Abstract
Staphylococcus epidermidis is the predominant cause of recalcitrant biofilm-associated infections, which are often highly resistant to antibiotics. Thus, the use of physico-chemical agents for disinfection offers a more effective approach to the control of S. epidermidis biofilm infections. However, the underlying tolerance mechanisms employed by S. epidermidis biofilm against these physico-chemical disinfectants remain largely unknown. The expression of a σB-dependent gene, alkaline shock protein 23 (asp23) and catalase activity by S. epidermidis biofilm and planktonic cells exposed to heat (50 °C), 0.8 M sodium chloride (NaCl), 5 mM sodium hypochlorite (NaOCl) or 50 μM hydrogen peroxide (H2O2) for 60 minutes were compared. Significantly higher asp23 expression levels were observed in biofilms exposed to 50 °C, 5 mM NaOCl or 50 μM H2O2 compared to the corresponding planktonic cells (p < 0.05). Conversely, asp23 expression levels in biofilm and planktonic cells exposed to 0.8 M NaCl were not significantly different (p > 0.05). Further, biofilms exposed to 50 °C, 0.8 M NaCl, 5 mM NaOCl or 50 μM H2O2 exhibited significantly higher catalase activity than the planktonic cells (p < 0.05). These results suggest that activities of σB and catalase may be involved in the tolerance of S. epidermidis biofilm against physico-chemical disinfection.
Introduction
Staphylococcus epidermidis biofilm is the predominant cause of primary bacteremia1 and medical implant device infections2. S. epidermidis biofilm is highly resistant to antibiotics and host immunity1. Hence, treatment of S. epidermidis biofilm-based infections often entails removal and replacement of the infected device, increasing morbidity and cost1. Effective control of S. epidermidis biofilm-associated infections is further hampered by the relatively high tendency of bacteria to develop resistance to antibiotics3. Consequently, the use of effective physico-chemical disinfection procedures to control biofilm-forming bacteria, such as S. epidermidis, is necessary4 in both domestic and healthcare settings, where bacterial biofilms are frequently encountered5,6. Nevertheless, the underlying tolerance mechanisms employed by S. epidermidis biofilm against physico-chemical disinfection remain largely unknown.
Multiple mechanisms have been proposed to account for the relatively high tolerance of biofilms to antimicrobials. These include, reduced diffusion of antimicrobials through the biofilm matrix7, neutralization of the antimicrobials by the biofilm matrix components8, physiological heterogeneity conferred by the three-dimensional biofilm structure9, higher expression of specific protective molecules10, and the presence of a subpopulation of highly resistant cells (persisters)9. However, these mechanisms only partially account for the high tolerance of biofilms against few antibiotics e.g. ciprofloxacin2,7,11, but not tolerance against commonly used physico-chemical disinfectants, such as heat, NaCl, NaOCl or H2O2.
The alternative factor B (σB), a sub-unit of RNA polymerase12, has been implicated in the tolerance of planktonic forms of different bacterial species against several stress agents, including heat13–16, osmotic stress15–18, H2O213,14 or antibiotics19,20 exposure. Although σB is essential in S. epidermidis biofilm formation, maturation and stability21, its role in the tolerance of S. epidermidis biofilm against heat, NaCl, NaOCl or H2O2 exposure has not been determined. The sigma B gene expression is dependent on both a σA and σB promoter22,23. On the contrary, alkaline shock protein 23 (asp23) gene expression is almost exclusively transcribed from σB-dependent promoters, making it a good marker for σB activity in S. epidermidis22,24.
Catalase enzyme has been linked with the survival of planktonic forms of different bacterial species against osmotic stress25,26, H2O226,27 and nitric oxide28 exposure. Despite S. epidermidis being a catalase-producing bacterium29, the role of catalase in the tolerance of S. epidermidis biofilm against heat, NaCl, NaOCl or H2O2-exposure has not been fully investigated.
Therefore, to better understand the mechanisms of tolerance of S. epidermidis biofilm against physico-chemical disinfection, the present study compared the expression of a σB-dependent gene, asp23 and catalase activity of S. epidermidis biofilm and planktonic cells in response to heat, NaCl, NaOCl or H2O2 exposure.
