Surveillance of circulating microbial populations is critical for monitoring the performance of a molecular diagnostic test. In this study, we characterized 31 isolates of Streptococcus agalactiae (group B Streptococcus [GBS]) from several geographic locations in the United States and Ireland that contain deletions in or adjacent to the region of the chromosome that encodes the hemolysin gene cfb, the region targeted by the Xpert GBS and GBS LB assays.
KEYWORDS: Streptococcus agalactiae, group B streptococcus, whole-genome sequencing
ABSTRACT
Surveillance of circulating microbial populations is critical for monitoring the performance of a molecular diagnostic test. In this study, we characterized 31 isolates of Streptococcus agalactiae (group B Streptococcus [GBS]) from several geographic locations in the United States and Ireland that contain deletions in or adjacent to the region of the chromosome that encodes the hemolysin gene cfb, the region targeted by the Xpert GBS and GBS LB assays. PCR-negative, culture-positive isolates were recognized during verification studies of the Xpert GBS assay in 12 laboratories between 2012 and 2018. Whole-genome sequencing of 15 GBS isolates from 11 laboratories revealed four unique deletions of chromosomal DNA ranging from 181 bp to 49 kb. Prospective surveillance studies demonstrated that the prevalence of GBS isolates containing deletions in the convenience sample was <1% in three geographic locations but 7% in a fourth location. Among the 15 isolates with chromosomal deletions, multiple pulsed-field gel electrophoresis types were identified, one of which appears to be broadly dispersed across the United States.
INTRODUCTION
Rates of early-onset neonatal group B streptococcal (EO-GBS) disease have fallen in the United States since the Centers for Disease Control and Prevention published the recommendation in 1996 to culture vaginal/rectal specimens from pregnant women at 35 to 37 weeks of gestation for detection of Streptococcus agalactiae (i.e., GBS) (1, 2). Women whose cultures yielded GBS are presumed to be colonized with the organism and subsequently are given intrapartum antimicrobial prophylaxis to prevent development of disease in the newborn (3, 4). The guidelines have been revised several times since then, with the most current version adding the use of nucleic acid amplification tests (NAATs) to detect GBS in Lim broth (LB) specimens or directly from intrapartum vaginal/rectal swab specimens, in addition to standard culture methods for detection of GBS from enrichment broths (5). These measures have been effective, with current rates of EO-GBS disease in the United States at record lows (i.e., 0.3 per 1,000 live births) (6). Several U.S. Food and Drug Administration (FDA)-cleared NAATs can be used to detect GBS in enrichment broth, and at least one FDA-cleared test can be used directly on vaginal/rectal swabs without broth enrichment for intrapartum testing (7–13). Intrapartum testing results are critical for women who present in labor prior to 35 weeks, who failed to have the surveillance cultures performed before time of delivery, or who do not have other indications for antimicrobial therapy to prevent EO-GBS disease (10). Intrapartum testing to detect GBS colonization is also used in many regions of the world in lieu of antepartum surveillance testing (14–16). In many laboratories, NAATs have replaced subcultures of enrichment broths to agar media because of NAATs’ improved workflow efficiency and its enhanced sensitivity, especially for the detection of nonhemolytic GBS strains or strains that do not produce CAMP factor, which may go undetected by culture (8, 17). A recent study by Gendrin et al. indicates that nonhemolytic strains of GBS still remain virulent, thus detecting them is important for guiding antimicrobial prophylaxis (17). One limitation of molecular diagnostic methods is the failure to detect organisms that have lost the target nucleic acid sequences either through deletion or mutation. Continuous surveillance of circulating microbial populations for the emergence of mutant strains is critical for maintaining the accuracy of molecular tests.
Xpert GBS (US-IVD, for in vitro diagnostic medical device) is a molecular in vitro test designed for intrapartum GBS screening of vaginal/rectal swab specimens, while Xpert GBS LB (US-IVD, for in vitro diagnostic use) is a molecular in vitro test intended to detect GBS from Lim broth-enriched vaginal/rectal swab specimens in antepartum women. Surveillance and laboratory verification studies conducted over the last 6 years have identified several specimens that were negative by either the Xpert GBS or Xpert GBS LB tests but yielded GBS isolates in culture. Whole-genome sequence analysis of a representative subset of the isolates revealed deletions of chromosomal DNA that included the target sequences of the Xpert GBS and Xpert GBS LB assays. Characterization of the chromosomal deletions present in these isolates and the results of several prospective surveillance studies to determine the prevalence of strains containing deletions are presented here.
MATERIALS AND METHODS
Characterization of isolates.
