Hepatitis C virus (HCV) RNA quantitation is the primary method by which active HCV infections are identified and the response to direct-acting antiviral therapy is monitored. This study describes the evaluation of the Aptima HCV Quant Dx assay (Aptima HCV) performed on the Panther system.
KEYWORDS: dried blood spots, hepatitis C virus, quantitative methods
ABSTRACT
Hepatitis C virus (HCV) RNA quantitation is the primary method by which active HCV infections are identified and the response to direct-acting antiviral therapy is monitored. This study describes the evaluation of the Aptima HCV Quant Dx assay (Aptima HCV) performed on the Panther system. The clinical performance of Aptima HCV was compared to that of the Cobas AmpliPrep/Cobas TaqMan HCV test v2.0 (CAP/CTM). Overall agreement was 84.9% (186/219) with a kappa statistic of 0.755 (standard error, 0.037; 95% confidence interval [CI], 0.682 to 0.828). Passing-Bablok regression of log10 IU/ml values revealed a regression line of Y = 1.163 × X − 0.991 (95% CI of the slope, 1.103 to 1.221, and intercept, −1.341 to −0.642). The 95% lower limit of detection (LLOD) for Aptima HCV on dried blood spot (DBS) samples was calculated to be 2.43 log10 IU/ml (267 IU/ml; 95% CI, 2.31 to 2.73 log10 IU/ml [204 to 540 IU/ml]). A comparison of Aptima HCV testing on paired DBS and serum specimens collected from patients at the time of routine blood collection for CAP/CTM demonstrated an overall agreement of 90.1% (82/91) with a kappa statistic of 0.657 (standard error, 0.101; 95% CI, 0.458 to 0.855). In conclusion, Aptima HCV provides a suitable alternative for HCV RNA testing on serum and DBS samples.
INTRODUCTION
Despite the increased availability of direct-acting antivirals (DAAs) capable of curing hepatitis C virus (HCV) infections, this blood-borne virus remains a significant global health problem. The World Health Organization estimates that there were 71 million people worldwide living with HCV infections in 2015, 1.75 million new infections that year, and 399,000 deaths related to HCV-associated chronic liver disease and hepatocellular carcinoma (1). In the United States, the number of HCV cases increased 3.5-fold between 2010 and 2016, due at least in part to the ongoing epidemic of injection drug use (2, 3). In 2016, the World Health Assembly endorsed the Global Health Sector Strategy (GHSS) on viral hepatitis, which calls for the elimination of viral hepatitis as a public health threat by 2030 (1). The goals of this strategy include reducing new infections by 90% and mortality by 65%. Critical to the success of the GHSS are laboratory testing for the diagnosis of viral hepatitis and monitoring the response to therapy.
The first step of the recommended HCV diagnostic algorithm involves serologic testing for anti-HCV antibodies. However, since 15% to 45% of infected persons spontaneously clear the virus within 6 months of infection without any treatment, antibody-positive individuals are subsequently tested for HCV RNA to identify active HCV infections (4, 5). Furthermore, HCV RNA quantitation by nucleic acid amplification testing (NAAT) is recommended prior to the start of therapy, after 4 weeks of therapy, and 12 weeks after the completion of therapy (6). While serum or plasma are the specimens of choice for HCV NAAT, whole-blood dried blood spot (DBS) specimens may be considered to facilitate access to testing in settings which lack laboratory facilities or the provision for timely delivery of specimens to a laboratory (7).
A recently developed NAAT for HCV RNA is the Aptima HCV Quant Dx assay (Aptima HCV) performed on the fully automated Panther system (both from Hologic Inc., San Diego, CA). This assay relies on real-time transcription-mediated amplification (TMA) for HCV RNA detection and quantitation. Previous work has demonstrated the utility of Aptima HIV on the Panther system for quantitation of HIV-1 from DBS (8), and in this study, the performance characteristics of Aptima HCV is investigated using both serum specimens and DBSs.
MATERIALS AND METHODS
Ethics statement.
This study was reviewed and approved by the Institutional Review Board of Stanford University (protocol 40509).
Aptima HCV Quant Dx assay.
