Serratia marcescens is an opportunistic bacterial pathogen. It is notorious for its increasing antimicrobial resistance and its potential to cause outbreaks of colonization and infections, predominantly in neonatal intensive care units (NICUs).
KEYWORDS: Serratia marcescens, WGS, bionumerics, molecular typing, neonatal intensive care, outbreak management, wgMLST
ABSTRACT
Serratia marcescens is an opportunistic bacterial pathogen. It is notorious for its increasing antimicrobial resistance and its potential to cause outbreaks of colonization and infections, predominantly in neonatal intensive care units (NICUs). There, its spread requires rapid infection control response. To understand its spread, detailed molecular typing is key. We present a whole-genome multilocus sequence typing (wgMLST) method for S. marcescens. Using a set of 299 publicly available whole-genome sequences (WGS), we developed an initial wgMLST system consisting of 9,377 gene loci. This included 1,455 loci occurring in all reference genomes and 7,922 accessory loci. This closed system was validated using three geographically diverse collections of S. marcescens consisting of 111 clinical isolates implicated in nosocomial dissemination events in three hospitals. The validation procedure showed a full match between epidemiological data and the wgMLST analyses. We set the cutoff value for epidemiological (non)relatedness at 20 different alleles, though for the majority of outbreak-clustered isolates, this difference was limited to 4 alleles. This shows that the wgMLST system for S. marcescens provides prospects for successful future monitoring for the epidemiological containment of this opportunistic pathogen.
INTRODUCTION
The new gold standard in microbial epidemiology is genome sequencing. The use of whole-genome (draft) sequences (WGS) to compare bacterial isolates in detail, and to delineate their spread, is based on either the detection of single nucleotide variants or polymorphisms (SNVs or SNPs, respectively) or on the assessment of overall gene content, including allelic differences between strains, by whole-genome multilocus sequence typing (wgMLST) (1–4). Both methods have their advantages and disadvantages. Where SNP analysis may have a higher intrinsic discriminatory power (since it covers coding and noncoding regions) and better resolves the ancestral relationship between lineages, wgMLST usually provides a more stable, generically applicable system, with results that are easier to translate into relevant epidemiological differences between isolates. wgMLST schemes have been developed for a multitude of microbial organisms, with the main driver being the development of a universal “typing language” (5–7). This will facilitate the monitoring of local institutional spread of certain pathogens but will also extend into regional, national, international, and possibly even global monitoring for the dissemination of given bacterial strain types (8–10). This will aid communication in international public health management and should in the end lead to early recognition of the emergence and spread of pathogenic microbial strains. Furthermore, this is of importance in the current era of multidrug-resistant bacteria and their global dispersal promoted by human travelling, international patient transfer, nosocomial transmission, and excessive use of antimicrobials.