Results
Optimal concentrations of physico-chemical disinfectants for analysis of S. epidermidis biofilm tolerance mechanisms
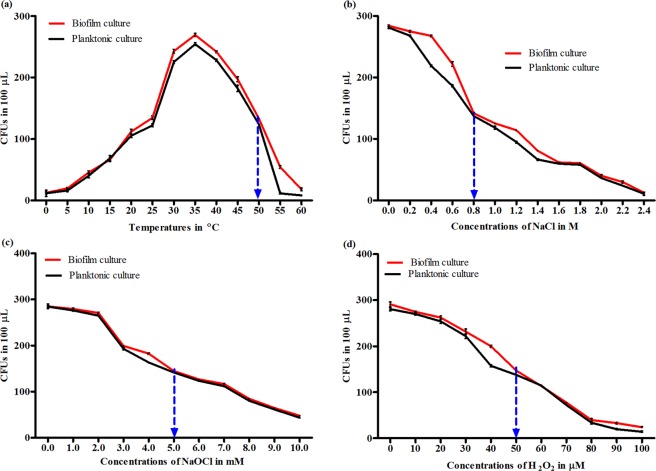
When subjected to increasing temperatures/concentrations of the physico-chemical disinfectants for 60 minutes, the S. epidermidis biofilm and planktonic cells were considerably stressed (growth reduced by almost 2-fold with reference to the highest CFU values) at 50 °C, 0.8 M NaCl, 5 mM NaOCl or 50 μM H2O2 (Fig. 1). Thus, the above temperature/concentrations of the physico-chemical disinfectants were considered optimal for analysis of asp23 expression and catalase activity in S. epidermidis biofilm and planktonic cells.
Figure 1.
The growth of S. epidermidis cells exposed to increasing physico-chemical disinfectant concentrations. The growth curves for a single mixture of S. epidermidis biofilm or planktonic culture subjected to increasing temperatures (a) or concentrations of NaCl (b), NaOCl (c) or H2O2 (d) for 60 minutes. The plots at each physico-chemical disinfectant concentration depict the mean ± standard deviation of three independent experiments with three technical replicates. The blue dotted arrow represents the optimal temperature/concentration at which both S. epidermidis biofilm and planktonic cells were considerably stressed (growth reduced by almost 2-fold with reference to the highest CFU value).
Asp23 expression is upregulated more in heat-exposed biofilm compared to planktonic cells
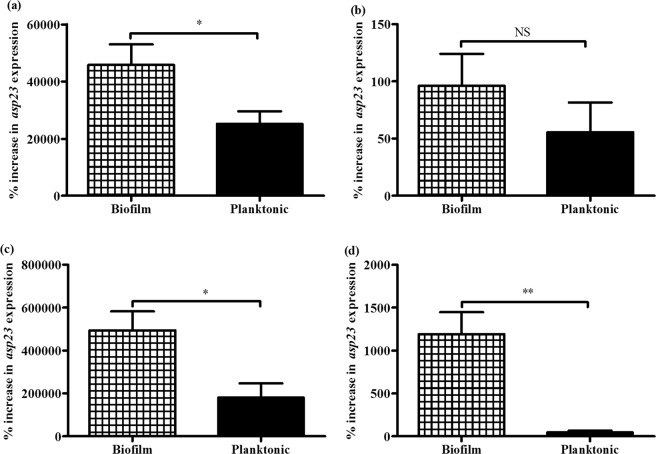
The S. epidermidis biofilm cells exposed to 50 °C temperature for 60 minutes had a significantly higher expression of a σB-dependent gene, asp23 compared to the corresponding planktonic cells (p = 0.0259; Fig. 2a). Further, the S. epidermidis biofilm or planktonic cells exposed to 50 °C temperature for 60 minutes exhibited significantly increased asp23 expressions than their respective cells exposed to 25 °C temperature (controls) (p < 0.0001; Table 2). Taken together, these results indicated that 50 °C-exposure enhances σB activity in both S. epidermidis biofilm and planktonic cells, but with significantly higher activity levels in the biofilm cells.
Figure 2.
Effects of physico-chemical disinfectant-exposure on asp23 expression by S. epidermidis cells. The percent increase in asp23 expression of pairs of S. epidermidis biofilm (n = 10) and planktonic (n = 10) samples exposed to 50 °C (a), 0.8 M NaCl (b), 5 mM NaOCl (c) or 50 μM H2O2 (d) for 60 minutes. For each sample, three repeats of asp23 expression measurements were performed. Bars represent the mean ± SEM. Statistical difference between biofilm and planktonic cells was evaluated using paired t-test (NS, p > 0.05; *p < 0.05; **p < 0.01).
