Voxilaprevir is a direct-acting antiviral agent (DAA) that targets the NS3/4A protease of hepatitis C virus (HCV). High sequence diversity of HCV and inadequate drug exposure during unsuccessful treatment may lead to the accumulation of variants with reduced susceptibility to DAAs, including NS3/4A protease inhibitors such as voxilaprevir.
KEYWORDS: clinical isolates, direct-acting antivirals, hepatitis C virus, pangenotypic, protease inhibitor, resistance, resistance-associated substitutions, voxilaprevir
ABSTRACT
Voxilaprevir is a direct-acting antiviral agent (DAA) that targets the NS3/4A protease of hepatitis C virus (HCV). High sequence diversity of HCV and inadequate drug exposure during unsuccessful treatment may lead to the accumulation of variants with reduced susceptibility to DAAs, including NS3/4A protease inhibitors such as voxilaprevir. The voxilaprevir susceptibility of clinical and laboratory strains of HCV was assessed. The NS3 protease regions of viruses belonging to 6 genotypes and 29 subtypes from 345 DAA-naive or -experienced (including protease inhibitor) patients and 344 genotype 1 to 6 replicons bearing engineered NS3 resistance-associated substitutions (RASs) were tested in transient-transfection assays. The median voxilaprevir 50% effective concentration against NS3 from protease inhibitor-naive patient samples ranged from 0.38 nM for genotype 1 to 5.8 nM for genotype 3. Voxilaprevir susceptibilities of HCV replicons with NS3 RASs were dependent on subtype background and the type and number of substitutions introduced. The majority of RASs known to confer resistance to other protease inhibitors had little to no impact on voxilaprevir susceptibility, except A156L, T, or V in genotype 1 to 4 which conferred >100-fold reductions but exhibited low replication capacity in most genotypes. These data support the use of voxilaprevir in combination with other DAAs in DAA-naive and DAA-experienced patients infected with any subtype of HCV.
INTRODUCTION
Chronic hepatitis C virus (HCV) infection is a worldwide health problem causing significant death and morbidity (1); the global prevalence of HCV was estimated to be 1% in 2015, corresponding to 71.1 million individuals with chronic HCV infection (2, 3). In recent years, tremendous resources have been directed toward discovery and development of novel direct-acting antiviral agents (DAAs) to treat HCV infection, including inhibitors of the NS3/4A protease. Seven protease inhibitors (PIs) have been approved in the United States and Europe for HCV treatment, namely, telaprevir (TPR) (4, 5), boceprevir (BOC) (6, 7), simeprevir (SIM) (8, 9), paritaprevir/r (10), grazoprevir (GZR) (11), voxilaprevir (VOX) (12, 13), and glecaprevir (GLE) (14). Voxilaprevir (15) and GLE (16) are the most recently approved, pangenotypic PIs with improved potency and a high barrier to resistance in combination with other DAAs.
Similar to other RNA-dependent RNA polymerases, the HCV NS5B polymerase has low fidelity, which, combined with the high replication rate of the virus, results in high genetic variability and adaptability (17). Naturally occurring variants of HCV with reduced susceptibility to inhibitors of NS3/4A protease, the NS5A protein, and the NS5B polymerase have been described (18, 19) and may affect treatment outcome (20). For example, the presence of the NS3 Q80K polymorphism at baseline is associated with reduced sustained viral response (SVR) rates in some patients treated with regimens, including SIM (8, 9, 21). Additionally, inadequate drug exposure during unsuccessful treatment may lead to the selection of viral variants with resistance-associated substitutions (RASs) with reduced susceptibility to DAAs.
Specific substitutions in NS3 have been associated with resistance to NS3/4A PIs. Mutations at position 156 confer reduced susceptibility to all known PIs, due to disruption of the drug binding site. However, the viral fitness of A156 variants is significantly lower than that of other resistant variants, and thus, they are not frequently observed in HCV-infected patients (22). RASs in NS3 at positions 155 and 156 were initially described as signature mutations for TPR and BOC (23, 24). More-recently approved PIs, including the macrocyclic drugs SIM and asunaprevir (ASU), have resistance profiles that are similar to TPR and BOC and additionally select for mutations at position 168 (25, 26). The macrocyclic PI GZR has decreased interactions with arginine 155, leading to lower impact on susceptibility of R155K (27, 28).
Voxilaprevir is a potent pangenotypic HCV NS3/4A PI with in vitro 50% effective concentration (EC50) values ranging from 0.33 nM to 6.1 nM in cells stably transfected with HCV replicons of genotypes (GT) 1 through 6, including subtypes 1a, 1b, 2a, 2b, 3a, 4a, 5a, 6a, and 6e (15). VOX susceptibility is reduced by less than 5-fold by nearly all RASs tested at several amino acid positions in GT 1 to 4, except R155W and A156V or T, which conferred a >38-fold reduction in VOX susceptibility (15). In a phase 1 clinical study of VOX monotherapy, A156T and V emerged in 5 patients with GT 1 (subtypes 1a and 1b) but not other GTs (15).
In this study, we characterized the resistance profile of VOX using three sets of HCV replicons, constructed using (i) 345 patient-derived NS3 protease regions from patients enrolled in the sofosbuvir/velpatasvir/VOX clinical development program, (ii) 49 synthesized NS3 protease regions containing RASs observed at low frequencies in clinical isolates, and (iii) 344 mutant replicons with RASs introduced by site-directed mutagenesis. The relationship between the presence of RASs and in vitro susceptibility to VOX and other antivirals in HCV from clinical isolates was investigated.
MATERIALS AND METHODS
Clinical specimens.
Plasma specimens were collected from 345 patients before initiation of treatment in the following clinical studies: GS-US-338-1121 (15, 29), GS-US-337-1468 (30, 31), GS-US-367-1168 (32), GS-US-367-1169 (33), GS-US-367-1871 (34), GS-US-342-1138 (35), GS-US-367-1170 (14), GS-US-367-1171 (14), GS-US-367-1172 (36), and GS-US-367-1173 (36). These patients were infected with HCV encompassing 6 GTs and 29 subtypes. In several of these clinical studies, prior exposure to DAAs (including PIs other than VOX) was permitted.
All studies were conducted in accordance with the Declaration of Helsinki, Good Clinical Practice guidelines, and local regulatory requirements. All patients provided written informed consent.
Compounds.
VOX (GS-9857), velpatasvir (VEL; GS-5816), ledipasvir (LDV; GS-5885), and sofosbuvir (SOF; GS-7977) were synthesized by Gilead Sciences, Inc. (Foster City, CA, USA).
Sequence analysis.
Deep sequencing was performed by DDL Diagnostic Laboratory (Rijswijk, The Netherlands) using the MiSeq platform (Illumina, San Diego, CA). Internally developed software (Gilead Sciences) was used to process and align sequencing data. The presence of RASs was established by comparison with the following wild-type reference sequences: H77 (GenBank accession number AF009606) for subtype 1a (37), Con1 (GenBank accession number AJ238799) for 1b (38), JFH1 (GenBank accession number AB047639) for 2a and other genotype 2 subtypes other than 2b (39), MD2b10 (GenBank accession number AY232748) for 2b (40), S52 (GenBank accession number GU814263) (subtype 3a) for GT 3 (40), ED43 (GenBank accession number GU814265) (subtype 4a) for GT 4 (40), SA13 (GenBank accession number AF064490) for 5a (41), and EUHK2 (GenBank accession number Y12083) (subtype 6a) for GT 6 (42).
HCV GT and subtype were determined by an analysis of NS3, NS5A, and NS5B sequences with the Basic Local Alignment Search Tool (BLAST) against a panel of standard subtype reference sequences recommended by the U.S. FDA.
NS3 RASs were defined as substitutions previously shown to confer reduced susceptibility (>2.5-fold change in EC50 compared with a GT specific reference) to any approved PI in a replicon model or that emerged in patients with virologic failure at the time of relapse. RASs include the following substitutions (using the Con1 subtype 1b as the reference amino acid): V36A/G/M/L/M, Q41R, F43L/S, T54A/C/G/S, V55A/I, Q80K/R/L, S122R, R155C/G/K/M/T/Q/S/W, A156F/G/N/P/T/V/S, D168A/E/F/G/H/I/N/K/L/P/V/T/Y, and V170A/T/L/F/V.
Generation of subtype 1b, 2a, 2b, and 6a NS3 replicon vectors.
