Abstract
The Gram-negative marine propylene-assimilating bacterium, strain PE-TB08W, was isolated from surface seawater. A structural gene analysis using the 16S rRNA gene showed 96, 94, and 95% similarities to Halioglobus species, Haliea sp. ETY-M, and Haliea sp. ETY-NAG, respectively. A phylogenetic tree analysis showed that strain PE-TB08W belonged to the EG19 (Chromatocurvus)-Congregibacter-Haliea cluster within the Halieaceae (formerly Alteromonadaceae) family. Thus, strain PE-TB08W was characterized as a newly isolated Halieaceae bacterium; we suggest that this strain belongs to a new genus. Other bacterial characteristics were investigated and revealed that strain PE-TB08W assimilated propylene, n-butane, 1-butene, propanol, and 1-butanol (C3 and C4 gaseous hydrocarbons and primary alcohols), but not various other alcohols, including methane, ethane, ethylene, propane, and i-butane. The putative alkene monooxygenase (amo) gene in this strain was a soluble methane monooxygenase-type (sMMO) gene that is ubiquitous in alkene-assimilating bacteria for the initial oxidation of alkenes. In addition, two epoxide carboxylase systems containing epoxyalkane, the co-enzyme M transferase (EaCoMT) gene, and the co-enzyme M biosynthesis gene, were found in the upstream region of the sMMO gene cluster. Both of these genes were similar to those in Xanthobacter autotrophicus Py2 and were inductively expressed by propylene. These results have a significant impact on the genetic relationship between terrestrial and marine alkene-assimilating bacteria.
Keywords: propylene assimilation, Halieaceae family, marine bacterium, alkene monooxygenase, EaCoMT
Non-methane hydrocarbons (NMHCs) exist naturally in the environment from sources such as vegetation and fuel combustion, and play an important role in chemical production and carbon circulation in the atmosphere. Since NMHCs are highly reactive, they provide a sink for hydroxyl radicals and play a crucial role in the production and destruction of ozone in the troposphere, influencing the photochemical reactions that occur in the atmosphere (11). NMHCs, particularly short-chain alkenes such as ethylene and propylene, are emitted into the environment not only by plants and microorganisms, but also by other natural and artificial sources. Therefore, the microbial degradation of short-chain NMHCs is critically important for global carbon circulation.
Propylene metabolism pathways in microorganisms are characterized by three properties that are distinct from ethylene metabolism pathways: (1) Ethylene is converted to epoxyethane by alkene monooxygenase, which then produces 2-hydroxyethyl-CoM by epoxyalkane:co-enzyme M transferase (EaCoMT); 2-hydroxyethyl-CoM is converted to 2-ketoethyl-CoM by an alcohol dehydrogenase from the short-chain dehydrogenase/reductase (SDR) family (25). In contrast, propylene is converted to the two (S)- and (R)-epoxypropane enantiomers by alkene monooxygenase, which are then converted to their respective enantiomers of 2-hydroxypropyl-CoM using two types of 2-hydroxypropyl-CoM dehydrogenases (19, 32). The two enantiomers of 2-hydroxypropyl-CoM are converted to 2-ketopropyl-CoM by the R- and S-forms of 2-hydroxypropyl-CoM dehydrogenase (1, 6, 33). (2) The second difference is that the EaCoMT genes of ethylene-assimilating bacteria are located independently in the upstream region of alkene monooxygenase gene clusters (8), which contain the operon. The EaCoMT genes of propylene-assimilating bacteria are contained in an epoxide carboxylase system with 2-hydroxypropyl-CoM dehydrogenase and 2-ketopropyl-CoM oxidoreductase/carboxylase (6, 17). (3) In ethylene metabolism, 2-ketoethyl-CoM is finally converted to acetyl-CoA by CoM reductase/carboxylase, a bifunctional alcohol/aldehyde dehydrogenase, and CoA transferase. Ethylene-assimilating bacteria possess a CoA-transferase/synthetase cluster near an alkene monooxygenase (25). In propylene metabolism, 2-ketopropyl-CoM is converted to acetoacetate or acetone by 2-ketopropyl-CoM oxidoreductase/carboxylase; therefore, a CoA-transferase/synthetase is not involved in propylene metabolism and the associated genes are not located near the alkene monooxygenase gene cluster. While these findings have been limited to soil bacteria, they show that the lineage of each monooxygenase subunit does not differ between species.
In surface seawater, NMHCs are produced as a by-product of the photochemical transformation of dissolved organic matter (29), but are also produced by micro- and macroalgae, photosynthetic bacteria, and cyanobacteria (5). Therefore, the concentrations of NMHCs in surface seawater are supersaturated, and are at higher levels than in the atmosphere (29), suggesting that carbon circulation in the sea surface, as well as in the terrestrial environment, primarily occurs via the activities of marine microorganisms. We previously isolated an alkene-assimilating bacterium and two ethylene-assimilating bacteria (Haliea spp.) from seawater, revealing the initial monooxygenase genes from both strains, which are different from other known alkene-assimilating bacteria (37). Until now, alkene-assimilating marine bacteria have only been reported in our isolates (37), and there is currently a lack of information on marine propylene-assimilating bacteria because no official reports on these organisms or their metabolic capabilities exist. Investigations on the bacterial and genetic characteristics of propylene-assimilating marine bacteria are important for elucidating microbiological dynamics and understanding genetic evolution between terrestrial and marine bacteria.
In the present study, we isolated propylene-assimilating bacteria from seawater and revealed their biological characteristics. Degradation properties were also examined. The complete nucleotide sequences of the putative propylene monooxygenase gene cluster and its related genes involved in epoxide metabolism were elucidated and compared with amo and its related genes from other known alkene-assimilating bacteria.
Materials and Methods
Bacterial strains and media
Propylene-assimilating bacteria were grown in −C (minus C) medium (100 mg of NH4NO3, 10 mg of KH2PO4, 2.5 mg of Fe[III]EDTA, 2.75 mg of vitamin B12, 0.5 mg of biotin, 100 mg of thiamine-HCl, 74.4 mg of Na2EDTA, 0.25 mg of CuSO4·5H2O, 5.75 mg of ZnSO4·7H2O, 4.55 mg of MnCl2·4H2O, 0.6 mg of CoCl2·6H2O, and 0.27 mg of [NH4]6Mo7O24·4H2O in 1 L of filtered seawater, pH 8.0) with propylene as the sole carbon source. Since propylene is a gas, it was provided by replacing 50% of the air in culture vessels. Plate cultures were made using −C containing 1% gellan gum. Cultures were incubated at 20°C. Escherichia coli strains were grown in Luria-Bertani (LB) medium containing 1% polypeptone, 0.5% yeast extract, and 1% NaCl supplemented with ampicillin (100 μg mL−1) when necessary.
Sampling and isolation
Surface seawater was collected from Tokyo Bay, Japan in 120-mL glass vials and cultured in −C liquid medium with propylene. After growth was observed in liquid medium, propylene-assimilating bacteria were purified by serial dilutions with liquid medium and cultured in 96-well plates with propylene. This isolation procedure was repeated five times, at which point the purity of the isolated strain was confirmed by microscopic observations. A transmission electron microscopy (TEM) analysis of purified strains, which were cultured at 22°C for 20 d, was performed by negative staining using JEM-2000EX (Japan Electron Optics Laboratory, Tokyo, Japan). Assessments of DNA G+C contents, cellular fatty acid profiles, and ubiquinones were performed by TechnoSuruga Laboratory (Shizuoka, Japan). Enzyme activity experiments were performed with API ZYM (BioMérieux, Lyon, France) according to the manufacturer’s instructions.
