Infectious diseases remain a global threat contributing to excess morbidity and death annually, with the persistent potential for destabilizing pandemics. Improved understanding of the pathogenesis of bacteria, viruses, fungi, and parasites, along with rapid diagnosis and treatment of human infections, is essential for improving infectious disease outcomes worldwide.
KEYWORDS: CRISPR-Cas, diagnostics, infectious diseases, therapeutics
ABSTRACT
Infectious diseases remain a global threat contributing to excess morbidity and death annually, with the persistent potential for destabilizing pandemics. Improved understanding of the pathogenesis of bacteria, viruses, fungi, and parasites, along with rapid diagnosis and treatment of human infections, is essential for improving infectious disease outcomes worldwide. Genomic loci in bacteria and archaea, termed clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins, function as an adaptive immune system for prokaryotes, protecting them against foreign invaders. CRISPR-Cas9 technology is now routinely applied for efficient gene editing, contributing to advances in biomedical science. In the past decade, improved understanding of other diverse CRISPR-Cas systems has expanded CRISPR applications, including in the field of infectious diseases. In this review, we summarize the biology of CRISPR-Cas systems and discuss existing and emerging applications to evaluate mechanisms of host-pathogen interactions, to develop accurate and portable diagnostic tests, and to advance the prevention and treatment of infectious diseases.
INTRODUCTION
Improved understanding of the structure and function of clustered regularly interspaced short palindromic repeats (CRISPR) and the accompanying CRISPR-associated (Cas) proteins has resulted in rapidly expanding research and clinical applications. The CRISPR locus was first identified in 1987, when a genetic structure containing 5 highly homologous repeats of 29 nucleotides separated by 32-nucleotide spacers was discovered in Escherichia coli (1). Years later, it was learned that spacer sequences derived from invading mobile genetic elements (MGEs), including bacteriophages, plasmids, and transposons, form the basis of adaptive and heritable immunity in bacteria and archaea (2). A subsequent sentinel discovery in CRISPR-Cas biology was that Cas9 nucleases, guided by CRISPR RNA (crRNA) and trans-activating RNA (tracrRNA) complexes, induce blunt double-stranded breaks in DNA (3, 4). CRISPR-Cas9 technology has since revolutionized the field of gene editing in basic sciences and clinical medicine (5). The structure and function of many additional CRISPR-Cas systems have now come to light, with rapidly diversifying applications of CRISPR technology. In this review, we discuss current knowledge regarding the biology of diverse CRISPR-Cas systems, highlight key features of CRISPR-Cas9 technology, and summarize existing and emerging applications related to infectious disease pathogenesis, diagnosis, and treatment.
CRISPR-CAS BIOLOGY
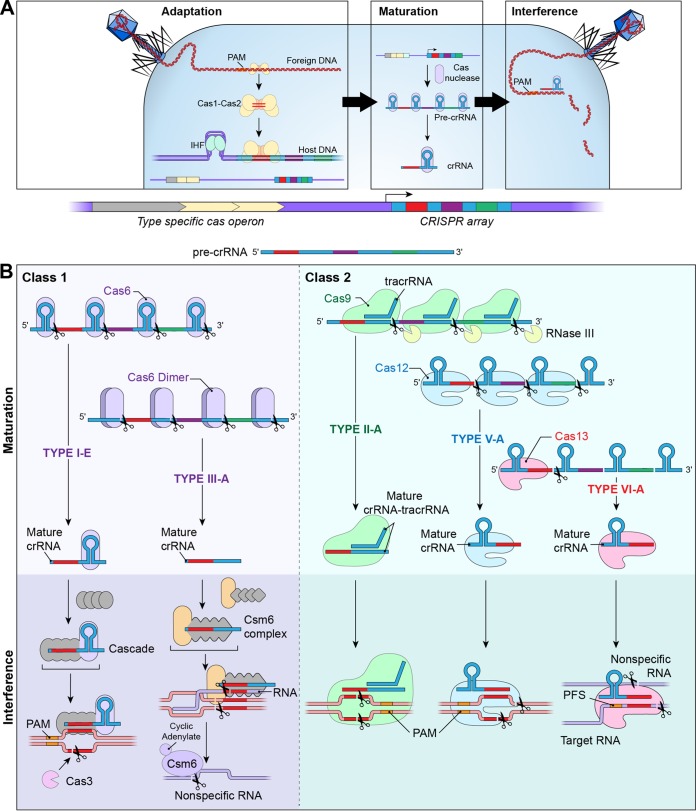
The defining feature of CRISPR-Cas systems is that they provide prokaryotes with heritable adaptive immunity against foreign genetic elements. The CRISPR genomic locus serves as a memory storage unit in which nucleic acid spacer sequences derived from invading genetic elements are sequestered; the sequences are later recalled to guide Cas proteins for targeted elimination of foreign invaders. At the molecular level, CRISPR-Cas systems function though the processes of adaption, crRNA maturation, and interference with significant biological diversity across systems (Fig. 1). Excellent comprehensive reviews on this topic have been published (6, 7), and salient features are summarized here. Two classes of CRISPR-Cas systems have been described to date, including six types and multiple subtypes. Class 1 CRISPR-Cas systems include types I, III, and IV, which utilize an interference machinery composed of multiple Cas proteins, while class 2 systems, including types II, V, and VI, employ a single Cas protein for interference.
FIG 1.
Biological diversity of the CRISPR-Cas system. (A) The CRISPR-Cas defense mechanism has three stages, namely, adaptation, maturation, and interference. In the type 1-E system, adaption begins with the recognition of foreign DNA and an associated PAM by the Cas1-Cas2 complex. The host protein IHF, which is not part of the Cas operon, bends the host DNA, allowing the Cas1-Cas2 complex to integrate the newly acquired spacer sequence. During maturation, pre-crRNA transcribed from the CRISPR array is generated and processed into individual mature crRNAs. In the final step of interference, mature crRNAs guide type-specific Cas nucleases to their respective target nucleic acid sequences, resulting in cleavage of foreign DNA. (B) Maturation in the class 1 systems of type I-E and type III-A is carried out by Cas6, which cleaves the pre-crRNA into mature crRNA. In the type 1-E system, a Cas6 monomer remains bound to the crRNA; in the type III-A system, a Cas6 dimer dissociates from the crRNA. Maturation in class 2 systems is performed by the same protein as utilized in interference. In type II-A systems, maturation requires binding of a tracrRNA to the pre-crRNA, forming a tracrRNA-crRNA complex. Cas9 binds the tracrRNA-crRNA complex, and cleavage of the pre-crRNA to mature crRNA is performed by RNase III. Maturation is carried out by Cas12 in the type V-A system and by Cas13 in the type VI-A system. Interference in class 1 systems is mediated by a multiprotein complex, while a single nuclease mediates interference in class 2 systems. In the type I-E system, Cascade binds to a mature crRNA and the complex recognizes the protospacer and an associated PAM sequence on foreign DNA. Cascade binding generates conformational changes in target DNA, resulting in the R-loop structure and recruitment of Cas3 to cleave the nonbound strand of target DNA. In the type III-A system, the Csm complex binds to mature crRNA. This complex then recognizes target nucleic acid sequences, resulting in cleavage of ssRNA and dsDNA. DNA cleavage results in cyclic adenylate production, which activates Csm6 cleavage of nonspecific RNA. In the type II-A system, interference is mediated by Cas9 bound to mature crRNA-tracrRNA. The resulting complex recognizes the PAM on target DNA, resulting in double-stranded cleavage with blunt ends. The type V-A system utilizes Cas12 bound to mature crRNA to recognize foreign DNA and the associated PAM, inducing double-stranded cleavage with staggered ends. Type VI systems employ Cas13, which, along with its mature crRNA, recognizes target ssRNA with its associated PFS. Binding to ssRNA initiates cleavage of the target ssRNA, along with indiscriminate cleavage of nonspecific ssRNA.
