincoated 30 μm PDMS layer, resulting in bonding of both layers after subsequent baking for 2 h at 80 °C. Holes for the fluidic inlet and outlet were punched with a 1.5 mm biopsy puncher (Integra Miltex, York, PA, USA), and the PDMS chip was finally bonded to a cleaned microscopy coverslip (Menzel, Braunschweig, Germany) after exposure to oxygen plasma for 120 s in a plasma cleaner (PDC-32G, Harrick Plasma, USA) at maximum power (18 W). The detailed description of the silicon master fabrication, chip design and fabrication as well as the CAD-file for the corresponding masks including all geometries is available in the ESI† (Fig. S1–S3 and file SF1).
Peptide synthesis
Peptides – melittin (GIGAVLKVLTTGLPALISWIKRKRQQ-NH2), aurein 1.2 and aurein 2.2 (GLFDIIKKIAESF-NH2 and GLFDIVKKVVGALGSL-NH2, respectively) – were synthesized with a Symphony robotic solid phase peptide synthesizer (Protein Technologies Inc., Tucson, AZ, USA) using the 9-fluorenylmethyloxycarbonyl (Fmoc) Merrifield synthesis procedure in dimethylformamide (DMF; Honeywell Specialty Chemicals, Seelze, Germany).36 A 10-fold excess of Fmoc-protected amino acids (Protein Technologies Inc., Tucson, AZ, USA) was used relative to the Fmoc-Rink-amide-MBHA resin loading (MBHA: methylbenzhydrylamine; Protein Technologies Inc., Tucson, AZ, USA). Coupling was performed with a ratio of 1 : 1 : 2 amino acid :HCTU:NMM in DMF (HCTU: 1H-benzotriazolium 1-[bis(dimethylamino)methylene]-5-chloro-hexafluorophosphate (1-),3-oxide; AAPPTEC, Louisville, KY, USA; NMM: 4-methylmorpholine, Thermo Fisher Scientific, Waltham, MA, USA). Fmoc deprotection was performed with 20% pyrrolidine (Thermo Fisher Acros Organics, Geel, Belgium) solution in DMF, and peptide cleavage from the resin was achieved with a TFA (2,2,2-trifluoroacetic acid, Thermo Fisher Scientific, Waltham, MA, USA) solution containing 2.5% triisopropylsilane (TIPS; Sigma-Aldrich, St. Louis, MO, USA) and 2.5% distilled water. The peptides were precipitated using diisopropyl ether stabilized with 2,6-di-tert-butyl-4-methylphenol (Merck KGaA, Darmstadt, Germany) and purified to >90% UV190nm on a preparative Prominence LCMS instrument (LC-20A, Shimadzu, Kyoto, Japan) using a reverse phase C18, 5 μM 150 × 21 mm column (Macherey-Nagel GmbH & Co. KG, Düren, Germany), with a linear gradient of 5 to 70% acetonitrile (Merck KGaA, Darmstadt, Germany) in water with 0.1% formic acid (Sigma Aldrich, Steinheim, Germany) and a flow rate of 24.5 ml min−1.
Cell culture and calcein staining
MCF-7 human breast adenocarcinoma cells were grown in Dulbecco's Modified Eagle Media (DMEM) with 4.5 g L−1 d-glucose supplemented with 10% foetal bovine serum, 1% l-glutamine and 1% penicillin–streptomycin at 37 °C in a 5% CO2 atmosphere (all Thermo Fisher Scientific, Waltham, MA, USA). For experiments, the cells were trypsinized (Trypsin-EDTA, Thermo Fisher Scientific, Waltham, MA, USA) and harvested at a concentration of 106 cells per mL in DMEM.
For visualization, cells were stained with calcein-AM (Thermo Fisher Scientific, Waltham, MA, USA). Calcein-AM stock solutions were prepared in dimethyl sulfoxide (DMSO; AppliChem, Darmstadt, Germany) at a concentration of 1 mM. For staining, 1 mL of the cell suspension (106 cells mL−1) was re-suspended in completed DMEM and incubated with a final concentration of 1 μM calcein-AM for 30 min at 37 °C. Once the non-fluorescent calcein-AM has permeated across the cell membrane, it is converted to green-fluorescent calcein through the hydrolysis of the acetoxymethyl ester by intracellular esterases such that it accumulates in the cytosol (including the mitochondria). The fluorescent calcein molecules can no longer permeate across the membrane and hence can be used as an indicator for membranolysis. After staining, cells were washed two times with completed DMEM by centrifugation at 500g for 5 min and subsequently filtered using a 20 μM gravity filter (Partec GmbH, Jettingen-Scheppach, Germany). Cell samples were stored at 37 °C at constant rotation on a MACSmix tube rotator (12 rpm, Miltenyi Biotec, Bergisch Gladbach, Germany) until use for a maximum of 2 h.
