Abstract
Background
Hepatitis C virus (HCV) is currently classified into 8 genotypes and 86 subtypes. The objective of this study was to characterize novel HCV subtypes and to investigate the impact of subtypes on treatment outcome.
Methods
Full-genome sequencing was performed on HCV plasma samples with <85% sequence homology of NS3, NS5A, and/or NS5B to HCV genotype (GT) 1–8 reference strains.
Results
A total of 14 653 patients with GT1–6 HCV infection were enrolled in clinical studies of sofosbuvir-based regimens. For the majority of the patients, a specific subtype could be assigned based on a close genetic relationship to previously described subtypes. However, for 19 patients, novel subtypes were identified with <85% homology compared with previously described subtypes. These novel subtypes had the following genotypes: 9 in GT2, 5 in GT4, 2 in GT6, and 1 each in GT1, GT3, and GT5. Despite the presence of polymorphisms at resistance-associated substitution positions, 18 of the 19 patients treated with sofosbuvir-containing therapy achieved SVR12.
Conclusions
Nineteen novel HCV subtypes were identified, suggesting an even greater genetic diversity of HCV subtypes than previously recognized.
Keywords: direct-acting antivirals (DAAs), phylogenetic analysis, resistance-associated substitutions (RAS), sofosbuvir, velpatasvir, voxilaprevir
Hepatitis C virus (HCV) has a high degree of genetic diversity, and the number of HCV genotypes and subtypes continues to increase. Until recently, HCV was classified into 7 distinct genotypes that differed by >30% at the nucleotide level [1]. A novel HCV genotype, genotype 8, was recently identified in 4 epidemiologically unlinked patients from the state of Punjab in India, which forms a distinct phylogenetic group from previously described sequences [2]. Genotypes are further divided into subtypes with a sequence divergence of >15% [3]. To date, 86 confirmed HCV subtypes have been described [4], with possibly even more genotypes and subtypes to be identified [5]. HCV genotypes 1, 2, and 3 are circulating worldwide, although with variable distribution in different geographical areas [6]. HCV genotype 1 is the most prevalent (46%) genotype globally. HCV subtypes 1a and 1b are predominant in North America, Europe, and Australia, whereas in Japan, 73% of HCV-infected individuals have subtype 1b infection. Genotype 3 is the second most prevalent (30%) genotype worldwide, primarily distributed in South Asia, and represents a disproportionately high distribution among people who inject drugs (PWIDs), regardless of geography. Infections with HCV genotype 4 are mainly found in Africa and the Middle East, and genotype 5 and genotype 6 are confined to Southern Africa and Southeast Asia, respectively [7]. Genotypes 1, 2, 3, 4, and 6 are comprised of multiple subtypes and display a high degree of genetic variability. In 2006, HCV genotype 7a was identified in a patient originating from the Democratic Republic of Congo. Subsequently, another patient from the same region was identified as being infected with genotype 7b [3, 8]. Genotype 5, as well as the recently described genotype 8, have 1 subtype each described to date.
In recent years, tremendous efforts have been directed toward discovering and developing novel direct-acting antivirals (DAAs) to treat HCV infection [9–15]. Due to the high genetic diversity of HCV and its potential to adapt quickly to different environments, the goal has been the clinical development of DAAs that are safe and highly effective across all genotypes and subtypes. The pan-genotypic NS5B HCV inhibitor sofosbuvir (SOF), when used in combination with other agents, has demonstrated high efficacy in patients infected with genotypes 1–6 and has also been effective in all 6 patients with genotype 7 or 8 [2, 16, 17]. Recently, SOF in combination with the NS5A inhibitor velpatasvir (VEL) showed high rates of sustained virologic response (SVR) among both previously treated and untreated patients infected with HCV genotypes 1–6 [17–19], including those with compensated cirrhosis [18]. Furthermore, a fixed-dose combination of pangenotypic SOF/VEL and the protease inhibitor voxilaprevir (VOX) provided high rates of SVR among patients across HCV genotypes in whom treatment with a DAA regimen had previously failed [20].
Given the high genetic diversity of HCV both at the genotype and subtype levels, there is a need to characterize the novel subtypes and to understand the potential impact of novel subtypes on treatment outcome. More than 14 000 HCV-infected patients have been enrolled in global clinical studies of sofosbuvir-based regimens. Here, extensive analysis of viral diversity and sequence variation across genotypes was performed to identify uncharacterized subtypes and the effect on treatment outcome.
METHODS
Clinical Samples
All patients included in these analyses have provided informed consent in writing, and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki (most recently updated in 2013), as reflected in a priori approval by the appropriate institutional review committee. All the patients received sofosbuvir-based regimens as part of Gilead global studies.
HCV Amplicon Amplification and Deep Sequencing
HCV RNA was determined at a central laboratory using the COBAS AmpliPrep/COBAS TaqMan HCV Quantitative Test, version 2.0 (Roche Molecular Diagnostics, Pleasanton, CA), with a lower limit of quantitation of 15 IU/mL. Amplification and sequencing of full-length NS3, NS5A, and NS5B were performed on plasma samples with HCV RNA ≥1000 IU/mL for all patients at baseline. For patients experiencing virologic failure, HCV amplicon amplification and sequencing were performed at the postbaseline time points as previously described [21]. Briefly, amplification and deep sequencing were performed by the DDL Diagnostic Laboratory (Rijswijk, the Netherlands) using the MiSeq deep sequencing platform (Illumina, San Diego, CA). Each gene was amplified using genotype-specific proprietary amplification primers and standard reverse transcription polymerase chain reaction technology. Library preparation, multiplexing, and deep sequencing were then performed. Illumina-based deep sequencing chemistry with paired ends was used with read lengths of 150 bases. Internally developed software was used to generate consensus sequences for each sample with inclusion of mixtures of amino acids, when present, between 15% and 85%. All aligned reads were then translated in-frame, and changes from a reference sequence were determined. Prevalence of resistance-associated subtitutions (RAS) was evaluated at 15% sequencing assay cutoffs.
