SUMMARY
Genome-wide studies identified genetic variants linked to neurologic diseases. Environmental factors also play important roles, but no methods are available for their comprehensive investigation. We developed an approach that combines genomic data, screens in a novel zebrafish model, computational modeling, perturbation studies and multiple sclerosis (MS) patient samples to evaluate the effects of environmental exposures on CNS inflammation. We found that the herbicide Linuron amplifies astrocyte pro-inflammatory activities by activating signaling via sigma receptor 1, inositol requiring enzyme-1α (IRE1α) and X-box binding protein 1 (XBP1). Indeed, astrocyte-specific shRNA- and CRISPR/Cas9-driven gene inactivation combined with RNA-Seq, ATAC-Seq, ChIP-Seq and the study of patient samples suggest that IRE1α/XBP1 signaling promotes CNS inflammation in experimental autoimmune encephalomyelitis (EAE) and potentially MS. In summary, these studies define environmental mechanisms that control astrocyte pathogenic activities and establish a multidisciplinary approach for the systematic investigation of the effects of environmental exposures in neurologic disorders.
In Brief
An environmental trigger, the herbicide Linuron, boosts astrocyte pathogenic activities in the context of CNS inflammation by activating IRE1alpha/XBP1 signaling in mice and potentially, multiple sclerosis patients.
Graphical Abstract
INTRODUCTION
Genetic and environmental factors contribute to the pathogenesis of neurologic diseases (Al-Chalabi and Hardiman, 2013; Chin-Chan et al., 2015; Kamel, 2013; Olsson et al., 2017; Rappaport and Smith, 2010; Tshala-Katumbay et al., 2015). Extensive genomic studies have identified genetic determinants that affect neurologic disorders. Environmental factors affecting disease pathogenesis have also been identified. For example, microbial metabolites and changes in melatonin levels driven by seasonal variations in night length affect central nervous system (CNS) inflammation (Farez et al., 2015; Rothhammer et al., 2018; Rothhammer et al., 2016). However, there is still an unmet need for methods to systematically evaluate the effects of environmental factors on CNS pathology and the mechanisms involved.
Astrocytes are CNS-resident cells that play important roles in tissue development and homeostasis, while they can also contribute to the pathogenesis of neurologic disorders (Liddelow et al., 2017; Mayo et al., 2014; Rothhammer and Quintana, 2015). Indeed, astrocytes and microglia control CNS inflammation and neurodegeneration (Colonna and Butovsky, 2017; Wheeler and Quintana, 2018). Accordingly, genetic variants linked to neurologic disorders modulate astrocyte and microglia activity (Raj et al., 2014; Sims et al., 2017). We and others recently showed that commensal bacteria metabolites regulate microglia and astrocyte responses, suggesting that environmental factors modulate CNS-resident cells to contribute to the pathogenesis of neurologic diseases (Erny et al., 2015; Rothhammer et al., 2018; Rothhammer et al., 2016; Sampson et al., 2016; Thion et al., 2018). However, the identity of most of these environmental factors and the mechanisms involved are still unknown.
Here we describe a multidisciplinary approach to identify environmental factors that boost astrocyte-driven inflammation, and define novel pathways involved in the regulation of astrocyte pro-inflammatory activities.
RESULTS
Induction of glial nos2a expression in a zebrafish model of CNS inflammation
Genetic and small molecule zebrafish screens have provided important insights in multiple biological processes (Jain et al., 2016; Li et al., 2015). To exploit the advantages offered by zebrafish for the study of neurologic disease, we developed a model of CNS inflammation based on the treatment of zebrafish embryos with pro-inflammatory E. coli K12 lipopolysaccharide (LPS) in combination with cuprizone, an inducer of demyelination (Matsushima and Morell, 2006) (Figure 1A). LPS/cuprizone treatment led to a reduction in myelin protein zero (mpz) expression (Figure 1B), recapitulating similar observations made in murine models of cuprizone-induced demyelination (Liu et al., 2010).
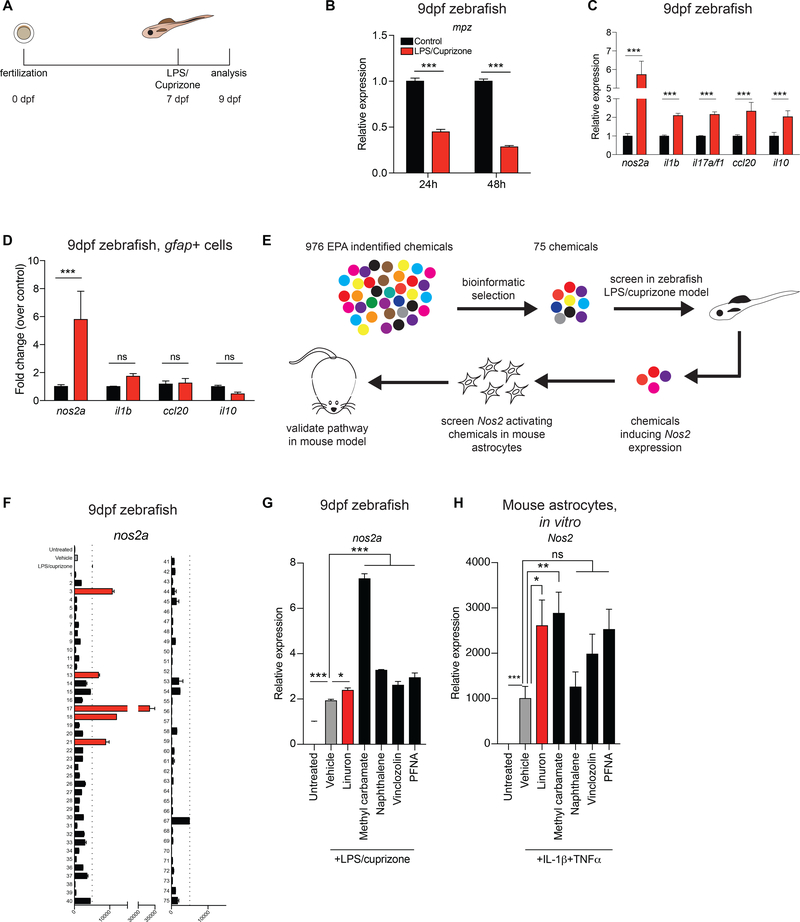
Figure 1. Induction of glial nos2a expression in a zebrafish model of CNS inflammation.
A) Zebrafish neuroinflammation model. B) qPCR of mpz expression in zebrafish. n=2 per condition per timepoint. Two-way ANOVA, Bonferroni post-test. C) qPCR analysis 48h after treatment. n=4 per condition, n=3 for il17a/f1 in LPS/cuprizone. Two-way ANOVA, Bonferroni post-test. D) qPCR in EGFP+ cells from gfap::egfp fish. n=4 per condition. Two-way ANOVA, Bonferroni post-test. E) Environmental chemical screen flowchart. F) qPCR of nos2a expression in response to environmental chemicals (see Table S1). Red bars indicate increase over baseline (dashed line). n=2 per condition. G) qPCR of nos2a expression from F. n=3 per condition. One-way ANOVA, Holm-Sidak post-test relative to vehicle. H) qPCR of Nos2 expression in neonatal primary mouse astrocytes treated for 24h. Control, n=8; Vehicle, n=8; Linuron, n=6; PFNA, n=6; Vinclozolin, n=9; Methyl carbamate, n=6; Naphthalene, n=4. One-way ANOVA, Bonferroni post-test relative to vehicle on ln-normalized data. See also Table S1.
Cuprizone-induced demyelination in mice is associated with the activation of CNS-resident glial cells such as astrocytes and microglia (Matsushima and Morell, 2006). Indeed, we found that LPS/cuprizone treatment induced the expression of genes associated with inflammation such as nos2a, il1b, il17a/f1, ccl20 and il10 (Figure 1C). To focus on the response of astrocyte-related cells to LPS/cuprizone treatment we used Tg(gfap::egfp) zebrafish in which enhanced green fluorescent protein (EGFP) expression is driven by the glial fibrillary acidic protein (gfap) promoter (Bernardos and Raymond, 2006). This reporter line labels zebrafish radial glia, which provide a model to study signaling pathways functional in astrocytes and other CNS-resident mammalian cells (Hui et al., 2010; Lyons and Talbot, 2014). The analysis of EGFP+ (Gfap+) cells showed that LPS/cuprizone treatment upregulated the expression of nos2a (Figure 1D), the zebrafish orthologue of inducible nitric oxide synthase (iNOS) which has been linked to astrocyte pro-inflammatory and neurodegenerative activities (Rabinovich et al., 2016; Sorbara et al., 2014). Thus, LPS/cuprizone induces a radial glia phenotype that resembles disease-promoting astrocytes in neurologic disorders.
Environmental exposures boost astrocyte pathogenicity
Environmental factors impact the pathogenesis of neurologic diseases, but methods for their systematic investigation are still needed (Al-Chalabi and Hardiman, 2013; Chin-Chan et al., 2015; Kamel, 2013; Olsson et al., 2017; Rappaport and Smith, 2010; Tshala-Katumbay et al., 2015). We used the zebrafish model of LPS/cuprizone-induced inflammation to systematically evaluate the effects of environmental chemicals on disease-promoting astrocyte functions (Figure 1E). In these studies we used the ToxCast chemical inventory established by the U.S. Environmental Protection Agency (EPA), which includes a broad collection of representative chemicals from multiple sources ranging from industrial and consumer products to food additives (Sipes et al., 2013). First, we performed a bioinformatic analysis of the ToxCast chemical inventory to identify compounds that modulate signaling pathways previously linked to neurodegeneration and inflammation in genetic and functional studies. Specifically, we focused on the identification of compounds that could potentially modify signaling associated with AHR (Rothhammer et al., 2016), DR5 (Aktas et al., 2005), IFNγ (Zamvil and Steinman, 2003), TGFβ1 (Shull et al., 1992), IL-1α (Liddelow et al., 2017), IL-6 (Glass et al., 2010), STAT3 (Anderson et al., 2016) and JAK3 (Levine et al., 2007). Our bioinformatic screening of 976 molecules in the ToxCast database identified 75 candidate compounds, which were then evaluated for their effects on nos2a expression in LPS/cuprizone-treated zebrafish (Figure 1F, Figure S1A, Table S1). Out of these 75 candidates, 5 compounds increased nos2a expression in LPS/cuprizone-treated zebrafish (Figures 1F-G).
To evaluate the relevance of these findings for the modulation of CNS inflammation in mammals, we analyzed the effect of these compounds on Nos2 expression in primary murine astrocytes activated in vitro with interleukin 1β (IL-1β) and tumor necrosis factor α (TNFα). These cytokines induce iNOS expression in astrocytes in vitro (Hewett et al., 1994) and contribute to CNS inflammation in vivo (Lock et al., 2002). We found that the herbicide Linuron and Methyl carbamate, which is used by the textile, polymer and pharmaceutical industries, boost Nos2 expression in murine astrocytes activated in vitro with pro-inflammatory IL-1β and TNFα (Figure 1H). Thus, in the context of inflammation, Methyl carbamate and Linuron boost evolutionarily conserved signaling pathways in mouse astrocytes that induce a disease-promoting phenotype.
Linuron boosts UPR activation to amplify astrocyte pathogenic activities
Linuron was recently banned in Europe because of its potential risk to mammals (Authority, 2016). Thus, we focused our studies on the mechanisms by which Linuron modulates astrocyte function; the study of these mechanisms may identify signaling pathways that control astrocyte responses. The bioinformatic analysis of the EPA gene expression database identified multiple interactions of Linuron with the unfolded protein response (UPR) (Figure 2A, Table S2) (Bettigole and Glimcher, 2015). The UPR is activated by three endoplasmic reticulum (ER)-bound transmembrane receptors: protein kinase RNA-like ER kinase (PERK), activating transcription factor 6 (ATF6) and IRE1α (Bettigole and Glimcher, 2015). Our bioinformatic analysis identified two candidate pathways for the activation of the UPR by Linuron: 1) PERK signaling, leading to the activation of CCAAT/enhancer-binding protein homologous protein (CHOP, encoded by Ddit3), and 2) IRE1α signaling, leading to the phosphorylation of IRE1α which removes a stop codon-containing intron in the Xbp1 transcript and enables the translation of a functional full-length XBP1 transcription factor (Calfon et al., 2002; Yoshida et al., 2001).
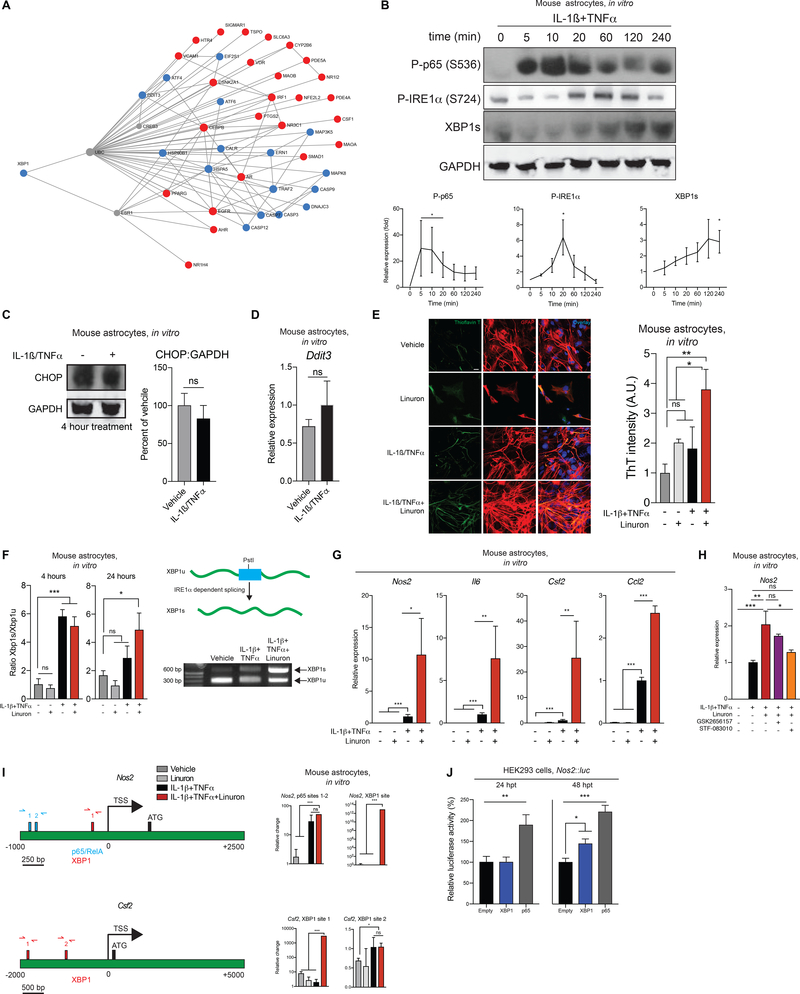
Figure 2. Environmental chemicals boost astrocyte pathogenicity.
A) Pathway analysis of ToxCast-identified genes for Linuron and the unfolded protein response. B) Western blot and quantification for treated neonatal astrocytes. n=3 P-p65, n=3 P-IRE1α, n=4 XBP1. Kruskal-Wallis non-parametric ANOVA, uncorrected Dunn’s post-test. C) Western blot. n=4 per condition. Unpaired two-tailed t-test. D) qPCR. n=6 per condition. Unpaired two-tailed t-test. E) Thioflavin T (ThT) fluorescence in neonatal astrocytes treated for 24 hours. n=5–6 images per condition from N=3 cultures. One-way ANOVA, Dunnett post-test. F) (Left) Quantification of Xbp1 splicing. (Right-top) Schematic of Xbp1 splicing assay. (Right-bottom) Splicing assay gel. n=5–6 per condition at 4 hours; n=9 per condition at 24 hours except Linuron (n=3). One-way ANOVA, Bonferroni post-test. G) qPCR in neonatal astrocytes after 24h treatment. n=6–9 per condition, n=4 for Csf2 Linuron condition. One-way ANOVA, Bonferroni post-test, performed on ln-normalized data for Nos2, Il6, and Csf2. H) qPCR of Nos2 expression in neonatal primary mouse astrocytes. n=6 per condition, n=3 for GSK2656157 condition. One-way ANOVA, Holm-Sidak post-test. I) (Left) Predicted XBP1 and p65 binding sites in the murine Nos2 and Csf2 promoters. (Right) ChIP-qPCR of XBP1 and p65 recruitment to Nos2 and Csf2 promoters in neonatal primary mouse astrocytes. n=5 for p65 vehicle, n=2–3 for Linuron, n=6 otherwise. Kruskal-Wallis non-parametric ANOVA, uncorrected Dunn’s test (Csf2, XBP1 site 2) or one-sample t-test (otherwise). J) HEK293 luciferase reporter assay at the indicated hours post transfection (hpt). n=5–6 per group. One-way ANOVA, Dunnett post-test. See also Figure S1 and Table S2.
We first analyzed the UPR in astrocytes activated with IL-1β and TNFα, pro-inflammatory cytokines considered important contributors to MS and EAE pathogenesis (Lin and Edelson, 2017; Valentin-Torres et al., 2016). Astrocyte treatment with IL-1β and TNFα activated the transcription factor NF-κB (Figure 2B), associated with astrocyte pro-inflammatory and neurotoxic activities (Liddelow and Barres, 2017). Moreover, IL-1β and TNFα triggered IRE1α phosphorylation which was highest 20–120 minutes after activation, preceding the increase in full length (active) XBP1 detected at 240 minutes. Of note, astrocyte activation with IL-1β and TNFα did not alter CHOP expression, ruling out a major contribution of PERK in cytokine-induced UPR activation in astrocytes (Figures 2B-D). Taken together, these findings suggest that pro-inflammatory stimuli relevant to MS activate IRE1α-XBP1 signaling in astrocytes.
We then investigated the effects of Linuron on the UPR and the pro-inflammatory activities of astrocytes. Linuron synergized with IL-1β and TNFα to induce unfolded protein accumulation in astrocytes (Figure 2E) and Xbp1 splicing (Figure 2F) up to 24 hours after activation, when spliced Xbp1 levels are decreased in cytokine-only treated astrocytes. Linuron also boosted the expression of pro-inflammatory genes in astrocytes such as Nos2, Il6, Csf2 and Ccl2 (Figure 2G). However, XBP1 activation was not induced by PFNA, methyl carbamate or other stimuli such as serum/glucose deprivation or extracellular ATP, suggesting specificity in the stressors that activate this pathway in astrocytes (Figures S1B-E).
