Abstract
Purpose:
Triple-negative breast cancers (TNBC) are often resistant to treatment with ionizing radiation (IR). We sought to investigate whether pharmacologic inhibition of Chk1 kinase, which is commonly overexpressed in TNBC, preferentially sensitizes TNBC cells to IR.
Methods:
Ten breast cancer cell lines were screened with small molecule inhibitors against Chk1 and other kinases. Chk1 inhibition was also tested in isogenic KRAS mutant or wild-type cancer cells. Cellular radiosensitization was measured by short-term and clonogenic survival assays and by staining for the DNA double-strand break (DSB) marker γ-H2AX. Radiosensitization was also assessed in breast cancer biopsies using an ex vivo assay. Aurora B kinase-dependent mitosis-like chromatin condensation, a marker of radioresistance, was detected using a specific antibody against co-localized phosphorylation of serine 10 and trimethylation of lysine 9 on histone 3 (H3K9me3/S10p). Expression of CHEK1 and associated genes was evaluated in TNBC and lung adenocarcinoma.
Results:
Inhibition of Chk1 kinase preferentially radiosensitized TNBC cells in vitro and in patient biopsies. Interestingly, TNBC cells displayed lower numbers of IR-induced DSBs than non-TNBC cells, correlating with their observed radioresistance. We found that Chk1 suppressed IR-induced DSBs in these cells, which was dependent on H3K9me3/S10p – a chromatin mark previously found to indicate radioresistance in KRAS mutant cancers. Accordingly, the effects of Chk1 inhibition in TNBC were reproduced in KRAS mutant but not wild-type cells. We also observed co-expression of genes in this Chk1 chromatin pathway in TNBC and KRAS mutant lung cancers.
Conclusions:
Chk1 promotes an unexpected, common phenotype of chromatin-dependent DSB suppression in radioresistant TNBC and KRAS mutant cancer cells, providing a direction for future investigations into overcoming the treatment resistance of TNBC.
Keywords: Triple-negative breast cancer, radioresistance, radiosensitization, Chk1, DNA double-strand breaks
Introduction
Triple-negative breast cancer (TNBC) constitutes approximately 15–20% of all invasive breast cancers and is considered the most aggressive of the breast cancer subtypes [1, 2]. It is classically defined by the absence of cellular expression of three receptors: estrogen, progesterone, and HER2/neu and it is often characterized by high tumor grade, an elevated rate of lymph node metastases, and a pattern of early, often distant recurrences [3]. Gene expression profiling studies have further stratified TNBC’s, characterizing them into a few molecular categories, including basal-like, mesenchymal, immune-enriched, and luminal androgen subtypes, with data supporting both unique response rates to systemic therapy as well as differences in survival by subgroup [4, 5].
Due to the aggressive nature of TNBC, patients often receive multimodality therapy including surgery, chemotherapy, and radiation. Yet, local tumor failure rates in this population remain higher than in other breast cancer subtypes, with 5-year failure rates as high as 10–20% [6–8]. This higher rate of local failure among TNBC patients is often attributed to an inherent radioresistance of TNBCs despite the presence of BRCA-like DNA repair defects [2, 9]. However, the mechanisms of radioresistance are poorly understood.
Molecular targets to overcome the treatment resistance of TNBC have remained elusive. TNBCs commonly overexpress the epidermal growth factor receptor (EGFR) [10]. However, early-phase clinical trials have failed to demonstrate significant activity of EGFR-targeted monoclonal antibodies or tyrosine kinase inhibitors and combinations with radiation treatment are poorly studied [11]. Other studies have shown that Chk1 is overexpressed and may be a therapeutic target in TNBC [12, 13]. Chk1 is a serine/threonine-specific kinase, encoded by CHEK1, which regulates the DNA damage response, homologous recombination repair (HRR), and several cell cycle checkpoints [14]. Preclinical data suggest that Chk1 inhibition is an effective radiosensitization strategy, particularly in tumors with non-functional p53 [15].
We, therefore, set out to investigate the radiosensitizing properties of a clinically relevant Chk1 inhibitor in a panel of triple-negative and hormone receptor-positive breast cancer cell lines. We report an unexpected chromatin-dependent suppression of DNA double-strand breaks (DSBs) by Chk1 which correlates with the radioresistance of TNBC cells. Interestingly, TNBC shares this phenotype with KRAS mutated cancer, which yields potentially novel therapeutic targets to overcome the treatment resistance of TNBC.