EC50 determination with MTT assay
Determination of the half effective concentration (EC50) of melittin, aurein 1.2, and aurein 2.2 was performed by a bulk 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, as described elsewhere.37 Briefly, 104 cells per well were seeded in a 96-well plate and left to adhere and grow for 24 hours. They were then treated with the peptide in a dilution series for 24 hours in order to determine the EC50. The supernatant was removed and the cells were incubated for 1 hour with a 0.5 mg mL−1 MTT solution (Sigma Aldrich, Steinheim, Germany). The supernatant was removed again and 100 μL of dimethyl sulfoxide (DMSO, Fisher Scientific, UK) were added into each well to resuspend the formazan crystals. The absorbance at 540 nm was measured to determine the cell survival under each of the conditions. The resultant sigmoidal curves were analysed with the drc R package using a log-logistic model with 3 fixed parameters to obtain the EC50 values.38
Chip and device operation
All fluids were supplied with a neMESYS syringe pump (Cetoni GmbH, Korbußen, Germany) using a 1 mL Primo syringe (BD, Franklin Lakes, NJ, USA) at flow rates of 0.1–20 μl min−1. The pressure valves embedded in the PDMS chip were pressurized to 2 bar to deflect the membrane and completely enclose the microfluidic chambers (ESI† Fig. S4 and video SV1). Measurement of the enclosed volume of the individual microchambers revealed a volume of 93 pL (ESI† Fig. S5). To reduce the adhesion of the cells to the channel walls, the chips were blocked with 1% BSA solution in physiological PBS (BSA: bovine serum albumin; PBS: phosphate buffered saline; both Sigma-Aldrich, St. Louis, MO, USA) for 30 min. The inlet of the BSA-coated chip was connected to the syringe pump and the outlet was coupled to a waste container using 1/16″ PTFE-tubing (0.86 mm inner diameter; PKM SA, Lyss, Switzerland). The chip was flushed for 10 min at 10 μL min−1 with completed DMEM. Then, 20 μL of the cell suspension were flushed into the chip at a flow rate of 10 μL min−1 and captured in the mechanical traps. After rinsing the chip with 10 μL of medium at the same flow rate, the pneumatic valves were closed and the cells were isolated in individual chambers. After flushing the chip with the ACP solution at 10 μL min−1 for 5 min, the fluid flow was reduced to 0.1 μL min−1. Peptides were diluted to the final experimental concentration in complete medium prior to administration. Upon opening the pneumatic valves, the cells of interest were exposed to the ACP and its effect was continuously monitored by fluorescence microscopy.
For optical analysis, two inverted microscopes were used and a custom heating stage kept the cells constantly at 37 °C. In contrast to a large incubation chamber, the custom heating stage simplifies the connection to external equipment and allows migration between different microscopes (ESI† Fig. S6). Cell viability was tested on- and off-chip every 60 min for a total time period of 6 h, which included the loading of calcein-AM. For off-chip tests, a 20 μL cell sample was evaluated in a Neubauer chamber (Brand GmbH & Co. KG, Wertheim, Germany) using 100 nM propidium iodide (PI; Thermo Fisher Scientific, Waltham, MA, USA) staining. On-chip viability tests were conducted on three different chips using a PI loading protocol adapted from Krämer et al.39 Long-term experiments were conducted on a setup comprising an XY-stage (HLD117IX, PRIOR Scientific GmbH, Jena, Germany), a LED-illumination system (pE-300, CoolLED Ltd., Andover, England) and an Andor Zyla camera (Andor Technology Ltd., Belfast, Ireland). All devices were controlled with the YouScope software package.40 The exposure time of the camera was set to 100 ms at 4 × 4 binning and a series of 24 images with 10× magnification was taken to image the entire chip (40 s per series). To generate statistically robust data, extrinsic variances were excluded by using an automated fluorescence microscope and standardized protocols for data evaluation.