The Aptima HCV Quant Dx assay (Aptima HCV) was performed on the automated Panther System (Hologic Inc., San Diego, CA). Aptima HCV is U.S. Food and Drug Administration (FDA) approved for the confirmation of active HCV infection and for monitoring HCV viral load using serum and plasma. This assay involves three steps: (i) target capture, (ii) amplification of the HCV target in the 5′ untranslated region (UTR) by transcription-mediated amplification (TMA), and (iii) real-time amplicon detection using fluorescently labeled probes. Quantitation is achieved by monitoring the time taken to reach a fluorescence threshold value, which is inversely proportional to the starting HCV concentration. Each reaction contains an internal calibrator/internal control (IC) that is used to account for the effects of inhibition and controls for variations in specimen processing and TMA. A single positive calibrator is run in triplicates along with one replicate each of the low-positive, high-positive, and negative controls for every new reagent kit loaded onto the Panther system. The calibrator and controls are then valid for 24 h. Aptima specimen aliquot tubes require a minimum input volume of 0.7 ml, and 0.5 ml is processed by the Panther system. The Panther system reports values for results ≥10 and ≤100,000,000 IU/ml (1.0 to 8.0 log10 IU/ml), and low-level positives that fall below this range are reported as detected <10 IU/ml (<1.0 log10 IU/ml).
For the testing of dried blood spots (DBSs), a single DBS was rehydrated in 1.0 ml of sample transport medium provided by Hologic and incubated at room temperature while rocking for 30 min. The tubes were centrifuged at 10,000 rpm for 1 min, and the supernatant was transferred to Aptima specimen aliquot tubes before loading into the Panther system. DBS testing using Aptima HCV is for research use only (RUO).
The results reported by the Panther are based on liquid calibrators that use 500 µl of sample. To estimate the HCV RNA IU/ml from this calibration, a conversion factor needs to be applied to account for the dilution of the starting samples and other factors. The HCV RNA IU/ml result obtained from the Panther instrument from a DBS was multiplied by a factor of 24.63. This value was obtained using the following equation: (volume of sample transport media in microliters)/[(volume of whole blood collected on DBS in microliters) × (1 − estimated hematocrit)] (8). The hematocrit value was estimated to be 42% based on the mean adult value from the National Health and Nutrition Examination Survey (NHANES) (9).
LLOD and precision analysis for DBS samples.
To establish the lower limit of detection (LLOD) and precision on DBS samples, DBSs were prepared from whole blood spiked with HCV using the HCV RNA AccuSpan Linearity panel (SeraCare, Milford, MA). EDTA whole blood was obtained from healthy blood donors at the Stanford Blood Center (Palo Alto, CA). The virus load was adjusted to 3.70 log10 (5,000), 3.00 log10 (1,000), 2.70 log10 (500), 2.40 log10 (250), and 2.10 log10 (125) IU/ml, and 70 µl spiked whole blood was spotted onto Whatman 903 sample collection cards (GE Healthcare Life Sciences, Marlborough, MA). DBSs were dried inside the biosafety cabinet for 30 min and stored at room temperature until testing. Leftover HCV-spiked whole blood was centrifuged at 2,500 rpm for 5 min to separate plasma. These plasma samples were run side by side with the DBS to verify the virus load. Ten spots were tested for each concentration per day for 3 separate days.
Clinical specimens.
A total of 93 DBSs were prepared at Stanford Health Care (SHC) blood draw locations at the time of routine blood collection for serum HCV NAAT. Each spot was composed of approximately 70 µl of whole blood spotted on Whatman 903 sample collection cards. DBSs were stored at room temperature until testing. The matching serum specimens were obtained from the Clinical Virology Laboratory archives at −80°C. The rehydrated DBSs and their matching serum samples were tested by Aptima HCV on the same day.
To evaluate specificity using serum as the specimen type, a total of 205 HCV‐seronegative specimens were tested by Aptima HCV. These were selected from clinical specimens that were submitted to the SHC Special Chemistry and Immunology Laboratory between May and June 2017 and tested negative for serum anti-HCV antibodies on the Architect i2000SR (Abbott, Abbott Park, IL).
