Molecular techniques in fungal detection and identification represent an efficient complementary diagnostic tool which is increasingly used to overcome limitations of routinely used culture techniques. The aim of this study was to characterize Candida sp.
KEYWORDS: Candida, biofilms, capillary electrophoresis, fungi, panfungal PCR, polyfungal sample, ureteral stents, urinary catheters
ABSTRACT
Molecular techniques in fungal detection and identification represent an efficient complementary diagnostic tool which is increasingly used to overcome limitations of routinely used culture techniques. The aim of this study was to characterize Candida sp. representation in samples from urine, urinary catheter, and ureteral stent biofilm using ITS2 ribosomal DNA (rDNA) amplification followed by fluorescent capillary electrophoresis (f-ITS2-PCR-CE) and to compare the results with those obtained by culture. A total of 419 samples were analyzed, and 106 (25.2%) were found positive, out of which 17 (16%) were polyfungal. The positivity rate did not differ between samples from catheters and stents (23.6% versus 20.9%) or between catheter and stent corresponding urine samples (40.2% versus 30.2%). Ten different Candida species were detected, with Candida parapsilosis (31.4%), Candida albicans (26.5%), and Candida tropicalis (12.4%) predominating. f-ITS2-PCR-CE was evaluated as substantially less time-consuming and 8.3 times more sensitive than the routinely applied culture technique with 1 µl of urine/sonicated fluid inoculated, detecting 67 (19.9%) versus 8 (2.4%) positive samples out of 337 initially analyzed samples. The culture sensitivity considerably improved to 1.7 times lower than that of f-ITS2-PCR-CE after the inoculation volume was increased to 100 µl in the additional 82 samples. Moreover, the molecular technique, unlike routine cultivation, enabled precise pathogen composition determination in polymicrobial samples. In conclusion, the f-ITS2-PCR-CE method was shown to be a quick and efficient tool for culture-independent detection and identification of fungi in urinary tract-related samples, demonstrating a higher sensitivity than culture.
INTRODUCTION
Ureteral stents and urinary catheters are valuable tools in urological practice although they offering, like all synthetic medical intracavity devices, an ideal surface for microbial colonization (1). They can be colonized by a wide range of microorganisms, including both bacteria and fungi, usually forming a community or multispecies biofilm. Colonization usually does not affect the patient's clinical status in any significant way. On the other hand, it can be an important source of infection, and biofilms may be critical for clinical infection development (2). Colonization has gained importance mainly due to an increasing number of immunocompromised patients who are particularly susceptible to infections. Candida spp. are the most frequently detected fungal agents in urinary tract samples. However, in many instances, a clinical laboratory report indicates that candiduria represents colonization or procurement specimen contamination rather than an invasive infection (3).
For many years, Candida albicans has been a predominant fungal species isolated from the urinary tract. With the increasing access and use of fluconazole, non-albicans Candida (NAC) species have emerged, and they can even dominate C. albicans in some patients (3). However, the most frequently occurring fungus is still C. albicans, followed by Candida glabrata, Candida tropicalis, and Candida krusei (4).
Traditionally, diagnosing Candida urinary tract infections has relied on clinical signs and symptoms and on microbiological urine culture, which is still the recommended method of choice (5). Species identification is based mostly on phenotypic features and is usually time-consuming as classical diagnostic work flow takes up to several days (6). Cultivation-independent approaches based on DNA isolation are superior to cultures in rapidity and in identifying species which cannot be easily grown and/or distinguished by a routine phenotypic approach. In order to characterize polyfungal samples, several molecular techniques, employed mainly for environmental material testing in the field of molecular ecology, can be applied. These community profiling techniques include, for instance, denaturing/temperature gradient gel electrophoresis (DGGE/TGGE), single-strand conformation polymorphism (SSCP), terminal restriction fragment length polymorphism (T-RFLP), cloning (7), or fluorescent capillary electrophoresis (CE) (8). The last method, based on utilizing the variability of the ITS2 ribosomal DNA (rDNA), has been proven to be superior to classical electrophoresis for analyzing DNA due to its higher separation efficiency, speed, and higher sensitivity and the fact that there is no need for sequencing (8, 9).
Therefore, the aim of this study was to characterize Candida occurrence and species prevalence in the urinary catheter and ureteral stent biofilm and in urine samples using fluorescent ITS2 rDNA amplicon detection by capillary electrophoresis (f-ITS2-PCR-CE) and to validate this approach using a comprehensive set of clinical samples.
MATERIALS AND METHODS
Determining reference polymicrobial samples.
A combination of different concentrations (as the number of CFU/milliliter) of seven clinical Candida species, C. albicans, C. glabrata, C. tropicalis, Candida parapsilosis, C. krusei, Candida kefyr, and Candida lusitaniae, were used to determine the ability of capillary electrophoresis to evaluate more than one fungal strain in a sample. Therefore, two fungal species were mixed in equal ratios or in 1:10, 1:100, and 1:1,000 ratios in triplicates; DNA was extracted as described below, and samples were evaluated by f-ITS2-PCR-CE. Accurate cell concentration was determined by assessing the CFU count on Sabouraud agar plates (Himedia, India) supplemented with vancomycin (5 mg) and amikacine (20 mg/liter) after 48 h of cultivation at 37°C.
Sample collection and clinical material preparation.