Table 2.
Effect of physico-chemical disinfectant-exposure on asp23 expressions by S. epidermidis cells.
| Type of cell | Mean ± SEM of asp23 expressions in response to physico-chemical disinfectants exposures | |||
|---|---|---|---|---|
| Heat | NaCl | NaOCl | H2O2 | |
| Biofilm (n = 10) | ||||
| Unexposed | 0.0178 ± 0.0021 | 0.0062 ± 0.0012 | 0.0286 ± 0.0051 | 0.0078 ± 0.0010 |
| Exposed | 7.4780 ± 0.9350 | 0.0127 ± 0.0032 | 112.11 ± 16.720 | 0.0869 ± 0.0161 |
| p < 0.0001 | p = 0.0203 | p < 0.0001 | p = 0.0002 | |
| Planktonic (n = 10) | ||||
| Unexposed | 0.0204 ± 0.0017 | 0.0052 ± 0.0011 | 0.0425 ± 0.0085 | 0.0141 ± 0.0019 |
| Exposed | 4.7510 ± 0.6449 | 0.0081 ± 0.0028 | 56.474 ± 16.37 | 0.0220 ± 0.0055 |
| p < 0.0001 | p = 0.1831 | p = 0.0073 | p = 0.0725 | |
The asp23 expression levels of S. epidermidis biofilm and planktonic cells exposed to 50 °C, 0.8 M NaCl, 5 mM NaOCl or 50 μM H2O2 for 60 minutes and the unexposed controls. For each sample, three repeats of asp23 expression measurements were performed. Boldface represent a statistically significant difference between the physico-chemical disinfectant-exposed cells and the unexposed controls as determined by paired-t test (p < 0.05).
NaCl exposure enhances asp23 expression in biofilm cells, but not in planktonic cells
When exposed to 0.8 M NaCl for 60 minutes, the expression levels of asp23 between S. epidermidis biofilm and planktonic cells were not significantly different (p = 0.4029; Fig. 2b). However, ap23 expression by 0.8 M NaCl-treated S. epidermidis biofilm cells was significantly higher compared to that of the untreated controls (p = 0.0203; Table 2). These findings indicated that 0.8 M NaCl exposure specifically enhances σB activity in S. epidermidis biofilm cells, but not in the corresponding planktonic cells.
NaOCl or H2O2 exposure elevates asp23 expression to higher extent in biofilm cells than in planktonic cells
The S. epidermidis biofilm or planktonic cells treated with 5 mM NaOCl for 60 minutes exhibited significantly increased asp23 expressions than their respective unexposed controls (p < 0.05; Table 2). Further analysis showed that asp23 expression was significantly higher in S. epidermidis biofilm cells compared to the corresponding planktonic cells upon exposure to 5 mM NaOCl for 60 minutes (p = 0.0109; Fig. 2c). Moreover, S. epidermidis biofilm cells exhibited significantly elevated asp23 expression levels compared to the corresponding planktonic cells upon exposure to 50 μM H2O2 for 60 minutes (p = 0.0020; Fig. 2d). Although the asp23 expressions by S. epidermidis planktonic cells upon 50 μM H2O2-exposure was not significantly different compared to their respective unexposed controls (p = 0.0725; Table 2), the ap23 expression by 50 μM H2O2-treated S. epidermidis biofilm cells was significantly higher than in the unexposed controls (p = 0.0002; Table 2). Taken together, these results indicated that 5 mM NaOCl and 50 μM H2O2-exposure significantly enhances σB activity in S. epidermidis biofilm cells than in the corresponding planktonic cells.
Heat or NaCl-exposed biofilm cells produce higher amounts of catalase than the planktonic cells
In this study, using a simple visual assay, we quantified catalase activity by measuring the trapped oxygen (O2) gas, which is visualized as foam. The O2 gas generated during the H2O2-catalase reaction in the test tubes and how the measurements were done is shown in Fig. 3.
Figure 3.

Images of the height of foam. The test tubes show the foam formed during H2O2-catalase reaction. Test tubes that formed well spread foam as shown in test tubes labeled 1–4 were measured in cm as illustrated in test tube labeled 3 (readings were taken at the bottom of the lower and upper meniscus). Test tubes that did not form a well spread foam as presented in test tube labeled C were not quantified.