A total of 28 S. agalactiae (GBS) isolates and 5 Lim broth specimens that failed to yield a positive result with the Xpert GBS or GBS LB tests (Cepheid, Sunnyvale, CA) were received by a central laboratory for analysis between 2012 and 2018 (Table 1). The isolates and specimens were retested with Xpert GBS LB, and all gave negative results with the exception of two isolates, which were subsequently excluded from the study. Specimens and isolates were subcultured onto 5% sheep blood agar (Hardy Diagnostics, Santa Maria, CA) and a selective agar with supplements to enhance beta hemolysis (GBS Detect; Hardy Diagnostics). Suspect colonies were Gram stained, tested for catalase production and production of CAMP factor, and tested for Lancefield group B antigen (Hardy Diagnostics StrepPro typing kit). A total of 31 Xpert GBS-negative S. agalactiae isolates were identified. Fifteen isolates representing broad geographic diversity and distinctive colonial morphologies were selected for genotypic analysis.
TABLE 1.
Group B streptococci used in this study and their observed phenotypes and genotypesa
| Strain | Yr received | Location | CAMP factor | MLST typeb | PFGE cluster | 181-bp deletion | 522-bp deletion | 7-kb deletion | 49-kb deletion |
|---|---|---|---|---|---|---|---|---|---|
| 15902 | 2018 | AK | + | ST-860 | A1 | + | |||
| 14141 | 2016 | LA (lab A) | + | ST-860 | A1 | + | |||
| 11133 | 2013 | ND | + | ST-860 | A1 | + | |||
| 15827 | 2018 | WI (lab C) | + | ST-860 | A1 | + | |||
| 14984 | 2017 | LA (lab B) | + | ST-860 | A2 | + | |||
| 14053 | 2015 | LA (lab A) | + | ST-860 | A3 | + | |||
| 14140 | 2016 | LA (lab A) | + | ST-860 | A3 | + | |||
| 15829 | 2018 | WY | + | ST-23 | B1 | + | |||
| 15830 | 2018 | WY | + | ST-23 | B2 | + | |||
| 11513 | 2014 | CO | + | ST-19 | E1 | + | |||
| 10072 | 2012 | IA | – | ST-22 | D1 | + | |||
| 10201 | 2012 | IA | – | Unknown* | D2 | + | |||
| 15844 | 2018 | KS | – | ST-19 | C1 | + | |||
| 15594 | 2017 | Ireland | – | ST-19 | C1 | + | |||
| 16396 | 2018 | MO | – | ST-19 | C1 | + | |||
| 14065 | 2015 | LA (lab A) | + | A1 | |||||
| 14313 | 2016 | LA (lab A) | + | A1 | |||||
| 14314 | 2016 | LA (lab A) | + | A1 | |||||
| 14327 | 2016 | LA (lab A) | + | A1 | |||||
| 14328 | 2016 | LA (lab A) | + | A1 | |||||
| 14330 | 2016 | LA (lab A) | + | A1 | |||||
| 14392 | 2016 | LA (lab A) | + | A1 | |||||
| 14312 | 2016 | LA (lab A) | + | A2 | |||||
| 14329 | 2016 | LA (lab A) | + | A2 | |||||
| 14331 | 2016 | LA (lab A) | + | A2 | |||||
| 14333 | 2016 | LA (lab A) | + | A2 | |||||
| 14391 | 2016 | LA (lab A) | + | A2 | |||||
| 14315 | 2016 | LA (lab A) | + | A4 | |||||
| 14326 | 2016 | LA (lab A) | + | A5 | |||||
| 14927 | 2017 | OH | + | B3 | |||||
| 14393 | 2016 | LA (lab A) | – | F1 |
+, Presence; –, absence. For the locations, U.S. state abbreviations are given where applicable. lab A, laboratory A.
Only 15 isolates were sequenced and typed using MLST. *, Unknown sequence type (ST) but very closely related to ST-22.
Pulsed-field gel electrophoresis.
SmaI restriction fragments of 31 Xpert GBS-negative S. agalactiae isolates were analyzed by pulsed-field gel electrophoresis (PFGE) as previously described (18) with the following modifications. Lysozyme and mutanolysin (Sigma-Aldrich, St. Louis, MO; 1 mg/ml and 50 U/ml final concentrations, respectively) were used for lysis of agarose-encased cells, followed by overnight incubation with proteinase K (Sigma-Aldrich; 0.1 µg/ml) and subsequent washes (TE buffer; 10 mM Tris, 1 mM EDTA [pH 8.0]) prior to restriction enzyme digestion.
Whole-genome sequencing.
Pure cultures of GBS grown overnight at 35°C in Lim broth (Hardy Diagnostics) were used for DNA extraction using a Sigma-Aldrich GenElute bacterial genomic DNA kit according to the manufacturer’s instructions. Concentrations of DNA were determined by the UV light absorbance method using the NanoPhotometer system (Implen, Munich, Germany). Sequencing libraries were prepared from extracted genomic DNA using either Kapa HyperPrep (Kapa Biosystems/Roche, Basel, Switzerland) or Nextera XT (Illumina, San Diego, CA) kits. The resultant libraries were sequenced on a MiSeq sequencer (Illumina), using V3 reagent chemistry with 301-cycle paired-end reads. Libraries were quantified using qPCR (Kapa Biosystems/Roche) or ddPCR (Bio-Rad, Hercules, CA). All procedures were performed in accordance with manufacturer protocols.