To determine agreement with routine NAAT testing, 219 serum specimens from 76 patients were tested by Aptima HCV. These were selected from clinical specimens that were submitted to the SHC Clinical Virology Laboratory between April 2016 and June 2017 for HCV NAAT using the Cobas AmpliPrep/Cobas TaqMan HCV test v2.0 (CAP/CTM) (Roche Molecular Systems, Pleasanton, CA). All specimens were collected as serum and stored at −80°C prior to testing. Archived samples were tested by Aptima HCV and the results were compared to previous results by CAP/CTM.
Statistical analysis.
The 95% LLOD was calculated using probit analysis in R using the generalized linear models (glm) package (10). Precision was analyzed in Microsoft Excel using the formula described by Chesher (11). Passing-Bablok and Deming regression analyses, as well as the Bland-Altman plot were generated in R using the mcr package (https://cran.r-project.org/web/packages/mcr/index.html).
RESULTS
Aptima HCV
(i) Serum. The specificity of Aptima HCV was 99.0% (203/205) when testing anti-HCV antibody-negative serum. The two HCV RNA-positive samples were detected below the quantifiable range, 1.0 log10 IU/ml (<10 IU/ml).
Two hundred nineteen archived serum samples from HCV-infected patients were tested for HCV RNA using Aptima HCV and the results were compared to those obtained through routine testing via CAP/CTM. Aptima HCV sensitivity (positive agreement) and specificity (negative agreement) were 92.3% (119/129; 95% confidence interval [CI] 86.2% to 96.2%) and 87.8% (79/90; 95% CI, 79.2% to 93.7%), respectively, with CAP/CTM as the reference.
Overall agreement was 84.9% (186/219) with a kappa statistic of 0.755 (standard error, 0.037; 95% CI, 0.682 to 0.828), indicating substantial agreement (Table 1). Ten samples that were detected at <1.0 log10 IU/ml (<10 IU/ml) by Aptima HCV were not detected by CAP/CTM. Similarly, 8 samples that were detected at <1.18 log10 IU/ml (<15 IU/ml) by CAP/CTM were not detected by Aptima HCV. In addition, there were 10 samples that were detected at <1.0 log10 IU/ml (<10 IU/ml) by Aptima HCV that were quantitated by CAP/CTM (mean, 1.46 log10 IU/ml; standard deviation [SD], 0.16), as well as 3 samples that were not detected by Aptima HCV that were quantitated by CAP/CTM (mean, 1.35 log10 IU/ml; SD, 0.08). Finally, there were 2 samples that were detected at <1.18 log10 IU/ml (<15 IU/ml) by CAP/CTM that were quantitated by Aptima HCV (mean, 1.02 log10 IU/ml; SD, 0.02).
TABLE 1.
Comparison of Aptima and CAP/CTM for detection of HCV RNA in clinical serum samples
| CAP/CTM result | No. of samples with indicated Aptima result |
Total | ||
|---|---|---|---|---|
| Not detected | <10 IU/ml | Quantitated | ||
| Not detected | 79 | 10 | 0 | 89 |
| <15 IU/mL | 8 | 17 | 2 | 27 |
| Quantitated | 3 | 10 | 90 | 103 |
| Total | 90 | 37 | 92 | 219 |
To investigate the quantitative agreement between Aptima HCV and CAP/CTM, the log10 concentrations of the 87 clinical samples quantifiable by both methods were plotted against one another and Passing-Bablok regression was performed (Fig. 1A). This analysis resulted in a regression line of Y = 1.163 × X − 0.991. The 95% confidence intervals of the slope (1.103 to 1.221) and intercept (−1.341 to −0.642) do not include 1 and zero, respectively, indicating that Aptima HCV shows positive proportional bias and negative systematic bias compared to CAP/CTM. Deming regression resulted in a similar regression line (Y = 1.120 × X − 0.775) and similar 95% confidence intervals of the slope (1.068 to 1.172) and intercept (−1.081 to −0.470). Next, the differences in log10 concentrations were plotted against the average values to generate a Bland-Altman plot (Fig. 1B). This analysis revealed a bias of −0.181 log10 IU/ml (Aptima − CAP/CTM) and 95% limits of agreement of −0.984 to 0.619.
FIG 1.