Genotype Distribution Among Patients in Clinical Trials
HCV genotypes were determined using the VERSANT HCV Genotype 2.0 assay (LiPA) or the Abbott RealTime HCV Genotype II assay (Abbott, IL).
Subtyping and HCV Full-Genome Sequencing
For each patient sample, HCV subtyping was based on amplicon deep sequencing, described above. A subset of patients with <85% homology to previously described HCV subtypes or with different subtypes assigned across the multiple sequencing targets were subsequently sequenced by full–HCV genome sequencing. HCV RNA was isolated from 200 µL of plasma using the QIAamp MinElute Virus spin kit (Qiagen, Hilden, Germany), and full–HCV genome sequencing was performed at the DDL Diagnostic Laboratory (Rijswijk, the Netherlands) as previously described [22]. Briefly, RNA was reverse-transcribed and amplified using the Ovation RNA-Seq V2 system (NuGEN, San Carlos, CA). Double-stranded DNA was generated and amplified using single-primer isothermal linear amplification according to the the manufacturer’s protocol. Amplified products were fragmented using the Covaris system (Covaris, Inc., Woburn, MA), and paired-end libraries were created for each sample using Ovation Ultralow DR Multiplex Systems (NuGEN) following the manufacturer’s instructions. Internally developed software (Gilead Sciences) was used to process and align sequencing data. Assignment of HCV subtype was performed by BLAST analysis of NS3, NS5A, and/or NS5B deep sequencing to genotype 1–7 reference strains (86 subtypes and 7 genotypes).
Phylogenetic Analysis of HCV Full-Genome Sequences
Full-genome consensus sequences were aligned to HCV reference sets obtained from the International Committee on Taxonomy of Viruses website containing the complete genomes of all 86 confirmed HCV subtypes and previously described unassigned subtypes (https://talk.ictvonline.org/ictv_wikis/flaviviridae/w/sg_flavi/56/hcv-classification). Maximum likelihood phylogenetic trees were inferred using GARLi (version 2.0) software, which optimizes the substitution model iteratively [23]. Default parameters were used. The confidence of the branches was assessed by the approximate likelihood ratio test [24] using PhyML 3.0. [25]. Phylogenetic trees were visualized using FigTree (version 1.3.1).
Analysis of Subtype Recombinants
The generated full-genome sequences were analyzed by BootScan in SimPlot, version 3.5.1, using default settings (Step: 20 bp; GapString: On; Reps: 100: Kimura, 2-parameter; T/t: 2.0; Neighbor-Joining). Individual analysis was performed for each sample at the nucleotide level, using previously described confirmed and unassigned subtypes of the corresponding sample to identify possible recombination between subtypes.
Resistance-Associated Substitutions Definitions
RAS were defined as substitutions that confer reduced susceptibility to any approved DAA inhibitor with >2.5-fold change compared with a genotype 1a reference (HCV1a H77 NC AF009606; NS3 RAS: V36A/G/I/L/M/T, Q41R/H/K, F43L/S/V, T54A/C/G/S, V55A/I, Y56H, Q80K/L/R, S122D/N/R, R155any, A156any, D168any, and I170A/T/V; NS5A RAS: K24A/E/G/N/R, M28A/G/T/V, Q30any, L31I/F/M/V, P32L, S38F, H58D/L/N, A92K/P/T, and Y93any; NS5B NI RAS: L159F, E237G, S282any, L320F, and V321A).
Phenotypic Analysis and Drug Susceptibility Assay
Phenotypic analysis was performed on new subtypes of clinical isolates to investigate in vitro susceptibility to sofosbuvir, velpatasvir, and voxilaprevir as previously described [26, 27]. Briefly, a patient specimen-derived HCV NS3, NS5A, and NS5B coding sequence was inserted into a chimeric replicon vector. RNA was transcribed from the vector in vitro and was transfected by electroporation into Huh-7 cell lines. Transfected cells were cultured in 96-well microplates, and serially diluted sofosbuvir, velpatasvir, and voxilaprevir were then added to the cells. For all assays, relative light unit signals were obtained 4 and 96 hours after transfection. Seventy-two hours after compound addition, luciferase signal was measured using the Promega Renilla-GLO Luciferase Assay kit or the Promega Renilla Luciferase Assay kit according to the manufacturer’s instructions (Promega). Inhibitor susceptibility was determined by evaluating 3 replicates. Intra-assay and interassay variations were each approximately 2–3-fold.