Adaptation.
During adaptation, foreign genetic elements are recognized, protospacer sequences are selected and processed, and the spacer is integrated into the CRISPR array (Fig. 1A, adaptation). In order for the CRISPR-Cas machinery to avoid autoimmunity and destruction of its own CRISPR array, it must reliably distinguish between foreign and self DNA. In the well-described CRISPR-Cas type I and II systems, recognition of a protospacer-adjacent motif (PAM) aids in distinguishing self from foreign genetic elements. In the subtype I-E system, the Cas1-Cas2 complex recognizes a compatible PAM sequence, cleaves foreign DNA, and adjusts protospacer size for integration into the CRISPR array. New spacer integration occurs immediately following the AT-rich leader sequence of the CRISPR array, resulting in a sequential timeline of foreign invaders. In the subtype I-E system, a CRISPR-independent protein, integrated host factor (IHF), bends DNA so that the Cas1-Cas2 complex can recognize the CRISPR array leader sequence and correctly position the spacer, acting as an integrase complex. In the subtype II-A system, Cas9, Csn2, and tracrRNA are also required, beyond Cas1-Cas2, for spacer acquisition (8). Recognition of the leader sequence in the subtype II-A system is IHF independent, and Cas1-Cas2 directly recognizes a leader-anchoring site (LAS) to orient the spacer correctly.
crRNA maturation.
Generation of mature crRNA begins with transcription of pre-crRNA, which is initiated within the leader sequence preceding the CRISPR array and results in multiple repeat and spacer segments. These segments are cleaved into individual mature crRNAs that guide Cas proteins to their foreign target (Fig. 1A, maturation). In class 1 type I and III systems, Cas6 enzymes cleave repeat segments within the pre-crRNA, yielding mature crRNA (Fig. 1B, class 1, maturation). Cas5d functions in place of Cas6 in the subtype I-C system and possibly in subtype III-C and III-D systems (11). In most type I systems, Cas6 remains bound to crRNA, serving as a building block for the interference complex. Subtypes I-A and I-B are exceptions, in which Cas6 becomes unbound following cleavage of the repeat sequence. In type III systems, Cas6 is a dimer that also becomes unbound. The mechanism of crRNA maturation in type IV systems has not been characterized. In class 2 systems, crRNA processing is accomplished using the same Cas proteins employed in interference and, in some cases, non-Cas proteins (Fig. 1B, class 2, maturation) . In subtype II-A and B systems, Cas9 binds to a crRNA-tracrRNA complex and recruits RNase III, a host protein, to cleave the pre-crRNA repeat segment (4). tracrRNA is required for crRNA maturation in type II and subtype V-B systems but is not required in other class 2 systems. In type V and VI CRISPR-Cas systems, both crRNA processing and interference are accomplished by Cas12 and Cas13 proteins, respectively.
Interference.
Interference is the most well-studied aspect of CRISPR-Cas systems, and improved understanding of CRISPR-Cas systems’ interference machinery, including DNA versus RNA targeting and specific versus nonspecific nucleic acid cleavage, has contributed to diversified emerging applications of CRISPR technology.
(i) Class 1 interference machinery.
The interference machinery of class 1 CRISPR-Cas systems consists of multiprotein complexes (Fig. 1B, class 1, interference). The CRISPR-Cas type I interference machinery is termed CRISPR-associated complex for antiviral defense (Cascade) (13). While Cascade components vary across type I subtypes (I-A to I-F and I-U), constant features include PAM recognition on foreign targets, Cas6- or Cas5-mediated crRNA binding to target DNA, a Cas7 backbone, R-loop stabilization, and target cleavage by Cas3. The subtype I-E interference complex has been well characterized and is composed of Cas5 and Cas6 bound to the 5′ and 3′ repeat portions of the crRNA, respectively, 6 centrally located Cas7 proteins that form the backbone, a large Cas8 subunit that mediates PAM recognition and initiates the unwinding of foreign DNA, and 2 small Cas11 subunits that form and stabilize the R-loop structure. Conformational changes in Cas8 and Cas11 subunits allow Cas3 recruitment and cleavage of the nonbound DNA strand within the R-loop (14). The structure and function of the interference machinery of other type I subtypes have not been fully elucidated.
The type III interference complex is Cascade like; however, cleavage of foreign DNA in this system depends on binding of the interference complex to RNA transcribed from foreign DNA. Interference complexes of subtypes III-A and III-B, termed Csm and Cmr, respectively, are composed of Cas5 bound to the 5′ repeat end of mature crRNA, a Cas7-family protein backbone, and Cas10 and Cas11 large and small subunits, respectively. Recognition of a PAM or RNA PAM (present on transcribed RNA) is required by some but not all type III systems, where other tools for self versus nonself discrimination are employed. Once the interference complex is bound, Cas7 cleaves the single-stranded RNA (ssRNA) transcript at regular intervals and Cas10 cleaves target DNA (15, 16). It was recently observed that DNA cleavage by Cas10 in the subtype III-A Csm complex triggers production of secondary messengers (cyclic adenylates) that activate robust nonspecific RNA cleavage by Csm6, a Cas-associated RNase. Little is known about subtype III-C and III-D and type IV interference mechanisms.
(ii) Class 2 interference machinery.
Class 2 CRISPR-Cas systems are different from class 1 systems, in that interference is performed with a single nuclease instead of a protein complex (Fig. 1B, class 2, interference). The defining features of the type II system are the bilobed Cas9 protein and the requirement for tracrRNA along with crRNA to guide the Cas9 nuclease (6). tracrRNA binds to complementary sequences of repeat regions of pre-crRNA, and the resulting complex recruits Cas9 (4). Cas9 recognizes PAM sequences on target DNA, and the crRNA-tracrRNA complex pairs with cDNA, resulting in Cas9 double-stranded cleavage of target DNA and leaving blunt ends (3). Subtypes of the type II system include II-A, II-B, and II-C, which are differentiated based on size and sequence variability in the Cas9 gene.
