Abstract
The poor outcomes in infant acute lymphoblastic leukemia (ALL) necessitate new treatments. Here we discover that EIF4E protein is elevated in most cases of infant ALL and test EIF4E targeting by the repurposed antiviral agent ribavirin, which has anticancer properties through EIF4E inhibition, as a potential treatment. We find that ribavirin treatment of actively dividing infant ALL cells on bone marrow stromal cells (BMSCs) at clinically achievable concentrations causes robust proliferation inhibition in proportion with EIF4E expression. Further, we find that ribavirin treatment of KMT2A-rearranged (KMT2A-R) infant ALL cells and the KMT2A-AFF1 cell line RS4:11 inhibits EIF4E, leading to decreases in oncogenic EIF4E-regulated cell growth and survival proteins. In ribavirin-sensitive KMT2A-R infant ALL cells and RS4:11 cells, EIF4E-regulated proteins with reduced levels of expression following ribavirin treatment include MYC, MCL1, NBN, BCL2 and BIRC5. Ribavirin-treated RS4:11 cells exhibit impaired EIF4E-dependent nuclear to cytoplasmic export and/or translation of the corresponding mRNAs, as well as reduced phosphorylation of the p-AKT1, p-EIF4EBP1, p-RPS6 and p-EIF4E signaling proteins. This leads to an S-phase cell cycle arrest in RS4:11 cells corresponding to the decreased proliferation. Ribavirin causes nuclear EIF4E to re-localize to the cytoplasm in KMT2A-AFF1 infant ALL and RS4:11 cells, providing further evidence for EIF4E inhibition. Ribavirin slows increases in peripheral blasts in KMT2A-R infant ALL xenograft-bearing mice. Ribavirin cooperates with chemotherapy, particularly L-asparaginase, in reducing live KMT2A-AFF1 infant ALL cells in BMSC co-cultures. This work establishes that EIF4E is broadly elevated across infant ALL and that clinically relevant ribavirin exposures have preclinical activity and effectively inhibit EIF4E in KMT2A-R cases, suggesting promise in EIF4E targeting using ribavirin as a means of treatment.
Keywords: Acute lymphoblastic leukemia; Infant, KMT2A; EIF4E; Ribavirin; Obatoclax
Introduction:
Leukemia causes the most cancer-related deaths in the first year of life and its incidence is increasing (SEER13 1992–2013) (1). Event-free survival in infant ALL, the commonest leukemia in infants <1 year old, is ~45% (2–6). KMT2A (Lysine Methyltransferase 2A; MLL) gene rearrangement, which is present in 70–80% of cases, is associated with dismal outcomes (7–9). Even in KMT2A-germline (KMT2A-G) cases, outcomes are inferior compared to ALL in children >1 year old. Molecularly tailored treatments for infant ALL are essential because intensive chemotherapy with or without hematopoietic stem cell transplantation has not improved these outcomes (2–6).
One opportunity to discover novel treatments is to identify correlations between in vitro cytotoxicity and gene expression profiles (10, 11). Here, while analyzing an infant ALL microarray dataset (12) for factors associated with resistance to the previously studied BCL2 family inhibitor obatoclax mesylate (GeminX Pharmaceuticals; now an indirect, wholly owned subsidiary of Teva Pharmaceutical Industries Ltd.) (13), we uncover a distinctive gene expression signature with up-regulation of eIF4/p70S6K pathway signaling. This leads us to investigate expression of the eukaryotic translation initiation factor EIF4E and its role in infant ALL, and to test ribavirin, a known EIF4E inhibitor (14), as a novel treatment.
EIF4E is over-expressed in many cancers including adult leukemias and lymphomas (14–19). EIF4E has two well established functions: 1) to mediate nuclear to cytoplasmic mRNA export, and 2) to increase translation efficiency of transcripts containing particular RNA elements (14, 16, 20–22). EIF4E associates with over 3000 mRNAs in the nucleus and regulates nuclear export and translation of many mRNAs (e.g. MYC, BCL2, MCL1 and others) important to oncogenesis (16, 19, 23–25). eIF4/p70S6K signaling, in which EIF4E is a key factor, and the interrelated PI3K/AKT1/mTOR pathway are central to cell growth, proliferation, metabolism and survival (26). These pathways intersect at the TORC1 complex, which phosphorylates EIF4EBP1 and p70S6K. When dephosphorylated, EIF4EBP1 binds to EIF4E and suppresses translation initiation (27). NBN, a known nuclear to cytoplasmic mRNA export target of EIF4E, is particularly important because NBN is an upstream activator of PI3K/AKT1/mTOR signaling, underscoring the significance of EIF4E in cell proliferation and survival signaling (28). The nuclear mRNA cargos of EIF4E extend to DNA methyltransferases and epigenetic regulators (19).
The guanosine analog ribavirin is an FDA-approved anti-viral agent for respiratory syncytial virus and hepatitis C (29, 30), but it also targets EIF4E (14). Ribavirin binds directly to EIF4E and competitively inhibits EIF4E binding to the 5’m7G cap of mRNA, thus inhibiting EIF4E’s nuclear to cytoplasmic mRNA export and translation functions (14, 16, 20–22). Ribavirin has been used to target EIF4E for clinical benefit in adult AML (15, 18), lymphoma (NCT03585725), metastatic breast cancer (NCT01056757), oropharyngeal cancer (31) (NCT01721525), prostate cancer (32, 33) and other solid tumors (NCT01309490). In adult AML, elevated EIF4E signifies addiction/dependence and predicts ribavirin sensitivity (15–18), and EIF4E inhibition by ribavirin correlates with clinical responses including remissions (15, 18). Although previously unstudied in pediatric cancers, as antiviral treatment ribavirin caused minimal complications in neonates and infants (34, 35), suggesting an interesting therapeutic option to investigate for infant ALL.
Results:
Elevated expression of translational signaling in obatoclax-resistant infant ALL
We reported that obatoclax treatment of infant ALL cells triggers a disease-specific triple apoptosis/necroptosis/autophagy killing mechanism (13). Forty-seven of the reported cases from the COG P9407 trial (6) had been studied both by MTT assays of obatoclax (13) and diagnostic gene expression profiling (GEO number GSE68720) (12). The gene expression profiles in these 47 cases were analyzed as a function of obatoclax EC50 in order to understand the mechanisms underlying obatoclax resistance and discover new drug targets.
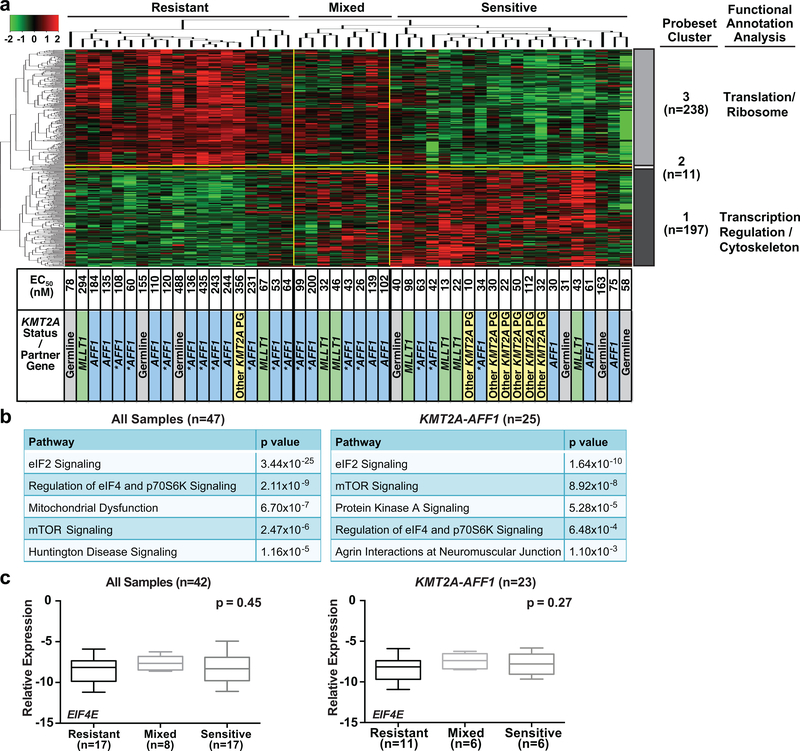
By Spearman test, log2 expression for 446 probesets correlated with obatoclax EC50 (p≤0.001). The probesets segregated into two major clusters, Gene Cluster 3 (238 probesets) and Gene Cluster 1 (197 probesets), and a third smaller 11-probeset cluster (Gene Cluster 2) (Figure 1a; Table S1). The two major probeset clusters separated the cases into three groups of 19 obatoclax-resistant cases with generally high EC50s (>176 nM; i.e. Cmax achieved at recommended adult phase II dose), 20 obatoclax-sensitive cases with generally low EC50s (<176 nM), and 8 cases with a mix of high and low obatoclax EC50s (Figure 1a). High expression of genes encoding translation/ribosomal proteins (Gene Cluster 3) but low expression of transcriptional regulatory/cytoskeleton genes (Gene Cluster 1) predicted obatoclax resistance (Tables 1, S2). Accordingly, by Ingenuity Pathway Analysis (IPA), obatoclax resistance correlated with three pathways important in translational control: the eIF4/p70S6K pathway and interrelated mTOR and eIF2 pathways (26, 36) (Figures 1b left, S1). The correlation with the eIF4/p70S6K pathway is not surprising because mRNA export and translation of the anti-apoptotic targets of obatoclax, BCL2 and MCL1, are EIF4E dependent (19, 37, 38). Phosphorylation of the EIF4E inhibitor EIF4EBP1 and P70S6K by the TORC1 complex up-regulates translation of EIF4E targets (27). The eIF2 pathway chaperones the initiator Met-tRNA to the ribosome and mediates AUG translation start site recognition (39). This suggests that obatoclax resistance and, more broadly, resistance to cell death in infant ALL are related to translation. For this reason, and because the pivotal translation regulator in the eIF4/p70S6K pathway EIF4E has established roles in cancer and ribavirin targets EIF4E (20–22), EIF4E was prioritized for our studies.
Figure 1. Correlation of basal gene expression with obatoclax response in diagnostic infant ALL samples from COG P9407 trial.
(a) Heatmap illustrating correlation between basal gene expression and 72 h single-agent EC50s of obatoclax from MTT assays in 47 diagnostic infant ALL samples. Note two major probeset clusters partitioning cases into resistant, sensitive and mixed groups in which EC50s were, in general, high (>176 nM; i.e. Cmax achieved at recommended adult phase II dose) (left), low (<176 nM) (right), or mix of high and low (middle). Functional annotation of genes in clusters is summarized (far right). Asterisks indicate cases with elevated HOXA gene expression pattern.
(b) Correlations determined by IPA between obatoclax EC50 and canonical signaling pathways in all 47 (left) and 25 KMT2A-AFF1 cases (right), respectively.
(c) Independent QPCR confirmation of lack of difference in EIF4E transcript levels between three sample groups with differing obatoclax sensitivities in 42 available cases from microarray (left), and in 23 of these 42 cases that were KMT2A-AFF1 (right). Horizontal lines represent median; bars, range.
Table 1.
Functional annotation analysis of probesets in gene clusters correlated with obatoclax EC50
| Category | Probesets (n) | Description | Probesets (n) by Functional Cluster |
Probesets (%) by Functional Cluster |
|||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | All | |||
| 1 | 159 | Translation | 43 | 0 | 116 | 21.8 | 0.0 | 48.7 | 35.7 |
| 2 | 35 | Ribosome; rRNA processing | 3 | 0 | 32 | 1.5 | 0.0 | 13.4 | 7.8 |
| 3 | 70 | Lumen | 20 | 1 | 49 | 10.2 | 9.1 | 20.6 | 15.7 |
| 4 | 44 | Mitochondria | 5 | 0 | 39 | 2.5 | 0.0 | 16.4 | 9.9 |
| 5 | 14 | Translation/Elongation | 0 | 0 | 14 | 0.0 | 0.0 | 5.9 | 3.1 |
| 6 | 57 | Mitochondria | 12 | 0 | 45 | 6.1 | 0.0 | 18.9 | 12.8 |
| 7 | 22 | Translation initiation/Regulation | 0 | 0 | 22 | 0.0 | 0.0 | 9.2 | 4.9 |
| 8 | 22 | Glycolysis | 7 | 0 | 15 | 3.6 | 0.0 | 6.3 | 4.9 |
| 9 | 39 | Cytoskeleton | 22 | 0 | 17 | 11.2 | 0.0 | 7.1 | 8.7 |
| 10 | 11 | Hexose/Alcohol biosynthetic process | 2 | 0 | 9 | 1.0 | 0.0 | 3.8 | 2.5 |
| 11 | 11 | Protein folding | 1 | 0 | 10 | 0.5 | 0.0 | 4.2 | 2.5 |
| 12 | 12 | NAD(P)-binding | 3 | 0 | 9 | 1.5 | 0.0 | 3.8 | 2.7 |
| 13 | 24 | 6 | 0 | 18 | 3.0 | 0.0 | 7.6 | 5.4 | |
| 14 | 4 | 2 | 0 | 2 | 1.0 | 0.0 | 0.8 | 0.9 | |
| 15 | 16 | 6 | 1 | 9 | 3.0 | 9.1 | 3.8 | 3.6 | |
| 16 | 13 | Pleckstrin homology domain | 11 | 0 | 2 | 5.6 | 0.0 | 0.8 | 2.9 |
| 17 | 26 | 3 | 0 | 23 | 1.5 | 0.0 | 9.7 | 5.8 | |
| 18 | 7 | 4 | 0 | 3 | 2.0 | 0.0 | 1.3 | 1.6 | |
| 19 | 48 | Transcription regulation | 32 | 0 | 16 | 16.2 | 0.0 | 6.7 | 10.8 |
| 20 | 9 | 8 | 0 | 1 | 4.1 | 0.0 | 0.4 | 2.0 | |
| All | 446 | 197 | 11 | 238 | |||||
Although expression of the entire eIF4/p70S6K pathway was highly enriched in obatoclax-resistant cases (p=2.11×10−9), the differentially expressed probesets (Table S1) did not include EIF4E (p=0.4), and EIF4E expression itself was not correlated with resistance (r=0.12) (Figure S1). In the KMT2A-AFF1 cases, which account for ~50% of KMT2A-R infant ALL, IPA also showed significant up-regulation of eIF4/p70S6K signaling (Figures 1b right, S2). Spearman test indicated moderate correlation between EIF4E expression and obatoclax resistance (r=0.4) and borderline significance (p=0.06) (Figure S2).