Assemblies were generated from fastq sequence files using a5-miseq, a software pipeline that performs a series of operations that include adapter trimming, quality control, error correction, assembly, and scaffolding (19). The software was used with default settings. Sequence alignments were made using BWA, an alignment program designed for processing large numbers of sequencing reads (20). Reference strain 2603V/R (GenBank accession number AE009948.1) (21) was used for alignment and subsequent visualizations and comparisons. Small deletions were determined using Sprites (22). Large deletions were detected using cnD (23). Phylogenetic analysis was performed on the assemblies, including a reference strain using parsnp from the Harvest suite (24). The resulting trees were visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Multilocus sequence typing.
Assemblies were uploaded to the Center for Genomic Epidemiology’s MLST 2.0 web tool for multilocus sequence typing (MLST) analysis (25).
Surveillance studies.
(i) Laboratory A. Between November 2015 and January 2016, a total of 145 Lim broth specimens were randomly selected from among 1,350 specimens sent for GBS screening cultures and tested with Xpert GBS LB. Four GBS culture-positive Xpert GBS LB-negative samples were identified. Laboratory A evaluated an additional 210 GBS culture-positive Lim broth samples collected between April 2016 and May 2016 from the same laboratory. Clinician-collected vaginal or vaginal/rectal CultureSwabs EZII (BBL, Cockeysville, MD) were submitted to the clinical laboratory for culture within 24 h of collection. Upon receipt by the laboratory, both swabs were placed into Lim broth (Remel, San Diego, CA) and incubated at 35°C. After 18 to 24 h of incubation, an aliquot of the Lim broths was inoculated onto Trypticase soy agar with 5% sheep blood plates (BAP; Remel) and CHROMID Strepto B plates (bioMérieux, Marcy-l’Étoile, France) and incubated at 35 to 37°C in ambient air. Plates were examined daily for 2 days for beta-hemolytic colonies on the BAP and/or pink-red colonies on the chromogenic agar. Suspected colonies were confirmed by a negative catalase test (BBL) and a positive latex agglutination (PathoDxtra Strep Group B Latex; Remel). All culture-positive Lim broths were tested with the Xpert GBS-LB assay, according to the product insert. Samples with discordant culture and PCR results were submitted to the central laboratory for evaluation.
(ii) Laboratory B. Between January and November 2017 physician-collected vaginal/rectal ESwabs (Copan, Murrieta, CA) submitted for prenatal screening culture were inoculated onto a series of plates, including a BAP and Columbia CNA agar with sheep blood (BBL) for better detection of Gram-positive organisms, as well as a Lim broth (BBL), for subsequent Xpert GBS-LB PCR testing. Plates were incubated at 35 to 37°C and examined at 18 and 40 h for significant growth. Suspicious colonies were identified by matrix-assisted laser desorption ionization--time of flight (MALDI-TOF) analysis (Vitek MS; bioMérieux) or by latex agglutination (PathoDx reagents; Remel). No discrepancies were identified between the culture results and PCR results.
(iii) Laboratory C. Initial Xpert GBS LB testing of Lim broth specimens showed that only 1 of 125 specimens was Xpert GBS negative but culture positive. The laboratory evaluated an additional 100 broth specimens to determine the prevalence of false-negative Xpert GBS LB results. Physician-collected vaginal/rectal CultureSwabs (BBL) were sent to the microbiology laboratory from women undergoing prenatal screening for routine GBS testing. Patient swabs were transported to the laboratory at room temperature, within 24 h of collection. On arrival in the laboratory, swabs were enriched in Lim broth (LB; Remel) for 18 to 24 h at 35 to 37°C. Postenrichment samples were tested with the Xpert GBS LB assay according to the manufacturer’s instructions. Between 1 May 2018 and 22 May 2018, 100 enriched LB patient specimens (one swab per patient) determined to be negative by the Xpert GBS LB assay were streaked onto prewarmed CHROMagar Strep B plates (Northeast Laboratory Services, Waterville, ME) within 1 h of PCR results becoming available. Freshly streaked CHROMagar Strep B plates were incubated in ambient air at 37°C for 18 to 24 h. After incubation, the plates were reviewed for mauve-colored colonies. If mauve-colored colonies were found, the original LB specimen was streaked onto blood agar plates (Remel) and incubated overnight at 35 to 37°C in 5% CO2. Morphologically suspicious colonies were identified by MALDI-TOF analysis (Bruker, MA).
