Key Points
Question
Are MUC16 mutations associated with tumor mutation load and prognosis in gastric cancer?
Findings
In this analysis of 437 samples from The Cancer Genome Atlas and 256 samples from an Asian cohort of patients with gastric cancer, MUC16 mutations were significantly associated with greater tumor mutation load and better outcomes among gastric cancer samples in The Cancer Genome Atlas cohort. These findings were independently validated in the Asian cohort.
Meaning
MUC16 mutations appear to be associated with tumor mutation load and can be used to stratify patients with gastric cancer into prognostically distinct groups.
Abstract
Importance
MUC16, which encodes cancer antigen 125 (CA-125), is frequently mutated in gastric cancer (GC); however, its association with tumor mutation load (TML) and outcome in patients with GC has not been established, to date.
Objective
To investigate whether MUC16 mutations are associated with TML and prognosis in patients with GC.
Design, Setting, and Participants
Statistical analysis of genomic data from 437 GC samples obtained from The Cancer Genome Atlas (TCGA) and 256 samples from an Asian cohort. Both cohorts contained data of patients with GC involved in previous genomic studies. Data were obtained from TCGA on September 3, 2017, and from the Asian cohort on March 5, 2013, and analyzed from September 3 to December 1, 2017. The TCGA cohort was used as a discovery set and the Asian cohort as a validation set. Kaplan-Meier survival analysis and multivariate Cox and logistic regression models were applied. Regression models addressed confounding factors; Bayesian variant nonnegative matrix factorization was used to extract mutational signatures. The MutSigCV algorithm was used to identify significantly mutated genes.
Main Outcomes and Measures
Primary outcomes were mutation frequency, overall survival, and TML, calculated using Kaplan-Meier survival analysis, odds ratios (ORs), and significance of signaling pathways.
Results
MUC16 was mutated in 168 of 437 (38.4%) of the GC samples from the TCGA cohort and in 57 of 256 (22.3%) from the Asian cohort. In both cohorts, GC samples with MUC16 mutations exhibited significantly greater TML than those without MUC16 mutations (median mutation counts: TCGA cohort, 264 with MUC16 mutation vs 115 without; Asian cohort, 134 with MUC16 mutation vs 74 without; Wilcoxon rank sum test, both P < .001). This association was independent of mutations in POLE and BRCA1/2 and mutational signatures in the TCGA cohort (OR, 1.87; 95% CI, 1.49-2.36; P < .001) and the Asian cohort (OR, 1.69; 95% CI, 1.25-2.29; P < .001). MUC16 mutations were significantly associated with better prognosis in both cohorts (median overall survival, 46.9 [95% CI, 26.4-NA (not available)] vs 26.7 [95% CI, 20.2-43.1] months; log-rank test, P = .007 [TCGA cohort] and not calculable [the median overall survival of patients with GC and MUC16 mutations could not be calculated because more than half the patients in the group were alive] vs 36.8 months; P = .04 [Asian cohort]). The association remained statistically significant after controlling for age, sex, TNM stage, mutations in POLE and BRCA1/2, and mutational signatures (hazard ratio, 0.61 [95% CI, 0.42-0.89]; log rank test, P = .01). Immune response and cell cycle regulation circuits were among the top altered signaling pathways in samples with MUC16 mutations (normalized enrichment score, 1.70 [95% CI, 1.57-1.79] and 2.04 [95% CI, 1.90-2.18]; adjusted P < .001). The prognostic significance of MUC16 mutation identified in the TCGA cohort was validated in the Asian cohort.
Conclusions and Relevance
These findings indicate that MUC16 mutations may be associated with higher TML, better survival outcomes, and immune response and cell cycle pathways. These findings may be immediately applicable for guiding immunotherapy treatment for patients with GC.
This analysis of genomic data from The Cancer Genome Atlas and an Asian cohort investigates associations between MUC16 mutation, tumor mutation load, and survival in patients with gastric cancer.