Identification of polyhydroxybutyrate (PHB)
The extraction of PHB from the isolate PE-TB08W was performed according to the method described by Law and Slepecky (20). To detect PHB, methyl esterification was performed using methanol dehydration and sulfuric acid after chloroform extraction. Putative PHB was extracted by chloroform from PE-TB08W cells grown on propylene. The combined organic phase was dried with NaSO4 and concentrated in vacuo to obtain crude extracts. The PHB obtained was added to anhydrous methanol and concentrated sulfuric acid (1:1), and the mixture was incubated at 160°C for 3 h. After gently cooling the sample, the same volume of hexane was added. The hexane layer containing crotonate methyl ester was analyzed by TRACE GC ULTRA gas chromatography coupled to a DSQ II mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA).
Characteristics of growth
Growth temperature
The temperature range for growth was estimated by growing the isolates in liquid −C medium with propylene at 4, 10, 20, 28, 37, and 45°C.
Assimilation of gaseous hydrocarbons
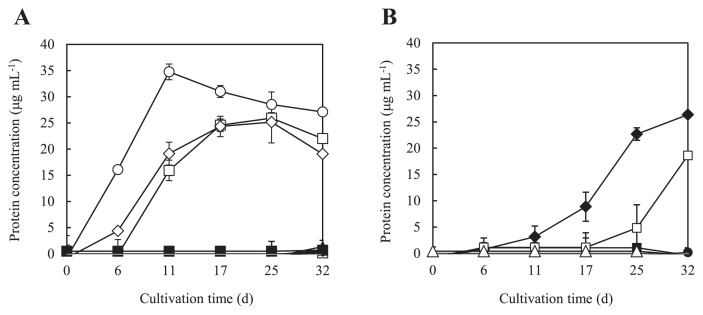
Using 120-mL vials, 100 μL of a 100-fold dilution from each pre-culture was inoculated into 25 mL of −C liquid medium and supplied with various gas hydrocarbons or alcohols. The vials were sealed with Teflon-lined rubber septa and then incubated at 20°C for 32 d. Methane, ethane, propane, n-butane, i-butane, ethylene, propylene, 1-butene, 2-butene, methanol, ethanol, propanol, 1-butanol, 2-butanol, and acetone were added as carbon sources at 50%. Only pentane was added at 0.2%. Except for the propylene culture, growing cells attached to the inner walls of the culture vessels in the liquid medium. Protein concentrations were used as a growth indicator and assessed by the Lowry method (2) using bovine serum albumin as a standard.
Effects of salt concentrations
To test the salt dependence of growth, 0, 0.26, 2.6, 13, 26, 52, 79, or 132 g of NaCl was added to 1 L of modified SOW (artificial sea water) medium containing 1.54 g CaCl2·2H2O, 100 mg KBr, 3 mg KF, 700 mg KCl, 30 mg H3BO3, 4.09 g K2SO4, 200 mg KHCO3, 17 mg SrCl2·6H2O, and 11.1 g MgCl2·6H2O in 1 L of distilled water. The isolates were cultured in these media with propylene as a carbon source at 20°C.
Genetic manipulations
The isolation of total DNA from PE-TB08W and DNA manipulations for E. coli were performed according to the standard protocols by Sambrook et al. (30). The 16S rRNA gene coding sequence was amplified using total DNA as a template with the primers 9F (7) and 1510R (41). PCR amplification was performed in a Bio-Rad S1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA) using Ex Taq DNA polymerase (Takara, Kyoto, Japan). PCR conditions were as described previously (37). The amplified fragment of the 16S rRNA gene (approximately 1.5 kb) was cloned into the T-vector pMD20 (Takara), and then transformed into E. coli DH5α as the host strain. DNA sequencing was conducted by cycle sequencing using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, CA, USA), and was performed using the ABI PRISMTM 310NT genetic analyzer (Applied Biosystems). Sequencing data were analyzed by GENETYX-MAC software ver. 15 (Genetyx Corporation, Tokyo, Japan). Southern blot hybridization was performed according to the method of Sambrook et al. (30). DNA probes were labeled with digoxigenin-11-dUTP (Roche Diagnostics, Mannheim, Germany). Hybridized DNAs were detected by nitrotetrazolium blue chloride (NBT) in combination with 5-bromo-4-chloro-indolyl phosphate (BCIP) solution using an enzyme-linked immunosorbent assay (ELISA).
Sequences of putative alkene monooxygenase (amo) gene clusters and their flanking regions
All primers used in the present study are listed in Table S1. Degenerate primers were designed from eight other known alkene-assimilating bacteria. The amoC gene was obtained by PCR amplification using the forward primer, degenerate-amoC-F3, and the reverse primer, degenerate-amoC-R4. Amplification from part of the amoA gene to the amoC gene containing the complete amoB gene was accomplished using the forward primer, degenerate-amoA-F2, and the reverse primer, PE-amoCR1. Amplification of the amoD gene was accomplished using the forward primer, PE-amoD-F1, and the reverse primer, PE-etnD-R5. Finally, amplification from part of the amoA gene to the amoD gene, containing the complete amoB and amoC genes, was obtained using the forward primer, PE-seq-F1, and the reverse primer, PED-2R4. The EaCoMT gene was amplified with the forward primer CoM-F1d and the reverse primer CoM-R4d, and the resulting fragments were sequenced. The areas encompassing the upstream region of the amoA gene and downstream region of the amoD gene, including the flanking region with the partial EaCoMT gene, were obtained using the LA in vitro Cloning Kit (Takara Bio, Otsu, Japan). Genomic DNA was digested with the proper restriction enzymes and ligated with the cassette DNAs provided in the kit. PCR was performed according to the manufacturer’s instructions using primers that were designed based on the sequence data obtained in this study in combination with the cassette-specific primers provided in the kit.
Construction of EaCoMT expression vectors and epoxy alkane conversion
The EaCoMT gene coding sequence was PCR-amplified using total DNA as the template with the primers PE-CoM-Nde and PE-CoM-Bam. The amplified fragment was digested with NdeI and BamHI and cloned into pET15b(+) (Novagen, Madison, WI) to obtain pSUPE-15E. pSUPE-15E clones were transformed into E. coli BL21(DE3) and cultivated in LB liquid medium containing 100 μg mL−1 ampicillin at 28°C for 6 h. They were cultured for a further 16 h after the addition of IPTG (1 mM).
The culture broth of E. coli BL21(DE3) harboring pSUPE-15E was harvested by centrifugation at 8,000 rpm and washed once with 50 mM PPB (50 mM potassium dihydrogen phosphate and 50 mM dipotassium hydrogen phosphate, pH 8.0). After centrifugation, the supernatant was discarded, and the cell pellet was resuspended in 50 mM PPB containing 10% (w/v) glycerol at approximately one-tenth of the broth volume. The cell suspension was subjected to ultrasonication (Digital Sonifier; Branson Ultrasonics Corporation, USA). The cell-free extract was collected by centrifugation at 15,000 rpm at 4°C for 15 min. The EaCoMT assay and GC analysis were performed according to the method of Boyd et al. (3). Protein concentrations were assessed according to Bradford (4) using bovine serum albumin as a standard.