The microarray data were independently confirmed by QPCR analysis of 42 cases with available samples from the microarray (23 KMT2A-AFF1; 6 KMT2A-MLLT1; 6 other KMT2A-R; 7 KMT2A-G) (Figures 1c, S3). The results demonstrated that EIF4E expression was not different between the three groups showing differing obatoclax sensitivities (Figure 1c). The similar EIF4E mRNA expression by microarray analysis and QPCR irrespective of obatoclax response yet entire eIF4/p70S6K pathway enrichment in obatoclax-resistant cases raised the possibilities that EIF4E transcript levels did not differ because they were elevated in all three groups, and that EIF4E protein may have been similarly elevated. Additionally, EIF4E is regulated post-transcriptionally (40), and the lack of differences in EIF4E mRNA expression by obatoclax response did not preclude protein elevation.
EIF4E protein is frequently elevated across infant ALL and childhood pre-B ALL
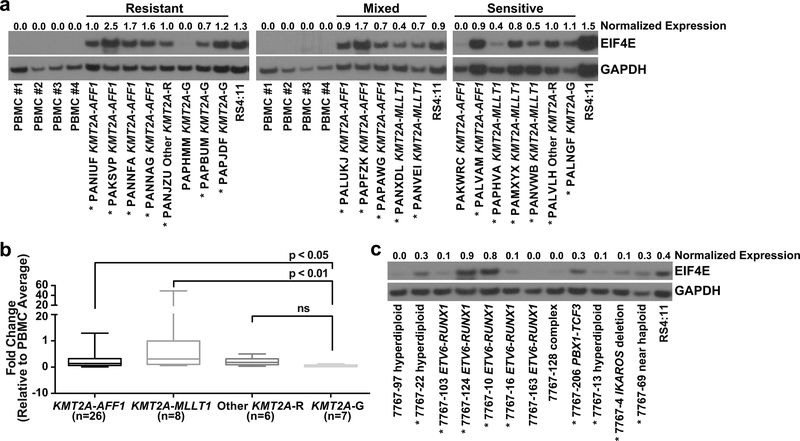
Given the above results, we used Western blot analysis to ask whether EIF4E protein is broadly elevated across infant ALL. Diagnostic infant ALL cells were studied for EIF4E protein levels compared to control healthy adult peripheral blood mononuclear cells (PBMCs), a typical control for EIF4E Western blot analysis (15, 17, 18, 41, 42). This control was utilized because, whereas infant ALL cells have a pre-B immunophenotype (9), B-cells do not migrate into circulation until more mature (after IG gene rearrangement) (43). EIF4E levels in PBMCs were compared to CD34+ cells (Figure S4), an alternative control for EIF4E Western blot analysis (17). The greater or equal normalized EIF4E levels in PBMCs compared to CD34+ cells (Figure S4) gave assurance that comparing blasts to PBMCs was informative.
Indeed, EIF4E protein levels proved highly elevated and similar across samples from the three groups showing differing obatoclax sensitivities. Thus, the lack of difference by obatoclax response in the microarray likely reflected a global elevation in EIF4E expression. Overall, compared to PBMCs, EIF4E protein was elevated in 18/20 cases (7/8 obatoclax-resistant, 5/5 mixed, 6/7 obatoclax-sensitive; 15/16 KMT2A-R, 3/4 KMT2A-G) (Figure 2a).
Figure 2. Basal EIF4E expression in diagnostic infant and pediatric ALL cells.
(a) Western blot analysis of basal EIF4E expression in available diagnostic infant ALL samples from microarray in Figure 1a and healthy adult PBMC controls. Microarray clusters by obatoclax response correspond to Figure 1a. Note elevated EIF4E protein (normalized to GAPDH) in 18 of 20 infant ALL samples compared to PBMC controls. Also note that proportions of cases with EIF4E elevation does not differ between three groups of samples with differing obatoclax sensitivities. EIF4E levels normalized to GAPDH are shown at top of blot. RS4:11 cells with elevated EIF4E are shown as a reference. Experiment was repeated twice on the same samples.
(b) QPCR analysis of EIF4E mRNA expression in infant ALL genetic subsets (42 COG P9407 samples from microarray, 5 samples from CHOP). Plots show normalized expression relative to 6 healthy adult PMBC controls expressed as 2-ΔΔCT for all samples by genetic subset. p-values indicate significance; ns, not significant. Horizontal lines represent median 2-ΔΔCT; bars, range.
(c) Western blot analysis of EIF4E in diagnostic samples from 12 childhood cases of pre-B ALL. Note elevated EIF4E in 9 cases (asterisks). Cytogenetic abnormalities are indicated for each sample. Complex karyotype included structural 1q, 3q, 4q and 7p abnormalities; high resolution SNP array showed 9p13.2 deletion including part of PAX3 locus, homozygous 7q34 and 14q11.2 TCR loci deletions, and 16q22.1 deletion including part of CTCF locus. EIF4E levels normalized to GAPDH are shown at top of blot. Due to sample limitations experiment was done once.
Protein samples from cases in the microarray were limited. Therefore, we compared infant ALL genetic subsets for EIF4E mRNA expression. QPCR detected higher EIF4E transcript levels in KMT2A-AFF1 (p<0.05) and KMT2A-MLLT1 (p<0.01) than KMT2A-G cases (Figure 2b). Even though transcript and protein expression may not correlate directly, these data demonstrate that, while EIF4E transcript levels in infant ALL do not differ by obatoclax response, they differ by genetic subset. Additional Western blot analysis showed high-level EIF4E in 9/12 representative cases of pre-B ALL (all KMT2A-G) from children >1 year old (Figure 2c), suggesting that EIF4E protein is frequently elevated and that EIF4E protein status has wider importance beyond the infant cases.
Single-agent ribavirin has robust anti-proliferative activity against infant ALL
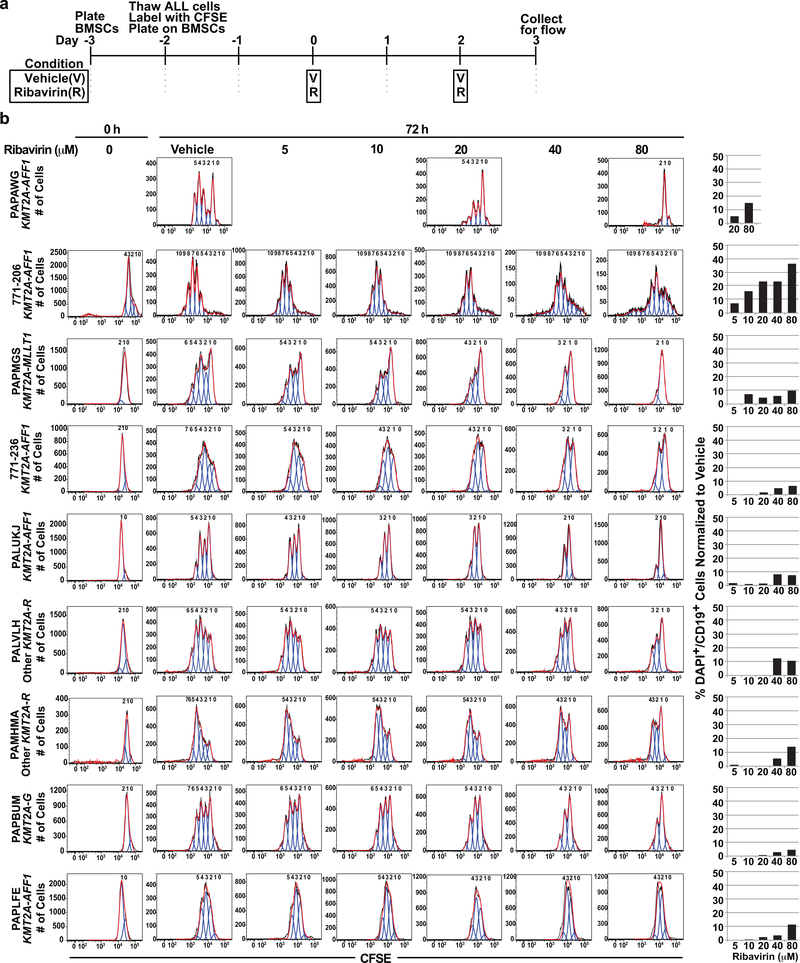
The frequent EIF4E protein elevation across infant ALL predicted ribavirin sensitivity. The importance of EIF4E targets in proliferation/cell cycle and survival regulation (16, 23–25) led us to investigate cell proliferation and survival after ribavirin treatment in diagnostic leukemia blasts from 9 cases of infant ALL (8 KMT2A-R; 1 KMT2A-G). The blasts were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE) for tracing cell divisions (44) and then co-cultured on BMSCs (45) ×48 h to surmount inherently limited proliferation of primary cells on plastic before 72 h of either vehicle or ribavirin treatment (Figure 3a). The number of cell divisions and percentage of DAPI+CD19+ cells, respectively, were used to quantify proliferation inhibition and cell death after treatment.
Figure 3. Effects of ribavirin on cell proliferation and survival in proliferating primary infant ALL cells co-cultured on BMSCs.
(a) Timeline for plating irradiated BMSCs, and plating and treatment of primary infant ALL cells with vehicle or ribavirin on the stromal layer. Time 0 represents the start of treatment.
(b) Analysis of proliferation and cell death in vehicle and ribavirin-treated infant ALL cells. Histograms (left) show modeled cell divisions based on CFSE intensity in viable DAPI-CD19+ cells. Peaks to left of parental peak representing cell divisions at time 0 and at 72 h after vehicle treatment indicate that all 9 samples underwent multiple cell divisions. Note dose-dependent decreases in numbers of modeled cell divisions at 72 h with ribavirin treatment indicative of proliferation inhibition in 6/8 KMT2A-R cases (top 6 rows). The histograms are representative of two intra-experimental replicates per condition. Graphs (right) show percentages of DAPI+CD19+ (dead) cells after 72 h treatment. The percentages represent average values for two intra-experimental replicates per condition. Due to the number of primary cells required (5×105 per replicate per condition) and primary sample limitations, experiment was done once. Note modest cell death compared to the marked proliferation inhibition.
By CFSE intensity (44), all 9 cases underwent multiple cell divisions (range 1–4) by the start of treatment (time 0; 48 h after plating). We found fewer cell divisions after 72 h of ribavirin treatment than after 72 h of vehicle treatment in 6/8 KMT2A-R cases, indicating marked dose-dependent reductions in proliferation (Figure 3b left). Additionally, ribavirin caused modest increases in cell death over vehicle compared to the robust proliferation inhibition (Figure 3 right). This mainly anti-proliferative activity is a known property of ribavirin and many effective molecularly targeted agents (22, 46–48).
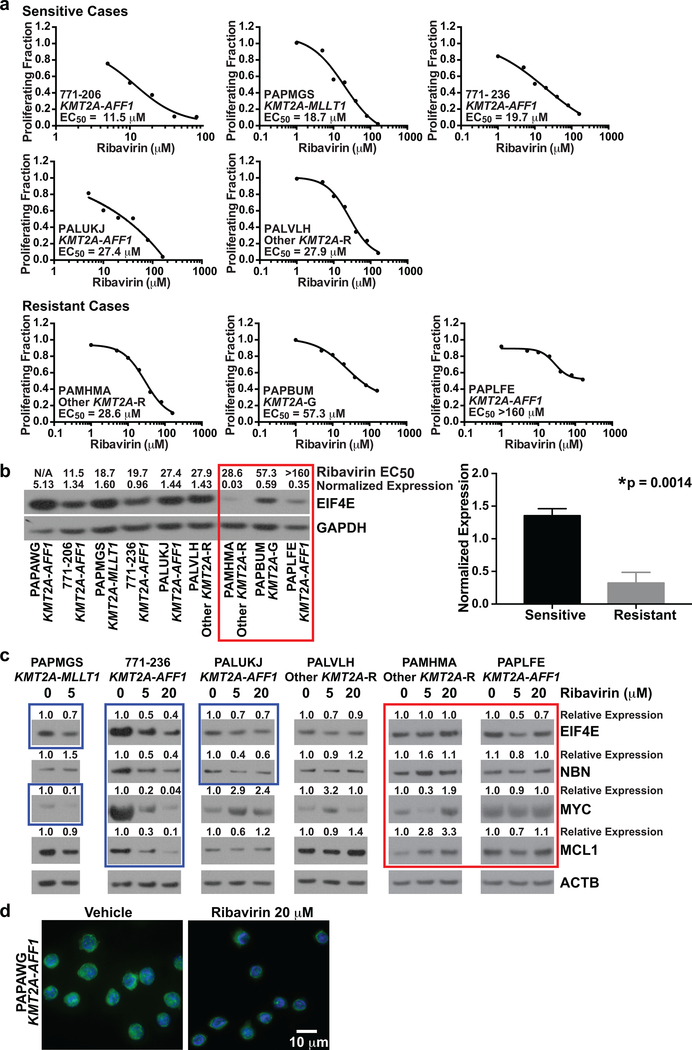
There was ample material in 8 of these 9 cases to measure CFSE intensity at multiple concentrations and determine EC50s. Steady-state ribavirin concentrations of 20–34 μM were reported in a neonate during intravenous antiviral treatment (49). The median ribavirin Cmax in adult AML trials was 14.8 μM (range 5–36 μM) as enteral monotherapy and 28 μM (range 7–76 μM) with low-dose cytarabine (15, 18). Using an EC50 cut-off of <28 μM, 5/8 cases (3/4 KMT2A-AFF1; 1/1 KMT2A-MLLT1; 1/2 other KMT2A-R; 0/1 KMT2A-G) were ribavirin-sensitive (Figure 4a). These results indicate that clinically relevant ribavirin exposures are cytostatic against a majority of cases of KMT2A-R infant ALL. Although ribavirin caused proliferation inhibition in the three other cases, the EC50s were not within the cut-off defining sensitivity (Figures 3b, 4a bottom).