Serratia marcescens is a bacterial pathogen for which no wgMLST scheme has yet been defined. S. marcescens is notorious for its pathogenicity in plants (11) but also in preterm neonates (12, 13). Therefore, setting up a robust epidemiological wgMLST typing scheme is essential for monitoring and interrupting outbreaks in neonatal intensive care units (NICUs) as well as other medical settings. In addition, S. marcescens is capable of efficiently acquiring multiple resistance determinants (that are unreliable epidemiological markers), which adds to its clinical relevance (14–18). We have developed a proprietary wgMLST toolbox for S. marcescens based on publicly available WGS data. We have validated the scheme using epidemiologically related isolates collected during recent outbreaks of colonization and infection in NICUs in both Dutch and German teaching hospitals.
MATERIALS AND METHODS
Strains.
Clinical S. marcescens isolates were obtained from three different institutions in Groningen (The Netherlands; n = 41), Cologne (Germany; n = 19), and Freiburg (Germany; n = 51).
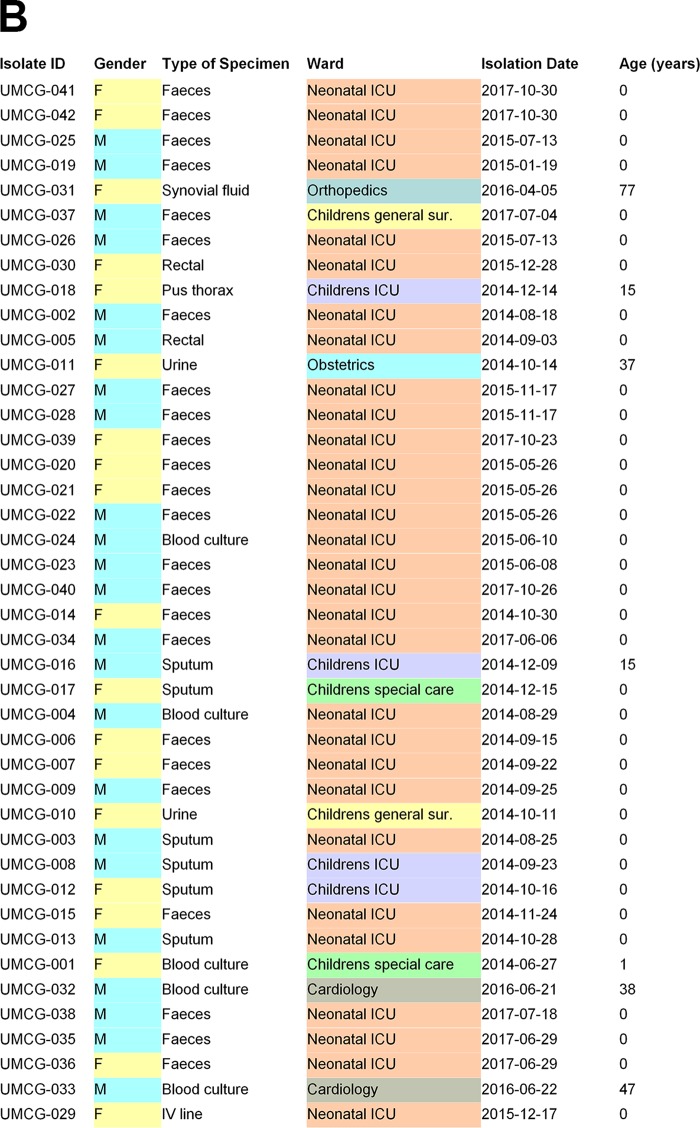
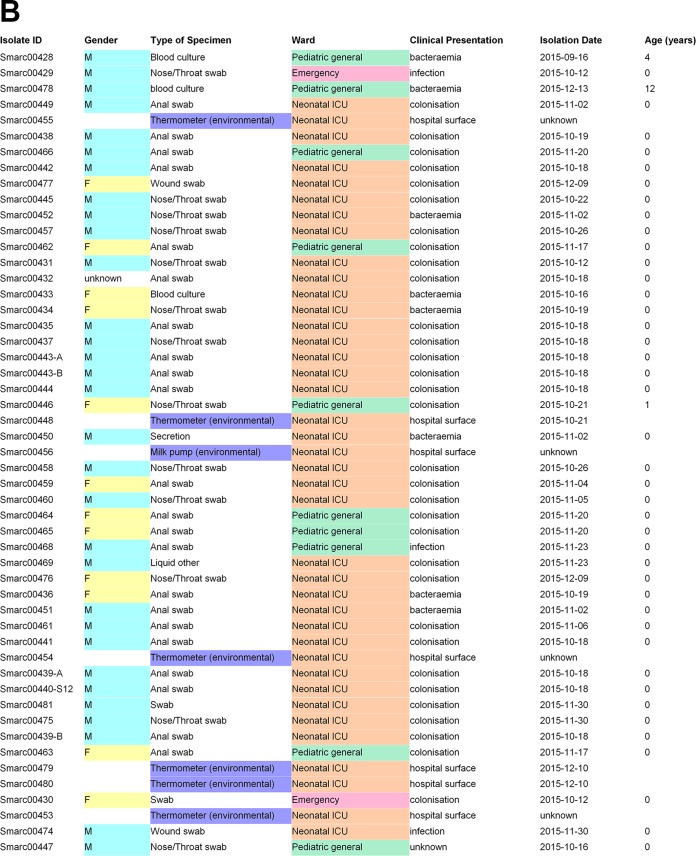
The 41 isolates from the University Medical Center Groningen were obtained between 2014 and 2017 from 38 patients, of which 4 were adults in nonpediatric wards (2 in cardiology, 1 in orthopedics, and 1 in obstetrics), 2 were from children >12 years of age in the pediatric intensive care unit (PICU), 1 from a child >18 months of age, and the others from children <6 months of age either on the pediatric special care unit (n = 1), the pediatric general surgery ward (n = 2), the PICU (n = 2), or the NICU (n = 26). From three patients, two isolates were sequenced. In one case, in addition to a positive culture from a rectal swab of the patient, an isolate was also cultured from the intravenous line, but this isolate appeared to be Serratia liquefaciens, originally misidentified as S. marcescens by conventional diagnostic methods. All other isolates were cultured from patients in the NICU using growth-based microbiology technology (see Fig. 1 for additional details on strain origin). The 19 isolates from Cologne were isolated between 2014 and 2017 and all originated from NICUs, PICUs, and general wards. The ages of the patients varied between 4 days and 11 months. The collection of isolates consisted of 5 epidemiologically related transmission clusters and 2 singleton isolates (see Fig. 2 for additional details). The 51 isolates from Freiburg mostly originated from the local NICU (n = 39), with patient ages varying between 0 and 12 weeks. Seven environmental isolates were included for comparative reasons and to gauge the relevance of environmental spread. For several patients (A to H, n = 8) multiple isolates were included in order to define basic levels of intrapatient variability of S. marcescens (see Fig. 3 for additional details).
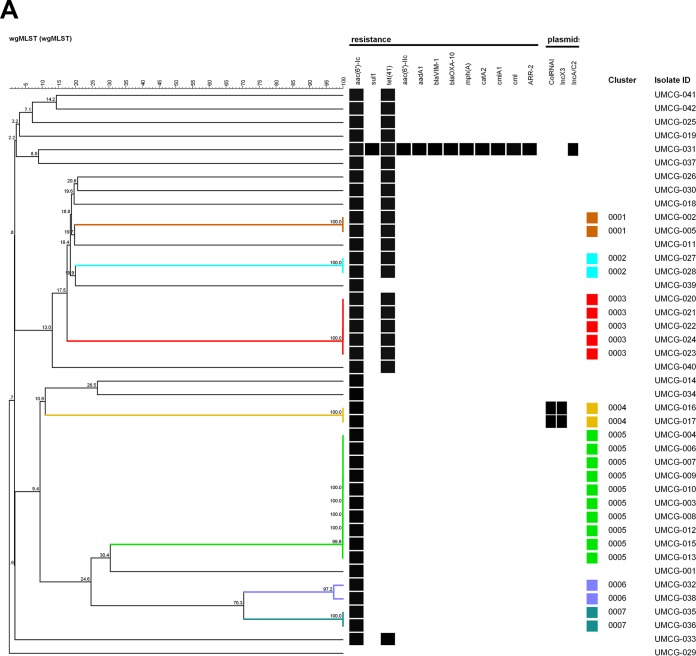
FIG 1.
UPGMA tree of the pan-genomic allelic profiles (n = 25) derived for S. marcescens isolates from the University Medical Center Groningen, The Netherlands. (A) Outbreaks and transfer events identified prior to our study (clusters 0001 to 0007) are highlighted by color. (B) Relevant microbiological, host-associated, and environmental metadata. The UPGMA tree was built using a similarity coefficient based on categorical values expressed as percentages. Isolate UMCG-029, located at the bottom of the tree, represents S. liquefaciens, a species only sharing approximately 2,900 loci with the S. marcescens wgMLST scheme, as opposed to 4,300 loci that are typically detected in S. marcescens.