Material and Methods
Cell Lines
The breast cancer cell lines BT-20, BT-549, EFM-19, MCF-7, MDA-MB-157, MDA-MB-231, MDA-MB-361, MDA-MB-436, MDA-MB-468, T-47D were purchased from ATCC. NCI-H1703 (KRAS mutant vs wild-type), DLD-1 (KRAS G13D/wt) and DWT7 (del/wt) were previously described [16]. MCF-7, MDA-MB-157, MDA-MB-231, MDA-MB-361, MDA-MB-436, MDA-MB-468 were cultivated in DMEM, BT-549, EFM-19, T-47D, NCI-H1703 in RPMI, DLD-1 and DWT7 in McCoy media, and BT-20 in Eagle’s Minimum Essential Medium, supplemented with 10% BGS, 2mM glutamine, 100 U/ml penicillin and 10% HEPES (all Sigma-Aldrich). All cell lines were cultured in a humidified incubator at 37°C and 5% CO2 and passaged for < 3 months after thawing a given frozen vial. All cell lines were tested mycoplasma free prior to the experiments (MycoAlert, Lonza) and none was ever treated for mycoplasma throughout the experiments. For 3D culture of tumor spheres, ~10,000 cells/well were grown in black round bottom polystyrene ultra-low attachment microplates (Corning) using serum-free medium composed of DMEM (Sigma-Aldrich), basic fibroblast growth factor (bFGF), and EGF (20 ng/mL each, Sigma-Aldrich), and B27 supplement (Life Technologies).
Tumor tissues
Tumor tissues were derived from untreated breast cancer patients under a protocol approved by the Institutional Review Board. Tumor samples were processed for ex-vivo foci analysis adapting a previously published protocol for breast cancer [17]. Dispensable tumor tissue not needed for pathological diagnosis were placed in RMPI medium within about 30 minutes of resection. Within 30–60 minutes the samples arrived in the laboratory and specimens were divided into samples of approximately 5 mm size. Samples were mock treated or exposed to 100 nm LY2603618 for 24 hours and irradiated with 6 Gy radiation, followed by snap freezing at 30 minutes.
Treatments
Ionizing radiation was administered using a Siemens Stabilipan 2 X-ray generator operated at 250 kVp and 12 mA, at a dose rate of 1.8 Gy/minute. LY2603618, VE-821 and KU-55933 were dissolved in dimethyl sulfoxide (DMSO) (LC Laboratories). Drugs were added 1 hour prior to irradiation and maintained for the duration of the respective experiment.
Cell proliferation and survival assays
Clonogenic cell survival assays and short-term radiosensitization experiments were performed as described [16, 18]. Short-term radiosensitization was assessed 5 days post-irradiation using the CellTiter-Glo (CTG) luminescence assay (Promega).
Immunofluorescence microscopy
Staining and visualization of γ-H2AX foci or trimethyl (Lys9)-phospho(ser10)-Histone H3 in vitro and in vivo were performed as described [19]. Exponentially growing cells were plated into 8-well chamber slides and treated with LY2602618 for 1 hour prior to irradiation. After 0.5 or 24 hours, cells were fixed for 15 minutes with 4% paraformaldehyde at room temperature. Subsequently, permeabilization was achieved using Triton X-100 (Sigma-Aldrich) in phosphate-buffered saline. Following blocking with 10% serum for 1 hour at room temperature, cells were incubated for 2 hours at 37°C with anti-γ-H2AX (1:100 dilution, #4411-PC-100 from Trevigen, Gaithersburg, USA) or anti-trimethyl (Lys9)-phospho(ser10)-Histone H3 (EMD Millipore 08–509) respectively. This was followed by incubation with species-specific Alexa-488-conjugated secondary antibody (Pierce #31583 or Molecular Probes #A-21441). All slides were counterstained with 40,6-diamidino-2-phenylindole (DAPI) and visualized by fluorescence microscopy (Olympus BX51). At least 150 nuclei were evaluated for each data point.
Gene expression analysis
To assess expression differences in genes of interest between breast cancer patients with/without TN status or lung cancer patients with/without mutant KRAS, we leveraged the Oncomine database [20]. We extracted fold-changes in gene expression along with T-statistics. We corrected all p-values for multiple hypothesis testing with false-discovery rate [21].