Figure 4. Dependencies of ribavirin response and EIF4E target modulation in infant ALL cells on basal EIF4E expression.
(a) Proliferating fractions plotted as inhibitory sigmoid Emax models to estimate ribavirin EC50s in 8 of 9 cases with sufficient material for ribavirin treatment at multiple concentrations (c.f. Figure 3b, all cases except PAPAWG). An EC50 cut-off of <28 μM, based on clinical Cmax in adult AML trial, was used to define sensitivity.
(b) Western blot analysis of primary infant ALL samples from cases in Figure 3 showing correlation between ribavirin response and basal eIF4E expression (left). Ribavirin EC50s are above the lanes for all samples where determined. N/A indicates EC50 not determined because material was not available (Case PAPAWG). EIF4E levels normalized to GAPDH are shown at top of blot. Note greater ribavirin sensitivity (lower EC50s) in samples with higher EIF4E expression. Red box indicates resistant cases. Experiment was performed once due to sample limitations. Normalized EIF4E levels in sensitive vs. resistant cases in the Western blot are compared in bar graph (right). Bars represent standard error. Note significantly higher EIF4E levels in ribavirin-sensitive than ribavirin-resistant cases.
(c) Western blot analysis showing abrogation of expression of MYC, MCL1 and/or NBN target proteins in EIF4E regulon by ribavirin treatment of infant ALL cells at clinically relevant concentrations. Note the greater protein reductions in the sensitive cases, which also show more abundant basal EIF4E expression. Band intensities normalized to ATCB and then made relative to vehicle for each protein are shown at top of blots. Blue boxes show decreased proteins after ribavirin treatment in the sensitive cases. Also note decreases in EIF4E in sensitive cases indicated by blue boxes. Red box indicates lack of consistent protein down-regulation by ribavirin treatment in resistant cases. Experiment was performed once due to sample limitations.
(d) Re-localization of nuclear EIF4E to the cytoplasm in ribavirin-treated infant ALL cells. The cells are splenocytes from secondary xenografts expanded in NSG mice created from Case PAPAWG. The primary cells responded to single-agent ribavirin on BMSCs (Figure 3b, top). The cells were treated for 72 h with vehicle or ribavirin at 20 μM. DAPI-labeled DNA in nucleus is blue; EIF4E labeled with anti-EIF4E-FITC is green. Image shows change in EIF4E nuclear localization to the cytoplasm in most cells in the field after ribavirin treatment compared to the nuclear localization of EIF4E in vehicle-treated cells. Experiment was performed once, but same result was seen in RS4:11 cell line (c.f. Figure 5g).
Elevated EIF4E protein in infant ALL correlates with ribavirin sensitivity
Like adult AML (15–18), the cases of infant ALL with higher EIF4E protein proved more ribavirin-sensitive (Figure 4b left). In the 8 cases where EC50s were determined, basal EIF4E expression was higher in the ribavirin-sensitive (EC50 <28 μM) than resistant cases (p=0.0014) (Figure 4b right). NBN is a known EIF4E nuclear export target and MYC and MCL1 are EIFE4-regulated at both nuclear mRNA export and translation levels (16, 19, 23–25, 37, 38). By Western blot analysis, clinically achievable ribavirin exposures (5 μM; 20 μM) ×72 h caused decreased NBN, MYC, and/or MCL1 protein levels in 3/4 ribavirin-sensitive (EC50 <28 μM) cases. In case PALVH with a more borderline EC50 (27.9 μM), the levels of these proteins did not change with ribavirin treatment. However, sample limitations allowed only few targets to be studied and precluded analysis of other factors (e.g. ribavirin uptake). Ribavirin did not cause corresponding dose-dependent changes in NBN, MYC or MCL1 levels in ribavirin-resistant (EC50 >28 μM) cases (Figure 4c).
Clinical ribavirin responses in adult AML are associated with reduced EIF4E (15, 18). Accordingly, Western blot analysis detected decreases in EIF4E after ribavirin treatment in 3/4 of the sensitive cases (Figure 4c). Similar to adult AML where ribavirin causes nuclear-to-cytoplasmic EIF4E re-localization (15, 18), in the ribavirin-responsive KMT2A-AFF1 case with insufficient primary material to determine EC50 (PAPAWG), ribavirin (20 μM×72 h) caused EIF4E to re-localize to the cytoplasm (Figure 4d), signifying molecular targeting of EIF4E.
Differences in EIF4E levels and drug uptake modulate ribavirin responsiveness in KMT2A-R ALL cell lines
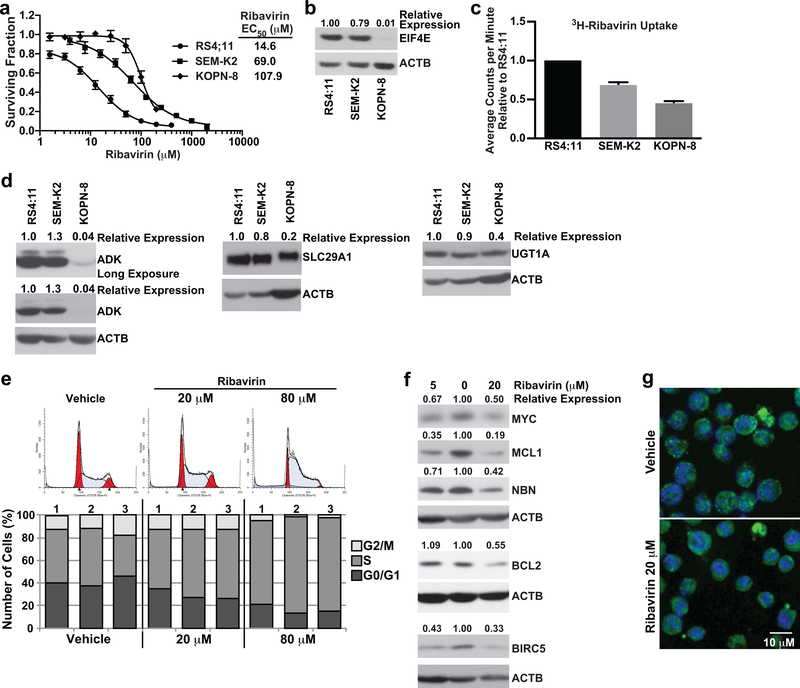
The ribavirin response in KMT2A-R ALL was further studied using cell lines. Consistent with dependence of ribavirin sensitivity on elevated EIF4E (15–18), by EC50s from MTT assays (RS4:11, 14.6 μM; SEM-K2, 69.0 μM; KOPN-8, 107.9 μM) (Figure 5a) and Western blot analysis (Figure 5b) the most ribavirin-sensitive cell line RS4:11 displayed the highest EIF4E expression. In contrast, KOPN-8 had the lowest EIF4E expression and the highest EC50 (Figure 5a,b).
Figure 5. Determinants of single-agent ribavirin activity in KMT2A-R ALL cell lines and functional effects of ribavirin on RS4:11 cells.
(a) MTT assay data represented as surviving fraction plots after treatment of KMT2A-R cell lines × 72 h with increasing ribavirin concentrations. Plots are mean values from 10 experiments for RS4:11, 8 experiments for SEM-K2 and 3 experiments for KOPN-8 cells. Each experiment included 3–6 replicates/condition. Error bars show standard error. Note lower ribavirin EC50 within clinically relevant concentrations for RS4:11 cells.
(b) Western blot showing correlation between ribavirin sensitivity (lower EC50) and high-level basal EIF4E expression. EIF4E levels normalized to ACTB and then made relative to RS4:11 are shown at top of blot. The blot is representative of data reproduced in 3 separate experiments. Note that the most ribavirin-sensitive cell line RS4:11 expresses the most EIF4E and KOPN-8 with highest EC50 has the least EIF4E expression.
(c) Bar graph of relative 3H-ribavirin uptake after 8 h treatment of KMT2A-R cell lines with 3H-ribavirin (RS4:11 cell line set to 1). Note that KOPN-8 with highest EC50 has the lowest uptake, and RS4:11 with lowest EC50 has the highest uptake (c.f. Figure 5a). Experiment performed twice with 3 intra-experimental replicates per experiment. Black bars indicate standard deviation.
(d) Western blot analysis of basal expression of ADK, SLC29A1 and UGT1A proteins. Band intensities for each protein normalized to ACTB and then made relative to RS4:11 cells are at top of blots. Blots are representative of two independent experiments. Note similar expression of ADK, SLC29A1 and UGT1A in RS4:11 cells compared to SEM-K2 cells, and lower expression of all three proteins in KOPN-8 cells.
(e) Analysis of cell cycle in ribavirin-treated RS4:11 cells. Representative flow cytometric analysis of propidium iodide labeling showing modeled cell cycle distribution after treatment × 24 h with vehicle or ribavirin at indicated concentrations (top). The histogram is representative of 3 separate experiments. Bar graphs (bottom) show percentages of cells in G0/G1, S, and G2/M in the 3 experiments. Note dose-dependent increase in cells in S phase indicative of cell cycle arrest with ribavirin treatment.
(f) Western blot analysis of EIF4E targets in ribavirin-treated RS4:11 cells. Cells were treated for 72 h with vehicle or ribavirin at 5 μM and 20 μM. Vehicle-treated cells are in center lanes. Band intensities normalized to ACTB and then made relative to vehicle for each protein are at top of blots. Experiments were done 3–5 times except BCL2 and BIRC5 done twice for 5 μM ribavirin. The blots show down-regulation of MYC, MCL1, NBN, BCL2 and BIRC5.
(g) Change in EIF4E subcellular distribution in ribavirin-treated RS4:11 cells. Confocal images show nuclear to cytoplasmic shift in most cells in population after 72 h ribavirin treatment compared to the nuclear EIF4E location in most cells following vehicle treatment. DAPI labeled DNA in the nucleus is blue and EIF4E labeled with anti-eIF4E-FITC is green. The images are representative of 3 separate experiments.
To identify factors modulating sensitivity besides EIF4E expression, these cell lines were studied for differences in 3H-ribavirin uptake and basal expression of ADK (Adenosine kinase) and SLC29A1 (ENT1; Equilibrative nucleoside transporter 1), reduced levels of which are linked to drug resistance in some cases of adult AML (42). After 8 h of exposure, 3H-ribavirin uptake was greatest in RS4:11 cells and least in KOPN-8 (Figure 5c). SEM-K2 cells displayed 30% less ribavirin uptake consistent with the higher ribavirin EC50 of this cell line compared to RS4:11 (Figure 5c). ADK and SLC29A1 expression was similar in RS4:11 and SEM-K2 but markedly reduced in KOPN-8 compared to both other cell lines (Figure 5d). Because ribavirin phosphorylation by ADK promotes cellular retention (50) and SLC29A1 is necessary for ribavirin transport (51), these findings suggest low-level ADK and SLC29A1 and resultant net reduction in drug uptake as additional causes for ribavirin resistance in KOPN-8. The GLI1-driven resistance factor UGT1A catalyzes ribavirin glucuronidation. The KOPN-8 cells showed lower-level expression of UGT1A (Figure 5d), indicating that up-regulation of the GLI1/UGT1A axis, which causes ribavirin resistance in some cases of adult AML (42), was not a resistance factor in this cell line.
Given that RS4:11 cells showed elevated EIF4E and an EC50 in range of clinically achievable plasma levels, these cells were prioritized for further studies. Particularly for KOPN-8 with the highest EC50, least EIF4E, and impaired ribavirin uptake (Figure 5a-d), the high ribavirin concentrations required for growth inhibition would likely cause off-target effects that would be difficult to interpret.
EIF4E expression is critical for driving expression of ALL-relevant cell growth and survival proteins
Given the proliferation inhibition in ribavirin-treated infant ALL cells (Figure 3b), it was of interest to determine how cell cycle was affected. Modeling cell cycle in ribavirin-treated RS4:11 cells revealed a marked S-phase arrest (Figure 5e) as the basis for the proliferation inhibition. Therefore, we next examined modulation of EIF4E targets important for leukemia cell proliferation and survival (16, 19, 23–25, 37, 38, 52). By Western blot analysis, ribavirin treatment of RS4:11 cells for 72 h at clinically relevant concentrations (5 μM; 20 μM) caused decreases in MYC, MCL1 and NBN protein levels similar to ribavirin-sensitive infant ALL cells, and decreases in the anti-apoptotic proteins BCL2 and BIRC5 (SURVIVIN) (Figure 5f). The reductions in these proteins are consistent with the inhibition of cell growth and survival seen with ribavirin treatment in both infant ALL and RS4:11 cells (Figure 3b, 5e). Ribavirin also caused EIF4E re-localization from the nucleus to the cytoplasm in RS4:11 cells (Figure 5g) similar to the KMT2A-AFF1 infant ALL cells (Figure 4d).