RESULTS
HCV Genotype and Subtype Assignment of Patients Enrolled in HCV Clinical Trials
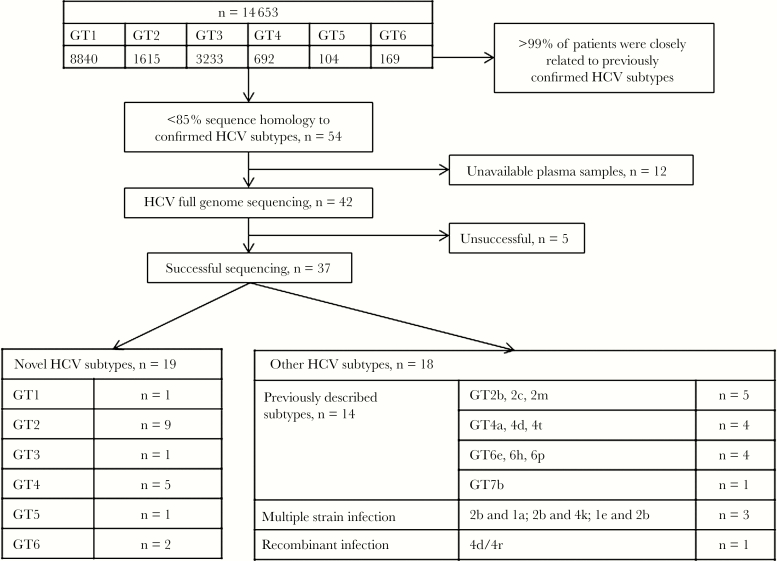
Across SOF-based clinical studies, 14 653 patients infected with HCV genotype 1–6 were enrolled. Determination of HCV subtype was performed at baseline for each patient using NS3, NS5A, and/or NS5B deep sequencing. For more than 99.5% of patients, the HCV subtype of the patient was closely related to 1 of the 86 subtypes currently confirmed for HCV. Of the patients included in the analysis, 8840, 1615, 3233, 692, 104, and 169 patients were classified at screening by commercial assays and/or amplicon sequencing as infected with HCV genotypes 1–6, respectively (Figure 1). However, in <0.5% of patients (54 of 14 653), the subtype showed <85% sequence homology to previously confirmed subtypes or discordant subtyping results across multiple targets. To determine the HCV subtype for these patients, full–HCV genome sequencing was performed on available baseline plasma samples (42 of 54 patients) (Table 1, Figure 1).
Figure 1.
Overview of hepatitis C virus (HCV) genotype and subtype assignment of patients enrolled in HCV clinical trials. Abbreviation: GT, genotype.
Table 1.
Genotyping Results Using Different Assays
| Patient | Subtype by Lipa/ Trugene or Abbott RT-PCR Assay | Amplicon Deep Sequencing | Full-Genome Sequencing | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS3 | NS5A | NS5B | Subtype | Nt Homology, % | |||||||||||
| nt | H omology, % | aa | Homology, % | nt | Homology, % | aa | Homology, % | nt | Homology, % | aa | Homology, % | ||||
| 1 | 2a/2c | ND | NA | ND | NA | 2j | 79 | 2i | 84 | 2e | 86 | 2d | 92 | Novel GT2 subtype | 82 (2c, 2r, 2d, 2) |
| 2 | 2a/2c | ND | NA | ND | NA | 2d | 79 | 2j | 85 | 2e | 86 | 2k | 91 | Novel GT2 subtype | 82 (2) |
| 3 | 2 | ND | NA | ND | NA | 2e | 82 | 2j | 86 | 2e | 88 | 2e | 94 | Novel GT2 subtype | 85 (2) |
| 4 | 2 | 2a | 82 | 2c | 92 | 2q | 78 | 2j | 80 | 2e | 83 | 2d | 89 | Novel GT2 subtype | 82 (2) |
| 5 | 2a/2c | 2q | 86 | 2c | 96 | 2q | 81 | 2k | 86 | 2k | 88 | 2k | 93 | Novel GT2 subtype | 84 (2k, 2q) |
| 6 | 2a/2c | 2a | 81 | 2c | 94 | 2a | 79 | 2a | 83 | 2a | 86 | 2a | 92 | Novel GT2 subtype | 82 (2a, 2) |
| 7 | 2b/2j | 2i | 75 | 2j | 87 | No hit | 0 | 2i | 77 | 2r | 80 | 2r | 86 | Novel GT2 subtype | 83 (2l) |
| 8 | 2l | 2e | 75 | 2i | 89 | No hit | 0 | 2b | 75 | 2r | 79 | 2r | 85 | Novel GT2 subtype | 81 (2l) |
| 9 | 2 | ND | NA | ND | NA | 2d | 80 | 2e | 85 | 2k | 86 | 2d | 92 | Novel GT2 subtype | 83 (2c, 2q) |
| 10 | 4 | 4a | 83 | 4a | 94 | 4v | 79 | 4v | 85 | 4l | 87 | 4l | 92 | Novel GT4 subtype | 82 (4v, 4l) |
| 11 | 4a/4c | 4a | 88 | 4a | 97 | 4a | 86 | 4a | 92 | 4c | 91 | 4c | 96 | Novel GT4 subtype | 86 (4a, 4) |
| 12 | 4l/4t | 4t | 83 | 4t | 94 | 4a | 79 | 4t | 85 | 4p | 87 | 4l | 93 | Novel GT4 subtype | 84 (4p) |
| 13 | 4m/4v | 4o | 82 | 4v | 95 | 4v | 79 | 4a | 85 | 4l | 86 | 4m | 93 | Novel GT4 subtype | 82 (4v, 4q, 4l, 4) |
| 14 | 4a/4c | 4c | 84 | ND | ND | 4v | 81 | ND | ND | ND | ND | 4a | 89 | Novel GT4 subtype | 84 (4) |
| 15 | 6c/6q | 6d | 78 | 6c | 94 | 6p | 75 | 6q | 87 | 6q | 86 | 6q | 92 | Novel GT6 subtype | 84 (6q) |
| 16 | 1b | ND | ND | ND | ND | 6w | 72 | 6g | 84 | No hit | 0 | 6g | 88 | Novel GT6 subtype | 80 (6q) |
| 17 | 1b | 1a | 79 | 1h | 91 | 1c | 78 | 1c | 84 | 1c | 86 | 1c | 92 | Novel GT1 subtype | 81 (1c, 1) |
| 18 | 3f | NA | NA | NA | NA | 3i | 79 | 3g | 87 | 3i | 87 | 3i | 94 | Novel GT3 subtype | 82 (3i) |
| 19 | 5a | NA | NA | NA | NA | 1b | 74 | 5a | 84 | 4v | 71 | 5a | 85 | Novel GT5 subtype | 78 (5a) |
Homology, in percentage, was calculated by BLASTN analysis for NS3, NS5A, and NS5B. The subtype with the highest match is shown. BLASTN was performed on the sequences generated by full–HCV genome sequencing; the query nucleotide length was 6567 for GT2, 9334 for GT3, 6576 for GT4, 4098 for GT6, 7270 for GT1, and 4095 for GT5.