Cas12 is the characteristic protein of the type V CRISPR-Cas system, which includes subtypes V-A, V-B, and V-C, with effector proteins 12a, 12b, and 12c, respectively. tracrRNA is required for 12b activity but not 12a activity; the requirements of 12c need further characterization (17). Following PAM recognition by the crRNA-Cas12 complex, target double-stranded DNA (dsDNA) is cleaved in a staggered fashion, leaving 5- to 7-nucleotide overhangs (17, 18). Type VI CRISPR-Cas systems do not require tracrRNA, and they employ a Cas13 nuclease that has characteristic higher eukaryotic and prokaryotic nucleotide (HEPN)-binding domains. Similar to type III systems, type VI systems target ssRNA. The crRNA-Cas13 complex recognizes a protospacer flanking site (PFS) adjacent to the complementary spacer on the 3′ end of ssRNA for Cas13a and a PFS on the 5′ and 3′ ends of the protospacer for Cas13b (20). Binding of Cas13 to the PFS and the target induces cleavage of both target and nonspecific RNA within the protein’s two HEPN-binding domains. Indiscriminate ssRNA cleavage by Cas13 resembles type III Csm6 activity (21).
CRISPR-CAS9 TECHNOLOGY OVERVIEW
CRISPR-Cas9 biotechnology is now extensively applied across multiple disciplines, including basic sciences, food/crop development, fuel generation, drug development, and human genome engineering (22). Naturally occurring crRNA-tracrRNA complexes of type II CRISPR systems may be synthetically engineered into chimeric single-guide RNAs (sgRNAs), which are programable fusion molecules (23). The development of guide RNAs (gRNAs), whether employed as a single fusion molecule (typically termed sgRNA) or as individual crRNA and tracrRNA components, has allowed the use of CRISPR-Cas9 for genome editing and high-throughput screening of genotype-phenotype relationships in a broad array of cell lines.
Genetic engineering with CRISPR-Cas9.
Since the first discoveries, CRISPR-Cas9 gene-editing applications have expanded rapidly. Unlike transcription activator-like effector nucleases (TALENs) and zinc finger nucleases, which require protein engineering to achieve their desired effects, the CRISPR-Cas9 system relies on sgRNAs to direct Cas proteins to target DNA cleavage sites (24). Cleavage of target dsDNA is followed by DNA repair by one of two host-mediated mechanisms, either nonhomologous end joining (NHEJ) or homology-directed repair (HDR). NHEJ is error prone, resulting in random indel and frameshift mutations that often result in loss of protein function (24). In contrast, HDR allows precise genetic modifications and is the basis for most CRISPR-Cas9-mediated gene editing. In this case, a donor repair template with homology to the area of the dsDNA break is introduced and serves as a template for precise edits (24).
Use of sgRNA libraries for screening of genotype-phenotype relationships.
CRISPR technology has been applied as a screening strategy to induce genetic mutations to evaluate gene function. CRISPR libraries include thousands of pooled plasmids, each containing multiple sgRNAs targeting genes of interest. Cells are treated with pooled libraries, resulting in distinct mutant cell populations (25). These populations are screened by positive or negative selection for a phenotype (typically survival) or gene of interest. This technology has been applied to many different human and nonhuman cell lines (26, 27).
Use of deactivated Cas9 to regulate gene transcription.
Deactivated Cas9 (dCas9) has also been used to upregulate or downregulate gene transcription (25). When dCas9 is used to activate gene expression, this is called CRISPR activation; when it is used to repress gene expression, it is termed CRISPR interference. One approach to CRISPR activation is to fuse dCas9 to VP6, a transcriptional activator. When this complex binds to the promoter region of a gene, transcription is activated. Repression of gene transcription or CRISPR interference can be achieved through binding of dCas9 itself to the promoter region and presumed steric hinderance or through fusion of dCas9 to the Krueppel-associated box domain, a transcriptional repressor (25).
INFECTIOUS DISEASE APPLICATIONS
Improved understanding of CRISPR-Cas biology has resulted in expanded applications in the field of infectious diseases. CRISPR technology provides tools that promise to clarify fundamental host and microbe interactions, to aid in the development of rapid and accurate diagnostics, and to advance the prevention and treatment of infectious diseases.
Understanding host-pathogen interactions.
Understanding mechanisms by which bacteria, viruses, fungi, and parasites induce human disease is essential to guide optimal clinical care and rational design of targeted therapies and vaccines. CRISPR-Cas9-based gene editing has been used across diverse pathogens to inform gene and protein contributions to molecular pathogenesis (28). Winter et al. employed a CRISPR-Cas9 sgRNA library to elucidate the mechanism by which α-hemolysin, a Staphylococcus aureus virulence factor, induces cytotoxicity (29). These experiments identified three genes (SYS1, ARFRP1, and TSPAN14) that posttranscriptionally regulate a disintegrin and metalloproteinase domain-containing protein 10 (ADAM-10), reducing its level on cell surfaces and thus decreasing α-hemolysin binding and toxicity. In a genome-wide screen of West Nile virus (WNV) using a CRISPR sgRNA library, Ma et al. identified seven genes (EMC2, EMC3, SEL1L, DERL2, UBE2G2, UBE2J1, and HRD1) that, when inactivated, protected against WNV-induced neuronal cell death (30). These genes are part of the endoplasmic reticulum-associated protein degradation pathway, leading the authors to conclude that this pathway mediates WNV-induced cytopathology and might provide a target for novel drug development.
The genomes of filamentous fungi are difficult to manipulate, due to low genome-editing efficiency (31). To address this problem, Nodvig et al. modified the Cas9 gene of Streptococcus pyogenes to include a 3′ simian virus 40 nuclear localization sequence and modified the sgRNA promoter to be more compatible with fungi (32). Using this system, the authors introduced RNA-guided mutations in the alleles of six Aspergillus species, supporting the utility of CRISPR technology for exploration of fungal biology. Parasites, including Toxoplasma gondii and Trypanosoma cruzi, the etiological agents of toxoplasmosis and Chagas disease, respectively, have also been investigated using CRISPR technology. CRISPR-Cas9 was employed by Sidik et al. in a high-throughput analysis of T. gondii gene function (33) and by Lander et al. to silence the T. cruzi gene encoding GP72, which is required for flagellar attachment and paraflagellar rod (PFR) proteins 1 and 2 (34). In the latter experiment, the authors demonstrated that PFR proteins 1 and 2 are required for flagellar attachment to the cell body and parasite motility. The aforementioned examples emphasize the breadth of CRISPR applications for evaluating host-pathogen interactions among diverse organisms.
Developing diagnostics for infectious diseases.