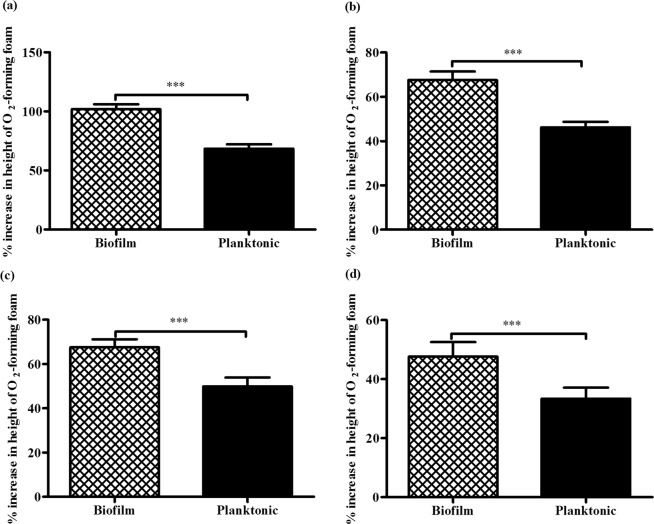
The S. epidermidis biofilm cells exposed to 50 °C for 60 minutes had a significantly higher height of foam compared to the corresponding planktonic cells (p < 0.0001; Fig. 4a). Moreover, a significant increase in height of foam was observed in S. epidermidis biofilm cells exposed to 0.8 M NaCl for 60 minutes compared to the analogous planktonic cells (p < 0.0001; Fig. 4b). Further, S. epidermidis biofilm or planktonic cells exposed to 50 °C or 0.8 M NaCl for 60 minutes exhibited significantly higher heights of foam than their respective unexposed controls (p < 0.0001; Table 3). Taken together, these findings implied that both S. epidermidis biofilm and planktonic cells enhance catalase release in response to 50 °C or 0.8 M NaCl exposure, but with significantly higher release by the biofilm cells.
Figure 4.
Effects of physico-chemical disinfectant-exposure on catalase release by S. epidermidis cells. The percent increase in height of O2-forming foam for pairs of S. epidermidis biofilm (n = 62) and planktonic (n = 62) samples exposed to 50 °C (a), 0.8 M NaCl (b), 5 mM NaOCl (c) or 50 μM H2O2 (d) for 60 minutes. For each sample, three repeats of catalase activity measurements were performed. Bars represent the mean ± SEM. Statistical difference between biofilm and planktonic cells was determined by paired t-test (*p < 0.05; ***p < 0.001).
Table 3.
Impact of physico-chemical disinfectant-exposure on catalase activity in S. epidermidis cells.
| Type of cell | Mean ± SEM of height of O2-forming foam in response to physico-chemical disinfectant exposure | |||
|---|---|---|---|---|
| Heat | NaCl | NaOCl | H2O2 | |
| Biofilm (n = 62) | ||||
| Unexposed | 0.2258 ± 0.0052 | 0.2226 ± 0.0064 | 0.2242 ± 0.0053 | 0.2226 ± 0.0064 |
| Exposed | 0.4479 ± 0.0071 | 0.3640 ± 0.0059 | 0.3672 ± 0.0068 | 0.3151 ± 0.0067 |
| p < 0.0001 | p < 0.0001 | p < 0.0001 | p < 0.0001 | |
| Planktonic (n = 62) | ||||
| Unexposed | 0.2505 ± 0.0056 | 0.2516 ± 0.0057 | 0.2511 ± 0.0057 | 0.2522 ± 0.0061 |
| Exposed | 0.4129 ± 0.0068 | 0.3602 ± 0.0058 | 0.3651 ± 0.0067 | 0.3258 ± 0.0070 |
| p < 0.0001 | p < 0.0001 | p < 0.0001 | p < 0.0001 | |
The height of O2-forming foam of S. epidermidis biofilm and planktonic cells exposed to 50 °C, 0.8 M NaCl, 5 mM NaOCl or 50 μM H2O2 for 60 minutes and the unexposed controls. Statistical differences between physico-chemical disinfectant-exposed cells and the unexposed controls were determined using paired t-test (all the differences were significant; p < 0.05).