To investigate the mechanism by which Linuron and the UPR control pro-inflammatory gene expression in astrocytes we studied the effects of PERK (GSK2656157) and IRE1α (STF-083010) inhibitors on Nos2 expression (Keestra-Gounder et al., 2016). IRE1α inhibition suppressed Nos2 expression upregulation induced by cytokines and potentiated by Linuron (Figure 2H); PERK inhibition had no effect. Moreover, Linuron boosted XBP1 recruitment to the regulatory regions of genes associated with astrocyte pathogenic activities such as Nos2 (linked to neurotoxicity (Hewett et al., 1994)) and Csf2 (linked to microglia activation (Becher et al., 2016)) (Figure 2I). Indeed, XBP1 transactivated the Nos2 promoter in reporter assays to similar levels as NF-κB p65 (Figure 2J), a known driver of Nos2 expression (Mayo et al., 2014). Taken together, these findings suggest that IRE1α-XBP1 signaling drives pro-inflammatory astrocyte responses, and that Linuron boosts these pathogenic activities by enhancing the UPR.
Linuron boosts IRE1α-XBP1 activation via Sigmar1
To identify the mechanism by which Linuron boosts XBP1 activation, we searched the EPA ToxCast inventory for molecules that interact with Linuron and are linked to the UPR. We identified the ligand-activated ER chaperone Sigmar1 as a candidate molecule mediating the boost of the UPR by Linuron (Figure 3A) (Sipes et al., 2013; Watanabe et al., 2016). Interestingly, Sigmar1 is reported to enhance the UPR by stabilizing active phosphorylated IRE1α (P-IRE1α) (Hayashi and Su, 2007; Mori et al., 2013). Thus, we hypothesized that Linuron boosts XBP1-driven pro-inflammatory gene expression in astrocytes by stabilizing P-IRE1α through a Sigmar1-dependent mechanism. In support of this hypothesis, Linuron increased the interaction of Sigmar1 with IRE1α in co-immunoprecipitation studies; this increased Sigmar1:IRE1α interaction was lost upon deletion of the Sigmar1 ligand biding domain (Figure 3B).
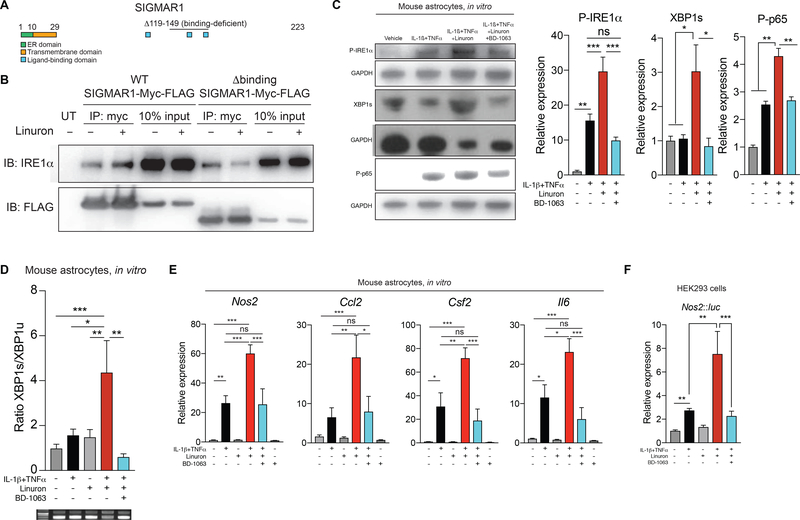
Figure 3. Linuron boosts IRE1α/XBP1 signaling via Sigmar1.
A) Schematic of Sigmar1 protein domains. B) Co-immunoprecipitation of Sigmar1 and IRE1α in HEK293 cells. C) Western blot for NF-κB, P-IRE1α and XBP1 activation in neonatal murine astrocytes treated for 1 hour. n=3–7 per condition. One-way ANOVA, Sidak post-test. D) Xbp1 splicing in murine astrocytes activated for 24 hours (top). Representative images of agarose Xbp1 splicing gels (bottom). n=4–9 per condition. One-way ANOVA, Dunnett post-test. E) qPCR in neonatal murine astrocytes activated for 24 hours. n=7–16 per condition; n=4 for BD-1063 alone. One-way ANOVA, Tukey post-test. F) HEK293 luciferase reporter assay. n=4 per condition. One-way ANOVA, Tukey post-test on ln-normalized data.
We then investigated the role of Sigmar1 in the enhancement of XBP1-driven pro-inflammatory gene expression in astrocytes by Linuron. Linuron boosted IRE1α activation induced in astrocytes by IL-1β and TNFα 1 hour after stimulation; this boost in IRE1α activation was suppressed when the Sigmar1-specific inhibitor BD-1063 was used (Figure 3C). In agreement with the increased IRE1α activation, Linuron increased XBP1 activation 1 hour after stimulation with IL-1β and TNFα, at a timepoint when XBP1 activation is not yet detected at the protein level in cytokine-only treated astrocytes (Figure 3C, compare with Figure 2B). Sigmar1 inhibition suppressed this boost in XBP1 and NF-κB activation induced by Linuron and cytokine activation (Figure 3C). Similarly, Sigmar1 inhibition abrogated the stabilization of spliced Xbp1 transcripts induced by Linuron 24 hours after treatment with IL-1β and TNFα, a timepoint at which Xbp1 activation returns to baseline levels in cytokine-only treated astrocytes (Figure 3D, compare with Figure 2F). Consequently, Sigmar1 inhibition suppressed the synergism detected between IL-1β, TNFα and Linuron in inducing the expression of Nos2, Csf2, Ccl2 and Il6 expression in astrocytes and Nos2 promoter activation (Figures 3E-F). Collectively, these findings suggest that Linuron enhances the interaction of Sigmar1 with P-IRE1α to promote its stabilization, thereby boosting the IRE1α-XBP1 driven expression of genes that contribute to astrocyte pathogenic activities in CNS inflammation and neurodegeneration.
Genomic control of disease-promoting astrocyte activities by IRE1α-XBP1 signaling
Astrocytes play important roles in the pathogenesis of MS and EAE (Locatelli et al., 2018; Mayo et al., 2014; Rothhammer et al., 2018; Rothhammer et al., 2016; Rothhammer and Quintana, 2015). Our studies on the mechanism of action of Linuron on astrocytes identified IRE1α-XBP1 signaling as a candidate regulator of astrocyte pathogenic activities. Thus, we investigated the role of IRE1α-XBP1 signaling in astrocytes in the EAE model of MS.
We detected increased expression of the XBP1 target gene Edem1 (Lee et al., 2003) in astrocytes isolated 22 days after EAE induction (Figure 4A). Ddit3 and Atf6 expression were unaffected, suggesting that PERK and ATF6 are not major UPR activators in astrocytes during EAE. Indeed, IRE1α blockade with STF-083010 suppressed Nos2, Csf2, and Edem1 expression induced by IL-1β and TNFα in adult murine astrocytes in culture, supporting a role for IRE1α in inducing XBP1-driven pro-inflammatory responses in adult astrocytes (Figure S1F).
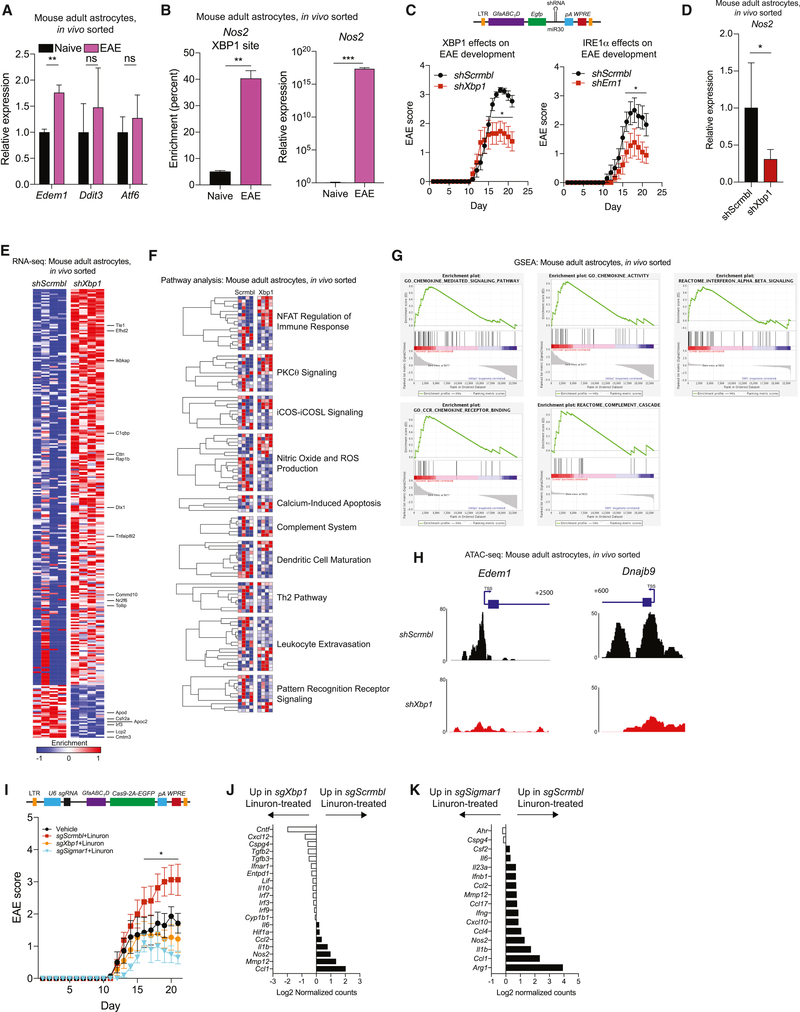
Figure 4. Genomic control of disease-promoting astrocyte activities by IRE1α-XBP1 signaling.
A) qPCR in sorted adult astrocytes from naïve and EAE mice. n=3 per group. Unpaired two-tailed t-test per gene. B) qPCR of Nos2 expression (left) or XBP1 binding to the Nos2 promoter by ChIP (right) in sorted adult astrocytes from naïve and EAE mice. n=3 naïve, n=8 EAE (qPCR) or n=2 (ChIP) per group. Unpaired two-tailed t-test on data ln-normalized (qPCR) or not (ChIP). C) Schematic of shRNA lentiviral vector and EAE development in transduced mice. n=13 shScrmbl (left), n=13 shXbp1, n=10 shScrmbl (right), n=9 shErn1. Two-way repeated measures ANOVA, Holm-Sidak post-test. LTR=long terminal repeats, GfaABC1D=Gfap promoter, EGFP=enhanced green fluorescent protein, pA=polyadenylation signal, WPRE=woodchuck hepatitis virus post-transcriptional regulatory element. D) qPCR of Nos2 expression in EAE astrocytes. n=9 per condition. Unpaired non-parametric Kolmogorov-Smirnov t-test. E) RNA-seq analysis of astrocytes from EAE mice. Columns are n=4 biological replicates. Genes with p<0.05 are shown. F) Pathway analysis of astrocyte signaling in Xbp1 knockdown. G) Gene set enrichment analysis for astrocyte RNA-seq. H) ATAC-seq peaks in astrocytes. n=4 shScrmbl, n=3 shXbp1. Scale bars show read density. Transcriptional schematics shown above plots. I) EAE scores. n=7 Vehicle, n=16 sgScrmbl, n=9 sgXbp1, n=10 sgSigmar1. Two-way repeated measures ANOVA, Holm-Sidak post-test. U6=human U6 promoter, sgRNA=single guide RNA, 2A=self-cleaving peptide. J-K) Astrocyte Nanostring analyses from I. RNA pooled from n=4 mice (sgScrmbl) or n=7 mice (sgXbp1) in J, n=5 mice per group in K. See also Figures S2–S5 and Tables S3–S6.
Chromatin immunoprecipitation (ChIP) analyses detected increased XBP1 recruitment to the Nos2 promoter concomitant with Nos2 expression in astrocytes during EAE (Figure 4B), suggesting that XBP1 promotes the expression of iNOS and other molecules that contribute to astrocyte pro-inflammatory activities. Indeed, the analysis of astrocytes during EAE by whole-genome ChIP followed by sequencing (ChIP-seq) detected XBP1 recruitment to its target genes Edem1 and Dnajb9 (Lee et al., 2003), and also to genes linked with immune activation such as Csf2 (Figure S2, Table S3). Taken together, these findings suggest that IRE1α-XBP1 signaling drives disease-promoting genomic programs in astrocytes during EAE.
To evaluate the role of IRE1α-XBP1 signaling in astrocytes during EAE we performed perturbation studies in which we knocked down Xbp1 and Ern1 (coding for IRE1α) in astrocytes using lentivirus-delivered small hairpin RNAs (shRNAs) expressed under the control of the Gfap promoter (Figure S3A-C). The knock down of Xbp1 or Ern1 in astrocytes ameliorated EAE (Figure 4C), but did not affect the T-cell response, nor the number of astrocytes or microglia (Figure S3D-E). Similarly, Xbp1 or Ern1 inactivation in astrocytes by CRISPR/Cas9-based deletion also ameliorated EAE (Figure S4A-C). However, inactivation of Eif2ak3 (coding for PERK) in astrocytes did not significantly affect EAE, suggesting that PERK signaling is not a major driver of astrocyte pathogenic activities in this experimental paradigm (Figure S4D-E).
The analysis of the transcriptional profile of astrocytes by qPCR and RNA-seq showed that Xbp1 knock down decreased Nos2 expression and the activation of inflammation-associated pathways (Figures 4D-G, Tables S4–S5). Further support for a pro-inflammatory role of XBP1 in astrocytes during EAE was provided by studies using the assay for transposase-accessible chromatin in combination with high-throughput sequencing (ATAC-seq) (Buenrostro et al., 2013). Following Xbp1 knock down we detected decreased chromatin accessibility in the XBP1-driven target genes Edem1 and Dnajb9, and in multiple pathways associated with inflammation (Figure 4H and Table S6). Collectively, these findings demonstrate that IRE1α-XBP1 signaling drives astrocyte-intrinsic pro-inflammatory activities during EAE.
Finally, we investigated the effect of boosting IRE1α-XBP1 signaling by Linuron on EAE. Linuron administration worsened EAE; this worsening was suppressed when Xbp1 or Sigmar1 were inactivated in astrocytes by CRISPR/Cas9-based deletion (Figure 4I, S4B, S5A). Xbp1 and Sigmar1 inactivation also suppressed the upregulation of pro-inflammatory gene expression induced by Linuron in astrocytes (Figure 4J-K), but did not affect the number of astrocytes, microglia, or T cells in the CNS (Figure S5B-C). Hence, Linuron acts via Sigmar1 to drive IRE1α-XBP1-dependent mechanisms in astrocytes that promote CNS inflammation during EAE.
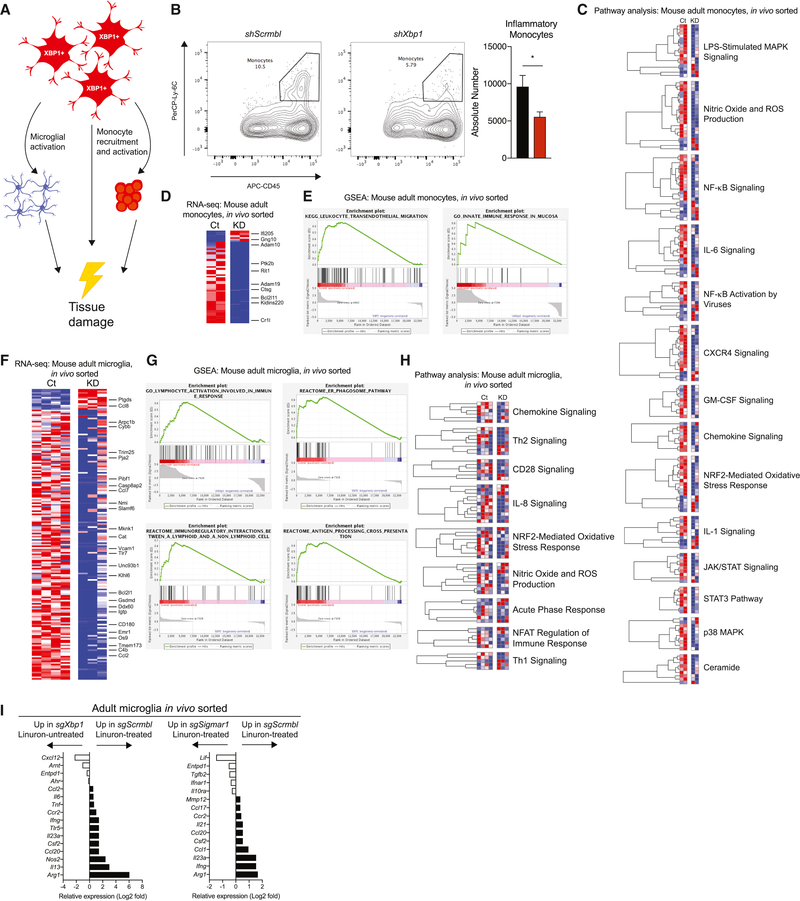
XBP1 in astrocytes controls monocytes and microglia during EAE
Astrocytes control microglial activation and the recruitment of inflammatory monocytes to the CNS (Rothhammer and Quintana, 2015; Wheeler and Quintana, 2018). Indeed, microglia and inflammatory monocytes are dominant cell types encountered in MS lesions, their presence and activation correlates with the emergence of tissue damage, and the suppression of monocyte recruitment to the CNS interferes with disease progression in MS experimental animal models (Ajami et al., 2011; Dendrou et al., 2015; Huitinga et al., 1990). Thus, we investigated the effects of XBP1 signaling in astrocytes on the response of monocytes and microglia during EAE (Figure 5A). Xbp1 knock down in astrocytes decreased the recruitment of peripheral inflammatory monocytes to the CNS and their expression of genes associated with inflammation (e.g. NF-κB) (Figures 5B-E). In addition, Xbp1 knock down in astrocytes decreased innate immune signaling and the expression of chemokines and cytokines in microglia (Figures 5F-H).
Figure 5. XBP1 in astrocytes controls monocytes and microglia during EAE.
A) Model of XBP1+ astrocyte pro-inflammatory signaling in the CNS. B) Flow cytometry plots (left) and quantification (right) of pro-inflammatory monocytes in CNS during EAE. n=10 per group. Unpaired two-tailed t-test. C) Pathway analysis of monocyte RNA-seq data. Columns are n=2 biological replicates. D) Heatmap of monocyte RNA-seq data (p<0.05). E) Gene set enrichment analysis of monocyte pathways. F) Heatmap of microglia RNA-seq data (p<0.05). n=4 shScrmbl, n=3 shXbp1. (G and H) Gene set enrichment analysis and pathway analysis of microglia RNA-seq. Ct=Gfap::shScrmbl, KD=Gfap::shXbp1. I) Nanostring analysis of EAE microglia from 4I. RNA pooled from n=5 mice, n=7 for sgXbp1. See also Tables S4–S5.