Rapid and accurate diagnostic tests facilitate early recognition and treatment of infectious diseases, allowing improved clinical care and timely implementation of infection control and other public health measures to limit disease spread. An ideal rapid diagnostic test would be sensitive and specific, easy to perform and to interpret, and portable and affordable, so that it might be used in diverse clinical settings, including resource-limited areas. CRISPR-Cas biology has contributed to important advances in the development of rapid and accurate infectious disease diagnostics.
(i) Diagnostics using CRISPR-Cas9.
CRISPR-Cas9 has been employed by multiple investigators developing infectious disease diagnostics. Pardee et al. combined nucleic acid sequence-based amplification (NASBA), an isothermal amplification technique, with CRISPR-Cas9 to distinguish accurately between closely related Zika virus strains in vitro and in a macaque model (35). The investigators appended a synthetic trigger sequence to NASBA-amplified viral RNA and used a sgRNA-Cas9 complex to cleave the resulting dsDNA. The presence or absence of a strain-specific PAM resulted in truncated or full-length DNA fragments upon Cas9 cleavage. Full-length strands, but not truncated strands, activated the trigger switch, which induced a color change on a paper disc, allowing reliable strain differentiation.
In another application, Muller et al. combined CRISPR-Cas9 with optical DNA mapping to identify bacterial antibiotic resistance genes (36). In this application, a guide RNA (gRNA)-Cas9 complex binds and cleaves specific nucleic acid sequences of plasmids containing resistance genes and a fluorescent dye (YOYO-1) and netropsin independently bind DNA based on AT-rich regions selectively, resulting in an emission intensity unique to each DNA segment, like a bar code. Using this assay, the authors were able to distinguish between plasmids producing different extended-spectrum β-lactamases (ESBLs), including cefotaxime 15 (CTX-M-15) and CTX-M-14 and carbapenemases including Klebsiella pneumoniae carbapenemase (KPC) and New Delhi metallo-β-lactamase 1 (NDM-1). Addition of multiple crRNAs allowed detection of numerous resistance genes in the same reaction.
Guk et al. coupled CRISPR-Cas9 with DNA fluorescent in situ hybridization (FISH) to detect methicillin-resistant Staphylococcus aureus (MRSA) (37). This method utilizes a dCas9 system in which a sgRNA-dCas9 complex coupled with a SYBR green I fluorescent probe recognizes the mecA gene of MRSA. While the complex recognizes the target DNA sequence, dCas9 does not induce DNA cleavage, thus making it suitable for detection by FISH. This assay can detect MRSA at a concentration of 10 CFU/ml and can distinguish between S. aureus isolates with and without the mecA gene.
(ii) CRISPR-Cas12- and CRISPR-Cas13-based applications.
More recent applications of CRISPR for the development of infectious disease diagnostics take advantage of collateral cleavage induced by Cas12 and Cas13 nucleases. The first of these platforms is termed specific high-sensitivity enzymatic reporter unlocking (SHERLOCK), which combines isothermal recombinase polymerase amplification (RPA) or reverse transcription (RT)-RPA with Cas13a cleavage (38). A crRNA-Cas13a complex binds and cleaves target nucleic acid with high specificity. Nontarget RNA coupled to a fluorescent reporter is also cleaved when the crRNA-Cas13a complex binds target nucleic acid, providing a fluorescent signal for pathogen detection. Using SHERLOCK, Gootenberg et al. differentiated between closely related Zika virus strains and Dengue viruses, identified E. coli and Pseudomonas aeruginosa, and distinguished between K. pneumoniae isolates with different resistance genes (KPC and NDM) (38). The feature that makes this technology attractive is its ability to discriminate single nucleotide changes.
A similar technique, termed DNA endonuclease-targeted CRISPR trans reporter (DETECTR), combines isothermal RPA with Cas12a enzymatic activity (19). In this assay, binding of the crRNA-Cas12a complex to target DNA induces indiscriminate cleavage of ssDNA that is coupled to a fluorescent reporter. Chen et al. used DETECTR to distinguish between human papillomavirus 16 (HPV16) and HPV18 in crude DNA extracts from cultured human cells and from clinical samples (19). Crude extracts from 25 human anal swab samples were tested using DETECTR, and the results were compared to those of an approved PCR-based assay. Following DNA extraction, DETECTR correctly identified HPV16 and HPV18 from clinical specimens within 1 h, with results for 25/25 and 23/25 samples, respectively, agreeing with PCR results.
Gootenberg et al. introduced SHERLOCKv2 with improvements that included single-reaction multiplexing with orthogonal CRISPR enzymes, quantitative target detection, enhanced sensitivity, and portability (39). Detection of up to four targets was achieved by combining multiple prescreened Cas13 nucleases and a Cas12 nuclease with nucleic acid-fluorescent reporter complexes that provided signal detection at different wavelengths. Quantitative detection was achieved by optimizing RPA primer concentrations so that sample input and signal intensity closely correlated across a broad range of sample concentrations. Enhanced sensitivity was achieved by adding Csm6 to increase cleavage of off-target ssRNA coupled to a fluorescent reporter. Portability of the assay was enhanced by replacing the fluorescence readout with streptavidin-based detection on a lateral flow paper test strip.
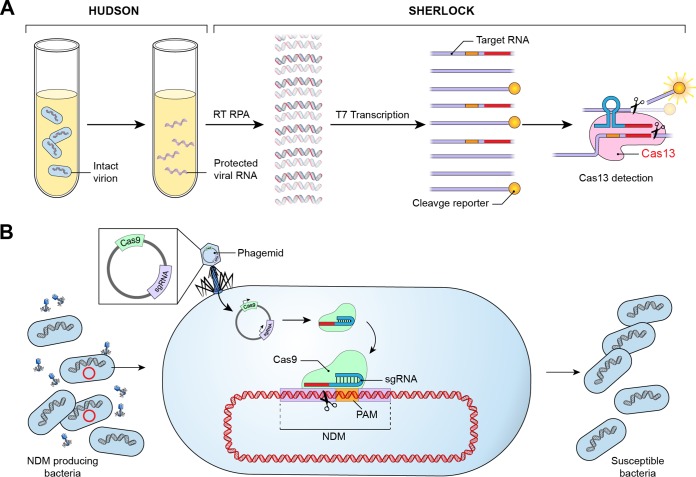
In a further advance, Myhrvold et al. combined SHERLOCK with heating unextracted diagnostic samples to obliterate nucleases (HUDSON) (40), which eliminates the need for nucleic acid extraction and allows pathogen detection directly from bodily fluids (Fig. 2A). In HUDSON, heat and chemical reduction inactivate high levels of nucleases present in body fluids, followed by lysis of viral particles, releasing nucleic acids into solution. HUDSON combined with SHERLOCK allowed highly sensitive detection of dengue virus in patient samples of whole blood, serum, and saliva within 2 h. The authors also demonstrated assay specificity and adaptability by distinguishing between four Dengue virus serotypes and developing an assay for detection of six common HIV reverse transcriptase mutations within 1 week.