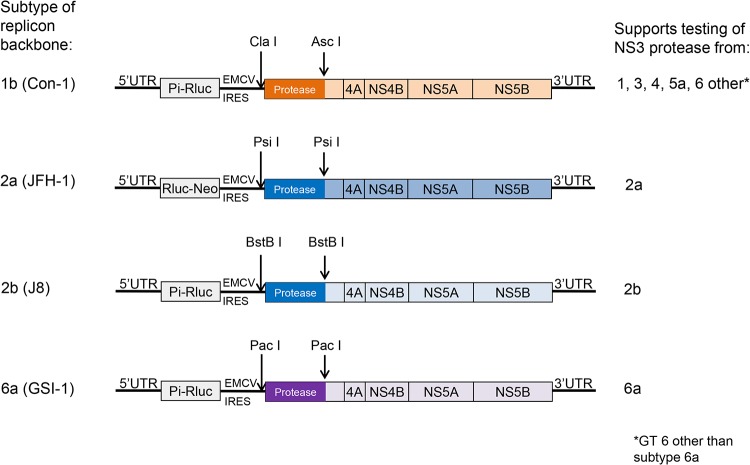
Replicons derived from subtype 1b (Con1, GenBank accession number AJ238799) (38), 2a (JFH1, GenBank accession number AB047639) (43), 2b (J8, GenBank accession number D10988), and 6a (GSI6a-1, GenBank accession number MF683840) (44) were designed and used as backbones for inserting amplified patient-derived sequences corresponding to the NS3 protease region (for a detailed description see Supplemental Material). The subtype 1b NS3 replicon vector that was described previously (45) was modified by the addition of two restriction enzyme sites, namely, ClaI (position 2856) and AscI (position 3159) (Fig. 1). A frameshift was introduced by deleting 1 nucleotide in NS3 between the ClaI and AscI sites. This frameshift disrupts the replication of the vector, unless a desired NS3 fragment is inserted in-frame. The subtype 2a (JFH-1 strain), 2b (J8), and 6a (GSI6a-1) NS3 replicon vectors were generated similarly using various restriction enzyme sites designed for patient sequence transfer (Fig. 1). The replicon vectors were completely sequenced to confirm the presence of the desired mutations and absence of any unintended mutations. Mutations introduced to create the restriction sites and frameshifts were reverted back to wild-type sequence during the patient sequence transfer process (see below).
FIG 1.
NS3 replicon vectors for susceptibility testing of GT 1 through 6 patient samples. Restriction sites used for transfer of the NS3 protease region from patient-derived viruses are indicated by arrows. UTR, untranslated region; Pi-Rluc, poliovirus internal ribosome entry site (IRES)-Renilla luciferase; Rluc-Neo, Renilla luciferase-aminoglycoside phosphotransferase (neomycin resistance) fusion protein; EMCV IRES, internal ribosome entry site from encephalomyocarditis virus.
Construction of chimeric replicons carrying NS3 protease regions from lab strains.
The PCR-amplified NS3 protease regions from lab strains representing GT 1 to 6 were inserted into replicon vectors of various subtypes using the In-Fusion HD EcoDry cloning kit (Clontech, Mountain View, CA) according to the supplier’s instructions. As part of the In-Fusion procedure, the primers were designed to revert mutations that created the vector restriction sites and frameshifts. A single clone for each subtype was selected and sequenced to verify that it matched the original cDNA.
Construction of chimeric replicons carrying NS3 protease regions from clinical isolates.
Total RNA isolation from patient plasma samples, NS3 cDNA synthesis, and PCR amplification were performed by DDL Diagnostic Laboratory (Rijswijk, The Netherlands). A second PCR amplification of the first 181 amino acids of NS3 protease region was carried out using semisubtype-specific PCR primers (see Supplemental Material) and the high fidelity PCR master kit (Roche Diagnostics, Dallas, USA). PCR products were purified, and 50 ng of insert DNA was recombined with 200 ng of vector DNA (of a compatible subtype, see Results) using the In-Fusion HD EcoDry cloning kit (Clontech, Mountain View, CA) according to the supplier’s instructions. The plasmid DNA was isolated using midi kits (Qiagen, Germantown, MD).
RASs that were detected at low frequency (1% to 10%) in clinical isolates by deep sequencing but not already represented in the panel of 347 site-directed mutants (see below) were introduced into replicons using either a cDNA derived from the same clinical isolate or the subtype-specific reference sequence as the backbone. Forty-nine such mutants were constructed by short fragment gene synthesis (see Table S1 in the supplemental material). The NS3 region of each newly generated chimeric replicon was sequenced to confirm that no unintended mutations were introduced.
Generation of NS3 chimeric replicons with single and multiple mutations.
A panel of 344 single and multiple RASs was created by site-directed mutagenesis of subtype 1a, 1b, 2a, 2b, 3a, 4a, or 6a nonchimeric replicons or the 5a/1b NS3 chimera. RASs were selected for inclusion in the panel based on (i) prior demonstration of impact on susceptibility to one or more HCV PIs, (ii) observed in PI-naive patients at known resistance-associated variant positions in the Gilead database, (iii) observed in patients who failed prior PI treatment to any HCV PI, or (iv) single or multiple substitutions that were observed in in vitro resistance selection studies to VOX.
Phenotypic susceptibility testing.
Transient transfections were performed as previously described (15, 46, 47). Briefly, RNA was synthesized from the linearized DNA using the Promega T7 RiboMAX express large scale RNA production system (Madison, WI) according to the supplier’s instructions. Replicon RNA was transfected into “cured” Huh-7 cells following the method of Lohmann et al. (38) in the presence of a range in drug concentration. Luciferase activity in the absence of drug was measured 96 hours after transfection and was used as an indicator of replication capacity. EC50 values were calculated using Prism version 6 (GraphPad, La Jolla, CA) by nonlinear regression analysis. The mean EC50 was calculated from at least 2 experiments. The fold change in EC50 for site-directed mutants was calculated as the ratio to the corresponding subtype parental replicon EC50.
RESULTS
Compatibility of NS3 protease regions from clinical isolates from GT 1 through 6 with subtype 1b, 2a, 2b, and 6a replicon vectors.
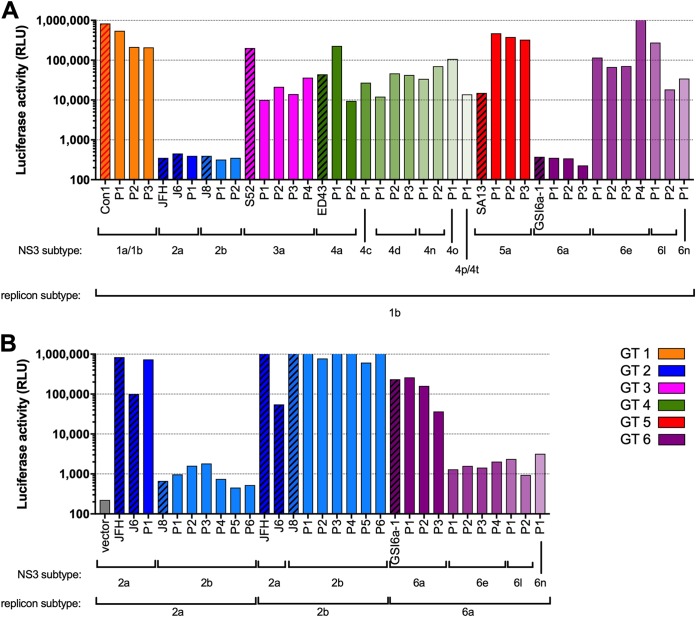
The replication of chimeric replicons containing NS3 protease regions from various subtypes of GT 1 through 6, as measured by the levels of luciferase activity in the absence of inhibitors, was tested to confirm the compatibility between the NS3 protease region and replicon vector of different subtypes (Fig. 2). The replication of subtype 1b-based chimeric replicons was sufficient to enable phenotypic testing of a sampling of patient-derived NS3 protease regions from subtypes 1a, 1b, 3a, 5a, and multiple subtypes of GT 4 and 6 (Fig. 2A). However, replicons containing the NS3 protease region from subtypes 2a, 2b, and 6a did not replicate in the subtype 1b backbone (Fig. 2A); therefore, new replicon vectors derived from subtypes 2a, 2b, and 6a were generated (see Materials and Methods and Fig. 1). High replication was observed for chimeric replicons containing NS3 protease regions from both lab strains and patient isolates using subtype-matched replicon vectors (Fig. 2B). Based on these results, the subtype 1b-based replicon vector was used for all subsequent testing of NS3 from subtypes other than 2a, 2b, or 6a, for which the subtype-matched vector was used.