(iv) Central laboratory testing. An additional 300 deidentified Lim broth specimens from laboratories in New Jersey, Texas, and Louisiana (100 specimens each) were sent to the central laboratory for testing using the phenotypic methods described above (see “Characterization of isolates”) and using the Xpert GBS LB assay according to the product insert.
A schematic representation of the study is shown in Fig. S1 in the supplemental material.
RESULTS
Surveillance studies.
Overall, of the 145 specimens tested from laboratory A during the evaluation from November 2015 to January 2016, 40 were both Xpert GBS LB positive and culture positive, 101 were determined to be negative by both tests, and four were determined to be positive by the culture method but negative by Xpert GBS LB. Of the 210 additional culture-positive samples tested between April and May 2016, 16 were culture positive but negative by Xpert GBS LB. Twenty GBS isolates were sent to the central laboratory, but only 18 were confirmed to be Xpert GBS test negative; the other two were positive with Xpert GBS LB after retesting. The prevalence of Xpert GBS LB-negative isolates among the first set of culture-positive specimens was 7.1% (18/254) (see Fig. S1 in the supplemental material).
Among the 100 Xpert GBS-negative specimens collected between January and July 2017 from laboratory B, none were culture positive for GBS (0% prevalence). In addition, a retrospective chart review was undertaken of the results of 150 GBS-positive cultures collected between January and November 2017 that showed GBS that also had undergone Xpert GBS LB testing. A single discrepant result where the corresponding Xpert GBS LB result was negative was identified. Thus, the prevalence of Xpert GBS LB-negative isolates among culture-positive specimens was 0.7% (1/150) in the first set and 0.4% (1/250) overall.
From 1 May 2018 to 22 May 2018, 100 vaginal/rectal specimens were received and tested by laboratory C. All were reported as negative for GBS by Xpert GBS LB. All 100 specimens were also culture negative for GBS (0% prevalence).
Of the 300 Lim broth specimens received from New Jersey, Texas, and Louisiana by the central laboratory, 54 Lim broth samples were determined to be positive by Xpert GBS LB and grew GBS in culture, whereas 229 were negative by both methods. Seventeen specimens were positive by Xpert GBS-LB but culture negative, likely due to the age of the Lim broth samples. There were no culture-positive/Xpert GBS LB-negative results.
Pulsed-field gel electrophoresis.
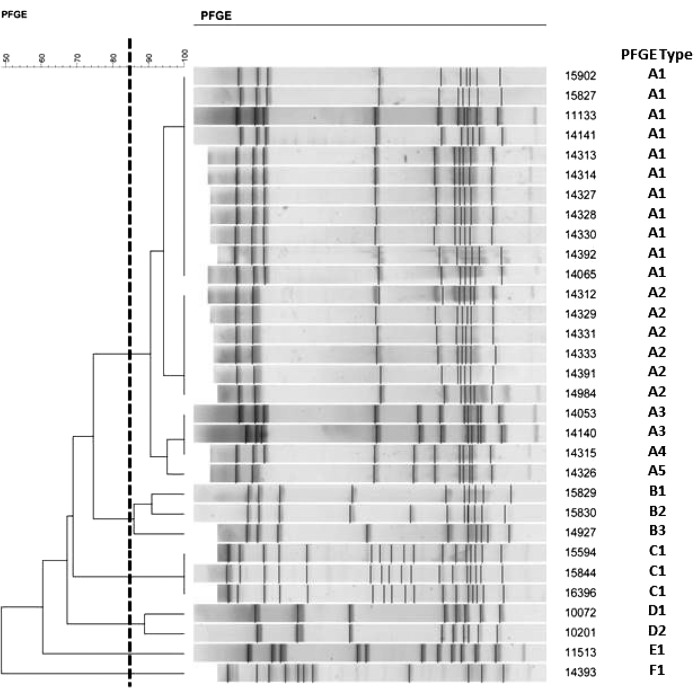
The results of PFGE analysis of 31 Xpert-negative GBS strains are shown in Fig. 1 and Table 1. Using ≥85% similarity as a threshold for similarity (26), there were three clusters of related isolates (A1 through A3) from Alaska, Louisiana, North Dakota, and Wisconsin that were highly related by PFGE and shared MLST sequence type ST-860. Two additional sets of isolates (B1 and C1) from Wyoming and Iowa, respectively, were unrelated to each other and to pattern “A” isolates by PFGE and MLST (Table 1).
FIG 1.
PFGE analysis results of 31 GBS strains not detected by Xpert GBS.
Whole-genome sequencing.