In line with our previous observations, CRISPR/Cas9-based deletion of Xbp1 or Sigmar1 in astrocytes suppressed the Linuron-induced upregulation of pro-inflammatory gene expression in microglia (Figure 5I). Thus, IRE1α-XBP1 signaling in astrocytes controls the pro-inflammatory response of microglia and monocytes during EAE and its exacerbation by Linuron.
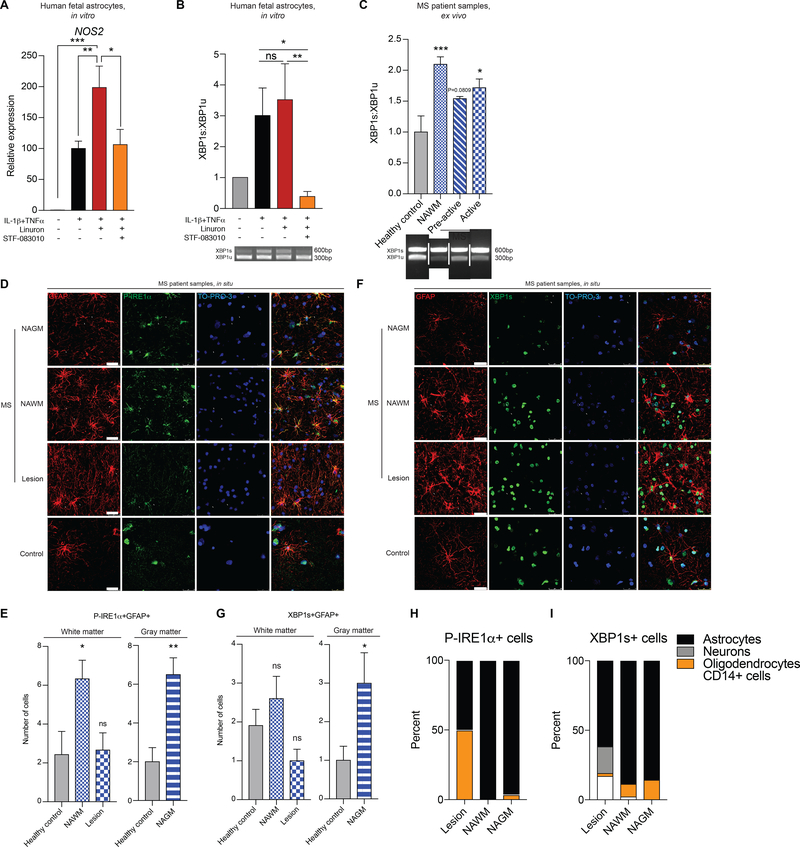
IRE1α-XBP1 signaling in astrocytes is upregulated in MS
Finally, we evaluated the effects of the UPR in human astrocytes. Activation with TNFα and IL-1β induced NOS2 expression in primary human astrocytes (Figure 6A), as well as XBP1 activation as determined by the increased abundance of the spliced XBP1 transcript (Figure 6B). Linuron boosted NOS2 expression in human astrocytes; this boost was blocked with the IRE1α inhibitor STF-083010 (Figures 6A-B).
Figure 6. IRE1α-XBP1 signaling in astrocytes is upregulated in MS.
A) qPCR of NOS2 in human fetal astrocytes treated for 48 hours. n=3 per condition. One-way ANOVA, Fisher’s LSD test. B) (Top) Quantification of XBP1 splicing. (Bottom) Gel of XBP1 splicing assay. n=3 per condition. One-way ANOVA, Bonferroni post-test on ln-transformed data. C) XBP1 splicing of MS patient tissue as quantification (top) and gel (bottom). n=3 healthy controls (HCs), n=4 otherwise. One-way ANOVA, Bonferroni post-test relative to control. pre-active=pre-active lesion, active=active lesion. White bars indicated spliced lanes. (D-F), Immunostaining of astrocyte colocalization with P-IRE1α (D-E) and XBP1s (F-G) in MS patients and HCs. NAGM=normally appearing gray matter. In (E) n=7 HC WM, n=6 NAWM, n=3 lesion, n=8 HC GM, n=8 NAGM images from N=3 MS patients and N=4 healthy controls. One-way ANOVA, Dunnett post-test relative to HC (left). Unpaired two-tailed t-test (right). In (G) n=11 HC WM, n=15 NAWM, n=10 lesion, n=6 HC GM, n=10 GM images from N=4 MS patients (NAWM, NAGM), N=3 MS patients (lesion), and N=2 HCs. One-way ANOVA, Dunnett post-test relative to HCs (left). Unpaired two-tailed t-test with Welch’s correction (right). (H-I) Quantification of P-IRE1α+ and XBP1s+ cells by cell type and MS pathology. n=30–54 cells per condition. See also Figure S6.
To evaluate the potential contribution of IRE1α-XBP1 signaling in astrocytes to human neurologic disease, we analyzed CNS samples from MS patients. When compared with healthy control tissue, MS samples from lesions and normal appearing white matter (NAWM) presented increased XBP1 activation as indicated by the abundance of spliced XBP1 transcripts (Figure 6C). Immunofluorescence studies detected increased astrocyte-specific IRE1α activation in NAWM and normal appearing gray matter (NAGM) from MS samples (Figures 6D-E, S6A); neurons, oligodendrocytes and CD14+ cells (microglia and recruited monocytes) showed little or no P-IRE1α (Figure S6A). Moreover, using an antibody specific for the spliced, active form of XBP1 (XBP1s), we detected increased abundance of active XBP1s in astrocytes from MS samples in NAWM and NAGM (Figures 6F-G, Figures S6B-C).
Interestingly, although we had detected increased XBP1 activation when we analyzed MS lesions by PCR in bulk (Figure 6C), we did not detect increased P-IRE1α or XBP1s in astrocytes located in MS lesions. These findings suggested that in MS lesions XBP1 is activated in other cell types besides astrocytes. Indeed, while in NAWM and NAGM astrocytes were the most abundant P-IRE1α+ and XBP1s+ cells, in MS lesions we also detected a significant contribution of neurons, oligodendrocytes, and CD14+ monocytes and microglia (Figures 6H-I). Collectively, these findings suggest that IRE1α-XBP1 signaling in astrocytes may contribute to MS pathology.
DISCUSSION
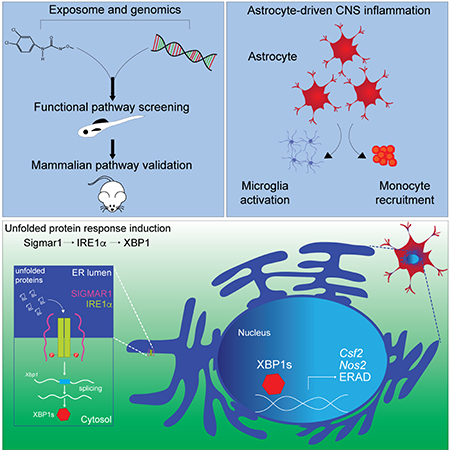
The exposome is defined as the totality of environmental exposures of an individual in a lifetime (Wild, 2005). In this study, we used a novel approach that combines new zebrafish models, bioinformatics, genomics and in vivo cell-specific genetic perturbations to systematically evaluate the effects of a wide range of common environmental chemicals on CNS inflammation and the molecular mechanisms involved. Our studies identified IRE1α-XBP1 signaling as a driver of genomic programs that contribute to disease-promoting astrocyte activities; IRE1α-XBP1 signaling and consequently, astrocyte pathogenic activities in EAE, were boosted by Linuron. Together with previous reports on the effects of commensal bacteria-derived metabolites (Erny et al., 2015; Rothhammer et al., 2018; Rothhammer et al., 2016; Sampson et al., 2016) and changes in melatonin levels induced by seasonal variations in day length (Farez et al., 2015), our findings support the need for systematic investigation of the effects of the exposome on neurologic diseases. These studies have the potential to identify unknown signaling pathways involved in the control of CNS inflammation and neurodegeneration, and also environmental factors that modulate their activation. Future studies should also evaluate additional variables such as the developmental stage at the time of exposure, a timely concern based on recent reports on the effects of the pesticide chlorpyrifos on CNS development (Rauh, 2018). In addition, the approach described in this work provides a cost-effective platform to screen for therapeutic compounds that suppress pathogenic activities of CNS-resident cells in their physiologic context.
We defined a role for Sigmar1 in the activation of IRE1α-XBP1 driven pathogenic activities in astrocytes by Linuron. Sigmar1 is an ER membrane receptor which controls multiple biological processes including the cellular response to stress and apoptosis (Schmidt et al., 2016). Interestingly, Sigmar1 has been linked to the pathology of neurologic disorders such as amyotrophic lateral sclerosis (ALS) (Al-Saif et al., 2011; Tsai et al., 2014). Indeed, environmental factors contribute to ALS pathogenesis and are considered a potential cause for the sudden increase in ALS incidence detected among young Gulf War veterans (Haley, 2003; Horner et al., 2003). In this context, our findings suggest that Sigmar1 activation in astrocytes by environmental chemicals may also participate in the pathogenesis of neurologic diseases such as ALS.
Endogenous Sigmar1 agonists have also been identified, including tryptaminergic trace amines, dehydroepiandrosterone and pregnenolone (Fontanilla et al., 2009). The regulation of Sigmar1 by endogenous ligands suggests its participation in astrocyte adaptation to the local CNS microenvironment. Interestingly, the ligand-binding cavity in Sigmar1 shows remarkable plasticity in ligand recognition, suggesting that structurally diverse agonists modulate Sigmar1 signaling under physiologic conditions (Schmidt et al., 2016). In this sense, Sigmar1 resembles the aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor activated by diverse ligands ranging from pollutants to microbial metabolites. AHR has been shown to control astrocytes in CNS inflammation, and is considered a candidate therapeutic target for inflammation and cancer (Gutiérrez-Vázquez and Quintana, 2018; Wheeler et al., 2017). Similarly, Sigmar1 may provide a target for the therapeutic modulation of astrocyte responses in MS and other neurologic disorders.
Our studies identify IRE1α-XBP1 signaling as a driver of disease-promoting astrocyte pathogenic activities during CNS inflammation. The UPR has been previously linked to the pathogenesis of neurologic disorders, particularly because of its role within neurons and oligodendrocytes (Hetz and Saxena, 2017). However, the role of IRE1α-XBP1 signaling as a driver of genomic programs controlling astrocyte pro-inflammatory activities was previously unknown. Our detection of IRE1α-XBP1 activation in MS tissue samples suggests that the UPR in astrocytes contributes to neurodegeneration and disease pathogenesis. Interestingly, PERK activation in oligodendrocytes is reported to suppress EAE (Lin et al., 2007). Based on the diverse biological roles of the UPR (Bettigole and Glimcher, 2015), these seemingly conflicting results likely reflect downstream cell-specific UPR functions associated with the specific pathways leading to its activation (e.g. IRE1α vs PERK), the adaptation of different cell types to inflammation (e.g. astrocytes vs oligodendrocytes) and the effects that UPR modulators may have on the inflammatory response (e.g. suppression of pathogenic T cells). Indeed, our data rule out major roles for PERK or ATF6 in astrocyte UPR activation during EAE. Given the heterogeneity of astrocytes in vivo (Ben Haim and Rowitch, 2017; Khakh and Sofroniew, 2015), IRE1α-XBP1 signaling may be associated to specific populations such as C3+ neurotoxic astrocytes (Liddelow et al., 2017). These factors should be considered for the design of therapeutic approaches targeting the UPR in neurologic disorders.
Microglia and inflammatory monocytes are important contributors to MS pathology (Ajami et al., 2011; Dendrou et al., 2015; Huitinga et al., 1990). Our data support a role for IRE1α-XBP1 signaling in astrocytes as a driver of genomic programs that control not only astrocyte-intrinsic disease-promoting activities, but also microglia and inflammatory monocytes. Indeed, astrocytes are a source of GM-CSF and CCL-2, involved in microglial activation and monocyte recruitment, respectively (Wheeler and Quintana, 2018). Moreover, recent reports identify astrocytes as a dominant source of signals that polarize myeloid cells in the CNS (Locatelli et al., 2018). Thus, although much attention has focused on the microglial control of astrocyte activation (Liddelow et al., 2017; Rothhammer et al., 2018), our findings highlight the role of astrocytes in the control of microglia and monocytes and the need to further characterize the molecular mechanisms involved.
Their intrinsic pathogenic activities and their ability to control microglia and monocytes, suggest an important role for astrocytes in MS progression, a disease process thought to be driven by CNS resident cells (Baecher-Allan et al., 2018; Thompson et al., 2018). Interestingly, IRE1α/XBP1 activation in NAWM and NAGM was predominantly detected in astrocytes. White and gray matter pathology contribute to disease progression in MS, but little is known about the mechanisms involved (Baecher-Allan et al., 2018; Calabrese et al., 2015; Ellwardt et al., 2018; Thompson et al., 2018). Taken together, these findings suggest that IRE1α/XBP1 signaling in astrocytes participates in pathogenic processes relevant for MS progression. However, since IRE1α/XBP1 activation was also detected in neurons, oligodendrocytes, microglia and recruited monocytes in MS lesions, this pathway may play additional pathological roles in other cell types besides astrocytes. Indeed, additional pathways such as PERK may trigger the UPR in other CNS-resident cells with cell-specific contributions to disease pathology, while IRE1α may also activate XBP1-independent pro-inflammatory mechanisms (Bettigole and Glimcher, 2015).
In conclusion, we describe a novel approach for the systematic investigation of the effects of environmental factors on neuroinflammation. This approach uncovered mechanisms that operate in astrocytes to control CNS inflammation and neurodegeneration, as well as environmental exposures that regulate them. This strategy may guide future epidemiologic studies on the effects of the environment on neurologic diseases, while identifying molecular mechanisms that control CNS pathology and potential therapeutic targets for MS and other neurologic diseases.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Francisco J. Quintana (fquintana@bwh.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals.
Mice
Adult Mus musculus males, females, and postnatal pups on a C57Bl/6J background were obtained from the Jackson Laboratory. B6.Cg-Tg(Gfap-cre)73.12Mvs/J mice (#012886, The Jackson Laboratory) used previously (Rothhammer et al., 2016) were crossed with B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J mice (#007909, The Jackson Laboratory) to generate Gfap(Cre/+);TdTomato(f/+) mice. Both male and female pups were sacrificed between P0-P3 for harvesting and culturing astrocytes and sex was assumed to be balanced and differences were not analyzed. Mice were kept in a pathogen-free facility at the Hale Building for Transformative Medicine at Brigham and Women’s Hospital in accordance with the IACUC guidelines, fed ad libitum on a 14/10-hour light/dark cycle. Mice were healthy and checked daily by veterinary staff. 8–10 week old mice were used for stereotactic injection and EAE induction. Mice were both drug and test naïve and not involved in previous procedures. All mice were at least doubly housed. All procedures were reviewed and approved under the IACUC guidelines at Brigham and Women’s Hospital.
Zebrafish
Danio rerio embryos, larvae, and adult AB* or Tg(gfap::egfp) (Bernardos and Raymond, 2006) fish were raised under standard conditions in a 28–30C system with a 14/10 hrs light/dark cycle. Until 7dpf, larvae were kept in E3 water (5 mM NaCl, 170 μM KCl, 433 μM CaCl2, 675 μM MgSO4, 80 μM HEPES, pH=7.5). Both male and female fish were used but sex was not checked in the experiments. All experiments were conducted in system water. Zebrafish were both drug and test naïve and not involved in previous procedures. All fish were group housed in tanks. All procedures were reviewed and approved under the IACUC guidelines at Brigham and Women’s Hospital.
LPS/Cuprizone treatment
7dpf larvae were exposed to freshly prepared cuprizone (#C9012, Sigma-Aldrich, Chemical name: Bis(cycohexanone)oxaldihydrazone) dissolved in E3 water (12.5 μg/mL) by warming to 42C and LPS-EK (#tlrl-eklps, InvivoGen) dissolved in E3 water (150 μg/mL) for 48h.
EAE
All mice used were WT animals on the C57Bl/6 background. EAE was induced with 25 μg of MOG35–55 (#110582, Genemed Synthesis Inc.) mixed with complete Freund’s Adjuvant at a ratio of 1:1 (v/v at a concentration of 5 mg/mL. Mice received 2 subcutaneous injections of 100 μL each of the MOG/CFA mix. Mice then received a single intraperitoneal injection of pertussis toxin (#180, List Biological Laboratories) at a concentration of 2 ng/μL in 200 μL of PBS. Mice received a second injection of pertussis toxin at the same concentration two days after the initial EAE induction. Mice were monitored and scored daily thereafter. EAE clinical scores were defined as follows: 0 – no signs, 1 – fully limp tail, 2 – hindlimb weakness, 3 – hindlimb paralysis, 4 – forelimb paralysis, 5 – moribund, as described previously (Mayo et al., 2014; Rothhammer et al., 2016). For studies of the effects of Linuron on EAE, mice were injected intraperitoneally with 100 mg/kg of Linuron dissolved in corn oil (#C8267–500ML, Sigma-Aldrich) at a concentration of 12.5 mg/mL on alternating days starting at day 6 post-EAE induction. Sex differences were not analyzed but only a single sex was used within any set of EAE experiments. Mice were randomly assigned to treatment groups.
MS tissue
For tissue used in qPCR experiments, brain tissue was from untreated individuals with clinically diagnosed and neuropathologically confirmed MS, as well as from healthy controls (Alvarez et al., 2015; Mayo et al., 2014; Rothhammer et al., 2016). Patient sex is unknown. White matter MS tissue samples were selected as previously described by analysis of tight junctions and adherens junctions near blood vessels (Alvarez et al., 2011). All individuals with MS and healthy controls, or their next of kin, had given informed consent for autopsy and use of their brain tissue for research purposes. Ethical approval was given before autopsy (University of Montreal Health Centre ethical approval: SL05.022 and SL05.023 and BH07.001). For tissue used in immunofluorescence experiments, human brain tissue was obtained from patients diagnosed with clinical and neuropathological MS according to the revised 2010 McDonald’s criteria (Polman et al., 2011). Tissue samples were collected from healthy donors (2 females, 2 males) and MS patients (2 females, 2 males) with full ethical approval and informed consent as approved by the local ethics committee (University of Montreal Health Centre ethical approval: HD04.046 and BH07.001). Autopsy samples were preserved and lesions classified using Luxol fast blue/haematoxylin & eosin staining and Oil Red O staining as previously published (Kuhlmann et al., 2017). Paraffin-embedded brain sections were obtained from 4 MS patients (2 females, 2 males) and 3 healthy controls (1 male, 1 female, 1 unknown) (Neuropathology Department of the Notre Dame Hospital). Samples were allocated into individual groups based on clinical diagnosis of MS according to criteria specified above. Healthy control samples were individuals who died of non-neurological traumatic injury. Sex differences were not analyzed but sex was balanced within all analyses. Sample size for MS patient sample studies was determined based on past analyses of similar comparisons (Mayo et al., 2014; Rothhammer et al., 2018; Rothhammer et al., 2016).