FIG 2.
Luciferase activity of replicon constructs containing lab strain (hatched fill) or patient-derived (solid fill) NS3 protease regions. (A) Genotype 1b (Con1) vector backbone. (B) Genotype 2a, 2b, and 6a backbones. The background luciferase activity of the NS3 frameshift (inactive) vector is shown in a gray bar.
Subgenomic, chimeric replicon constructs containing NS3 from subtypes 1a (H77), 3a (S52), and 4a (ED43) in the subtype 1b (Con1) backbone had similar VOX EC50 (1.8, 6.4, and 0.82 nM, respectively) compared with the native (nonchimeric) NS3 to NS5B replicons for the corresponding subtype (1.8, 4.2, and 1.1 nM, respectively). This result indicates that a mismatch between the subtype of the NS3 region and the replicon vector does not impact VOX susceptibility.
VOX susceptibility of GT 1 through 6 HCV clinical isolates from PI-naive patients.
The NS3 protease regions (amino acid 1 to 181) from 345 clinical isolates were transferred to replicon vectors and tested in the transient-transfection system. Among those, luciferase activity from 332 replicons was sufficient for VOX susceptibility determination, including NS3 from 6 GTs (for details, see Table S2 in the supplemental material), and 81 DAA-experienced (but PI-naive) patients.
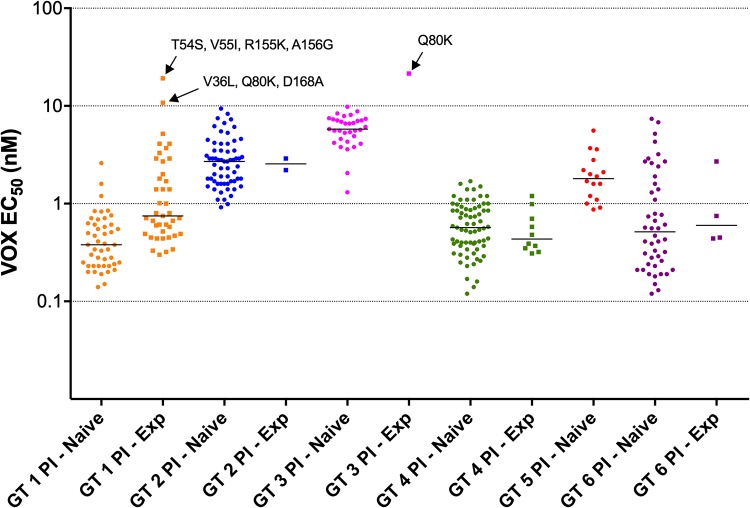
The median VOX EC50 values for the 274 clinical isolates belonging to PI-naive patients infected with GT 1, GT 2, GT 3, GT 4, GT 5, and GT 6 were 0.38, 2.7, 5.8, 0.57, 1.8, and 0.52 nM, respectively (Fig. 3; see Table S3 in the supplemental material). For comparison, the VOX EC50 of replicon constructs with lab strain NS3 from subtypes 1a, 1b, 2a, 2b, 3a, 4a, 5a, and 6a were 3.9 nM, 3.3 nM, 3.7 nM, 6.6 nM, 6.1 nM, 2.9 nM, 1.9 nM, and 3.0 nM, respectively. The ratio between the 95th and 5th percentiles of VOX EC50 values among GT 1 to 5 ranged from 2.9- to 7-fold, compared with 31.6-fold for GT 6.
FIG 3.
VOX susceptibility (EC50, nM) of replicons containing GT 1 to 6 patient NS3 protease regions. Samples are grouped by genotype and whether or not the patient had previously been treated with PIs (squares) or not (circles). Median EC50 for each group is shown by the horizontal bars. There were no subtype 5a, PI-experienced patients. The RASs present in 3 samples with EC50 of >10 nM are indicated (all RASs present in >90% of sequence reads).
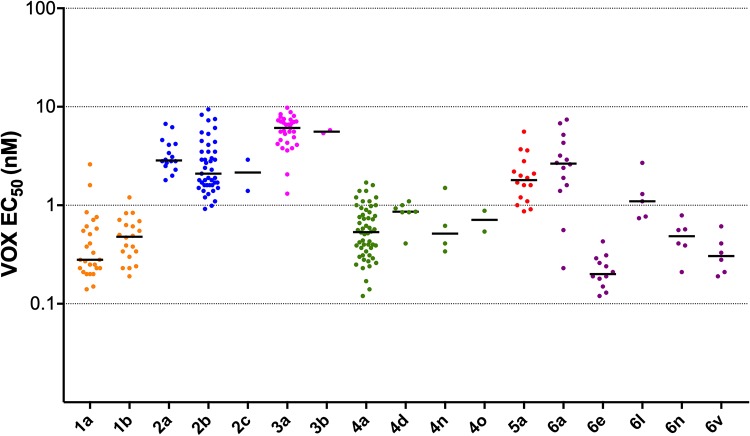
VOX EC50 values grouped by subtype are shown in Fig. 4 for each subtype with at least 2 patients. Median VOX EC50 ranged from 0.2 nM (subtype 6e, n = 12) to 6.1 nM (subtype 3a). The maximum VOX EC50 observed was 9.8 nM.
FIG 4.
VOX susceptibility (EC50, nM) of replicons containing GT 1 to 6 patient NS3 protease regions from PI-naive patients. Median EC50 for each group is shown by the horizontal bars. Samples are grouped by subtype. Results are shown for subtypes with at least 2 data points.
It is possible that associations between elevated VOX EC50 and the presence of RASs might be obscured because the prevalence of RASs is low within a particular sample. However, we used deep sequencing mutation prevalence data to focus on the 15 samples from PI-naive patients that contained a substitution at position 155, 156, or 168 in at least 50% of sequencing reads. The median VOX EC50 for these samples was 2.1 nM, ranging from 0.38 (a subtype 1a sample with Q80K and R155K) to 8.8 nM (a subtype 3a sample with Q168K).
VOX susceptibility of GT 1 through 6 HCV clinical isolates from PI-experienced patients.
VOX EC50 was determined from 58 replicons containing the NS3 protease region from PI-experienced patients. The median VOX EC50 values for these samples, grouped by genotype (for groups with more than 2 results each), were 0.75, 0.44, and 0.60 nM for GT 1, 4 and 6, respectively (Fig. 3; Table S3). Three samples had EC50 over 10 nM, namely, a subtype 1a sample with changes at positions 155 and 156 (T54S/V55I/R155K/A156G; EC50, 19 nM), a subtype 1a sample with D168A (V36L/Q80K/D168A; EC50, 11 nM), and a subtype 3a sample with Q80K as the only RAS (EC50, 22 nM) (Fig. 3; Table S2). Since HCV containing NS3 RASs often has impaired fitness (48–50) and the timing of sampling in PI-experienced patients with respect to when they stopped their prior therapy was variable, many NS3 protease regions from PI-experienced patients did not contain detectable RASs at conserved sites. In total, 23 clinical isolates from PI-experienced patients had NS3 with one or more NS3 RAS at positions 155, 156, or 168 in at least 50% of sequencing reads, including R155K, A156G, or D168A, E or V in various combinations with each other, or other NS3 RASs. VOX EC50 among these samples ranged from 0.3 to 19 nM. Other than the 2 subtype 1a outliers described above, the next highest EC50 was 5.2 nM (Table 1).
TABLE 1.