Urinary catheters (C; n = 127), ureteral stents (S; n = 67), and corresponding urine samples (U-C, n = 101; U-S; n = 64), were collected from 161 patients (125 males and 36 females; median ages, 76.0 and 65.5 years, respectively) hospitalized in the Department of Urology, St. Anne's University Hospital, Brno, Czech Republic, during the years 2012 to 2014, irrespective of patient diagnosis or underlying disease, as performed previously by others (10), except for the consecutive sampling approach in our study (n = 196). Thus, both infectious and noninfectious samples were included. In total, 419 samples were analyzed (Table 1). Catheter removal was based on a urologist’s decision. The study was approved by the Ethics Committee of the St. Anne’s University Hospital in Brno; no informed consent was required because neither human cells nor human tissues were processed, and no procedure in addition to standard care was performed.
TABLE 1.
Detection of fungi using f-ITS2-PCR-CEa
| Materialb | No. (%) of samples |
||||
|---|---|---|---|---|---|
| Total analyzed | Negative | Positive | Monofungalc | Polyfungalc | |
| C | 127 | 97 (76.4) | 30 (23.6) | 26 (86.7) | 4 (13.3) |
| U-C | 101 | 60 (59.4) | 41 (40.6) | 30 (73.2) | 11 (26.8) |
| Sd | 127 | 111 (87.4) | 16 (12.6) | 16 (100) | 0 (0) |
| U-S | 64 | 45 (70.3) | 19 (29.7) | 17 (89.5) | 2 (10.5) |
| Total | 419 | 313 (74.8) | 106 (25.2) | 89 (84) | 17 (16) |
The table shows an overview of molecular detection in clinical samples. C. robusta detected in sonication fluids (material C and S) was evaluated as a contaminant and was excluded from the results. These results also include three non-Candida fungal species, identified by sequencing as Geotrichum candidum, Rhodotorula rubra, and Cryptococcus neoformans, in 2 U-C, 1 U-C, and 1 polyfungal U-C sample, respectively. Candida was detected in 8 samples (3 C. parapsilosis, 3 C. albicans, 1 C. tropicalis, and 1 C. famata isolate) out of 29 and 2 (C. albicans) out of 3 catheter and stent samples without a corresponding urine sample, respectively. In 3 U-C cases without a corresponding catheter sample, no Candida was detected.
C, urinary catheter; U-C, urine from patients with catheter; S, ureteral stent; U-S, urine from patients with stent.
Percentage of positive samples.
Data for the stent category are based on the results of 127 unique samples. In the disjunction of results of proximal and distal tip samples, there were 14 (20.9%) Candida sp. positive stents; no stent was polyfungal.
Stents and catheters were aseptically removed from the body; 5-cm-long tips (both the proximal and distal parts of the stents and the distal part of the catheters) were snipped off and placed into sterile tubes with 5 ml of brain heart infusion (BHI) medium for sonication. The sonication procedure was based on a previously described protocol (11) and consisted of two 5-min sonications interspaced by 2 min of vortexing. In parallel, urine samples obtained through the catheter or stent before their removal were also treated aseptically. Urine collection was impossible in 29 and 3 sampling cases of catheters and stents, respectively, because of patients’ oligo-anuria at the time of collection. In 3 U-C cases, urine without a catheter was collected. Sonicates and urine samples were subsequently cultured and, in parallel, used for direct DNA extraction.
Phenotypic identification.
The suspension (1 μl in 337 samples and 100 μl in an additional 82 samples) was inoculated to blood agar (with 7% of sterile sheep blood; Columbia Blood Agar Base, Oxoid, United Kingdom), UriSelect 4 (Bio-Rad, France), and Sabouraud agar (Himedia, India) supplemented with vancomycin (5 mg) and amikacine (20 mg/liter) and cultured for 48 h and with Sabouraud agar for 1 week at 37°C to isolate individual strains. All isolated strains were identified at the species/genus level using ChromAgar Candida (CHROMagar, France), conventional biochemical tests (Micro-LA-tests; Lachema, Czech Republic; API bioMérieux, France). Culture results were provided in a quantified manner as a number of CFU per urinary catheter or urinary stent and urine sample.
To verify ambiguous results, matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) analysis, not easily available for routine use at the time of sample collection, was performed. A MALDI Biotyper with FlexControl, version 3.4, software (Bruker Daltonik) was used according to the manufacturer’s instructions. The manufacturer-recommended cutoff scores were used for identification, with scores of ≥2.000 indicating identification to the species level, scores between 1.700 and 1.999 indicating identification to the genus level, and scores of <1.700 indicating no identification.
DNA extraction.
The genomic DNA from clinical specimens was extracted by a QIAamp DNA Blood Mini kit (Qiagen, Germany), according to the manufacturer's instructions with the following modification. Briefly, 2 ml of the urine sample and 1 ml of the catheter or stent sonicate were centrifuged at 23,000 rpm for 20 min and at 14,000 rpm for 10 min, respectively. The supernatant was removed, and the sediment was incubated with 50 μl of lysis buffer (500 mM EDTA, pH 8.0, 1 M Tris, pH 8.0, 43.2 μl/ml Triton X-100), 20 μl of lysozyme (180 mg/ml; Sigma-Aldrich, USA), and 20 μl of lyticase (5 U/μl; Sigma-Aldrich, USA) for 60 min at 37°C and treated according to the manufacturer’s instructions. DNA samples were stored at −20°C. Sterile DNA-free water (Qiagen, Germany) was used as a specimen to check for possible contamination during the extraction step.
PCR amplification by using a fluorescently labeled primer (f-ITS2-PCR).