RNA manipulations
PE-TB08W isolates were grown for one week in −C liquid medium with 50% propylene and n-butane in the gas phase and 0.3% 1-butanol in the medium; cells were cultivated at 20°C for ten d. Total RNA was extracted using the RNeasy Mini Kit (Qiagen K. K., Tokyo, Japan) according to the manufacturer’s instructions. Trace amounts of DNA were removed with RQ1 RNase-Free DNase (Promega, Madison, WI, USA). A reverse transcription-polymerase chain reaction (RT-PCR) was performed with OneStep RT-PCR Kit Ver. 2 (Takara, Tokyo, Japan). RT-PCR without the reverse transcription step was used as a control and was performed using Ex Taq DNA polymerase (Takara). Reverse transcription and cDNA synthesis were performed at 50°C for 30 min and 94°C for 2 min, followed by 25 cycles of 96°C for 30 s, 58°C for 30 s, and 72°C for 1 min.
Phylogenetic analysis
Genetic analyses were conducted using the 16S rRNA gene of strain PE-TB08W, which has been deposited in GenBank. Homology searches were conducted using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). Sequences were aligned with the CLUSTALW ver. 1.83 program. Phylogenetic trees were constructed with TreeViewX software using the neighbor-joining method. A bootstrap analysis with 100 trial replications was performed to assess the reliability of clustering patterns.
Genbank accession
The gene accession number assigned to the 16S rRNA gene of the strain PE-TB08W is AB728559. The gene accession number for the ~20-kb DNA sequence containing the putative amo gene cluster and adjacent alkene metabolism-related genes is AB728560.
Results and Discussion
Isolation and characterization of the propylene-assimilating bacterium, strain PE-TB08W
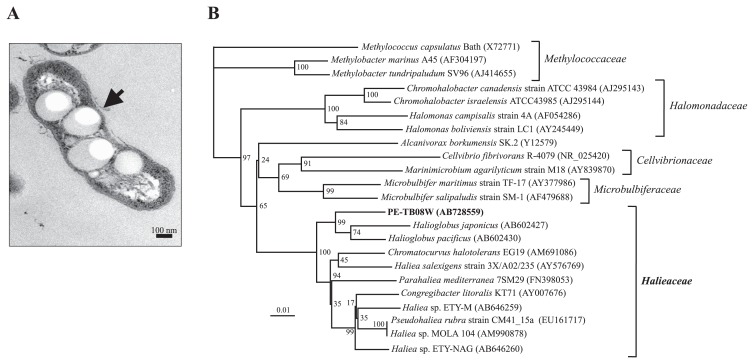
We isolated one propylene-assimilating bacterium, strain PE-TB08W, from the surface seawater of Tokyo Bay, Japan using an enrichment culturing with 50% propylene as the carbon source. This strain was Gram-negative and formed cream-colored colonies on the −C-gellan gum plate with propylene, but rarely grew on Marine Agar 2216 (MA) plates. Using the TEM analysis, we observed the cell type and dimensions for this isolate as a non-flagellated, short rod that was 0.3–0.4 μm in width and 1.2–1.4 μm in length (Fig. 1A). The TEM analysis also showed the low electron density structure of the cell membrane and revealed the accumulation of PHB. Notably, trans- and cis-crotonic acids resulted in the methyl-esterification of PHB (27) and were detected by the GC/MS analysis (data not shown). The optimum growth temperature was 20°C, and optimum salinity range was 26.4 g L−1, similar to Haliea sp. ETY-NAG (37). Cellular fatty acids were composed of four primary fatty acids: C18:1ω7c, C10:0 3OH, C16:1ω7c or C15:0 iso 2OH, and C16:0, similar to Haliea sp. ETY-M (37). The quinone was ubiquinone Q-8, as observed in other Haliea species. The DNA G+C content was 55.1 mol%, which was lower than 57.8, 63.0, and 55.8–65.2 mol% observed in Congregibacter litoralis KT71 (34), Chromatocurvus halotolerans EG19 (10, 35), and other Haliea species (21, 23, 37–39), respectively. The Halieaceae family was re-classified from the Alteromonadaceae family in 2015 and belongs to the order Cellvibrionales (36). Halieaceae family bacteria are gammaproteobacteria, Gram-negative, aerobic, and marine, and have often been isolated from coastal, open, and deep-sea waters (15). Other Halieaceae family bacteria also grow in the presence of NaCl under aerobic conditions and produce pigments; however, the reported colors differ. Park et al. recently reported that two Halioglobus species, H. japonicus and H. pacificus (Gram-negative gammaproteobacteria), were isolated from the seawater of the Northwestern Pacific Ocean (28). Their salinity range, growth temperature, and quinone systems were very similar to our PE-TB08W isolate. The characteristics of PE-TB08W were compared with other bacteria from the Halieaceae family in Table 1.
Fig. 1.
(A) Transmission electron microscopy analysis of strain PE-TB08W. The arrow indicates accumulated PHB. (B) Phylogenetic tree of 16S rRNA genes constructed using a neighbor-joining algorithm. Methylococcus capsulatus served as the outgroup. The numbers on the right are GenBank accession numbers, and the scale bar indicates 0.01 substitutions per 100 base positions. Bootstrap values from 100 trials are listed at the tree nodes. Bacterial families were examined by “List of Prokaryotic names with Standing in Nomenclature” (LPSN; http://www.bacterio.net).
Table 1.
Comparison of PE-TB08W and other Halieaceae family bacteria.