Patient-derived NS3 samples with RAS at positions 155, 156, or 168
| Subtype | RAS(s) (%) | Additional resistance-associated polymorphisms (%) | VOX EC50 (nM) (mean ± SD) |
|---|---|---|---|
| 1a | V36L (>99), R155K (99.0) | Q80N (>99) | 3.3 ± 1.1 |
| 1a | Q80K (>99), R155K (>99) | 2.0 ± 0.03 | |
| 1a | V36M (>99), R155K (>99), I170V (57.9) | 4.1 ± 0.95 | |
| 1a | D168A (95.5), I170T (1.1), I170V (3.7) | 2.7 ± 0.94 | |
| 1a | Q80K (>99), R155K (84.4) | 1.8 ± 0.51 | |
| 1a | V36M (>99), R155K (>99) | 5.2 ± 0.10 | |
| 1a | R155K (>99) | 0.30 ± 0.08 | |
| 1a | Q80K (>99), D168E (>99) | 0.71 ± 0.25 | |
| 1a | T54S (>99), V55I (16.2), R155K (81.3) | S122G (1.1) | 0.60a |
| 1a | T54S (>99), V55I (94.4), R155K (98.1), A156G (>99) | 19 ± 0.66 | |
| 1a | Q80L (>99), D168E (57.4) | 0.80 ± 0.25 | |
| 1a | V55A (>99), Q80R (92.8), D168E (98.4), I170V (98.3) | 1.4 ± 0.15 | |
| 1a | Q80K (>99), D168E (51.4), I170V (>99) | 4.1 ± 0.84 | |
| 1a | S122R (>99), R155N (11.2), D168E (>99) | 0.47 ± 0.04 | |
| 1a | Q80K (>99), D168E (>99) | 1.0 ± 0.04 | |
| 1a | V36L (>99), Q80K (>99), D168A (>99) | 11 ± 0.63 | |
| 1a | V36L (>99), R155K (>99), I170V (>99) | 0.67 ± 0.13 | |
| 1b | Q80L (>99), D168V (>99) | 0.45 ± 0.19 | |
| 1b | Q80R (94.4), D168E (>99) | 2.9 ± 1.3 | |
| 1b | D168E (>99) | V170I (>99) | 1.0 ± 0.04 |
| 1b | F43L (1.3), Q80R (31.6), R155L (1.0), D168E (98.3) | 1.4 ± 0.26 | |
| 4d | R155Q (1.4), D168V (91.9) | 0.58 ± 0.19 | |
| 6a | Y56H (10.7), L80K (98.2), D168E (>99) | 2.7 ± 0.53 |
EC50 value derived from a single experiment.
VOX susceptibility in a panel of gene-synthesized NS3 constructs containing multiple mutations observed in VOX clinical studies.
The NS3 protease region from several patient samples (both PI-naive and PI-experienced) contained one or more RAS in a low proportion of sequence reads. Phenotypic assays may underestimate the impact of RASs if their prevalence in the pool of replicon constructs derived from that sample is low; the threshold is thought to be variable depending on the magnitude of the effect and the fitness of the mutant compared with drug-susceptible variants (51). To evaluate the VOX susceptibility of the RASs present at low frequency in clinical isolates, 49 NS3 genes containing different patterns of RASs were synthesized and cloned into the replicon vectors of the appropriate subtype (Table S1). Thirty-six of the 49 replicon constructs replicated well enough to enable VOX EC50 determination in the transient-transfection susceptibility assay (Table 2). The fold change in EC50 (FC) was calculated by comparison to the corresponding parental replicon containing the lab strain NS3 protease region of the same subtype or a clone from the same patient.
TABLE 2.
Synthetic constructs with NS3 RAS observed in patient samples
| Subtype | Backbone | RAS(s) detected by deep sequencing (%) | NS3 SDM in construct | VOX EC50-fold change (mean ± SD) |
|---|---|---|---|---|
| 1a | H77 | R155K (65.7), I170V (>99) | R155K/I170V | 1.2 ± 0.3 |
| 1a | H77 | Q41K (1.5), Q80K (>99) | Q41K/Q80K | 2.8 ± 1.2 |
| 1a | H77 | F43L (1.1), Q80K (>99) | F43L/Q80K | 3.4 ± 0.62 |
| 1a | H77 | S122G (>99), R155K (3.0) | S122G/R155K | 6.2 ± 0.95 |
| 1a | H77 | Q80L (>99), D168E (57.4) | Q80L/D168E | 11 ± 2.2 |
| 1a | H77 | Q80K (98.7), D168Y (1.3) | Q80K/D168Y | 117 ± 13 |
| 1a | H77 | V55A (>99), D168E (3.6), I170V (1.5) | V55A/D168E/I170V | 1.5 ± 0.25 |
| 1a | H77 | Q41H (1.2), Q80K (>99), S122N (13.%) | Q41H/Q80K/S122N | 1.9 ± 0.33 |
| 1a | H77 | Q41H (10.1), Q80K (>99), S122G (4.9) | Q41H/Q80K/S122G | 2.0 ± 0.78 |
| 1a | H77 | V36M (>99), R155K (>99), I170V (57.9) | V36M/R155K/I170V | 3.6 ± 0.53 |
| 1a | H77 | S122G (3.0), R155K (29.2), I170V (65.6) | S122G/R155K/I170V | 5.2 ± 0.94 |
| 1a | H77 | T54S (86.8), V55I (87.3), Q80L (85.8) | T54S/V55I/Q80L | 7.0 ± 1.5 |
| 1a | H77 | Q80K (>99), D168E (51.4), I170V (>99) | Q80K/D168E/I170V | 8.0 ± 3.4 |
| 1a | H77 | S122R (>99), R155N (11.2), D168E (>99) | S122R/R155N/D168E | 11 ± 0.09 |
| 1a | H77 | T54S (>99), Y56H (1.6), I170V (>99) | T54S/Y56H/I170V | 11 ± 2.2 |
| 1a | H77 | T54S (29.5), V55A (3.6), Q80R (2.0), I170V (1.2) | T54S/V55A/Q80R/I170V | 1.3 ± 0.04 |
| 1a | H77 | V55A (>99), Q80R (92.8), D168E (98.4), I170V (98.3) | V55A/Q80R/D168E/I170V | 4.9 ± 0.06 |
| 1a | H77 | T54S (>99), V55I (16.2), S122G (1.1), R155K (81.3) | T54S/V55I/S122G/R155K | 5.9 ± 0.45 |
| 1a | H77 | T54S (>99), S122G (1.3), R155K (>99), I170V (1.1) | T54S/S122G/R155K/I170V | 9.2 ± 3.2 |
| 1a | H77 | Q41K (2.5), F43L (1.3), Q80K (98.9), D168Y (1.7) | Q41K/F43L/Q80K/D168Y | 233 ± 66 |
| 1a | H77 | T54S (>99), V55I (64.0), S122G (3.2), R155K (37.6), I170V (7.3) | T54S/V55I/S122G/R155K/I170V | 4.1 ± 0.95 |
| 1b | Con1 | D168N (1.0), V170I (>99) | D168N/V170I | 0.7 ± 0.01 |
| 1b | Con1 | Y56F (>99), D168Y (1.2) | Y56F/D168Y | 0.8 ± 0.03 |
| 1b | Con1 | Y56F (>99), Q80R (87.9) | Y56F/Q80R | 1.1 ± 0.41 |
| 1b | Con1 | Y56F (>99), V170I (95.9), V170T (2.1) | Y56F/V170T | 1.1 ± 0.09 |
| 1b | Con1 | Y56F (>99), V170I (95.9), V170T (2.1) | Y56F/V170I | 1.6 ± 0.48 |
| 1b | Con1 | Y56F (>99), S122N (1.0) | Y56F/S122N | 3.4 ± 0.62 |
| 1b | Con1 | Y56F (98.9), S122N (71.1), S122T (28.1), V170I (>99) | Y56F/S122T/V170I | 1.7 ± 0.02 |
| 1b | Con1 | Y56F (98.9), S122N (71.1), S122T (28.1), V170I (>99) | Y56F/S122N/V170I | 3.3 ± 0.36 |
| 2b | MD2b-1 | Y56F (>99), R122G (1.4) | Y56F/R122G | 0.9 ± 0.41 |
| 2b | MD2b-1 | L36M (1.6), F43L (3.1), G80V (1.9), G80W (2.1), R122L (1.5), R155M (2.3), R155S (2.1), D168Y (7.4), L175I (2.4) | L36M/F43L/G80W/R122L/R155M/D168Y/L175I | 0.8 ± 0.07 |
| 4a | Patient | F43S (1.7), Q80K (1.0), D168Y (1.1) | F43S/Q80K/D168Y | 1.0 ± 0.06 |
| 4d | Patient | R155Q (1.4), D168V (91.9) | R155Q/D168V | 1.0 ± 0.38 |
| 6a | Patient | L80K (98.2), D168E (>99) | L80K/D168E | 1.1a |
| 6a | Patient | V55A (1.1), L80K (>99), S122N (98.5) | V55A/L80K/S122N | 2.0 ± 0.04 |
EC50 value derived from a single experiment.