Human fetal astrocytes
Human fetal astrocytes were isolated as previously described (Jack et al., 2005; Rothhammer et al., 2016) from human CNS tissue from fetuses at 17–23 weeks of gestation that were obtained from the Birth Defects Research Laboratory at the University of Washington Dept. of Pediatrics (Project Number: 5R24HD000836–51) following Canadian Institutes of Health Research-approved guidelines. The sex of the human fetal astrocytes used is unknown.
METHOD DETAILS
Zebrafish flow cytometry
Cells were sorted from Tg(gfap:gfp) fish as described (Manoli and Driever, 2012). Briefly, larvae were anesthetized on ice, homogenized in FACS buffer (1% BSA, 2 mM EDTA pH=8.0, in 1X PBS), and filtered through a 70 μm cell strainer (#22–363-548, Fisher Scientific). Cells were sorted with a FACSAria IIU (BD Biosciences) directly into TRIzol LS (#10296010, Thermo Fisher Scientific) or FACS buffer for RNA analysis and processed according to the manufacturer’s protocol.
RNA isolation from zebrafish
Zebrafish RNA was isolated with TRIzol LS, followed by RNA purification with Direct-zol RNA kit (#R2050, Zymo Research) or Qiagen RNeasy kit (#74106, Qiagen), according to the manufacturer’s protocol. cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (#4368813, Life Technologies). Gene expression was measured by qPCR using Taqman Fast Universal PCR Master Mix (#4367846, Life Technologies). Taqman probes used in this study are: actb1 (Dr03432610_m1), ccl20 (Dr03431608_m1), il1b (Dr03114369_m1), il10 (Dr03103209_m1), il17a/f1 (Dr03096843_g1), mpz (Dr03131914_m1) and nos2a (Dr03124753_m1). The results were normalized to actb1 and all probes used in this study were from Life Technologies.
EPA ToxCast library screen in zebrafish
Test compounds were added to fish water together with LPS/cuprizone. After 48h, the larvae were sacrificed, RNA isolated and gene expression analyzed. Compounds listed in Table S1 were used at 0.2 μM (Compounds 1–40) and 20 μM (Compounds 41–75), dissolved in DMSO.
Primary astrocyte cultures
Brains of mice aged P0-P3 were dissected into PBS on ice. Cortices were discarded and the brain parenchyma were pooled, centrifuged at 500xg for 10 minutes at 4C and resuspended in 0.25% Trypsin-EDTA (#25200–072, Thermo Fisher Scientific) at 37C for 10 minutes. DNase I (#90083, Thermo Fisher Scientific) was then added to the solution (1 mg/mL), and the brains were digested for 10 more minutes. Trypsin was neutralized by adding DMEM/F12+GlutaMAX (#10565018, Thermo Fisher Scientific) supplemented with 10% FBS (#10438026, Thermo Fisher Scientific) and 1% penicillin/streptomycin (#15140148, Thermo Fisher Scientific), and cells were passed through a 70 μm cell strainer. Cells were centrifuged at 500xg for 10 minutes at 4C, resuspended in DMEM/F12+GlutaMAX with 10% FBS/1% penicillin/streptomycin and cultured in T-75 flasks (#353136, Falcon) at 37C in a humidified incubator with 5% CO2, for 7–10 days until confluency was reached. Astrocytes were shaken for 30 minutes at 180 rpm, the media was changed, then astrocytes were shaken for at least 2 hours at 220 rpm and media was changed again. Medium was replaced every 2–3 days. For microglial cultures, after the first round of shaking, detached cells were replated into a fresh 10-cm dish and cultured at 37C in a humidified incubator with 5% CO2.
Primary astrocyte pharmacological studies
Compound treatment was performed for 24 hours with compounds diluted in DMEM/F12+GlutaMAX that was supplemented with 10% FBS and 1% penicillin/streptomycin. Compounds used in these studies are: 0.2–20 μM linuron (#36141, Sigma-Aldrich, 20 mM stock in EtOH), 20 μM methyl carbamate (#246352, Sigma-Aldrich, 20 mM stock in EtOH), 20 μM naphthalene (#147141, Sigma-Aldrich, 20 mM stock in EtOH), 20 μM vinclozolin (#45705, Sigma-Aldrich, 20 mM stock in EtOH), 20 μM PFNA (#394459, Sigma-Aldrich, 20 mM stock in EtOH), 10 μM ATP (#P0756, NEB, 1:1000), 50 μM BD1063 (#SML0276, Sigma-Aldrich, 50 mM stock in PBS), 100 ng/mL IL-1ß (#401-ML-005, R&D Systems, 100 μg/mL stock in PBS), 50 ng/mL TNFα (#410-MT-010, R&D Systems, 100 μg/mL stock in PBS), 1 μM GSK2656157 (#504651, Calbiochem, 1 mM stock in EtOH), and 50 μM STF083010 (#SML0409, Sigma-Aldrich, 50 mM stock in DMSO).
RNA isolation from cultured mouse astrocytes
Astrocytes were lysed in Buffer RLT (Qiagen) and RNA was isolated from cultured astrocytes using the Qiagen RNeasy Mini kit (#74106, Qiagen). cDNA was transcribed using the High-Capacity cDNA Reverse Transcription Kit (#4368813, Life Technologies). Gene expression was then measured by qPCR using Taqman Fast Universal PCR Master Mix (#4367846, Life Technologies). Taqman probes used in this study are: Ccl2 (Mm00441242_m1), Gapdh (Mm99999915_g1), Il6 (Mm00446190_m1), Csf2 (Mm01290062_m1), Nos2 (Mm00440502_m1), Ddit3 (Mm01135937_g1), Xbp1 (Mm00457357_m1), Atf6 (Mm01295319_m1) and Edem1 (Mm00551797_m1). qPCR data were analyzed by the ddCt method by normalizing the expression of each gene for each replicate to Gapdh and then to the control group.
Western blot
Protein lysates were prepared by lysing astrocytes with boiling 1X Laemmli buffer (#BP-111R, Boston BioProducts) followed by boiling at 95C for 5 minutes. SDS-PAGE was performed largely as described using Bolt 4–12% Bis-Tris Plus gradient gels (#NW04125BOX, Invitrogen). Western blotting was performed by transferring proteins onto a PVDF membrane (#IPVH15150, Millipore) in 1X NuPAGE buffer (#NP00061, Thermo Fisher Scientific). Membranes were blocked in 10% milk (#M0841, Lab Scientific) in TBS-T (#IBB-180–2L, Boston BioProducts) or alternatively blocked in Odyssey Blocking Buffer (TBS) (#927–50000, LI-COR Biosciences). Primary antibodies used in this study are: mouse anti-XBP-1 (#sc-8015, Santa Cruz, 1:500), rabbit anti-P-IRE1a (Ser724) (#NB100–2323, Novus Biologicals, 1:1000), mouse anti-CHOP (#2895T, Cell Signaling Technology, 1:1000), rabbit anti-P-p65 (Ser536) (#3033S, Cell Signaling Technology, 1:1000), and rabbit anti-GAPDH (#2118S, Cell Signaling Technology, 1:1000). Secondary antibodies used in this study are: anti-mouse IgG-HRP conjugate (#7076S, Cell Signaling Technology, 1:1000), anti-rabbit IgG-HRP conjugate (#7074S, Cell Signaling Technology, 1:1000), IRDye 800CW goat anti-mouse IgG (H+L) (#925–32210, LI-COR Biosciences, 1:10000), and IRDye 800CW goat anti-rabbit IgG (H+L) (#925–32211, LI-COR Biosciences, 1:10000). HRP-conjugated blots were developed using SuperSignal West Femto Maximum Sensitivity Substrate (#34095, Thermo Fisher Scientific) and CL-XPosure Film (#34090, Thermo Fisher Scientific). Some HRP-conjugated blots were developed using the KwikQuant imaging system (Kindle Biosciences). Film was developed using a M35A X-OMAT Film Processor (Kodak). Alternatively, IRDye-conjugated blots were imaged using an Odyssey Imaging System (LI-COR Biosciences). For quantification, each protein was normalized to GAPDH. For time course analysis, time=0 for each time course experiment was set to 1 and fold change was measured.
Co-Immunoprecipitation
HEK293 cells were transfected with human WT IRE1α (a gift from Fumihiko Urano, Addgene plasmid #20744) (Lipson et al., 2008), Sigmar1-myc-FLAG transcript variant 1 (#RC201206, Origene), or Sigmar1-myc-FLAG transcript variant 2 (#RC214331, Origene) using Lipofectamine 2000 according to the manufacturer’s protocol. Four days post-transfection, cells were stimulated with 20 μM Linuron. 16 hours later, cells were lysed in ice cold non-denaturing lysis buffer (20 mM Tris-HCl pH=8.0, 137 mM NaCl, 1% NP-40, 2 mM EDTA, recipe obtained from Abcam) supplemented with protease inhibitor cocktail (#5871, Cell Signaling, 1:100 working concentration) and phosphatase inhibitor cocktail (#5870, Cell Signaling, 1:100 working concentration). Immunoprecipitation for c-myc was performed for 4 hours at 4C using the following kit (#23620, Thermo Fisher Scientific). SDS-PAGE was then performed as described above. The following antibodies were used: Mouse anti-FLAG M2 (#F1804–200UG, Sigma-Aldrich, 1:1000) and Rabbit anti-IRE1α (14C10) (#3294S, Cell Signaling, 1:1000).
Xbp1 splicing analysis
Mouse astrocyte cDNA was amplified using Phusion HF PCR Master Mix (#F548L, Thermo Fisher Scientific) using the following primers: forward: 5’-AAACAGAGTAGCAGCGCAGACTGC-3’ and reverse: 5’-GGATCTCTAAAACTAGAGGCTTGGTG-3’ to generate a 598 bp fragment containing a unique PstI restriction enzyme site (Calfon et al., 2002). For human astrocyte studies, the XBP1 transcript was amplified using the primers: forward: 5’-GCGCTGAGGAGGAAACTGAAAAAC-3’ and reverse: 5’-CAGAGGGTATCTCTAAGACTAG-3’ to generate a 621 bp fragment containing a unique PstI restriction enzyme site.
For human and mouse studies the PCR amplified fragment was purified using a QIAquick PCR Purification Kit (#28104, Qiagen) and digested using PstI-HF and DpnI in CutSmart Buffer (R3140S, R0176S, New England Biolabs) for 30 minutes at 37C. The cut PCR products were then run on a 0.8% agarose gel for analysis. Densitometry was performed using FIJI (Schindelin et al., 2012) by comparing the ratio of spliced versus unspliced transcript.
In silico promoter analysis
The M. musculus Nos2 and Csf2 genomic sequences were obtained using Ensembl (Yates et al., 2016). The DNA sequences ~2000 bp upstream of the protein coding transcripts Nos2–201 and Csf2–201 were analyzed. p65/RelA transcription factor binding sites were identified using Mulan (Ovcharenko et al., 2005). XBP1 DNA binding sites were defined as those possessing the XBP1 consensus binding sequence, 5’-acgt-3’ (Kanemoto et al., 2005).
Chromatin immunoprecipitation (ChIP)
Approximately 1.2 million astrocytes were exposed to the indicated treatments followed by cell preparation according to the ChIP-IT Express Enzymatic Shearing and ChIP protocol (#53009, Active Motif). Briefly, cells were fixed in 1% formaldehyde for 10 minutes with gentle agitation, washed in 1X PBS, washed for 5 minutes in 1X glycine Stop-Fix solution in PBS, and scraped in 1X PBS supplemented with 500 μM PMSF. Cells were pelleted, nuclei were isolated, and chromatin was sheared using the Enzymatic Shearing Cocktail (Active Motif) for 10 minutes at 37C with vortexing every 2 minutes. Sheared chromatin was immunoprecipitated according to the Active Motif protocol overnight at 4C with rotation. The next day, the protein-bound magnetic beads were washed 1X with ChIP buffer 1, 1X with ChIP buffer 2, and 1X with 1X TE. Cross-links were reversed in 100 μL of 0.1% SDS and 300 mM NaCl in 1X TE at 63C for 4–5 hours, as described (van Galen et al., 2016). DNA was purified using QIAquick PCR Purification Kit (#28104, Qiagen). qPCR was performed using Fast SYBR Green Master Mix (#4385612, Thermo Fisher Scientific). Anti-IgG immunoprecipitation and 10% input were used as controls. Antibodies used in this study are: mouse anti-XBP1 antibody (#sc-8015X, Santa Cruz Biotechnology, 1:100), rabbit anti-p65 (#8242S, Cell Signaling Technology), and rabbit IgG polyclonal isotype control (ChIP grade) (#ab171870, Abcam, 1:100). PCR primers were developed by using Primer3 (Untergasser et al., 2012) to generate amplicons of approximately 150 bp. Data were analyzed by either ddCt as above or percent enrichment relative to input. Percent enrichment was calculated by: Adjusted input: Ct(input)-log2(input fraction−1). Percent enrichment: 100*2adjusted input-Ct(sample). Primer sequences used for this analysis are: Nos2-Xbp1-forward 5’-AGCTGCAAGCCAGGGTATGT-3’, Nos2-Xbp1-reverse 5’-CTGTGGTGTATCCTCATGCAA-3’, Nos2-p65-forward 5’-CACAGACTAGGAGT GTCCATCA-3’, Nos2-p65-reverse 5’-TATACCCCTCCAGGCTCTGC-3’, Csf2-Xbp1–1-forward 5’-TAGGAGAAAGATACATATCTACCCACA-3’, Csf2-Xbp1–1-reverse 5’-GGCAGGGTGGTGGCTAAG-3’, Csf2-Xbp1–2-forward 5’-GAGCTTCTGGAGAGGGAGGT-3’, Csf2-Xbp1–2-reverse 5’-AATAACCAGGCACGCACAC-3’.
Intracranial injection of lentivirus
Lentiviral constructs used in this study were generated by modifying the pLenti-GFAP::EGFP-shRNA vector constructed previously (Yan et al., 2012) and used by our lab previously (Mayo et al., 2014; Rothhammer et al., 2018; Rothhammer et al., 2016), which contains a truncated form of the human GFAP promoter, gfa2, (GfaABC1D) (Lee et al., 2008) driving the expression of EGFP and an shRNA. For the scrambled shRNA hairpin sequence, 5’-GCGCGATAGCGCTAATAATTT- TAGTGAAGCCACAGATGTA-AAATTATTAGCGCTATCGCGC-3’ (#SHC016, Sigma-Aldrich) was used. For the shXbp1 hairpin sequence, 5’- GGTTGAGAACCAGGAGTTAAG-TAGTGAAGCCACAGATGTA-CTTAACTCCTGGTTCTCAACC-3’ (#TRCN0000232018, Thermo-Fisher Scientific) was used. For the shErn1 hairpin sequence, (#TRCN0000356311, Thermo-Fisher Scientific) 5’-GGAATCCTCTACATGGGTAAA-TAGTGAAGCCACAGATGTA-TTTACCCATGTAGAGGATTCC-3’ was used. Sequences were obtained using the Broad Institute’s GPP Web Portal. Lentiviral constructs were grown in NEB Stable Cells (#C3040, New England Biolabs) at 30C and DNA was prepared using QIAprep Spin Miniprep Kit (#2710, Qiagen). Lentiviral plasmids were transfected into HEK293FT cells (the sex of these cells is unknown) according to the ViraPower Lentiviral Packaging Mix protocol (#K497500, Thermo Fisher Scientific). Lentiviruses were packaged with pLP1, pLP2, and pseudotyped with pLP/VSVG. Lentivirus was collected and concentrated using Lenti-X Concentrator (#631231, Clontech) overnight at 4C followed by centrifugation according to the manufacturer’s protocol and resuspension in 1/100–1/500 of the original volume in 1X PBS. Viral titer was determined by using the qPCR Lentivirus Titration Kit (#LV900, Applied Biological Materials). Titers for lentiviruses used were on the order of 108 IU/mL.
Delivery of lentiviruses via intracerebroventricular (ICV) injection was performed largely as described previously (Rothhammer et al., 2016). Briefly, mice were anesthetized using 1–3% isoflurane mixed with oxygen. Their heads were shaved and cleaned followed by a medial incision of the skin to expose the skull. The ventricles were targeted bilaterally using the coordinates: +/− 1.0 (lateral), −0.44 (posterior), −2.2 (ventral) relative to Bregma. Mice were injected with approximately 107 total IU of lentivirus delivered by two 10 μL injections using a 25 μL Hamilton syringe (#20787, Sigma-Aldrich) on a Stereotaxic Alignment System (#1900, Kopf), sutured, and permitted to recover in a separate clean cage. Mice were permitted to recover for between 4–7 days before induction of EAE.
CRISPR/Cas9 gene deletion and validation
A fragment encoding a U6-sgRNA derived from lentiCRISPR v2 (a gift from Feng Zhang, Addgene plasmid #52961) (Sanjana et al., 2014) and the ABC1D gfa2 GFAP promoter (Lee et al., 2008) (both purchased from Genewiz) was substituted for the EFS promoter into the lentiCas9-EGFP backbone (a gift from Phil Sharp and Feng Zhang, Addgene plasmid #63592) (Chen et al., 2015). sgRNA sequences were designed using a combination of the Broad Institute’s sgRNA GPP Web Portal (portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design), Synthego (https://design.synthego.com/#/validate), and cross-referenced with activity-optimized sequences contained within the Addgene library #1000000096 (a gift from David Sabatini and Eric Lander) (Wang et al., 2017). Lentiviruses were produced and injected as described above. sgRNAs used in this study were: sgXbp1: 5’-gGGACACGCTGGATCCTGACG-3’, sgErn1: 5’-gACATCCTGAGATACGGTGGT-3’, sgSigmar1: 5’-gGAATGCCGTGGGCCGCGGGA-3’, sgEif2ak3: 5’-gCTTTGAACTTCGGTATATTC-3’, and sgScramble: 5’-gGCACTACCAGAGCTAACTCA-3’ (sequence from #GE100003, Origene).