| Characteristics | Strains | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| PE-TB08W | Halioglobus pacificus S1-72T | Haliea salexigens 3X/A02/235T | Haliea sp. ETY-M | Chromocurvus halotolerans EG19T | Congregibacter litoralis KT71T | |
| Isolation source | Surface seawater | Surface seawater | Surface seawater | Surface seawater | Spring run-off stream | Seawater (depth of 8 m) |
| Cell morphology | Short Rods | Coccus | Straight rods | Short Rods | Pleomorphic | Pleomorphic |
| Cell dimensions (mm) | 0.4–0.45×1.2–1.3 | 0.3×0.5 | 0.3–0.7×1.3–1.9 | 0.4–0.45×1.2–1.3 | 0.7×1.5–3.0 | 0.5–4.5×0.4–0.7 |
| Colony color (agar medium) | Cream (−C) | N.A. | Cream (MA) | Purple (5VM) | Pale pinkish-purple (Medium A) | Cream (MA) Pale yellow to orange-red (Under certain conditions) |
| Growth of nutrient agar (MA) | − | + | + | − | N.A. | + |
| Flagella | − | − | + (1 polar) | − | N.A. | + (1–2 polar) |
| DNA G+C content (mol%) | 55.1 | 59.4 | 61.4 | 65.2 | 63.0 | 57.8 |
| Optimum growth temperature (°C) | 20 | 20–25 | 25–30 | 30 | 37 | 28 |
| Optimum salt concentration (g L−1) | 26.4 | 20 | 40 | 13.2 | 40 | 20 |
| PHB accumulation | + | N.A. | N.A. | − | N.A. | N.A. |
| Quinone | Ubiquinone Q-8 | Ubiquinone Q-8 | Ubiquinone Q-8 | Ubiquinone Q-8 | Ubiquinone Q-8 | Ubiquinone Q-8 |
| Major fatty acid compositions | C18:1ω7c, C16:1ω7c or C15:0 iso 2OH, C16:0, C14:0, C10:0 3OH | C18:1ω7c, C16:1ω7c, C17:1ω8c, C11:0 | C18:1ω7c, C17:1ω8c, C16:1ω7c, C17:0 | C18:1ω7c, C16:1ω7c or C15:0 iso 2OH, C16:0, C14:0, C10:0 3OH | N.A. | C18:1ω7c, C16:1ω7c, C16:0, C10:0 3OH |
| Gas hydrocarbon utilization of | ||||||
| Ethylene | − | N.A. | N.A. | + | N.A. | N.A. |
| Propylene | + | N.A. | N.A. | − | N.A. | N.A. |
| 1-butene | + | N.A. | N.A. | − | N.A. | N.A. |
| 2-butene | − | N.A. | N.A. | − | N.A. | N.A. |
| Methane | − | N.A. | N.A. | − | N.A. | N.A. |
| Ethane | − | N.A. | N.A. | − | N.A. | N.A. |
| Propane | − | N.A. | N.A. | − | N.A. | N.A. |
| n-butane | + | N.A. | N.A. | − | N.A. | N.A. |
| i-butane | − | N.A. | N.A. | − | N.A. | N.A. |
| Alcohol utilization of | ||||||
| Methanol | − | N.A. | N.A. | − | — | — |
| Ethanol | − | N.A. | N.A. | + | — | — |
| 1-propanol | + | N.A. | N.A. | (+) | N.A. | N.A. |
| 2-propanol | − | N.A. | N.A. | − | N.A. | N.A. |
| 1-butanol | + | N.A. | N.A. | − | N.A. | N.A. |
| 2-butanol | − | N.A. | N.A. | − | N.A. | N.A. |
| Pentane | − | N.A. | N.A. | − | N.A. | N.A. |
MA, marine agar 2216; +, positive; −, negative; N.A., data not available.
A 16S rRNA gene sequence analysis was performed to examine the phylogeny of this strain. The sequence analysis showed 96, 94, and 95% similarities to Halioglobus species, Haliea sp. ETY-M, and Haliea sp. ETY-NAG, respectively. The phylogenetic tree showed that strain PE-TB08W belonged to the EG19 (Chromatocurvus)-Congregibacter-Haliea cluster (10) in the Halieaceae family (36), which are members of the OM60/NOR5 clade (35); however, PE-TB08W formed a new branch near the Halioglobus species (Fig. 1B). Based on these results, PE-TB08W was assumed to be a new species or even a new genus within the Halieaceae family of bacteria.
PE-TB08W assimilated C3 and C4 gaseous hydrocarbons and primary alcohols
The assimilation of gaseous hydrocarbons and alcohols was examined. PE-TB08W assimilated n-butane and 1-butene as gaseous hydrocarbons, as well as propylene (Fig. 2A), but not other alkenes. The most well-known propylene-assimilating bacteria are Rhodococcus rhodochrous B-276 and Xanthobacter autotrophicus Py2. R. rhodochrous B-276 has been shown to grow on ethylene, propylene, propane, 1-butene, and butane, but not on methane or ethane (12). On the other hand, X. autotrophicus Py2 has been shown to utilize ethylene and propene, but not alkanes, such as ethane, propane, or butane (40). Propylene-utilizing Mycobacterium strains, such as Mycobacterium sp. M156, were shown to grow on propylene and 1-butene, but not ethylene or saturated alkanes (42). It is noteworthy that gaseous hydrocarbon assimilation patterns differed among alkene-assimilating bacteria. The genetic characterization of these bacteria differs from species to species. The alkene monooxygenase gene components of the CMNR group, including R. rhodochrous B-276, comprise four open reading frames, while the alkene monooxygenase gene components of X. autotrophicus Py2 comprise six open reading frames (43) (see below). Some strains of bacteria contain several types of monooxygenases, such as Mycobacterium chubuense NBB4, which is able to grow on alkenes (ethene, propene, butene) and alkanes (ethane, propane, butane, pentane, hexane, heptane, octane, and hexadecane) (9).
Fig. 2.
Growth curves of strain PE-TB08W using various growth substrates. The longitudinal axis indicates protein concentrations (μg mL−1) using the Lowry method. The horizontal axis indicates cultivation times (d). Results are plotted as the mean values obtained for at least two experiments. Error bars represent the range of individual points. (A) Assimilation of gaseous hydrocarbons: (●), Methane; (■), Ethane; (▲), Propane; (◆), Ethylene; (○), Propylene; (□), n-butane; (△), i-butane; (⋄), 1-butene; (▼), cis-2-butene; (▽), trans-2-butene; (×), control (no gaseous hydrocarbon substrate added). (B) Assimilation of alcohols: (●), Methanol; (■), Ethanol; (▲), Acetone; (◆), 1-propanol; (○), 2-propanol; (□), 1-butanol; (△), 2-butanol; (⋄), Pentane; (×), control (not alcohol added as a substrate).
Regarding the assimilation of alcohols, PE-TB08W grew well on 1-propanol and 1-butanol, but not on methanol, ethanol, acetone, 2-propanol, 2-butanol, or pentane (Fig. 2B). X. autotrophicus Py2 was able to utilize various alcohols, including 1-propanol and 1-butanol (40), while R. rhodochrous B-276 grew on 1,2-propanediol (12). Within the former Alteromonadaceae family, Microbulbifer strains were not able to utilize all alcohols, while Marinimicrobium strains were able to utilize methanol, ethanol, 1-propanol, 2-propanol, and 1-butanol (14). Haliea spp. ETY-M and ETY-NAG were only able to grow on ethanol (37). With the exception of strains ETY-M and ETY-NAG, H. salexigens (38), H. mediterranea (23), and Pseudohaliea rubra (formerly Haliea rubra) (35, 39), all other known members of the Haliea genus were unable to assimilate ethylene and propylene (37). However, it is notable that the assimilation of gaseous hydrocarbons has not been reported in the Halieaceae family, except for Haliea sp. ETY-M and ETY-NAG. These results suggest that differences in gaseous hydrocarbons and alcohol-assimilation patterns are related to those in the genes and gene organization in each microorganism.