The internal transcribed fungal genomic rDNA spacer region ITS2 was amplified using the previously described panfungal primers UNF1 (5′-GCATCGATGAAGAACGTAGC-3′) and UNF2 (5′-AACTATACGAATTCAAGTCGCC-3′). Due to the final amplicon detection with capillary electrophoresis, a UNF1 primer 5′ fluorescently labeled with a 6-carboxyfluorescein (6-FAM) dye was used. PCR was performed in a total volume of 20 µl of HotStarTaq Master Mix (Qiagen, Germany) with 0.5 μM of each primer and 2 mM final Mg2+ concentration, using cycling conditions as described previously (8). A negative control of both DNA extraction (see above) and amplification (sterile DNA-free-water as a template) was used in each PCR run.
Capillary electrophoresis (f-ITS2-PCR-CE).
FAM-labeled ITS2-PCR amplicons were separated and detected in terms of their size using capillary electrophoresis on an ABI Prism 3130 Avant Genetic Analyser (Life Technologies, USA). The instrument was adjusted according to the manufacturer's instructions.
One microliter of PCR amplicon was mixed with 0.25 µl of a GeneScan 500 6-carboxy-X-rhodamine (ROX) dye size standard (Life Technologies, USA) and 9.75 µl of highly deionized (HiDi) formamide (Life Technologies, USA). After 2 min of denaturation at 95°C and quick freezing at −80°C, the samples were injected into a 36-cm capillary column containing the POP-7 (Life Technologies, USA) high-performance polymer. Electrophoretic parameters were set at 16-s injection times, 1.2-kV injection voltages, 15-kV electrophoresis voltages, and 60°C oven temperature. Finally, PCR product lengths were analyzed using GeneMapper, version 4.1, software. A successful analysis was derived from the red peaks being assigned to the ROX-labeled size standard GeneScan 500 (Life Technologies, USA).
The fungi present in each sample were evaluated by comparing ITS2 lengths with our previously prepared internal laboratory database of the reference strain ITS2 lengths. Identification was considered successful if the analyzed sample length appeared within a 0.3-nucleotide (nt) interval of the respective reference strain’s length (8).
Data interpretation and statistical analysis.
For positivity rate evaluation and data set characteristics (Table 1), proximal and distal stent tip samples were handled as independent ones. For Candida sp. prevalence evaluation and statistics addressing clinical material, a disjunction of proximal and distal stent tip results was applied. A statistical analysis employing a two-tailed Fischer's exact test was performed to test categorical data. Nonparametric analysis of variance (ANOVA) was used to test mutually species prevalence in catheter- and stent-related urine samples and in catheters and stents separately.
ITS2 sequencing.
The representative ITS2 rDNA amplicons were separated on a 2% agarose gel, excised, purified using a QIAquick Gel Extraction kit (Qiagen, Germany) according to the manufacturer's instructions, and sequenced as described previously (12).
RESULTS
Evaluation of artificial polyfungal samples.
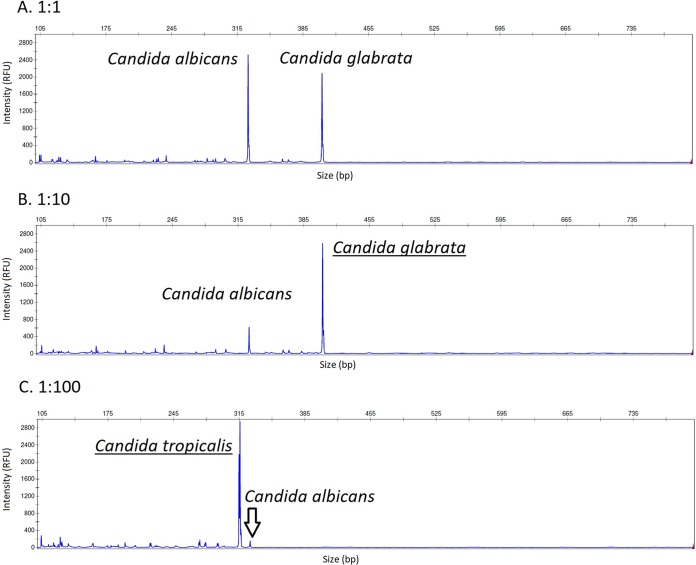
At first, we examined cultivation-independent f-ITS2-PCR-CE performance in detecting individual Candida sp. in cell suspensions containing various mixed populations of two different species. Each sample was assessed three times. The results showed that both fungal species were detected in all samples if present in equal or 10-fold different concentrations, while only the major species was identified if the concentration difference was 100-fold or higher (Fig. 1).
FIG 1.
f-ITS2-PCR-CE analysis of mixed Candida sp. populations with different CFU ratios at the beginning of DNA extraction. It was possible to differentiate two Candida spp. if present in equal concentrations (A) or 10-fold-different concentrations (B). Only the major species was reliably identified if its concentration was 100-fold higher (C). Major species are underlined in panels B and C. RFU, relative fluorescence units. The range of the x axis is set to 100 to 800 bp.
Analysis of clinical specimens.