Results
Preferential radiosensitization of TNBC cells by Chk1 inhibition
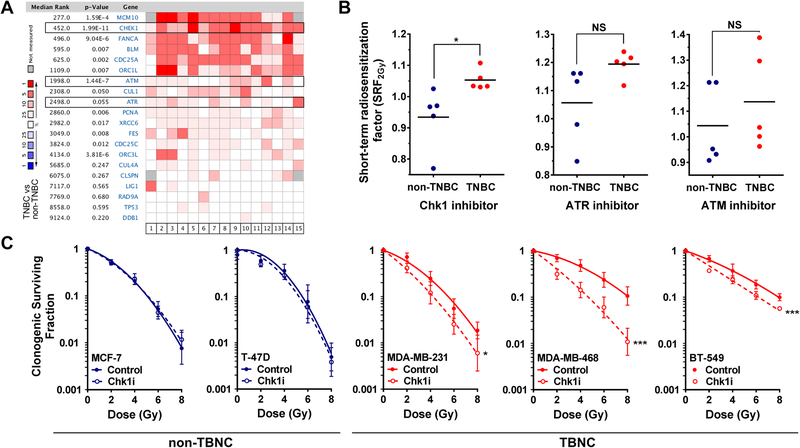
In a meta-analysis of 15 clinical databases, we observed CHEK1 to be upregulated to a higher degree in TNBC than its upstream kinases ATR and ATM (Fig. 1A), prompting us to assess these three kinases as targets for radiosensitization of TNBC cells. In a panel of five TNBC and five non-TNBC cell lines, only pharmacological inhibition of Chk1 led to preferential radiosensitization of TNBC cells using a validated screening assay (Fig. 1B) (Suppl. Fig. S1A,B) [18]. In contrast, ATM and ATR inhibitors caused varying degrees of radiosensitization in both subsets of cell lines (Fig. 1B). Thus, expression of CHEK1 and its importance for radioresistance of TNBC cells is distinct from ATR as well as ATM. The preferential Chk1-dependent sensitization of radioresistant TNBC cell lines was confirmed with two additional assays (Fig. 1C, Suppl. Fig. S2A). Compared to non-TNBC cells, TNBC cells were also somewhat more sensitive to Chk1 inhibitor alone (Suppl. Fig. S2B).
Fig. 1. Preferential radiosensitization of TNBC cells by Chk1 inhibition.
A, Meta-analysis across 15 published clinical data bases to illustrate expression patterns of triple-negative breast cancer (TNBC) and non-TNBC cell lines for a panel of genes associated with Chk1.
B, Comparison of short-term radiosensitization factors (SRF2Gy) for selective inhibitors of Chk1 (LY2603618, 100 nM), ATR-(VE-821, 2.5 μM), and ATM-(KU-55933, 2.5 μM) in TNBC and non-TNBC cell lines. Each data point represents averages of at least 3 biological repeats and horizontal lines indicate the mean. Statistical comparisons were performed with the unpaired T-test (two-tailed).
C, Clonogenic survival of non-triple-negative (MCF-7, T-47D) and triple-negative cell lines (MDA-MB-231, MDA-MB-468, BT-549) after single dose irradiation with or without LY2603618 (100 nM) treatment initiated 1 hour before irradiation. Statistical comparisons were based on 3 independent repeat experiments and carried out using the F-test (GraphPad Prism 6). * p≤0.05, ** p ≤0.01, *** p ≤0.001
Chk1 suppresses ionizing radiation (IR)-induced DSBs in TNBC cells in vitro and in tumor biopsies
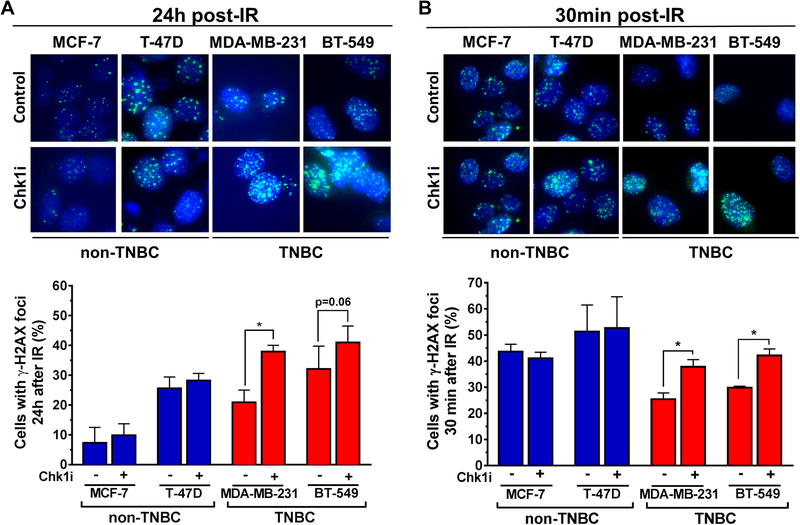
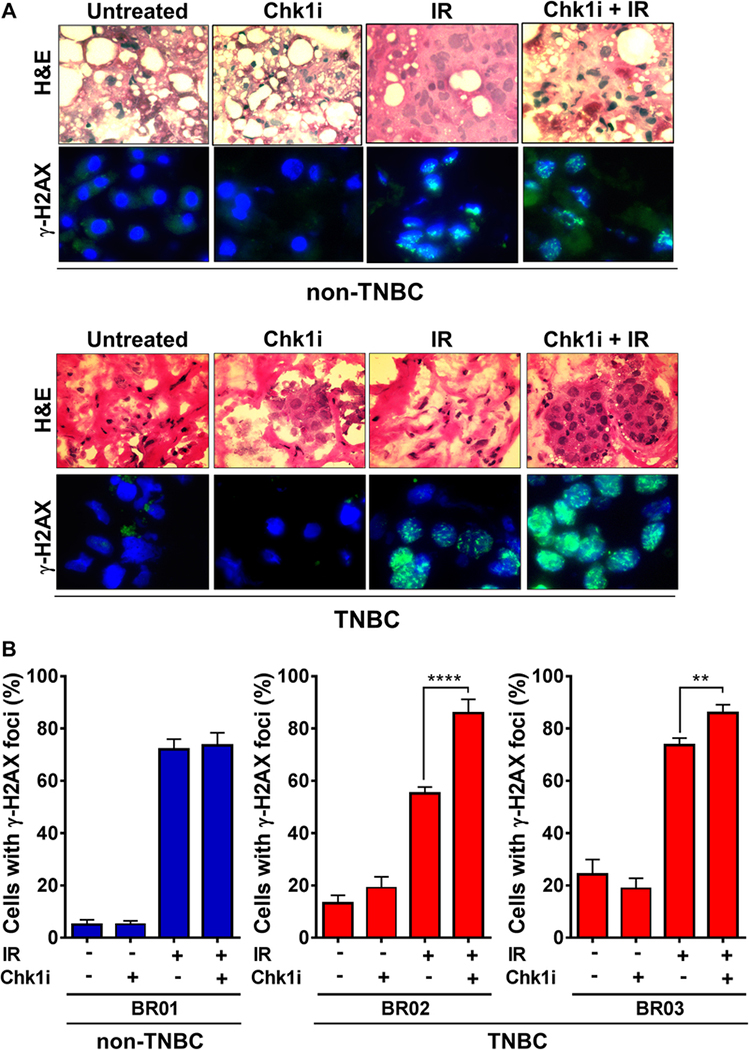