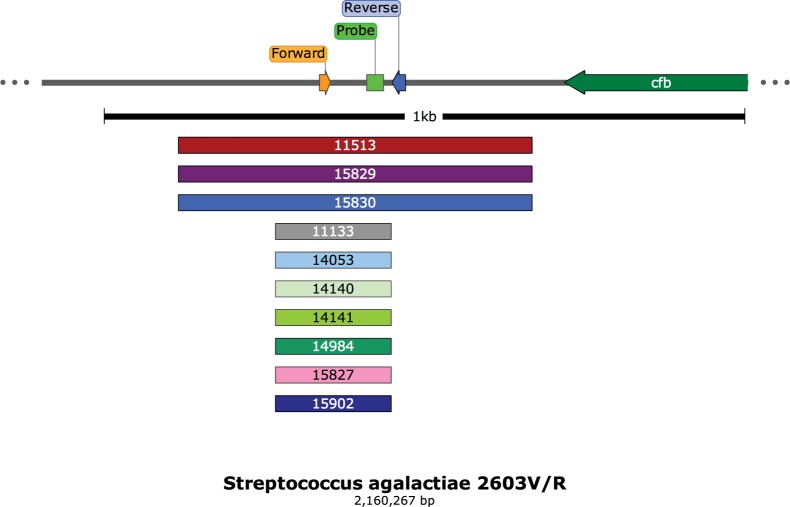
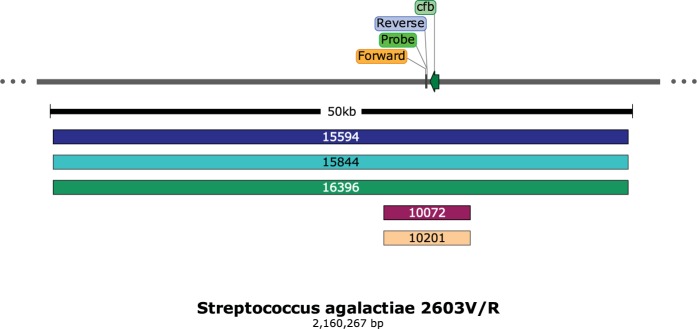
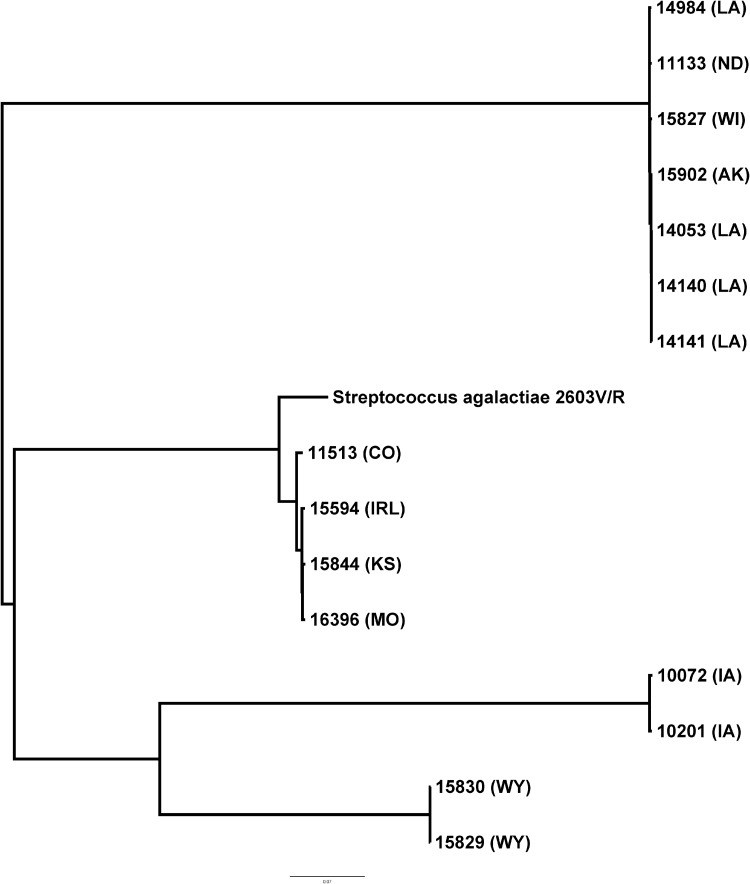
By whole-genome sequence analysis, 10 isolates from diverse geographical regions of the United States showed one of two small deletions that removed nucleotide sequences from one or both of the primer and the probe sites used in the Xpert GBS tests (Xpert GBS and Xpert GBS-LB), resulting in a lack of target amplification (Fig. 2). Strains 11133, 14053, 14140, 14141, 14984, 15827, and 15902 shared the same 181-bp deletion, while strains 11513, 15829, and 15830 shared the same 552-bp deletion at the primer and probe sites (Fig. 2). Five additional strains had larger chromosomal deletions that remove the sites for both primer and probe binding (Fig. 3); four isolates are from different geographical regions of the United States, and one isolate is from Ireland. Strains 15594, 15844, and 16396 share the same 49-kb deletion, while strains 10072 and 10201 have a 7-kb deletion (Table 1). The sequences of the isolates appear to differ in other areas of the genome as well, but many of them, although collected over at least a 2-year period, are highly related by core genome single-nucleotide polymorphism phylogenetic analysis (Fig. 4).