In contrast to these experiments, high-magnification image series of individual cells were taken with a 100× oil immersion objective on an Olympus IX71 microscope equipped with a SpectraX LED illumination system (Lumencor, Beaverton, OR, USA) and an Andor iXon Ultra EMCCD camera (Andor Technology Ltd., Belfast, Ireland). Exposure times were set to 50 ms at an imaging frequency of 10 Hz and an EM gain of 100.
Post-processing of long-term imaging data
At each time point during long-term imaging experiments, 24 fluorescence images at different locations were collected to monitor all microchambers. Each series was stitched to one large image using the stitching plug-in of the software Fiji.41,42 These large images were then combined into one time-lapse series of the entire chip. The captured cells were automatically detected (analyse particles plug-in) and the fluorescence intensities were extracted (ESI† Fig. S7).43 Finally, the single cell data was sorted and the results were visualized with Matlab (version R2015a, MathWorks, USA).
Results and discussion
Cell handling and on-chip viability
To exclude any negative influence from the measurement setup on cell viability, MCF-7 cells were first incubated in completed DMEM at 106 cells per mL off-chip at 37 °C under continuous rotation. Viability was tested every 60 min and found to be constant at 92 ± 1% over more than 6 hours. On-chip incubation of the cells for 6 hours with constant medium supply at 0.1 μL min−1 yielded a comparable cell viability of 91 ± 2%. This confirms that the measurement setup, including the heating stage, ensures viability of the MCF-7 cells over the experimental time period (ESI† Fig. S6A).
After mounting the chip onto the heating stage and the inverted fluorescence microscope, the cell trapping was tested by introducing 20 μL of stained cells (106 cells mL−1) into the chip. On average, more than 200 individual microchambers were occupied by a single MCF-7 cell. Due to the aggregating nature of MCF-7 cells, most of the remaining chambers were not empty, but occupied by multiple cell aggregates (ESI† Fig. S7) and therefore excluded from the final data evaluation process.
Monitoring single cells at high temporal and spatial resolution
Calcein-AM was used to monitor the individual time-resolved cell viability in all experiments performed on-chip. When the cell loses its membrane integrity, i.e. ACPs induce membrane disruption, the dye leaks out of the cell and the fluorescence signal intensity decreases, which indicates cell death.
First, we monitored the response of single cells to melittin, aurein 1.2 and aurein 2.2 at a fixed concentration of 50 μM over a time period of 100 s. This concentration was chosen because the half effective concentrations of aurein peptides have been shown to range between 1.5 and 100 μM.14 Upon ACP administration, the valves were opened and the effect of the ACP on an isolated single cell was observed at 100× magnification (Fig. 2). Due to the integration of eight independent valves, up to eight experiments could be performed sequentially on one chip.
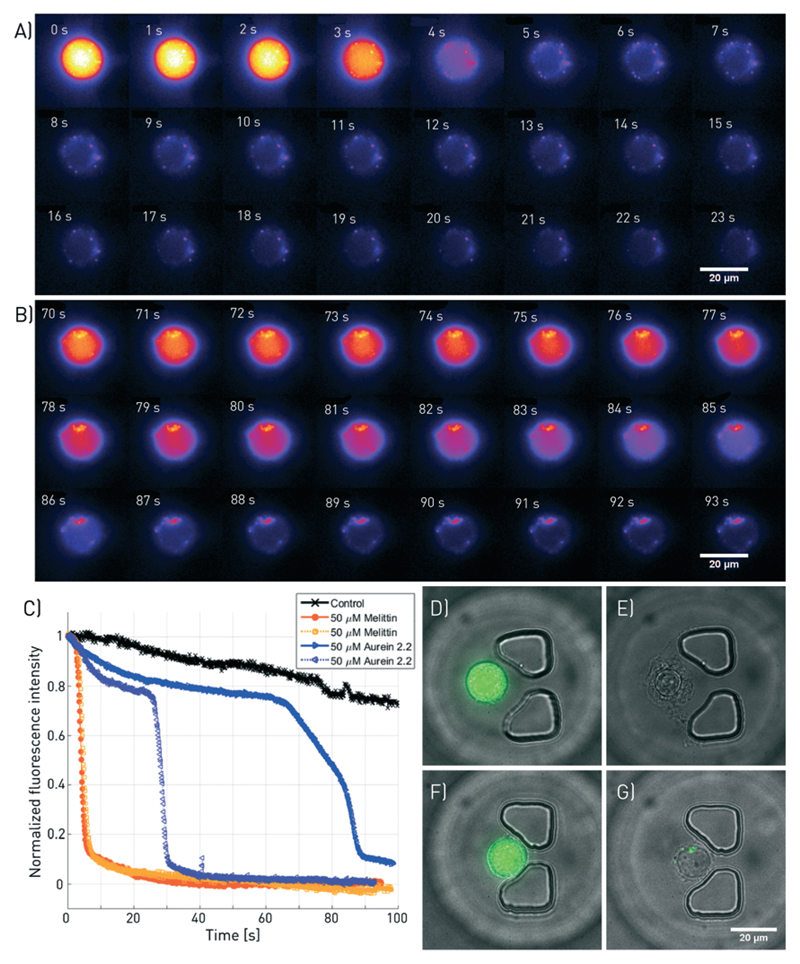
Fig. 2.