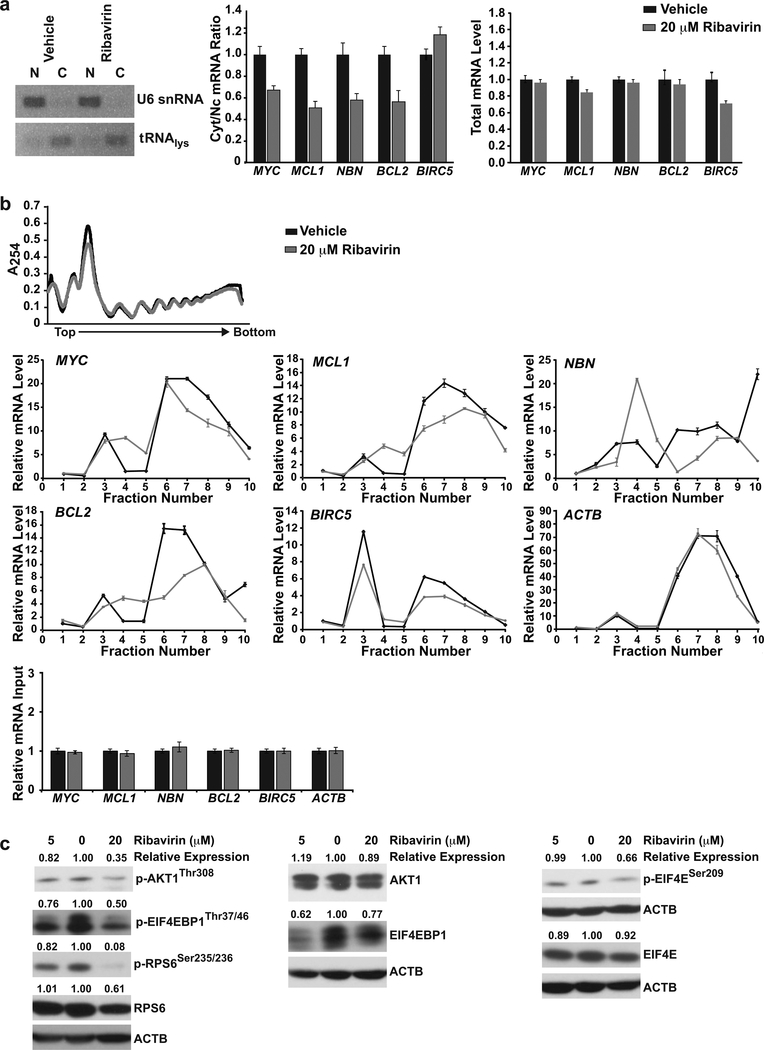
Ribavirin inhibits mRNA export and translation of EIF4E targets in RS4:11 cells
The decreases in EIF4E-regulated oncogenic cell growth and survival proteins led us to investigate ribavirin inhibition of nuclear to cytoplasmic mRNA export and translation. mRNA export was determined by nuclear and cytoplasmic fractionation and monitoring the mRNAs by QPCR. Semi-quantitative RT-PCR analysis of the nuclear and cytoplasmic markers U6 snRNA and tRNAlys, respectively, confirmed the quality of fractionation (Figure 6a, left). Ribavirin treatment (20 μM × 72 h) reduced mRNA export as evidenced by decreased cytoplasmic:nuclear ratios compared to vehicle for MYC, MCL1, NBN and BCL2, all of which are known mRNA export targets. The cytoplasmic:nuclear ratio for BIRC5, an EIF4E translation initiation target only (Figure 6a, middle) and all relevant total mRNA levels (Figure 6a, right) were unchanged by the treatment as expected.
Figure 6. Inhibition of EIF4E-mediated nuclear to cytoplasmic mRNA export and translation, and PI3K/AKT1/mTOR and eIF4/p70S6K pathway inhibition in ribavirin-treated RS4:11 cells.
(a) Analysis of cytoplasmic:nuclear ratios of EIF4E target mRNAs. (a, left) Verification of cytoplasmic and nuclear mRNA fractionation by semi-quantitative RT-PCR analysis of tRNAlys (cytoplasmic marker) and U6 snRNA (nuclear marker). Ethidium bromide stained agarose gel shows tRNAlys product in cytoplasmic but not nuclear fraction, and product for U6 snRNA in nuclear but not cytoplasmic fraction. (a, middle) Cytoplasmic:nuclear mRNA ratios of EIF4E target transcripts in RS4:11 cells determined by QPCR after treatment with vehicle or ribavirin (20 μM) for 72h. The data show decreased export of MYC, MCL1, NBN and BCL2 but not BIRC5 mRNAs. (a, right) Total mRNA levels for transcripts analyzed in the same experiment. Data were normalized to ACTB and shown relative to vehicle (set to 1). Experiment was carried out three independent times.
(b) Polysomal profiling of RS4:11 cells after treatment with vehicle or ribavirin (20 μM) for 72h. (b, top) Unchanged total polysomal profiles with ribavirin treatment. (b, middle) Polysomal profiling of known EIF4E translation targets. The data show shifts of MYC, MCL1, BCL2 and BIRC5 mRNAs and also NBN mRNA from higher to lower polysomal fractions (right side in plots) with ribavirin treatment. ACTB mRNA was analyzed as negative control. Fractions are shown relative to monosome fraction of vehicle control (Fraction 1). (b, bottom) Input levels of analyzed transcripts normalized to UBC and shown relative to vehicle (set to 1). Experiments were carried out two independent times.
(c) Western blot analysis of PI3K/AKT1/mTOR and eIF4/p70S6K signaling in ribavirin-treated RS4:11 cells. Cells were treated for 72 h with vehicle or ribavirin at 5 μM and 20 μM. Vehicle-treated cells are in center lanes. Band intensities normalized to ACTB and then made relative to vehicle for each protein are at top of blots. The blots show decreased phospho-AKT1Thr308 but not AKT1 total protein; decreased phospho-EIF4EBP1Thr37/46 with only modestly decreased EIF4EBP1 total protein; >90% decrease in phospho-RPS6Ser235/236 with only modestly decreased RPS6 total protein, and decreased phospho–EIF4ESer209 but not EIF4E total protein. Experiments were done at least 3 independent times. The same blot was used for BCL2 (Figure 5f) and EIF4E, and the same ACTB panel is shown in both figures.
To determine whether translation efficiency of these targets was affected, mRNA content on polysomes as a function of ribavirin treatment was quantified by QPCR. Total polysomal profiling indicated that ribavirin did not alter polysome formation (Figure 6b, top), which is consistent with previous studies in other cell lines using siRNA or ribavirin for EIF4E depletion (19, 22). Compared to vehicle, ribavirin caused the known EIF4E translation targets MYC, MCL1, BCL2 and BIRC5 to be depleted from the higher molecular-weight fractions, indicative of reduced translation (Figure 6b, middle). Additionally, ribavirin reduced translation of NBN, not studied before for EIF4E regulation at the level of translation (Figure 6b, middle). By contrast, the negative control (non-EIF4E target) mRNA ACTB did not show reduced translation (Figure 6b, middle) and, further, RNA input was not altered by the treatment (Figure 6b, bottom).
These data are consistent with the known mechanism of action of ribavirin as a 5’m7G cap competitor that inhibits EIF4E binding to its target mRNAs and thus prevents their nuclear export and translation (14, 16, 20–22). Mechanistically, they suggest that the observed decreased protein levels (Figure 5e) are a consequence of ribavirin effects on EIF4E-mediated mRNA export and translation. Our discovery that ribavirin decreases NBN translation indicates that NBN is a new EIF4E translation initiation target besides a known EIF4E nuclear export target. Importantly, the biological effects of ribavirin in RS4:11 cells occur at the same concentration as NBN mRNA export and translation inhibition. This finding is significant because of the pivotal role of NBN in PI3K/AKT1/mTOR signaling activation.
Ribavirin inhibits PI3K/AKT1/mTOR and eIF4/p70S6K signaling in RS4:11 cells
NBN, via PI3K, activates AKT1 and downstream EIF4EBP1 and RPS6 signaling. Previously, EIF4E inhibition in fibroblasts using ribavirin or siRNA reduced NBN nuclear to cytoplasmic export and AKT1 signaling (25, 28, 53). Therefore, given the NBN inhibition in ribavirin-sensitive infant ALL and RS4:11 cells (Figure 4c, 5f, 6a, 6b), we investigated RS4:11 cells for altered signaling by AKT1 and downstream effectors as a possible mechanism behind the decreased proliferation (Figures 3b, 5e). Consistent with this predicted mechanism, by Western blot analysis, ribavirin-treated (5 μM or 20 μM × 72 h) RS4:11 cells displayed a dose-dependent decrease in phospho-AKT1Thr308, which phosphorylates mTOR in the TORC1 complex (26). The reduced phospho-AKT1Thr308 was not due to a decrease in AKT1 total protein (Figure 6c). Additionally, ribavirin treatment led to a dose-dependent decrease in phospho-EIF4EBP1Thr37/46 (Figure 6c). EIF4EBPI total protein was decreased but only modestly by ribavirin treatment (Figure 6c). The reduced phospho-AKT1Thr308 and phospho-EIF4EBP1Thr37/46 were confirmed by phosphoflow cytometry (Figure S5). Finally, also consistent with effects of NBN on PI3K/AKT1/mTOR signaling, ribavirin caused >90% abrogation of phospho-RPS6Ser235/236, which is immediately downstream of p70S6K in the mTOR pathway (26), with only moderate decrease (40%) in RPS6 total protein (Figure 6c). All of these effects of ribavirin on RS4:11 cells including its effects on NBN occurred in the same concentration range as the EC50 (14.6 μM; Figure 5a) and, thus, they are consistent with inhibition of signaling by AKT1 and its downstream effectors underpinning the biological effects of ribavirin treatment.
Also in regards to signaling, EIF4E phosphorylation by MNK1 promotes EIF4E-mediated mRNA translation (37, 54, 55). Ribavirin treatment down-regulated phospho-EIF4E in primary AML cells (41), a Ph+ ALL cell line (56) and a breast cancer cell line xenograft (57). By Western blot analysis, RS4:11 cells showed a 30% decrease in phospho–EIF4ESer209 after ribavirin treatment (20 μM × 72 h) while EIF4E total protein levels were unchanged (Figure 6c). Thus, similar to other cell lines, ribavirin inhibits phospho–EIF4ESer209 in RS4:11 cells. Taken together, the data indicate that the effects of ribavirin on RS4:11 cell proliferation are paralleled by inhibition of central proliferation signaling pathways.
Ribavirin has in vivo activity against KMT2A-R infant ALL
Ribavirin monotherapy was evaluated in NSG mouse models with established xenografts from three cases of primary KMT2A-R infant ALL (PAPAWG, PAPMGS, 771–206), each of which was ribavirin-responsive when co-cultured on BMSCs (Figure 3b) and had elevated EIF4E (Figure 4b). The increase in median peripheral blast count was significantly slower over time in the ribavirin arm compared to vehicle in one of the KMT2A-AFF1 cases (PAPAWG; Figure S6a). Mice with KMT2A-MLLT1 xenografts showed heterogeneous responses (PAPMGS; 3 of 5 responders; Figure S6b asterisks). Ribavirin was ineffective against the second KMT2A-AFF1 case tested (771–206; Figure S6c). Interestingly, this case was the most rapidly proliferating and had the lowest ribavirin EC50 on BMSCs (Figures 3b, 4a). This suggests that although the cells were ribavirin-sensitive, their in vivo proliferation surpassed the anti-proliferative effects of ribavirin treatment. These results indicate in vivo activity of ribavirin monotherapy against some cases of KMT2A-R infant ALL, but limited cases have been studied.
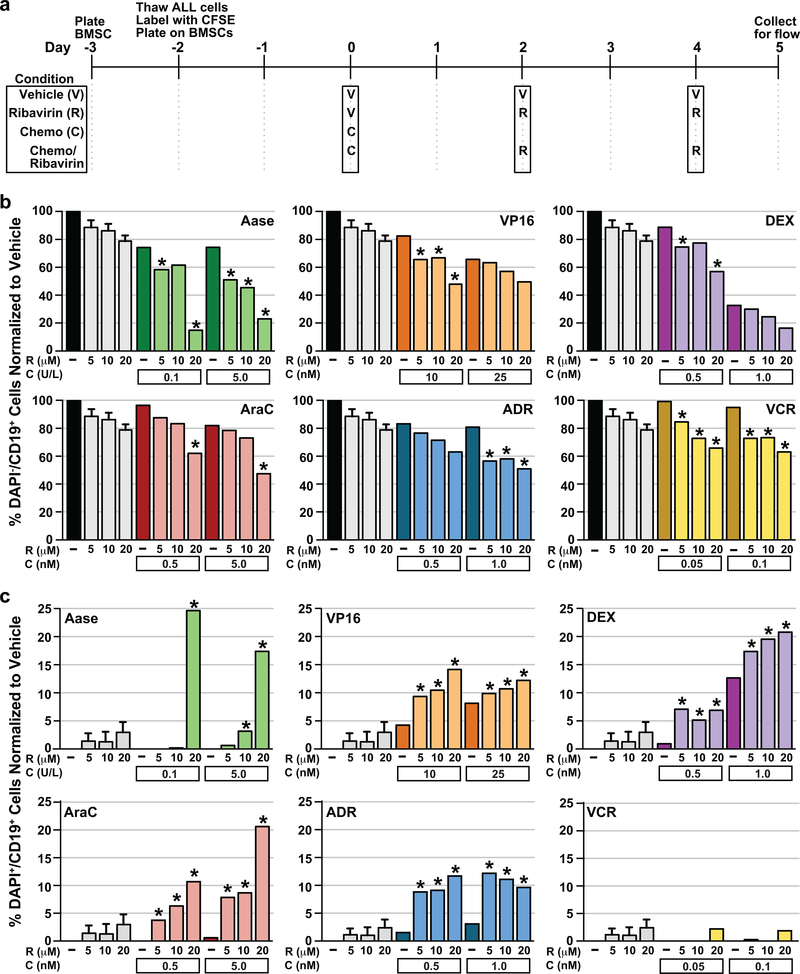
Cooperative effects of ribavirin against KMT2A-AFF1 infant ALL in chemotherapy combinations
Combination effects of ribavirin with clinically relevant concentrations of cytotoxic chemotherapies used for infant ALL (58–65) were tested against the KMT2A-AFF1 case in which ribavirin showed in vivo activity (PAPAWG). Due to the large cell numbers needed (~20–30×106/combination), the cells were expanded in NSG mice before co-culture on BMSCs. The chemotherapy was sequenced before ribavirin (Figure 7a) because chemotherapy is most effective in dividing cells (66–68) whereas ribavirin is anti-proliferative (Figure 3b) and arrests the cell cycle (Figure 5e). Live (DAPI-CD19+) cells reflecting the starting number of cells, proliferation and survival altogether, and dead (DAPI+CD19+) cells were counted. Cell divisions were not modeled because prolonged culture attenuates CFSE intensity (44). Decreases in DAPI-CD19+ cells exceeding the sum of decreases after single-agent treatments indicated dose-dependent cooperative interactions between ribavirin and L-asparaginase, cytarabine, dexamethasone, etoposide, doxorubicin and vincristine in reducing live cell numbers (Figure 7b asterisks).