FIG 2.
Diagnostic and therapeutic applications of CRISPR-Cas. (A) A CRISPR-based rapid diagnostic test combines HUDSON with SHERLOCK, allowing rapid detection of viral nucleic acid directly from body fluids. HUDSON employs heat and chemical reduction to inactivate high levels of nucleases present in body fluids and lyses viral particles, releasing viral nucleic acids into solution. SHERLOCK integrates isothermal nucleic acid amplification by RT-RPA with T7-dependent transcription and detection of target RNA through Cas13-mediated cleavage of nonspecific RNA coupled to a fluorescent reporter. (B) CRISPR-based therapeutics include the potential to remove antibiotic resistance plasmids from drug-resistant bacteria. NDM-producing bacteria are transfected with a phagemid containing Cas9 and a sgRNA targeting a spacer in NDM. The sgRNA-Cas9 complex cleaves the NDM gene, resulting in bacteria that are antibiotic susceptible.
Emerging therapeutic applications.
Antibiotic-resistant bacteria cause an estimated 2 million infections and 23,000 deaths annually in the United States, and the pipeline for new antibiotics is running dry (41). Since its inception, the global human immunodeficiency virus (HIV) epidemic has claimed an estimated 35 million lives, and a similar number of individuals remain persistently infected today (42). Drug-resistant bacteria and persistent viral infections, including HIV and hepatitis B virus (HBV), are primary targets of emerging CRISPR-based infectious disease therapeutics (Table 1).
TABLE 1.
Potential targeted therapeutics for CRISPR-Cas systems
| Organism | CRISPR system | Gene target | Finding | Reference |
|---|---|---|---|---|
| E. coli, S. enterica | Type I-E (Cas3) | ftsA, asd, msbA, nusB, fucP, ogr, groL, arpA, PentC, phoH, PppsR | Selective removal of individual strains in pure and mixed cultures | Gomaa et al. (46) |
| EHEC, E. coli | Type II (Cas9) | eae, blaSHV-18, blaNDM-1, gyrA | Phagemid targeted to eae in vitro resulted in 20-fold reduction in EHEC and in vivo improved survival rates in Galleria mellonella; targeting of SHV-18 and NDM-1 was bactericidal | Citorik et al. (47) |
| E. coli | Type I-E (Cas3) | blaNDM-1, blaCTX-M-15 | Eliminated antibiotic resistance plasmids, leading to enrichment of antibiotic-sensitive bacteria using CRISPR-Cas along with temperate and lytic bacteriophages | Yosef et al. (48) |
| E. coli | Type II (Cas9) | blaTEM, blaSHV | Restored β-lactam sensitivity in ESBL-producing E. coli | Kim et al. (49) |
| S. aureus | Type II (Cas9) | aph-3, mecA | Phagemid delivery to selectively kill virulent and avirulent Staphylococcus aureus and to eliminate mecA gene without killing host | Bikard et al. (50) |
| HIV-1 | Type II (Cas9) | LTR U3 region | Inactivated gene expression and replication in infected T cells and microglial cells and use of multiplex gRNAs to prevent HIV-1 infection in TZM-bI-cells | Hu et al. (51) |
| HIV-1 | Type II (Cas9) | LTR, gag, pol | Excision of chronic HIV-1 in spleen, lungs, heart, colon, and brain from humanized mice with AAV vector | Yin et al. (53) |
| HIV-1 | Type II (Cas9) | LTR | Removal of HIV-1 DNA from human peripheral blood mononuclear cells engrafted in humanized mice with multiplex of gRNAs delivered with lentivirus vector | Bella et al. (54) |
| HSV-1 | Type II (Cas9) | Essential: UL15, UL27, UL29, UL30, UL36, UL37, UL42, UL5, UL52, UL8, UL54, UL9; nonessential: US3, US8 | Simultaneous targeting of essential genes with 2 gRNAs was superior to that with 1 gRNA in clearing HSV-1-infected cells | van Diemen et al. (55) |
| HSV-1 | Type II (Cas9) | ICP0, ICP4, ICP27 | Limited HSV-1 infection cycle in human oligodendroglioma cells | Roehm et al. (56) |
| EBV | Type II (Cas9) | EBNA-1, OriP | Targeting of Burkitt lymphoma cells infected with EBV with 2 gRNAs against EBNA-1 resulted in 95% loss of EBV genome | van Diemen et al. (55) |
| CMV | Type II (Cas9) | Essential: UL44, UL54, UL57, UL70, UL105, UL86, UL84; nonessential: US6, US7, US11 | Targeting of essential genes led to inhibition of CMV replication and targeting of nonessential genes did not | van Diemen et al. (55) |
| HBV | Type II (Cas9) | Repeat region of integrated genome | Removal of integrated and episomally located HBV genome from HBV cell line | Li et al. (58) |
| HBV | Type II (Cas9) | S open reading frame | Inactivation of cccDNA of HBV in hNTCP-HepG2 cells with AAV vector | Scott et al. (59) |
| JCV | Type II (Cas9) | T-antigen | Suppressed JCV replication in HJC-2 cells using lentivirus vector delivery | Wollebo et al. (60) |
| HPV | Type II (Cas9) | E6, E7 | Targeting of E6 and E7 with gRNA delivered by lentivirus vector resulted in death of HPV-transformed cells | Kennedy et al. (61) |
(i) Rationale for use of CRISPR-Cas systems to target bacterial resistance.
While not all bacteria employ CRISPR-Cas systems, increasing evidence supports the role of these systems in preventing the acquisition of genomic elements that confer antibiotic resistance, raising the possibility that bacteria’s own defenses might be therapeutically applied against them. Aydin et al. showed that the presence of a type I-F CRISPR system in E. coli was associated with antibiotic susceptibility (43), and Price et al. showed that Enterococcus faecalis strains with a deletion in the Cas9 gene were more likely to acquire resistance elements through conjugation (44). Additionally, in an in vivo mouse model, plasmid transfer was less effective in E. faecalis strains with intact CRISPR-Cas systems than in those in which CRISPR-Cas function was impaired by mutagenesis (44).
In the setting of human bacterial infections, broad-spectrum antibiotic use, often initiated empirically, applies pressure for the development of resistant bacteria. Recent in vitro data suggest that exposure to broad-spectrum antibiotics may suppress CRISPR-Cas activity, aiding bacterial acquisition of antibiotic resistance elements. Lin et al. observed that exposure to the broad-spectrum antibiotic imipenem suppressed the CRISPR-Cas activity in K. pneumoniae by stimulating the transcriptional repressor histone-like nucleoid structuring protein (H-NS) (45). These data support the biological fitness of bacteria adapting to external stimuli and the complex interplay between clinical interventions and consequent microbial responses. Improved understanding of how CRISPR-Cas systems function at baseline and under variable conditions will guide the rational use of this technology as it expands into the clinic.