Fluorescence microscopy of melittin and aurein 2.2-induced membranolysis at high temporal and spatial resolution. A subset of the images taken at a frequency of 10 Hz and 100× magnification are shown for A) 50 μM melittin and B) 50 μM aurein 2.2. C) Fluorescence intensity decrease of four individual cells treated with melittin and aurein 2.2 (2 each) as well as a negative control without ACP. D) to G) Overlaid bright-field and fluorescence images of the cells, taken before and after the treatment. D and E correspond to the cell shown in A, and F and G depict the cell presented in B. The corresponding videos can be found in ESI† SV2.
Melittin induced rapid cell death within 4.2 ± 0.7 s of exposure. The time of cell death was defined as the point at which the fluorescence signal intensity of a cell decreased below 50% of the initial intensity (Fig. 2A and C). In contrast, cells treated with aurein 2.2 at identical concentrations died notably slower (Fig. 2B and C). As some of the cells resisted the treatment for the time of observation, no mean for the time of death can be given. The findings are in accordance with the EC50 values obtained from bulk experiments, which are 1.83 ± 0.08 μM and 35 ± 3 μM, respectively (ESI† Fig. S8), suggesting considerable shorter time scales until membrane disruption for melittin compared to aurein 2.2 at identical concentrations. A change in cell morphology such as the formation of bulges and blebs was visible in bright-field microscopy (Fig. 2E and G). Furthermore, the leakage of calcein from the cells indicated cell death via membrane disruption for both peptides. A video comparing the membranolysis induced by the two peptides is available in ESI† SV2.
Aurein 1.2 is the least active among the tested peptides and causes slower cell death than the other two peptides, which makes high temporal resolution monitoring of the membranolysis process impossible due to bleaching of the calcein. A correlation with the EC50 value obtained from measurements off-chip (EC50 = 44.7 ± 2.7 μM, ESI† Fig. S8) to on-chip data was therefore not feasible. The bleaching data in ESI† Fig. S9 provides more detailed information. For experiments lasting much longer than 100 s, the excitation light intensity or exposure time had to be reduced to avoid fast bleaching. Therefore, for long-term experiments, the cells were exposed to light only for 100 ms at each measurement time point. Here, the bleaching was much slower and we could clearly determine the time of the membrane lysis characterized by the fast drop in the calcein fluorescence (ESI† Fig. S9). This reduced excitation was used for the following parallel single-cell analysis.
High-throughput analysis at the single-cell level
The inherent heterogeneity of cellular responses and the study of less active peptides require the analysis of a larger number of cells with reasonable temporal and spatial resolution. This challenge was addressed by imaging all 612 microfluidic chambers every two minutes on an automated fluorescence microscope setup. Acquiring all images necessary for the stitching of one entire chip image took less than 40 s using a 10× objective. This type of experiment is complementary to high-resolution imaging of single cells as it gives insight into the cellular response over longer periods of time, minimizes the light exposure and avoids fast bleaching of the cells.