Transfections and luciferase assays
HEK-293 cells (ATCC, #CRL-1573) were grown in DMEM/F12+GlutaMAX supplemented with 10% FBS, 1% penicillin/streptomycin, and 500 μg/mL Geneticin (#10131035, Thermo Fisher Scientific). Cells were transfected using Fugene-HD Transfection Reagent (#E2311, Promega). For knockdown validation studies, GL261 cells (#CRL-1887, ATCC) were transfected using Lipofectamine 2000 (#11668019, Thermo Fisher Scientific) with the indicated constructs and assayed for expression 48 hours post-transfection. GL261 cells were grown in the media described above supplemented with 100 ng/mL Geneticin (#10131035, Thermo Fisher Scientific). Constructs used were: pGL2-NOS2Promoter-Luciferase (a gift from Charles Lowenstein, Addgene plasmid #19296) (Lowenstein et al., 1993), pcDNA3-RelA-cFlag (a gift from Stephen Smale, Addgene plasmid #20012) (Sanjabi et al., 2005), pFLAG.XBP1.p.CMV2 (a gift from David Ron, Addgene plasmid #21833) (Calfon et al., 2002), and FLAG-HA-pcDNA3.1 (a gift from Adam Antebi, Addgene plasmid #52535) (Horn et al., 2014), and pRL-CMV-Renilla (#E2261, Promega). Luciferase activity was analyzed either 24 or 48 h post-transfection using the Dual Luciferase Reporter System (#E1960, Promega) and normalized to Renilla luciferase activity. For experiments testing pharmacological induction of Nos2, chemicals were used at the following doses: 20 μM Linuron, 50 ng/mL TNFα, 100 ng/mL IL-1ß, and 50 μM BD-1063. All cells were grown in a humidified incubator at 37C with 5% CO2. Cells lines were purchased from and authenticated by the ATCC. The sex of HEK293 and GL261 cells is unknown.
Immunostaining
Mice were intracardially perfused with ice cold 1X PBS followed by ice cold 4% PFA. Brains were harvested, post-fixed in 4% PFA overnight at 4C, followed by dehydration in 30% sucrose for 2 days at 4C. Brains were then frozen in OCT (#4583, Sakura) and 30 μm sections were obtained by cryostat on SuperFrost Plus slides (#22–037-246, Fisher Scientific). A hydrophobic barrier was drawn (#H-4000, Vector Laboratories) and sections were washed 3X for 5 minutes with 0.3% Triton X-100 in PBS (PBS-T). Sections were blocked with 5% donkey serum (#D9663, Sigma-Aldrich) in 0.3% PBS-T at RT for 30 minutes. Sections were then incubated with primary antibodies diluted in blocking buffer overnight at 4C. Following primary antibody incubation, sections were washed 3X with 0.3% PBS-T and incubated with secondary antibodies diluted in blocking buffer for 2 hours at RT. Following secondary incubation, sections were washed 3X with 0.3% PBS-T, dried, and coverslips were mounted using Fluoromount-G with DAPI (#0100–20, SouthernBiotech). Primary antibodies used in this study were: mouse anti-GFAP (#MAB360, Millipore, 1:500), chicken anti-GFP (#ab13970, Abcam, 1:1000), mouse anti-Sigma receptor 1 (B-5) (#sc-137075, Santa Cruz Biotechnology, 1:100), rabbit anti-P-IRE1α (#NB100–2323, Novus Biologicals, 1:100), rabbit anti-PERK (#ab217322, Abcam, 1:100), and rabbit anti-XBP1s (#619502, Biolegend, 1:100). Secondary antibodies used in this study were: Alexa Fluor 647 donkey anti-mouse (#ab150107, Abcam), donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed, Alexa Fluor 568 (#A10042, Life Technologies), Rhodamine Red-X-AffiniPure Fab Fragment donkey anti-mouse IgG (H+L) (#715–297-003, Jackson ImmunoResearch, 2 mg/mL stock), and Goat anti-Chicken IgY (H+L), Alexa Fluor 488 (#A11039, Life Technologies) all at 1:500 working dilution. Dual labeling using mouse primary antibodies was accomplished by incubating with a single primary antibody on Day 1, staining with the anti-mouse Fab fragment on Day 2, washing 6X with PBS-T, followed by incubation with the remainder of primary and secondary antibodies as described above.
For immunocytochemistry, astrocytes were plated on poly-D-lysine and laminin (100 μg/mL) coated coverslips. They were fixed in 4% PFA, washed 3X with 0.3% PBS-T, blocked for 30 minutes in 5% donkey serum in 0.3% PBS-T, and incubated with primary antibody overnight at 4C. The next day, cells were washed 3X with 0.3% PBS-T and samples were incubated with secondary antibodies overnight. Primary antibody used was mouse anti-GFAP (#MAB360, Millipore, 1:500), and the secondary antibodies used were thioflavin T (#596200, Millipore, 5 μM working dilution from 5 mM stock diluted in EtOH) and rhodamine donkey anti-mouse IgG Fab fragment (H+L) (#715–297-003, Jackson Immunoresearch, 1:500 at 0.5 mg/mL).
Confocal imaging and analysis
Sections were imaged on a Zeiss LSM710 confocal using a 20X objective. For all imaging analyses, regions were randomly chosen based on DAPI fluorescence. Imaging was performed using the LSM710 smart setup parameters with each channel acquired in sequence to minimize crosstalk between channels to essentially zero. Image analysis for Thioflavin T experiments was performed using FIJI by quantifying total GFP channel fluorescence for each acquired section and subtracting background. Quantification of XBP1, Sigmar1, P-IRE1α, and PERK staining in brain sections was performed by first quantifying the number of positive cells and then counting the number of those cells that are GFAP+ with a visible cell body in the DAPI channel.
Human primary astrocyte culture and RNA purification
Human cells were plated at 20,000 cells/mL into 12 well plates. Once cells were 80% confluent, they were treated under the indicated conditions. Astrocytes were treated with vehicle (1X PBS at 1:1000 in culture medium), 20 μM linuron (#36141, Sigma-Aldrich, 20 mM stock in EtOH), 100 ng/mL IL-1ß (#401-ML-005, R&D Systems, 100 μg/mL stock in PBS), 50 ng/mL TNFα (#410-MT-010, R&D Systems, 100 μg/mL stock in PBS), and 50 μM STF083010 (#SML0409, Sigma-Aldrich, 50 mM stock in DMSO). RNA was isolated from human astrocytes by lysing with Buffer RLT (Qiagen) and was purified in the same manner as mouse RNA followed by RT-PCR, which was performed in the same manner as mouse samples. Human probes used in this study were: EDEM1 (Hs00976004_m1), NOS2 (Hs01075529_m1), and GAPDH (Hs02758991_g1).
Immunostaining of frozen MS tissue
Frozen brain tissue from 4 MS patients and 4 healthy control individuals was cut into 7 μm thick sections, air dried, and fixed in ice-cold acetone for 10 minutes. Sections were delipidised in 70% ethanol for 5 minutes, followed by blocking of non-specific binding with 10% donkey serum (#D9663, Sigma-Aldrich). Rabbit anti-human P-IRE1α (#NB100–2323, Novus Biologicals, 1:50) was incubated with mouse anti-human GFAP-Cy3 (#C9205, Sigma-Aldrich, 1:50), mouse anti-human NeuN (#MAB377, Millipore, 1:50), mouse anti-human CNPase (#MAB326, Millipore, 1:100) or mouse anti-human CD14 (#555397, BD Biosciences, 1:10) in blocking buffer overnight at 4C. The next day slides were washed with 0.05% PBS-Tween and incubated with a mixture of donkey anti-rabbit Alexa Fluor 488 (#R37118, Life Technologies, 1:400) and donkey anti-mouse-Cy3 (#715–165-151, Jackson ImmunoResearch, 1:200) or donkey anti-rabbit-Rhodamine Red X (#711–295-152, Jackson ImmunoResearch, 1:400) and donkey anti-mouse-Alexa Fluor 488 (Jackson ImmunoResearch, 1:400) for 40 minutes at room temperature. Sections were mounted in Mowiol (#81381, Sigma-Aldrich) containing TOPRO-3 (#T3605, Invitrogen). In secondary only controls, primary antibodies were omitted to control for non-specific binding.
Immunostaining of paraffin-embedded MS tissue
4 μm thick paraffin embedded brain sections from 4 MS patients and 3 healthy controls were deparaffinised, washed in PBS, and treated with heat-induced antigen retrieval in Tris-EDTA pH=9. Endogenous avidin/biotin was blocked using an avidin-biotin blocking kit (#004303, Life Technologies) and non-specific binding was further blocked with 10% Goat serum (#G9023, Sigma). Mouse anti-human XBP1s (#MABC521, Millipore, 1:50) was incubated in blocking buffer with 0.1% Triton-X100 overnight at 4C. The next day slides were washed with 1% PBS-Triton-X100 and incubated with goat anti-mouse biotin (#0433, Dako, 1:500), followed by blocking of endogenous peroxidase using 0.3% H2O2 for 15 min. Sections were washed and incubated with streptavidin-HRP (#554066, BD Biosciences, 1:1000) for 40 minutes at room temperature and the signal developed with Tyramide-FITC (#B40953, Life Technologies). After extensive washing, sections were incubated with mouse anti-human GFAP-Cy3 (#C9205, Sigma, 1:50) in blocking buffer for 1 hour at room temperature. Sections were washed again and mounted in Mowiol containing TOPRO-3. In secondary only controls, primary antibodies were omitted to control for non-specific binding. Additional primary antibodies used are: rabbit anti-human NOGO-A (#ab62024, Abcam, 1:200), rabbit anti-human CD14 (#ab133335, Abcam, 1:500), chicken anti-human MAP2 (#ab5392, Abcam, 1:500). Additional secondary antibodies used are: Goat anti-rabbit Cy3 (#111–165-003, Jackson Immunoresearch, 1:300), Goat anti-chicken Alexa 555 (#A21437, Invitrogen, 1:400).
Image acquisition of MS tissue
Images were acquired as z-stacks using a Leica SP5 confocal microscope with Leica LAS AF software and processed using FIJI and LAS X. All acquisition settings were kept the same between replicates. P-IRE1α+ and XBP1s+ cells were quantified per field of view in GFAP+, NeuN+, CNPase+ and CD14+ cell bodies.
Isolation of cells from adult mouse CNS
Astrocytes were isolated by flow cytometry as described (Mayo et al., 2014; Rothhammer et al., 2016). Briefly, mice were perfused with 1X PBS and the brain was isolated into digestion solution (0.05% Trypsin (#25200–072, Thermo Fisher Scientific), 1 mM EDTA pH=8.0, in HBSS (#14175, Thermo Fisher Scientific)) and finely chopped with a razor blade. The CNS was incubated at 37C for 10–40 minutes and filtered through a 70 μm cell strainer. Cells were pelleted at 500xg at 4C for 10 minutes in a centrifuge followed by suspension of the pellet in 33% Percoll TM (#17–5445-01, GE Healthcare) in 1X PBS. The suspension was centrifuged at 500xg at 4C for 25 minutes with slow acceleration and deceleration settings to separate myelin and intact cells. Pelleted intact cells were washed with 1X PBS and then stained with antibodies for flow cytometry.
Adult mouse astrocyte cultures
Brains of Gfap(Cre/+);TdTomato(f/+) mice were dissected similar to the described protocol (Foo, 2013). A single cell suspension was obtained and cells were sorted according to TdTomato fluorescence. Sorted cells were then cultured in poly-L-lysine coated wells in serum-free DMEM/F12 media supplemented with 5 ng/mL heparin-binding EGF-like growth factor (#E4643–50UG, Sigma-Aldrich). Cells were cultured until confluent and then treated with the indicated compounds for 24 hours.
Flow cytometry
Cells were stained in the dark on ice for 15 minutes with flow cytometry antibodies. Cells were then washed once with 1X PBS and resuspended in 1X PBS for sorting as described previously (Mayo et al., 2014; Rothhammer et al., 2016). Antibodies used in this study were: PE anti-mouse CD45R/B220 (#553089, BD Biosciences, 1:100), PE anti-mouse TER-119 (#116207, Biolegend, 1:100), PE anti-O4 (#FAB1326P, R&D Systems, 1:100), PE anti-CD105 (#12–1051-82, eBioscience, 1:100), PE anti-CD140a (#12–1401-81, eBioscience, 1:100), PE anti-Ly-6G (#127608, Biolegend, 1:100), PerCP anti-Ly-6C (#128028, Biolegend, 1:100), APC anti-CD45 (#17–0451-83, eBioscience, 1:100), APC-Cy7 anti-CD11c (#561241, BD Biosciences, 1:100), and PE-Cy7 or FITC anti-CD11b (#25–0112-82, #11–0112-85, eBioscience, 1:100). All cells were gated on the following parameters: CD105negCD140anegO4negTer119negLy-6GnegCD45Rneg. Astrocytes were subsequently gated on: CD11bnegCD45negLy-6CnegCD11cneg. Microglia were subsequently gated on: CD11bhighCD45lowLy-6Clow. Pro-inflammatory monocytes were subsequently gated on: CD11bhighCD45highLy-6Chigh. Compensation was performed on single-stained samples of cells and an unstained control.
FACS analysis of T cells
Single cell suspensions from the CNS and from the spleen were stimulated using 500 ng/mL PMA (phorbol 12-myristate 13-acetate) (#P1585, Sigma-Aldrich), 500 ng/mL ionomycin (#I9657, Sigma-Aldrich), and GolgiSTOP (#554724, BD Biosciences, 1:1000) diluted in T-cell culture medium (RPMI (#11875119, Life Technologies) containing 10% FBS, 1% penicillin/streptomycin, 50 μM ß-mercaptoethanol (#M6250, Sigma-Aldrich), and 1% Non-Essential Amino Acids (#11140050, Life Technologies)) for 4 hours. Following stimulation, T cells were washed with 1X PBS, centrifuged, and incubated with antibodies against surface markers, using a live/dead cell marker for 15 minutes on ice. Cells were then washed once with 1X PBS followed by use of a fixation and intracellular antibody labeling kit (#00–5523, eBioscience). Antibodies used were: eFluor 450 anti-CD3 (#48–0032-82, eBioscience, 1:100), FITC anti-CD4 (#100510, BioLegend, 1:50), 405 Aqua LIVE/DEAD cell stain kit (#L34966, Thermo Fisher Scientific, 1:400), APC-Cy7 anti-IFNγ (#561479, BD Biosciences, 1:100), PE anti-IL-17a (#12–7177-81, eBioscience, 1:100), APC anti-IL-10 (#505010, BioLegend, 1:100), PerCP-Cy5.5 anti-FoxP3 (#45–5773-82, eBioscience, 1:100). Compensation was performed on single-stained samples and an unstained control. Gating of the CNS was performed on 5,000 live CD3+/CD4+ cells and gating for the spleen was performed on 10,000 live CD3+ CD4+ cells. FACS was performed on an LSRII (BD Biosciences).
nCounter gene expression.
50 ng of total RNA was hybridized with reporter and capture probes in custom-made astrocyte-targeted nCounter gene expression code sets according to the manufacturer’s instructions (NanoString Technologies), as previously described (Rothhammer et al., 2016). Data were analyzed using nSolver Analysis software.
RNA-seq
Astrocytes, microglia, and pro-inflammatory monocytes were lysed and RNA isolated using the Qiagen RNeasy Mini kit (#74106, Qiagen) with on-column DNase I digestion (#79254, Qiagen). RNA was cleaned using Agencourt RNA Clean XP magnetic beads (#A63987, Beckman Coulter) according to the manufacturer’s protocol and concentrated using a Savant Speedvac Concentrator (#DNA120, Thermo Fisher Scientific). RNA was suspended in 10 μL of nuclease free water and sequenced using 3’ Digital Gene Expression (Soumillon et al., 2014) by the Broad Technology Labs and the Broad Genomics Platform. Processed RNA-Seq data was filtered, removing genes with low read counts. Read counts were normalized using TMM normalization and CPM (counts per million) were calculated to create a matrix of normalized expression values. Differential binding in astrocytes, microglia, and monocytes were performed using R (v3.3.3) and Limma (v3.34.1) (Ritchie et al., 2015). A p-value <0.05 was used to determine differential genes. Heatmaps were generated with GENE-E program of the Broad Institute. Enrichment plots for RNA-Seq data were generated using GSEA (Subramanian et al., 2005) using molecular signatures for canonical pathways: KEGG/Reactome/Biocarta (c2.cp.all), Gene ontology (c5.cp.all), and Hallmark (h.all) in astrocytes, microglia, and monocytes individually. Default settings were used. Pathway analysis was performed using IPA (Qiagen).
ATAC-seq
Sequencing libraries were prepared largely as described previously (Buenrostro et al., 2013; Buenrostro et al., 2015). After isolation of nuclei, transposition was performed using the kit (#FC-121–1030, Illumina) according to the Buenrostro protocol. DNA was then amplified using NEBNext High Fidelity 2X PCR Master Mix (M0541S, New England Biolabs) for 5 cycles. DNA quantity was then measured using a Viia 7 Real-Time PCR System (Thermo Fisher Scientific) and the number of cycles required to achieve 1/3 of maximal SYBR Green fluorescence was determined and libraries were amplified accordingly. TruSeq adaptors (universal: Ad1_noMX and barcoded: Ad2.1-Ad2.7) were used according to the Buenrostro protocol. Libraries were purified using MiniElute PCR Purification Kit (#28006, Qiagen) followed by double sided AMPure XP bead purification (#A63881, Beckman Coulter) to remove primer dimers and large DNA fragments. Libraries were analyzed on an Agilent 2200 High Sensitivity DNA TapeStation in the Harvard Biopolymers Facility. Libraries were sequenced by Genewiz on an Illumina HiSeq by 2×150 bp paired end sequencing. Mean quality score for the data was 32.17 with QC≥30 of 68.21% and an average of 40,000,000 reads per sample.
Paired-end ATAC-Seq reads were first trimmed from 150bp to 50bp with Fastx (v0.0.13) fastx_trimmer and then aligned against the GRCm39/mm10 mouse genome assembly with Bowtie (v2.3.0) (Langmead and Salzberg, 2012) in local mode and a maximum fragment size of 2000. Alignments were filtered with SAMtools (v1.3) to exclude reads with mapping quality <30, not properly paired, aligned to mitochondrial genome, and/or aligned to ENCODE blacklist regions. PCR duplicates were not present in the dataset. Alignments with an insertion size of >100bp were removed to enrich for nucleosome-free reads. ATAC-Seq peaks were called for each replicate using MACS2, using --format BAMPE and --keep-dup all. Differential peaks were determined using DiffBind, 600bp peak intervals (300 up- and down-stream of peak summits). Differential peaks were then selected with FDR<0.1 and were annotated to gene symbols using Bedtool (v2.26.0) closest. For signal tracks, ATAC-Seq was processed in similar manner as ChIP-Seq but used a window size of 10bp.