We next examined the effect of Chk1 inhibition on residual (at 24 hours post IR) as well as induced (at 30 minutes) DSB using the established DSB marker γ-H2AX in irradiated TNBC and non-TNBC cell lines. Chk1 inhibition not only increased the number of residual DSB in TNBC cells but unexpectedly also the number of DSBs induced by IR (Fig. 2A,B). The magnitude of effect was very similar for these endpoints, i.e., in the order of a ~1.5-fold increase of cells with DSBs upon Chk1 inhibition. In contrast, there was no effect of Chk1 inhibition on DSB numbers in non-TNBC cells. Of note, because Chk1 inhibitor is only added 1 hour prior to IR, there was not sufficient time for any Chk1-dependent cell cycle disturbances to affect the γ-H2AX readout. To confirm the in vitro findings in patient samples, we utilized a previously employed ex vivo assay where fresh tumor biopsies from breast cancer patients are subjected to IR and drug treatments in the laboratory [17]. Again, we observed that Chk1 inhibitor treatment increased induced DSB numbers preferentially in TNBC cells (Fig. 3A,B).
Figure 2. Chk1 suppresses the induction of DSB in TNBC cells.
A, Non-TNBC (MCF-7, T-47-D) and TNBC (MDA-MB-231, BT-549) cell lines were analyzed 24 hours following irradiation with 6 Gy ± LY2603618 treatment (100 nM) initiated 1 hour prior to irradiation. Upper panel, representative images of γ-H2AX immunofluorescence staining (40X). Lower panel, percentage of cells with ≥ 20 foci/nucleus at 24 hours post-irradiation.
B, Analogous comparison of γ-H2AX signal 30 minutes following irradiation with 1 Gy ± LY2603618 treatment (100 nM) initiated 1 hour before irradiation. Bars represent mean ± standard error based on 3 biological repeats. Statistical comparison was performed with the t test.
Figure 3. Chk1 suppresses the induction of DSB in TNBC patient tumor biopsies.
A, Representative images of ex vivo γ-H2AX foci in breast cancer patient tumor biopsies. Tumor biopsies were incubated under standard cell culture conditions and exposed to LY2603618 (100 nM) or mock treatment for 24h. Specimens were snap frozen 30 minutes after 1 Gy irradiation or mock treatment and analyzed by immunofluorescence microscopy and H&E staining.
B, Percentage of cells with LY2603618-induced γ-H2AX foci at 30 minutes post-irradiation in breast cancer explants. Bars represent mean ± standard error based on 8–10 random images and 200–400 nuclei per data point.
Chk1-dependent radiosensitization correlates with mitosis-like chromatin condensation
The observed suppression of IR-induced DSB by Chk1 inhibition and the magnitude of effect were reminiscent of a previously observed role of the EGFR in KRAS mutant cancer cells [19]. In that study, we uncovered a pathway consisting of EGFR, PKCα, and Aurora B kinase, which promotes mitosis-like chromatin condensation in interphase cells and protects against IR-induced DSB specifically in KRAS mutant cancer cells. As a result, KRAS mutant cells displayed lower numbers of DSB than KRAS wild-type cells following IR treatment [19].