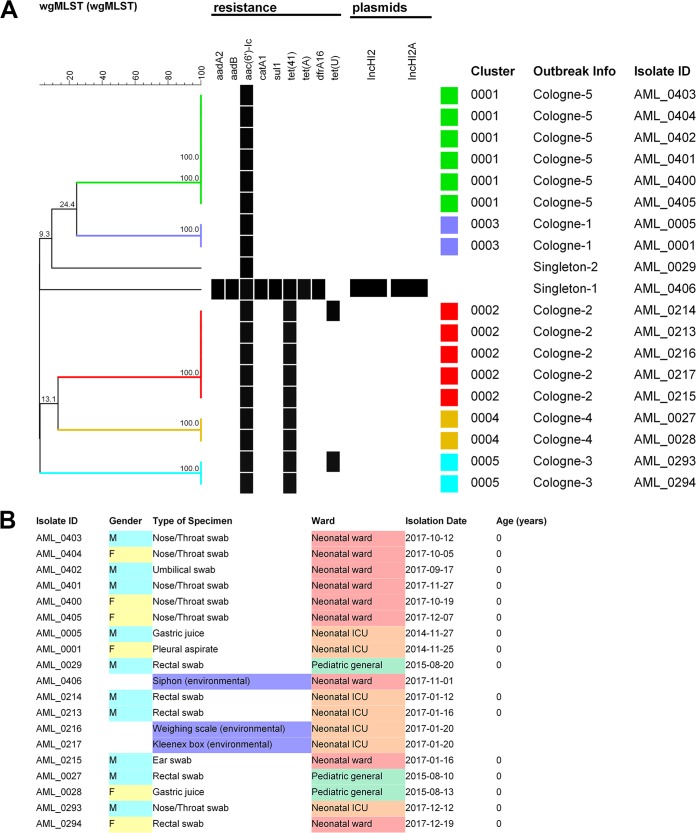
FIG 2.
UPGMA tree of the pan-genomic allelic profiles (n = 7) derived for S. marcescens isolates from the Institute for Medical Microbiology, Immunology and Hygiene at the University of Cologne, Germany. (A) Outbreaks and transfer events (Cologne-1 to Cologne-5) identified prior to our study are highlighted by color. (B) Relevant microbiological, host-associated, and environmental metadata. The UPGMA tree was built using a similarity coefficient based on categorical values expressed as percentages. Isolates originating from inanimate surfaces are highlighted in blue.
FIG 3.
UPGMA tree of the pan-genomic allelic profiles (n = 4) derived for S. marcescens isolates from the University Hospital of Freiburg, Germany. (A) A single major outbreak event generated all strains except four (red and nonboxed). (B) Relevant microbiological, host-associated, and environmental metadata. The UPGMA tree was built using a similarity coefficient based on categorical values expressed as percentages. Note that in this case, multiple isolates were included for 8 different individuals. Isolates originating from inanimate surfaces are highlighted in blue.
Isolates were either directly processed or stored at −80°C in glycerol-containing medium until they were cultured for DNA isolation and genome sequencing. In addition to the WGS data, clinical and epidemiological data were included. Metadata included, but were not limited to, isolation dates, outbreak associations, patients’ sex and age, type (and outcome) of infections, specimen types submitted for microbiological analyses, location of the ward, and whether local typing data obtained previously were available.
DNA isolation.
DNA was extracted using the Ultraclean Microbial DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA, USA) or the MagAttract HMW DNA isolation kit according to the manufacturers’ instructions (Qiagen, Hilden, Germany) and quantified using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific Inc, Waltham, MA, USA) and/or the Qubit dsDNA HS assay (Thermo Fisher Scientific GmbH, Schwerte, Germany).
Genome sequencing.
DNA libraries were prepared using the Nextera XT library preparation kit and the Nextera XT v2 index kit (Illumina, San Diego, CA, USA). The library was sequenced on a MiSeq, using the reagent kit v2 generating 250-bp paired-end reads. Tables S1A to C in the supplemental material disclose the quality parameters for the sequences determined. All WGS included met the required quality criteria.
Development of the wgMLST scheme.
A scheme for wgMLST of S. marcescens was developed using publicly available WGS data for this species (June 2017) and will be made commercially available through a plugin in BioNumerics (Applied Maths NV, Sint-Martens-Latem, Belgium). The scheme is intended to facilitate the detection of subtype- or outbreak-specific markers. Using a selection of 299 annotated, publicly available reference genomes which were assumed to capture the diversity within S. marcescens, a pan-genomic scheme with high discriminatory power was developed (see Table S2 for a list of all WGS included). Starting from the reference genomes, our scheme creation procedure used a sampling-based multireciprocal BLAST procedure to determine those sets of alleles that make up the stable loci in the pan-genome. A per-locus allele assessment procedure then determined the central prototype allele and thus the definition of the locus. The wgMLST scheme for S. marcescens was tested, validated, and approved by epidemiological and microbiological analyses using information on the strain collections from Groningen, Cologne, and Freiburg.
Bioinformatics analyses.
De novo genome assembly for all WGS was performed using SPAdes 3.7.1. All de novo calculations were run on the cloud-based calculation engine that comes with BioNumerics 7.6.3. wgMLST analysis was also performed using the BioNumerics cloud-based calculation engine. Alleles were identified by both an assembly-free k-mer–based approach using the raw reads and an assembly-based BLAST approach. Identification was performed against the S. marcescens wgMLST database in BioNumerics. Categorical coefficients were used for defining similarity levels, and unweighted pair group method with arithmetic mean (UPGMA) was used as the clustering algorithm. Minimum spanning trees (MST) were constructed using the wgMLST allelic profiles as input data. The sizes of the nodes were chosen proportional to the numbers of isolates in the nodes (i.e., isolates with the same allelic profiles). Branch lengths reflect the numbers of allele differences between the isolates in the connected nodes.
Accession number(s).
All primary sequences from WGS were deposited in the public domain (ENA project numbers PRJEB28358 and PRJEB28681).
RESULTS
A new system for wgMLST for S. marcescens.