Abbreviations: aa, amino acid; GT, genotype; HCV, hepatitis C virus; nt, nucleotide; RT-PCR, reverse transcriptase polymerase chain reaction.
Identification of Novel HCV Subtypes by Phylogenetic Analysis
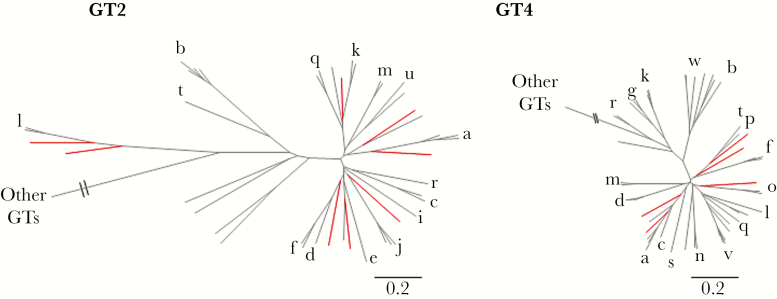
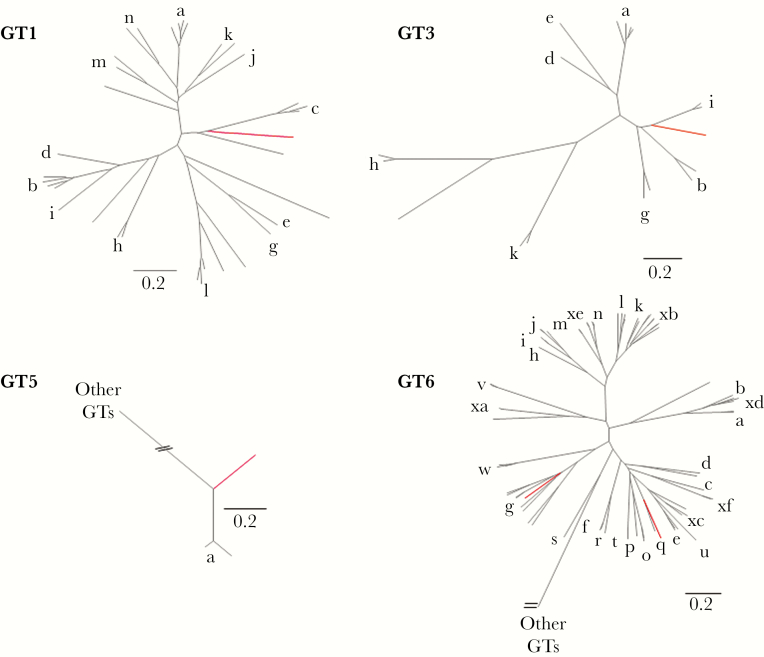
Of the 42 patients who underwent sequencing of the complete HCV genome and phylogenetic analysis, 14 patients were assigned to known HCV subtypes, 4 patients had multiple strains or recombinant infection, sequencing was unsuccessful for 5 patients, and novel subtypes were identified for the remaining 19 patients. Of the 19 patients with novel subtypes, 9 patients were classified as having a novel subtype within genotype 2, 5 patients with novel genotype 4 subtypes, 2 patients with novel genotype 6 subtypes, and 1 each for genotypes 1, 3, and 5 (Figure 1). These novel subtypes showed an absence of recombination break-point using SimPlot analysis (data not shown). For genotype 2, the identified subtypes were distinct from the 15 confirmed and 8 unassigned subtypes previously described for genotype 2 (Figure 2). The sequence of 1 of the 9 patients was shorter, and a separate phylogenetic tree generated consistent results (data not shown). Compared with genotype 2, genotype 4 has a higher genetic diversity, with 18 confirmed and 10 unassigned subtypes. Five patients with novel genotype 4 subtypes were identified, which were phylogenetically distinct from each other and from previously confirmed and unassigned genotype 4 subtypes (Figure 2). One patient with a novel genotype 1 subtype was identified, which was phylogenetically distinct from the 13 confirmed and 7 unassigned genotype 1 subtypes previously described (Figure 3). In addition to the novel genotype 1 subtype, 1 patient with novel genotype 3 subtype was identified, which was phylogenetically distinct from the 8 confirmed subtypes and 1 unassigned subtype previously described (Figure 3). Furthermore, 1 patient with novel genotype 5 subtype was identified. This subtype was distinct from 5a, which is the only subtype confirmed for genotype 5 (Figure 3). In contrast to genotype 5, genotype 6 is the most diverse of all HCV genotypes, with 29 confirmed and 21 unassigned subtypes. Two patients with novel genotype 6 subtypes were identified, which were phylogenetically distinct from each other and from previously confirmed and unassigned genotype 6 subtypes (Figure 3).
Figure 2.