The median FC values among the synthetic NS3 constructs with 2 (n = 15), 3 (n = 14), 4 (n = 5), or >4 (n = 2) RASs were 1.2-, 2.6-, 5.9-, and 2.4-fold, respectively. Eighteen of them had an FC of >2.5 and only 2 had an FC of >20; all constructs with these elevated FC belonged to subtype 1a. One of them had Q80K and D168Y (117-fold), while the second had Q41H/F43L/Q80K/D168Y (233-fold) (Table 2). Four other constructs also had D168Y but had an FC of ≤1.0 (Y56F/D168Y in subtype 1b, L36M/F43L/G80W/R122L/R155M/D168Y/L175I in 2b, and F43S/Q80K/D168Y in 4a, lab strain or patient clone).
VOX susceptibility of a panel of replicons with engineered NS3 RASs.
A panel of 344 NS3 mutants, including 233 single, 86 double, and 25 triple mutants was generated by site-directed mutagenesis in replicons derived from GT 1 to 6. Of the 344 mutants, 284 mutants replicated well enough to enable susceptibility determination, including 189 single, 73 double, and 22 triple mutants. Most (172 of 189, 91.0%) of the single mutants demonstrated ≤2.5-fold (133 of 189, 70.4%) or 2.5- to 20-fold (39 of 189, 20.6%) reductions in VOX susceptibility (Table 3). Patterns of RASs that conferred 20- to 100-fold reduction in EC50 included R155W, D168K, L, or R in GT 1 and some combinations of 2 RASs, including D168A, E, or H in subtypes 5a and 6a (Table 3). Combinations of RASs that conferred a >100-fold reduction in EC50 included A156L, T, or V in multiple subtypes or Y56H/Q168R in subtype 3a (see Table S5 and S6 in the supplemental material).
TABLE 3.
Levels of resistance conferred by GT 1 NS3 single mutants
| Subtype | Mutant by fold-change category: |
|||
|---|---|---|---|---|
| ≤2.5 | 2.5–20 | 20–100 | >100 | |
| 1a | V36A/I/L/M, Q41H/K/L, F43L, T54A/C/G/S, V55A/I, Y56F/H, Q80K/L/M/N/R, V107I, S122A/C/G/N/P/R/T/V, I132V, R155A/K/T, A156G/S, V158A/I, A166S, D168E/G/H/N/S/Y, I170F/T/V, L175M | V36G, Q41R, F43S, R155G, D168A/F/I/T/V | R155W, D168K/L/R | A156L/T |
| 1b | V36I/A/S, V55A, Y56F, Q80K/L/M/R, V107I, S122G/N/T, R155K, V158I, A166S, D168A/E/G, V170I/T, M175L | V36A/M, S122D, A156S, D168V/Y, V170A | R155W | A156T/V |
| 2a | L36M, Q41V, V55S, Y56F, G80E/T, K122I/R/T, A156G, A166S, D168E/K/S/V | F43V | A156T | A156L/V |
| 2b | V55I, Y56F, A166S, D168Y | None | None | None |
| 3a | K26R, Q41R, T54A, Q80R, S122A/C/F/T/Y, R155K, Q168H/K/R, V170I | Q41K, Q80K, L175M | None | A156T/V |
| 4a | Q41H, T54S, T122A/N/S/V, R155C/K, A166S, D168K, V170I/L | Q41R, D168E/T/V | None | A156L/T/V |
| 5a | T122A/G/V, A166S, D168E/V, I170V | D168A/H/K/R/Y | None | None |
| 6a | V36A, V55A, L80K/L/Q/R, S122A/D/G/N/T, D168E/V/Y, I170V | Q41K, Q41R, Y56H, D168A, D168H | None | None |
The following substitutions, when introduced on their own, did not result in significant reductions in VOX susceptibility (≤2.5-fold change) in any subtype: 54A/C/G/S, 55A/I/S, 56F, 80E/L/M/N/Q/R/T, 107I, 122A/C/F/G/I/N/P/S/T/V/Y, 132V, 155A/C/K/T, 156G, 158A/I, 166S, 168G/S, and 170F/I/L/T/V. Substitutions at positions 36 (A/D/G/I/L/M), 41 (H/K/L/R/V), 43 (L/S/V), 56 (H), 80 (K), 155 (G/W), 122 (D), 156 (S), 168 (A/H/I/T/V/Y), 170 (A), and 175 (M) conferred ≤20-fold reduced susceptibility to VOX. Substitutions at positions 155 (W), 156 (T), and 168 (K/L/R) conferred 20- to 100-fold reduced susceptibility to VOX in some subtype backbones. Lastly, variants with RASs at position 156 (A156L/T/V) demonstrated >100-fold reduced susceptibility to VOX (Table 3). Of the 13 mutants with >100-fold reductions in susceptibility, 12 contained A156L, T, or V in subtypes 1a, 1b, 2a, 3a, or 4a; the remaining one had Y56H/Q168R in subtype 3a. These replicons had replication capacity ranging from 0.4% to 53% in genotypes 1, 2, and 3 (Table S5).
Susceptibility of replicons with NS3 RASs to other classes of DAA.
Subtype 1a and 1b replicons bearing NS3 RASs at positions 36, 43, 54, 55, 80, 122, 155, 156, or 168 were tested for cross-resistance to the nucleoside NS5B inhibitor SOF and the NS5A inhibitors LDV and VEL. Nine of the 38 RASs tested had VOX EC50 FC of >2.5 and 3 had VOX EC50 FC of >10. No cross-resistance (FC, <1.5) was observed for SOF, LDV, or VEL (see Table S7 in the supplemental material).
DISCUSSION
This study describes the in vitro VOX susceptibility of replicons successfully constructed from NS3 protease regions from 332 patients and 284 engineered mutant replicons as measured in transient-transfection assays. Samples tested covered 6 GTs and 29 subtypes of HCV and over 50 RASs in various combinations. To test all these samples, four replicon vectors were engineered. NS3 protease regions derived from clinical isolates with subtype 1a, 1b, 3a, 3b, 5a, and multiple subtypes of GT 4 and 6 (other than 6a) were compatible with the GT 1b replicon, while NS3 protease regions derived from clinical samples with subtype 2a, 2b, and 6a required the use of subtype-matched replicon vectors. VOX demonstrated potent (EC50, <10 nM) antiviral activity across a diverse range of NS3 RAS and HCV clinical isolates, including those with multiple RASs and patients previously treated with a PI.
Susceptibility of replicons containing NS3 from three PI-experienced patients (two with subtype 1a, 1 with subtype 3a) demonstrated relatively high VOX EC50 (>10 nM). In addition, synthetic constructs based on RASs observed in two other subtype 1a patients at low frequency also had elevated VOX EC50. Each NS3 protease region had a different complement of RASs, and only Q80K and D168Y were represented more than once. One sequence had both R155K and A156G, both of which are at positions that can have strong effects on susceptibility to other PIs. However, neither RAS, when introduced in isolation in a subtype 1a replicon, caused significant reduction in VOX susceptibility (FC, <1.5). Similarly, D168A or Y imparted no to small effects (FC, <4) except in a subtype 5a backbone (8.1- to 13.5-fold). The subtype 3a sample from a PI-experienced patient with an isolated Q80K had a relatively elevated EC50 (21.5 nM). The analogous substitution L80K was present in all subtype 6a samples, most of which had VOX EC50 values in the 1 to 10 nM range. The site-directed mutant containing this RAS in the subtype 6a replicon did not display reduced VOX susceptibility compared with the parental replicon (fold change, 0.5). When present as the only RAS in GT 1 patient samples or site-directed mutant replicons, Q80K was not associated with elevated EC50 (<1 nM; FC, <1.0). The Q80K RAS in subtype 3a had a modest (3.5-fold) impact on VOX susceptibility. Taken together, these results indicate a high context-dependence (i.e., influenced strongly by both subtype and presence of other RASs) of the effect of particular RAS on VOX susceptibility. This conclusion is consistent with previous studies of the effect of various RASs on in vitro susceptibility to VOX (15) and other DAAs (52, 53).