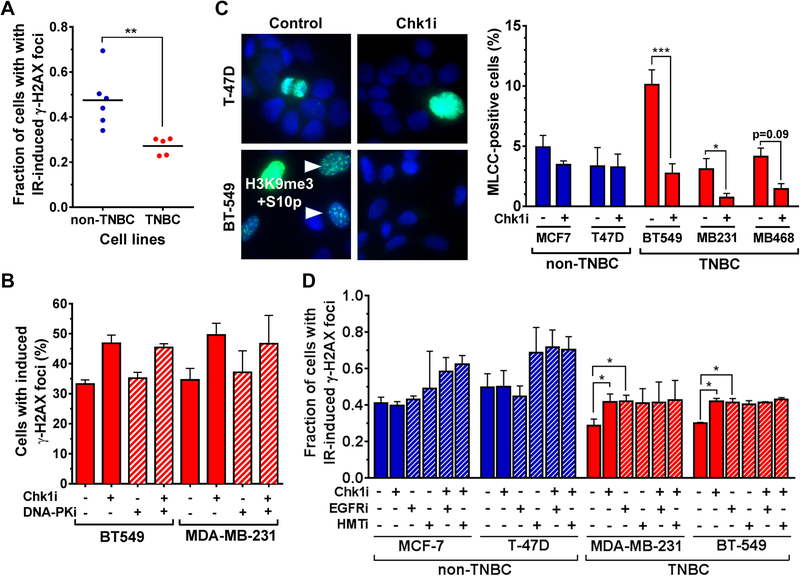
Similarly, TNBC cells clearly exhibited less induced DSBs than non-TBNC directly after irradiation which correlated with their enhanced radioresistance (Fig. 4A). To address the possibility that the suppression of DSB induction in TNBC cells by Chk1 was dependent on chromatin structure, similar to KRAS mutant cancer cells, we first confirmed that pharmacological blockade of non-homologous end-joining (NHEJ), which operates within minutes after irradiation, did not influence the effects of Chk1 inhibition (Fig. 4B). HRR was not thought to affect DSBs that early after their induction because this repair pathway operates much slower than NHEJ, and we have not found altered HRR in TNBC cell lines (Suppl. Fig S3).
Figure 4. Chk1-dependent radiosensitization correlates with mitosis-like chromatin condensation.
A, Left, comparison of percentage with ≥ 20 IR-induced γ-H2AX foci of non-TNBC and TNBC cell lines 30 minutes following irradiation with 1 Gy. Statistical comparison was performed with the t test.
B, Percentage of TNBC cell lines (BT-549 and MDA-MB-231) with ≥ 20 IR-induced γ-H2AX foci ± LY2603618 and ± DNA-PKcs inhibitor NU7026 (10 μM) initiated 1 hour prior to irradiation.
C, Left panel, representative immunofluorescence images showing co-localized phospho-H3S10 and H3K9me3 marking mitosis-like chromatin condensation (MLCC) [19]. Arrows, punctate interphase-like staining pattern. Diffuse nuclear staining consistent with metaphase. Right panel, percentage of cells with punctuated mitosis-like chromatin staining pattern 30 minutes after Chk1 inhibitor (Chk1i) LY2603618 (100 nM) or mock treatment. Bars represent mean ± standard error based on 3 independent repeats.
D, Fraction of non-TNBC (MCF-7, T-47D) and TNBC (MDA-MB-231, BT-549) cells with ≥20 IRinduced γ-H2AX foci ± LY2603618 (100 nM), ± erlotinib (2 μM), and ± histone-methyltransferase (HMT) inhibitor Chaetocin (100 nM). Bars represent mean ± standard error based on 3 independent repeats.
We then assayed for Aurora B-dependent mitosis-like chromatin condensation using a highly specific antibody against co-localized serine 10 phosphorylation and lysine 9 trimethylation on histone 3 (H3K9me3/S10p), as described previously [16, 19]. Fig. 4C, left panel, illustrates the distinct granular staining pattern of the binary H3K9me3/S10p modification that is observable in interphase cells. Strikingly, incubation with Chk1 inhibitor for 1 hour resulted in a reduction of this staining signal in the TNBC cell lines but not the non-TNBC cell lines (Fig. 4C, right panel). Further, analogously to our findings in KRAS mutant cancer cells, EGFR inhibition mirrored, and was epistatic with, the increase in DSBs seen with Chk1 inhibition (Fig. 4D). Next, we treated cells with a histone methyl-transferase inhibitor to block H3K9me3 formation and hence abrogate the putative target for Chk1. Analogous to Chk1 and EGFR inhibition, this treatment increased DSBs, and no additional increase was seen in Chk1 inhibitor co-treated cells, suggesting an epistatic effect (Fig. 4D).