Nucleotide sequence of a 20-kb region containing genes related to alkene assimilation
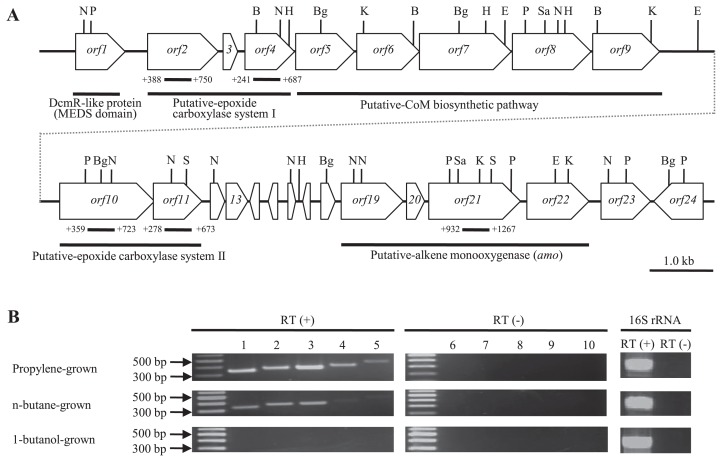
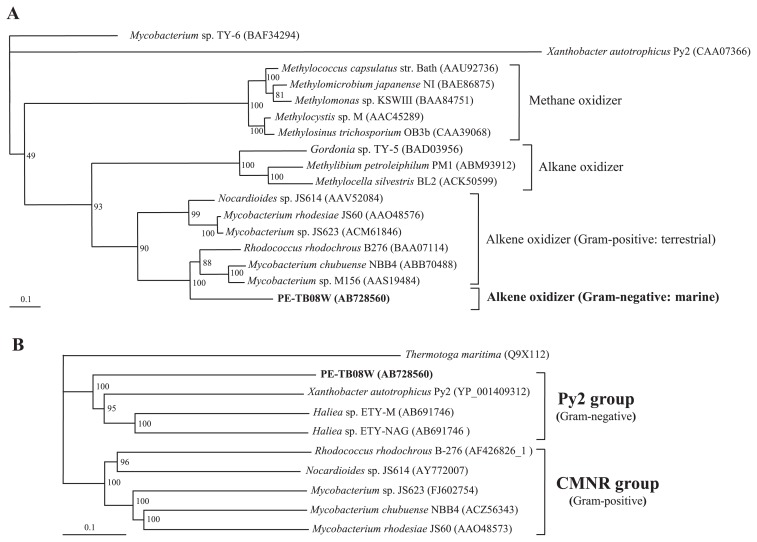
Nearly all alkene-assimilating bacteria possess an initial monooxygenase that is a soluble methane monooxygenase-like (sMMO-like) gene, except for the marine Haliea spp. ETY-M and ETY-NAG (37), which possess a pmo-like gene. To detect this gene in PE-TB08W, we performed Southern blot hybridization using the pmoA-like gene from ETY-M and ETY-NAG as a probe and PCR with the universal primer for pmoA, which was designed from the pmoA-like genes of ETY-M and ETY-NAG. However, no pmo-like genes were detected in PE-TB08W total DNA (data not shown). In order to elucidate the sMMO-like gene sequences that are possessed extensively by alkene-assimilating bacteria, degenerate primers were designed from amoC, which encodes the alpha subunit of other known alkene-assimilating bacteria. We were ultimately able to sequence ~20 kb of the flanking region of the amoC-like gene by genome walking, resulting in a complete amo-like gene cluster and its various functional genes. These regions contained 24 open reading frames, as shown in Fig. 3A. The 20-kb region contained a putative alkene monooxygenase (amo) gene cluster (ORF19–ORF22), which is similar to other known alkene-assimilating bacteria, such as the CMNR group. The phylogenetic tree analysis showed that the deduced amino acid sequence of amoC formed a new branch near the monooxygenases of alkene-assimilating terrestrial bacteria (Fig. 4A). In addition to finding this putative amo gene cluster, two putative epoxide carboxylase systems were located around the clusters and were shown to contain epoxyalkane:co-enzyme M transferase (EaCoMT) (ORF2–ORF4 and ORF10–ORF11), a co-enzyme M biosynthetic gene cluster (ORF5–ORF9), and other genes (ORF1, ORF12–18, and ORF23–ORF24). Each deduced amino acid in these genes, except for in the putative amo gene cluster, was very similar to its counterpart in X. autotrophicus Py2, while the order in the putative amo gene cluster was the same as in other known alkene-assimilating bacteria. Predicted gene products and database similarities are shown in Table S2.
Fig. 3.
Gene organization of approximately 20 kb containing the amo gene cluster and its flanking region in PE-TB08W (A) N, NdeI; P, PstI; B, BamHI; H, HindIII; Bg, BglII; E, EcoRI; Sa, SacI; S, SalI; K, KpnI. Black lines indicate the region amplified by RT-PCR. Numbers on the side indicate positions from the start codon of each gene. (B) RT-PCR analysis. Transcriptional analysis of the amoC, EaCoMT, 2-KPCC, S-HPCDH, and R-HPCDH genes. The upper panel shows the transcription of each mRNA in propylene-grown cells. The middle panel shows the transcription of each mRNA in n-butane-grown cells. The lower panel shows the transcription of each mRNA in 1-butanol-grown cells. Lane 1, amoC; lane 2, EaCoMT gene; lane 3, 2-KPCC gene; lane 4, S-HPCDH gene; lane 5, R-HPCDH gene; lanes 6–10, non-RT corresponding to lanes 1–5, respectively.
Fig. 4.
Phylogenetic tree of deduced amino acid sequences of alkene monooxygenase α-subunits (A) and EaCoMT genes (B) using the neighbor-joining algorithm. Numbers on the right are accession numbers in the database. The scale bar indicates 0.1 substitutions per 100 base positions. Bootstrap values from 100 trials are listed at the tree nodes. In the phylogenetic tree of EaCoMT, Thermotoga maritima (Q9X112) served as an outgroup.
A single copy of the EaCoMT gene exists in the PE-TB08W genome
EaCoMT genes have been found in many alkene-assimilating bacteria, such as the CMNR group and X. autotrophicus Py2 (6, 8, 24, 43). When we searched for homologous EaCoMT genes using a Southern blot hybridization analysis (data not shown), a single copy of a putative EaCoMT gene (ORF2) existed in the epoxide carboxylase system (ORF2–ORF4) of PE-TB08W. The deduced amino acid sequence showed 66, 63, and 62% similarities to the EaCoMTs of X. autotrophicus Py2, Haliea sp. ETY-M, and Haliea sp. ETY-NAG, respectively. The phylogenetic tree showed that the putative EaCoMT gene of PE-TB08W was the most closely related to the 2-hydroxypropyl-CoM lyase of X. autotrophicus strain Py2 (Fig. 4B), and together with Haliea spp. ETY-M and ETY-NAG constituted the EaCoMT Py2 group.
The EaCoMT gene plays a critical role in alkene assimilation (22) because EaCoMT adds co-enzyme M to epoxyalkane during alkene degradation. To examine the function of EaCoMT, we constructed the expression plasmid pSUPE-15E and examined the conversion of various epoxy alkane compounds (epoxypropane, epoxyethane, and epoxybutane) using the recombinant EaCoMT. Table 2 shows the residual epoxyalkane from the cell-free extract conversion. While a crude extract from E. coli harboring pET15b(+) did not decrease the amount of any epoxyalkane, the recombinant EaCoMT showed conversion activity against some epoxyalkanes. Epoxypropane, a mixture of chiral epoxides ((R)- and (S)-epoxypropane), decreased by 70% from that of the control following the addition of recombinant EaCoMT, while (R)- and (S)-epoxypropane were decreased by 30.2 and 61.0%, respectively, from that of the control. Similar to epoxypropane, epoxyethane was markedly decreased (15.8%) by the addition of the recombinant EaCoMT.
Table 2.