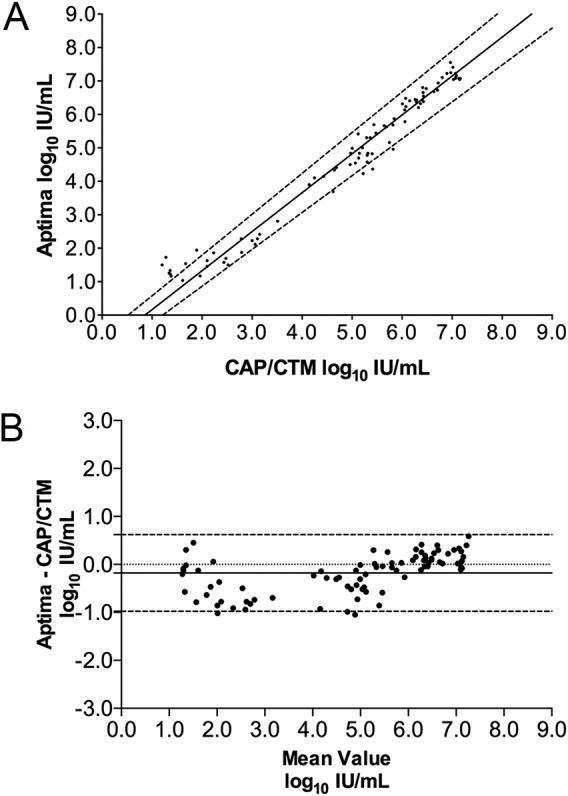
Quantitative comparison of clinical serum specimens tested by the Aptima HCV Quant Dx assay (Aptima) and Cobas AmpliPrep/Cobas TaqMan HCV test v2.0 (CAP/CTM). (A) Passing-Bablok regression analysis of 87 clinical specimens tested by both Aptima HCV and CAP/CTM. The regression line (solid line) and 95% confidence intervals (dashed lines) are displayed. (B) Bland-Altman plot comparing Aptima with CAP/CTM. The bias (solid line) was −0.181 log10 IU/ml. The 95% limits of agreement (dashed lines) and zero line (dotted black line) are also shown.
(ii) DBS.
The 95% LLOD for Aptima HCV on DBS samples was calculated to be 2.43 log10 IU/ml (267 IU/ml; 95% CI, 2.31 to 2.92 log10 IU/ml [204 to 840 IU/ml]). The within-run, between-day, and within-laboratory (total) imprecision levels (i.e., standard deviations) were 0.13 log10, 0.04 log10, and 0.13 log10 IU/ml, respectively, for DBSs containing a nominal HCV concentration of 3.7 log10 IU/ml (5,000 IU/ml) (mean observed concentration, 3.24 log10 IU/ml), the lowest concentration at which all replicates were within the quantifiable range of the assay.
Ninety-three DBSs were tested by Aptima HCV with concurrently collected serum. Overall agreement was 90.1% (82/91) with a kappa statistic of 0.657 (standard error, 0.101; 95% CI, 0.458 to 0.855), indicating substantial agreement (Table 2). Five samples that were detected at <1.0 log10 IU/ml (<10 IU/ml) in serum were not detected in DBSs. Similarly, 3 samples that were detected at <2.43 log10 IU/ml (<270 IU/ml) in DBSs were not detected in serum. In addition, one sample detected at <1.0 log10 IU/ml (<10 IU/ml) in serum was detected at <2.43 log10 IU/ml (<270 IU/ml) in DBS, and one sample detected at 1.45 log10 IU/ml (28 IU/ml) in serum was detected at <2.43 log10 IU/ml (<270 IU/ml) in DBS. For samples quantifiable in both matrices, the mean difference (serum − DBS) was 0.85 log10 IU/ml (95% CI, 0.19 to 1.50).
TABLE 2.
Comparison of Aptima HCV from paired DBS and serum
| Result in serum | No. of pairs with indicated result in DBS |
Total | ||
|---|---|---|---|---|
| Not detected | <270 IU/ml | ≥270 IU/ml | ||
| Not Detected | 72 | 3 | 0 | 75 |
| <10 IU/mL | 5 | 1 | 0 | 6 |
| ≥10 IU/mL | 0 | 1 | 9 | 10 |
| Total | 77 | 5 | 9 | 91 |
DISCUSSION
This study compared the performance characteristics of the Aptima HCV Quant Dx assay performed on the Panther system with the Cobas AmpliPrep/Cobas TaqMan HCV test v2.0 for the quantitation of HCV RNA in serum. The Aptima HCV DBS protocol was then investigated using HCV-spiked whole blood and paired serum and DBS specimens from patients infected with HCV.