Introduction
Gastric adenocarcinoma (herein referred to as gastric cancer [GC]) is the leading cause of cancer-related death worldwide. Despite progress in Helicobacter pylori eradication and early cancer screening, the 5-year survival rate for GC remains 29.6% worldwide.1
Gastric cancer is genomically heterogeneous, with varying tumor mutation loads (TMLs). Recent studies have shown that GC samples with microsatellite instability–high (MSI-H) or POLE (OMIM 174762) mutations had DNA mismatch repair (MMR) signatures and higher TMLs.2,3 Tumor mutation load is an important determinant in molecular subtyping of GC in The Cancer Genome Atlas (TCGA). With the use of GC samples in TCGA, 4 molecular subtypes have been identified, each defined by distinct genomic characteristics.3 Previous studies of GC showed that clonal complexity and driver mutation patterns were associated with survival.2,4 Recent advances in immunotherapy show that MMR-deficient tumors are more sensitive to immune checkpoint blockade, irrespective of tissue of origin.5
MUC16 is a type I transmembrane mucin protein with 3 components: a C-terminal domain, a tandem repeat region, and an extracellular N-terminal section.6,7 Cancer antigen 125 (CA-125), used to monitor disease progression in ovarian cancer, is part of the tandem repeat domain.7
MUC16 (OMIM 606154) is one of the most frequently mutated genes in GC; however, its associations with TML and prognosis remain unclear. In this study, we investigated whether MUC16 mutations are associated with TML and prognosis in patients with GC.
Methods
Genomic Data of GC
Somatic mutation and gene expression data for 437 GC samples in the TCGA were downloaded from Genome Data Commons (https://portal.gdc.cancer.gov). For the Asian cohort, clinical and somatic mutation data were obtained from a previous study.2 The Asian cohort contained data from 256 patients with GC, comprising 78 patients from northern China (1 sample from this group had no mutation in its exomic region and was excluded),4 100 from Hong Kong,8 49 from South Korea,9 and 30 from Japan.10 Gene expression data for the Asian cohort are not available, and survival data were only available for the 78 patients from northern China. We did not include esophageal adenocarcinoma in our study because it differs substantially from GC with respect to mutational signatures, driver mutations (eg, TP53 mutation was present in 140 of 171 esophageal adenocarcinoma samples [81.9%] vs 165 of 347 GC samples [47.6%]; χ2 test, P < .001), and genomic ploidy (genomic doubling event was present in 153 of 365 GC samples [41.9%] vs 97 of 163 esophageal adenocarcinoma samples [59.5%]; χ2 test, P < .001). This study was approved by the Tianjin Medical University Cancer Institute and Hospital Institutional Review Board, which waived additional informed consent because all data used in this study were obtained from public databases. Participants in the original genomic studies provided informed consent.
Mutational Signature Extraction
We used SignatureAnalyzer11 (https://software.broadinstitute.org/cancer/cga/Home) to extract mutational signatures by combining somatic mutation data from the TCGA and Asian cohorts rather than by extracting signatures in each cohort separately. SignatureAnalyzer uses Bayesian-based nonnegative matrix factorization that automatically determines the optimal number of mutational signatures. The Bayesian nonnegative matrix factorization method exploits a shrinkage or automatic relevance determination technique by iteratively pruning components that do not contribute to explanation of final mutation portraits. SignatureAnalyzer factorized the mutational portrait matrix A into 2 nonnegative matrices, W and H (ie, A equals approximately W × H), with W representing mutational signatures and H representing mutational activities. The number of columns of matrix W is the number of mutational signatures. The rows of matrix A are the 96 mutational contexts, and its columns are the 693 GC samples of both cohorts. The 96 mutational contexts are derived from combinations of 6 mutational types (ie, C > A, C > G, C > T, T > A, T > C, and T > G) and their 5′ and 3′ adjacent bases. The pruning process is performed by introducing weight parameter λk, which is associated with the kth column of W and the kth row of H. During inference, the columns and rows of irrelevant components rapidly shrink to zero as λk approaches the optimal number of signatures, which is the number of nonzero columns of matrix W.12 Mutational signatures were annotated by calculating cosine similarity against 21 independently validated mutational signatures in the Catalogue of Somatic Mutations in Cancer13 and by manual review.