Oxidation of various epoxyalkanes by recombinant EaCoMT in PE-TB08W
| Substrates | Relative residual substrates (%) | |
|---|---|---|
|
| ||
| Control | Recombinant EaCoMT | |
| Epoxypropane | 89.4±1.11 | 62.6±1.39 |
| (R)-(+)-epoxypropane | 92.8±0.12 | 28.0±0.73 |
| (S)-(+)-epoxypropane | 90.2±0.54 | 55.1±0.41 |
| Epoxyethane | 96.2±0.22 | 15.2±0.21 |
| 1,2-epoxybutane | 100.4±0.20 | 99.6±0.20 |
| cis-2,3-epoxybutane | 91.9±0.25 | 78.7±0.10 |
| trans-2,3-epoxybutane | 105.9±0.07 | 96.9±0.24 |
Epoxypropane contains (R)-(+)-epoxypropane and (S)-(−)-epoxypropane. Since epoxyethane is a liquid, 1.2 mL (5 μmol) of epoxyethane was injected into a vial. One-hundred-milliliter samples were injected into a gas chromatographic system. Data are mean values in duplicate represented as means±standard deviations.
In contrast to the above epoxy compounds, 1,2-epoxybutane; cis-2,3-epoxybutane; and trans-2,3-epoxybutane did not show a marked decrease. In the cases of ethylene-assimilating bacteria, their EaCoMT were able to decrease epoxyethane, but not epoxypropane (Suzuki, submitted). These results suggest that the EaCoMT of PE-TB08W exhibits epoxy compound conversion activity, which plays a key role in propylene metabolism.
Transcriptional analysis and localization of genes related to alkene metabolism
To examine whether the putative amo genes and epoxide carboxylase systems of strain PE-TB08W were expressed inductively or constitutively by propylene and if they functioned in the oxidation of propylene, we performed a transcriptional analysis using reverse transcription-PCR. Growth substrates were used for propylene, n-butane, and 1-butanol. Total mRNAs were isolated from propylene-grown and n-butane-grown cells at ten d and were isolated from 1-butanol-grown cells at 25 d according to the growth of each (Fig. 2A and B). The indicators for each gene cluster used were the amoC, EaCoMT, 2-ketopropyl-CoM oxidoreductase/carboxylase (2-KPCC), (R)-2-hydroxypropyl-CoM dehydrogenase (R-HPCDH), and (S)-2-hydroxypropyl-CoM dehydrogenase (S-HPCDH) genes. When propylene-grown cells were used, mRNAs were detected for all genes. These results suggest that all the putative amo genes and epoxide carboxylase systems were inductively expressed in the presence of propylene, indicating that they function in the oxidation of propylene. In the putative amo gene cluster, a putative sigma54-like promoter sequence (24) (GGCCCCCAGGTCG AGCAGC; boldface indicates −24 and −12 elements) existed in a 193-bp region upstream of the amoA start codon, while a putative stem-loop sequence (ATGATGAGCCTCCTCGGA ACACAGCAT; boldface indicates stem sequences) existed in the region downstream of amoD. A putative Shine-Dalgarno sequence was identified in the upstream region of each putative amo gene. This result also suggests that putative amo genes constitute an operon structure. In addition, putative amo genes may be expressed separately from epoxide carboxylase systems via some regulatory systems.
Very thin bands were detected in S-HPCDH and R-HPCDH (Fig. 3B; lanes 4 and 5) in n-butane-grown cells; however, it is unlikely that these genes are involved in n-butane assimilation because they act on their respective enantiomers of 2-hydroxypropyl-CoM in propylene metabolism (19, 32). S-HPCDH and R-HPCDH are a part of putative epoxide carboxylase system I and putative epoxide carboxylase system II, respectively, and are present in the region downstream with EaCoMT and 2-KPCC (Fig. 3A). Since EaCoMT and 2-KPCC were transcribed in n-butane-grown cells, S-HPCDH and R-HPCDH appear to constitute an operon with EaCoMT and 2-KPCC, respectively, because promoter-like sequences were not found in their upstream regions. Kotani et al. reported that the propane hydroxylase genes in Gordonia sp. TY-5 and Mycobacterium sp. TY-6 comprised the following four components: (1) a hydroxylase large subunit, (2) a reductase, (3) a hydroxylase small subunit, and (4) a coupling protein (18). Thus, the putative amo genes in PE-TB08W may also function in butane assimilation. PE-TB08W was positive for n-butane assimilation, whereas X. autotrophicus Py2 did not have the ability to assimilate alkanes. A recent study reported that Mycobacterium chubuense NBB4 possesses a pmo-like gene homolog (pHMO), and the heterologous expression of pHMO led to a decrease in ethane, propane, and butane (9). Nocardioides sp. CF8 also possesses a pmo-like gene homolog (pBMO) in addition to alkane hydroxylase genes (31). However, no pmo-like genes were detected in PE-TB08W total DNA. In contrast, no mRNAs were detected for any of the indicator genes in cells grown on 1-butanol (Fig. 3B) with any of the growth substrates used, suggesting that neither putative epoxide carboxylase systems nor putative amo genes were involved in 1-butanol metabolism.
To examine whether the putative amo genes and epoxide carboxylase systems are essential for propylene assimilation, we attempted to express the putative amo genes and disrupt the putative amo genes and epoxide carboxylase systems; however, we were unsuccessful. The putative amo gene was expressed in the presence of propylene as the sole carbon and energy source in RT-PCR. In addition, putative amo genes constitute an operon and are present near the regions of epoxide carboxylase systems and the putative CoM biosynthetic pathway. These results strongly suggest that the putative amo gene functions as an alkene monooxygenase of PE-TB08W.
Positional relationship between the putative amo gene cluster and EaCoMT gene
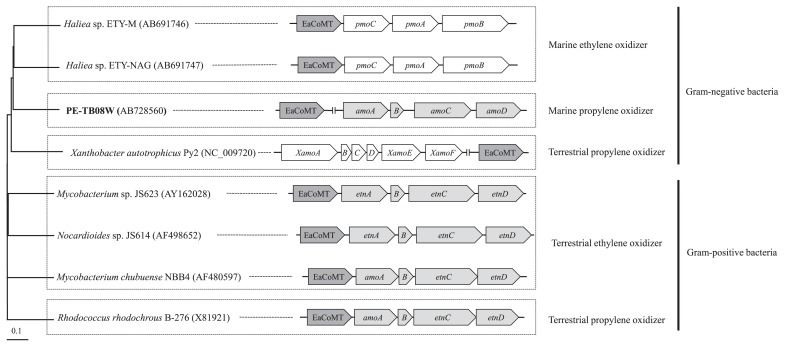
Observations of sequence data, assimilation data, and transcriptional data from the PE-TB08W isolate gradually clarified the order of the EaCoMT/amo genes in the relationship between terrestrial and marine alkene-assimilating bacteria. The location of the EaCoMT gene differed among ethylene-and propylene-assimilating bacteria (Fig. 5). The ethylene-assimilating terrestrial bacteria, Nocardioides sp. JS614 and Mycobacterium strain JS60, possess EaCoMT genes immediately upstream of their alkene monooxygenase gene clusters (8, 16). In contrast, XecA of X. autotrophicus strain Py2 is part of the epoxide carboxylase system that does not form an operon. Although the EaCoMT gene of Mycobacterium strain JS60 (8) is co-transcribed with the amo gene cluster, the EaCoMT gene of marine ethylene-assimilating Haliea sp. ETY-M is co-transcribed with the emo gene cluster (Suzuki, submitted). These findings explicitly suggest a difference in genetic diversity between terrestrial and marine gaseous hydrocarbon-assimilating bacteria.
Fig. 5.