Phylogenetic analysis of genotype 2 and genotype 4 subtypes. Individual maximum likelihood trees were inferred for each genotype, including all previously described confirmed and unassigned subtypes. Previously described subtypes are shown in black, with a corresponding letter for the confirmed subtypes and no letter for the unassigned subtypes. The novel subtypes are shown in red. For genotype 2, 15 confirmed and 8 unassigned subtypes previously described were included. For genotype 4, the 18 confirmed and 10 unassigned subtypes were included in the tree.
Figure 3.
Phylogenetic analysis of genotype 1, 3, 5, and 6 subtypes. Individual maximum likelihood trees were inferred for each genotype, including all previously described confirmed and unassigned subtypes. Previously described subtypes are shown in black, with a corresponding letter for the confirmed subtypes and no letter for the unassigned subtypes. The novel subtypes are shown in red. For GT1, the 13 confirmed and 7 unassigned subtypes previously described were included. For GT3, the 1 unassigned and 8 confirmed subtypes previously described were included. For GT5, the single confirmed subtype was included. For GT6, the 29 confirmed and 21 unassigned subtypes were included in the tree.
Characteristics and Demographics of Patients With Novel HCV Subtypes
Of the 19 patients infected with novel HCV subtypes, 12 patients were enrolled in France (Table 2). Of the remaining 7 patients, 2 patients each were enrolled in Great Britain and the United States, 1 patient was enrolled in each of Canada, India, and Taiwan. The overrepresentation of patients from France was particularly apparent for patients with novel genotype 2 and genotype 4 subtypes, where 6 of 9 patients and 4 of 5 patients, respectively, were from France. Of the patients from France with novel genotype 2 subtypes, the majority were black (4 of 6). There was a difference in gender between the subtypes, as the majority of patients with novel genotype 2 subtypes were male (6 of 9), but for the other novel subtypes, the majority were female (8 of 10).
Table 2.
Characterization of Patients With Novel HCV Subtypes
| Patient | Country/Age/ Gender/Race | Prior Treatment Experience | Cirrhosis | Viral Load, IU/mL | Subtype by Full-Genome Sequencing | Treatment | SVR |
|---|---|---|---|---|---|---|---|
| 1 | GBR/37/M/BL/NH | TN | No | 9 560 000 | Novel GT2 subtype | SOF/VEL 12 wk | Yes |
| 2 | FRA/50/M/BL/NH | TN | No | 4 320 000 | Novel GT2 subtype | SOF/VEL 12 wk | Yes |
| 3 | FRA/69/M/WH/NH | TE (PEG-IFN+RBV) | No | 5 440 000 | Novel GT2 subtype | SOF/VEL 12 wk | Yes |
| 4 | FRA/59/F/BL/NH | TN | No | 3 870 000 | Novel GT2 subtype | SOF/VEL 12 wk | Yes |
| 5 | FRA/72/M/BL/NH | TE (PEG-IFN+RBV) | No | 26 200 000 | Novel GT2 subtype | SOF/VEL 12 wk | Yes |
| 6 | USA/75/F/WH/H | TN | No | 1 590 000 | Novel GT2 subtype | SOF/VEL 12 wk | Yes |
| 7 | FRA/72/M/WH/NH | Unknown | Yes | 2 920 000 | Novel GT2 subtype | SOF/VEL/VOX 12 wk | Yes |
| 8 | USA/73/F/WH/NH | TN | No | 325 000 | Novel GT2 subtype | SOF/VEL/VOX 8 wk | Yes |
| 9 | FRA/62/M/BL/NH | TE (PEG-IFN+RBV) | No | 514 000 | Novel GT2 subtype | SOF/VEL 12 wk | Yes |
| 10 | FRA/42/F/BL/NH | TN | No | 137 000 | Novel GT4 subtype | SOF/VEL 12 wk | Yes |
| 11 | CAN/43/F/BL/NH | TN | No | 119 000 | Novel GT4 subtype | SOF/VEL/VOX 8 wk | Yes |
| 12 | FRA/66/M/WH/NH | TN | No | 4 130 000 | Novel GT4 subtype | SOF/VEL 12 wk | Yes |
| 13 | FRA/64/F/BL/NH | TE (PEG-IFN+RBV) | No | 1 590 000 | Novel GT4 subtype | SOF/VEL 12 wk | Yes |
| 14 | FRA/62/F/AS/NH | TN | No | 1 110 000 | Novel GT4 subtype | SOF/VEL 12 wk | Yes |
| 15 | FRA/60/F/AS/NH | TE (PEG-IFN+RBV) | No | 17 100 000 | Novel GT6 subtype | SOF/VEL/VOX 8 wk | Yes |
| 16 | TWN/62/F/AS/NH | Unknown | No | 3 060 000 | Novel GT6 subtype | LDV/SOF 12 wk | Yes |
| 17 | GBR/58/M/BL/NH | TE (PEG-IFN+RBV) | Yes | 584 000 | Novel GT1 subtype | SOF/VEL 12 wk | No |
| 18 | IND/39/M/AS/ND | TN | No | 2 200 000 | Novel GT3 subtype | SOF+RBV 24 wk | Yes |
| 19 | FRA/58/F/WH/NH | TE (LDV/SOF) | No | 222 000 | Novel GT5 subtype | SOF/VEL/VOX 12 wk | Yes |
Abbreviations: AS, Asian; BL, black; CAN, Canada; F, female; FRA, France; GBR, Great Britain; H, Hispanic; IND, India; LDV, ledipasvir; M, male; NH, non-Hispanic; PEG-IFN+RBV, pegylated interferon with ribavirin; RBV, ribavirin; SOF, sofosbuvir; SVR, sustained virologic response; TE, treatment experienced; TN, treatment naïve; TWN, Taiwan; VEL, velpatasvir; VOX, voxilaprevir; WH, white.