Figure 7. Anti-leukemia effects of ribavirin against KMT2A-AFF1 infant ALL in cytotoxic chemotherapy combinations.
(a) Timeline for plating irradiated BMSCs, and co-culture and treatment of infant ALL cells with vehicle, chemotherapy/ribavirin combination, single-agent chemotherapy or single-agent ribavirin on the stromal layer. Time 0 represents start of treatment.
(b) Percentages of live (DAPI-CD19+) cells remaining normalized to vehicle after chemotherapy/ribavirin combination, single-agent chemotherapy or single-agent ribavirin treatment. Chemotherapy drugs tested in the combinations are at top right in plots. Asterisks indicate greater decreases in DAPI-CD19+ cells with the combination compared to the sum of decreases with the relevant single-agent treatments, consistent with cooperative interactions with every ribavirin/chemotherapy combination.
(c) Percentages of dead (DAPI+CD19+) cells normalized to vehicle after chemotherapy/ribavirin combination, single-agent chemotherapy or single-agent ribavirin treatment. Chemotherapy drugs are at top left in plots. Asterisks indicate greater proportions of DAPI+CD19+ cells with the combination compared to the sum of decreases with the relevant single-agent treatments, consistent with cooperative ALL cell killing by every combination tested with exception of vincristine/ribavirin.
(b, c) The data plotted are averages for 2 intra-experimental replicates per condition from a single experiment for each of the 6 single-agent chemotherapies and each chemotherapy/ribavirin combination. For single-agent ribavirin, the data plotted are averages for 2 intra-experimental replicates per condition from all 6 experiments. Error bars represent standard error. Abbreviations: Aase, L-asparaginase; AraC, cytarabine; DEX, dexamethasone; VP16, etoposide; ADR, doxorubicin; VCR, vincristine.
Although these chemotherapies are cytotoxic (46), the counting occurred five days after chemotherapy exposure due to the sequential schedule (Figure 7a), and only modest cell death was detected, likely because of clearance of additional dead cells as debris prior to analysis. Still, each ribavirin/chemotherapy combination except ribavirin/vincristine caused cooperative cytotoxicity and more DAPI+CD19+ cells than the sum of dead cells after the respective single-agent treatments (Figure 7c asterisks). Therefore, ribavirin augments activity of a broad spectrum of chemotherapies used for infant ALL. The greatest cooperativity occurred with ribavirin/L-asparaginase where live (DAPI-CD19+) cells decreased to as low as 14.9% (Figure 7b), and combining the single agents at concentrations with low/no cytotoxicity alone caused 24.6% cell killing (DAPI+CD19+ cells) (Figure 7c).
Discussion:
This work provides the first evidence that EIF4E inhibition by ribavirin has activity in KMT2A-R infant ALL, an orphan disease with exceedingly poor survival. Whereas conventional treatments have significant complications, enteral ribavirin has been well tolerated in two adult AML trials (15, 18), oropharyngeal (31) and prostate cancer trials (32, 33), and in standard of care therapy for hepatitis C in children (30). Furthermore, extensive aerosolized usage for respiratory syncytial virus proved that ribavirin is safe in neonates and infants (34, 35). In this study, single-agent ribavirin caused pronounced proliferation inhibition and modest cytotoxicity against actively dividing infant ALL cells on BMSCs. This in vitro response after short duration (72 h) ribavirin treatment was primarily cytostatic. Cytostasis does not preclude therapeutic benefit when molecularly targeted agents are given as continuous monotherapy (46). In fact, targeted agents including ribavirin are not purely cytostatic and cytotoxicity ensues after prolonged cytostasis (15, 18, 46). Ribavirin is not plasma protein bound (69, 70), allowing comparisons between effective in vitro concentrations and patient plasma concentrations (15, 18). The EC50 cut-off (<28 μM) for in vitro sensitivity derived from median Cmax values in adult AML trials in which oral ribavirin showed activity (15, 18) identified 5/7 cases of KMT2A-R infant ALL (~70%) as ribavirin-sensitive. Furthermore, ribavirin efficacy was observed in KMT2A-R infant ALL xenograft-bearing mice and ribavirin enhanced activity of chemotherapy.
Consistent with ribavirin sensitivity, we discover that infant ALL and RS4:11 cells have elevated EIF4E levels. In RS4:11 cells we uncover that the EIF4E elevation, which in turn drives mRNA export and translation of central EIF4E cell proliferation and survival targets, is suppressed by ribavirin treatment. Similar to the nuclear to cytoplasmic re-localization first reported with ribavirin treatment in AML (15, 18) and later discovered to be due to impaired Importin 8-mediated nuclear import of EIF4E after EIF4E-mediated mRNA export (71), we also observe that ribavirin causes EIF4E re-localization to the cytoplasm in KMT2A-AFF1 infant ALL and RS4:11 cells. This EIF4E re-localization and ability of ribavirin to act as a 5’m7G cap competitor triggers reduced levels of directly regulated EIF4E targets (e.g. MYC, NBN, BCL2, MCL1, BIRC5), thereby underpinning the biological effects of ribavirin treatment.
In regards to obatoclax, it is interesting that elevated eIF4/p70S6K pathway signaling correlated with resistance. The fact that EIF4E drives BCL2 and MCL1 expression stitches together the associations between elevations in this pathway and related pathways and high obatoclax EC50. In this way, our studies explain a mechanism for at least one type of obatoclax resistance and, more broadly, resistance to apoptosis. Although obatoclax is no longer clinically available, they also suggest a means to overcome this form of resistance in infant ALL using ribavirin treatment.
We also find some KMT2A-R cell lines that are insensitive to ribavirin treatment. Pharmacological resistance is observed in AML (15, 18, 42) and head and neck cancer models (42). As in AML (15, 18, 42), we find that lower-level EIF4E protein is associated with lower sensitivity. While our infant ALL microarray data (12) do not indicate differential GLI1 expression and we did not observe resistance due to elevated UGTA1 glucuronidation enzyme, we did observe reduced 3H-ribavirin uptake likely due to lower ADK and SLC29A1 expression. Current clinical trials are addressing these resistance mechanisms by excluding patients with low EIF4E, ADK or SLC29A1 expression and by using GLI1 inhibition as required (42); such strategies could also be considered for pediatric trials.
Evidence we observe that clinically relevant ribavirin exposures inhibit EIF4E in KMT2A-R infant ALL includes decreases in known EIF4E-regulated proteins (NBN, MYC, MCL1) and EIF4E re-localization to the cytoplasm. This evidence is augmented by mechanistic studies of ribavirin-sensitive RS4:11 cells, which have high EIF4E and robust ribavirin uptake. These cells display reductions in nuclear to cytoplasmic export and/or translation of EIF4E-regulated mRNAs and, as a consequence, decreases in the relevant proteins (MYC, MCL1, NBN, BCL2, BIRC5) with ribavirin treatment. Relatedly, signaling phospho-proteins in the PI3K/AKT1/mTOR and eIF4/p70S6K pathways critical to translation (phospho-AKT1, phospho-EIF4EBP1, phospho-RPS6, phospho-EIF4E) are reduced by ribavirin treatment. In particular, our study brings new knowledge that the EIF4E target NBN, previously only known to be EIF4E-regulated at the nuclear export level, is also EIF4E-regulated at the translation level. The dual inhibition of NBN nuclear export and translation by ribavirin, in turn, down-regulates PI3K/AKT1/mTOR signaling downstream of NBN and is thus important to the observed cell proliferation and survival inhibition because NBN promotes AKT1 activation (25, 28, 53). These results indicate that the functional anti-leukemia effects of ribavirin in infant ALL occur by down-regulation of EIF4E activity and resultant decreases in PI3K/AKT1/mTOR and intersecting eIF4/p70S6K pathway signaling.
Our results generally are concordant with reported functional effects of EIF4E inhibition by ribavirin in multiple other cancers. For instance, AKT1 de-phosphorylation, which results from down-regulation of the direct EIF4E target transcript NBN, also was reported after ribavirin treatment in a glioblastoma cell line (72), breast cancer cell lines (53), hepatocellular carcinoma (73), oral squamous cell carcinoma (74), Ph+ ALL and CML (56) and in AML in patients (15). Additionally, we find decreases in MYC levels with ribavirin treatment in infant ALL and RS4:11 cells, and ribavirin caused MYC levels to decrease in primary AML cells (41), a diffuse large B-cell lymphoma cell line (19), colon and cervical cancer (75) and hepatocellular carcinoma (73). The decreased MYC is of interest because of protean roles of MYC in up-regulating EIF4E (76, 77) and ribosome biogenesis (e.g. RPS6) gene transcription (78, 79). Furthermore, the reduced microRNA let-7b known to occur in KMT2A-R ALL causes MYC overexpression (80), MYC activates EIF4E transcription (76, 77) and EIF4E increases MYC in a positive-feedback loop (81). Therefore, in the infant ALL and RS4:11 cells where ribavirin affected MYC it is possible that the decreased MYC contributed to decreased RPS6 and EIF4E levels. However, experiments to rescue the effects of ribavirin treatment or depletion experiments were not undertaken because, given the multiple altered EIF4E targets and larger EIF4E regulon (16, 19, 23–25), forced expression or deletion of limited targets would be unlikely to capture the broad roles of EIF4E. These results clearly demonstrate the multi-faceted importance of EIF4E to infant ALL pathobiology.
Single-agent enteral ribavirin delayed the increase in peripheral leukemia cell burden in xenograft models of one case each of KMT2A-AFF1 and KMT2A-MLLT1 infant ALL, consistent with the proliferation inhibition by ribavirin treatment in these cases when grown on BMSCs. In the third case tested in the mouse, ribavirin did not reduce peripheral blasts despite the low EC50 on BMSCs and elevated EIF4E. Although the dosing utilized (40 mg/kg oral gavage/d) potentially could achieve clinically meaningful concentrations in mouse plasma (82), it also was conservative (83–85). Further, the mice were sacrificed at 14 days, whereas in humans it takes four weeks for ribavirin to reach steady state concentrations (70), and absolute bioavailability is 40–50% with ribavirin oral dosing (86). Better efficacy including a reduction in tumor size by 50% in 10 days was observed in rapidly growing diffuse large B-cell lymphoma xenograft models with intraperitoneal ribavirin dosing at 80 mg/kg/d. Therefore, optimal mouse plasma levels may not have been attained before the mice were sacrificed. Interestingly, in breast cancer xenograft models, ribavirin resistance emerged as early as 25 days (57). Thus, more rapidly growing, resistant populations may have emerged early in our experiment. These considerations will be important future directions to advance this agent. Nonetheless, the observed in vivo responses to ribavirin, even as monotherapy, are significant because infant ALL is recalcitrant to current treatments.
To improve on these results, we tested combinations of ribavirin with chemotherapies used for infant ALL. Indeed, cooperative in vitro activity occurred between ribavirin and idarubicin, cytarabine or azacytidine in adult AML (41). We found broad cooperative ribavirin/chemotherapy interactions in reducing live KMT2A-AFF1 infant ALL cells on BMSCs. Additionally, except with vincristine, cooperative cell killing occurred with every combination. Cooperation between ribavirin and chemotherapy enabling effective dose reductions is highly desirable for infant ALL because chemotherapy is ineffective and has severe complications (2–6, 87–89). The potential of ribavirin to sensitize infant ALL to corticosteroids and L-asparaginase is particularly significant because, by in vitro drug resistance profiling, infant ALL is highly resistant to these agents compared to common/pre-B ALL in children (89). The greatest cooperation occurred with ribavirin/L-asparaginase. Amino acid deprivation responses to asparagine depletion by L-asparaginase increase EIF2S1 (eIF2α) phosphorylation, which inactivates EIF2S1, decreases TORC1 signaling and suppresses translation (90, 91). The ribavirin/L-asparaginase cooperation may suggest specific benefit to co-targeting two different nodes in translational control with this combination. The ribavirin-treated RS4:11 cells showed a S-phase arrest in the cell cycle. This differs from the G0/G1 arrest caused by ribavirin in NIH 3T3 (22), melanoma (47) and glioblastoma (48, 92) cell lines and G2/M arrest in a renal carcinoma cell line (93). These apparent cell context-dependent cytokinetic perturbations have implications about combining ribavirin with chemotherapies that affect cytokinetics or are cell cycle-dependent.