ChIP-seq
Approximately 400,000 cells were fixed, nuclei collected, and chromatin isolated using the ChIP-IT Express Enzymatic Shearing Kit (#53009, Active Motif). Chromatin complexes were immunoprecipitated using a mouse anti-XBP1 antibody (#sc-8015X, Santa Cruz Biotechnology, 1:100). Cross-links were reversed in the following buffer: (1% SDS, 0.1M NaHCO3 in H2O) by heating at 65C for 8–12 hours. DNA was purified using a QIAquick PCR Purification Kit (#28104, Qiagen). DNA was analyzed using an Agilent 2200 High Sensitivity DNA TapeStation in the Harvard Biopolymers Facility. DNA libraries were then prepared for sequencing using the NEBNext Ultra II DNA Library Prep Kit (#E7645S, New England Biolabs) and sequencing adaptors were ligated using NEBNext Multiplex Oligos for Illumina (#E7500S, E7335S) as described. ChIPed DNA was amplified by using 13 cycles according to the NEBNext protocol. DNA libraries were not size selected but primer dimers were removed via purification using Agencourt AMPure XP Beads (#A63881, Beckman Coulter). DNA libraries consisting of ChIPed DNA and input DNA for each sample were sequenced on an Illumina HiSeq using 2×150 paired end sequencing. Mean quality score for the data was 36.79 with QC≥30 of 86.35% and an average of 42,000,000 reads per sample.
Paired-end ChiP-Seq reads were aligned using BWA mem (v.0.7.8) (Li and Durbin, 2009) against the GRCm39/mm10 mouse genome assembly with default settings. PCR duplicates were not present in the dataset. Alignments were filtered with SAMtools (v1.3) (Li et al., 2009) to exclude reads with mapping quality <30, not properly paired, aligned to mitochondrial genome, and aligned to ENCODE blacklist regions (Consortium, 2012). For peak calling, MACS2 callpeak (v2.1.1) (Zhang et al., 2008) were called on individual replicates for each ChIP (treatment) and Input (control) pair, using p-value<0.01. Differential peak binding was accessed with Bioconductor DiffBind (v. 2.6.1) (Stark, 2011) using 800bp peak intervals (400bp up- and down-stream of summits). The consensus peak dataset was found within the DiffBind framework, where merged peaks overlap >=1bp. Differential peaks were then selected with FDR<0.1 and were annotated to gene symbols using Bedtools (v2.26.0) (Quinlan and Hall, 2010) closest. To generate signal tracks, the fold enrichment for each replicate was calculated using MACS2 bdcmp. Regions of interest were then binned into 100bp windows with Bedtools makeWindows for smoothing and Bedtools map calculated the coverage for each bin in every replicate. The coverage for both groups (Naïve and EAE) were pooled by summing the total reads and density plots were create using Sushi R package (v1.16.0) (Phanstiel, 2015) to visualize the genomic regions.
Toxcast Pathway Analysis
ER stress genes and bioactivity data were downloaded from EPA’s Toxcast database (Dix et al., 2007). Datasets were parsed for Linuron (C330552) in all active and tested chemical-assay combinations. AC50 was transformed to −log10(AC50/10000000) to anchor AC50 values to 0. Dataset was split between activated and inhibited activity based on signal direction of assay readout for targets. Since chemicals are run in multiple-concentrations for different time-points, the same target can appear more than once for the same assay. These AC50 values were averaged. Heatmaps were generated using R software. ER-stress response network diagram was created with NetworkAnalyst (Xia et al., 2015) using settings for zero-order network (direct interactions) and StringDB database default parameters. Data from Toxcast was further analyzed for assays involved in ER stress response. XBP1, ATF6, Jun/Fos, and Creb3 were the only target genes found to be tested in Toxcast with available data. IPA was used to generate the ER-Stress response diagram.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical tests, comparisons, and sample sizes are included in the Figure Legends. All data are shown as mean±SEM. In all cases, ***p<0.001, **p<0.01, *p<0.05, ns=not statistically significant, p>0.05. All statistical tests were performed using Prism 7 (GraphPad). In all cases, except imaging analysis of mouse and human samples, the stated “n” value indicates a biological replicate. In imaging analyses, lowercase “n” indicates the number of images analyzed and upper case “N” indicates the number of biological replicates (independent wells, mice, or patients). In no case does “n” represent a technical replicate. No statistical methods were used to pre-determine sample sizes. Outliers were formally excluded using Grubbs’ test (Prism, www.graphpad.com/quickcalcs/Grubbs1.cfm) or PCA (RNA-seq data) and not more than one outlier was excluded per group. Blinding was not performed in this study. All samples were randomly allocated into different treatment groups. Fig. 3B (Sigmar1 co-IP) and Fig. S4D (sgEif2ak3 EAE) were performed once. All other in vitro and in vivo experiments were replicated at least once, and all attempts at replication were successful.
DATA AND SOFTWARE AVAILABILITY
A SuperSeries page has been created in the GEO database with the accession number GSE122119. All ATAC-seq, ChIP-seq, and RNA-seq data have been deposited in the GEO database under this SuperSeries with the accession numbers GSE121923, GSE121935, and GSE122113, respectively.
Supplementary Material
Figure S1. XBP1 activation by stressors different from Linuron, Related to Figure 1 and Figure 2. A) nos2a expression in vehicle or Linuron treated zebrafish. n=4 replicates per condition with at least 10 fish per group. Unpaired two-tailed t-test. B) Xbp1 splicing in human fetal astrocytes exposed to different stressors. n=4 per condition. One-way ANOVA, Dunnett post-test. C) Xbp1 splicing in mouse neonatal astrocytes treated with multiple stressors for 4 hours. n=3–6 per condition. One-way ANOVA, Tukey post-test. D) Xbp1 splicing in mouse neonatal astrocytes treated with PFNA or Methyl Carbamate. n=3 per condition. Two-way ANOVA, Sidak post-test. E) Xbp1 splicing in neonatal mouse microglia in culture. n=4 per condition. One-way ANOVA, Dunnett post-test. F) Effect of IRE1α/XBP1 signaling inhibition in sorted adult murine TdTomato+ astrocytes. n=3 per condition. One-way ANOVA, Tukey post-test. ns=not significant. ***p<0.001, **p<0.01, *p<0.05. Data shown as mean ± SEM.
Table S4. Effects of Xbp1 knock down on the transcriptional response of astrocytes in EAE, Related to Figure 4 and Figure 5. Genes detected as differentially modulated by RNA-seq with p<0.05 are listed. logFC measured as ratio of expression in shXbp1 condition compared to shScrmbl condition.
Table S5. Effects of Xbp1 knock down in astrocytes on the transcriptional response of microglia and CNS-recruited monocytes in EAE, Related to Figure 4 and Figure 5. Genes detected as differentially modulated by RNA-seq with p<0.05 are listed. logFC measured as ratio of expression in shXbp1 condition compared to shScrmbl condition.
Table S6. Effects of Xbp1 knockdown in astrocytes on chromatin accessibility, Related to Figure 4. Ingenuity pathway analysis of XBP1 targets identified by ATAC-seq. Pathways with p<0.05 are listed.
Table S7. List of oligonucleotides used. Related to Key Resources Table.
Figure S2. XBP1 ChIP-seq pathway analysis, Related to Figure 4. A) Table of statistically significant XBP1-driven pathways in astrocytes from EAE mice compared to naïve mice. Table generated by Ingenuity Pathway Analysis. B) XBP1 ChIP-seq density plots for Edem1, Dnajb9, and Csf2 genomic loci in astrocytes isolated from naïve and EAE mice, normalized to input DNA. n=3 EAE, n=2 naïve. Scale bars show read density. Schematics of transcriptional regulation shown above density plots.
Figure S3. Evaluation of shRNA-based knockdown, Related to Figure 4. A) XBP1 expression in glial cells determined by western blot. n=4 for shXbp1, n=2 for WT. One-sample t-test. B) IRE1α expression in glial cells determined by Western blot. n=3 per condition. One-sample t-test. C) Xbp1 expression in astrocytes, microglia, or monocytes. n=5 replicates per condition. Unpaired two-tailed t-test. D-E) FACS analysis of astrocytes, microglia, and T cells isolated from mice undergoing EAE transduced with Gfap-driven shXbp1 or non-targeting lentiviruses. T cells isolated from the CNS shown in (D) or the spleen shown in (E). n=10 per condition for astrocytes and microglia, n=3 per condition for T cells. Unpaired two-tailed t-test. **p<0.01, *p<0.05. ns=not significant. Data shown as mean ± SEM.
Figure S4. UPR perturbation during EAE, Related to Figure 4. A) EAE clinical scores of mice in which Xbp1 and Ern1 were inactivated using CRISPR/Cas9. n=14 sgScrmbl, n=14 sgErn1, and n=12 sgXbp1. Two-way repeated measures ANOVA, Holm-Sidak post-test. B) Quantification of XBP1+ astrocytes in EAE mice following CRISPR/Cas9-based Xbp1 inactivation. n=8–9 sections from N=3 brains per genotype. One-way ANOVA, Tukey post-test. C) sgErn1 knockdown efficiency in astrocytes. n=4–5 sections from N=3 mice. Unpaired two-tailed t-test. D) EAE development in control or sgEif2ak3 (sgPerk)-treated mice. n=4–5 mice per group. Two-way repeated-measures ANOVA, Holm-Sidak post-test. E) Quantification of sgEif2ak3 knockdown efficiency. n=4–6 images from N=3 mice per group. Unpaired two-tailed t-test. ***p<0.001, **p<0.01, *p<0.05, ns=not significant. Data shown as mean ± SEM.
Figure S5. Effects of Sigmar1 inactivation and Linuron on EAE, Related to Figure 4. A) Quantification of sgSigmar1 knockdown validation in astrocytes. n=8–9 images from N=3 mice per condition. Unpaired two-tailed t-test. B) T-cell subsets, astrocytes and microglia in mice treated with sgScrmbl or sgXbp1 Gfap-driven lentiviruses and Linuron or vehicle. n=3 per group (T cells) and n=4–5 per group (otherwise). One-way ANOVA, Tukey post-test. C) T cells, astrocytes and microglia in mice treated with sgScrmbl or sgSigmar1 Gfap-driven lentiviruses and Linuron. n=3 per group (T cells) and n=5 per group (otherwise). Unpaired two-tailed t-test. *p<0.05, ns=not significant. Data shown as mean ± SEM.
Figure S6. Cell-type specific analysis of IRE1α-XBP1s activation in MS, Related to Figure 6. A) IRE1α activation in neurons, oligodendrocytes, and CD14+ cells in MS samples. Images total n=17 for astrocytes, n=9 for neurons, n=8 for oligodendrocytes, and n=3 for CD14+ cells, from N=4 (astrocytes) or N=2 (neurons, oligodendrocytes, and CD14+ cells) MS patients. Cells analyzed per cell type are: n=85 NeuN+, n=109 CNPase+, n=29 CD14+, n=118 GFAP+. ND=not detected. Fisher’s exact test between positive and negative cells compared to astrocytes. B) Analysis of XBP1s expression. n=20–38 sections per cell type. N=4 MS patients per cell type analyzed. Cells analyzed per cell type are: n=60 MAP2+, n=70 NOGOA+, n=59 CD14+, n=212 GFAP+. Fisher’s exact test between positive and negative cells compared to astrocytes. C) Control immunostaining analysis validating antibody target specificity. ***p<0.001. Data shown as mean ± SEM.
Table S1. Candidate chemicals tested in zebrafish screen, Related to Figure 1. Chemicals analyzed in Fig. 1F.
Table S2. Molecular targets of Linuron, Related to Figure 2. Ingenuity pathway analysis of Linuron molecular targets. Pathways with p<0.05 are listed.
Table S3. XBP1 targets in astrocytes during EAE, Related to Figure 4. Ingenuity pathway analysis of XBP1 targets identified by ChIP-seq. Pathways with p<0.05 are listed.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-XBP-1 | Santa Cruz | Cat#sc-8015; RRID:AB_628449 |
| Rabbit anti-P-IRE1a (Ser724) | Novus Biologicals | Cat#NB100–2323; RRID:AB_10145203 |
| Mouse anti-CHOP | Cell Signaling | Cat#2895T; RRID:AB_2089254 |
| Rabbit anti-P-p65 | Cell Signaling | Cat#3033S; RRID:AB_331284 |
| Rabbit anti-GAPDH | Cell Signaling | Cat#2118S; RRID:AB_561053 |
| Anti-mouse IgG-HRP conjugate | Cell Signaling | Cat#7076S; RRID:AB_330924 |
| Anti-rabbit IgG-HRP conjugate | Cell Signaling | Cat#7074S; RRID:AB_2099233 |
| IRDye 800CW goat anti-mouse IgG (H+L) | LI-COR | Cat#925–32210; RRID:AB_2687825 |
| IRDye 800CW goat anti-rabbit IgG (H+L) | LI-COR | Cat#925–32211; RRID:AB_2651127 |
| Mouse anti-FLAG M2 | Sigma-Aldrich | Cat#F1804–200UG; RRID:AB_262044 |
| Rabbit anti-IRE1a (14C10) | Cell Signaling | Cat#3294S; RRID:AB_823545 |
| Mouse anti-XBP1 antibody | Santa Cruz | Cat#sc-8015X; RRID:AB_628449 |
| Rabbit anti-p65 | Cell Signaling | Cat#8242S; RRID:AB_10859369 |
| Rabbit IgG polyclonal isotype control | Abcam | Cat#ab171870; RRID:AB_2687657 |
| Mouse anti-GFAP | Millipore | Cat#MAB360; RRID:AB_11212597 |
| Chicken anti-GFP | Abcam | Cat#ab13970; RRID:AB_300798 |
| Mouse anti-Sigma receptor 1 (B-5) | Santa Cruz | Cat#sc-137075; RRID:AB_2285870 |
| Rabbit anti-PERK | Abcam | Cat#ab217322 |
| Rabbit anti-XBP1s | Biolegend | Cat#619502; RRID:AB_315908 |
| Alexa Fluor 647 donkey anti-mouse | Abcam | Cat#ab150107 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed, Alexa Fluor 568 | Life Technologies | Cat#A10042; RRID:AB_2534017 |
| Rhodamine Red-X-AffiniPure Fab Fragment donkey anti-mouse IgG (H+L) | Jackson ImmunoResearch | Cat#715–297-003; RRID:AB_2340836 |
| Goat anti-Chicken IgY (H+L), Alexa Fluor 488 | Life Technologies | Cat#A11039; RRID:AB_2534096 |
| Thioflavin T | Millipore | Cat#596200 |
| Mouse anti-human GFAP-Cy3 | Sigma-Aldrich | Cat#C9205; RRID:AB_476889 |
| Mouse anti-human NeuN | Millipore | Cat#MAB377; RRID:AB_2298772 |
| Mouse anti-human CNPase | Millipore | Cat#MAB326; RRID:AB_2082608 |
| Mouse anti-human CD14 | BD Biosciences | Cat#555397; RRID:AB_395798 |
| Donkey anti-rabbit Alexa Fluor 488 | Life Technologies | Cat#R37118; RRID:AB_2556546 |
| Donkey anti-mouse-Cy3 | Jackson Immunoresearch | Cat#715–165-151; RRID:AB_2315777 |
| Donkey anti-rabbit-Rhodamine Red X | Jackson Immunoresearch | Cat#711–295-152; RRID:AB_2340613 |
| Donkey anti-mouse-Alexa Fluor 488 | Jackson Immunoresearch | Cat#715–545-150; RRID:AB_2340846 |
| Mouse anti-human XBP1s | Millipore | Cat#MABC521 |
| Goat anti-mouse biotin | Dako | Cat#0433; RRID:AB_2687905 |
| Streptavidin-HRP | BD Biosciences | Cat#554066 |
| Tyramide-FITC | Life Technologies | Cat#B40953 |
| Rabbit anti-human NOGO-A | Abcam | Cat#ab62024; RRID:AB_956171 |
| Rabbit anti-human CD14 | Abcam | Cat#ab133335 |
| Chicken anti-human MAP2 | Abcam | Cat#ab5392; RRID:AB_2138153 |
| Goat anti-rabbit Cy3 | Jackson Immunoresearch | Cat#111–165-003; RRID:AB_2338000 |
| Goat anti-chicken Alexa 555 | Invitrogen | Cat#A21437; RRID:AB_2535858 |
| PE anti-mouse CD45R/B220 | BD Biosciences | Cat#553089; RRID:AB_394619 |
| PE anti-mouse TER-119 | Biolegend | Cat#116207; RRID:AB_313708 |
| PE anti-O4 | R&D | Cat#FAB1326P; RRID:AB_664169 |
| PE anti-CD105 | eBioscience | Cat#12–1051-82; RRID:AB_657524 |
| PE anti-CD140a | eBioscience | Cat#12–1401-81; RRID:AB_657615 |
| PE anti-Ly-6G | Biolegend | Cat#127608; RRID:AB_2129141 |
| PerCP anti-Ly-6C | Biolegend | Cat#128028; RRID:AB_10897805 |
| APC anti-CD45 | eBioscience | Cat#17–0451-83; RRID:AB_469393 |
| APC-Cy7 anti-CD11c | BD Biosciences | Cat#561241; RRID:AB_10611727 |
| PE-Cy7 anti-CD11b | eBioscience | Cat#25–0112-82; RRID:AB_469588 |
| FITC anti-CD11b | eBioscience | Cat#11–0112-85; RRID:AB_464936 |
| eFluor 450 anti-CD3 | eBioscience | Cat#48–0032-82; RRID:AB_1272193 |
| FITC anti-CD4 | BioLegend | Cat#100510; RRID:AB_312713 |
| 405 Aqua LIVE/DEAD cell stain kit | Thermo Fisher | Cat#L34966 |
| APC-Cy7 anti-IFNg | BD Biosciences | Cat#561479; RRID:AB_10898181 |
| PE anti-IL-17a | eBioscience | Cat#12–7177-81; RRID:AB_763582 |
| APC anti-IL-10 | BioLegend | Cat#505010; RRID:AB_315364 |
| PerCP-Cy5.5 anti-FoxP3 | eBioscience | Cat#45–5773-82; RRID:AB_914351 |
| Bacterial and Virus Strains | ||
| NEB Stable E. Coli | NEB | Cat#C3040 |
| Biological Samples | ||
| Human fetal astrocytes | University of Washington Dept. of Pediatrics, Birth Defects Research Laboratory | N/A |
| Human pathology samples | University of Montreal Health Centre | N/A |
| Paraffin-embedded brain sections | Neuropathology Dept. of the Notre Dame Hospital | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cuprizone | Sigma | Cat#C9012 |
| LPS-EK | InvivoGen | Cat#tlrl-eklps |
| TRIzol LS | Thermo Fisher | Cat#10296010 |
| Linuron | Sigma-Aldrich | Cat#36141 |
| Methyl carbamate | Sigma-Aldrich | Cat#246352 |
| Naphthalene | Sigma-Aldrich | Cat#147141 |
| Vinclozolin | Sigma-Aldrich | Cat#45705 |
| PFNA | Sigma-Aldrich | Cat#394459 |
| ATP | NEB | Cat#P0756 |
| BD1063 | Sigma-Aldrich | Cat#SML0276 |
| IL-1b | R&D | Cat#401-ML-005 |
| TNFa | R&D | Cat#410-MT-010 |
| GSK2656157 | Calbiochem | Cat#504651 |
| STF083010 | Sigma-Aldrich | Cat#SML0409 |
| Pertussis toxin | List Biological Laboratories | Cat#180 |
| MOG 35–55 | Genemed Synthesis Inc | Cat#110582 |
| Heparin-binding EGF-like growth factor | Sigma-Aldrich | Cat#E4643–50UG |