MUC16 Mutations vs TML
Because mutations in BRCA1/2 (OMIM 113705 and OMIM 600185, respectively) and POLE and MMR deficiency increase mutation rates in the cancer genome,3 we used a multivariate regression model to analyze associations between MUC16 mutation and TML by including them as confounding factors. Tumor mutation load is defined as log2 transformation of mutation rate per megabase. The extracted MMR mutational signatures were treated as binary variables (ie, 0 and 1) in the multivariate model according to the principle used in a previous study: a signature was considered significant if it contributed to more than 100 substitutions or more than 25% of total mutations.13 We used stan_lm from the R package rstanarm, version 2.13.1 (https://cran.r-project.org/web/packages/rstanarm/index.html) to perform multivariate regression analyses.
Significantly Mutated Genes
We used the MutSigCV algorithm14 to define significantly mutated genes (SMGs) in GC samples with and without MUC16 mutations. Before performing MutSigCV analysis, we removed GC samples with substantial MMR signatures (>100 substitutions or >25% of total mutations) to avoid skewing the results. An additional procedure was performed to identify expressed SMGs in TCGA data15 and an encyclopedia of cell lines16; a gene was considered to be expressed if it had 3 or more reads in 75% or more of the samples, as described in a 2013 study by Kandoth et al.15
Gene Set Enrichment Analysis
As in the analysis of SMGs, we first removed samples with significant MMR signatures and mutations in BRCA1/2 and POLE. The R packages limma17 and edgeR18 were used to evaluate differential expression of each gene in GC samples with and without MUC16 mutations. Specifically, read counts of gene expression data were downloaded from Genomic Data Commons (https://gdc.cancer.gov) and normalized by calcNormFactors in R package edgeR, and then fed to lmFit and eBayes functions in the R limma package. The differential expression statistics obtained from the eBayes function were used as input to perform gene set enrichment analysis for a list of cell-signaling pathways downloaded from MSigDB.19 The fast gene set enrichment analysis algorithm20 implemented in the Bioconductor R package fgsea was used. The P value was calculated based on 1 million permutations.
Prognosis
Kaplan-Meier survival and multivariate Cox regression analyses implemented in the R package survival were used to analyze associations between MUC16 mutations and survival. The log-rank test was used to determine significant differences of survival curves stratified by MUC16 mutations. A 2-sided P < .05 was considered statistically significant. Median overall survival time and 95% CIs are reported where relevant.
Results
TCGA Cohort
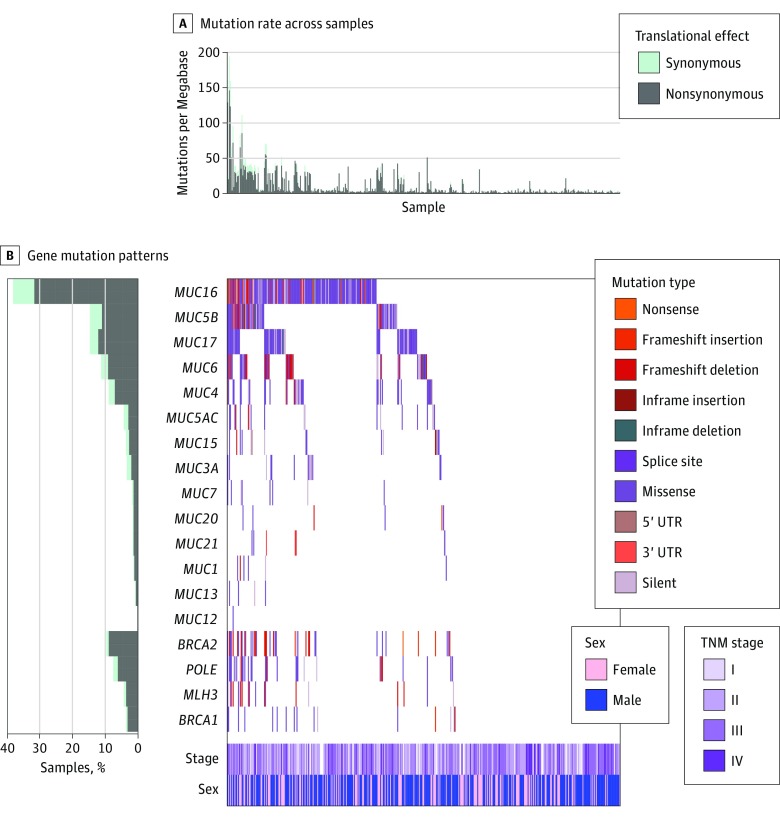
Of the 437 patients in the TCGA cohort, 280 (64.1%) were male, and the median (IQR) age was 67.6 (15.3) years. MUC16 was one of the most frequently mutated genes in the TCGA cohort, accounting for 168 of 437 patients (38.4%). Gastric cancer samples with MUC16 mutations had higher TMLs than samples without MUC16 mutation (Figure 1A). Of the GC samples with MUC16 mutations, 73 of 165 (44.2%) also harbored mutations in genes related to maintenance of genomic integrity, DNA replication proofreading, and MMR, such as BRCA1/2, POLE, and MLH3 (Figure 1B). The mutational associations between MUC16 and its family members are shown in Figure 1B.