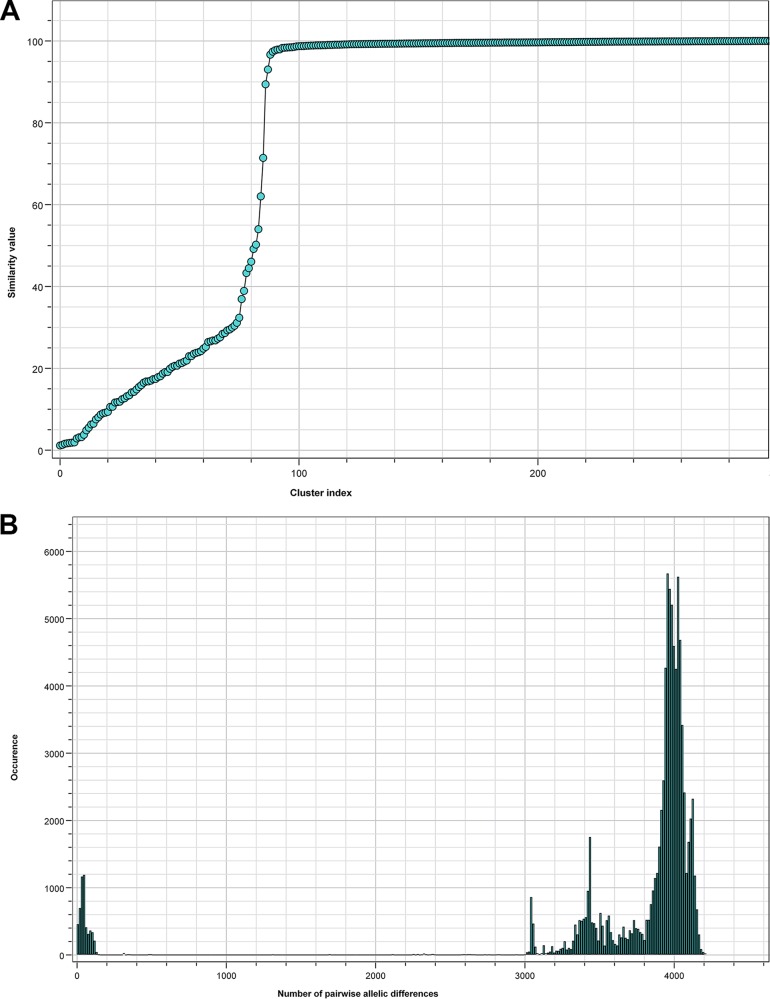
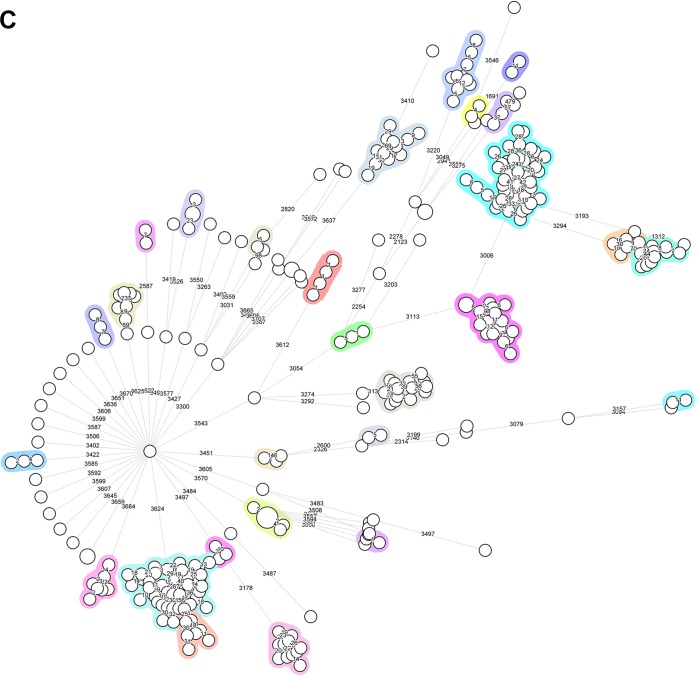
In total, 299 reference genome sequences were included while building the wgMLST scheme. These displayed a conformity between 85% and 97% after constructing the scheme and showed an average of 95% global coverage of the included loci. The scheme was validated in August 2017 on the basis of 373 sequence read archives (SRAs), which included all Illumina data sets publicly available as of 28 August 2017. In this way, a total of 9,377 loci were added to the scheme, including 1,455 loci which were present in all references and 7,922 accessory loci. The wgMLST scheme had high discriminatory power and allowed for the detection of markers specific for S. marcescens subtypes or outbreak strains, thus enabling powerful classification and outbreak definition (Fig. 4C). The two allele detection procedures (either assembly based or assembly free) performed fast and reliable allele calling for cluster detection. Figure 4A indicates the diversity within the reference genome set and provides an overview of the numbers of clusters as a function of the similarity cutoff value, indicating the presence of both distant and highly related isolates in the reference set of 299 strains. Figure 4B depicts the number of pairwise allelic differences and the frequency of their occurrence, peaking at approximately 4,000 allelic differences given the current wgMLST scheme complexity. Figure 4C shows a global perspective of the genomic diversity among the references used to build the wgMLST scheme, where all circles identify distinct wgMLST types (as also semiquantified by the numbers of allelic differences quantified on the branches) and the colored blocks identify isolates of more closely related and sometimes indistinguishable genomic sequences. This confirms our assumption that the genome sequences obtained from the public domain show significant levels of diversity, allowing them to serve as a reference of genomic variability. Overall, the quality parameters indicate that the scheme covers the diversity within the species and provides sufficient resolving power for distinguishing even closely related bacterial isolates. Finally, it seems that the population structure of S. marcescens is largely genetically diverse with many singletons present. However, there seem to be indications for the successful expansion of clones (Fig. 4C, colored circles).
FIG 4.
Review of quality parameters for the S. marcescens-specific whole-genome sequences used to construct the wgMLST reference database. (A) Correlation between numbers of clusters and similarity cutoff values for the founding S. marcescens wgMLST database. The cluster index was based on the average numbers of alleles being different between closely related strain pairs. The analysis was performed using all WGS listed in Table S2 in the supplemental material. (B) Correlation between the numbers of pairwise allelic differences and their frequency of occurrence. (C) Minimum spanning tree based on the pan-genomic allelic profiles of 299 S. marcescens isolates, representing the reference set used to create the wgMLST database. Colors highlight closely related isolates, numbers of allelic differences are indicated on the lines connecting the various types.