Relationships between 16S rRNA, alkene monooxygenase, and EaCoMT genes from various alkene-assimilating bacteria. Phylogenetic tree of 16S rRNA genes constructed using a neighbor-joining dendrogram. The scale bar indicates 0.1 substitutions per 100 base positions. Numbers in parentheses on the right of the strain name are accession numbers in the database. Components of the EaCoMT and alkene monooxygenase gene cluster are represented by different colors: EaCoMT gene, gray; soluble methane monooxygenase-type (amo or etn) genes, light gray; xamo and particulate methane monooxygenase (pmo)-type genes, white.
It is particularly noteworthy that EaCoMT and other gene clusters from PE-TB08W belong to the Py2 group, whereas the putative amo genes were more closely related to the CMNR group. Furthermore, the amo gene cluster and EaCoMT phylogeny did not reflect 16s rRNA gene phylogeny and the location of the EaCoMT gene differed from species to species (Fig. 5). There were seven truncated genes (orf12-18) reading in the opposite direction between the putative epoxide carboxylase system and amo gene cluster of PE-TB08W (Fig. 3A). Although each of the amo genes in PE-TB08W and their order were more similar to those in the CMNR group than in X. autotrophicus Py2, the epoxide carboxylase system is more closely related to the latter. Based on these results, we speculated that the gene arrangements in strain PE-TB08W were constituted with non-homologous recombination or horizontal gene transfer to enable efficient propylene assimilation in the marine environment; genetic transformation within a host genome occurs rapidly in this environment (26). Our supposition is also supported by the existence of the transposase/integrase gene and truncated genes that were similar to those in Pseudoalteromonas haloplanktis and the genus Shewanella, both of which are marine microorganisms (13).
Alkene-assimilating marine bacteria belonging to the Halieaceae family have only been isolated in our laboratory. Among this family of bacteria, the assimilation of other gaseous hydrocarbons, related genes, and similar genetic characterizations could be common to each microorganism. Future studies to examine further gaseous hydrocarbon assimilation and additional common genes within members of the Halieaceae family will be important for elucidating microbial dynamics together with genetic relationships between terrestrial and marine bacteria groups. The isolation of many other gaseous hydrocarbon-assimilating marine bacteria is greatly desired.
Supplementary Information
Acknowledgements
We thank Dr. Toshiya Shigeno (Tsukuba Environmental Microorganism Institute, Japan) and Dr. Ken-ichiro Suzuki (Tokyo University of Agriculture) for their helpful discussions. We also thank Ms. Seira Furumatsu and Ms. Aya Akiba for their assistance with genetic analyses and bacterial characterization. This research was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan, under the project supporting the formation of research centers in private universities.
References
- 1.Allen J.R., Clark D.D., Krum J.G., Ensign S.A. A role for coenzyme M (2-mercaptoethanesulfonic acid) in a bacterial pathway of aliphatic epoxide carboxylation. Proc Natl Acad Sci USA. 1999;96:8432–8437. doi: 10.1073/pnas.96.15.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bensadoun A., Weinstein D. Assay of proteins in the presence of interfering materials. Anal Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 3.Boyd J.M., Clark D.D., Kofoed M.A., Ensign S.A. Mechanism of inhibition of aliphatic epoxide carboxylation by the coenzyme M analog 2-bromoethanesulfonate. J Biol Chem. 2010;285:25232–25242. doi: 10.1074/jbc.M110.144410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 5.Broadgate W.J., Malin G., Küpper F.C., Thompson A., Liss P.S. Isoprene and other non-methane hydrocarbons from seaweeds: A source of reactive hydrocarbons to the atmosphere. Mar Chem. 2004;88:61–73. [Google Scholar]
- 6.Broberg C.A., Clark D.D. Shotgun proteomics of Xanthobacter autotrophicus Py2 reveals proteins specific to growth on propylene. Arch Microbiol. 2010;192:945–957. doi: 10.1007/s00203-010-0623-3. [DOI] [PubMed] [Google Scholar]
- 7.Brosius J., Dult T.J., Sleeter D.D., Noller H.F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 8.Coleman N.V., Spain J.C. Epoxyalkane: Coenzyme M transferase in the ethene and vinyl chloride biodegradation pathways of Mycobacterium strain JS60. J Bacteriol. 2003;185:5536–5545. doi: 10.1128/JB.185.18.5536-5545.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman N.V., Le N.B., Ly M.A., Ogawa H.F., McCarl V., Wilson N.L., Holmes A.J. Hydrocarbon monooxygenase in Mycobacterium: recombinant expression of a member of the ammonia monooxygenase superfamily. ISME J. 2012;6:171–182. doi: 10.1038/ismej.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csotonyi J.T., Stackebrandt E., Swiderski J., Schumann P., Yurkov V. Chromocurvus halotolerans gen. nov., sp. nov., a gammaproteobacterial obligately aerobic anoxygenic phototroph, isolated from a Canadian hypersaline spring. Arch Microbiol. 2011;193:573–582. doi: 10.1007/s00203-011-0698-5. [DOI] [PubMed] [Google Scholar]
- 11.Donahue N.M., Prinn R.G. Nonmethane hydrocarbon chemistry in the remote marine boundary layer. J Geophys Res. 1990;95:18387–18411. [Google Scholar]
- 12.Furuhashi K., Taoka A., Uchida S., Karube I., Suzuki S. Production of 1,2-epoxyalkanes from 1-alkenes by Nocardia corallina B-276. Eur J Appl Microbiol Biotechnol. 1981;12:39–45. [Google Scholar]
- 13.Gauthier G., Gauthier M., Christen R. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int J Syst Bacteriol. 1995;45:755–761. doi: 10.1099/00207713-45-4-755. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez J., Mayer F., Moran M., Hodson R., Whitman W. Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium geor-giense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int J Syst Evol Microbiol. 1997;47:369–376. doi: 10.1099/00207713-47-2-369. [DOI] [PubMed] [Google Scholar]
- 15.Ivanova E.P., Mikhailov V.V. A new family, Alteromonadaceae fam. nov., including marine proteobacteria of the genera Alteromonas, Pseudoalteromonas, Idiomarina and Colwellia. Microbiology. 2001;70:15–23. [PubMed] [Google Scholar]
- 16.Jin Y.O., Cheung S., Coleman N.V., Mattes T.E. Association of missense mutations in epoxyalkane coenzyme M transferase with adaptation of Mycobacterium sp. strain JS623 to growth on vinyl chloride. Appl Environ Microbiol. 2010;76:3413–3419. doi: 10.1128/AEM.01320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kofoed M.A., Wampler D.A., Pandey A.S., Peters J.W., Ensign S.A. Roles of the redox-active disulfide and histidine residues forming a catalytic dyad in reactions catalyzed by 2-ketopropyl coenzyme M oxidoreductase/carboxylase. J Bacteriol. 2011;193:4904–4913. doi: 10.1128/JB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotani T., Kawashima Y., Yurimoto H., Kato N., Sakai Y. Gene structure and regulation of alkane monooxygenases in propane-utilizing Mycobacterium sp. TY-6 and Pseudonocardia sp. TY-7. J Biosci Bioeng. 2006;102:184–192. doi: 10.1263/jbb.102.184. [DOI] [PubMed] [Google Scholar]