NaOCl or H2O2-exposed biofilm cells release more catalase than the planktonic cells
The S. epidermidis biofilm cells subjected to 5 mM NaOCl for 60 minutes had a significantly higher height of foam compared to the corresponding planktonic cells (p < 0.0001; Fig. 4c). Similarly, a significantly higher height of foam was observed in S. epidermidis biofilm cells exposed to 50 μM H2O2 for 60 minutes compared to the planktonic cells (p < 0.0001; Fig. 4d). Further, S. epidermidis biofilm or planktonic cells exposed to 5 mM NaOCl or 50 μM H2O2 for 60 minutes exhibited significantly higher heights of foam than their respective unexposed controls (p < 0.0001; Table 3). Taken together, these results indicated that 5 mM NaOCl or 50 μM H2O2-exposure stimulates catalase release by S. epidermidis biofilm and planktonic cells, with significantly higher release by the biofilm cells.
Discussion
The mechanisms underlying the tolerance of S. epidermidis biofilm against physico-chemical disinfection remain largely unknown. Therefore, in this study, the activities of σB and catalase were evaluated as potential tolerance mechanisms employed by S. epidermidis biofilm against heat, NaCl, NaOCl or H2O2-exposure.
Results presented showed that σB activity is enhanced in both S. epidermidis biofilm and planktonic cells in response to 50 °C-exposure, but with significantly higher activities in the biofilm cells. The present finding is in agreement with previous reports on B. cereus15 and B. subtilis16. However, the previous studies only focused on the planktonic forms of the Bacillus species. Microbial metabolic activities are temperature-dependent30. Considering that σB regulates bacterial metabolism31 and that planktonic cells have higher metabolic activity than biofilm32, it was expected that planktonic cells would have a higher σB activity than the corresponding biofilm cells. However, the observed nearly 2-fold higher σB activity by S. epidermidis biofilm compared to the planktonic cells suggests that the tolerance of biofilm against heat exposure might be more dependent on σB activity.
Further, we showed that 0.8 M NaCl-exposure enhances σB activity in the S. epidermidis biofilm cells, but not in the planktonic cells. Our finding that σB activities between 0.8 M NaCl-treated planktonic cells and their untreated controls were not statistically different contradicts previous reports on different bacterial species14,15,17,18. Considering that NaCl-exposure response regulatory mechanisms in different bacterial species may not follow common patterns33, the discrepancy between the present and previous outcomes could be attributed to the different regulatory patterns in planktonic cells of S. epidermidis and the previously studied bacterial species. Although we did not observe a significant difference in σB activity between NaCl-exposed S. epidermidis planktonic cells and the unexposed controls, some low level of σB activity was detected. Perhaps the low levels of σB activity detected in the planktonic cells could be as a result of the basal σB activity22, and not necessarily as a NaCl-exposure response mechanism. A possible explanation for the observed significantly elevated σB activity levels in biofilm cells, but not in planktonic cells is that tolerance of S. epidermidis biofilm against NaCl-exposure might be more specifically dependent on σB activity.
Further, our data showed that 5 mM NaOCl-exposure upregulates σB activity in both S. epidermidis biofilm and planktonic cells, but with significantly higher activities in the biofilm cells. So far, there are no direct reports on the effect of NaOCl exposure on σB activity. NaOCl-exposure exerts its anti-bacterial effects by targeting several metabolic processes, such as DNA synthesis, adenosine triphosphate synthesis etc.34. The same metabolic processes are regulated by σB activity31. Considering that planktonic cells have higher metabolic activity than the corresponding biofilm32, planktonic cells should have exhibited higher σB activity in response to NaOCl exposure. However, we observed the opposite, suggesting that tolerance of S. epidermidis biofilm against NaOCl exposure is more dependent on the σB activity.
Furthermore, we revealed that 50 μM H2O2 exposure enhances σB activity in S. epidermidis biofilm cells, but not in the planktonic cells. The present findings concur with a previous report, showing that 50 μM H2O2 has a limited effect on σB expression by B. cereus cells15. Moreover, the present finding agrees with a previous report in which 60 μM H2O2 was shown not to affect σB expression in S. aureus cells14. However, the previous reports only focused on the planktonic forms of B. cereus and S. aureus. It is probable that no σB expressions were detected in the 60 μM H2O2-exposed to S. aureus14 because the study employed northern blot analysis, which is insensitive and only detects large gene expressions35. Taking into the findings of the present and previous studies, it appears that σB activity by planktonic cells is largely independent of H2O2-exposure. The low levels of σB activity detactable in planktonic cells in the present and previous studies could be probably be related to the basal σB activity22, but not necessarily as a H2O2-stress response mechanism. A more plausible explanation for the observed significantly higher σB activities in the biofilm cells, but not in planktonic cells is that σB has a more significant role in the S. epidermidis biofilm’s tolerance against H2O2-exposure.