(ii) Targeting pathogenic and drug-resistant bacteria.
It has been proposed that CRISPR technology might be used to develop selective and titratable antimicrobials to eliminate pathogenic bacteria. Gomaa et al. were among the first investigators to prove this concept in vitro, using a subtype I-E CRISPR-Cas system to selectively kill individual strains of E. coli and Salmonella enterica in pure and mixed culture experiments (46). Citorik et al. used RNA-guided Cas9 delivered by phagemid (a plasmid packaged in phage capsids) to eliminate the eae gene of enterohemorrhagic E. coli (EHEC), which encodes a virulence factor for bacterial adhesion to host epithelial cells (47). Targeting eae in vitro resulted in a 20-fold reduction in bacterial counts, while targeting eae in EHEC-infected Galleria mellonella larvae resulted in improved survival rates, compared with off-target-treated and chloramphenicol-treated controls. The authors also successfully targeted and eliminated the β-lactamase genes sulfhydryl variable 18 (SHV-18) and NDM-1 of E. coli, individually or combined in a multiplexed system that resulted in bacterial death.
Elimination of drug-resistant genetic elements and restoration of antibiotic susceptibility while maintaining bacterial viability, to limit disruption to the normal microbiota, has also been explored in vitro using CRISPR technology (Fig. 2B). To remove resistance elements without killing host bacteria, Yosef et al. employed temperate phages to deliver a subtype I-E CRISPR-Cas system that eliminated plasmids encoding NDM-1 and CTM-X-15 and provided protection against lytic phages, while not killing the E. coli host (48). Following challenge with lytic phages, drug-resistant bacteria were eliminated, thus enriching for antibiotic-sensitive bacteria. Similarly, Kim et al. used a CRISPR-Cas9-based strategy termed resensitization to antibiotics from resistance (ReSAFR) to restore β-lactam antibiotic activity in ESBL-producing E. coli (49). The authors targeted separate conserved sequences among multiple TEM- and SHV-type ESBL E. coli strains. Targeting these sequences restored susceptibility to ampicillin and ceftazidime.
These concepts have been similarly applied to Gram-positive bacteria. Bikard et al. used RNA-guided Cas9 delivered by phagemid to selectively kill virulent but not avirulent S. aureus strains and to eliminate plasmids carrying the mecA methicillin resistance gene without killing the host bacteria (50). The studies outlined here provide preliminary evidence that CRISPR-Cas systems might be used to therapeutically target specific bacteria to treat acute infections, to decolonize patients harboring resistant organisms, or to otherwise reshape the human microbiome in a beneficial fashion.
(iii) Targeting persistent viral infection.
Following initial infection, many viral pathogens establish persistent infection by integrating their genome into human chromosomal DNA or maintaining it episomally within host cells. Viral pathogens that cause persistent infection include but are not limited to HIV, hepatitis viruses, herpesviruses, and papillomaviruses. In recent years, CRISPR technology has been used to reduce or to eliminate persistent viral infections in vitro and in animal models, raising new hope for cures for latent and chronic viral infections.
Following acute HIV infection, proviral DNA becomes integrated into host cells, resulting in chronic infection despite antiretroviral therapy. In vitro applications of CRISPR technology have been used to prevent or to eliminate HIV infection. Hu et al. prevented de novo HIV-1 infection in TZM-bI cells expressing a gRNA-Cas9 construct targeting long terminal repeat (LTR) sequences of HIV (51). With this construct, the authors also inactivated HIV gene expression in infected microglial cells and T cells. Similar constructs targeting HIV LTR, gag, and env genes have been used to eliminate HIV proviral DNA from multiple other cell lines (52).
Yin et al. eliminated HIV proviral DNA from neural progenitor cells using multiple sgRNAs and S. aureus Cas9 delivered by an adeno-associated virus (AAV) vector (53). This work was extended into a humanized bone marrow/liver/thymus mouse model of chronic HIV infection, in which a quadruplex sgRNA-S. aureus Cas9 system packaged into an AAV vector that was administered by intravenous infusion excised proviral DNA from animals’ spleen, lung, heart, colon, and brain tissues (53). In similar work, Bella et al. used CRISPR technology delivered by a lentivirus vector to eliminate HIV proviral DNA from infected human peripheral blood mononuclear cells in a transgenic mouse model (54).
Similar to applications for HIV, CRISPR has been used to prevent and to eliminate herpes simplex virus 1 (HSV-1) infections in vitro. van Diemen et al. used gRNAs targeting 12 different essential genes and 2 nonessential genes to reduce replication of HSV-1 in Vero cells. Interestingly, targeting of multiple genes simultaneously resulted in greater efficiency of HSV-1 clearance, compared with targeting of single genes (55). Roehm et al. limited HSV-1 infection and suppressed viral replication in human oligodendroglioma cells using a CRISPR-Cas9 system to introduce indels into gene targets important for viral replication (56). Other herpesviruses that have been targeted by CRISPR in vitro include Epstein-Barr virus (EBV), which predisposes individuals to certain lymphomas and nasopharyngeal cancers, and human cytomegalovirus (CMV), which causes severe disease when acquired congenitally or by immunocompromised hosts. van Diemen et al. used a lentivirus vector to deliver a CRISPR-Cas9 system and two gRNAs targeting EBV nuclear antigen 1 (EBNA-1), achieving 95% clearance of EBV genomes in latently infected Burkitt lymphoma cells (55). These findings raise the possibility that CRISPR-based therapies might someday be used to eradicate persistent EBV infections from tissues and to prevent the development of EBV-associated malignancies. The authors also used gRNAs targeting essential and nonessential genes for CMV replication and observed that targeting of essential genes decreased replication, while targeting of nonessential genes had no effect, despite effective Cas-9 editing. Of note, however, while multiple gRNAs led to CMV suppression for up to 11 days, viral escape mutants emerged, highlighting potential challenges with CRISPR-based therapies (55).
More than 250 million people are infected with HBV worldwide, resulting in an estimated 887,000 deaths each year (57). While an effective HBV vaccine exists, a cure remains elusive. Li et al. used CRISPR-Cas9 to remove a full-length 3,175-bp HBV DNA fragment that was chromosomally integrated and episomally located in chronically infected cells (58), increasing the potential for complete HBV eradication. Scott et al. used a single-stranded AAV vector to deliver sgRNA targeting the S open reading frame of HBV and S. aureus Cas9. With this system, they inactivated covalently closed circular DNA (cccDNA) of HBV-infected hNTCP-HepG2 cells (59). JC virus (JCV) and HPV have also been targeted in vitro using CRISPR technology. Wollebo et al. used a CRISPR-Cas9 system with gRNA targeting the JCV T-antigen. Using a lentivirus vector with a doxycycline-inducible Cas9 gene, the authors transduced HJC-2 cells and successfully eliminated T-antigen expression (60). Kennedy et al. targeted HPV-16 and HPV-18 with gRNAs against E6 and E7 oncogenes in conjunction with CRISPR-Cas9, to inactivate these oncogenes in HPV-transformed cells (61). Interestingly, E6 inactivation led to increased p53 expression, while inactivation of E7 resulted in increased Rb expression, each resulting in cell death and highlighting a potential role in the treatment of HPV-associated malignancy (61).