These data suggested that Chk1 modulates an Aurora B kinase specific chromatin signal outside of mitosis. Indeed, Aurora B protein expression was observed in the G1 phase of TNBC cells and pharmacological inhibition of Aurora B kinase increased DSBs throughout interphase (Suppl. Fig. 4A, B). Interestingly, in addition to previously reported EGFR and CHEK1, AURBK was also more highly expressed in TNBC cells lines compared to receptor-positive cell lines, which is consistent with the hypothesized functional interaction of these genes (Fig. S5).
Effects of Chk1 inhibition in KRAS mutant cancer cells mirror TNBC findings
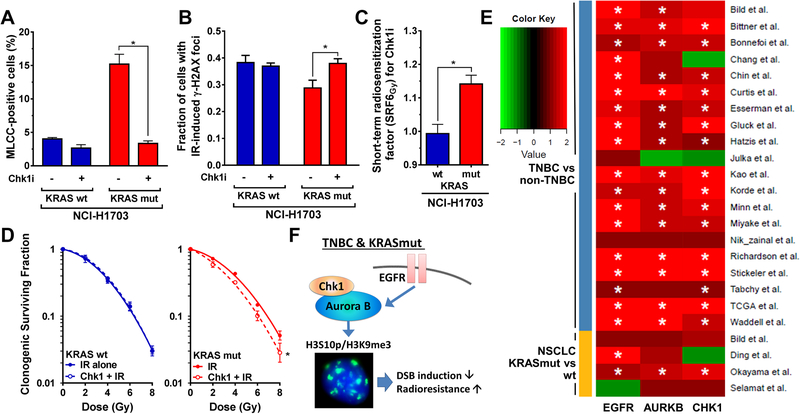
To further investigate phenotypical similarities between the triple-negative and KRAS mutant states, we assessed the effects of Chk1 inhibition in isogenic KRAS mutant and wild-type cancer cells. In KRAS mutant cells, there was a more pronounced H3K9me3/S10p signal compared to wild-type cells, and this was sensitive to Chk1 inhibition (Fig. 5A). Conversely, upon Chk1 inhibition the number of IR-induced DSBs was increased in KRAS mutant but not wild-type cells (Fig. 5B). As predicted, Chk1 inhibition specifically sensitized KRAS mutant cells to IR (Fig. 5C,D). Lastly, because the H3K9me3/S10p modification is dependent on Aurora B kinase activity [19] and in light of our in vitro findings (Fig. 4), we explored the expression of CHEK1 and AURBK as well as EGFR in breast and lung cancers (Fig. 5E). Strikingly, overexpression of these genes in TNBC and KRAS mutant cancers was clearly correlated. Taken together, the data indicate that a Chk1-dependent pathway of DSB suppression is associated with radioresistance in both TNBC and KRAS mutant cancer (model in Fig. 5F, see Discussion).
Figure 5. Effects of Chk1 inhibition in KRAS mutant cancer cells.
A, Percentage of cells with mitosis-like chromatin condensation (MLCC) staining pattern in isogenic NCI-H1703 lung cancer cells with or without mutant KRAS expression 30 minutes after LY2603618 (100 nM) or mock treatment. In all panels, bars represent mean with standard error based on at least 3 repeats. Statistical comparisons were performed with the t test.
B, Fraction of NCI-H1703 cells with or without mutant KRAS expression with ≥ 20 IR-induced γ-H2AX foci as a function of LY2603618 (100 nM) treatment.
C, Short-term radiosensitization factors (SRF6Gy) for LY2603618 (100 nM) for isogenic NCI-H1703 cells grown as 3D sphere to augment the difference between wild-type (wt) and mutant (mut) KRAS [16].
D, Clonogenic survival of isogenic DLD-1 cells harboring a mutant KRAS or the DWT7 derivative line with mutant KRAS allele deleted after single dose irradiation with or without LY2603618 (100 nM) treatment initiated 1 hour before irradiation. Statistical comparison was carried out using the F-test.
E, Expression of selected genes in the hypothesized Chk1 pathway of chromatin control. Asterisks indicate significance of a t-test at a level of q < 0.05, where q is the correct p-value based on the false-discovery-rate.
F, Model of Chk1-dependent mitosis-like chromatin condensation (MLCC), which represses DSB induction and promotes radioresistance of TNBC and KRAS mutant tumor cells. Aurora B and EGFR were previously implicated in supporting MLCC expression [19].