Further results of this study demonstrated that 50 °C-exposure enhances catalase release by S. epidermidis biofilm and planktonic cells, but with higher release by the biofilm cells. The present finding is consistent with two previous reports on a fungus Aspergillus nidulans36 and Rhodobacter sphaeroides37. However, the previous reports only focused on the planktonic forms of the two organisms. Taking into account that temperatures above 37 °C increases the porosity of S. epidermidis biofilm38, a probable explanation for the higher catalase release by the biofilms could be related to the increased porosity of the S. epidermidis biofilm by the 50 °C-exposure thus availing more catalase for quantification. Alternatively, catalase release is a metabolic process, which is expected to be higher in planktonic cells compared to the biofilm32; however, S. epidermidis biofilm exhibited higher catalase activity in response to 50 °C-exposure suggesting a more important role of catalase in biofilm’s tolerance against heat-exposure.
Further, we showed that 0.8 M NaCl exposure stimulates catalase release by both S. epidermidis biofilm and planktonic cells, but with significantly higher release by the biofilm cells. Our observation that 0.8 M NaCl-treated planktonic cells released higher catalase than the untreated controls is in line with previous reports on different bacterial species25,26. Additional analyses demonstrated that 5 mM NaOCl or 50 μM H2O2-exposure enhances catalase release by S. epidermidis biofilm and planktonic cells, but with significantly more production by the biofilm cells. Our observations that NaOCl or H2O2-exposed planktonic cells produce significantly higher catalase than the unexposed controls are in agreement with previous reports on different bacterial species37,39. However, a contradicting observation has been reported for P. aeruginosa exposed to H2O240,41. The differences in the exposure durations prior to catalase measurements could be responsible for the observed discrepancy between the results of the present and the two previous studies. From the present findings, it appears that catalase activity is involved in both S. epidermidis biofilm and planktonic cells’ tolerance against NaCl, NaOCl or H2O2-exposure. However, considering that catalase production is a metabolic activity, we expected planktonic cells to show significantly higher catalase activity than the biofilm32 per unit viable cells. We, instead, observed the opposite, suggesting that catalase might be involved to a greater extent in the tolerance of S. epidermidis biofilm against NaCl, NaOCl or H2O2-exposure.
In conclusion, we observed that 50 °C, 0.8 M NaCl, 5 mM NaOCl or 50 μM H2O2-exposed S. epidermidis biofilm cells significantly enhance σB and catalase activities more than the corresponding planktonic cells, suggesting that both σB and catalase activities might be having a greater contribution in the tolerance of S. epidermidis biofilm against the physico-chemical disinfectants than in the planktonic cells. Therefore, σB and /or catalase could be explored further as promising targets for the development of more potent anti-staphylococcal biofilm eradication approaches. Nevertheless, further studies incorporating σB and catalase mutants or promoter reporters are required to reach a more definite conclusion regarding the dependence of S. epidermidis biofilm on the activities of σB and /or catalase enzyme for survival against the physico-chemical disinfectants.
Methods
Sample collection, bacterial isolates and growth conditions
Skin swab samples were collected from outpatients at Kisumu County Referral Hospital (KCRH), Kenya in accordance with relevant guidelines and regulations and research approved by the Maseno University Ethics Review Committee (Reference number: MSU/DRPI/MUERC/000187/15). Further, permission to recruit outpatients was granted by the KCRH. Written informed consent was obtained from participants before recruitment. Collection of the skin swabs and S. epidermidis isolation on mannitol salt agar (HiMedia Laboratories Pvt. Limited, Nashik, India) were conducted as described elsewhere42. The S. epidermidis isolates were further identified using routine microbiologic methods namely, Gram staining, catalase, coagulase and novobiocin sensitivity tests and grown on tryptic soy agar (TSA; HiMedia Laboratories Pvt. Limited, Mumbai, India) at 37 °C overnight. Staphylococcus aureus American Type Culture Collection (ATCC) 29213, a good former of mature biofilm within 24 hours43, was used as a reference control strain. In this study, analogous planktonic cells served as control samples.