Phase 3 clinical trials of coformulated SOF-VEL-VOX demonstrated high rates of SVR (96% to 98%) among patients who were previously treated with DAA regimens, including PIs (14, 36). Among the small number of patients who failed treatment, emergence of resistance was uncommon and none of the virologic failures had emergent NS3 RASs, including the substitutions at position 156 shown here to have large effects on VOX susceptibility. No RAS or combination of RASs present before treatment was associated with reduced SVR rates, nor was the detection of RASs known to confer up to 100-fold resistance in vitro (54). As expected, we found no impact of NS3 RASs, including several with detectable impact on NS3 susceptibility to VOX, on susceptibility to SOF, VEL, or LDV.
In summary, our studies demonstrate potent antiviral activity of VOX in a collection of 332 HCV GT 1 through 6 clinical samples from DAA-naive and -experienced patients treated in the SOF-VEL-VOX phase 3 program, 36 synthesized NS3 genes, and 284 RAS-containing replicons, many of which are known to display large reductions in susceptibility to other PIs. VOX susceptibility among PI-experienced patient isolates was only 5-fold higher on average than that of PI-naive patient isolates. No cross-resistance of NS3 RASs to LDV, VEL, or SOF was observed. The results of this study are consistent with the resistance analyses of the SOF-VEL-VOX phase 3 clinical program, which demonstrated that the regimen has a high barrier to development of DAA resistance and good efficacy without regard to the presence of NS3 RAS (54, 55).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Gilead Sciences, Inc.
Technical writing (manuscript editing and table and figure formatting) was provided by Data First Consulting, Inc. We gratefully acknowledge the patients who participated in the study, the investigators, nursing staff, and research support staff involved in the study, and members of the project teams at Gilead Sciences.
All authors are employees of Gilead Sciences.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01844-18.
[This article was published on 28 March 2019 with a byline that lacked Julia Lu. The byline was updated in the current version posted on 22 April 2020.]
REFERENCES
- 1.Cooke GS, Lemoine M, Thursz M, Gore C, Swan T, Kamarulzaman A, DuCros P, Ford N. 2013. Viral hepatitis and the global burden of disease: a need to regroup. J Viral Hepat 20:600–601. doi: 10.1111/jvh.12123. [DOI] [PubMed] [Google Scholar]
- 2.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. 2013. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 3.Polaris Observatory HCV Collaborators. 2017. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol 2:161–176. doi: 10.1016/S2468-1253(16)30181-9. [DOI] [PubMed] [Google Scholar]
- 4.McHutchison JG, Manns MP, Muir AJ, Terrault NA, Jacobson IM, Afdhal NH, Heathcote EJ, Zeuzem S, Reesink HW, Garg J, Bsharat M, George S, Kauffman RS, Adda N, Di Bisceglie AM, Team PS. 2010. Telaprevir for previously treated chronic HCV infection. N Engl J Med 362:1292–1303. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 5.Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R, George J, Rizzetto M, Shouval D, Sola R, Terg RA, Yoshida EM, Adda N, Bengtsson L, Sankoh AJ, Kieffer TL, George S, Kauffman RS, Zeuzem S, Team AS. 2011. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med 364:2405–2416. doi: 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- 6.Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, Poordad F, Goodman ZD, Sings HL, Boparai N, Burroughs M, Brass CA, Albrecht JK, Esteban R, for the HCV RESPOND-2 Investigators. 2011. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med 364:1207–1217. doi: 10.1056/NEJMoa1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poordad F, McCone J Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, DiNubile MJ, Sniukiene V, Brass CA, Albrecht JK, Bronowicki JP, Investigators S-. 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med 364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forns X, Lawitz E, Zeuzem S, Gane E, Bronowicki JP, Andreone P, Horban A, Brown A, Peeters M, Lenz O, Ouwerkerk-Mahadevan S, Scott J, De La Rosa G, Kalmeijer R, Sinha R, Beumont-Mauviel M. 2014. Simeprevir with peginterferon and ribavirin leads to high rates of SVR in patients with HCV genotype 1 who relapsed after previous therapy: a phase 3 trial. Gastroenterology 146:1669–1679. e1663. doi: 10.1053/j.gastro.2014.02.051. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson IM, Dore GJ, Foster GR, Fried MW, Radu M, Rafalsky VV, Moroz L, Craxi A, Peeters M, Lenz O, Ouwerkerk-Mahadevan S, De La Rosa G, Kalmeijer R, Scott J, Sinha R, Beumont-Mauviel M. 2014. Simeprevir with pegylated interferon alfa 2a plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-1): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet 384:403–413. doi: 10.1016/S0140-6736(14)60494-3. [DOI] [PubMed] [Google Scholar]
- 10.Lawitz E, Sullivan G, Rodriguez-Torres M, Bennett M, Poordad F, Kapoor M, Badri P, Campbell A, Rodrigues L Jr, Hu Y, Pilot-Matias T, Vilchez RA. 2015. Exploratory trial of ombitasvir and ABT-450/r with or without ribavirin for HCV genotype 1, 2, and 3 infection. J Infect 70:197–205. doi: 10.1016/j.jinf.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Kwo P, Gane EJ, Peng CY, Pearlman B, Vierling JM, Serfaty L, Buti M, Shafran S, Stryszak P, Lin L, Gress J, Black S, Dutko FJ, Robertson M, Wahl J, Lupinacci L, Barr E, Haber B. 2017. Effectiveness of elbasvir and grazoprevir combination, with or without ribavirin, for treatment-experienced patients with chronic hepatitis C infection. Gastroenterology 152:164–175.e164. doi: 10.1053/j.gastro.2016.09.045. [DOI] [PubMed] [Google Scholar]
- 12.Poordad F, Felizarta F, Asatryan A, Sulkowski MS, Reindollar RW, Landis CS, Gordon SC, Flamm SL, Fried MW, Bernstein DE, Lin CW, Liu R, Lovell SS, Ng TI, Kort J, Mensa FJ. 2017. Glecaprevir and pibrentasvir for 12 weeks for hepatitis C virus genotype 1 infection and prior direct-acting antiviral treatment. Hepatology 66:389–397. doi: 10.1002/hep.29081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poordad F, Pol S, Asatryan A, Buti M, Shaw D, Hézode C, Lalezari J, Felizarta F, Reindollar RW, Gordon SC, Pianko S, Fried MW, Bernstein DE, Gallant J, Lin C-W, Lei Y, Ng TI, Pilot-Matias T, Kort J, Mensa F. 2017. MAGELLAN-1, Part 2: glecaprevir and pibrentasvir for 12 or 16 weeks in patients with chronic hepatitis C virus genotype 1 or 4 and prior direct-acting antiviral treatment failure, abstr 422. Abstr The International Liver Congress, European Association for the Study of the Liver (EASL), Amsterdam, The Netherlands. [Google Scholar]
- 14.Bourliere M, Gordon SC, Flamm SL, Cooper CL, Ramji A, Tong M, Ravendhran N, Vierling JM, Tran TT, Pianko S, Bansal MB, de Ledinghen V, Hyland RH, Stamm LM, Dvory-Sobol H, Svarovskaia E, Zhang J, Huang KC, Subramanian GM, Brainard DM, McHutchison JG, Verna EC, Buggisch P, Landis CS, Younes ZH, Curry MP, Strasser SI, Schiff ER, Reddy KR, Manns MP, Kowdley KV, Zeuzem S; Polaris-4 Investigators. 2017. Sofosbuvir, velpatasvir, and voxilaprevir for previously treated HCV infection. N Engl J Med 376:2134–2146. doi: 10.1056/NEJMoa1613512. [DOI] [PubMed] [Google Scholar]