Resistance Analysis of Isolates From Patients With Novel HCV Subtypes
Baseline RAS in NS3, NS5A, and/or NS5B were observed in all but 1 of the novel subtypes. For the 9 patients with novel genotype 2 subtypes, none had NS3 RAS at baseline. For NS5A, T24S was observed at baseline in all patients as a single RAS or in combination with other substitutions: L31M, F28C, L31V, or C92T. For NS5B, M289L was observed in 2 patients (Table 3). The impact on SOF, VEL, and VOX susceptibility to the novel subtypes at baseline was assessed by phenotypic analysis of patient isolates. None of the novel genotype 2 subtypes showed any reduced susceptibility to VOX, with <2-fold EC50 change compared with susceptible wild-type (WT) virus. In 8 of the 9 patient isolates tested, no reduced susceptibility to VEL was observed. One patient isolate with T24S and L31M exhibited a 22-fold increased EC50 to VEL. None of the patient isolates replicated with NS5B in genotype 1b backbone (Table 3).
Table 3.
Phenotypic Analyses of Patient Isolates
| Patient | Virology Subtype | RAS in Patient Isolates | VOX EC50 FC From WT | VEL EC50 FC From WT | SOF EC50 FC From WT | ||
|---|---|---|---|---|---|---|---|
| NS3 | NS5A | NS5B | |||||
| 1 | Novel GT2 | ND | T24S | None | 0.58 | 1.2 | NR |
| 2 | Novel GT2 | ND | T24S L31M | None | 0.66 | 1.2 | NR |
| 3 | Novel GT2 | ND | T24S L31M | None | ND | 22.5 | NR |
| 4 | Novel GT2 | None | T24S F28C | None | 1.09 | 3.7 | NR |
| 5 | Novel GT2 | None | T24S L31M | None | 1.37 | 0.125 | NR |
| 6 | Novel GT2 | None | F28V L31V | M289L | 0.29 | 3.7 | NR |
| 7 | Novel GT2 | None | T24S L31M | M289L | 1.43 | 0.62 | NR |
| 8 | Novel GT2 | None | T24S L31M C92T | None | No data | ND | ND |
| 9 | Novel GT2 | NA | T24S L31M | None | Failed | Failed | NR |
| 10 | Novel GT4 | None | L30R | None | 1.42 | NR | NR |
| 11 | Novel GT4 | None | L30R | None | 0.36 | 2 | NR |
| 12 | Novel GT4 | None | L30R | E237G | 1 | 2 | NR |
| 13 | Novel GT4 | None | L28V L30H | None | 1.25 | 1 | NR |
| 14 | Novel GT4 | None | L30R | None | ND | ND | ND |
| 15 | Novel GT6 | None | F28M | M289L | 1.27 | 3.33 | NR |
| 16 | Novel GT6 | ND | None | None | ND | 0.67 | NR |
| 17 | Novel GT1 | I170V | Q30R L31M | None | ND | 1.6 | 1 |
| 18 | Novel GT3 | ND | A30K | None | ND | ND | ND |
| 19 | Novel GT5 | No FG coverage in NS3 | Q30S L31M T93S | None | ND | 340 | ND |
Abbreviations: FC, fold change; FG, full genome sequencing; GT, genotype; NA, not applicable due to low sequence coverage; ND, not done due to sample unavailability; NR, no replication; RAS, resistance-associated substitutions; SOF, sofosbuvir; VEL, velpatasvir; VOX, voxilaprevir; WT, wild-type.
None of the genotype 4 patients had NS3 RAS at baseline. For NS5A, L30R was observed in 3 patients and L30R/H was observed in 1 patient at baseline. For NS5B, E237G was observed in 1 patient. None of the patient isolates exhibited reduced susceptibility to VOX or VEL with <2-fold EC50 change compared with WT. None of the patient isolates with NS5B replicated in genotype 1b backbone (Table 3).
For the genotype 1 patient, I170V in NS3 and Q30R and L31M in NS5A were observed at baseline. No RAS were observed in NS5B. There was no reduced susceptibility observed to VEL or SOF in this patient isolate (Table 3).
For 1 genotype 3 patient, A30K in NS5A was observed at baseline. No RAS were observed in NS5B. The genotype 5 patient had A30K in NS5A at baseline, and no RAS were observed in NS5B. One genotype 6 patient had F28M in NS5A and M289L in NS5B at baseline. None of the genotype 6 patients had NS3 RAS at baseline. One patient had F28M in NS5A and M289L in NS5B at baseline.
Treatment Outcome for Patients Infected With Novel Subtypes
Overall, across patients with genotype 1–6 infection, 19 patients were identified with novel HCV subtypes. Prior treatment information was available for 17 of 19 patients; 10 were treatment naïve, and 7 were treatment experienced. For the patients with prior treatment experience, 6 had received prior pegylated interferon with ribavirin (PEG-IFN+RBV) treatment and 1 had received prior ledipasvir (LDV)/SOF treatment (Table 2). Following an SOF-containing regimen, 18 of 19 (95%) patients with novel subtypes achieved SVR. The GT5 patient with prior LDV/SOF treatment failure was re-treated with SOF/VEL/VOX for 12 weeks and achieved SVR. The only patient who did not achieve SVR was the patient with a novel genotype 1 subtype who had cirrhosis and received SOF/VEL for 12 weeks. This patient had previously failed PEG-IFN+RBV treatment. Of note, the 5 patient isolates exhibiting a >2-fold increase in VEL EC50 achieved SVR with SOF/VEL or SOF/VEL/VOX treatment (Table 3).