DISCUSSION
Prokaryotes have diverse CRISPR-Cas systems, which provide heritable adaptive immunity. As new insights into the composition and function of these systems come to light, clinical applications for CRISPR technology will continue to expand. Infectious diseases impose an enormous burden worldwide, and new tools are needed to study underlying mechanisms and to diagnose accurately and to treat infections in all settings. CRISPR-Cas9 technology is advancing the understanding of microbe-host interactions as not previously possible and is being applied to develop new diagnostics for infectious diseases, adding to the existing armamentarium. Perhaps the most exciting advance in CRISPR-based diagnostics for infectious diseases is the application of nonspecific nucleic acid cleavage observed in CRISPR-Cas type III, V, and VI systems to develop accurate and portable diagnostic tests. These emerging platforms will require careful validation against approved diagnostic methods and field testing to ensure end user functionality.
CRISPR-based therapies hold promise for the treatment of cancer and inherited disorders such as sickle cell disease and for the prevention and treatment of infectious diseases. The transition from preclinical observations to proven and approved therapies has not yet occurred, as no approved CRISPR-based therapies are available and only a limited number of early clinical trials are ongoing. Efficient targeted delivery of CRISPR technology in vivo, without significant on- or off-target toxicity, remains a challenge. Viral vectors, including adenoviruses and lentiviruses, may deliver a CRISPR construct to the target of interest, although concerns related to potential carcinogenesis and immunogenicity remain. Nonviral vectors, including lipid and polymer nanocarriers such as polyethylenimine, poly(l-lysine), polyamidoamine dendrimers, and chitosan, are also under investigation (62). Recent reports suggest that CRISPR-Cas9 can induce damage to the p53-mediated DNA repair mechanism, raising concerns because of the association between p53 mutations and cancer (63).
To date, early investigations into CRISPR-based therapies targeting infectious diseases have focused on prevention and treatment of pathogenic drug-resistant bacteria and persistent viral infections. This is good news because these infections, including infections with multidrug-resistant bacteria, HIV, and HBV, significantly contribute to the global disease burden. Challenges remain beyond safe and effective delivery of CRISPR-based therapies for infectious diseases. Bacterial and viral plasticity may result in genetic polymorphisms of gRNA targets, rendering CRISPR-based therapies ineffective. PAM sequence mutations have also been shown to allow phages to escape CRISPR-Cas systems (64, 65). Whether this can be addressed by packaging and delivering multiple gRNAs with diverse targets remains to be seen.
Moving forward with CRISPR-Cas systems as treatments in the realm of infectious disease will require standardized methods for safe treatment delivery. If successful, an antibiotic resistance decolonization strategy, in which patients who are colonized with carbapenemase-producing organisms are given an oral formulation of a targeted CRISPR-Cas system aimed at removing resistant organisms from the gastrointestinal tract, thus positively restructuring the human microbiome, could be imagined. Safe and effective CRISPR-based therapies for persistent viral infections would remarkably change the global landscape of infectious diseases. While these challenges are formidable, they are not insurmountable, suggesting that a future infectious disease community might routinely integrate CRISPR technology into daily practice.
ACKNOWLEDGMENTS
The Intramural Research Program of the National Institutes of Health (Clinical Center, Critical Care Medicine Department, and Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases) supported this work.
The content of this publication does not necessarily reflect the views or policies of the U.S. Department of Health and Human Services, and the mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. government.
REFERENCES
- 1.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. 1987. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169:5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishino Y, Krupovic M, Forterre P. 2018. History of CRISPR-Cas from encounter with a mysterious repeated sequence to genome editing technology. J Bacteriol 200:e00580-17. doi: 10.1128/JB.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 4.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hille F, Richter H, Wong SP, Bratovic M, Ressel S, Charpentier E. 2018. The biology of CRISPR-Cas: backward and forward. Cell 172:1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 7.Koonin EV, Makarova KS, Zhang F. 2017. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. 2015. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reference deleted.
- 10. Reference deleted.
- 11.Garside EL, Schellenberg MJ, Gesner EM, Bonanno JB, Sauder JM, Burley SK, Almo SC, Mehta G, MacMillan AM. 2012. Cas5d processes pre-crRNA and is a member of a larger family of CRISPR RNA endonucleases. RNA 18:2020–2028. doi: 10.1261/rna.033100.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reference deleted.
- 13.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao Y, Luo M, Hayes RP, Kim J, Ng S, Ding F, Liao M, Ke A. 2017. Structure basis for directional R-loop formation and substrate handover mechanisms in type I CRISPR-Cas system. Cell 170:48–60.e11. doi: 10.1016/j.cell.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staals RH, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, Sakamoto K, Suzuki T, Dohmae N, Yokoyama S, Schaap PJ, Urlaub H, Heck AJ, Nogales E, Doudna JA, Shinkai A, van der Oost J. 2014. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell 56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. 2015. Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell 161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. 2015. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell 60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. 2018. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. 2016. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.East-Seletsky A, O'Connell MR, Knight SC, Burstein D, Cate JH, Tjian R, Doudna JA. 2016. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang F, Doudna JA. 2017. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 25.Shalem O, Sanjana NE, Zhang F. 2015. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MC, Yusa K. 2014. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. 2014. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 28.Doerflinger M, Forsyth W, Ebert G, Pellegrini M, Herold MJ. 2017. CRISPR/Cas9: the ultimate weapon to battle infectious diseases? Cell Microbiol 19:e12693. doi: 10.1111/cmi.12693. [DOI] [PubMed] [Google Scholar]
- 29.Winter SV, Zychlinsky A, Bardoel BW. 2016. Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus α-hemolysin-mediated toxicity. Sci Rep 6:24242. doi: 10.1038/srep24242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma H, Dang Y, Wu Y, Jia G, Anaya E, Zhang J, Abraham S, Choi JG, Shi G, Qi L, Manjunath N, Wu H. 2015. A CRISPR-based screen identifies genes essential for West-Nile-virus-induced cell death. Cell Rep 12:673–683. doi: 10.1016/j.celrep.2015.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi TQ, Liu GN, Ji RY, Shi K, Song P, Ren LJ, Huang H, Ji XJ. 2017. CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art. Appl Microbiol Biotechnol 101:7435–7443. doi: 10.1007/s00253-017-8497-9. [DOI] [PubMed] [Google Scholar]
- 32.Nodvig CS, Nielsen JB, Kogle ME, Mortensen UH. 2015. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One 10:e0133085. doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sidik SM, Hackett CG, Tran F, Westwood NJ, Lourido S. 2014. Efficient genome engineering of Toxoplasma gondii using CRISPR/Cas9. PLoS One 9:e100450. doi: 10.1371/journal.pone.0100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lander N, Chiurillo MA, Vercesi AE, Docampo R. 2017. Endogenous C-terminal tagging by CRISPR/Cas9 in Trypanosoma cruzi. Bio Protoc 7:e2299. doi: 10.21769/BioProtoc.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O'Connor DH, Gehrke L, Collins JJ. 2016. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 36.Muller V, Rajer F, Frykholm K, Nyberg LK, Quaderi S, Fritzsche J, Kristiansson E, Ambjornsson T, Sandegren L, Westerlund F. 2016. Direct identification of antibiotic resistance genes on single plasmid molecules using CRISPR/Cas9 in combination with optical DNA mapping. Sci Rep 6:37938. doi: 10.1038/srep37938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guk K, Keem JO, Hwang SG, Kim H, Kang T, Lim EK, Jung J. 2017. A facile, rapid and sensitive detection of MRSA using a CRISPR-mediated DNA FISH method, antibody-like dCas9/sgRNA complex. Biosens Bioelectron 95:67–71. doi: 10.1016/j.bios.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 38.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F. 2017. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. 2018. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, Tan AL, Paul LM, Parham LA, Garcia KF, Barnes KG, Chak B, Mondini A, Nogueira ML, Isern S, Michael SF, Lorenzana I, Yozwiak NL, MacInnis BL, Bosch I, Gehrke L, Zhang F, Sabeti PC. 2018. Field-deployable viral diagnostics using CRISPR-Cas13. Science 360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf. [Google Scholar]
- 42.World Health Organization. 2018. Global Health Observatory (GHO) data: HIV/AIDS. http://www.who.int/gho/hiv/en.