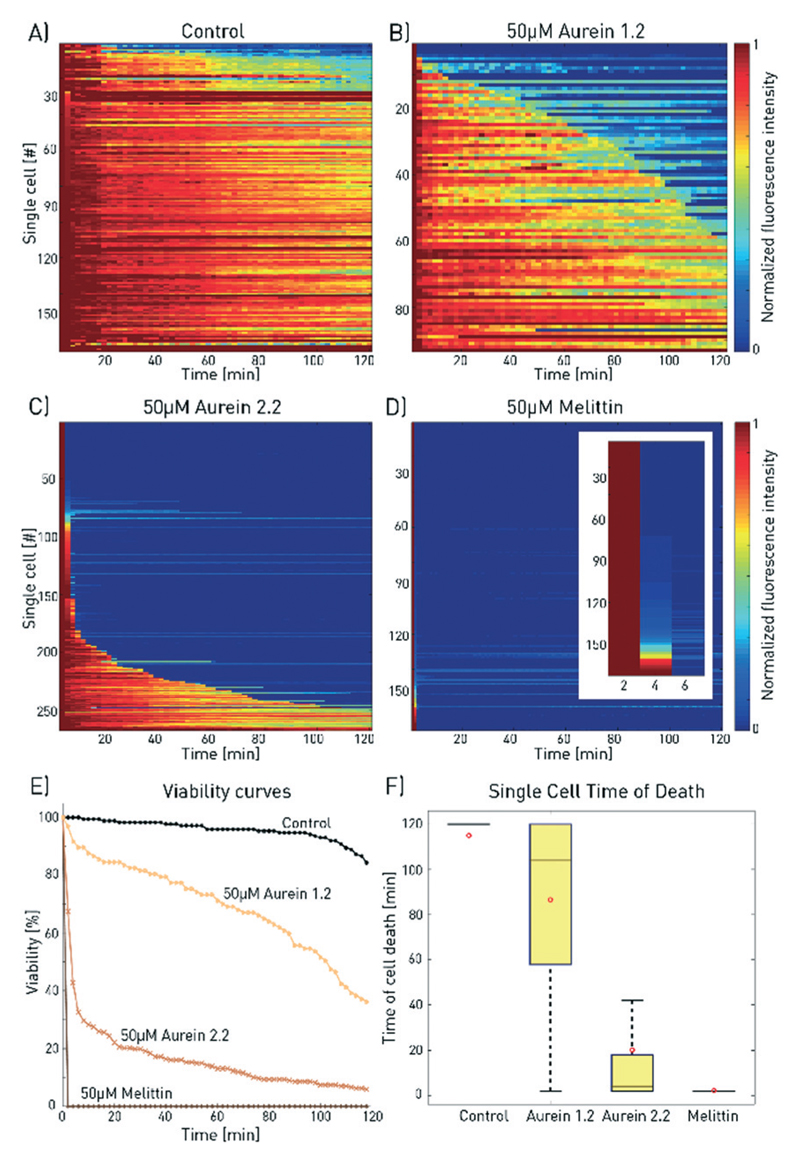
Fig. 3 shows the results for the population viability after treatment with melittin, aurein 2.2, and aurein 1.2 at a concentration of 50 μM, together with a control sample without an anticancer agent (the time series for aurein 2.2 is available in ESI† SV3). In accordance with previous findings, melittin was the fastest acting and most potent membranolytic peptide for MCF-7 cells with 100% membrane disruption on all investigated cells in less than 4 min. Aurein 2.2 and aurein 1.2 showed intermediate activities with 7% and 35% of the cells surviving after 2 h of incubation at 50 μM concentration, with large deviations in the time of death. Although these deviations are obscured in bulk analysis, they are a general cellular behaviour within each cell population and not artefacts of the measurement procedure. The mean time of death between three different experimental runs (different chip devices) was exemplarily determined for 50 μM aurein 2.2 to be 20.4 ± 2.0 min. This corresponds to an error of less than 10% and verifies reproducibility of the on-chip measurements.
Fig. 3.
Comparison of the effect of melittin, aurein 2.2 and aurein 1.2 on MCF-7 cells. The 2D-matrix plots show the results for A) a control experiment and for 50 μM B) aurein 1.2, C) aurein 2.2 and D) melittin (the inset shows a zoom for the first 6 min). Every horizontal line depicts the viability of a single cell over time, whereby cells are alive at high fluorescence intensity and dead at low fluorescence signal (total number of analysed cells: nA = 170, nB = 97, nC = 267, and nD = 364). E) The time until membrane disruption differs for each of the peptides as depicted by the combined single-cell measurements. F) For aurein 1.2, this time period is distributed much wider than those for aurein 2.2 and melittin, underlining the heterogeneous cell response. For improved visibility, outliers are not shown and mean values are marked by red circles.