Figure 1. Mutation Patterns of the Mucin Gene Family in Relation to Genes Associated With Genomic Instability in The Cancer Genome Atlas Cohort.
A, Mutation rates per megabase in synonymous and nonsynonymous mutations. B, Frequency of synonymous and nonsynonymous mutations, gene mutation patterns across each sample, and distribution of TNM stage by patient sex. UTR indicates untranslated region.
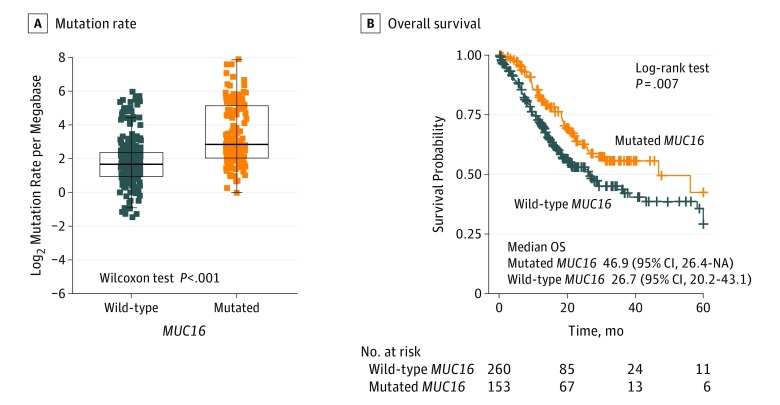
MUC16 Mutation Association With TML
Gastric cancer samples with MUC16 mutations had a significantly higher mutation rate (Figure 2A; Wilcoxon rank sum test, P < .001). Tumor mutation load is largely attributed to genomic instability, which is prevalent in GC. In these samples, we found 6 mutational signatures (eFigure 1 in Supplement 1), including those related to genomic instability. The numbers of somatic mutations attributed to each mutational signature varied considerably in each sample. Underlying associations with these 6 mutational signatures included defects in DNA proofreading owing to recurrent somatic mutations in POLE13 (signature 10, 8256 of 171 732 [4.8%]), overactivity of mRNA-editing enzyme APOBEC (signature 2, 18 669 of 171 732 [10.9%]), reflux of gastric acid (signature 17, 11 267 of 171 732 [6.6%]),21 age-related accumulation of C>T at cytosine-phosphate-guanine dinucleotide (signature 1, 71 816 of 171 732 [41.8%]) and defective MMR (signature 15, 41 769 of 171 732 [24.3%] and signature 21, 19 954 of 171 732 [11.6%]). Signature 21 significantly co-occurred with signature 15 (Fisher exact test, odds ratio [OR], 186; 95% CI, 45.8-1596.3; P < .001). Tumors with MSI-H and a substantial presence of signatures 15 or 21 had greater TML compared with tumors without these features, whereas for TML of tumors with MSI-H, the presence of signatures 15 and 21 was comparable (eFigure 2 in Supplement 1; median TML, 5.51 [95% CI, 2.54 to 7.78] vs 5.74 [95% CI, −0.22 to 7.23]; Kruskal-Wallis rank sum test, P = .18). Mutational activities of signatures 15 and 21 were significantly higher in MSI-H tumors than either MSI-low or MS-stable tumors (eFigure 3 in Supplement 1; MSI-H: 344.3 vs 7.8, MS-stable: 108.4 vs 2.7; Wilcoxon rank sum test, both P < .001). The mutational activity attributable to each mutational signature in each GC sample and variation of these mutational activities is shown in eFigure 4 in Supplement 1. A heat map depicting these 6 mutational signatures and Catalogue of Somatic Mutations in Cancer signatures is shown in eFigure 5 in Supplement 1.