Beyond EIF4E protein elevation, other features of infant ALL suggest EIF4E as a rational target. KMT2A-R infant ALL often has high-level HOXA9 mRNA, and KMT2A-AFF1 cases partition into high and low HOXA transcript clusters (12, 94–97). Because HOXA9 displaces the EIF4E repressor PRH/Hex from EIF4E and up-regulates EIF4E-mediated nuclear mRNA export and translation (98), this suggests that HOXA9 may cause further EIF4E dysregulation. In fact, 16/25 KMT2A-AFF1 cases in the microarray (Figure 1a, asterisks) fell into the high HOXA transcript cluster (12).
Our main focus was KMT2A-R infant ALL, but we also find high-level EIF4E expression to be more prevalent in KMT2A-G pediatric pre-B ALL compared to adult ALL (17). Additionally, in vitro ribavirin activity was reported against a pediatric Ph+ALL cell line (56). These findings suggest that ribavirin anti-leukemia activity merits broader testing.
Drug repurposing creates opportunities to accelerate progress with new agents for orphan diseases (99–102). This work establishes that clinically relevant ribavirin concentrations inhibit cell proliferation and survival signaling through EIF4E in KMT2A-R infant ALL, that ribavirin monotherapy has preclinical activity against KMT2A-R infant ALL in vivo, and that it is advantageous to combine ribavirin with mainstay chemotherapies used for infant ALL. These discoveries are critical preclinical steps towards advancing ribavirin in suitable combinations to achieve chemosensitization in KMT2A-R infant ALL. They now will form the basis for preclinical testing of ribavirin/chemotherapy combinations for in vivo efficacy.
Materials and methods:
This research was performed following tenants of the Declaration of Helsinki under CHOP IRB-approved protocol (IRB 01–002460), which uses materials from subjects recruited to CHOP parent protocol 94–000771, and de-identified samples from COG leukemia cell banks or CHOP IRB study 10–007767, all with consent for future research.
Samples and cell lines
Pre-treatment COG P9407 infant ALL samples (25 KMT2A-AFF1, 8 KMT2A-MLLT1, 7 other KMT2A-R, 7 KMT2A-G)(12, 13) and pre-treatment infant (3 KMT2A-AFF1, 2 KMT2A-MLLT1) and pediatric (12 KMT2A-G) ALL samples from the Children’s Hospital of Philadelphia (CHOP) were examined. Samples were chosen based on availability. RS4:11 (ATCC), SEM-K2 (kind gift of F. Cotter) and KOPN-8 (DSMZ Collection) cell lines were authenticated by profiling with the AmpFLSTR Identifiler PCR Amplification Kit (ThermoFisher) and screened for mycoplasma contamination using PCR. Cell lines were maintained (103, 104) and acclimated (13) as reported. Primary ALL cells and cryopreserved healthy adult PBMCs (Stem Cell Technologies or University of Pennsylvania Human Immunology Core) were acclimated overnight in RPMI-1640/20% serum substitute (BIT 9500; Stem Cell Technologies, Vancouver, Canada), 10ng/mL IL-7 and SCF before basal QPCR and Western blot analyses.
Bioinformatics analyses
Spearman test was used to identify correlations between log2 expression levels for each probeset from Affymetrix HG_U133 Plus2.0 arrays (GEO number GSE68720) and obatoclax EC50s.(12, 13) A heatmap of 446 probesets with significant correlations (p≤0.001) was generated by transforming expression levels into z-scores, computing Pearson correlation between probesets and between samples by complete linkage clustering, and plotting the data using R/gplots.
Functional annotation and gene ontology (GO) analysis was performed using DAVID (david.abcc.ncifcrf.gov). Overlap with genes in prebuilt lists was determined using default settings, OMIM annotation, protein functional domain and motif information, GO and pathway annotations. Genes with significant expression differences were used to identify GO groups. Significant terms (Benjamini multiple-testing corrected <0.1) were included.
Correlations between obatoclax EC50 and cell signaling pathways were determined by Ingenuity Pathway Analysis (IPA) of significant probesets (p<0.1).
QPCR analysis of primary samples
Random-primed cDNAs synthesized from 1 μg total RNA with a High-Capacity cDNA Reverse Transcription Kit were amplified using TaqMan™ Universal PCR Master Mix and EIF4E (Hs00854166_g1), EIF2A (Hs01045430_m1) or ACTB (Hs99999903_m1) primers (ThermoFisher Scientific, Waltham, MA, USA). There were 3 intra-experimental replicates per reaction.
Relative expression between groups by obatoclax response was compared by computing ΔCT values (avgCT[target] – avgCT [ACTB]).
For comparisons of normalized EIF4E expression in infant ALL KMT2A genetic subsets relative to PBMCs, average ΔCT values (avgCT[EIF4E] - avgCT[ACTB]) based on 3 intra-experimental replicates per reaction were used to calculate ΔΔCT (avgΔCT [sample] - meanΔCT [6 PBMC samples]) and 2-ΔΔCT for each sample.
Western blot analyses
Total protein lysates in RIPA buffer (103) were separated on NuPAGE gels (Invitrogen, Carlsbad, CA, USA), transferred onto PVDF membranes and analyzed using antibodies to EIF4E, NBN, EIF4EBP1, RPS6, AKT1, UGT1A, MCL1, MYC, GAPDH, ADK, SLC29A1 or ACTB (Table S3). RS4:11 cell lysates in Phospho-Lysis buffer (28) were studied using antibodies to phospho-AKT1Thr308, phospho-EIF4EBP1Thr37/46, phospho-RPS6Ser235/236 or phospho–EIF4ESer209 (Table S3).
Band intensities were quantified using ImageJ (v1.49) to normalize expression to loading control proteins (ACTB or GAPDH) and examine relative expression (105).
Assays of infant ALL cells on BMSCs
Immortalized BMSCs (BMSC-hTERT) (45) were irradiated with 2000 cGy, plated onto 24 well plates (1×105 cells/well) (day −3, Figure 3a), allowed to adhere overnight and washed with X-VIVO 10 media (Lonza, Portsmouth, NH, USA) before adding ALL cells (day −2, Figure 3a).
Thawed ALL cells were labeled with 1μM CFSE (eBioscience, San Diego, CA, USA), washed twice, co-cultured ×48 h on BMSC-hTERT (106) to allow proliferation, and then treated (starting at time 0, Figure 3a) with vehicle or ribavirin (Sigma Aldrich) × 72 h with ribavirin replenishment at 48 h (Borden, unpublished). Cells were labeled with anti-CD19-APC-AF750 (Invitrogen) and DAPI (Invitrogen) and analyzed on a LSR II cytometer (BD Biosciences, San Jose, CA, USA). Cell divisions based on CFSE staining were modeled and cell death (DAPI+CD19+ cells) quantified using FlowJo (Tree Star, Inc. v9.7.6).
Replication indices (RIs) calculated using FlowJo were used in the equation modified from Munson et al. (107) to determine proliferating fractions (PFs). Inhibitory sigmoid Emax models (1.0 top down to 0.0 bottom, variable slope) based on PFs were generated using GraphPad Prism.
ALL cells were isolated with CD19 MicroBeads using MACS (Miltenyi Biotec, Auburn, GA) for Western blot analysis.
ALL cells were expanded as secondary xenografts in NOD/SCID/Il2rγtm1wjl/SzJ (NSG) mice (Supplemental Methods) and splenic lymphoblasts co-cultured on BMSC-hTERT. CD19+ cells were purified by immunomagnetic positive selection using an EasySepTM kit (Stem Cell Technologies) to determine ribavirin effects on EIF4E subcellular distribution (Case PAPAWG).
Splenic lymphoblasts co-cultured on BMSC-hTERT (Case PAPAWG) were treated with vehicle, chemotherapy followed by ribavirin, or single-agent chemotherapy or ribavirin at times matched to those in the combinations (Figure 7a). BMSCs were plated on day −3 (24 h before adding ALL cells). ALL cells were added on day −2 and co-cultured on the stromal layer ×48 h before treatment to allow proliferation. The chemotherapy was L-asparaginase, cytarabine, dexamethasone, etoposide, doxorubicin or vincristine (Table S4). Ribavirin was replenished after 48 h. The cells were collected on day 5 of treatment, labeled with anti-CD19-APC-AF750 and DAPI, and counted using CountBrightTM Absolute Counting Beads (Molecular Probes, Eugene, OR, USA). Viable (DAPI-CD19+) and dead (DAPI+CD19+) cells were quantified. The ALL cells were labeled with CFSE before plating; however, cell divisions were not analyzed because of signal attenuation due to cell divisions in excess of the limit of resolvable generations after prolonged culture.
MTT assays
Cell lines were plated and MTT assays (ATCC) (13) performed after 72 h ribavirin treatment. Surviving fraction plots were generated and EC50s determined from inhibitory sigmoid Emax models.
3H-ribavirin uptake
Cells were incubated with 1 μM 3H-ribavirin (Table S4) for 8 h, lysed, and scintillation counting performed as described (42).
Cellular Fractionation and Export Assay
Fractionation protocol was followed as previously described (19). About 3 × 107 cells were collected and washed twice in ice cold PBS with centrifugation at 1,200 rpm for 3–5 min. The cell pellet was resuspended with slow pipetting in 1 mL of lysis buffer B (10 mM Tris pH 8.4, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP40, 1 mM DTT and 100 U/mL RNase Inhibitors (ThermoFisher Scientific). The lysate was centrifuged at 1000×g for 3 min at 4°C and supernatant (cytoplasmic fraction) was transferred into a fresh micro-centrifuge tube. The pellet (nuclear fraction) was resuspended in 1 mL of lysis buffer B and transferred to a round bottom, polypropylene tube, 1/10 volume (100 μL) of detergent stock (3.3% Sodium Deoxycholate and 6.6% Tween 40 in DEPC H20) was added with slow vortexing, and the tube was incubated on ice for 5 min. The suspension was transferred to a micro-centrifuge tube and centrifuged at 1,000×g for 3 min at 4°C. The supernatant (post-nuclear fraction) was removed and added to cytoplasmic fraction, and the pelleted-nuclear fraction rinsed in 1 mL of lysis buffer B and centrifuged at 1000×g for 3 min at 4°C.
RNA was extracted from the different fractions by adding TRIzol reagent (ThermoFisher Scientific) and processed using a DirectZol RNA Miniprep kit (Zymo Research, Irvine, CA). DNase treated RNAs were reversed transcribed using M-MLV Reverse Transcriptase (ThermoFisher Scientific, Cat# 28025013). QPCR assays were performed using listed primers (Table S5) and a SensiFAST SYBR Lo-ROX Kit (Bioline, London, UK) in an Applied Biosystems ViiA7 thermal cycler by the relative standard curve method (Applied Biosystems User Bulletin #2) according to described conditions (19). Semi-quantitative RT-PCR analysis of U6 snRNA and tRNAlys was performed using Taq DNA Polymerase with ThermoPol Reaction Buffer (NEB, Ipswich, MA, USA) and 30 cycles of 95°30 sec, 56°×30 sec, 68°×30 sec.
Polysomal profiling
Polysomal profiling was done as described (19). Briefly, cells were treated with cyclohexamide (100 μg/mL, Sigma Aldrich) for 10 min before harvesting and lysates were prepared using polysome lysis buffer (15mM Tris pH 7.4, 250 mM NaCl, 15 mM MgCl2, 1% Triton X-100, 100 μg/mL cyclohexamide, 1 mM DTT, 400 U/mL RNase inhibitors) with EDTA-free cOmplete protease inhibitors (Roche). Equal amounts (3–5 mg) of protein lysates were layered on a 20–50% linear sucrose gradient (20 and 50% sucrose solutions in 15 mM Tris pH 7.4, 15 mM MgCl2, 150 mM NaCl, 1 mM DTT, 100 μg/mL cyclohexamide, 20 U/mL RNase inhibitors), mixed on a Gradient Station IP (Biocomp) and centrifuged in a Beckman SW41Ti rotor at 92,000×g for 3h at 4˚C. Following centrifugation, polysomal fractions were collected by continuously monitoring and recording the A254 on a Gradient Station IP (Biocomp) attached to a UV-MII (GE Healthcare) spectrophotometer. RNAs were isolated from polysomal fractions using TRIzol reagent. RNAs from each fraction were measured using QPCR (Table S5).
Cell cycle analysis
Propidium iodide (Sigma-Aldrich) labeling was quantified on a LSR II flow cytometer.(108) Cell cycle was modeled using ModFit LTTM (Verity Software House, Topsham, ME, USA).
EIF4E subcellular distribution
Cells (6–12×104/sample) were smeared onto slides, dried at 37°C, fixed and permeabilized with 100% methanol at −20°C ×20 min, washed twice with PBS and blocked with 10% FBS/0.2% Tween-20/PBS at 37°C ×1 h. After labeling with 1:50 anti-EIF4E-FITC (BD Transduction) in 2% FBS/0.2% Tween-20/PBS at 37°C ×2 h, slides were mounted using Fluoro-Gel II with DAPI (Electron Microscopy Sciences, Hatfield, PA, USA).
Microscopy
DAPI and EIF4E images of primary cells were acquired with a 100X/1.30 oil UPlan Fluo objective using an Olympus IX71 Microscope equipped with Leica DP80 camera. DAPI and EIF4E images of RS4:11 cells were acquired with a Zeiss LSM710 confocal microscope using a 63X/1.4 oil Plan-Apochromat objective and Axiocam High Resolution Camera. Individual DAPI and EIF4E images were overlaid using Adobe Photoshop CS5 (v.12.0.4×64).