Strain characteristics and outbreak features.
It has to be stated that only one patient died as a consequence of S. marcescens colonization/infection. Also, the presence was mostly due to the colonization, and real infection was only apparent in a limited number of cases (Groningen, 9 of 38 patients [24%]; Cologne, 2/16 [13%]; Freiburg, 6/23 [26%] [one sample of unknown origin]). Overall, 22% patients had an infection.
Groningen outbreak analyses.
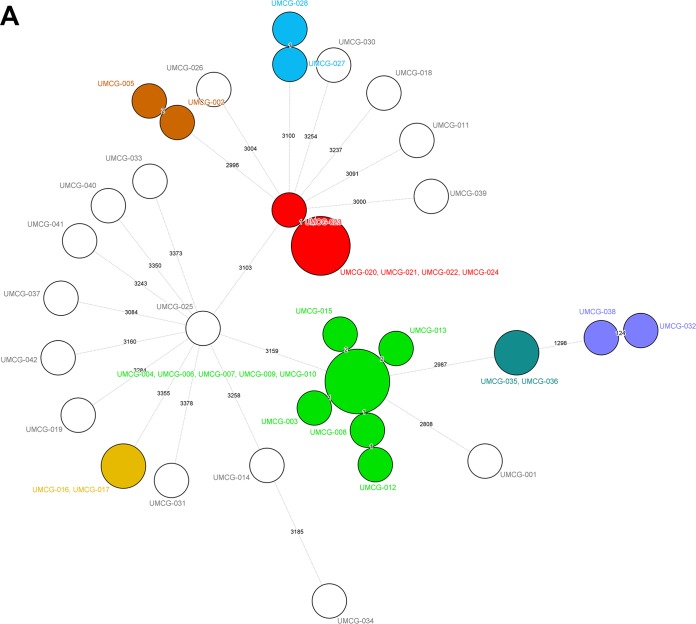
Forty-one clinical isolates were obtained from 38 patients in the University Medical Center Groningen (UMCG). The wgMLST analysis detected a small cluster of related isolates: five isolates obtained from three patients in May to June in 2015 (Fig. 1, cluster 0003). From one patient, two isolates from the rectal swab appeared to be 100% wgMLST identical, and from the other patient, the isolate found in the blood was identical to the one found in the rectal swab. In addition, a larger cluster was found containing isolates, all from different patients, from a protracted outbreak in August to November 2014 (Fig. 1, cluster 0005). The single invasive isolate that was isolated during this episode was indistinguishable from the other isolates. In addition, four suspected cases of single transmission events involving two patients were confirmed (Fig. 1, clusters 0001, 0002, 0004, 0006, and 0007). Hence, the clustering aligns very well with the prior epidemiological scenarios. The 0002 cluster contained two separate isolates from the same patient, showing the reproducibility of the method. All isolates contained the aminoglycoside resistance-associated gene aac(6′)-I-C, and approximately half of them contained the tetracycline resistance determinant Tet(41). A single multiresistant isolate was cultured from the synovial fluid of an elderly female nursed at the orthopedics department. The origin of this strain is not clear.
Cologne outbreak analyses.
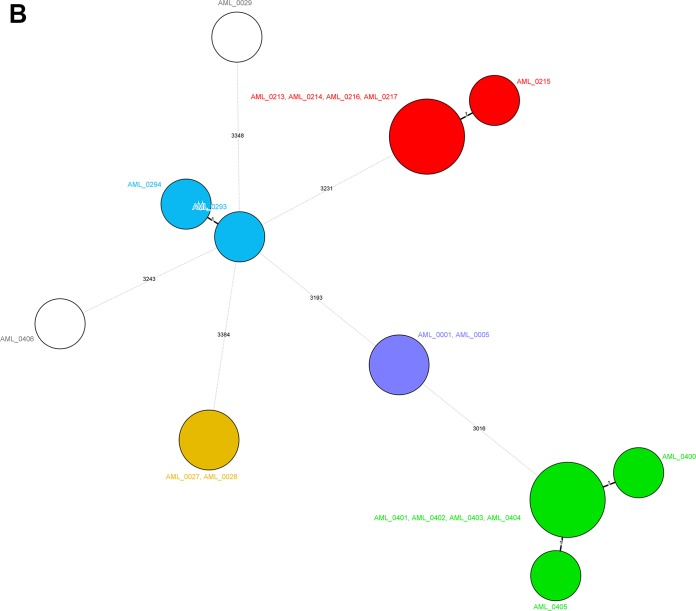
wgMLST analysis of the 19 isolates from the Cologne University hospital correctly defined the anticipated clustering and identified two main outbreak clusters and three cases where interpatient transfer was already suspected (Cologne-1 to Cologne-5). The two singleton isolates were separated from all of the other isolates. Figure 2 summarizes the overall data and sketches the outbreak scenarios, also showing that all related isolates were 100% identical at the wgMLST level. One of the singleton isolates contained at least 8 different resistance genes.
Freiburg outbreak analyses.
The collection of isolates derived from the laboratory in the Freiburg University hospital contained 47 of 51 isolates that were nearly indistinguishable by wgMLST (Fig. 3, green boxes), indicating a local outbreak which occurred in October and November 2015 involving 19 patients and 7 environmental isolates. Additionally, two isolates were identified (Fig. 3, red boxes) that were not distinguished by wgMLST, reflecting a single known transmission event of a different strain type outside the NICU. Most of the outbreak isolates were considered to represent colonization rather than infection or bacteremia (16/19 patients). All serial isolates obtained from individual patients were identical at the wgMLST level. Small differences were documented only in the cases of patients F and H but were within the boundaries of the epidemiological cutoff value. Finally, the environmental isolates all fell within the same outbreak category.
Minimum spanning trees.