Figure 2. Association of MUC16 Mutation With Tumor Mutation Load and Prognosis in The Cancer Genome Atlas Cohort.
A, Mutation rates of gastric cancer stratified by MUC16 mutation status. B, Kaplan-Meier survival analysis stratified by MUC16 mutation status. NA indicates not available.
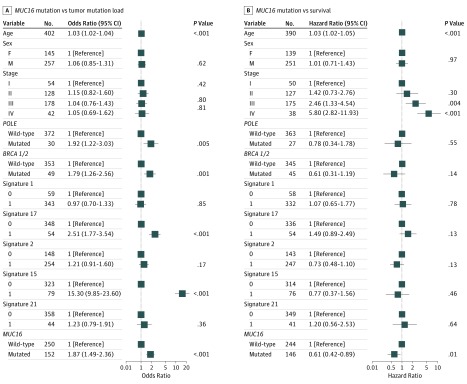
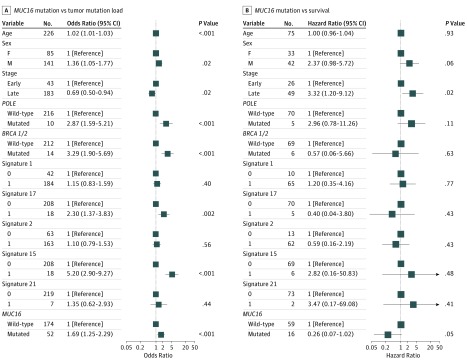
To rule out the possibility that associations between MUC16 mutations and TML were affected by these confounding factors, we included all mutational signatures (except signature 10) and mutations in BRCA1/2 and POLE in the multivariate model. Four GC samples showed a significant presence of signature 10 and 2 samples harbored somatic mutations in POLE. Associations between MUC16 mutations and TML remained statistically significant (OR, 1.87; 95% CI, 1.49-2.36; Wilcoxon rank sum test, P < .001) (Figure 3A).
Figure 3. Association of MUC16 Mutation With Higher Tumor Mutation Load and Survival Outcome in The Cancer Genome Atlas Cohort of Gastric Cancer Samples.
Data are adjusted for age, sex, TNM stage, mutational signatures, and mutations in BRCA1/2 and POLE. Square data markers indicate estimated odds ratios (A) and hazard ratios (B). Error bars represent 95% CIs.
Survival in TCGA Cohort
In Kaplan-Meier survival analysis, the MUC16 mutation was significantly associated with a better survival outcome in the TCGA cohort (Figure 2B; median overall survival, 46.9 [95% CI, 26.4-NA (not available)] vs 26.7 [95% CI, 20.2-43.1] months; log-rank test, P = .007). This association remained statistically significant after controlling for confounding factors such as age, sex, TNM stage, mutations in BRCA1/2 and POLE, and defective MMR signatures (hazard ratio, 0.61 [95% CI, 0.42-0.89]; log-rank test, P = .01) (Figure 3B).