FIG 2.
Schematic of the Xpert GBS primer/probe region adjacent to the cfb gene. Larger and smaller deletions (552 and 181 bp, respectively) are listed by strain number.
FIG 3.
Schematic of the Xpert GBS primer/probe region adjacent to the cfb gene. The strains below share larger deletions (49 and 7 kb, respectively) affecting primer/probe region, as well as the cfb gene.
FIG 4.
Phylogenetic analysis of the 15 GBS strains visualized using FigTree, with strain 2603V/R as a reference (GenBank accession number AE009948.1).
Multilocus sequence typing.
The results of multilocus sequence typing of 15 Xpert GBS-negative strains are shown in Table 1. With the exception of strain 11513, MLST types, the sizes of the deletions, and the PFGE clusters appear to correlate well.
DISCUSSION
Several FDA-cleared NAATs can be used to detect GBS in enrichment broths (such as Lim broth) or directly from vaginal/rectal swabs without broth enrichment during intrapartum testing (7–13, 27–29). The availability of rapid results from intrapartum testing is critical for women who either present in labor prior to 35 weeks, who failed to have the surveillance cultures performed before time of delivery, or who do not otherwise have indications for antimicrobial therapy to prevent early-onset disease in the neonate (10). However, one limitation of molecular diagnostic tests is the failure to detect the organisms targeted due to nucleotide changes or deletions in the primer or probe binding regions. Such false-negative results may go undetected unless culture is performed in parallel to recover isolates (30). Verification studies, such as those required by most accreditation agencies, are undertaken to ensure that the testing laboratory can produce accurate and reproducible results from any new test before reporting patient results. Verification studies of new molecular tests typically employ a legacy phenotypic method but may also use another molecular method for comparison to allow users to detect potential problems with the new test in the patient population served by the laboratory. It was during such a verification study that isolates with deletions in the cfb region that affected Xpert GBS results were first identified.
In this study, among the geographically diverse specimens tested, there was an overall concordance of 96.1% between the results of Xpert GBS and recovery of GBS by culture. Thirty-one specimens were Xpert GBS-negative but culture positive, more than half of which came from a single hospital and were shown by PFGE typing to be highly related. This may represent local clonal expansion, although the reasons for this are unclear. Of note, that same strain type was obtained from three other states. Surveillance studies of specimens collected in New Jersey, Texas, and Wisconsin and at a second hospital in Louisiana in an effort to understand how widespread strains with deletions in the target region may be did not identify additional Xpert GBS-negative strains of GBS.
To understand the mechanism responsible for the Xpert GBS negative results, whole-genome sequencing was undertaken among 15 isolates and revealed four types of chromosomal deletions, ranging from 181 to 49 kb. Large recombination events, driven by selective pressures, have been described as important evolutionary drivers of GBS diversity (31, 32). Nonetheless, small genetic changes, such as those reported here, can also contribute to phenotypic diversity and host adaptation among certain GBS serotypes (33). Some of the deletions observed in our study were large enough to include the entire cfb gene, resulting in phenotypically CAMP-negative GBS isolates. Since the CAMP test is often used for presumptive identification of GBS in culture, strains with chromosomal deletions affecting the cfb gene could have gone undetected, especially before the introduction of alternative phenotypic identification methods such as chromogenic and selective agars (34). None of the Xpert GBS-negative results reported in this study resulted in infections in either mothers or their infants.
In summary, four types of chromosomal deletions were noted in the region of the cfb gene in GBS isolates that resulted in negative Xpert GBS tests. These mutant strains encompassed four different MLST types and multiple PFGE types. Strains with deletions appear to be uncommon, and the overall performance of the assay remains within performance claims. One limitation of the study is that other locations with clonal expansion of strains with deletion may exist. Nonetheless, based on convenience samples from three geographically distinct locations, the data indicate that although these mutations are rare, there is a continuing need to monitor strains for modifications that may affect the accuracy of molecular detection devices.
Supplementary Material
ACKNOWLEDGMENTS
I.A.T., F.C.T., S.D., V.M.L., R.N.B., M.J.L., D.G., and E.J.B. are current or former employees of Cepheid. R.V.G. has received research funding from Cepheid. All other authors report no conflicts of interest.