Discussion
Triple-negative breast cancer comprises a challenging subset of tumors owing to often poor prognosis, resistance to standard treatments including radiation, and lack of targeted therapies to date. Here, we suggest that Chk1 kinase is a potential target for radiosensitization of TNBC cells (Fig. 1–3). Mechanistically, Chk1 inhibition increases the number of radiation-induced DSBs in a manner that is dependent on EGFR signaling and a mitosis-like chromatin signal known to be promoted by Aurora B kinase (Fig. 4) [19]. This phenotype bears a striking resemblance to a previously described radioresistance mechanism in KRAS mutant cancer cells (Fig. 5F) [16, 19]. In irradiated KRAS mutant cells, a non-canonical pathway including EGFR, PKCα, and Aurora B suppresses the induction of DSBs and promotes a cancer stem cell-like phenotype, thereby leading to radioresistance [16, 19]. Our current data suggest that Chk1 plays an additional role in this pathway in KRAS mutant cells (Fig. 5A-D).
The exact mechanisms by which Chk1 impacts radioresistance of TNBC cells remain to be elucidated. Chk1 has multiple functions in cell cycle control and HRR [14]. Interestingly, Chk1 also regulates the activity of Aurora B kinase in mitosis [22]. It is tempting to speculate that the chromatin modifying role of Aurora B in interphase cells is similarly modified by Chk1 kinase activity but more mechanistic study is needed. The difference between CHEK1 and ATR (Fig. 1A,B) and the observed co-expression of CHEK1 and AURBK genes in TNBC as well as KRAS mutated lung cancers is consistent with this notion (Fig. 5E).
Do additional commonalities exist between KRAS mutated cancers and TNBC? Mutations in KRAS are very rare in breast cancers, including TNBC [23]. However, increased KRAS activity has been reported in basal-type breast cancers compared to other subtypes, and TNBCs are enriched for the functional KRAS variant rs61764370 [24, 25]. Interestingly, in the context of inter-tumoral heterogeneity among KRAS mutant cancers, we recently linked co-mutations in the TP53 tumor suppressor to the radioresistant phenotype [16, 26]. Similarly, TP53 mutations are common in TNBC [23]. EGFR is typically overexpressed in TNBC, and we have implicated EGFR in the observed radioresistance phenotype in both TNBC and in KRAS mutated cancer cells. We have further identified important roles of not only EGFR but also PKCα in a cancer stem cell-like phenotype of KRAS mutated tumors [16, 18]. Consistent with this idea, the Weinberg lab reported that PKCα activity associates with stem-ness and TNBC [27].
Taken together, our findings reveal an unexpected, common phenotype of Chk1-mediated cellular radioresistance associated with triple-negative receptor status and mutated KRAS. Our data not only draw attention to cancer type-independent mechanisms of radioresistance but also raise the important question as to whether targeted treatment approaches in KRAS mutated cancers could be applied to TNBC as well. Interestingly, both TNBC and KRAS mutated lung cancer with TP53 co-mutations are promising targets for immunotherapy [28, 29]. Additional study will be needed to establish novel targeted therapy strategies for overcoming the treatment resistance and prognosis of these challenging tumor types.
Supplementary Material
Acknowledgements
This work was in part supported by the American Cancer Society 123420RSG-12-224-01-DMC (H. Willers), UK Wellcome Trust 102696 (C.H. Benes), the Werner-Otto-Stiftung, Hamburg (P.H. Dinkelborg), and the National Cancer Institute of the National Institutes of Health under Award Number U01CA220714 (H. Willers, C.H. Benes). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Compliance with Ethical Standards
All patient tumor samples were collected under a protocol approved by the institutional review board. The following authors declare: Benes, C.H. – funding from Novartis, Amgen, Araxes; Hong, T. S. – consultant/advisory role: Clinical Genomics, EMD Serono, funding from Novartis, Taiho; Juric, D. – consultant/advisory role: Novartis, EMD Serono, Eisai, Genentech. All other authors declare that they have no conflict of interest.