- 43.Aydin S, Personne Y, Newire E, Laverick R, Russell O, Roberts AP, Enne VI. 2017. Presence of type I-F CRISPR/Cas systems is associated with antimicrobial susceptibility in Escherichia coli. J Antimicrob Chemother 72:2213–2218. doi: 10.1093/jac/dkx137. [DOI] [PubMed] [Google Scholar]
- 44.Price VJ, Huo W, Sharifi A, Palmer KL. 2016. CRISPR-Cas and restriction-modification act additively against conjugative antibiotic resistance plasmid transfer in Enterococcus faecalis. mSphere 1:e00064-16. doi: 10.1128/mSphere.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin TL, Pan YJ, Hsieh PF, Hsu CR, Wu MC, Wang JT. 2016. Imipenem represses CRISPR-Cas interference of DNA acquisition through H-NS stimulation in Klebsiella pneumoniae. Sci Rep 6:31644. doi: 10.1038/srep31644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. 2014. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio 5:e00928-13. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Citorik RJ, Mimee M, Lu TK. 2014. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol 32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yosef I, Manor M, Kiro R, Qimron U. 2015. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci U S A 112:7267–7272. doi: 10.1073/pnas.1500107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim JS, Cho DH, Park M, Chung WJ, Shin D, Ko KS, Kweon DH. 2016. CRISPR/Cas9-mediated re-sensitization of antibiotic-resistant Escherichia coli harboring extended-spectrum β-lactamases. J Microbiol Biotechnol 26:394–401. doi: 10.4014/jmb.1508.08080. [DOI] [PubMed] [Google Scholar]
- 50.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. 2014. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol 32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J, Mo X, Khalili K. 2014. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci U S A 111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang G, Zhao N, Berkhout B, Das AT. 2018. CRISPR-Cas based antiviral strategies against HIV-1. Virus Res 244:321–332. doi: 10.1016/j.virusres.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 53.Yin C, Zhang T, Qu X, Zhang Y, Putatunda R, Xiao X, Li F, Xiao W, Zhao H, Dai S, Qin X, Mo X, Young WB, Khalili K, Hu W. 2017. In vivo excision of HIV-1 provirus by saCas9 and multiplex single-guide RNAs in animal models. Mol Ther 25:1168–1186. doi: 10.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bella R, Kaminski R, Mancuso P, Young W-B, Chen C, Sariyer R, Fischer T, Amini S, Ferrante P, Jacobson JM, Kashanchi F, Khalili K. 2018. Removal of HIV DNA by CRISPR from patient blood engrafts in humanized mice. Mol Ther Nucleic Acids 12:275–282. doi: 10.1016/j.omtn.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Diemen FR, Kruse EM, Hooykaas MJ, Bruggeling CE, Schurch AC, van Ham PM, Imhof SM, Nijhuis M, Wiertz EJ, Lebbink RJ. 2016. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog 12:e1005701. doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roehm PC, Shekarabi M, Wollebo HS, Bellizzi A, He L, Salkind J, Khalili K. 2016. Inhibition of HSV-1 replication by gene editing strategy. Sci Rep 6:23146. doi: 10.1038/srep23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.World Health Organization. 2018. Finding a cure for hepatitis B: are we close? http://www.who.int/hepatitis/news-events/hbv-cure-overview/en.
- 58.Li H, Sheng C, Wang S, Yang L, Liang Y, Huang Y, Liu H, Li P, Yang C, Yang X, Jia L, Xie J, Wang L, Hao R, Du X, Xu D, Zhou J, Li M, Sun Y, Tong Y, Li Q, Qiu S, Song H. 2017. Removal of integrated hepatitis B virus DNA using CRISPR-Cas9. Front Cell Infect Microbiol 7:91. doi: 10.3389/fcimb.2017.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott T, Moyo B, Nicholson S, Maepa MB, Watashi K, Ely A, Weinberg MS, Arbuthnot P. 2017. ssAAVs containing cassettes encoding SaCas9 and guides targeting hepatitis B virus inactivate replication of the virus in cultured cells. Sci Rep 7:7401. doi: 10.1038/s41598-017-07642-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wollebo HS, Bellizzi A, Kaminski R, Hu W, White MK, Khalili K. 2015. CRISPR/Cas9 system as an agent for eliminating polyomavirus JC infection. PLoS One 10:e0136046. doi: 10.1371/journal.pone.0136046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kennedy EM, Kornepati AV, Goldstein M, Bogerd HP, Poling BC, Whisnant AW, Kastan MB, Cullen BR. 2014. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol 88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li L, He ZY, Wei XW, Gao GP, Wei YQ. 2015. Challenges in CRISPR/CAS9 delivery: potential roles of nonviral vectors. Hum Gene Ther 26:452–462. doi: 10.1089/hum.2015.069. [DOI] [PubMed] [Google Scholar]
- 63.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. 2018. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 64.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bikard D, Barrangou R. 2017. Using CRISPR-Cas systems as antimicrobials. Curr Opin Microbiol 37:155–160. doi: 10.1016/j.mib.2017.08.005. [DOI] [PubMed] [Google Scholar]