This study was funded by Cepheid.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.02040-18.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2009. Trends in perinatal group B streptococcal disease–United States, 2000-2006. MMWR Morb Mortal Wkly Rep 58:109–112. [PubMed] [Google Scholar]
- 2.Verani JR, Schrag SJ. 2010. Group B streptococcal disease in infants: progress in prevention and continued challenges. Clin Perinatol 37:375–392. doi: 10.1016/j.clp.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Lin FY, Weisman LE, Azimi P, Young AE, Chang K, Cielo M, Moyer P, Troendle JF, Schneerson R, Robbins JB. 2011. Assessment of intrapartum antibiotic prophylaxis for the prevention of early-onset group B streptococcal disease. Pediatr Infect Dis J 30:759–763. doi: 10.1097/INF.0b013e31821dc76f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Dyke MK, Phares CR, Lynfield R, Thomas AR, Arnold KE, Craig AS, Mohle-Boetani J, Gershman K, Schaffner W, Petit S, Zansky SM, Morin CA, Spina NL, Wymore K, Harrison LH, Shutt KA, Bareta J, Bulens SN, Zell ER, Schuchat A, Schrag SJ. 2009. Evaluation of universal antenatal screening for group B Streptococcus. N Engl J Med 360:2626–2636. doi: 10.1056/NEJMoa0806820. [DOI] [PubMed] [Google Scholar]
- 5.Verani JR, McGee L, Schrag SJ, Division of Bacterial Diseases and Respiratory Diseases, Centers for Disease Control and Prevention. 2010. Prevention of perinatal group B streptococcal disease: revised guidelines from CDC, 2010. MMWR Recomm Rep 59:1–36. [PubMed] [Google Scholar]
- 6.Verani JR, Spina NL, Lynfield R, Schaffner W, Harrison LH, Holst A, Thomas S, Garcia JM, Scherzinger K, Aragon D, Petit S, Thompson J, Pasutti L, Carey R, McGee L, Weston E, Schrag SJ. 2014. Early-onset group B streptococcal disease in the United States: potential for further reduction. Obstet Gynecol 123:828–837. doi: 10.1097/AOG.0000000000000163. [DOI] [PubMed] [Google Scholar]
- 7.Alfa MJ, Sepehri S, De Gagne P, Helawa M, Sandhu G, Harding GK. 2010. Real-time PCR assay provides reliable assessment of intrapartum carriage of group B Streptococcus. J Clin Microbiol 48:3095–3099. doi: 10.1128/JCM.00594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchan BW, Faron ML, Fuller D, Davis TE, Mayne D, Ledeboer NA. 2015. Multicenter clinical evaluation of the Xpert GBS LB assay for detection of group B Streptococcus in prenatal screening specimens. J Clin Microbiol 53:443–448. doi: 10.1128/JCM.02598-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies HD, Miller MA, Faro S, Gregson D, Kehl SC, Jordan JA. 2004. Multicenter study of a rapid molecular-based assay for the diagnosis of group B Streptococcus colonization in pregnant women. Clin Infect Dis 39:1129–1135. doi: 10.1086/424518. [DOI] [PubMed] [Google Scholar]
- 10.El Helali N, Nguyen JC, Ly A, Giovangrandi Y, Trinquart L. 2009. Diagnostic accuracy of a rapid real-time polymerase chain reaction assay for universal intrapartum group B streptococcus screening. Clin Infect Dis 49:417–423. doi: 10.1086/600303. [DOI] [PubMed] [Google Scholar]
- 11.Miller SA, Deak E, Humphries R. 2015. Comparison of the AmpliVue, BD Max system, and Illumigene molecular assays for detection of group B Streptococcus in antenatal screening specimens. J Clin Microbiol 53:1938–1941. doi: 10.1128/JCM.00261-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couturier BA, Weight T, Elmer H, Schlaberg R. 2014. Antepartum screening for group B Streptococcus by three FDA-cleared molecular tests and effect of shortened enrichment culture on molecular detection rates. J Clin Microbiol 52:3429–3432. doi: 10.1128/JCM.01081-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silbert S, Rocchetti TT, Gostnell A, Kubasek C, Widen R. 2016. Detection of group B Streptococcus directly from collected ESwab samples by use of the BD Max GBS assay. J Clin Microbiol 54:1660–1663. doi: 10.1128/JCM.00445-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koppes DM, Vriends A, van Rijn M, van Heesewijk AD. 2017. Clinical value of polymerase chain reaction in detecting group B Streptococcus during labor. J Obstet Gynaecol Res 43:996–1000. doi: 10.1111/jog.13321. [DOI] [PubMed] [Google Scholar]
- 15.El Helali N, Giovangrandi Y, Guyot K, Chevet K, Gutmann L, Durand-Zaleski I. 2012. Cost and effectiveness of intrapartum group B streptococcus polymerase chain reaction screening for term deliveries. Obstet Gynecol 119:822–829. doi: 10.1097/AOG.0b013e31824b1461. [DOI] [PubMed] [Google Scholar]
- 16.Bjorklund V, Nieminen T, Ulander VM, Ahola T, Saxen H. 2017. Replacing risk-based early-onset-disease prevention with intrapartum group B Streptococcus PCR testing. J Matern Fetal Neonatal Med 30:368–373. doi: 10.3109/14767058.2016.1173030. [DOI] [PubMed] [Google Scholar]