DISCUSSION
Hepatitis C virus is a leading cause of chronic liver disease throughout the world [28]. HCV has a high degree of genetic diversity, and the number of recognized HCV genotypes and subtypes continues to increase [8, 29]. Genotype and subtype information is important for the understanding of the epidemiology of HCV and during the clinical development process to determine the breadth of activity for a given DAA and their combinations, which until recently has been limited to certain genotypes and/or prior treatment exposures. In this study, we identified 19 novel subtypes by analyzing >14 000 HCV-infected patients enrolled in clinical studies of SOF-containing regimens. Interestingly, novel subtypes were identified among all of the most common genotypes (1–6): 9 in genotype 2, 5 in genotype 4, 2 in genotype 6, and 1 each in genotypes 1, 3, and 5. Prior treatment experience information was available for 17 of the patients; 10 were treatment naïve, and 7 had prior treatment experience.
The majority of the patients identified with novel HCV subtypes were enrolled in France; notably, those patients with novel genotype 2 subtypes from France were mainly black men.
This probably reflects the fact that there is an important population diversity in France due to migration from the begining of the 20th century.
In general, a high prevalence of genotype 2 has been described in West Africa [30], and genotype 4 predominates in Central Sub-Saharan Africa and Southeast Asia [6]. However, for both genotype 2 and genotype 4 in these regions, information on the prevalence of specific subtypes is unknown, and it is possible that even more diverse subtypes are prevalent in these regions. The selection of HCV treatment regimens for patients in these regions requires the consideration of the high diversity and potential resistance variants that may impact some regimens. The patterns of diversity and novel subtypes described in this study may be the consequence of human population mobility, recent spread into new risk groups, or immigration from a potentially much older endemic circulation of HCV in Sub-Saharan Africa and Southeast Asia to Europe. Further investigation is needed to investigate the prevalence of these novel subtypes or potentially even additional subtypes in different geographic regions.
Of note, in this study, we identified 54 patients with <85% sequence homology to confirmed subtypes or discordant subtyping results across multiple targets. Of these patients, full-genome sequencing revealed that 14 patient sequences were assigned to previously confirmed HCV subtypes (ie, not novel subtypes). This suggests that partial sequencing information (eg, sequencing of NS3, NS5A, and/or NS5B) is not always sufficient to accurately assign a subtype to the sample, and additional sequence investigation may be required in the event of a viral relapse after DAA therapy.
Resistance analysis was performed to characterize the novel subtypes and the potential effect on treatment outcome. Overall, baseline RAS in NS3, NS5A, and/or NS5B were observed in all but 1 of the novel subtypes. For the majority of the patient isolates, no reduced susceptibility was observed to either VEL or VOX. For the NS5B isolates, the novel subtypes supported weak replication in genotype 1b backbone, and susceptibility to SOF could not be assessed. Despite the presence of RAS at baseline in these patients, 18 of 19 (95%) patients achieved SVR following an SOF-containing regimen, suggesting that SOF-containing regimens are effective against these novel subtypes. The response of these novel subtypes to other HCV regimens has not been previously described. There are now abundant clinical effectiveness data with SOF/VEL across these many genotypes and subtypes, which suggests that baseline genotyping is unnecessary, especially in resource-limited settings, and that a test-and-treat strategy based solely on the identification of active viral replication through HCV core antigen or dried blood spot technology may be sufficient to initiate therapy and monitor response. Taken together, across Gilead clinical studies, 19 novel HCV subtypes were identified, suggesting an even greater genetic diversity of HCV subtypes than previously recognized. Of these patients, 95% achieved SVR following an SOF-containing regimen.
Acknowledgments
Financial support. This work was supported by Gilead Sciences.
Potential conflicts of interest. Charlotte Hedskog, Bandita Parhy, Silvia Chang, Robert H. Hyland, Hongmei Mo, and Evguenia Svarovskaia are employees and stock holders of Gilead Sciences. Stephen D. Shafran has received research support from AbbVie, Bristol-Myers Squibb, Gilead, Janssen, and Merck and honoraria/speaker fees from Gilead, Merck, and Pfizer. Sergio M. Borgia has received research support from AbbVie, Gilead, and Merck and honoraria/speaker fees from AbbVie, Gilead, and Merck. Tarik Asselah is a speaker, consultant, and investigator for AbbVie, BMS, Janssen, Gilead, Roche, and Merck. Christophe Moreno was paid as speaker or advisor for Abbvie, Bayer, BMS, Gilead, and MSD. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Prior presentation. This work was previously presented in part as an oral presentation at the annual meeting of the American Association for the Study of Liver Disease (AASLD); October 2017; Washington DC.
References
- 1. Smith DB, Bukh J, Kuiken C, et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 2014; 59:318–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Borgia SM, Hedskog C, Parhy B, et al. Identification of a novel hepatitis C virus genotype from Punjab, India: expanding classification of hepatitis C virus into 8 genotypes. J Infect Dis 2018; 218:1722–9. [DOI] [PubMed] [Google Scholar]
- 3. Smith DB, Bukh J, Kuiken C, et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 2014; 59:318–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. International Committee on Taxonomy of Viruses (ICTV) https://talk.ictvonline.org/ictv_wikis/flaviviridae/w/sg_flavi/56/hcv-classification. Accessed June 2018.