Biofilm was formed as previously described44 with few modifications. Briefly, a single S. epidermidis colony, from a TSA plate, was inoculated into 2 mL of tryptic soy broth (TSB; HiMedia Laboratories Pvt. Limited, Mumbai, India) and incubated at 37 °C with shaking at 120 revolutions per min (rpm) overnight. To form biofilm culture, 100 µL of the overnight culture adjusted to ~1 × 109 colony-forming units (CFU)/mL was simultaneously inoculated into two polystyrene tubes containing 10 mL of fresh TSB supplemented with 1% glucose (Unilab Limited, Nairobi, Kenya), to enhance biofilm formation, and incubated at 37 °C with shaking at 120 rpm for 24 hours. Then, the spent medium in one of the tubes was removed and the biofilm was rinsed twice with 200 μL of 0.9% NaCl. Biofilm formation on the tube was verified by a qualitative tube method biofilm assay with 0.1% crystal violet staining as described in details elsewhere45. In case of strong biofilm formation in the first tube, the spent medium in the parallel second tube was carefully removed, and the biofilm was washed twice with 200 μL of 0.9% NaCl. Then, 1 mL of 0.9% NaCl was added to the tube and vortexed for 2 minutes to detach the biofilm cells (vortexing detaches S. epidermidis biofilm from surfaces46,47). The detached biofilm cell suspension was centrifuged at 10,000 rpm, 4 °C for 10 minutes. To grow planktonic cells, 100 µL of the overnight culture adjusted to ~1 × 109 CFU/mL was inoculated into 10 mL of fresh TSB in a polystyrene tube and incubated at 37 °C with shaking at 120 rpm for 18 hours. Then, the bacterial cells in suspension were centrifuged at 10,000 rpm, 4 °C for 10 minutes. The biofilm or planktonic cell pellets were suspended in 0.9% NaCl and adjusted to ~1 × 109 CFU/mL.
Optimal concentrations of physico-chemical disinfectants for analysis of the tolerance mechanisms
To minimize variability between samples, the optimal temperature/concentration of each of the physico-chemical disinfectants was determined using a single mixture of S. epidermidis biofilm or planktonic culture as previously described42 with some modifications. Briefly, 150 μL of ~1 × 109 CFU/mL of a single mixture of S. epidermidis biofilm or planktonic culture (prepared by mixing equivalent amount i.e. 150 μL of ~1 × 109 CFU/mL of S. epidermidis biofilm or planktonic culture drawn from six random biofilm samples or corresponding planktonic samples) was inoculated into 1.5 mL of increasing concentrations of NaCl, NaOCl (Supersleek, Nairobi, Kenya) or H2O2 (RFCL Limited, New Delhi, India) and exposed for 60 minutes. For heat exposure, tubes containing 1.5 mL of sterile distilled water were inoculated with 150 μL of ~1 × 109 CFU/mL of the single mixture of S. epidermidis biofilm or planktonic culture and exposed to increasing temperatures in a water bath for 60 minutes. The NaOCl and H2O2-exposed cultures were neutralized by 200 μL of 0.1% sodium thiosulphate. Whereas, NaCl and heat-exposed cultures were neutralized by 200 μL of sterile distilled water at 4 °C. To collect biofilm cells, the bacteria in suspension were discarded and the biofilm was gently rinsed once with 200 μL of 0.9% NaCl. One mL of 0.9% NaCl was added to the biofilm, vortexed for 2 minutes then centrifuged at 9,000 rpm for 8 minutes. Planktonic cells were collected by centrifuging the bacteria in suspension at 9,000 rpm for 8 minutes. The biofilm or planktonic cell pellets were suspended in 1 mL of sterile distilled water and CFUs enumerated on TSA as described elsewhere42. At each temperature/concentration of the physico-chemical disinfectant, three experiments were performed with three technical replicates. In this study, 50 °C, 0.8 M NaCl, 5 mM NaOCl or 50 μM H2O2 induced considerable stress to the biofilm and planktonic cells hence, used in the subsequent procedure.