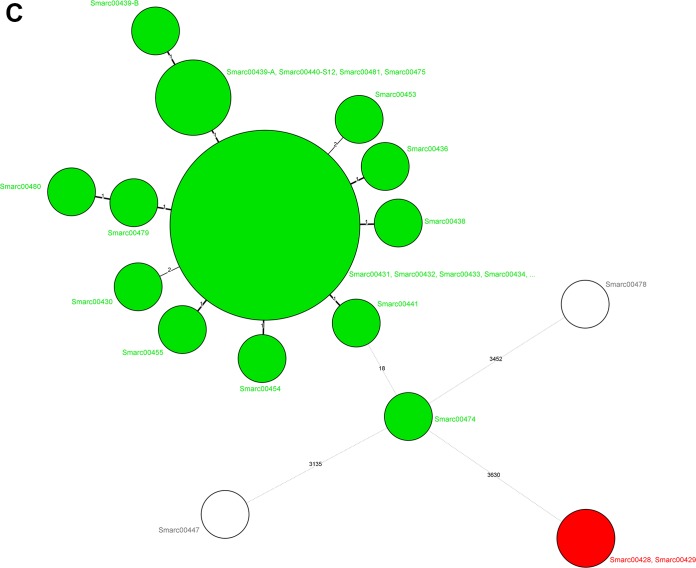
Figure 5 displays the minimum spanning trees for the three studies, and there is good concordance with the UPGMA trees in Fig. 1 and 3. The number of allele differences ranged between 0 and 4 for the epidemiologically defined strain clusters with two exceptions. There is only a single strain in the Freiburg cluster that differs by 18 alleles from its counterparts. This suggests that a cutoff value of <20 alleles would represent a conservative but useful estimate for transmission-related isolates, also given the significantly higher genetic distance between the nonrelated S. marcescens isolates. Figure 6 once more displays the robustness of the wgMLST scheme; although all WGS entries in the database were included, the strain clusters identified above remained unchanged.
FIG 5.
Minimum spanning trees for the S. marcescens isolates from Groningen (A), Cologne (B), and Freiburg (C) built from the pan-genomic allelic profiles. Colors of the circles identify the epidemiological clusters and cases of transmission. The values on the lines indicate the numbers of allelic differences between the connected isolates. Circle sizes are associated with the numbers of isolates per type. The figure implies that there are no clusters extending across hospitals. Color codes are specific for the three different panels and should not be compared between panels.
FIG 6.
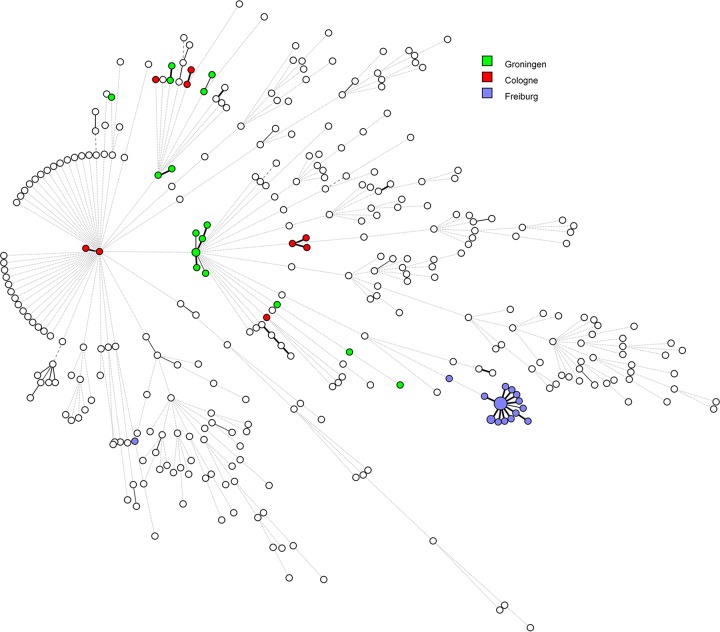
Overall genomic population structure of S. marcescens based on a combined analysis of our epidemiologically related isolates and the reference genomes that were used to construct the wgMLST scheme. Note the extended number of singletons and the occurrence of epidemic clones seemingly originating from several of such singletons. Green bullets represent isolates from Groningen, red ones the isolates from Cologne, and blue ones identify the isolates from Freiburg.
DISCUSSION
S. marcescens is a nosocomial pathogen of clinical importance, and both species identification and antimicrobial susceptibility testing are well covered in routine diagnostic clinical microbiology laboratories. However, epidemiological typing of S. marcescens is less developed, and for this reason, we developed a wgMLST scheme. The system allowed for the adequate recognition of clonally related organisms and for the detection of outbreak events. At the level of wgMLST, the numbers of changes between the most closely related isolates were <20 alleles (given the time frame during which our outbreak-related strains were captured), although a significant fraction of the closely related genomes only differed by 0 to 4 alleles. This latter level of resolution does not allow for detailed epidemiological tracing of spread from one patient to the other given the apparently low number of changes associated with such transfers. We performed a limited number of wgSNP analyses, and surprisingly, for the ten related isolates from Groningen, this did not increase the resolution. The number of SNPs encountered between the ten isolates ranged from zero to five, in the same range as the wgMLST variation and insufficient to decipher transmission of strains between patients (data not shown). Of note, a recent cgMLST study for Brucella melitensis revealed similar findings: epidemiological cutoff values for nonvariance were defined as <6 loci for wgMLST and <7 loci for wgSNP analyses, similar to what we document here for S. marcescens (19).