Independent Validation of MUC16 Mutations in the Asian Cohort
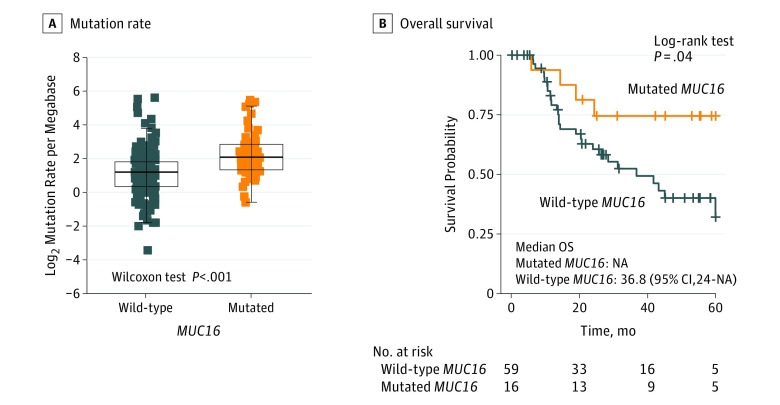
Of the 256 patients in the Asian cohort, 141 (55.1%) were male and median (IQR) age was 63 (17.8) years. MUC16 was also frequently mutated (57 of 256 patients [22.3%]) in the Asian cohort, as were BRCA1/2, POLE, and MLH3 (26 of 256 patients [10.2%] total for all 3). A significantly higher mutation count was also observed in GC samples with MUC16 mutations (mutation count, 134 vs 74; Wilcoxon rank sum test, P < .001) (eFigure 6A in Supplement 1; upper panel). The most prevalent mutational signatures included signature 1, which accounted for 11 401 of 30 115 total mutations (37.9%), and signature 2, which accounted for 7628 of 30 115 (25.3%). Mismatch repair signature 15 contributed to 4363 of 30 115) total mutations (14.5%) and MMR signature 21 contributed to 2158 of 30 115 (7.2%) (eFigure 6B and C in Supplement 1). Associations of mutations among the mucin gene family and BRCA1/2, POLE, and MLH3 are shown in the middle panel of eFigure 6A in Supplement 1. As in TCGA cohort, GC samples with MUC16 mutations had significantly more mutations than those without MUC16 mutation (TML, 2.1 vs 1.2 per megabase; log2 transformation of mutation count per megabase; Wilcoxon rank sum test, P < .001) (Figure 4A). The association of MUC16 mutations with higher TML remained statistically significant after controlling for age, sex, TNM stage, mutational signatures, and mutations in BRCA1/2 and POLE in the multivariate model (OR, 1.69; 95% CI, 1.25-2.29; P < .001) (Figure 5A). In Kaplan-Meier survival analyses, MUC16 mutations were significantly associated with better survival outcomes (Figure 4B; median overall survival, not calculable [the median overall survival of patients with GC and MUC16 mutations could not be calculated because more than half the patients in the group were alive] vs 36.8 months; log-rank test, P = .04). This association remained statistically significant after controlling for confounding factors such as age, sex, TNM stage, and mutational signatures (hazard ratio, 0.26 [95% CI, 0.07-1.02]; P = .05) (Figure 5B).
Figure 4. Association of MUC16 Mutation With Tumor Mutation Load and Prognosis in the Asian Cohort.
A, Mutation rates of gastric cancer stratified by MUC16 mutation status. B, Kaplan-Meier survival analysis stratified by MUC16 mutation status. NA indicates not available.
Figure 5. Association of MUC16 Mutation With Tumor Mutation Load and Survival Outcome in the Asian Cohort.
Data are adjusted for age, sex, TNM stage, mutational signature, and mutations in BRCA1/2 and POLE. Square data markers indicate estimated odds ratios (A) and hazard ratios (B). Error bars represent 95% CIs.
Significantly Mutated Genes and Pathways Associated With MUC16 Mutations
In this analysis, we excluded GC samples with significant MMR signatures and mutations in BRCA1/2 and POLE (see Methods). We performed SMG and gene set enrichment analyses for GC samples with and without MUC16 mutations, respectively. The SMG mutational landscapes of these 2 groups (eFigure 7 in Supplement 1) exhibited differential mutations in RPL22 (8 of 165 [4.8%] vs 4 of 428 [0.9%]; 2-sided P = .005), ACVR2A (10 of 165 [6.1%] vs 6 of 428 [1.4%]; P = .003), APC (21 of 165 [12.7%] vs 30 of 428 [7%]; P = .03), CDH1 (9 of 165 [5.5%] vs 53 of 428 [12.4%]; P = .02) and ELF3 (0 of 165 [0%] vs 12 of 428 [2.8%]; P = .02) (eFigure 8 in Supplement 1). Although mutation frequency for B2M was not statistically significant in GC samples with and without MUC16 mutations (4 of 165 [2.4%] vs 3 of 428 [0.7%]; P = .10), it was significant by the MutSigCV algorithm in the MUC16 mutant group. It was not significant in the MUC16 wild-type group (eFigure 7 in Supplement 1). B2M was associated with antigen presentation and cytolytic activity, and previously its mutation was associated with resistance to immune checkpoint blockade in melanoma.22 Signaling pathways involved in the immune system, cell cycle checkpoints, antigen processing, and DNA replication and repair were significantly altered in GC samples with MUC16 mutations compared with those without MUC16 mutations (normalized enrichment score, 1.70 [95% CI, 1.57-1.79] and 2.04 [95% CI, 1.90-2.18]; adjusted P < .001) (eFigure 9 in Supplement 1). Results of differential gene expression analysis are shown in the eTable in Supplement 2.