The specificity of Aptima HCV was observed to be 99.0% (203/205) when testing anti-HCV antibody-negative serum. Note that the aliquots used for specificity testing had been tested for anti-HCV antibodies on the Architect instrument prior to Aptima HCV, and the false positives were at very low virus loads, suggesting Architect cross contamination. Though sufficient serum was not available for additional molecular investigation, these results support the policy of requiring dedicated aliquots for hepatitis virus load testing.
Overall, Aptima HCV and CAP/CTM gave comparable quantitations of HCV RNA from serum. The evaluation of 219 clinical serum specimens revealed substantial classification agreement. Both Passing-Bablok and Deming regression analyses of the samples quantifiable by both methods indicated that Aptima HCV shows a negative systematic bias compared to CAP/CTM. Negative bias was also observed by Bland-Altman analysis, −0.181 log10 IU/ml. It is important to note, however, that these Aptima HCV serum values were compared to historical CAP/CTM virus loads, which may help explain the observed negative bias.
Several studies have compared Aptima HCV to a variety of HCV NAATs (12–14), including CAP/CTM (15–17). Consistent with the data presented here, there was reasonable agreement between the methods for both qualitative and quantitative results.
Importantly, the manuscript also demonstrates the utility of Aptima HCV for the detection of HCV RNA in DBSs, as has been shown in previous studies with other NAAT methods (18–30). Though Aptima HCV on DBSs was less analytically sensitive than serum testing and demonstrated a negative bias compared to quantitation in serum, these data are similar to those previously reported for DBSs tested for HCV RNA by CAP/CTM and Abbott m2000 (20, 27, 28). However, future studies will be required to compare Aptima HCV to other methods of HCV RNA detection from DBSs.
In addition, HCV RNA was detected at a low level in three DBSs, whereas concurrent serum was negative. Given the higher analytical sensitivity of Aptima HCV on serum, these results are unexpected and may represent false-positive DBSs, though sufficient remaining serum and DBS samples were not available for discrepancy analysis. Further studies detailing the specificity of Aptima HCV on DBS are warranted. Finally, additional work is needed to improve the Aptima HCV DBS conversion factor, which in this study, did not account for potential HCV RNA loss during DBS extraction.
Interestingly, the 2017 World Health Organization (WHO) testing guidelines indicate that qualitative NAATs can be used for both the detection of viremic HCV infection and the assessment of treatment response (5). The WHO reports that >95% of those individuals with chronic HCV infection have a virus load of >4.0 log10 IU/ml (>10,000 IU/ml) (5); furthermore, the minority of individuals with lower virus loads typically have HCV RNA levels of ≥3.0 log10 IU/ml (≥1,000 IU/ml) (31, 32). Consistent with these data, over the past 2 years (1 January 2017 to 1 January 2019) at our institution, 95.3% (402/422) of samples submitted for HCV RNA quantitation with reflex to HCV genotyping, primarily ordered for patients with chronic infection prior to therapy initiation, had virus loads of ≥4.0 log10 IU/ml (≥10,000 IU/ml), and 97.6% (412/422) had virus loads of ≥3.0 log10 IU/ml (≥1,000 IU/ml). Of the patients with virus loads <3.0 log10 IU/ml (<1,000 IU/ml), three had recently completed direct-acting antiviral therapy and two were immediately posttransplantation from an HCV+ donor. Similarly, virus loads in nonresponders and those that relapse after therapy are also ≥3.0 log10 IU/ml (≥1,000 IU/ml) (33, 34). Taken together, these data indicate that Aptima HCV on DBSs provides adequate analytical performance to be used for both the detection of viremic infection and therapeutic failure. Given the simple automated Panther DBS workflow, DBS testing using Aptima HCV is a promising option to meet the increased demand for HCV NAAT necessitated by the GHSS goal of eliminating viral hepatitis as a public health threat by 2030.
Limitations of this study include the limited diversity of HCV genotypes observed in this patient population, primarily genotypes 1a and 1b. Future studies will be necessary to confirm the performance of Aptima HCV on DBSs in a larger cohort of viremic patients with representation of all circulating HCV genotypes (35). Furthermore, future work would benefit from study designs that specifically select patients to evaluate Aptima HCV on DBSs to identify active infections as part of an HCV diagnostic algorithm and to identify virological failure in patients on DAAs.