It should be noted that with appropriate imaging techniques, the cell morphology including cell size can be evaluated and a relationship of morphological differences to drug resistance can be discovered. For the peptides investigated here, no such correlation between the cell size and viability was found (ESI† Fig. S11).
Aurein 2.2 single-cell dose–response curve
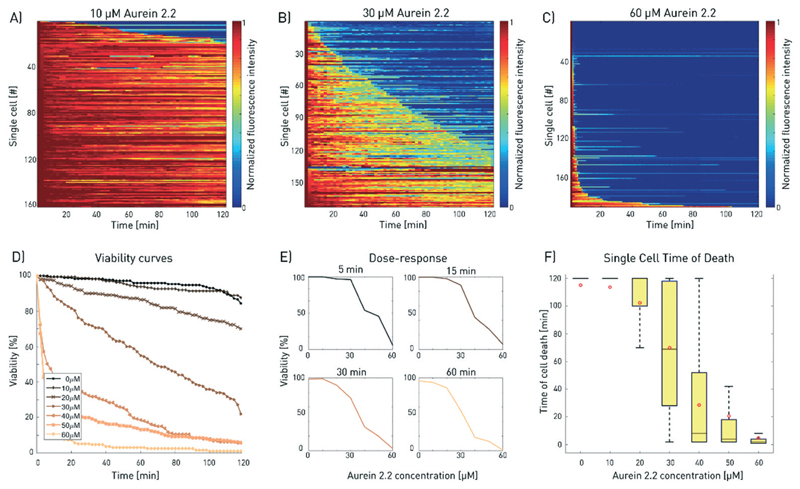
The effect of the peptide concentration on cell viability was further investigated. For this purpose, cells were trapped onchip and treated with aurein 2.2 at concentrations ranging from 0 to 60 μM (Fig. 4). Higher peptide doses resulted in faster membranolytic cell death, leaving virtually no surviving cells at the concentration of 60 μM. The large number of single cells measured for each peptide concentration allowed the derivation of population-based viability and dose–response curves (Fig. 4E and F) as well as tracking of individual cells of interest. The on-chip EC50 value for aurein 2.2 can be calculated at each time point during the experiment, which simplifies the comparison of the results with other studies that is often hampered by varying incubation times. After 30, 60 and 120 min of incubation, the EC50 values were found to be 36 ± 3 μM, 31 ± 3 μM and 24 ± 3 μM. Hence, these data reveal both time and dose dependencies of the drug efficacy on the cells. We would like to emphasise the fact that the onchip derived EC50 value matches well with the value from conventional bulk tests in well plates (ESI† Fig. S8C).
Fig. 4.
Dose–response analysis for aurein 2.2. A–C) Increasing aurein 2.2 concentrations induced faster cell death. The results for three different concentrations of aurein 2.2 are shown exemplarily (total number of analysed cells: nA = 163, nB = 173, nC = 190; for all 2D plots, refer to ESI† S10). The single-cell measurements were combined to plot D) the viability of the cell population over time for increasing doses and E) the drug response curves for selected time points during the measurement. F) The time of cell death is evaluated for increasing aurein 2.2 concentrations. Higher concentrations show faster mean cell death but broad variations are found for the intermediate concentrations (30 and 40 μM). The red dots depict the mean value of the corresponding measurements, outliers are not shown for better visibility. Differences between the mean and median are a consequence of the non-Gaussian distribution of the data. This results from the fact that the measurement duration was limited to 120 min.
Conclusions
We analysed the time-dependent effect of the ACPs melittin, aurein 1.2 and aurein 2.2 on MCF-7 cell populations at the single-cell level. ACP-specific membranolysis was monitored either for individual cells at high local and temporal resolution for up to 100 s or in a high-throughput mode over extended time periods for hundreds of single cells simultaneously for 2 h. Both modes revealed ACP-induced membrane disruption after strongly varying and ACP-specific time periods. These periods scale with EC50 values determined off-chip. We could further identify cells that are resistant to both aurein peptides under the set experimental conditions.