In total, 106 out of 419 (25.3%) of the C, U-C, S, and U-S samples were positive using cultivation-independent f-ITS2-PCR-CE (Table 1 provides a detailed description). Among the positive samples, 89 (84.0%) were monofungal; the remaining samples contained more than one fungal pathogen (Tables 1 to 3). With respect to different clinical materials, surprisingly, the positivity rate did not differ between samples from either the catheters and stents (23.6% versus 20.9%; P = 0.72) or between catheter- and stent-corresponding urine samples (40.2% versus 30.2%; P = 0.24), as has been reported in bacteria previously (13). No stent was found to be polyfungal. We detected 10 different fungal species, with 38 C. parapsilosis (31.4%), 32 C. albicans (26.5%), and 15 C. tropicalis (12.4%) strains predominating (Table 2). Correct species identification was confirmed by ITS2 sequencing in all representative Candida species (Table 2). Moreover, due to the absence of non-Candida species in our library, sequencing was also used to identify three detected non-Candida species: Geotrichum candidum was identified in 2 U-C samples, and Rhodotorula rubra and Cryptococcus neoformans were each identified in one U-C sample. No statistical difference was observed in species prevalences mutually between catheters and stents or between catheter- and stent-corresponding urine samples.
TABLE 2.
Summary of Candida species prevalencea
| Identified species | No. (%) of positive samplesb
|
||||
|---|---|---|---|---|---|
| C | U-C | Sc | U-S | Total | |
| C. parapsilosis | 14 (40) | 17 (34.7) | 1 (7.1) | 6 (28.6) | 38 (31.4) |
| C. albicans | 10 (28.6) | 8 (16.3) | 7 (50.0)d | 6 (28.6) | 32 (26.5) |
| C. tropicalis | 6 (17.1) | 7 (14.3) | 1 (7.1) | 1 (4.8) | 15 (12.4) |
| C. robusta | 0e | 8 (16.3) | 0e | 3 (14.3) | 11 (9.1) |
| C. krusei | 1 (2.9) | 1 (2.0) | 4 (28.6)d | 2 (9.5) | 9 (7.4) |
| C. famata | 2 (5.7) | 4 (8.2) | 1 (7.1) | 1 (4.8) | 8 (6.7) |
| C. lusitaniae | 1 (2.9) | 2 (4.1) | 0 | 0 | 3 (2.5) |
| C. glabrata | 1 (2.9) | 1 (2.0) | 0 | 0 | 2 (1.7) |
| C. lipolytica | 0 | 1 (2.0) | 0 | 1 (4.8) | 2 (1.7) |
| C. dubliniensis | 0 | 0 | 0 | 1 (4.8) | 1 (0.8) |
| Total | 35 | 49 | 14 | 21 | 121 |
The table shows the prevalence of Candida spp. detected by f-ITS2-PCR-CE in clinical samples, regardless of mono- or polyfungal infection. C. robusta detected in sonication fluids (materials C and S) was evaluated as a contaminant and was excluded from results. Candida was detected in 8 samples (3 C. parapsilosis, 3 C. albicans, 1 C. tropicalis, and 1 C. famata isolate) out of 29 and 2 samples (C. albicans) out of 3 catheter and stent samples without a corresponding urine sample, respectively. In 3 U-C cases without a corresponding catheter sample, no Candida was detected.
C, urinary catheter; U-C, urine from patients with catheter; S, ureteral stent; U-S, urine from patients with stent.
Results for stents represent a disjunction of results from stent proximal and distal tip samples.
In one case of C. albicans and C. krusei, respectively, representatives were detected in the proximal and distal tip samples.
C. robusta was excluded as a contaminant if detected in catheter or stent sonicate.
TABLE 3.
Characterization of polyfungal samples identified by f-ITS2-PCR-CEa
| Identified speciesb | No. (%) of positive samplesc |
||
|---|---|---|---|
| C | U-C | U-S | |
| C. albicans > C. parapsilosis | 1 | ||
| C. albicans > C. robusta | 1 | ||
| C. famata > C. lusitaniae | 1 | ||
| C. famata > C. robusta | 1 | ||
| C. krusei > C. albicans | 1 | ||
| C. lipolytica > C. tropicalis | 1 | ||
| C. parapsilosis > C. albicans | 1 | 1 | |
| C. parapsilosis > C. tropicalis | 2 | ||
| C. tropicalis > C. glabrata | 1 | 1 | |
| C. robusta > C. albicans | 1 | ||
| C. robusta > C. famata | 1 | ||
| C. parapsilosis > C. albicans | 1 | ||
| C. albicans > C. parapsilosis | 1 | ||
| C. famata > C. parapsilosis > C. lusitaniae | 1 | ||
| Total | 4 (23) | 11 (65) | 2 (12) |
The table shows the characterization of polyfungal samples identified by f-ITS2-PCR-CE. C. robusta detected in sonication fluids (materials C and S) was evaluated as a contaminant and was excluded from the results. C. albicans is in boldface, and polyfungal-tending NAC species (species with two or more times higher prevalence in polyfungal than mono-fungal samples) are underlined. No cooccurrence of C. albicans with polyfungal-tending NAC species is depicted.
Species in the same line are in order of decreasing signal.
C, urinary catheter; U-C, urine from patients with catheter; U-S, urine from patients with stent. No stent was found to be polyfungal.
We identified 17 polyfungal samples with 35 representatives (Tables 1 and 3). C. glabrata was identified exclusively in polyfungal samples. Candida famata, Candida lipolytica, and C. lusitaniae showed more than 2-fold-higher prevalence in poly- than monofungal samples. Interestingly, these four polyfungal-tending NAC species were detected exclusively together with other NAC species in six polyfungal samples. In seven polyfungal samples containing C. albicans, apart from the previously mentioned NAC species, such as C. parapsilosis, Candida robusta, or C. krusei, were detected. Conversely, C. krusei prevalence was more than three times higher in mono- than polyfungal samples, and Candida dubliniensis was detected exclusively in one monofungal sample. Unsurprisingly, the highest detection rate of more than one species in a sample was observed in 11 out of 41 positive catheter urine samples (26.8%).