- 19.Krishnakumar A.M., Sliwa D., Endrizzi J.A., Boyd E.S., Ensign S.A., Peters J.W. Getting a handle on the role of coenzyme M in alkene metabolism. Microbiol Mol Biol Rev. 2008;72:445–456. doi: 10.1128/MMBR.00005-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law J.H., Slepecky R.A. Assay of Poly-b-hydroxybutyric acid. J Bacteriol. 1960;82:33–36. doi: 10.1128/jb.82.1.33-36.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin C.Y., Zhang X.Y., Liu A., Liu C., Song X.Y., Su H.N., Qin Q.L., Xie B.B., Zhang Y.Z., Chen X.L. Haliea atlantica sp. nov., isolated from seawater, transfer of Haliea mediterranea to Parahaliea gen. nov. as Parahaliea mediterranea comb. nov. and emended description of the genus Haliea. Int J Syst Evol Microbiol. 2015;65:3413–3418. doi: 10.1099/ijsem.0.000431. [DOI] [PubMed] [Google Scholar]
- 22.Liu X., Mattes T.E. Epoxyalkane: coenzyme M transferase gene diversity and distribution in groundwater samples from chlorinated ethene contaminated sites. Appl Environ Microbiol. 2016;82:3269–3279. doi: 10.1128/AEM.00673-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucena T., Pascual J., Garay E., Arahal D.R., Macián M.C., Pujalte M.J. Haliea mediterranea sp. nov., a marine gammaproteobacterium. Int J Syst Evol Microbiol. 2010;60:1844–1848. doi: 10.1099/ijs.0.017061-0. [DOI] [PubMed] [Google Scholar]
- 24.Mattes T.E., Coleman N.V., Spain J.C., Gossett J.M. Physiological and molecular genetic analyses of vinyl chloride and ethene biodegradation in Nocardioides sp. strain JS614. Arch Microbiol. 2005;183:95–106. doi: 10.1007/s00203-004-0749-2. [DOI] [PubMed] [Google Scholar]
- 25.Mattes T.E., Alexander A.K., Coleman N.V. Aerobic biodegradation of the chloroethanes: pathways, enzymes, ecology, and evolution. FEMS Microbiol Rev. 2010;34:445–475. doi: 10.1111/j.1574-6976.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 26.McDaniel L.D., Young E., Delaney J., Ruhnau F., Ritchie K.B., Paul J.H. High frequency of horizontal gene transfer in the oceans. Science. 2010;330:50. doi: 10.1126/science.1192243. [DOI] [PubMed] [Google Scholar]
- 27.Monteil-Rivera F., Betancourt A., Van Tra H., Yezza A., Hawari J. Use of headspace solid-phase microextraction for the quantification of poly(3-hydroxybutyrate) in microbial cells. J Chromatogr A. 2007;1154:34–41. doi: 10.1016/j.chroma.2007.03.121. [DOI] [PubMed] [Google Scholar]
- 28.Park S., Yoshizawa S., Inomata K., Kogure K., Yokota A. Halioglobus japonicus gen. nov., sp. nov., and Halioglobus pacificus sp. nov., within the class Gammaproteobacteria, isolated from seawater. Int J Syst Evol Microbiol. 2011;62:1784–1789. doi: 10.1099/ijs.0.031443-0. [DOI] [PubMed] [Google Scholar]
- 29.Riemer D.D., Milne P.J., Zika R.G., Pos W.H. Photoproduction of nonmethane hydrocarbons (NMHCs) in seawater. Mar Chem. 2000;71:177–198. [Google Scholar]
- 30.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; NY: 2001. [Google Scholar]
- 31.Sayavedra-Soto L.A., Hamamura N., Liu C-W., Kimbrel J.A., Chang J.H., Arp D.J. The membrane-associated monooxygenase in the butane-oxidizing Gram-positive bacterium Nocardioides sp. strain CF8 is a novel member of the AMO/PMO family. Environ Microbiol Rep. 2011;3:390–396. doi: 10.1111/j.1758-2229.2010.00239.x. [DOI] [PubMed] [Google Scholar]
- 32.Shennan J.L. Utilisation of C2–C4 gaseous hydrocarbons and isoprene by microorganisms. J Chem Technol Biotechnol. 2006;81:237–256. [Google Scholar]
- 33.Sliwa D.A., Krishnakumar A.M., Peters J.W., Ensign S.A. Molecular basis for enantioselectivity in the (R)- and (S)-hydroxypropylthioethanesulfonate dehydrogenases, a unique pair of stereoselective short-chain dehydrogenases/reductases involved in aliphatic epoxide carboxylation. Biochemistry. 2010;49:3487–3498. doi: 10.1021/bi100294m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spring S., Lünsdorf H., Fuchs B.M., Tindall B.J. The photosynthetic apparatus and its regulation in the aerobic gammaproteobacterium Congregibacter litoralis gen. nov., sp. nov. PLoS One. 2009;4:e4866. doi: 10.1371/journal.pone.0004866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spring S., Riedel T., Spröer C., Yan S., Harder J., Fuchs B.M. Taxonomy and evolution of bacteriochlorophyll a-containing members of the OM60/NOR5 clade of marine gammaproteobacteria: description of Luminiphilus syltensis gen. nov., sp. nov., reclassification of Haliea rubra as Pseudohaliea rubra gen. nov., comb. nov., and emendation of Chromatocurvus halotolerans. BMC Microbiol. 2013;13:118. doi: 10.1186/1471-2180-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spring S., Scheuner C., Göker M., Klenk H.P. A taxonomic framework for emerging groups of ecologically important marine gammaproteobacteria based on the reconstruction of evolutionary relationships using genome-scale data. Front Microbiol. 2015;6:281. doi: 10.3389/fmicb.2015.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki T., Nakamura T., Fuse H. Isolation of two novel marine ethylene-assimilating bacteria, Haliea species ETY-M and ETY-NAG, containing particulate methane monooxygenase-like fenes. Microbes Environ. 2012;27:54–60. doi: 10.1264/jsme2.ME11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urios L., Intertaglia L., Lesongeur F., Lebaron P. Haliea salexigens gen. nov., sp. nov., a member of the Gammaproteobacteria from the Mediterranean Sea. Int J Syst Evol Microbiol. 2008;58:1233–1237. doi: 10.1099/ijs.0.65470-0. [DOI] [PubMed] [Google Scholar]
- 39.Urios L., Intertaglia L., Lesongeur F., Lebaron P. Haliea rubra sp. nov., a member of the Gammaproteobacteria from the Mediterranean Sea. Int J Syst Evol Microbiol. 2009;59:1188–1192. doi: 10.1099/ijs.0.002220-0. [DOI] [PubMed] [Google Scholar]
- 40.Van Ginkel C.G., de Bont J.A.M. Isolation and characterization of alkene-utilizing Xanthobacter spp. Arch Microbiol. 1986;145:403–407. [Google Scholar]
- 41.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woodland M.P., Matthews C.S., Leak D.J. Properties of a soluble propene monooxygenase from Mycobacterium sp. (strain M156) Arch Microbiol. 1994;163:231–234. [Google Scholar]
- 43.Zhou N.Y., Jenkins A., Chion C.K., Leak D.J. The alkene monooxygenase from Xanthobacter strain Py2 is closely related to aromatic monooxygenases and catalyzes aromatic monohydroxylation of benzene, toluene, and phenol. Appl Environ Microbiol. 1999;65:1589–1595. doi: 10.1128/aem.65.4.1589-1595.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.