| PMA (phorbol 12-myristate 13-acetate) | Sigma-Aldrich | Cat#P1585 |
| Ionomycin | Sigma-Aldrich | Cat#I9657 |
| GolgiSTOP | BD Biosciences | Cat#554724 |
| Geneticin | Thermo Fisher | Cat#10131035 |
| Critical Commercial Assays | ||
| Pierce c-Myc-Tag IP/Co-IP Kit | Thermo Fisher | Cat#23620 |
| QIAquick PCR Purification Kit | Qiagen | Cat#28104 |
| Taqman Fast Universal PCR Master Mix | Life Technologies | Cat#4367846 |
| Zebrafish: actb1 probe | Life Technologies | Cat#Dr03432610_m1 |
| Zebrafish: ccl20 probe | Life Technologies | Cat#Dr03431608_m1 |
| Zebrafish: il1b probe | Life Technologies | Cat#Dr03114369_m1 |
| Zebrafish: il10 probe | Life Technologies | Cat#Dr03103209_m1 |
| Zebrafish: il17a/f1 probe | Life Technologies | Cat#Dr03096843_g1 |
| Zebrafish: mpz probe | Life Technologies | Cat#Dr03131914_m1 |
| Zebrafish: nos2a probe | Life Technologies | Cat#Dr03124753_m1 |
| RNeasy Mini Kit | Qiagen | Cat#74106 |
| High-Capacity cDNA Reverse Transcription Kit | Life Technologies | Cat#4368813 |
| Mouse: Ccl2 probe | Life Technologies | Cat#Mm00441242_m1 |
| Mouse: Gapdh probe | Life Technologies | Cat#Mm99999915_g1 |
| Mouse: Il6 probe | Life Technologies | Cat#Mm00446190_m1 |
| Mouse: Csf2 probe | Life Technologies | Cat#Mm01290062_m1 |
| Mouse: Nos2 probe | Life Technologies | Cat#Mm00440502_m1 |
| Mouse: Ddit3 probe | Life Technologies | Cat#Mm01135937_g1 |
| Mouse: Xbp1 probe | Life Technologies | Cat#Mm00457357_m1 |
| Mouse: Edem1 probe | Life Technologies | Cat#Mm00551797_m1 |
| Mouse: Atf6 probe | Life Technologies | Cat#Mm01295319_m1 |
| SuperSignal West Femto Maximum Sensitivity Substrate | Thermo Fisher | Cat#34095 |
| Phusion HF PCR Master Mix | Thermo Fisher | Cat#F548L |
| PstI-HF | NEB | Cat#R3140S |
| DpnI | NEB | Cat#R0176S |
| ChIP-IT Express Enzymatic Shearing Kit | Active Motif | Cat#53009 |
| Fast SYBR Green Master Mix | Thermo Fisher | Cat#4385612 |
| ViraPower Lentiviral Packaging Mix | Thermo Fisher | Cat#K497500 |
| Lenti-X Concentrator | Clontech | Cat#631231 |
| qPCR Lentivirus Titration Kit | Applied Biological Materials | Cat#LV900 |
| Fugene-HD Transfection Reagent | Promega | Cat#E2311 |
| Lipofectamine 2000 | Thermo Fisher | Cat#11668019 |
| Dual Luciferase Reporter System | Promega | Cat#E1960 |
| Human: EDEM1 probe | Life Technologies | Cat#Hs00976004_m1 |
| Human: NOS2 probe | Life Technologies | Cat#Hs01075529_m1 |
| Human: GAPDH probe | Life Technologies | Cat#Hs02758991_g1 |
| Avidin-biotin blocking kit | Life Technologies | Cat#004303 |
| Intracellular antibody labeling kit | eBioscience | Cat#00–5523 |
| nCounter gene expression codesets | Nanostring Technologies | N/A |
| Agencourt RNA Clean XP magnetic beads | Beckman Coulter | Cat#A63987 |
| Nextera DNA Library Prep Kit | Illumina | Cat#FC-121–1030 |
| NEBNext High Fidelity 2X PCR Master Mix | NEB | Cat#M0541S |
| MiniElute PCR Purification Kit | Qiagen | Cat#28006 |
| NEBNext Ultra II DNA Library Prep Kit | NEB | Cat#E7645S |
| NEBNext Multiplex Oligos for Illumina | NEB | Cat#E7500S, Cat#E7335S |
| Deposited Data | ||
| ATAC-seq | This paper | GEO: GSE121923 |
| ChIP-seq | This paper | GEO: GSE121935 |
| RNA-seq | This paper | GEO: GSE122113 |
| Experimental Models: Cell Lines | ||
| Human: HEK293 cells | ATCC | Cat#CRL-1573 |
| Human: HEK293FT cells | Thermo Fisher | Cat#R70007 |
| Mouse: GL261 cells | ATCC | Cat#CVCL_Y003 |
| Experimental Models: Organisms/Strains | ||
| WT C57Bl/6 mice | Jackson Laboratory | JAX: 000664; RRID:MGI:5656552 |
| B6.Cg-Tg(Gfap-cre)73.12Mvs/J | Jackson Laboratory | JAX: 012886; RRID:IMSR_JAX:012886 |
| B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | Jackson Laboratory | JAX: 007909; RRID:IMSR_JAX:007909 |
| Tg(gfap::egfp) zebrafish | Bernardos and Raymond, 2006 | ZFIN: ZDB-TGCONSTRCT-070117–154 |
| AB* zebrafish | This paper | ZFIN: ZDB-GENO-960809–7; RRID:ZIRC_ZL1438 |
| Oligonucleotides | ||
| See Table S7 | ||
| Recombinant DNA | ||
| WT IRE1a | Lipson et al., 2008 | Addgene Cat#20744 |
| Sigmar1-myc-FLAG transcript variant 1 | Origene | Cat#RC201206 |
| Sigmar1-myc-FLAG transcript variant 2 | Origene | Cat#RC214331 |
| pLenti-Gfap-GFP-shScramble | This paper | N/A |
| pLenti-Gfap-GFP-shErn1 | This paper | N/A |
| pLenti-Gfap-GFP-shXbp1 | This paper | N/A |
| pLenti-U6-sgScramble-Gfap-Cas9–2A-GFP | This paper | N/A |
| pLenti-U6-sgXbp1-Gfap-Cas9–2A-GFP | This paper | N/A |
| pLenti-U6-sgErn1-Gfap-Cas9–2A-GFP | This paper | N/A |
| pLenti-U6-sgSigmar1-Gfap-Cas9–2A-GFP | This paper | N/A |
| pLenti-U6-sgEif2ak3-Gfap-Cas9–2A-GFP | This paper | N/A |
| pGL2-NOS2Promoter-Luciferase | Lowenstein et al., 1993 | Addgene Cat#19296 |
| pcDNA3-RelA-cFlag | Sanjabi et al., 2005 | Addgene Cat#20012 |
| pFLAG.XBP1.p.CMV2 | Calfon et al., 2002 | Addgene Cat#21833 |
| FLAG-HA-pcDNA3.1 | Horn et al., 2014 | Addgene Cat#52535 |
| pRL-CMV-Renilla | Promega | Cat#E2261 |
| Software and Algorithms | ||
| Prism 7 | GraphPad | N/A |
| FIJI | RRID:SCR_002285 | https://imagej.net/Welcome |
| Mulan | NCBI | https://mulan.dcode.org/ |
| Ensembl | RRID:SCR_008356; EBI/WTSI | https://useast.ensembl.org/index.html |
| Primer3 | RRID:SCR_003139 | http://bioinfo.ut.ee/primer3-0.4.0/ |
| R v3.3.3 | The R Foundation | https://www.r-project.org/ |
| Limma (v3.34.1) | Ritchie et al., 2015; RRID:SCR_010943 | https://www.bioconductor.org/install/ |
| Fastx (v0.0.13) | Gregory Hannon lab | http://hannonlab.cshl.edu/fastx_toolkit/ |
| Bowtie (v2.3.0) | Langmead and Salzberg, 2012; RRID:SCR_016368 | http://bowtie-bio.sourceforge.net/index.shtml |
| SAMtools (v1.3) | RRID:SCR_002105 | http://samtools.sourceforge.net/ |
| Bedtools (v2.26.0) | Quinlan and Hall, 2010; RRID:SCR_006646 | https://bedtools.readthedocs.io/en/latest/index.html |
| BWA mem (v.0.7.8) | Li and Durbin, 2009 | http://bio-bwa.sourceforge.net/ |
| MACS2 (v2.1.1) | Zhang et al., 2008 | https://github.com/taoliu/MACS |
| DiffBind (v. 2.6.1) | Stark, 2011; RRID:SCR_012918 | https://www.bioconductor.org/packages/devel/bioc/html/DiffBind.html |
| Sushi R package (v1.16.0) | Phanstiel, 2015 | https://bioconductor.org/packages/release/bioc/html/Sushi.html |
| EPA Toxcast database | Dix et al., 2007 | https://actor.epa.gov/dashboard/ |
| NetworkAnalyst | Xia et al., 2015 | https://www.networkanalyst.ca/ |
| Other | ||
Highlights.
A multidisciplinary approach to study effects of environmental factors on astrocytes.
Linuron boosts IRE1α-XBP1 signaling in astrocytes via Sigmar1
Astrocytic IRE1α-XBP1 signaling promotes CNS inflammation in EAE
IRE1α-XBP1 signaling is activated in astrocytes in multiple sclerosis
ACKNOWLEDGMENTS
This work was supported by grants NS102807, NS087867, ES02530, AI126880 and AI093903 from the National Institutes of Health, RSG-14–198-01-LIB from the American Cancer Society and RG4111A1 and JF2161-A-5 from the National Multiple Sclerosis Society to FJQ. FJQ and JPA received support from the International Progressive MS Alliance. MAW was supported by a training grant from the NIH and Dana-Farber Cancer Institute (T32CA207201), a post-doctoral fellowship from the NIH (F32NS101790), and a Traveling Neuroscience Fellowship from the Program in Neuroscience at Brigham and Women’s Hospital. RC was supported by a postdoctoral fellowship from the Swedish Research Council. Sequencing was carried out at the DNA Resource Core of Dana-Farber/Harvard Cancer Center (funded in part by NCI Cancer Center support grant 2P30CA006516–48). We thank all members of the Quintana laboratory for helpful advice and discussions, S. Adoro for advice regarding XBP1 and C. Lentucci for technical assistance. We thank D. Kozoriz and A. Chicoine for technical assistance with flow cytometry studies. We thank E. Buys, T. Pedulla and A. Cintolo for assistance with zebrafish husbandry. We thank Laurie Glimcher (Dana Farber Cancer Institute, Boston, USA) for advice and tools for the investigation of the UPR, and Kevin Crofton and Richard Judson (National Center for Computational Toxicology, US Environmental Protection Agency, Research Triangle Park, NC, USA) for the bioinformatic analysis of the ToxCast database.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing financial interests. The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ajami B, Bennett JL, Krieger C, McNagny KM, and Rossi FM (2011). Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat Neurosci 14, 1142–1149. [DOI] [PubMed] [Google Scholar]
- Aktas O, Smorodchenko A, Brocke S, Infante-Duarte C, Schulze Topphoff U, Vogt J, Prozorovski T, Meier S, Osmanova V, Pohl E, et al. (2005). Neuronal damage in autoimmune neuroinflammation mediated by the death ligand TRAIL. Neuron 46, 421–432. [DOI] [PubMed] [Google Scholar]
- Al-Chalabi A, and Hardiman O (2013). The epidemiology of ALS: a conspiracy of genes, environment and time. Nature reviews Neurology 9, 617–628. [DOI] [PubMed] [Google Scholar]
- Al-Saif A, Al-Mohanna F, and Bohlega S (2011). A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann Neurol 70, 913–919. [DOI] [PubMed] [Google Scholar]
- Alvarez JI, Dodelet-Devillers A, Kebir H, Ifergan I, Fabre PJ, Terouz S, Sabbagh M, Wosik K, Bourbonniere L, Bernard M, et al. (2011). The Hedgehog pathway promotes blood-brain barrier integrity and CNS immune quiescence. Science 334, 1727–1731. [DOI] [PubMed] [Google Scholar]
- Alvarez JI, Saint-Laurent O, Godschalk A, Terouz S, Briels C, Larouche S, Bourbonniere L, Larochelle C, and Prat A (2015). Focal disturbances in the blood-brain barrier are associated with formation of neuroinflammatory lesions. Neurobiol Dis 74, 14–24. [DOI] [PubMed] [Google Scholar]
- Anderson MA, Burda JE, Ren Y, Ao Y, O’Shea TM, Kawaguchi R, Coppola G, Khakh BS, Deming TJ, and Sofroniew MV (2016). Astrocyte scar formation aids central nervous system axon regeneration. Nature 532, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Authority EFS (2016). Peer review of the pesticide risk assessment of the active substance linuron. EFSA Journal 14. [Google Scholar]
- Baecher-Allan C, Kaskow BJ, and Weiner HL (2018). Multiple Sclerosis: Mechanisms and Immunotherapy. Neuron 97, 742–768. [DOI] [PubMed] [Google Scholar]
- Becher B, Tugues S, and Greter M (2016). GM-CSF: From Growth Factor to Central Mediator of Tissue Inflammation. Immunity 45, 963–973. [DOI] [PubMed] [Google Scholar]
- Ben Haim L, and Rowitch DH (2017). Functional diversity of astrocytes in neural circuit regulation. Nat Rev Neurosci 18, 31–41. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, and Raymond PA (2006). GFAP transgenic zebrafish. Gene Expr Patterns 6, 1007–1013. [DOI] [PubMed] [Google Scholar]
- Bettigole SE, and Glimcher LH (2015). Endoplasmic reticulum stress in immunity. Annu Rev Immunol 33, 107–138. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese M, Magliozzi R, Ciccarelli O, Geurts JJ, Reynolds R, and Martin R (2015). Exploring the origins of grey matter damage in multiple sclerosis. Nat Rev Neurosci 16, 147–158. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, and Ron D (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96. [DOI] [PubMed] [Google Scholar]
- Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, et al. (2015). Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin-Chan M, Navarro-Yepes J, and Quintanilla-Vega B (2015). Environmental pollutants as risk factors for neurodegenerative disorders: Alzheimer and Parkinson diseases. Frontiers in cellular neuroscience 9, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M, and Butovsky O (2017). Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu Rev Immunol 35, 441–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendrou CA, Fugger L, and Friese MA (2015). Immunopathology of multiple sclerosis. Nat Rev Immunol 15, 545–558. [DOI] [PubMed] [Google Scholar]
- Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, and Kavlock RJ (2007). The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol Sci 95, 5–12. [DOI] [PubMed] [Google Scholar]
- Ellwardt E, Pramanik G, Luchtman D, Novkovic T, Jubal ER, Vogt J, Arnoux I, Vogelaar CF, Mandal S, Schmalz M, et al. (2018). Maladaptive cortical hyperactivity upon recovery from experimental autoimmune encephalomyelitis. Nature Neuroscience 21, 1392–1403. [DOI] [PubMed] [Google Scholar]
- Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, et al. (2015). Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 18, 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farez MF, Mascanfroni ID, Mendez-Huergo SP, Yeste A, Murugaiyan G, Garo LP, Balbuena Aguirre ME, Patel B, Ysrraelit MC, Zhu C, et al. (2015). Melatonin Contributes to the Seasonality of Multiple Sclerosis Relapses. Cell 162, 1338–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanilla D, Johannessen M, Hajipour AR, Cozzi NV, Jackson MB, and Ruoho AE (2009). The hallucinogen N,N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator. Science (New York, NY) 323, 934–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo LC (2013). Purification of rat and mouse astrocytes by immunopanning. Cold Spring Harb Protoc 2013, 421–432. [DOI] [PubMed] [Google Scholar]
- Glass CK, Saijo K, Winner B, Marchetto MC, and Gage FH (2010). Mechanisms underlying inflammation in neurodegeneration. Cell 140, 918–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Vázquez C, and Quintana FJ (2018). Regulation of the Immune Response by the Aryl Hydrocarbon Receptor. Immunity 48, 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley RW (2003). Excess incidence of ALS in young Gulf War veterans. Neurology 61, 750–756. [DOI] [PubMed] [Google Scholar]
- Hayashi T, and Su TP (2007). Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell 131, 596–610. [DOI] [PubMed] [Google Scholar]
- Hetz C, and Saxena S (2017). ER stress and the unfolded protein response in neurodegeneration. Nat Rev Neurol 13, 477–491. [DOI] [PubMed] [Google Scholar]
- Hewett SJ, Csernansky CA, and Choi DW (1994). Selective potentiation of NMDA-induced neuronal injury following induction of astrocytic iNOS. Neuron 13, 487–494. [DOI] [PubMed] [Google Scholar]
- Horn M, Geisen C, Cermak L, Becker B, Nakamura S, Klein C, Pagano M, and Antebi A (2014). DRE-1/FBXO11-dependent degradation of BLMP-1/BLIMP-1 governs C. elegans developmental timing and maturation. Dev Cell 28, 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner RD, Kamins KG, Feussner JR, Grambow SC, Hoff-Lindquist J, Harati Y, Mitsumoto H, Pascuzzi R, Spencer PS, Tim R, et al. (2003). Occurrence of amyotrophic lateral sclerosis among Gulf War veterans. Neurology 61, 742–749. [DOI] [PubMed] [Google Scholar]
- Hui SP, Dutta A, and Ghosh S (2010). Cellular response after crush injury in adult zebrafish spinal cord. Dev Dyn 239, 2962–2979. [DOI] [PubMed] [Google Scholar]
- Huitinga I, van Rooijen N, de Groot CJ, Uitdehaag BM, and Dijkstra CD (1990). Suppression of experimental allergic encephalomyelitis in Lewis rats after elimination of macrophages. The Journal of experimental medicine 172, 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CS, Arbour N, Manusow J, Montgrain V, Blain M, McCrea E, Shapiro A, and Antel JP (2005). TLR Signaling Tailors Innate Immune Responses in Human Microglia and Astrocytes. The Journal of Immunology 175, 4320–4330. [DOI] [PubMed] [Google Scholar]
- Jain IH, Zazzeron L, Goli R, Alexa K, Schatzman-Bone S, Dhillon H, Goldberger O, Peng J, Shalem O, Sanjana NE, et al. (2016). Hypoxia as a therapy for mitochondrial disease. Science 352, 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel F (2013). Epidemiology. Paths from pesticides to Parkinson’s. Science (New York, NY) 341, 722–723. [DOI] [PubMed] [Google Scholar]
- Kanemoto S, Kondo S, Ogata M, Murakami T, Urano F, and Imaizumi K (2005). XBP1 activates the transcription of its target genes via an ACGT core sequence under ER stress. Biochem Biophys Res Commun 331, 1146–1153. [DOI] [PubMed] [Google Scholar]
- Keestra-Gounder AM, Byndloss MX, Seyffert N, Young BM, Chavez-Arroyo A, Tsai AY, Cevallos SA, Winter MG, Pham OH, Tiffany CR, et al. (2016). NOD1 and NOD2 signalling links ER stress with inflammation. Nature 532, 394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, and Sofroniew MV (2015). Diversity of astrocyte functions and phenotypes in neural circuits. Nature neuroscience 18, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann T, Ludwin S, Prat A, Antel J, Bruck W, and Lassmann H (2017). An updated histological classification system for multiple sclerosis lesions. Acta Neuropathol 133, 13–24. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, and Glimcher LH (2003). XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23, 7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Messing A, Su M, and Brenner M (2008). GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia 56, 481–493. [DOI] [PubMed] [Google Scholar]