- 15.Lawitz E, Yang JC, Stamm LM, Taylor JG, Cheng G, Brainard DM, Miller MD, Mo H, Dvory-Sobol H. 2018. Characterization of HCV resistance from a 3-day monotherapy study of voxilaprevir, a novel pangenotypic NS3/4A protease inhibitor. Antivir Ther 23:325–334. doi: 10.3851/IMP3202. [DOI] [PubMed] [Google Scholar]
- 16.Ng TI, Tripathi R, Reisch T, Lu L, Middleton T, Hopkins TA, Pithawalla R, Irvin M, Dekhtyar T, Krishnan P, Schnell G, Beyer J, McDaniel KF, Ma J, Wang G, Jiang LJ, Or YS, Kempf D, Pilot-Matias T, Collins C. 2018. In vitro antiviral activity and resistance profile of the next-generation hepatitis C Virus NS3/4A protease inhibitor glecaprevir. Antimicrob Agents Chemother 62:e01620-17. doi: 10.1128/AAC.01620-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 18.Bartels DJ, Sullivan JC, Zhang EZ, Tigges AM, Dorrian JL, De Meyer S, Takemoto D, Dondero E, Kwong AD, Picchio G, Kieffer TL. 2013. Hepatitis C virus variants with decreased sensitivity to direct-acting antivirals (DAAs) were rarely observed in DAA-naive patients prior to treatment. J Virol 87:1544–1553. doi: 10.1128/JVI.02294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alves R, Queiroz AT, Pessoa MG, da Silva EF, Mazo DF, Carrilho FJ, Carvalho-Filho RJ, de Carvalho IM. 2013. The presence of resistance mutations to protease and polymerase inhibitors in Hepatitis C virus sequences from the Los Alamos databank. J Viral Hepat 20:414–421. doi: 10.1111/jvh.12051. [DOI] [PubMed] [Google Scholar]
- 20.Sarrazin C, Dvory-Sobol H, Svarovskaia ES, Doehle BP, Pang PS, Chuang SM, Ma J, Ding X, Afdhal NH, Kowdley KV, Gane EJ, Lawitz E, Brainard DM, McHutchison JG, Miller MD, Mo H. 2016. Prevalence of resistance-associated substitutions in HCV NS5A, NS5B, or NS3 and outcomes of treatment with ledipasvir and sofosbuvir. Gastroenterology 151:501–512.e501. doi: 10.1053/j.gastro.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Lawitz E, Matusow G, DeJesus E, Yoshida EM, Felizarta F, Ghalib R, Godofsky E, Herring RW, Poleynard G, Sheikh A, Tobias H, Kugelmas M, Kalmeijer R, Peeters M, Lenz O, Fevery B, De La Rosa G, Scott J, Sinha R, Witek J. 2016. Simeprevir plus sofosbuvir in patients with chronic hepatitis C virus genotype 1 infection and cirrhosis: A phase 3 study (OPTIMIST-2). Hepatology 64:360–369. doi: 10.1002/hep.28422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong X, Chase R, Skelton A, Chen T, Wright-Minogue J, Malcolm BA. 2006. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res 70:28–38. doi: 10.1016/j.antiviral.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Forestier N, Susser S, Welker MW, Weegink CJ, Reesink HW, Zeuzem S, Sarrazin C. 2007. Telaprevir resistance mutations in patients with hepatitis C who relapsed after sequential therapy with telaprevir, peg-interferon alfa 2a and ribavirin, abstr 50 58th Annual Meeting of the American Society for the Study of Liver Diseases, Boston, MA. [Google Scholar]
- 24.Susser S, Welsch C, Wang Y, Zettler M, Domingues FS, Karey U, Hughes E, Ralston R, Tong X, Herrmann E, Zeuzem S, Sarrazin C. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718. doi: 10.1002/hep.23192. [DOI] [PubMed] [Google Scholar]
- 25.Lenz O, Verbinnen T, Lin TI, Vijgen L, Cummings MD, Lindberg J, Berke JM, Dehertogh P, Fransen E, Scholliers A, Vermeiren K, Ivens T, Raboisson P, Edlund M, Storm S, Vrang L, de Kock H, Fanning GC, Simmen KA. 2010. In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435. Antimicrob Agents Chemother 54:1878–1887. doi: 10.1128/AAC.01452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McPhee F, Friborg J, Levine S, Chen C, Falk P, Yu F, Hernandez D, Lee MS, Chaniewski S, Sheaffer AK, Pasquinelli C. 2012. Resistance analysis of the hepatitis C virus NS3 protease inhibitor asunaprevir. Antimicrob Agents Chemother 56:3670–3681. doi: 10.1128/AAC.00308-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romano KP, Ali A, Aydin C, Soumana D, Ozen A, Deveau LM, Silver C, Cao H, Newton A, Petropoulos CJ, Huang W, Schiffer CA. 2012. The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors. PLoS Pathog 8:e1002832. doi: 10.1371/journal.ppat.1002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summa V, Ludmerer SW, McCauley JA, Fandozzi C, Burlein C, Claudio G, Coleman PJ, Dimuzio JM, Ferrara M, Di Filippo M, Gates AT, Graham DJ, Harper S, Hazuda DJ, Huang Q, McHale C, Monteagudo E, Pucci V, Rowley M, Rudd MT, Soriano A, Stahlhut MW, Vacca JP, Olsen DB, Liverton NJ, Carroll SS. 2012. MK-5172, a selective inhibitor of hepatitis C virus NS3/4a protease with broad activity across genotypes and resistant variants. Antimicrob Agents Chemother 56:4161–4167. doi: 10.1128/AAC.00324-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Torres M, Glass S, Hill J, Freilich B, Hassman D, Di Bisceglie AM, Taylor JG, Kirby BJ, Dvory-Sobol H, Yang JC, An D, Stamm LM, Brainard DM, Kim S, Krefetz D, Smith W, Marbury T, Lawitz E. 2016. GS-9857 in patients with chronic hepatitis C virus genotype 1-4 infection: a randomized, double-blind, dose-ranging phase 1 study. J Viral Hepat 23:614–622. doi: 10.1111/jvh.12527. [DOI] [PubMed] [Google Scholar]
- 30.Gane EJ, Hyland RH, Yang Y, Svarovskaia E, Stamm LM, Brainard DM, McHutchison JG, Stedman CAM. 2017. Efficacy of ledipasvir plus sofosbuvir for 8 or 12 weeks in patients with hepatitis C virus genotype 2 infection. Gastroenterology 152:1366–1371. doi: 10.1053/j.gastro.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 31.Gane EJ, Schwabe C, Hyland RH, Yang Y, Svarovskaia E, Stamm LM, Brainard DM, McHutchison JG, Stedman CA. 2016. Efficacy of the combination of sofosbuvir, velpatasvir, and the NS3/4A protease inhibitor GS-9857 in treatment-naive or previously treated patients with hepatitis C virus genotype 1 or 3 infections. Gastroenterology 151:448–456.e441. doi: 10.1053/j.gastro.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 32.Lawitz E, Reau N, Hinestrosa F, Rabinovitz M, Schiff E, Sheikh A, Younes Z, Herring R Jr, Reddy KR, Tran T, Bennett M, Nahass R, Yang JC, Lu S, Dvory-Sobol H, Stamm LM, Brainard DM, McHutchison JG, Pearlman B, Shiffman M, Hawkins T, Curry M, Jacobson I. 2016. Efficacy of sofosbuvir, velpatasvir, and GS-9857 in patients with genotype 1 hepatitis C virus infection in an open-label, phase 2 trial. Gastroenterology 151:893–901.e891. doi: 10.1053/j.gastro.2016.07.039. [DOI] [PubMed] [Google Scholar]
- 33.Gane EJ, Kowdley KV, Pound D, Stedman CA, Davis M, Etzkorn K, Gordon SC, Bernstein D, Everson G, Rodriguez-Torres M, Tsai N, Khalid O, Yang JC, Lu S, Dvory-Sobol H, Stamm LM, Brainard DM, McHutchison JG, Tong M, Chung RT, Beavers K, Poulos JE, Kwo PY, Nguyen MH. 2016. Efficacy of sofosbuvir, velpatasvir, and GS-9857 in patients with hepatitis C virus genotype 2, 3, 4, or 6 infections in an open-label, phase 2 trial. Gastroenterology 151:902–909. doi: 10.1053/j.gastro.2016.07.038. [DOI] [PubMed] [Google Scholar]
- 34.Lawitz E, Poordad F, Wells J, Hyland RH, Yang Y, Dvory-Sobol H, Stamm LM, Brainard DM, McHutchison JG, Landaverde C, Gutierrez J. 2017. Sofosbuvir-velpatasvir-voxilaprevir with or without ribavirin in direct-acting antiviral-experienced patients with genotype 1 hepatitis C virus. Hepatology 65:1803–1809. doi: 10.1002/hep.29130. [DOI] [PubMed] [Google Scholar]