Next-generation sequencing (NGS) is becoming very popular in clinical microbiology (20, 21), but wgMLST for S. marcescens has not yet been described. WGS has been used to study S. marcescens virulence after wound infections and infection of snake bites (22). As well, WGS has been used to study the national dissemination of drug resistance elements and plasmids in S. marcescens throughout Germany, although only a limited number of isolates were subjected to NGS (16). The authors of that study also demonstrated that S. marcescens genomes can be used to generate genomic catalogues of antibiotic resistance genes. Relevant to our current clinical study is the work done by Iguchi et al. (23). These authors defined the WGS for two selected S. marcescens strains. Their analysis revealed a degree of genetic heterogeneity that our current study exploited. Iguchi et al. (23) already tried to define core and variable genes and used ways for defining the genetic distance between S. marcescens isolates. Most recently, Martineau and colleagues (24) used WGS to elucidate transmission patterns in a NICU in Montreal, Canada. WGS for ten clinical isolates were instrumental in resolving S. marcescens routes of spread in this setting. We were able to confirm their data using our wgMLST scheme (results not shown). In the examples brought forward by Martineau et al. (24), a single outbreak was analyzed, whereas we have now taken the method to a higher level, including the development of a dedicated wgMLST WGS database and an informatics tool for the semiautomated analysis of potential outbreak scenarios. With turnaround calculation times of less than 30 min per sample and simultaneous processing of up to 24 samples, high-powered wgMLST performance is guaranteed. Using BioNumerics and a cloud-based calculation engine provides a high-throughput environment that enables a fast and simple outbreak analysis of WGS data for S. marcescens. The calculation engine’s quality-controlled de novo assembly possibilities allow for rapid push-button assembly of WGS data without the need of local computing power. In short, even high-resolution typing needs optimal epidemiological data and cannot stand on its own. Although we here focus on patients in NICUs, it should be emphasized that genomic typing of S. marcescens will have wider implications, as these bacteria infect other risk groups as well (25, 26). We acknowledge the fact that we are not disclosing the precise methodology used for wgMLST scheme development, since this module will become available only in combination with BioNumerics.
In conclusion, all laboratory-run typing methods, wgMLST included, are valuable in the context of hospital-wide screening for pathogens but also for analyses of random clinical isolates (27, 28). wgMLST for S. marcescens has here been demonstrated to be a promising epidemiological typing support tool. In combination with tools for deciphering a genomic antibiogram and the presence of virulence genes, WGS by NGS may help trace and follow outbreaks, understand the acquisition and spread of resistance factors, and explain the disease-invoking potential for this not-to-be-underestimated human pathogen.
Supplementary Material
ACKNOWLEDGMENTS
This work was conducted in collaboration with the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Genomic and Molecular Diagnostics (ESGMD) and the ESCMID Study Group on Epidemiological Markers (ESGEM), Basel, Switzerland.
Alex van Belkum, Jill Dombrecht, Diederik Vanfleteren, and Katrien De Bruyne are employees of bioMérieux, a company designing, developing, and selling infectious disease diagnostics and hence have a business implication in this work. John W. A. Rossen consults for IDbyDNA. All other authors declare no conflicts of interest and have submitted the ICMJE form for disclosure of potential conflicts of interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed. No external financial support was provided for the studies presented herein.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01652-18.
REFERENCES
- 1.Sabat AJ, Budimir A, Nashev D, Sá-Leão R, van Dijl JM, Laurent F, Grundmann H, Friedrich AW, ESCMID Study Group of Epidemiological Markers (ESGEM). 2013. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill 18:20380. [DOI] [PubMed] [Google Scholar]
- 2.Goering RV, Köck R, Grundmann H, Werner G, Friedrich AW, ESCMID Study Group for Epidemiological Markers (ESGEM). 2013. From theory to practice: molecular strain typing for the clinical and public health setting. Euro Surveill 18:20383. [DOI] [PubMed] [Google Scholar]
- 3.Higgins PG, Prior K, Harmsen D, Seifert H. 2017. Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS One 12:e0179228. doi: 10.1371/journal.pone.0179228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossen JWA, Friedrich AW, Moran-Gilad J, ESCMID Study Group for Genomic and Molecular Diagnostics (ESGMD). 2018. Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin Microbiol Infect 24:355–360. doi: 10.1016/j.cmi.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Revez J, Zhang J, Schott T, Kivistö R, Rossi M, Hänninen ML. 2014. Genomic variation between Campylobacter jejuni isolates associated with milk-borne-disease outbreaks. J Clin Microbiol 52:2782–2786. doi: 10.1128/JCM.00931-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jolley KA, Maiden MC. 2014. Using multilocus sequence typing to study bacterial variation: prospects in the genomic era. Future Microbiol 9:623–630. doi: 10.2217/fmb.14.24. [DOI] [PubMed] [Google Scholar]