To be comprehensive, the most frequently identified species from urinary catheters and ureteral stents was Candida robusta (Saccharomyces cerevisiae) (data not shown). However, when samples with BHI medium as a negative control were analyzed, we observed that the species’ DNA had been present in the medium. Therefore, this species was evaluated as contamination coming from the BHI medium used to sonicate solid material samples. Thus, sonicates containing only S. cerevisiae were evaluated as negative, and S. cerevisiae was not considered an additional agent in any positive catheter or stent sample.
f-ITS2-PCR-CE comparison with phenotypic identification.
The same clinical samples were also analyzed by culture-dependent phenotypic identification, considered a gold standard. However, using this technique in a routine setting with just 1 μl of sonicate fluid/urine inoculation volume, we identified only eight strains (2.4% of the total number of 337 analyzed samples) belonging to four different species. Thus, an 8.3-fold-lower sensitivity than that with f-ITS2-PCR-CE (67 out of 337 samples positive, 19.9%) was achieved. C. albicans was found in four samples (50%), two catheters and two stents; C. krusei was identified in two stent samples (25%); and C. parapsilosis and C. tropicalis were each identified only once (12.5%), with both found in catheters. The fungal population reached 103, 104, and 105 CFU/ml, in 4, 3, and 1 positive sample, respectively.
In order to increase the sensitivity of classical culture techniques, since the culture positivity rate was lower than expected, 100 μl of sonication fluids and urine instead of the routinely used 1 μl was inoculated in an additional 82 clinical samples. Then, 23 samples (28.0%) were culture positive, while 39 (47.6%) were positive using f-ITS2-PCR-CE analysis (Table 4), indicating a much improved sensitivity, but culture sensitivity was still 1.7-fold lower than that of the molecular approach. An overall higher f-ITS2-PCR-CE positivity rate, regardless of the material type, in this additional sample set (compare 39.5%, 64.5%, 22.3%, and 50% of positive samples of additionally tested samples to 16.9%, 31.3%, 11.9%, and 28.3% positive samples of those initially tested in C, U-C, S, and U-S groups, respectively) is probably related to the different nature of samples because no difference was registered in the collection strategy or f-ITS2-PCR-CE analysis.
TABLE 4.
Comparison of culture and f-ITS2-PCR-CE results by using a higher inoculation volume for culture (100 μl)
| Result type | Culture method result |
f-ITS2-PCR-CE result |
No. (%) of positive samplesb
|
|||||
|---|---|---|---|---|---|---|---|---|
| Identificationa | No. (%) of samples tested | Identificationa | No. (%) of samples tested | C | U-C | S | U-S | |
| Concordant | Negative | 43 | Negative | 43 | 23 | 11 | 7 | 2 |
| C. tropicalis | 5 | C. tropicalis | 5 | 3 | 2 | |||
| C. albicans | 6 | C. albicans | 6 | 2 | 1 | 2 | 1 | |
| C. parapsilosis | 2 | C. parapsilosis | 2 | 1 | 1 | |||
| Partially concordant | C. albicans > Candida sp. | 2 | C. albicans | 2 | 1 | 1 | ||
| C. albicans > Candida sp. | 1 | C. albicans > C. parapsilosis | 1 | 1 | ||||
| Candida sp. | 2 | C. parapsilosis | 2 | 1 | 1 | |||
| Candida sp. | 1 | C. parapsilosis > C. albicans | 1 | 1 | ||||
| C. tropicalis | 2 | C. tropicalis > C. glabrata | 2 | 1 | 1 | |||
| C. albicans > C. glabrata | 1 | C. parapsilosis > C. albicans | 1 | 1 | ||||
| Rhodotorula rubra | 1 | Non-Candida sp.c | 1 | 1 | ||||
| Discrepant | Negative | 16 | C. albicans | 3 | 2 | 1 | ||
| C. famata | 2 | 1 | 1 | |||||
| C. lusitaniae | 1 | 1 | ||||||
| C. parapsilosis | 5 | 1 | 4 | |||||
| C. robusta | 1 | 1 | ||||||
| C. famata > C. lusitaniae | 1 | 1 | ||||||
| C. parapsilosis > C. tropicalis | 1 | 1 | ||||||
| C. robusta > C. albicans | 1 | 1 | ||||||
| C. robusta > C. famata | 1 | 1 | ||||||
| Total | 82 | 82 | 38 | 31 | 9 | 4 | ||
| Negative | 59 (72) | 43 (52) | 23 | 11 | 7 | 2 | ||
| Positive | 23 (28.0) | 39 (47.6) | 15 (39.5) | 20 (64.5) | 2 (22.3) | 2 (50) | ||
| Polyfungald | 4 (17) | 9 (23) | ||||||
Species in the same line are in order of decreasing signal.
C, urinary catheter; U-C, urine from patient with catheter; S, stent; U-S, urine from patient with stent.
Identified as Rhodotorula rubra by sequencing.
Number (percentage) of polyfungal samples out of positive samples.
Correspondingly, more polyfungal samples (17.4% of culture-positive samples) were found when a 100-μl inoculation volume was applied than with a 1-μl inoculation volume (0.0%), while 23.1% of positive samples were polyfungal using f-ITS2-PCR-CE. However, in only 1 of 4 polyfungal samples was the second pathogen identified to species level by culture (Table 4).