Statistical Analysis
Significance in QPCR experiments was determined by Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparison post-hoc tests in order to asses for gene expression as a continuous variable by samples categorized by drug response (Figures 1c, S3) or KMT2A genetic subset (Figure 2b). Box and whisker plots were created using GraphPad Prism (v6.0). Student’s t-test was used to analyze ribavirin-sensitive vs. resistant sample categories for a difference in basal normalized EIF4E expression (Figure 4b).
Supplementary Material
Acknowledgements
Childhood ALL samples provided by CHOP CCCR Biorepository.
C.A.F. is Joshua Kahan Endowed Chair in Pediatric Leukemia Research. S.P.H. is Jeffrey E. Perelman Distinguished Chair in the Department of Pediatrics at CHOP. K.L.B.B. holds a Canada Research Chair in Molecular Biology of the Cell Nucleus.
We thank Ed Lopata and Lisa McCoy for supporting our research on KMT2A-R leukemia in honor of their son Jesse.
Sources of Support: C.A.F., L.-S.W., J.S.B., C.L.W., M.D. and S.P.H. supported by Leukemia & Lymphoma Society SCOR 7372–07. C.A.F. supported by ASH Bridge Funding Program, CHOP Bridge Funding Program, Eagles Fly for Leukemia, and CHOP Hematologic Malignancies Research Fund. C.A.F. and S.K.T. supported by SU2C – St. Baldrick’s Pediatric Dream Team Translational Research Grant SU2C-AACR-DT1113 (SU2C is a program of Entertainment Industry Foundation administered by AACR). C.A.F. and D.T.T. supported by Cookies for Kids’ Cancer Foundation. S.K.T. supported by K08CA184418 and Rally Foundation for Childhood Cancer Research and was ALSF Scholar in Developmental Therapeutics. A.E.S. supported by ACS MRSG-12–215-01-LIB and Hyundai Hope on Wheels Scholar Award. M.P.C. supported by R01CA198089. K.L.B.B. funded by R01CA098571 and R01CA080728 and holds a Canada Research Chair. Other support provided by U10CA98413 (COG Statistical Center), U24CA114766 (COG Specimen Banking).
Footnotes
Conflicts of interest:
C.A.F. owns an unlicensed patent relevant to genetic classification of leukemia cases: Methods and Kits for Analysis of Chromosomal Rearrangements Associated with Leukemia. United States of America 6,368,791. 2002 April 09.
C.A.F. owns unlicensed, unrelated patents: United States of America Patent Numbers 6,174,684; and 8,642,265 B2. C.A.F. and L.-S.W. submitted an unrelated patent application: United States of America Patent Application 61/490,975.
References
- 1.Barrington-Trimis JL, Cockburn M, Metayer C, Gauderman WJ, Wiemels J, McKean-Cowdin R. Trends in childhood leukemia incidence over two decades from 1992 to 2013. Int J Cancer. 2017;140(5):1000–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunger SP, Lu X, Devidas M, Camitta BM, Gaynon PS, Winick NJ, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children’s oncology group. J Clin Oncol. 2012;30(14):1663–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biondi A, Rizzari C, Valsecchi MG, De Lorenzo P, Arico M, Basso G, et al. Role of treatment intensification in infants with acute lymphoblastic leukemia: results of two consecutive AIEOP studies. Haematologica. 2006;91(4):534–7. [PubMed] [Google Scholar]
- 4.Hilden JM, Dinndorf PA, Meerbaum SO, Sather H, Villaluna D, Heerema NA, et al. Analysis of prognostic factors of acute lymphoblastic leukemia in infants: report on CCG 1953 from the Children’s Oncology Group. Blood. 2006;108(2):441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pieters R, Schrappe M, De Lorenzo P, Hann I, De Rossi G, Felice M, et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet. 2007;370(9583):240–50. [DOI] [PubMed] [Google Scholar]
- 6.Dreyer ZE, Hilden JM, Jones TL, Devidas M, Winick NJ, Willman CL, et al. Intensified chemotherapy without SCT in infant ALL: results from COG P9407 (Cohort 3). Pediatr Blood Cancer. 2015;62(3):419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferster A, Bertrand Y, Benoit Y, Boilletot A, Behar C, Margueritte G, et al. Improved survival for acute lymphoblastic leukaemia in infancy: the experience of EORTC-Childhood Leukaemia Cooperative Group. Br J Haematol. 1994;86(2):284–90. [DOI] [PubMed] [Google Scholar]
- 8.Dordelmann M, Reiter A, Borkhardt A, Ludwig WD, Gotz N, Viehmann S, et al. Prednisone response is the strongest predictor of treatment outcome in infant acute lymphoblastic leukemia. Blood. 1999;94(4):1209–17. [PubMed] [Google Scholar]
- 9.Reaman GH, Sposto R, Sensel MG, Lange BJ, Feusner JH, Heerema NA, et al. Treatment outcome and prognostic factors for infants with acute lymphoblastic leukemia treated on two consecutive trials of the Children’s Cancer Group. J Clin Oncol. 1999;17(2):445–55. [DOI] [PubMed] [Google Scholar]
- 10.Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJ, Kazemier KM, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351(6):533–42. [DOI] [PubMed] [Google Scholar]
- 11.Cheok MH, Evans WE. Acute lymphoblastic leukaemia: a model for the pharmacogenomics of cancer therapy. Nat Rev Cancer. 2006;6(2):117–29. [DOI] [PubMed] [Google Scholar]
- 12.Kang H, Wilson CS, Harvey RC, Chen IM, Murphy MH, Atlas SR, et al. Gene expression profiles predictive of outcome and age in infant acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood. 2012;119(8):1872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urtishak KA, Edwards AY, Wang LS, Hudome A, Robinson BW, Barrett JS, et al. Potent obatoclax cytotoxicity and activation of triple death mode killing across infant acute lymphoblastic leukemia. Blood. 2013;121(14):2689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Culjkovic B, Borden KL. Understanding and Targeting the Eukaryotic Translation Initiation Factor eIF4E in Head and Neck Cancer. J Oncol. 2009;2009:981679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Assouline S, Culjkovic B, Cocolakis E, Rousseau C, Beslu N, Amri A, et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood. 2009;114(2):257–60. [DOI] [PubMed] [Google Scholar]
- 16.Borden KL, Culjkovic-Kraljacic B. Ribavirin as an anti-cancer therapy: acute myeloid leukemia and beyond? Leuk Lymphoma. 2010;51(10):1805–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Topisirovic I, Guzman ML, McConnell MJ, Licht JD, Culjkovic B, Neering SJ, et al. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol Cell Biol. 2003;23(24):8992–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Assouline S, Culjkovic-Kraljacic B, Bergeron J, Caplan S, Cocolakis E, Lambert C, et al. A phase I trial of ribavirin and low-dose cytarabine for the treatment of relapsed and refractory acute myeloid leukemia with elevated eIF4E. Haematologica. 2015;100(1):e7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Culjkovic-Kraljacic B, Fernando TM, Marullo R, Calvo-Vidal N, Verma A, Yang S, et al. Combinatorial targeting of nuclear export and translation of RNA inhibits aggressive B-cell lymphomas. Blood. 2016;127(7):858–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kentsis A, Volpon L, Topisirovic I, Soll CE, Culjkovic B, Shao L, et al. Further evidence that ribavirin interacts with eIF4E. RNA. 2005;11(12):1762–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volpon L, Osborne MJ, Zahreddine H, Romeo AA, Borden KL. Conformational changes induced in the eukaryotic translation initiation factor eIF4E by a clinically relevant inhibitor, ribavirin triphosphate. Biochem Biophys Res Commun. 2013;434(3):614–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004;101(52):18105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175(3):415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Culjkovic B, Topisirovic I, Borden KL. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle. 2007;6(1):65–9. [DOI] [PubMed] [Google Scholar]
- 25.Tan K, Culjkovic B, Amri A, Borden KL. Ribavirin targets eIF4E dependent Akt survival signaling. Biochem Biophys Res Commun. 2008;375(3):341–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett D, Brown VI, Grupp SA, Teachey DT. Targeting the PI3K/AKT/mTOR signaling axis in children with hematologic malignancies. Paediatr Drugs. 2012;14(5):299–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapuis N, Tamburini J, Green AS, Willems L, Bardet V, Park S, et al. Perspectives on inhibiting mTOR as a future treatment strategy for hematological malignancies. Leukemia. 2010;24(10):1686–99. [DOI] [PubMed] [Google Scholar]
- 28.Culjkovic B, Tan K, Orolicki S, Amri A, Meloche S, Borden KL. The eIF4E RNA regulon promotes the Akt signaling pathway. J Cell Biol. 2008;181(1):51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riner A, Chan-Tack KM, Murray JS. Original research: Intravenous ribavirin--review of the FDA’s Emergency Investigational New Drug Database (1997–2008) and literature review. Postgrad Med. 2009;121(3):139–46. [DOI] [PubMed] [Google Scholar]
- 30.Yang CHT, Yoo ER, Ahmed A. The Role of Direct-acting Antivirals in the Treatment of Children with Chronic Hepatitis C. J Clin Transl Hepatol. 2017;5(1):59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunn LA, Fury MG, Sherman EJ, Ho AA, Katabi N, Haque SS, et al. Phase I study of induction chemotherapy with afatinib, ribavirin, and weekly carboplatin and paclitaxel for stage IVA/IVB human papillomavirus-associated oropharyngeal squamous cell cancer. Head Neck. 2018;40(2):233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosaka T, Maeda T, Shinojima T, Nagata H, Mizuno R, Oya M. A clinical study to evaluate the efficacy and safety of docetaxel with ribavirin in patients with progressive castration resistant prostate cancer who have previously received docetaxel alone. J Clin Oncol. 2017;35(15_suppl):e14010–e. [Google Scholar]
- 33.Kosaka T, Shinojima T, Kikuchi K, Hagiwara S, Kojima S, Hongo H, et al. A phase 1/2a trial of docetaxel plus ribavirin for reprogramming efficacy in patients with progressive metastatic castration resistant prostate cancer who have previously received docetaxel alone: DRREEM trial. J Clin Oncol. 2018;36(6_suppl):329-. [Google Scholar]
- 34.Hall CB, McBride JT, Walsh EE, Bell DM, Gala CL, Hildreth S, et al. Aerosolized ribavirin treatment of infants with respiratory syncytial viral infection. A randomized double-blind study. N Engl J Med. 1983;308(24):1443–7. [DOI] [PubMed] [Google Scholar]
- 35.Krilov LR. Safety issues related to the administration of ribavirin. Pediatr Infect Dis J. 2002;21(5):479–81. [DOI] [PubMed] [Google Scholar]
- 36.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6(4):318–27. [DOI] [PubMed] [Google Scholar]
- 37.Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21(24):3232–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borden KL. The eukaryotic translation initiation factor eIF4E wears a “cap” for many occasions. Translation (Austin). 2016;4(2):e1220899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asano K, Phan L, Valasek L, Schoenfeld LW, Shalev A, Clayton J, et al. A multifactor complex of eIF1, eIF2, eIF3, eIF5, and tRNA(i)Met promotes initiation complex assembly and couples GTP hydrolysis to AUG recognition. Cold Spring Harb Symp Quant Biol. 2001;66:403–15. [DOI] [PubMed] [Google Scholar]
- 40.Topisirovic I, Siddiqui N, Orolicki S, Skrabanek LA, Tremblay M, Hoang T, et al. Stability of eukaryotic translation initiation factor 4E mRNA is regulated by HuR, and this activity is dysregulated in cancer. Mol Cell Biol. 2009;29(5):1152–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraljacic BC, Arguello M, Amri A, Cormack G, Borden K. Inhibition of eIF4E with ribavirin cooperates with common chemotherapies in primary acute myeloid leukemia specimens. Leukemia. 2011;25(7):1197–200. [DOI] [PubMed] [Google Scholar]
- 42.Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, Gendron P, Romeo AA, Morris SJ, et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature. 2014;511(7507):90–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanderson RD, Lalor P, Bernfield M. B lymphocytes express and lose syndecan at specific stages of differentiation. Cell Regul. 1989;1(1):27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roederer M Interpretation of cellular proliferation data: avoid the panglossian. Cytometry A. 2011;79(2):95–101. [DOI] [PubMed] [Google Scholar]
- 45.Mihara K, Imai C, Coustan-Smith E, Dome JS, Dominici M, Vanin E, et al. Development and functional characterization of human bone marrow mesenchymal cells immortalized by enforced expression of telomerase. Br J Haematol. 2003;120(5):846–9. [DOI] [PubMed] [Google Scholar]
- 46.Rixe O, Fojo T. Is cell death a critical end point for anticancer therapies or is cytostasis sufficient? Clin Cancer Res. 2007;13(24):7280–7. [DOI] [PubMed] [Google Scholar]
- 47.Vallee S, Fouchier F, Braguer D, Marvaldi J, Champion S. Ribavirin-induced resistance to heat shock, inhibition of the Ras-Raf-1 pathway and arrest in G(1). Eur J Pharmacol. 2000;404(1–2):49–62. [DOI] [PubMed] [Google Scholar]
- 48.Volpin F, Casaos J, Sesen J, Mangraviti A, Choi J, Gorelick N, et al. Use of an anti-viral drug, Ribavirin, as an anti-glioblastoma therapeutic. Oncogene. 2016. [DOI] [PubMed] [Google Scholar]