For realization of the present study, we designed a microfluidic device where mechanical cell traps kept the cells at a constant location to facilitate the quick and efficient exchange of aqueous solutions and automated image analysis. Further increase of the chamber number can be achieved easily. In contrast to bulk analyses, these experiments consumed only minimal quantities of the tested drugs and cells, reducing the costs per test. This is an advantage especially for drug candidates that are synthesized in small quantities for initial tests. The microchip further enables the performance of up to eight different experiments in parallel (different drugs, concentrations or cell types) or tests of multiple drug combinations. Using new scientific cameras with large fields of view and high pixel numbers will speed up the highthroughput measurements. Temporal resolution of few seconds to precisely monitor the effects of peptides with high activities will then be possible.
We envision our microfluidic platform to facilitate the parallel, systematic analysis of ACPs, other therapeutic agents, toxins or cell-affecting compounds at the single-cell level. Furthermore, the cell traps can be adapted to other cell types and bacteria.44 Depending on the application, transmission or fluorescence microscopy can be used and this setup is compatible with protein expression studies from single mammalian to single bacterial cells by means of on-chip immunoassays.44,45 Such an integration of single cell capture, drug response monitoring and analysis into one microfluidic chip can help to understand the underlying principles of drug resistance and aid the design and testing of targeted therapeutics.
Supplementary Material
† Electronic supplementary information (ESI) available. See DOI: 10.1039/c7lc00505a
Acknowledgements
The authors would like to acknowledge the support from Christoph Bärtschi and Christian Marro from the mechanical workshop and the FIRST cleanroom facility (ETH Zurich). We further thank Prof. Dr. Cornelia Halin at ETH Zurich for providing the MCF-7 cell line and cell culture facilities. Financial support by the European Research Council (ERC Consolidator Grant No. 681587 “HybCell” to P. S. D.) and the Swiss National Science Foundation (SNF Grants No. 200021_157190 and No. CRSII2_160699 to G. S.) is acknowledged.
Footnotes
G. Schneider: 0000-0001-6706-1084
P. S. Dittrich: 0000-0001-5359-8403
References
- 1.Altschuler SJ, Wu LF. Cell. 2010;141:559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marusyk A, Polyak K. Biochim Biophys Acta, Rev Cancer. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, Halushka MK, et al. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Kennecke H, Yerushalmi R, Woods R, Cheang MCU, Voduc D, Speers CH, Nielsen TO, Gelmon K. J Clin Oncol. 2010;28:3271–3277. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt F, Efferth T. Pharmaceuticals. 2016;9(2):33. doi: 10.3390/ph9020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrell RA, McGranahan N, Bartek J, Swanton C. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 7.Mader JS, Hoskin DW. Expert Opin Invest Drugs. 2006;15:933–946. doi: 10.1517/13543784.15.8.933. [DOI] [PubMed] [Google Scholar]
- 8.Riedl S, Zweytick D, Lohner K. Chem Phys Lipids. 2011;164:766–781. doi: 10.1016/j.chemphyslip.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schweizer F. Eur J Pharmacol. 2009;625:190–194. doi: 10.1016/j.ejphar.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 10.Gaspar D, Salomé Veiga A, Castanho MARB. Front Microbiol. 2013;4:1–16. doi: 10.3389/fmicb.2013.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terwilliger TC, Eisenberg D. J Biol Chem. 1988;257:6016–6022. [PubMed] [Google Scholar]
- 12.Kim S, Kim SS, Bang YJ, Kim SJ, Lee BJ. Peptides. 2003;24:945–953. doi: 10.1016/s0196-9781(03)00194-3. [DOI] [PubMed] [Google Scholar]
- 13.Asthana N, Yadav SP, Ghosh JK. J Biol Chem. 2004;279:55042–55050. doi: 10.1074/jbc.M408881200. [DOI] [PubMed] [Google Scholar]