When the consistency of identification between both methods was considered, 56 out of 82 samples (68.3%) were in full concordance (43 samples were concordantly negative), 10 samples (12.2%) were concordantly positive but discrepant or partially discrepant at species level identification, and the remaining 16 samples (19.5%) were culture negative but PCR positive (Table 4). Importantly, there was no sample that was culture positive and f-ITS2-PCR-CE negative.
DISCUSSION
We optimized the method for ITS2 rDNA length polymorphism analysis using fluorescent capillary electrophoresis (f-ITS2-PCR-CE) and evaluated the technique’s efficiency in our previous study (8), and we now proceeded to evaluate its applicability for urinary catheter, ureteral stent, and urine clinical samples. This technique enabled us to distinguish 26 out of 29 tested medically important Candida species. This method is usable for closely related species which are difficult to phenotypically distinguish, such as C. dubliniensis and C. albicans or Candida fabianii and Candida pelliculosa. Candida guilliermondii, Candida fermentati, and Candida carpophila were indistinguishable because of identical amplicon lengths. Amplicon length varied from 229 nucleotides (C. lipolytica) to 420 nucleotides (C. kefyr) (8).
Catheters and stents are valuable tools in urologic practice, but colonizing microorganisms might concurrently be a source of serious infection (1). To the best of our knowledge, here we provide the most comprehensive study focused on Candida identification in urinary catheters and ureteral stents, alongside corresponding urine samples, both by culture-dependent and culture-independent techniques. In addition, we compared the contribution of molecular culture-independent f-ITS2-PCR-CE for fungal detection and Candida sp. identification in clinical samples with culture-dependent phenotypic identification.
Candida species identification is mostly based on phenotypic features. Although urine culture is more time-consuming than the molecular-based methods and is not a very sensitive approach, it is considered a standard, widely used method to detect stent colonization (14, 15). In our previous study we reported f-ITS2-PCR-CE as a sensitive and efficient tool for culture-independent identification of a substantial number of clinically important Candida species that was also applicable to polyfungal specimens. Moreover, it was shown to be fast, making a difference of more than 3 h compared with the time required for panfungal PCR followed by sequencing, the method currently used in the clinical setting in our laboratory (8). This study confirmed the higher sensitivity of f-ITS2-PCR-CE than that of a culture approach even though the sensitivity of cultivation was substantially improved by using 100-fold-higher volume of sonicate or urine than was routinely applied. Using f-ITS2-PCR-CE, we detected considerably more fungal species than by cultivation followed by phenotypic identification (19.9% versus 2.4% and 47.6% versus 28.0%, when 1 μl and 100 μl of urine/sonicated fluid, respectively, were inoculated). The detection limit of a cultivation-dependent approach in this setting was roughly 103 CFU/ml. Thus, f-ITS2-PCR-CE had high sensitivity and could therefore be applied to rapidly detect fungi in clinical urological specimens.
Of note, the BHI medium used to sonicate different clinical materials was largely contaminated by C. robusta DNA (S. cerevisiae), identified by f-ITS2-PCR-CE not by cultivation. A possible explanation is the presence of yeast extract DNA in BHI broth after inactivation of the living cells. For this purpose, we suggest using another medium for sonication, such as Ringer solution or phosphate-buffered saline (PBS)/phosphate buffer, as reported previously by (10, 16).
Higher f-ITS2-PCR-CE sensitivity was probably also caused by detection of DNA fragments of floating or dead fungal cells and by the multicopy nature of ITS2 rDNA in the genome. Both of these probable causes raise the question of the clinical relevance of Candida sp. detection and identification.
Candida spp. are often considered contaminants, and verifying their presence does not prove clinical impact (4). On the other hand, they might be a source of serious infection, particularly in immunocompromised patients (17). Kauffman et al. (18) stated that Candida growth in urine represents a spectrum of states, including external perineal colonization, catheter infection, cystitis, or even secondary seeding from an undetected bloodstream infection. A debate exists about the threshold of microorganism concentration of 104 versus 105 CFU/ml in urine that may potentially be used as a criterion for treating candiduria (18). One of the limitations of culture-independent f-ITS2-PCR-CE analysis is that it is impossible to determine the cell concentration in the sample because of the difficulty of standardization (calibration) caused by different genomic ITS2 rDNA copy numbers in various fungal species. However, quantitation may be achieved by combining the f-ITS2-PCR-CE and cultivation-based approaches. Detecting nonliving cells still may be of diagnostic value, particularly in culture-negative samples analyzed from treated patients.
Numerous authors (19–22) reported that the most frequently occurring fungal microorganism in clinical materials is C. albicans, followed by C. glabrata, C. tropicalis, and C. krusei. We detected mostly C. parapsilosis (31.4%), C. albicans (26.5%), and C. tropicalis (12.4%), followed by C. robusta (9.1%) and other NAC species. The high prevalence of saprophytic urinary tract commensals (C. parapsilosis and C. robusta) is not surprising because our sampling strategy did not consider each patient´s diagnosis. f-ITS2-PCR-CE showed the capability to distinguish even between species of C. parapsilosis complex (8); therefore, we do not assume any misidentification. Biofilms are critical for developing clinical infection. C. albicans and C. parapsilosis, which were often identified in analyzed materials, have lower incidences of fluconazole, voriconazole, amphotericin B, and echinocandins resistance than other NAC species (23–26). As NAC specie resistance together with prevalence in infections is increasingly being reported (22), NAC species detection and identification must not be neglected. Of note, fungi identified in this study (including R. rubra and G. candidum) are common urinary tract pathogens or commensals.