References
- 1.Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F, Valero V, Lehmann BD, Pietenpol JA, Hortobagyi GN et al. : Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res 2013, 19(19):5533–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moran MS: Radiation therapy in the locoregional treatment of triple-negative breast cancer. Lancet Oncol 2015, 16(3):e113–122. [DOI] [PubMed] [Google Scholar]
- 3.Foulkes WD, Smith IE, Reis-Filho JS: Triple-negative breast cancer. N Engl J Med 2010, 363(20):1938–1948. [DOI] [PubMed] [Google Scholar]
- 4.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC et al. : Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res 2015, 21(7):1688–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA: Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011, 121(7):2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arvold ND, Taghian AG, Niemierko A, Abi Raad RF, Sreedhara M, Nguyen PL, Bellon JR, Wong JS, Smith BL, Harris JR: Age, breast cancer subtype approximation, and local recurrence after breast-conserving therapy. J Clin Oncol 2011, 29(29):3885–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jwa E, Shin KH, Kim JY, Park YH, Jung SY, Lee ES, Park IH, Lee KS, Ro J, Kim YJ et al. : Locoregional Recurrence by Tumor Biology in Breast Cancer Patients after Preoperative Chemotherapy and Breast Conservation Treatment. Cancer Res Treat 2016, 48(4):1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C, Wang S, Israel HP, Yan SX, Horowitz DP, Crockford S, Gidea-Addeo D, Clifford Chao KS, Kalinsky K, Connolly EP: Higher locoregional recurrence rate for triple-negative breast cancer following neoadjuvant chemotherapy, surgery and radiotherapy. Springerplus 2015, 4:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L: Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 2016, 13(11):674–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM: Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist 2013, 18(2):123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa R, Shah AN, Santa-Maria CA, Cruz MR, Mahalingam D, Carneiro BA, Chae YK, Cristofanilli M, Gradishar WJ, Giles FJ: Targeting Epidermal Growth Factor Receptor in triple negative breast cancer: New discoveries and practical insights for drug development. Cancer Treat Rev 2017, 53:111–119. [DOI] [PubMed] [Google Scholar]
- 12.Albiges L, Goubar A, Scott V, Vicier C, Lefebvre C, Alsafadi S, Commo F, Saghatchian M, Lazar V, Dessen P et al. : Chk1 as a new therapeutic target in triple-negative breast cancer. Breast 2014, 23(3):250–258. [DOI] [PubMed] [Google Scholar]
- 13.Ma CX, Cai S, Li S, Ryan CE, Guo Z, Schaiff WT, Lin L, Hoog J, Goiffon RJ, Prat A et al. : Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Invest 2012, 122(4):1541–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu Z, Oleinick NL, Zhang J: ATR/CHK1 inhibitors and cancer therapy. Radiother Oncol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan MA, Parsels LA, Zhao L, Parsels JD, Davis MA, Hassan MC, Arumugarajah S, Hylander-Gans L, Morosini D, Simeone DM et al. : Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res 2010, 70(12):4972–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, Han J, Marcar L, Black J, Liu Q, Li X, Nagulapalli K, Sequist LV, Mak RH, Benes CH et al. : Radiation Resistance in KRAS-Mutated Lung Cancer Is Enabled by Stem-like Properties Mediated by an Osteopontin-EGFR Pathway. Cancer Res 2017, 77(8):2018–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willers H, Gheorghiu L, Liu Q, Efstathiou JA, Wirth LJ, Krause M, von Neubeck C: DNA Damage Response Assessments in Human Tumor Samples Provide Functional Biomarkers of Radiosensitivity. Semin Radiat Oncol 2015, 25(4):237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, Wang M, Kern AM, Khaled S, Han J, Yeap BY, Hong TS, Settleman J, Benes CH, Held KD et al. : Adapting a drug screening platform to discover associations of molecular targeted radiosensitizers with genomic biomarkers. Mol Cancer Res 2015, 13(4):713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Kern AM, Hulskotter M, Greninger P, Singh A, Pan Y, Chowdhury D, Krause M, Baumann M, Benes CH et al. : EGFR-mediated chromatin condensation protects KRAS-mutant cancer cells against ionizing radiation. Cancer Res 2014, 74(10):2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM: ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hochberg Y, Benjamini Y: More powerful procedures for multiple significance testing. Stat Med 1990, 9(7):811–818. [DOI] [PubMed] [Google Scholar]
- 22.Petsalaki E, Akoumianaki T, Black EJ, Gillespie DA, Zachos G: Phosphorylation at serine 331 is required for Aurora B activation. J Cell Biol 2011, 195(3):449–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas N: Comprehensive molecular portraits of human breast tumours. Nature 2012, 490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim RK, Suh Y, Yoo KC, Cui YH, Kim H, Kim MJ, Gyu Kim I, Lee SJ: Activation of KRAS promotes the mesenchymal features of basal-type breast cancer. Exp Mol Med 2015, 47:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paranjape T, Heneghan H, Lindner R, Keane FK, Hoffman A, Hollestelle A, Dorairaj J, Geyda K, Pelletier C, Nallur S et al. : A 3’-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol 2011, 12(4):377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong TS, Wo JW, Borger DR, Yeap BY, McDonnell EI, Willers H, Blaszkowsky LS, Kwak EL, Allen JN, Clark JW et al. : Phase II study of proton-based stereotactic body radiation therapy for liver metastases: Importance of tumor genotype. J Natl Cancer Inst 2017, [DOI] [PubMed] [Google Scholar]
- 27.Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F, Wu ZJ, Krall JA, Bierie B, Guo W et al. : Protein kinase C alpha is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell 2013, 24(3):347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, Behrens C, Kadara H, Parra ER, Canales JR et al. : Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015, 5(8):860–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nanda R, Chow LQ, Dees EC, Berger R, Gupta S, Geva R, Pusztai L, Pathiraja K, Aktan G, Cheng JD et al. : Pembrolizumab in Patients With Advanced Triple-Negative Breast Cancer: Phase Ib KEYNOTE-012 Study. J Clin Oncol 2016, 34(21):2460–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.