- 14.Rozek T, Wegener KL, Bowie JH, Olver IN, Carver JA, Wallace JC, Tyler MJ. Eur J Biochem. 2000;267:5330–5341. doi: 10.1046/j.1432-1327.2000.01536.x. [DOI] [PubMed] [Google Scholar]
- 15.Heath JR, Ribas A, Mischel PS. Nat Rev Drug Discovery. 2015;15:204–216. doi: 10.1038/nrd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Junkin M, Tay S. Lab Chip. 2014;14:1246–1260. doi: 10.1039/c3lc51182k. [DOI] [PubMed] [Google Scholar]
- 17.Burton MG, Huang QM, Hossain MA, Wade JD, Palombo EA, Gee ML, Clayton AHA. Langmuir. 2016;32:6496–6505. doi: 10.1021/acs.langmuir.6b00596. [DOI] [PubMed] [Google Scholar]
- 18.Kao F-S, Pan Y-R, Hsu R-Q, Chen H-M. Biochim Biophys Acta. 2012;1818:2927–2935. doi: 10.1016/j.bbamem.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 19.Piyasena ME, Graves SW. Lab Chip. 2014;14:1044. doi: 10.1039/c3lc51152a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Léonard L, Chibane LB, Bouhedda BO, Degraeve P, Oulahal N. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armbrecht L, Dittrich PS. Anal Chem. 2016;89:2–21. doi: 10.1021/acs.analchem.6b04255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Carlo D, Wu LY, Lee LP. Lab Chip. 2006;6:1445. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 23.Hümmer D, Kurth F, Naredi-Rainer N, Dittrich PS. Lab Chip. 2015;16:447–458. doi: 10.1039/c5lc01314c. [DOI] [PubMed] [Google Scholar]
- 24.Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. Nat Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitz CHJ, Rowat AC, Köster S, Weitz DA. Lab Chip. 2009;9:44–49. doi: 10.1039/b809670h. [DOI] [PubMed] [Google Scholar]
- 27.Sauzade M, Brouzes E. Lab Chip. 2017;17:2186–2192. doi: 10.1039/c7lc00283a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer A, Pellaux R, Potot S, Becker K, Hohmann H-P, Panke S, Held M. Nat Chem. 2015;7:673–678. doi: 10.1038/nchem.2301. [DOI] [PubMed] [Google Scholar]
- 29.Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 30.White AK, Vaninsberghe M, Petriv OI, Hamidi M, Sikorski D, Marra MA, Piret J, Aparicio S, Hansen CL. Proc Natl Acad Sci U S A. 2011;108:13999–14004. doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorsen T, Maerkl SJ, Quake SR. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 32.Johnson-Chavarria EM, Agrawal U, Tanyeri M, Kuhlman TE, Schroeder CM. Lab Chip. 2014;14:2688–2697. doi: 10.1039/c4lc00057a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li XJ, Ling V, Li PCH. Anal Chem. 2008;80:4095–4102. doi: 10.1021/ac800231k. [DOI] [PubMed] [Google Scholar]
- 34.Chattopadhyay PK, Gierahn TM, Roederer M, Love JC. Nat Immunol. 2014;15:128–135. doi: 10.1038/ni.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eyer K, Kuhn P, Hanke C, Dittrich PS. Lab Chip. 2012;12:765–772. doi: 10.1039/c2lc20876h. [DOI] [PubMed] [Google Scholar]
- 36.Merrifield RB. J Am Chem Soc. 1963;85:2149–2154. [Google Scholar]
- 37.Douglas S, Hoskin DW, Hilchie AL. In: Therapeutic Peptides: Methods and Protocols. 1088th edn. Totowa NJ, editor. Humana Press; 2014. [Google Scholar]
- 38.Ritz C, Baty F, Streibig JC, Gerhard D. PLoS One. 2016;10:1–13. doi: 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krämer CEM, Wiechert W, Kohlheyer D. Sci Rep. 2016;6 doi: 10.1038/srep32104. 32104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lang M, Rudolf F, Stelling J. Current Protocols in Molecular Biology. Vol. 98 John Wiley & Sons, Inc.,; 2001. [Google Scholar]
- 41.Preibisch S, Saalfeld S, Tomancak P. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, et al. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abràmoff MD, Magalhães PJ, Ram SJ. Biophotonics Int. 2004;11:36–41. [Google Scholar]
- 44.Eyer K, Stratz S, Kuhn P, Dittrich PS. Anal Chem. 2013;85:3280–3287. doi: 10.1021/ac303628j. [DOI] [PubMed] [Google Scholar]
- 45.Stratz S, Eyer K, Kurth F, Dittrich PS. Anal Chem. 2014;86:12375–12381. doi: 10.1021/ac503766d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.