- 5. Widell A, Forslund O, Medstrand P. A divergent hepatitis C virus (HCV) strain may represent a new major HCV genotype (Genotype 8). In: 23rd International Symposium on Hepatitis C Virus and Related Viruses; October 11–15, 2016; Kyoto Japan. Abstract. [Google Scholar]
- 6. Messina JP, Humphreys I, Flaxman A, et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 2015; 61:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Asselah T, Hassanein T, Waked I, et al. Eliminating hepatitis C within low-income countries - the need to cure genotypes 4, 5, 6. J Hepatol 2017; 68:814–26. [DOI] [PubMed] [Google Scholar]
- 8. Schreiber J, McNally J, Chodavarapu K, et al. Treatment of a patient with genotype 7 hepatitis C virus infection with sofosbuvir and velpatasvir. Hepatology 2016; 64:983–5. [DOI] [PubMed] [Google Scholar]
- 9. De Francesco R, Carfí A. Advances in the development of new therapeutic agents targeting the NS3-4A serine protease or the NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Adv Drug Deliv Rev 2007; 59:1242–62. [DOI] [PubMed] [Google Scholar]
- 10. Hebner CM, Han B, Brendza KM, et al. The HCV non-nucleoside inhibitor tegobuvir utilizes a novel mechanism of action to inhibit NS5B polymerase function. PLoS One 2012; 7:e39163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Manns MP, Bourlière M, Benhamou Y, et al. Potency, safety, and pharmacokinetics of the NS3/4A protease inhibitor BI201335 in patients with chronic HCV genotype-1 infection. J Hepatol 2011; 54:1114–22. [DOI] [PubMed] [Google Scholar]
- 12. Manns M, Palmer M, Flisiak R, et al. A phase-2B trial to evaluate the safety, tolerability and efficacy of a caspase inhibitor, GS-9450, in adults failing PEG/RBV therapy for chronic HCV infection. J Hepatol 2011; 54:S55. [Google Scholar]
- 13. Lam AM, Espiritu C, Bansal S, et al. Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of hepatitis C virus. Antimicrob Agents Chemother 2012; 56:3359–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lam AM, Espiritu C, Bansal S, et al. Hepatitis C virus nucleotide inhibitors PSI-352938 and PSI-353661 exhibit a novel mechanism of resistance requiring multiple mutations within replicon RNA. J Virol 2011; 85:12334–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lam AM, Murakami E, Espiritu C, et al. PSI-7851, a pronucleotide of beta-D-2’-deoxy-2’-fluoro-2’-C-methyluridine monophosphate, is a potent and pan-genotype inhibitor of hepatitis C virus replication. Antimicrob Agents Chemother 2010; 54:3187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lam AM, Espiritu C, Bansal S, et al. Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of hepatitis C virus. Antimicrob Agents Chemother 2012; 56:3359–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeuzem S, Dusheiko GM, Salupere R, et al. ; VALENCE Investigators Sofosbuvir and ribavirin in HCV genotypes 2 and 3. N Engl J Med 2014; 370:1993–2001. [DOI] [PubMed] [Google Scholar]
- 18. Feld JJ, Jacobson IM, Hézode C, et al. ; ASTRAL-1 Investigators Sofosbuvir and velpatasvir for HCV genotype 1, 2, 4, 5, and 6 infection. N Engl J Med 2015; 373:2599–607. [DOI] [PubMed] [Google Scholar]
- 19. Abergel A, Metivier S, Samuel D, et al. Ledipasvir plus sofosbuvir for 12 weeks in patients with hepatitis C genotype 4 infection. Hepatology 2016; 64:1049–56. [DOI] [PubMed] [Google Scholar]
- 20. Bourlière M, Gordon SC, Flamm SL, et al. ; POLARIS-1 and POLARIS-4 Investigators Sofosbuvir, velpatasvir, and voxilaprevir for previously treated HCV infection. N Engl J Med 2017; 376:2134–46. [DOI] [PubMed] [Google Scholar]
- 21. Mizokami M, Dvory-Sobol H, Izumi N, et al. Resistance analyses of Japanese hepatitis C-infected patients receiving sofosbuvir or ledipasvir/sofosbuvir containing regimens in phase 3 studies. J Viral Hepat 2016; 23:780–8. [DOI] [PubMed] [Google Scholar]
- 22. Hedskog C, Chodavarapu K, Ku KS, et al. Genotype- and subtype-independent full-genome sequencing assay for hepatitis C virus. J Clin Microbiol 2015; 53:2049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zwickl DJ. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets Under the Maximum Likelihood Criterion. PhD dissertation, The University of Texas at Austin; 2006. [Google Scholar]
- 24. Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 2006; 55:539–52. [DOI] [PubMed] [Google Scholar]
- 25. Guindon S, Dufayard JF, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010; 59:307–21. [DOI] [PubMed] [Google Scholar]
- 26. Shih IH, Vliegen I, Peng B, et al. Mechanistic characterization of GS-9190 (tegobuvir), a novel nonnucleoside inhibitor of hepatitis C virus NS5B polymerase. Antimicrob Agents Chemother 2011; 55:4196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinson M, Yang H, Sun SC, et al. Novel hepatitis C virus reporter replicon cell lines enable efficient antiviral screening against genotype 1a. Antimicrob Agents Chemother 2010; 54:3099–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis 2005; 5:558–67. [DOI] [PubMed] [Google Scholar]
- 29. Hedskog C, Bhardwaj N, Chang S, et al. Identification of novel HCV genotype and subtypes in patients treated with sofosbuvir-based regimens. Paper presented at: AASLD; October 20–24, 2017; Washington DC. [Google Scholar]
- 30. Kassa E, Bane A, Kefene H. Common genotypes and treatment outcomes of HCV infection among ethiopian patients: a prospective study. Ethiop Med J 2016; 54:1–7. [PubMed] [Google Scholar]