- 7.Kingry LC, Rowe LA, Respicio-Kingry LB, Beard CB, Schriefer ME, Petersen JM. 2016. Whole genome multilocus sequence typing as an epidemiologic tool for Yersinia pestis. Diagn Microbiol Infect Dis 84:275–280. doi: 10.1016/j.diagmicrobio.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kluytmans-van den Bergh MF, Rossen JW, Bruijning-Verhagen PC, Bonten MJ, Friedrich AW, Vandenbroucke-Grauls CM, Willems RJ, Kluytmans JA. 2016. Whole-genome multilocus sequence typing of extended-spectrum-beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol 54:2919–2927. doi: 10.1128/JCM.01648-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiebig L, Kohl TA, Popovici O, Mühlenfeld M, Indra A, Homorodean D, Chiotan D, Richter E, Rüsch-Gerdes S, Schmidgruber B, Beckert P, Hauer B, Niemann S, Allerberger F, Haas W. 2017. A joint cross-border investigation of a cluster of multidrug-resistant tuberculosis in Austria, Romania and Germany in 2014 using classic, genotyping and whole genome sequencing methods: lessons learnt. Euro Surveill 22:30439. doi: 10.2807/1560-7917.ES.2017.22.2.30439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadon C, Van Walle I, Gerner-Smidt P, Campos J, Chinen I, Concepcion-Acevedo J, Gilpin B, Smith AM, Man Kam K, Perez E, Trees E, Kubota K, Takkinen J, Nielsen EM, Carleton H, FWD-NEXT Expert Panel. 2017. PulseNet International: vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro Surveill 22:30544. doi: 10.2807/1560-7917.ES.2017.22.23.30544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Besler KR, Little EL. 2017. Diversity of Serratia marcescens strains associated with cucurbit yellow vine disease in Georgia. Plant Dis 101:129–136. doi: 10.1094/PDIS-05-16-0618-RE. [DOI] [PubMed] [Google Scholar]
- 12.Mahlen SD. 2011. Serratia infections: from military experiments to current practice. Clin Microbiol Rev 24:755–791. doi: 10.1128/CMR.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dramowski A, Aucamp M, Bekker A, Mehtar S. 2017. Infectious disease exposures and outbreaks at a South African neonatal unit with review of neonatal outbreak epidemiology in Africa. Int J Infect Dis 57:79–85. doi: 10.1016/j.ijid.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 14.Batah R, Loucif L, Olaitan AO, Boutefnouchet N, Allag H, Rolain JM. 2015. Outbreak of Serratia marcescens coproducing ArmA and CTX-M-15 mediated high levels of resistance to aminoglycoside and extended-spectrum beta-lactamases, Algeria. Microb Drug Resist 21:470–476. doi: 10.1089/mdr.2014.0240. [DOI] [PubMed] [Google Scholar]
- 15.Gruber TM, Göttig S, Mark L, Christ S, Kempf VA, Wichelhaus TA, Hamprecht A. 2015. Pathogenicity of pan-drug-resistant Serratia marcescens harboring blaNDM-1. J Antimicrob Chemother 70:1026–1030. doi: 10.1093/jac/dku482. [DOI] [PubMed] [Google Scholar]
- 16.Wendel AF, Kaase M, Autenrieth IB, Peter S, Oberhettinger P, Rieber H, Pfeffer K, MacKenzie CR, Willmann M. 2017. Protracted regional dissemination of GIM-1-producing Serratia marcescens in western Germany. Antimicrob Agents Chemother 61:e01880-16. doi: 10.1128/AAC.01880-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mataseje LF, Boyd DA, Delport J, Hoang L, Imperial M, Lefebvre B, Kuhn M, Van Caeseele P, Willey BM, Mulvey MR. 2014. Serratia marcescens harbouring SME-type class A carbapenemases in Canada and the presence of blaSME on a novel genomic island, SmarGI1-1. J Antimicrob Chemother 69:1825–1829. doi: 10.1093/jac/dku040. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez C, Brengi S, Cáceres MA, Mochi S, Viñas MR, Rizza CA, Merletti G, Bru E, Assa JD, Raya RR, Centrón D. 2018. Successful management with fosfomycin + ceftazidime of an infection caused by multiple highly related subtypes of multidrug-resistant and extensively drug-resistant KPC-producing Serratia marcescens. Int J Antimicrob Agents 52:737–739. doi: 10.1016/j.ijantimicag.2018.07.020. [DOI] [PubMed] [Google Scholar]
- 19.Janowicz A, De Massis F, Ancora M, Cammà C, Patavino C, Battisti A, Prior K, Harmsen D, Scholz H, Zilli K, Sacchini L, Di Giannatale E, Garofolo G. 2018. Core genome multilocus sequence typing and single nucleotide polymorphism analysis in the epidemiology of Brucella melitensis infections. J Clin Microbiol 56:e00517-18. doi: 10.1128/JCM.00517-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westblade LF, van Belkum A, Grundhoff A, Weinstock GM, Pamer EG, Pallen MJ, Dunne WM Jr, 2016. Role of clinicogenomics in infectious disease diagnostics and public health microbiology. J Clin Microbiol 54:1686–1693. doi: 10.1128/JCM.02664-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunne WM Jr, Westblade LF, Ford B. 2012. Next-generation and whole-genome sequencing in the diagnostic clinical microbiology laboratory. Eur J Clin Microbiol Infect Dis 31:1719–1726. doi: 10.1007/s10096-012-1641-7. [DOI] [PubMed] [Google Scholar]
- 22.Huang YT, Cheng JF, Liu YT, Mao YC, Wu MS, Liu PY. 2018. Genome-based analysis of virulence determinants of a Serratia marcescens strain from soft tissues following a snake bite. Future Microbiol 13:331–343. doi: 10.2217/fmb-2017-0202. [DOI] [PubMed] [Google Scholar]
- 23.Iguchi A, Nagaya Y, Pradel E, Ooka T, Ogura Y, Katsura K, Kurokawa K, Oshima K, Hattori M, Parkhill J, Sebaihia M, Coulthurst SJ, Gotoh N, Thomson NR, Ewbank JJ, Hayashi T. 2014. Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol Evol 6:2096–2110. doi: 10.1093/gbe/evu160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martineau C, Li X, Lalancette C, Perreault T, Fournier E, Tremblay J, Gonzales M, Yergeau É, Quach C. 2018. Serratia marcescens outbreak in a neonatal intensive care unit: new insights from next-generation sequencing applications. J Clin Microbiol 56:e00235-18. doi: 10.1128/JCM.00235-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leng P, Huang WL, He T, Wang YZ, Zhang HN. 2015. Outbreak of Serratia marcescens postoperative infection traced to barbers and razors. J Hosp Infect 89:46–50. doi: 10.1016/j.jhin.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Us E, Kutlu HH, Tekeli A, Ocal D, Cirpan S, Memikoglu KO. 2017. Wound and soft tissue infections of Serratia marcescens in patients receiving wound care: a health care-associated outbreak. Am J Infect Control 45:443–447. doi: 10.1016/j.ajic.2016.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Dawczynski K, Proquitté H, Roedel J, Edel B, Pfeifer Y, Hoyer H, Dobermann H, Hagel S, Pletz MW. 2016. Intensified colonization screening according to the recommendations of the German Commission for Hospital Hygiene and Infectious Diseases Prevention (KRINKO): identification and containment of a Serratia marcescens outbreak in the neonatal intensive care unit, Jena, Germany, 2013-2014. Infection 44:739–746. doi: 10.1007/s15010-016-0922-y. [DOI] [PubMed] [Google Scholar]
- 28.Åttman E, Korhonen P, Tammela O, Vuento R, Aittoniemi J, Syrjänen J, Mattila E, Österblad M, Huttunen R. 2018. A Serratia marcescens outbreak in a neonatal intensive care unit was successfully managed by rapid hospital hygiene interventions and screening. Acta Paediatr 107:425–429. doi: 10.1111/apa.14132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.