Discussion
We analyzed 437 GC samples from the TCGA cohort and 256 GC samples from an Asian cohort for validation. MUC16 was frequently mutated in GC, and its mutation was associated with higher TML and better survival outcome. The association of MUC16 mutation with TML was independent of a significant presence of mutational signatures and of mutations in BRCA1/2 and POLE. Gastric cancer samples with MUC16 mutations were characterized by upregulation of signaling pathways involved in immune response, antigen processing, cell cycle checkpoints, and DNA replication and repair.
MUC16 is frequently mutated in multiple types of human cancer. Owing to its large size, it was often excluded from lists of significantly mutated genes.14 Nonetheless, MUC16 is known to modulate immune response to cancer.6 Our gene set enrichment analyses also indicated that immune response, cell cycle checkpoints, and DNA replication and repair were significantly altered in GC samples with MUC16 mutations. Therefore, therapeutic regimens to abrogate immune inhibition, such as immune checkpoint blockade, may be beneficial for patients with GC who have MUC16 mutations. Gastric cancer may develop other strategies to survive host immune attack, such as loss of antigen presentation via B2M mutation (eFigure 7 in Supplement 1), which has been associated with acquired resistance to anti–programmed death 1 immunotherapy in patients with melanoma.22
Limitations
Our study has several limitations. First, somatic mutation data of the Asian cohort were aggregated from 4 previous studies,4,8,9,10 and the tools used in analyzing sequencing data may have been different between these studies. This difference in sequencing could introduce bias in the final mutation list. Second, the number of samples with follow-up data in the Asian cohort was limited, which limits the ability to adjust for confounding factors. In the Asian cohort, TML was significantly lower in the TCGA cohort (1.4 vs 2.2 log2 transformation of mutation count per megabase; Wilcoxon rank sum test, P < .001). The proportion of GC samples with significant presence of signatures 15 and 21 (associated with MMR) is significantly lower than the TCGA cohort (signature 15: 7.8% vs 20.1%; signatures 21: 3.1% vs 11%; χ2 test, both P < .001). This is probably because there was a higher proportion of MSI-H samples in the TCGA cohort than in the Asian cohort (22% vs 10%; χ2 test, P < .001).
MUC16 is frequently mutated in many other human cancer types (the eTable in Supplement 1). MUC16 or CA-125 has been implicated in pancreatic, breast, lung, and bladder cancers. For instance, MUC16 is involved in inhibiting anticancer immune responses by binding to natural killer cells and acting as a barrier between natural killer cells and targeted cancer cells, thus preventing direct interaction between the natural killer cells and their targets.7 However, the mechanisms underlying the association between MUC16 mutations with better prognosis and higher TML are still unclear. The full implication of MUC16 or CA-125 in GC diagnosis and monitoring remains elusive and requires in-depth studies.
Conclusions
In 2 independent genomic data sets from TCGA and Asian cohorts, MUC16 mutations were associated with higher TML and improved outcome in patients with GC. This finding may have implications for prognostic prediction and therapeutic guidance for GC.
eFigure 1. Mutational Signatures Extracted From Gastric Cancer.
eFigure 2. Tumor Mutation Load Stratified by MSI Status, Signatures 15 and 21.