- 35.Feld JJ, Jacobson IM, Hezode C, Asselah T, Ruane PJ, Gruener N, Abergel A, Mangia A, Lai CL, Chan HL, Mazzotta F, Moreno C, Yoshida E, Shafran SD, Towner WJ, Tran TT, McNally J, Osinusi A, Svarovskaia E, Zhu Y, Brainard DM, McHutchison JG, Agarwal K, Zeuzem S; Astral-1 Investigators A. 2015. Sofosbuvir and velpatasvir for HCV genotype 1, 2, 4, 5, and 6 infection. N Engl J Med 373:2599–2607. doi: 10.1056/NEJMoa1512610. [DOI] [PubMed] [Google Scholar]
- 36.Jacobson IM, Lawitz E, Gane EJ, Willems BE, Ruane PJ, Nahass RG, Borgia SM, Shafran SD, Workowski KA, Pearlman B, Hyland RH, Stamm LM, Svarovskaia E, Dvory-Sobol H, Zhu Y, Subramanian GM, Brainard DM, McHutchison JG, Brau N, Berg T, Agarwal K, Bhandari BR, Davis M, Feld JJ, Dore GJ, Stedman CAM, Thompson AJ, Asselah T, Roberts SK, Foster GR. 2017. Efficacy of 8 weeks of sofosbuvir, velpatasvir, and voxilaprevir in patients with chronic HCV infection: 2 phase 3 randomized trials. Gastroenterology 153:113–122. doi: 10.1053/j.gastro.2017.03.047. [DOI] [PubMed] [Google Scholar]
- 37.Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 38.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 39.Kato T, Furusaka A, Miyamoto M, Date T, Yasui K, Hiramoto J, Nagayama K, Tanaka T, Wakita T. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J Med Virol 64:334–339. doi: 10.1002/jmv.1055. [DOI] [PubMed] [Google Scholar]
- 40.Gottwein JM, Scheel TK, Callendret B, Li YP, Eccleston HB, Engle RE, Govindarajan S, Satterfield W, Purcell RH, Walker CM, Bukh J. 2010. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J Virol 84:5277–5293. doi: 10.1128/JVI.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bukh J, Apgar CL, Engle R, Govindarajan S, Hegerich PA, Tellier R, Wong DC, Elkins R, Kew MC. 1998. Experimental infection of chimpanzees with hepatitis C virus of genotype 5a: genetic analysis of the virus and generation of a standardized challenge pool. J Infect Dis 178:1193–1197. doi: 10.1086/515683. [DOI] [PubMed] [Google Scholar]
- 42.Adams NJ, Chamberlain RW, Taylor LA, Davidson F, Lin CK, Elliott RM, Simmonds P. 1997. Complete coding sequence of hepatitis C virus genotype 6a. Biochem Biophys Res Commun 234:393–396. doi: 10.1006/bbrc.1997.6627. [DOI] [PubMed] [Google Scholar]
- 43.Cheng G, Chan K, Yang H, Corsa A, Pokrovskii M, Paulson M, Bahador G, Zhong W, Delaney W. 2011. Selection of clinically relevant protease inhibitor-resistant viruses using the genotype 2a hepatitis C virus infection system. Antimicrob Agents Chemother 55:2197–2205. doi: 10.1128/AAC.01382-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Camus G, Xu S, Han B, Lu J, Dvory-Sobol H, Yu M, Cheng G, Miller MD, Doehle BP, Mo H. 2018. Establishment of robust HCV genotype 4d, 5a, and 6a replicon systems. Virology 514:134–141. doi: 10.1016/j.virol.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Qi X, Bae A, Liu S, Yang H, Sun SC, Harris J, Delaney W, Miller M, Mo H. 2009. Development of a replicon-based phenotypic assay for assessing the drug susceptibilities of HCV NS3 protease genes from clinical isolates. Antiviral Res 81:166–173. doi: 10.1016/j.antiviral.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Shih I-h, Vliegen I, Peng B, Yang H, Hebner C, Paeshuyse J, Purstinger G, Fenaux M, Tian Y, Mabery E, Qi X, Bahador G, Paulson M, Lehman LS, Bondy S, Tse W, Reiser H, Lee WA, Schmitz U, Neyts J, Zhong W. 2011. Mechanistic characterization of GS-9190 (tegobuvir), a novel non-nucleoside inhibitor of hepatitis C virus NS5B polymerase. Antimicrob Agents Chemother 55:4196–4203. doi: 10.1128/AAC.00307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyles D, Dvory-Sobol H, Svarovskaia ES, Doehle BP, Martin R, Afdhal NH, Kowdley KV, Lawitz E, Brainard DM, Miller MD, Mo H, Gane EJ. 2017. Post-treatment resistance analysis of hepatitis C virus from phase II and III clinical trials of ledipasvir/sofosbuvir. J Hepatol 66:703–710. doi: 10.1016/j.jhep.2016.11.022. [DOI] [PubMed] [Google Scholar]
- 48.Susser S, Vermehren J, Forestier N, Welker MW, Grigorian N, Fuller C, Perner D, Zeuzem S, Sarrazin C. 2011. Analysis of long-term persistence of resistance mutations within the hepatitis C virus NS3 protease after treatment with telaprevir or boceprevir. J Clin Virol 52:321–327. doi: 10.1016/j.jcv.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan JC, De Meyer S, Bartels DJ, Dierynck I, Zhang EZ, Spanks J, Tigges AM, Ghys A, Dorrian J, Adda N, Martin EC, Beumont M, Jacobson IM, Sherman KE, Zeuzem S, Picchio G, Kieffer TL. 2013. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. Clin Infect Dis 57:221–229. doi: 10.1093/cid/cit226. [DOI] [PubMed] [Google Scholar]
- 50.Thomas XV, de Bruijne J, Sullivan JC, Kieffer TL, Ho CK, Rebers SP, de Vries M, Reesink HW, Weegink CJ, Molenkamp R, Schinkel J. 2012. Evaluation of persistence of resistant variants with ultra-deep pyrosequencing in chronic hepatitis C patients treated with telaprevir. PLoS One 7:e41191. doi: 10.1371/journal.pone.0041191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Underwood MR, Ross LL, Irlbeck DM, Gerondelis P, Rouse E, St Clair MH, Trinh L, Parkin N, Lanier E. 2009. Sensitivity of phenotypic susceptibility analyses for nonthymidine nucleoside analogues conferred by K65R or M184V in mixtures with wild-type HIV-1. J Infect Dis 199:84–88. doi: 10.1086/595296. [DOI] [PubMed] [Google Scholar]
- 52.Zhou S, Williford SE, McGivern DR, Burch CL, Hu F, Benzine T, Ingravallo P, Asante-Appiah E, Howe AYM, Swanstrom R, Lemon SM. 2018. Evolutionary pathways to NS5A inhibitor resistance in genotype 1 hepatitis C virus. Antiviral Res 158:45–51. doi: 10.1016/j.antiviral.2018.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu R, Curry S, McMonagle P, Yeh WW, Ludmerer SW, Jumes PA, Marshall WL, Kong S, Ingravallo P, Black S, Pak I, DiNubile MJ, Howe AY. 2015. Susceptibilities of genotype 1a, 1b, and 3 hepatitis C virus variants to the NS5A inhibitor elbasvir. Antimicrob Agents Chemother 59:6922–6929. doi: 10.1128/AAC.01390-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarrazin C, Cooper CL, Manns MP, Reddy KR, Kowdley KV, Roberts SK, Dvory-Sobol H, Svarovskia E, Martin R, Camus G, Doehle BP, Stamm LM, Hyland RH, Brainard DM, Mo H, Gordon SC, Bourliere M, Zeuzem S, Flamm SL. 2018. No impact of resistance-associated substitutions on the efficacy of sofosbuvir, velpatasvir, and voxilaprevir for 12weeks in HCV DAA-experienced patients. J Hepatol 69:1221–1230. doi: 10.1016/j.jhep.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 55.Wyles D, Thompson A, Lawitz E, Willems B, Gane EJ, Svarovskaia E, Dvory-Sobol H, Martin R, Camus G, Doehle BP, Stamm LM, Hyland RH, Brainard DM, Mo HM, Asselah T, Jacobson I, Foster GR, Roberts S. 2017. No impact of RASs on the high efficacy of SOF/VEL/VOX for 8 weeks in DAA-naïve patients: an integrated resistance analysis of the POLARIS-2 and POLARIS-3 studies, abstr THU-257. Abstr The International Liver Congress, European Association for the Study of the Liver (EASL), Amsterdam, The Netherlands. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.