- 49.Aebi C, Headrick CL, McCracken GH, Lindsay CA. Intravenous ribavirin therapy in a neonate with disseminated adenovirus infection undergoing extracorporeal membrane oxygenation: pharmacokinetics and clearance by hemofiltration. J Pediatr. 1997;130(4):612–5. [DOI] [PubMed] [Google Scholar]
- 50.Willis RC, Carson DA, Seegmiller JE. Adenosine kinase initiates the major route of ribavirin activation in a cultured human cell line. Proc Natl Acad Sci U S A. 1978;75(7):3042–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarvis SM, Thorn JA, Glue P. Ribavirin uptake by human erythrocytes and the involvement of nitrobenzylthioinosine-sensitive (es)-nucleoside transporters. Br J Pharmacol. 1998;123(8):1587–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mamane Y, Petroulakis E, Martineau Y, Sato TA, Larsson O, Rajasekhar VK, et al. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS One. 2007;2(2):e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersson F, Yau C, Dobocan MC, Culjkovic-Kraljacic B, Retrouvey H, Puckett R, et al. Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clin Cancer Res. 2011;17(9):2874–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Landon AL, Muniandy PA, Shetty AC, Lehrmann E, Volpon L, Houng S, et al. MNKs act as a regulatory switch for eIF4E1 and eIF4E3 driven mRNA translation in DLBCL. Nat Commun. 2014;5:5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Topisirovic I, Ruiz-Gutierrez M, Borden KL. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res. 2004;64(23):8639–42. [DOI] [PubMed] [Google Scholar]
- 56.Shi F, Len Y, Gong Y, Shi R, Yang X, Naren D, et al. Ribavirin Inhibits the Activity of mTOR/eIF4E, ERK/Mnk1/eIF4E Signaling Pathway and Synergizes with Tyrosine Kinase Inhibitor Imatinib to Impair Bcr-Abl Mediated Proliferation and Apoptosis in Ph+ Leukemia . PLoS One. 2015;10(8):e0136746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersson F, Del Rincon SV, Emond A, Huor B, Ngan E, Ng J, et al. Genetic and pharmacologic inhibition of eIF4E reduces breast cancer cell migration, invasion, and metastasis. Cancer Res. 2015;75(6):1102–12. [DOI] [PubMed] [Google Scholar]
- 58.Asselin BL, Whitin JC, Coppola DJ, Rupp IP, Sallan SE, Cohen HJ. Comparative pharmacokinetic studies of three asparaginase preparations. J Clin Oncol. 1993;11(9):1780–6. [DOI] [PubMed] [Google Scholar]
- 59.Frost BM, Eksborg S, Bjork O, Abrahamsson J, Behrendtz M, Castor A, et al. Pharmacokinetics of doxorubicin in children with acute lymphoblastic leukemia: multi-institutional collaborative study. Med Pediatr Oncol. 2002;38(5):329–37. [DOI] [PubMed] [Google Scholar]
- 60.Lowis SP, Pearson AD, Newell DR, Cole M. Etoposide pharmacokinetics in children: the development and prospective validation of a dosing equation. Cancer Res. 1993;53(20):4881–9. [PubMed] [Google Scholar]
- 61.Moore AS, Norris R, Price G, Nguyen T, Ni M, George R, et al. Vincristine pharmacodynamics and pharmacogenetics in children with cancer: a limited-sampling, population modelling approach. Journal of paediatrics and child health. 2011;47(12):875–82. [DOI] [PubMed] [Google Scholar]
- 62.Yang L, Panetta JC, Cai X, Yang W, Pei D, Cheng C, et al. Asparaginase may influence dexamethasone pharmacokinetics in acute lymphoblastic leukemia. J Clin Oncol. 2008;26(12):1932–9. [DOI] [PubMed] [Google Scholar]
- 63.Groninger E, Proost JH, de Graaf SS. Pharmacokinetic studies in children with cancer. Crit Rev Oncol Hematol. 2004;52(3):173–97. [DOI] [PubMed] [Google Scholar]
- 64.McLeod HL, Evans WE. Clinical pharmacokinetics and pharmacodynamics of epipodophyllotoxins. Cancer Surv. 1993;17:253–68. [PubMed] [Google Scholar]
- 65.McLeod HL, Relling MV, Crom WR, Silverstein K, Groom S, Rodman JH, et al. Disposition of antineoplastic agents in the very young child. The British journal of cancer Supplement. 1992;18:S23–9. [PMC free article] [PubMed] [Google Scholar]
- 66.Krishan A, Frei E 3rd. Effect of adriamycin on the cell cycle traverse and kinetics of cultured human lymphoblasts. Cancer Res. 1976;36(1):143–50. [PubMed] [Google Scholar]
- 67.Jedema I, Barge RM, Frankel AE, Willemze R, Falkenburg JH. Acute myeloid leukemia cells in G0 phase of the cell cycle that are unresponsive to conventional chemotherapy are sensitive to treatment with granulocyte-macrophage colony-stimulating factor/diphtheria toxin fusion proteins. Exp Hematol. 2004;32(2):188–94. [DOI] [PubMed] [Google Scholar]
- 68.van Leeuwen IM, Rao B, Sachweh MC, Lain S. An evaluation of small-molecule p53 activators as chemoprotectants ameliorating adverse effects of anticancer drugs in normal cells. Cell Cycle. 2012;11(9):1851–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laskin OL, Longstreth JA, Hart CC, Scavuzzo D, Kalman CM, Connor JD, et al. Ribavirin disposition in high-risk patients for acquired immunodeficiency syndrome. Clin Pharmacol Ther. 1987;41(5):546–55. [DOI] [PubMed] [Google Scholar]
- 70.Glue P The clinical pharmacology of ribavirin. Semin Liver Dis. 1999;19 Suppl 1:17- [PubMed] [Google Scholar]
- 71.Volpon L, Culjkovic-Kraljacic B, Osborne MJ, Ramteke A, Sun Q, Niesman A, et al. Importin 8 mediates m7G cap-sensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proc Natl Acad Sci U S A. 2016;113(19):5263–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Isakovic AM, Dulovic M, Markovic I, Kravic-Stevovic T, Bumbasirevic V, Trajkovic V, et al. Autophagy suppression sensitizes glioma cells to IMP dehydrogenase inhibition-induced apoptotic death. Exp Cell Res. 2017;350(1):32–40. [DOI] [PubMed] [Google Scholar]
- 73.Tan J, Ye J, Song M, Zhou M, Hu Y. Ribavirin augments doxorubicin’s efficacy in human hepatocellular carcinoma through inhibiting doxorubicin-induced eIF4E activation. J Biochem Mol Toxicol. 2018;32(1). [DOI] [PubMed] [Google Scholar]
- 74.Dai D, Chen H, Tang J, Tang Y. Inhibition of mTOR/eIF4E by anti-viral drug ribavirin effectively enhances the effects of paclitaxel in oral tongue squamous cell carcinoma. Biochem Biophys Res Commun. 2017;482(4):1259–64. [DOI] [PubMed] [Google Scholar]
- 75.Xi C, Wang L, Yu J, Ye H, Cao L, Gong Z. Inhibition of eukaryotic translation initiation factor 4E is effective against chemo-resistance in colon and cervical cancer. Biochem Biophys Res Commun. 2018. [DOI] [PubMed] [Google Scholar]
- 76.Rosenwald IB, Rhoads DB, Callanan LD, Isselbacher KJ, Schmidt EV. Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2 alpha in response to growth induction by c-myc. Proc Natl Acad Sci U S A. 1993;90(13):6175–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jones RM, Branda J, Johnston KA, Polymenis M, Gadd M, Rustgi A, et al. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol Cell Biol. 1996;16(9):4754–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20(6):1383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16(4):253–64. [DOI] [PubMed] [Google Scholar]
- 80.Schotte D, De Menezes RX, Akbari Moqadam F, Khankahdani LM, Lange-Turenhout E, Chen C, et al. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica. 2011;96(5):703–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin CJ, Malina A, Pelletier J. c-Myc and eIF4F constitute a feedforward loop that regulates cell growth: implications for anticancer therapy. Cancer research. 2009;69(19):7491–4. [DOI] [PubMed] [Google Scholar]
- 82.Endres CJ, Moss AM, Govindarajan R, Choi DS, Unadkat JD. The role of nucleoside transporters in the erythrocyte disposition and oral absorption of ribavirin in the wild-type and equilibrative nucleoside transporter 1−/− mice. J Pharmacol Exp Ther. 2009;331(1):287–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smee DF, Wandersee MK, Wong MH, Bailey KW, Sidwell RW. Treatment of mannan-enhanced influenza B virus infections in mice with oseltamivir, ribavirin and viramidine. Antivir Chem Chemother. 2004;15(5):261–8. [DOI] [PubMed] [Google Scholar]
- 84.Sidwell RW, Bailey KW, Wong MH, Barnard DL, Smee DF. In vitro and in vivo influenza virus-inhibitory effects of viramidine. Antiviral research. 2005;68(1):10–7. [DOI] [PubMed] [Google Scholar]
- 85.Khare GP, Sidwell RW, Witkowski JT, Simon LN, Robins RK. Suppression by 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide (virazole, ICN 1229) of influenza virus-induced infections in mice. Antimicrob Agents Chemother. 1973;3(4):517–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koren G, King S, Knowles S, Phillips E. Ribavirin in the treatment of SARS: A new trick for an old drug? CMAJ 2003;168(10):1289–92. [PMC free article] [PubMed] [Google Scholar]
- 87.Scully RE, Lipshultz SE. Anthracycline cardiotoxicity in long-term survivors of childhood cancer. Cardiovascular toxicology. 2007;7(2):122–8. [DOI] [PubMed] [Google Scholar]
- 88.Hijiya N, Ness KK, Ribeiro RC, Hudson MM. Acute leukemia as a secondary malignancy in children and adolescents: current findings and issues. Cancer. 2009;115(1):23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramakers-van Woerden NL, Beverloo HB, Veerman AJ, Camitta BM, Loonen AH, van Wering ER, et al. In vitro drug-resistance profile in infant acute lymphoblastic leukemia in relation to age, MLL rearrangements and immunophenotype. Leukemia. 2004;18(3):521–9. [DOI] [PubMed] [Google Scholar]
- 90.Bunpo P, Dudley A, Cundiff JK, Cavener DR, Wek RC, Anthony TG. GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent L-asparaginase. J Biol Chem. 2009;284(47):32742–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bunpo P, Cundiff JK, Reinert RB, Wek RC, Aldrich CJ, Anthony TG. The eIF2 kinase GCN2 is essential for the murine immune system to adapt to amino acid deprivation by asparaginase. The Journal of nutrition. 2010;140(11):2020–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ogino A, Sano E, Ochiai Y, Yamamuro S, Tashiro S, Yachi K, et al. Efficacy of ribavirin against malignant glioma cell lines. Oncology letters. 2014;8(6):2469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Teng L, Ding D, Chen Y, Dai H, Liu G, Qiao Z, et al. Anti-tumor effect of ribavirin in combination with interferon-alpha on renal cell carcinoma cell lines in vitro. Cancer cell international. 2014;14:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113(11):2375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Imamura T, Morimoto A, Takanashi M, Hibi S, Sugimoto T, Ishii E, et al. Frequent co-expression of HoxA9 and Meis1 genes in infant acute lymphoblastic leukaemia with MLL rearrangement. Br J Haematol. 2002;119(1):119–21. [DOI] [PubMed] [Google Scholar]
- 96.Trentin L, Giordan M, Dingermann T, Basso G, Te Kronnie G, Marschalek R. Two independent gene signatures in pediatric t(4;11) acute lymphoblastic leukemia patients. Eur J Haematol. 2009;83(5):406–19. [DOI] [PubMed] [Google Scholar]
- 97.Stam RW, Schneider P, Hagelstein JA, van der Linden MH, Stumpel DJ, de Menezes RX, et al. Gene expression profiling-based dissection of MLL translocated and MLL germline acute lymphoblastic leukemia in infants. Blood. 2010;115(14):2835–44. [DOI] [PubMed] [Google Scholar]
- 98.Topisirovic I, Kentsis A, Perez JM, Guzman ML, Jordan CT, Borden KL. Eukaryotic translation initiation factor 4E activity is modulated by HOXA9 at multiple levels. Mol Cell Biol. 2005;25(3):1100–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu K, Cote TR. Database identifies FDA-approved drugs with potential to be repurposed for treatment of orphan diseases. Brief Bioinform. 2011;12(4):341–5. [DOI] [PubMed] [Google Scholar]
- 100.Ekins S, Williams AJ, Krasowski MD, Freundlich JS. In silico repositioning of approved drugs for rare and neglected diseases. Drug Discov Today. 2011;16(7–8):298–310. [DOI] [PubMed] [Google Scholar]
- 101.Manara MC, Garofalo C, Ferrari S, Belfiore A, Scotlandi K. Designing novel therapies against sarcomas in the era of personalized medicine and economic crisis. Curr Pharm Des. 2013;19(30):5344–61. [DOI] [PubMed] [Google Scholar]
- 102.Pessetto ZY, Weir SJ, Sethi G, Broward MA, Godwin AK. Drug repurposing for gastrointestinal stromal tumor. Mol Cancer Ther. 2013;12(7):1299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Robinson BW, Behling KC, Gupta M, Zhang AY, Moore JS, Bantly AD, et al. Abundant anti-apoptotic BCL-2 is a molecular target in leukaemias with t(4;11) translocation. Br J Haematol. 2008;141(6):827–39. [DOI] [PubMed] [Google Scholar]
- 104.Matsuo Y, Drexler HG. Establishment and characterization of human B cell precursor-leukemia cell lines. Leuk Res. 1998;22(7):567–79. [DOI] [PubMed] [Google Scholar]
- 105.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Teachey DT, Obzut DA, Cooperman J, Fang J, Carroll M, Choi JK, et al. The mTOR inhibitor CCI-779 induces apoptosis and inhibits growth in preclinical models of primary adult human ALL. Blood. 2006;107(3):1149–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Munson ME. An improved technique for calculating relative response in cellular proliferation experiments. Cytometry A. 2010;77(10):909–10. [DOI] [PubMed] [Google Scholar]
- 108.Pozarowski P, Grabarek J, Darzynkiewicz Z. Flow cytometry of apoptosis. Curr Protoc Cytom. 2003;Chapter 7:Unit 7 19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.