In conclusion, we report the evaluation of Aptima HCV performed on the Panther system. This assay demonstrated comparable performance to that of CAP/CTM on serum specimens collected from a U.S. cohort. Furthermore, Aptima HCV showed sufficient analytical and clinical performance on DBSs to be considered by the WHO as a method to facilitate worldwide access to HCV RNA testing, particularly in areas where infrastructure challenges prevent the timely delivery of other, more labile, specimen types for virus load testing. In addition, the availability of Aptima tests for HIV-1 and hepatitis B virus (HBV) is essential given the global burden of HCV coinfections with these blood-borne viral pathogens (36–38). Serum and DBS testing using the Aptima HCV Quant Dx assay and other Aptima assays on the Panther system may provide the necessary performance and efficiency to contribute to the worldwide elimination of viral hepatitis.
ACKNOWLEDGMENTS
We thank the Stanford Health Care Clinical Virology and Special Chemistry and Immunology Laboratories for their continued hard work and dedication to patient care.
The financial support for this research was provided by Hologic, Inc.
The funders had no role in data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.World Health Organization. 2017. WHO global hepatitis report. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2018. Surveillance for viral hepatitis–United States, 2016. https://www.cdc.gov/hepatitis/statistics/2016surveillance/commentary.htm. Accessed 24 November 2018.
- 3.Zibbell JE, Iqbal K, Patel RC, Suryaprasad A, Sanders KJ, Moore-Moravian L, Serrecchia J, Blankenship S, Ward JW, Holtzman D, Centers for Disease Control and Prevention. 2015. Increases in hepatitis C virus infection related to injection drug use among persons aged ≤30 years - Kentucky, Tennessee, Virginia, and West Virginia, 2006–2012. MMWR Morb Mortal Wkly Rep 64:453–458. [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2013. Testing for HCV infection: an update of guidance for clinicians and laboratorians. MMWR Morb Mortal Wkly Rep 62:362–365. [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. 2017. WHO guidelines on hepatitis B and C testing. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.AASLD-IDSA. 2018. HCV testing and linkage to care. Recommendations for testing, managing, and treating hepatitis C. http://www.hcvguidelines.org/full-report/hcv-testing-and-linkage-care. Accessed 23 November 2018.
- 7.Easterbrook PJ, WHO Guidelines Development Group. 2016. Who to test and how to test for chronic hepatitis C infection - 2016 WHO testing guidance for low- and middle-income countries. J Hepatol 65:S46–S66. doi: 10.1016/j.jhep.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Sahoo MK, Varghese V, White E, Winslow M, Katzenstein DA, Shafer RW, Pinsky BA. 2016. Evaluation of the Aptima HIV-1 Quant Dx assay using plasma and dried blood spots. J Clin Microbiol 54:2597–2601. doi: 10.1128/JCM.01569-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacher DA, Barletta J, Hughes JP. 2012. Biological variation of hematology tests based on the 1999–2002 national health and nutrition examination survey. Natl Health Stat Report 2012:1–10. [PubMed] [Google Scholar]
- 10.Venables WN, Ripley BD, Venables WN. 2002. Modern applied statistics with S, 4th ed Springer, New York, NY. [Google Scholar]
- 11.Chesher D. 2008. Evaluating assay precision. Clin Biochem Rev 29:S23–S26. [PMC free article] [PubMed] [Google Scholar]