- Levine RL, Pardanani A, Tefferi A, and Gilliland DG (2007). Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat Rev Cancer 7, 673–683. [DOI] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Lahvic JL, Binder V, Pugach EK, Riley EB, Tamplin OJ, Panigrahy D, Bowman TV, Barrett FG, Heffner GC, et al. (2015). Epoxyeicosatrienoic acids enhance embryonic haematopoiesis and adult marrow engraftment. Nature 523, 468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, and Barres BA (2017). Reactive Astrocytes: Production, Function, and Therapeutic Potential. Immunity 46, 957–967. [DOI] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Munch AE, Chung WS, Peterson TC, et al. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, and Edelson BT (2017). New Insights into the Role of IL-1beta in Experimental Autoimmune Encephalomyelitis and Multiple Sclerosis. J Immunol 198, 4553–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Bailey SL, Ho H, Harding HP, Ron D, Miller SD, and Popko B (2007). The integrated stress response prevents demyelination by protecting oligodendrocytes against immune-mediated damage. J Clin Invest 117, 448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson KL, Ghosh R, and Urano F (2008). The role of IRE1alpha in the degradation of insulin mRNA in pancreatic beta-cells. PLoS One 3, e1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Belkadi A, Darnall L, Hu T, Drescher C, Cotleur AC, Padovani-Claudio D, He T, Choi K, Lane TE, et al. (2010). CXCR2-positive neutrophils are essential for cuprizoneinduced demyelination: relevance to multiple sclerosis. Nat Neurosci 13, 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli G, Theodorou D, Kendirli A, Jordão MJC, Staszewski O, Phulphagar K, Cantuti-Castelvetri L, Dagkalis A, Bessis A, Simons M, et al. (2018). Mononuclear phagocytes locally specify and adapt their phenotype in a multiple sclerosis model. Nature Neuroscience. [DOI] [PubMed] [Google Scholar]
- Lock C, Hermans G, Pedotti R, Brendolan A, Schadt E, Garren H, Langer-Gould A, Strober S, Cannella B, Allard J, et al. (2002). Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat Med 8, 500–508. [DOI] [PubMed] [Google Scholar]
- Lowenstein CJ, Alley EW, Raval P, Snowman AM, Snyder SH, Russell SW, and Murphy WJ (1993). Macrophage nitric oxide synthase gene: two upstream regions mediate induction by interferon gamma and lipopolysaccharide. Proceedings of the National Academy of Sciences 90, 9730–9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DA, and Talbot WS (2014). Glial cell development and function in zebrafish. Cold Spring Harb Perspect Biol 7, a020586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli M, and Driever W (2012). Fluorescence-activated cell sorting (FACS) of fluorescently tagged cells from zebrafish larvae for RNA isolation. Cold Spring Harb Protoc 2012. [DOI] [PubMed] [Google Scholar]
- Matsushima GK, and Morell P (2006). The Neurotoxicant, Cuprizone, as a Model to Study Demyelination and Remyelination in the Central Nervous System. Brain Pathology 11, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo L, Trauger SA, Blain M, Nadeau M, Patel B, Alvarez JI, Mascanfroni ID, Yeste A, Kivisakk P, Kallas K, et al. (2014). Regulation of astrocyte activation by glycolipids drives chronic CNS inflammation. Nature medicine 20, 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Hayashi T, Hayashi E, and Su TP (2013). Sigma-1 receptor chaperone at the ER-mitochondrion interface mediates the mitochondrion-ER-nucleus signaling for cellular survival. PLoS One 8, e76941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson T, Barcellos LF, and Alfredsson L (2017). Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nature reviews Neurology 13, 25–36. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Giardine BM, Hou M, Ma J, Hardison RC, Stubbs L, and Miller W (2005). Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res 15, 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel DH (2015). Sushi: Tools for visualizing genomics data. In R package version 1160.
- Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, Fujihara K, Havrdova E, Hutchinson M, Kappos L, et al. (2011). Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 69, 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich D, Yaniv SP, Alyagor I, and Schuldiner O (2016). Nitric Oxide as a Switching Mechanism between Axon Degeneration and Regrowth during Developmental Remodeling. Cell 164, 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, Feng T, Lee M, Asinovski N, Frohlich I, et al. (2014). Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM, and Smith MT (2010). Epidemiology. Environment and disease risks. Science 330, 460–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauh VA (2018). Polluting Developing Brains - EPA Failure on Chlorpyrifos. N Engl J Med 378, 1171–1174. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V, Borucki DM, Tjon EC, Takenaka MC, Chao C-C, Ardura-Fabregat A, de Lima KA, Gutiérrez-Vázquez C, Hewson P, Staszewski O, et al. (2018). Microglial control of astrocytes in response to microbial metabolites. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, Chao CC, Patel B, Yan R, Blain M, et al. (2016). Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nature medicine 22, 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V, and Quintana FJ (2015). Control of autoimmune CNS inflammation by astrocytes. Semin Immunopathol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, et al. (2016). Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 167, 1469–1480 e1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi S, Williams KJ, Saccani S, Zhou L, Hoffmann A, Ghosh G, Gerondakis S, Natoli G, and Smale ST (2005). A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev 19, 2138–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HR, Zheng S, Gurpinar E, Koehl A, Manglik A, and Kruse AC (2016). Crystal structure of the human sigma1 receptor. Nature 532, 527–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, et al. (1992). Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature 359, 693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, Kunkle BW, Boland A, Raybould R, Bis JC, et al. (2017). Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nature genetics 49, 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, and Knudsen TB (2013). Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol 26, 878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorbara CD, Wagner NE, Ladwig A, Nikic I, Merkler D, Kleele T, Marinkovic P, Naumann R, Godinho L, Bareyre FM, et al. (2014). Pervasive axonal transport deficits in multiple sclerosis models. Neuron 84, 1183–1190. [DOI] [PubMed] [Google Scholar]
- Soumillon M, Cacchiarelli D, Semrau S, van Oudenaarden A, and Mikkelsen TS (2014). Characterization of directed differentiation by high-throughput single-cell RNA-Seq. bioRxiv. [Google Scholar]
- Stark R, and Brown GD (2011). DiffBind: differential binding analysis of ChIP-Seq peak data (Bioconductor).
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thion MS, Low D, Silvin A, Chen J, Grisel P, Schulte-Schrepping J, Blecher R, Ulas T, Squarzoni P, Hoeffel G, et al. (2018). Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell 172, 500–516 e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Baranzini SE, Geurts J, Hemmer B, and Ciccarelli O (2018). Multiple sclerosis. The Lancet 391, 1622–1636. [DOI] [PubMed] [Google Scholar]
- Tsai SY, Pokrass MJ, Klauer NR, De Credico NE, and Su TP (2014). Sigma-1 receptor chaperones in neurodegenerative and psychiatric disorders. Expert Opin Ther Targets 18, 1461–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tshala-Katumbay D, Mwanza JC, Rohlman DS, Maestre G, and Oria RB (2015). A global perspective on the influence of environmental exposures on the nervous system. Nature 527, S187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, and Rozen SG (2012). Primer3--new capabilities and interfaces. Nucleic Acids Res 40, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Torres A, Savarin C, Hinton DR, Phares TW, Bergmann CC, and Stohlman SA (2016). Sustained TNF production by central nervous system infiltrating macrophages promotes progressive autoimmune encephalomyelitis. J Neuroinflammation 13, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Galen P, Viny AD, Ram O, Ryan RJ, Cotton MJ, Donohue L, Sievers C, Drier Y, Liau BB, Gillespie SM, et al. (2016). A Multiplexed System for Quantitative Comparisons of Chromatin Landscapes. Mol Cell 61, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, and Sabatini DM (2017). Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell 168, 890–903 e815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Ilieva H, Tamada H, Nomura H, Komine O, Endo F, Jin S, Mancias P, Kiyama H, and Yamanaka K (2016). Mitochondria-associated membrane collapse is a common pathomechanism in SIGMAR1- and SOD1-linked ALS. EMBO Mol Med 8, 1421–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MA, and Quintana FJ (2018). Regulation of Astrocyte Functions in Multiple Sclerosis Cold Spring Harb Perspect Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MA, Rothhammer V, and Quintana FJ (2017). Control of immune-mediated pathology via the aryl hydrocarbon receptor. J Biol Chem 292, 12383–12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CP (2005). Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev 14, 1847–1850. [DOI] [PubMed] [Google Scholar]
- Xia J, Gill EE, and Hancock RE (2015). NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 10, 823–844. [DOI] [PubMed] [Google Scholar]
- Yan Y, Ding X, Li K, Ciric B, Wu S, Xu H, Gran B, Rostami A, and Zhang GX (2012). CNS-specific therapy for ongoing EAE by silencing IL-17 pathway in astrocytes. Mol Ther 20, 1338–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, et al. (2016). Ensembl 2016. Nucleic Acids Res 44, D710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, and Mori K (2001). XBP1 mRNA Is Induced by ATF6 and Spliced by IRE1 in Response to ER Stress to Produce a Highly Active Transcription Factor. Cell 107, 881–891. [DOI] [PubMed] [Google Scholar]
- Zamvil SS, and Steinman L (2003). Diverse Targets for Intervention during Inflammatory and Neurodegenerative Phases of Multiple Sclerosis. Neuron 38, 685–688. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. XBP1 activation by stressors different from Linuron, Related to Figure 1 and Figure 2. A) nos2a expression in vehicle or Linuron treated zebrafish. n=4 replicates per condition with at least 10 fish per group. Unpaired two-tailed t-test. B) Xbp1 splicing in human fetal astrocytes exposed to different stressors. n=4 per condition. One-way ANOVA, Dunnett post-test. C) Xbp1 splicing in mouse neonatal astrocytes treated with multiple stressors for 4 hours. n=3–6 per condition. One-way ANOVA, Tukey post-test. D) Xbp1 splicing in mouse neonatal astrocytes treated with PFNA or Methyl Carbamate. n=3 per condition. Two-way ANOVA, Sidak post-test. E) Xbp1 splicing in neonatal mouse microglia in culture. n=4 per condition. One-way ANOVA, Dunnett post-test. F) Effect of IRE1α/XBP1 signaling inhibition in sorted adult murine TdTomato+ astrocytes. n=3 per condition. One-way ANOVA, Tukey post-test. ns=not significant. ***p<0.001, **p<0.01, *p<0.05. Data shown as mean ± SEM.
Table S4. Effects of Xbp1 knock down on the transcriptional response of astrocytes in EAE, Related to Figure 4 and Figure 5. Genes detected as differentially modulated by RNA-seq with p<0.05 are listed. logFC measured as ratio of expression in shXbp1 condition compared to shScrmbl condition.
Table S5. Effects of Xbp1 knock down in astrocytes on the transcriptional response of microglia and CNS-recruited monocytes in EAE, Related to Figure 4 and Figure 5. Genes detected as differentially modulated by RNA-seq with p<0.05 are listed. logFC measured as ratio of expression in shXbp1 condition compared to shScrmbl condition.
Table S6. Effects of Xbp1 knockdown in astrocytes on chromatin accessibility, Related to Figure 4. Ingenuity pathway analysis of XBP1 targets identified by ATAC-seq. Pathways with p<0.05 are listed.
Table S7. List of oligonucleotides used. Related to Key Resources Table.
Figure S2. XBP1 ChIP-seq pathway analysis, Related to Figure 4. A) Table of statistically significant XBP1-driven pathways in astrocytes from EAE mice compared to naïve mice. Table generated by Ingenuity Pathway Analysis. B) XBP1 ChIP-seq density plots for Edem1, Dnajb9, and Csf2 genomic loci in astrocytes isolated from naïve and EAE mice, normalized to input DNA. n=3 EAE, n=2 naïve. Scale bars show read density. Schematics of transcriptional regulation shown above density plots.
Figure S3. Evaluation of shRNA-based knockdown, Related to Figure 4. A) XBP1 expression in glial cells determined by western blot. n=4 for shXbp1, n=2 for WT. One-sample t-test. B) IRE1α expression in glial cells determined by Western blot. n=3 per condition. One-sample t-test. C) Xbp1 expression in astrocytes, microglia, or monocytes. n=5 replicates per condition. Unpaired two-tailed t-test. D-E) FACS analysis of astrocytes, microglia, and T cells isolated from mice undergoing EAE transduced with Gfap-driven shXbp1 or non-targeting lentiviruses. T cells isolated from the CNS shown in (D) or the spleen shown in (E). n=10 per condition for astrocytes and microglia, n=3 per condition for T cells. Unpaired two-tailed t-test. **p<0.01, *p<0.05. ns=not significant. Data shown as mean ± SEM.
Figure S4. UPR perturbation during EAE, Related to Figure 4. A) EAE clinical scores of mice in which Xbp1 and Ern1 were inactivated using CRISPR/Cas9. n=14 sgScrmbl, n=14 sgErn1, and n=12 sgXbp1. Two-way repeated measures ANOVA, Holm-Sidak post-test. B) Quantification of XBP1+ astrocytes in EAE mice following CRISPR/Cas9-based Xbp1 inactivation. n=8–9 sections from N=3 brains per genotype. One-way ANOVA, Tukey post-test. C) sgErn1 knockdown efficiency in astrocytes. n=4–5 sections from N=3 mice. Unpaired two-tailed t-test. D) EAE development in control or sgEif2ak3 (sgPerk)-treated mice. n=4–5 mice per group. Two-way repeated-measures ANOVA, Holm-Sidak post-test. E) Quantification of sgEif2ak3 knockdown efficiency. n=4–6 images from N=3 mice per group. Unpaired two-tailed t-test. ***p<0.001, **p<0.01, *p<0.05, ns=not significant. Data shown as mean ± SEM.
Figure S5. Effects of Sigmar1 inactivation and Linuron on EAE, Related to Figure 4. A) Quantification of sgSigmar1 knockdown validation in astrocytes. n=8–9 images from N=3 mice per condition. Unpaired two-tailed t-test. B) T-cell subsets, astrocytes and microglia in mice treated with sgScrmbl or sgXbp1 Gfap-driven lentiviruses and Linuron or vehicle. n=3 per group (T cells) and n=4–5 per group (otherwise). One-way ANOVA, Tukey post-test. C) T cells, astrocytes and microglia in mice treated with sgScrmbl or sgSigmar1 Gfap-driven lentiviruses and Linuron. n=3 per group (T cells) and n=5 per group (otherwise). Unpaired two-tailed t-test. *p<0.05, ns=not significant. Data shown as mean ± SEM.
Figure S6. Cell-type specific analysis of IRE1α-XBP1s activation in MS, Related to Figure 6. A) IRE1α activation in neurons, oligodendrocytes, and CD14+ cells in MS samples. Images total n=17 for astrocytes, n=9 for neurons, n=8 for oligodendrocytes, and n=3 for CD14+ cells, from N=4 (astrocytes) or N=2 (neurons, oligodendrocytes, and CD14+ cells) MS patients. Cells analyzed per cell type are: n=85 NeuN+, n=109 CNPase+, n=29 CD14+, n=118 GFAP+. ND=not detected. Fisher’s exact test between positive and negative cells compared to astrocytes. B) Analysis of XBP1s expression. n=20–38 sections per cell type. N=4 MS patients per cell type analyzed. Cells analyzed per cell type are: n=60 MAP2+, n=70 NOGOA+, n=59 CD14+, n=212 GFAP+. Fisher’s exact test between positive and negative cells compared to astrocytes. C) Control immunostaining analysis validating antibody target specificity. ***p<0.001. Data shown as mean ± SEM.
Table S1. Candidate chemicals tested in zebrafish screen, Related to Figure 1. Chemicals analyzed in Fig. 1F.
Table S2. Molecular targets of Linuron, Related to Figure 2. Ingenuity pathway analysis of Linuron molecular targets. Pathways with p<0.05 are listed.
Table S3. XBP1 targets in astrocytes during EAE, Related to Figure 4. Ingenuity pathway analysis of XBP1 targets identified by ChIP-seq. Pathways with p<0.05 are listed.
Data Availability Statement
A SuperSeries page has been created in the GEO database with the accession number GSE122119. All ATAC-seq, ChIP-seq, and RNA-seq data have been deposited in the GEO database under this SuperSeries with the accession numbers GSE121923, GSE121935, and GSE122113, respectively.