We did not detect C. glabrata on stents and catheters or in urine samples, except in two polyfungal samples (Tables 2 and 3), although its higher adherence to epithelial cells and silicone than other NAC species and C. parapsilosis or C. tropicalis, respectively, was reported (22, 23). In contrast to these findings, Shin et al. (27) reported lower C. glabrata biofilm-forming capabilities than other NAC species after culture in nutritionally rich medium.
In addition, unlike cultivation, f-ITS2-PCR-CE enabled more efficient polyfungal sample composition analysis. Although the culture-based polyfungal sample detection rate improved by increasing the inoculation volume, the capacity to distinguish individual pathogens in complex samples remained low; both strains were identified to the species level in just 1 out of 4 samples. Wolcott et al. (28) stated that it is important to identify all of the species present in biofilm because minor microbial constituents can provide a multitude of different advantages to their neighbors, including increased virulence. However, it should be pointed out that f-ITS2-PCR-CE does not allow minor species identification in polymicrobial samples present in a 100-fold-lower concentration than major species (Fig. 1).
We recognized groups of Candida spp. according to poly- or monofungal sample inclinations. C. glabrata, C. famata, C. lipolytica, and C. lusitaniae can be suspected as polyfungal-tending species because they were present in polyfungal samples more often or even exclusively. Of note, these four NAC species were not detected together with C. albicans in any case. Mutually opposed higher C. krusei prevalence in stents and higher C. parapsilosis prevalence in catheters (Table 2) could imply that C. krusei has an inhibitory effect on C. parapsilosis, a phenomenon well described in cases of C. krusei and C. albicans (29, 30). The possible inhibitory effect of C. krusei on other NAC species is supported by a 3-fold-higher prevalence in monofungal than in polyfungal samples. This hypothesis needs to be supported by further investigations.
In this study, we provided an example of the previously reported f-ITS2-PCR-CE method used in urinary tract-associated samples. This technique was found to be more sensitive and more specific than routine culture both in mono- and polyfungal samples. We suggest further improvement of f-ITS2-PCR-CE by prospectively broadening the in-house database of non-Candida fungi distinguishable by the unique ITS2 length and including an internal amplification control.
Further studies are desired to clarify and identify the Candida sp. colonizers, their mutual relationships, and involvement in infection.
ACKNOWLEDGMENTS
This work was supported by research grants from the Ministry of Health of the Czech Republic (grant number 16-31593 A) and the Ministry of Education, Youth and Sports (grant number MUNI/A/0925/2017).
We acknowledge Lenka Suchánková-Křupková for her technical support.
REFERENCES
- 1.Lojanapiwat B. 2006. Colonization of internal ureteral stent and bacteriuria. World J Urol 24:681–683. doi: 10.1007/s00345-006-0135-6. [DOI] [PubMed] [Google Scholar]
- 2.Fanning S, Mitchell AP. 2012. Fungal biofilms. PLoS Pathog 8:e1002585. doi: 10.1371/journal.ppat.1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher JF, Sobel JD, Kauffman CA, Newman CA. 2011. Candida urinary tract infections—treatment. Clin Infect Dis 52:S457–S466. doi: 10.1093/cid/cir112. [DOI] [PubMed] [Google Scholar]
- 4.Sobel JD, Fisher JF, Kauffman CA, Newman CA. 2011. Candida urinary tract infections—epidemiology. Clin Infect Dis 52:S433–S436. doi: 10.1093/cid/cir109. [DOI] [PubMed] [Google Scholar]
- 5.Hooton TM, Bradley SF, Cardenas DD, Colgan R, Geerlings SE, Rice JC, Saint S, Schaeffer AJ, Tambayh PA, Tenke P, Nicolle LE. 2010. Diagnosis, prevention, and treatment of catheter-associated urinary tract infection in adults: 2009 International Clinical Practice Guidelines from the Infectious Diseases Society of America. Clin Infect Dis 50:625–663. doi: 10.1086/650482. [DOI] [PubMed] [Google Scholar]
- 6.Idelevich EA, Grunewald CM, Wüllenweber J, Becker K. 2014. Rapid identification and susceptibility testing of Candida spp. from positive blood cultures by combination of direct MALDI-TOF mass spectrometry and direct inoculation of Vitek 2. PLoS One 9:e114834. doi: 10.1371/journal.pone.0114834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson IC, Cairney JWG. 2004. Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ Microbiol 6:769–779. doi: 10.1111/j.1462-2920.2004.00675.x. [DOI] [PubMed] [Google Scholar]
- 8.Obručová H, Tihelková R, Kotásková I, Růžička F, Holá V, Němcová E, Freiberger T. 2016. Evaluation of fluorescent capillary electrophoresis for rapid identification of Candida fungal infections. J Clin Microbiol 54:1295–1303. doi: 10.1128/JCM.00118-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu CY, Zhang J, Xu X, Chen J. 2011. Prediction of DNA separation by capillary electrophoresis with polymer additives. J Chromatogr Sci 49:310–315. doi: 10.1093/chrsci/49.4.310. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Moser C, Al-Soud WA, Sørensen S, Høiby N, Nielsen PH, Thomsen TR. 2012. Culture-Dependent and -Independent Investigations of Microbial Diversity on Urinary Catheters. J Clin Microbiol 50:3901–3908. doi: 10.1128/JCM.01237-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holá V, Ruzicka F, Horka M. 2010. Microbial diversity in biofilm infections of the urinary tract with the use of sonication techniques. FEMS Immunol Med Microbiol 59:525–528. doi: 10.1111/j.1574-695X.2010.00703.x. [DOI] [PubMed] [Google Scholar]