- 12.Garbuglia AR, Bibbo A, Sciamanna R, Pisciotta M, Capobianchi MR. 2017. Performance evaluation of the Aptima HCV Quant Dx assay for hepatitis C virus (HCV) RNA detection and quantification in comparison to the Abbott RealTime HCV assay. J Clin Virol 92:1–6. doi: 10.1016/j.jcv.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Schalasta G, Speicher A, Borner A, Enders M. 2016. Performance of the new Aptima HCV Quant Dx assay in comparison to the Cobas TaqMan HCV2 test for use with the high pure system in detection and quantification of hepatitis C virus RNA in plasma or serum. J Clin Microbiol 54:1101–1107. doi: 10.1128/JCM.03236-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.May S, Adamska E, Tang JW. 2018. Evaluating the Aptima HIV-1 Quant Dx, HCV Quant Dx and HBV quant assays against the Abbott HIV-1, HCV and HBV RealTime assays. J Clin Virol 106:7–10. doi: 10.1016/j.jcv.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 15.Worlock A, Blair D, Hunsicker M, Le-Nguyen T, Motta C, Nguyen C, Papachristou E, Pham J, Williams A, Vi M, Vinluan B, Hatzakis A. 2017. Analytical characteristics and comparative evaluation of Aptima HCV Quant Dx assay with the Abbott RealTime HCV assay and Roche Cobas AmpliPrep/Cobas TaqMan HCV quantitative test v2.0. Virol J 14:66. doi: 10.1186/s12985-017-0727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schonning K, Pedersen MS, Johansen K, Landt B, Nielsen LG, Weis N, Westh H. 2017. Analytical and clinical performance of the Hologic Aptima HCV Quant Dx assay for the quantification of HCV RNA in plasma samples. J Virol Methods 248:159–165. doi: 10.1016/j.jviromet.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Chevaliez S, Dubernet F, Dauvillier C, Hezode C, Pawlotsky JM. 2017. The new Aptima HCV Quant Dx Real-time TMA assay accurately quantifies hepatitis C virus genotype 1-6 RNA. J Clin Virol 91:5–11. doi: 10.1016/j.jcv.2017.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Solmone M, Girardi E, Costa F, Pucillo L, Ippolito G, Capobianchi MR. 2002. Simple and reliable method for detection and genotyping of hepatitis C virus RNA in dried blood spots stored at room temperature. J Clin Microbiol 40:3512–3514. doi: 10.1128/JCM.40.9.3512-3514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Crignis E, Re MC, Cimatti L, Zecchi L, Gibellini D. 2010. HIV-1 and HCV detection in dried blood spots by SYBR green multiplex real-time RT-PCR. J Virol Methods 165:51–56. doi: 10.1016/j.jviromet.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 20.Tuaillon E, Mondain AM, Meroueh F, Ottomani L, Picot MC, Nagot N, Van de Perre P, Ducos J. 2010. Dried blood spot for hepatitis C virus serology and molecular testing. Hepatology 51:752–758. doi: 10.1002/hep.23407. [DOI] [PubMed] [Google Scholar]
- 21.Bennett S, Gunson RN, McAllister GE, Hutchinson SJ, Goldberg DJ, Cameron SO, Carman WF. 2012. Detection of hepatitis C virus RNA in dried blood spots. J Clin Virol 54:106–109. doi: 10.1016/j.jcv.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Santos C, Reis A, Dos Santos CV, Damas C, Silva MH, Viana MV, Ferraz ML, Carnauba D, El-Far F, Serra F, Diaz RS. 2012. The use of real-time PCR to detect hepatitis C virus RNA in dried blood spots from Brazilian patients infected chronically. J Virol Methods 179:17–20. doi: 10.1016/j.jviromet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Ross RS, Stambouli O, Gruner N, Marcus U, Cai W, Zhang W, Zimmermann R, Roggendorf M. 2013. Detection of infections with hepatitis B virus, hepatitis C virus, and human immunodeficiency virus by analyses of dried blood spots–performance characteristics of the ARCHITECT system and two commercial assays for nucleic acid amplification. Virol J 10:72. doi: 10.1186/1743-422X-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dokubo EK, Evans J, Winkelman V, Cyrus S, Tobler LH, Asher A, Briceno A, Page K. 2014. Comparison of hepatitis C virus RNA and antibody detection in dried blood spots and plasma specimens. J Clin Virol 59:223–227. doi: 10.1016/j.jcv.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tejada-Strop A, Drobeniuc J, Mixson-Hayden T, Forbi JC, Le NT, Li L, Mei J, Terrault N, Kamili S. 2015. Disparate detection outcomes for anti-HCV IgG and HCV RNA in dried blood spots. J Virol Methods 212:66–70. doi: 10.1016/j.jviromet.