eFigure 3. Mutational Activities of Signatures 15 and 21 Stratified by MSI Status
eFigure 4. Mutational Activity of Each Signature in Each TCGA Sample (A) and Total Contribution of Each Signature (B)
eFigure 5. Cosine Similarity Between Six Extracted Mutational Signatures Versus 21 COSMIC Signatures
eFigure 6. Mutation Patterns of Mucin Gene Family in Relation to Genes Associated With Genomic Instability (eg, BRCA1/2, POLE and MLH3) in the Asian Cohort
eFigure 7. SMG Mutation Landscape Stratified by MUC16 Mutation
eFigure 8. Mutation Frequencies of SMGs Stratified by MUC16 Mutation
eFigure 9. Gene Set Enrichment Plots of Top Up-Regulated Signaling Pathways
eTable. MUC16 Mutation Frequency Among Human Cancer Types Downloaded From cBioPortal (http://www.cbioportal.org)
eTable. Differential Gene Expression Analysis Result.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359-E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Li X, Wu WK, Xing R, et al. Distinct subtypes of gastric cancer defined by molecular characterization include novel mutational signatures with prognostic capability. Cancer Res. 2016;76(7):1724-1732. doi: 10.1158/0008-5472.CAN-15-2443 [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202-209. doi: 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen K, Yang D, Li X, et al. Mutational landscape of gastric adenocarcinoma in Chinese: implications for prognosis and therapy. Proc Natl Acad Sci U S A. 2015;112(4):1107-1112. doi: 10.1073/pnas.1422640112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409-413. doi: 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suh H, Valle S, Morris DL. Targeting MUC16 in cancer therapy. Chemotherapy: Open Access. 2017;06:6-10. [Google Scholar]
- 7.Felder M, Kapur A, Gonzalez-Bosquet J, et al. MUC16 (CA125): tumor biomarker to cancer therapy, a work in progress. Mol Cancer. 2014;13:129. doi: 10.1186/1476-4598-13-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46(6):573-582. doi: 10.1038/ng.2983 [DOI] [PubMed] [Google Scholar]
- 9.Wong SS, Kim KM, Ting JC, et al. Genomic landscape and genetic heterogeneity in gastric adenocarcinoma revealed by whole-genome sequencing. Nat Commun. 2014;5:5477. doi: 10.1038/ncomms6477 [DOI] [PubMed] [Google Scholar]
- 10.Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46(6):583-587. doi: 10.1038/ng.2984 [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Mouw KW, Polak P, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet. 2016;48(6):600-606. doi: 10.1038/ng.3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan VY, Févotte C. Automatic relevance determination in nonnegative matrix factorization with the β-divergence. IEEE Trans Pattern Anal Mach Intell. 2013;35(7):1592-1605. doi: 10.1109/TPAMI.2012.240 [DOI] [PubMed] [Google Scholar]
- 13.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. ; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain . Signatures of mutational processes in human cancer [published correction appears in Nature. 2013;502(7470):258]. Nature. 2013;500(7463):415-421. doi: 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214-218. doi: 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333-339. doi: 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klijn C, Durinck S, Stawiski EW, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33(3):306-312. doi: 10.1038/nbt.3080 [DOI] [PubMed] [Google Scholar]
- 17.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139-140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sergushichev A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. Preprint. Published online June 20, 2016. bioRxiv. doi: 10.1101/060012 [DOI] [Google Scholar]
- 21.Dulak AM, Stojanov P, Peng S, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45(5):478-486. doi: 10.1038/ng.2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375(9):819-829. doi: 10.1056/NEJMoa1604958 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. Mutational Signatures Extracted From Gastric Cancer.
eFigure 2. Tumor Mutation Load Stratified by MSI Status, Signatures 15 and 21.
eFigure 3. Mutational Activities of Signatures 15 and 21 Stratified by MSI Status
eFigure 4. Mutational Activity of Each Signature in Each TCGA Sample (A) and Total Contribution of Each Signature (B)
eFigure 5. Cosine Similarity Between Six Extracted Mutational Signatures Versus 21 COSMIC Signatures
eFigure 6. Mutation Patterns of Mucin Gene Family in Relation to Genes Associated With Genomic Instability (eg, BRCA1/2, POLE and MLH3) in the Asian Cohort
eFigure 7. SMG Mutation Landscape Stratified by MUC16 Mutation
eFigure 8. Mutation Frequencies of SMGs Stratified by MUC16 Mutation
eFigure 9. Gene Set Enrichment Plots of Top Up-Regulated Signaling Pathways
eTable. MUC16 Mutation Frequency Among Human Cancer Types Downloaded From cBioPortal (http://www.cbioportal.org)
eTable. Differential Gene Expression Analysis Result.