- 12.Nemcova E, Cernochova M, Ruzicka F, Malisova B, Freiberger T, Nemec P. 2015. Rapid identification of medically important Candida isolates using high resolution melting analysis. PLoS One 10:e0116940. doi: 10.1371/journal.pone.0116940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maki DG, Tambyah PA. 2001. Engineering out the risk for infection with urinary catheters. Emerg Infect Dis 7:342–347. doi: 10.3201/eid0702.700342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riedl CR, Plas E, Hübner WA, Zimmerl H, Ulrich W, Pflüger H. 1999. Bacterial colonization of ureteral stents. Eur Urol 36:53–59. doi: 10.1159/000019927. [DOI] [PubMed] [Google Scholar]
- 15.Lifshitz DA, Winkler HZ, Gross M, Sulkes J, Baniel J, Livne PM. 1999. Predictive value of urinary cultures in assessment of microbial colonization of ureteral stents. J Endourol 13:735–738. doi: 10.1089/end.1999.13.735. [DOI] [PubMed] [Google Scholar]
- 16.Zhai Z, Li H, Qin A, Liu G, Liu X, Wu C, Li H, Zhu Z, Qu X, Dai K. 2014. Meta-analysis of sonication fluid samples from prosthetic components for diagnosis of infection after total joint arthroplasty. J Clin Microbiol 52:1730–1736. doi: 10.1128/JCM.03138-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes RA. 2008. Early diagnosis of fungal infection in immunocompromised patients. J Antimicrob Chemother 61:i3–i6. doi: 10.1093/jac/dkm424. [DOI] [PubMed] [Google Scholar]
- 18.Kauffman CA, Vazquez JA, Sobel JD, Gallis HA, McKinsey DS, Karchmer AW, Sugar AM, Sharkey PK, Wise GJ, Mangi R, Mosher A, Lee JY, Dismukes WE. 2000. Prospective Multicenter Surveillance Study of Funguria in Hospitalized Patients. The National Institute for Allergy and Infectious Diseases (NIAID) Mycoses Study Group. Clin Infect Dis 30:14–18. doi: 10.1086/313583. [DOI] [PubMed] [Google Scholar]
- 19.Dariane C, Cornu JN, Esteve E, Cordel H, Egrot C, Traxer O, Haab F. 2015. Infections fongiques et matériel urétéral: quelle prise en charge? Prog Urol 25:306–311. doi: 10.1016/j.purol.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Weinstein RA, Lundstrom T, Sobel J. 2001. Nosocomial candiduria: a review. Clin Infect Dis 32:1602–1607. doi: 10.1086/320531. [DOI] [PubMed] [Google Scholar]
- 21.Sobel JD, Vazquez JA. 1999. Fungal infections of the urinary tract. World J Urol 17:410–414. doi: 10.1007/s003450050167. [DOI] [PubMed] [Google Scholar]
- 22.Padawer D, Pastukh N, Nitzan O, Labay K, Aharon I, Brodsky D, Glyatman T, Peretz A. 2015. Catheter-associated candiduria: risk factors, medical interventions, and antifungal susceptibility. Am J Infect Control 43:e19–e22. doi: 10.1016/j.ajic.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Berkow EL, Lockhart SR. 2017. Fluconazole resistance in Candida species: a current perspective. Infect Drug Resist 10:237–245. doi: 10.2147/IDR.S118892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perea S, Patterson TF, Patterson TF. 2002. Antifungal resistance in pathogenic fungi. Clin Infect Dis 35:1073–1080. doi: 10.1086/344058. [DOI] [PubMed] [Google Scholar]
- 25.Mandras N, Tullio V, Allizond V, Scalas D, Banche G, Roana J, Robbiano F, Fucale G, Malabaila A, Cuffini AM, Carlone N. 2009. In vitro activities of fluconazole and voriconazole against clinical isolates of Candida spp. determined by disk diffusion testing in Turin, Italy. Antimicrob Agents Chemother 53:1657–1659. doi: 10.1128/AAC.00998-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perlin DS. 2015. Echinocandin resistance in Candida. Clin Infect Dis 61:S612–S617. doi: 10.1093/cid/civ791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin JH, Kee SJ, Shin MG, Kim SH, Shin DH, Lee SK, Suh SP, Ryang DW. 2002. Biofilm production by isolates of Candida species recovered from nonneutropenic patients: comparison of bloodstream isolates with isolates from other sources. J Clin Microbiol 40:1244–1248. doi: 10.1128/JCM.40.4.1244-1248.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolcott R, Costerton JW, Raoult D, Cutler SJ. 2013. The polymicrobial nature of biofilm infection. Clin Microbiol Infect 19:107–112. doi: 10.1111/j.1469-0691.2012.04001.x. [DOI] [PubMed] [Google Scholar]
- 29.Rossoni RD, Barbosa JO, Vilela SFG, dos Santos JD, de Barros PP, Prata MC, de A, Anbinder AL, Fuchs BB, Jorge AOC, Mylonakis E, Junqueira JC. 2015. Competitive interactions between C. albicans, C. glabrata and C. krusei during biofilm formation and development of experimental candidiasis. PLoS One 10:e0131700. doi: 10.1371/journal.pone.0131700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos JD. d, Piva E, Vilela SFG, Jorge AOC, Junqueira JC. 2016. Mixed biofilms formed by C. albicans and non-albicans species: a study of microbial interactions. Braz Oral Res 30:S1806-83242016000100232. doi: 10.1590/1807-3107BOR-2016.vol30.0023. [DOI] [PubMed] [Google Scholar]