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 26.Mossner BK, Staugaard B, Jensen J, Lillevang ST, Christensen PB, Holm DK. 2016. Dried blood spots, valid screening for viral hepatitis and human immunodeficiency virus in real-life. World J Gastroenterol 22:7604–7612. doi: 10.3748/wjg.v22.i33.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soulier A, Poiteau L, Rosa I, Hezode C, Roudot-Thoraval F, Pawlotsky JM, Chevaliez S. 2016. Dried blood spots: a tool to ensure broad access to hepatitis C screening, diagnosis, and treatment monitoring. J Infect Dis 213:1087–1095. doi: 10.1093/infdis/jiv423. [DOI] [PubMed] [Google Scholar]
- 28.Marins EG, Bodinaidu K, Lin M, Deforest A. 2017. Evaluation of the Cobas AmpliPrep/Cobas TaqMan HCV test v2.0 for HCV viral load monitoring using dried blood spot specimens. J Virol Methods 247:77–80. doi: 10.1016/j.jviromet.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 29.Greenman J, Roberts T, Cohn J, Messac L. 2015. Dried blood spot in the genotyping, quantification and storage of HCV RNA: a systematic literature review. J Viral Hepat 22:353–361. doi: 10.1111/jvh.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lange B, Roberts T, Cohn J, Greenman J, Camp J, Ishizaki A, Messac L, Tuaillon E, van de Perre P, Pichler C, Denkinger CM, Easterbrook P. 2017. Diagnostic accuracy of detection and quantification of HBV-DNA and HCV-RNA using dried blood spot (DBS) samples - a systematic review and meta-analysis. BMC Infect Dis 17:693. doi: 10.1186/s12879-017-2776-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hajarizadeh B, Grady B, Page K, Kim AY, McGovern BH, Cox AL, Rice TM, Sacks-Davis R, Bruneau J, Morris M, Amin J, Schinkel J, Applegate T, Maher L, Hellard M, Lloyd AR, Prins M, Dore GJ, Grebely J, InC3 Study Group. 2015. Patterns of hepatitis C virus RNA levels during acute infection: the InC3 study. PLoS One 10:e0122232. doi: 10.1371/journal.pone.0122232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGovern BH, Birch CE, Bowen MJ, Reyor LL, Nagami EH, Chung RT, Kim AY. 2009. Improving the diagnosis of acute hepatitis C virus infection with expanded viral load criteria. Clin Infect Dis 49:1051–1060. doi: 10.1086/605561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swain MG, Lai MY, Shiffman ML, Cooksley WG, Zeuzem S, Dieterich DT, Abergel A, Pessoa MG, Lin A, Tietz A, Connell EV, Diago M. 2010. A sustained virologic response is durable in patients with chronic hepatitis C treated with peginterferon alfa-2a and ribavirin. Gastroenterology 139:1593–1601. doi: 10.1053/j.gastro.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Manns MP, Pockros PJ, Norkrans G, Smith CI, Morgan TR, Haussinger D, Shiffman ML, Hadziyannis SJ, Schmidt WN, Jacobson IM, Barcena R, Schiff ER, Shaikh OS, Bacon B, Marcellin P, Deng W, Esteban-Mur R, Poynard T, Pedicone LD, Brass CA, Albrecht JK, Gordon SC. 2013. Long-term clearance of hepatitis C virus following interferon alpha-2b or peginterferon alpha-2b, alone or in combination with ribavirin. J Viral Hepat 20:524–529. doi: 10.1111/jvh.12074. [DOI] [PubMed] [Google Scholar]
- 35.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platt L, Easterbrook P, Gower E, McDonald B, Sabin K, McGowan C, Yanny I, Razavi H, Vickerman P. 2016. Prevalence and burden of HCV co-infection in people living with HIV: a global systematic review and meta-analysis. Lancet Infect Dis 16:797–808. doi: 10.1016/S1473-3099(15)00485-5. [DOI] [PubMed] [Google Scholar]
- 37.Mavilia MG, Wu GY. 2018. HBV-HCV coinfection: viral interactions, management, and viral reactivation. J Clin Transl Hepatol 6:296–305. doi: 10.14218/JCTH.2018.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konstantinou D, Deutsch M. 2015. The spectrum of HBV/HCV coinfection: epidemiology, clinical characteristics, viral interactions and management. Ann Gastroenterol 28:221–228. [PMC free article] [PubMed] [Google Scholar]