- 17.Gendrin C, Vornhagen J, Armistead B, Singh P, Whidbey C, Merillat S, Knupp D, Parker R, Rogers LM, Quach P, Iyer LM, Aravind L, Manning SD, Aronoff DM, Rajagopal L. 2018. A nonhemolytic group B Streptococcus strain exhibits hypervirulence. J Infect Dis 217:983–987. doi: 10.1093/infdis/jix646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goering RV. 2010. Pulsed field gel electrophoresis: a review of application and interpretation in the molecular epidemiology of infectious disease. Infect Genet Evol 10:866–875. doi: 10.1016/j.meegid.2010.07.023. [DOI] [PubMed] [Google Scholar]
- 19.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tettelin H, Masignani V, Cieslewicz MJ, Eisen JA, Peterson S, Wessels MR, Paulsen IT, Nelson KE, Margarit I, Read TD, Madoff LC, Wolf AM, Beanan MJ, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Kolonay JF, Madupu R, Lewis MR, Radune D, Fedorova NB, Scanlan D, Khouri H, Mulligan S, Carty HA, Cline RT, Van Aken SE, Gill J, Scarselli M, Mora M, Iacobini ET, Brettoni C, Galli G, Mariani M, Vegni F, Maione D, Rinaudo D, Rappuoli R, Telford JL, Kasper DL, Grandi G, Fraser CM. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc Natl Acad Sci U S A 99:12391–12396. doi: 10.1073/pnas.182380799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, Wang J, Luo J, Ding X, Zhong J, Wang J, Wu FX, Pan Y. 2016. Sprites: detection of deletions from sequencing data by re-aligning split reads. Bioinformatics 32:1788–1796. doi: 10.1093/bioinformatics/btw053. [DOI] [PubMed] [Google Scholar]
- 23.Simpson JT, McIntyre RE, Adams DJ, Durbin R. 2010. Copy number variant detection in inbred strains from short read sequence data. Bioinformatics 26:565–567. doi: 10.1093/bioinformatics/btp693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover FC, Arbeit RD, Goering RV. 1997. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. Infect Control Hosp Epidemiol 18:426–439. doi: 10.2307/30141252. [DOI] [PubMed] [Google Scholar]
- 27.Bourgeois-Nicolaos N, Cordier AG, Guillet-Caruba C, Casanova F, Benachi A, Doucet-Populaire F. 2013. Evaluation of the Cepheid Xpert GBS assay for rapid detection of group B streptococci in amniotic fluids from pregnant women with premature rupture of membranes. J Clin Microbiol 51:1305–1306. doi: 10.1128/JCM.03356-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Church DL, Baxter H, Lloyd T, Miller B, Gregson DB. 2011. Evaluation of the Xpert® group B Streptococcus real-time polymerase chain reaction assay compared to StrepB carrot broth for the rapid intrapartum detection of group B streptococcus colonization. Diagn Microbiol Infect Dis 69:460–462. doi: 10.1016/j.diagmicrobio.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Ke D, Bergeron MG. 2001. Molecular methods for rapid detection of group B streptococci. Expert Rev Mol Diagn 1:175–181. doi: 10.1586/14737159.1.2.175. [DOI] [PubMed] [Google Scholar]
- 30.Yang S, Rothman RE. 2004. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis 4:337–348. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brochet M, Rusniok C, Couve E, Dramsi S, Poyart C, Trieu-Cuot P, Kunst F, Glaser P. 2008. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc Natl Acad Sci U S A 105:15961–15966. doi: 10.1073/pnas.0803654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Da Cunha V, Davies MR, Douarre PE, Rosinski-Chupin I, Margarit I, Spinali S, Perkins T, Lechat P, Dmytruk N, Sauvage E, Ma L, Romi B, Tichit M, Lopez-Sanchez MJ, Descorps-Declere S, Souche E, Buchrieser C, Trieu-Cuot P, Moszer I, Clermont D, Maione D, Bouchier C, McMillan DJ, Parkhill J, Telford JL, Dougan G, Walker MJ, Holden MT, Poyart C, Glaser P. 2015. Corrigendum: Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat Commun 6:6108. doi: 10.1038/ncomms7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flores AR, Galloway-Pena J, Sahasrabhojane P, Saldana M, Yao H, Su X, Ajami NJ, Holder ME, Petrosino JF, Thompson E, Margarit YRI, Rosini R, Grandi G, Horstmann N, Teatero S, McGeer A, Fittipaldi N, Rappuoli R, Baker CJ, Shelburne SA. 2015. Sequence type 1 group B Streptococcus, an emerging cause of invasive disease in adults, evolves by small genetic changes. Proc Natl Acad Sci U S A 112:6431–6436. doi: 10.1073/pnas.1504725112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosa-Fraile M, Spellerberg B. 2017. Reliable detection of group B Streptococcus in the clinical laboratory. J Clin Microbiol 55:2590–2598. doi: 10.1128/JCM.00582-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.