Exposure of biofilm and planktonic cells to optimal physico-chemical disinfectants
S. epidermidis biofilm (n = 62) and planktonic (n = 62) samples were exposed to the optimal temperature/concentration of the disinfectants as previously described48 with some modifications. Briefly, 200 μL of ~1 × 109 CFU/mL of biofilm or planktonic suspension were inoculated into 1100 μL of TSB adjusted to 0.8 M NaCl, 5 mM NaOCl or 50 μM H2O2, vortexed for 2 minutes and incubated at 37 °C with shaking at 80 rpm for 60 minutes. For heat exposure, 200 μL of biofilm or planktonic suspension adjusted to ~1 × 109 CFU/mL were inoculated into 1100 μL of TSB alone and transferred to a water bath at 50 °C for 60 minutes. The effects of the disinfectants were neutralized as in the preceding section. Untreated controls were set up by inoculating 200 μL of biofilm or planktonic suspension adjusted to ~1 × 109 CFU/mL into 1100 μL of TSB alone and incubated at 37 °C for 60 minutes. A similar set up incubated at 25 °C for 60 minutes served as a control for heat exposure. After incubation, the biofilm or planktonic cells were collected as described in the preceding section. For gene expression analysis, the biofilm or planktonic cells were suspended in 0.9% NaCl, adjusted to ~1 × 109 CFU/mL and immediately transferred into an equal volume of a 1:1 mixture of ice-cold acetone and ethanol, then kept at −80 °C for at least 20 minutes. For catalase activity analysis, viable biofilm and planktonic cells were enumerated in duplicate on TSA as described elsewhere46 and adjusted to 1 × 107 cells/mL.
Quantification of asp23 expression
Total RNA was isolated from the disinfectant-exposed S. epidermidis biofilm (n = 10) and planktonic (n = 10) samples and the unexposed controls as previously described49. Genomic DNA was degraded using DNase I (New England Biolabs, Ipswich, England) following the manufacturer’s instructions. The purity and concentration of RNA were determined as previously described50. RNA samples with an OD280/OD260 ratio between 1.8 and 2.2 were used for complementary DNA (cDNA) synthesis. Two μg of RNA was reverse transcribed into cDNA using ProtoScript® First Strand cDNA Synthesis kit (New England Biolabs, Ipswich, England) following the manufacturer’s instructions. Quantitative-PCR (qPCR) was performed on a Rotor-Gene Q real-time thermal cycler (Qiagen, Hilden, Germany) as previously described51 with few modifications. Briefly, each 20 μL of qPCR reaction mixture contained 10 μL of Lunar Universal qPCR mastermix (New England Biolabs, Ipswich, England), 0.5 μL each of the forward and reverse primers (Inqaba Biotechnical, Pretoria, South Africa) specific for 16S rRNA (reference) and asp23 (target) genes (Table 1), 7 μL of nuclease-free water and 2 μL of cDNA template. The thermal cycling conditions were as follows: 1 minute at 95 °C, followed by 45 cycles of 15 seconds at 95 °C, 30 seconds at 60 °C. A control lacking the M-MuLV enzyme mix was included in each reaction. The asp23 expression level was determined using the EfficiencyΔCt method, where, ΔCt = Ct (reference gene)−Ct (target gene)50 for the disinfectant-exposed samples and their unexposed controls. All primers had an efficiency of ~100%; hence, the real efficiency i.e. 2 was substituted in the EfficiencyΔCt formula. For each isolated RNA, three repeats of asp23 expression measurements were performed. For each sample, asp23 expression level was expressed as percentage change by 100 × [{asp23 expression(exposed cells) − asp23 expression(unexposed control)}/asp23 expression(unexposed control)].
Table 1.
Primers used for qPCR amplification.
| Target gene | Set | Sequence (5′–3′) | Reference |
|---|---|---|---|
| S. epidermidis | |||
| asp23 | Forward | CAGCAGCTTGTTTTTCTCCA | 21 |
| Reverse | CATGAAAGGTGGCTTCACAG | ||
| 16S rRNA | Forward | GGGCTACACACGTGCTACAA | 44 |
| Reverse | GTACAAGACCCGGGAACGTA | ||
| S. aureus ATCC 29213 | |||
| asp23 | Forward | TCGCTGCACGTGAAGTTAAA | 53 |
| Reverse | CAGCAGCTTGTTTTTCACCA | ||
| 16S rRNA | Forward | GTAGGTGGCAAGCGTTATCC | 54 |
| Reverse | CGCACATCAGCGTCAG | ||
Quantification of catalase activity
Catalase activities of the disinfectant-exposed S. epidermidis biofilm (n = 62) and planktonic (n = 62) samples and the unexposed controls were determined by a simple visual assay as previously described52 with few modifications. Briefly, 100 μL of ~1 × 107 of viable biofilm or planktonic cells/mL was transferred to a Pyrex test tube (1.3 centimetre (cm) diameter by 10 cm height). Then, 100 μL each of 1% Triton X-100 (Rohm and Haas, Philadelphia, USA) and 30% H2O2 were added, mixed thoroughly and incubated at 25 °C. Once reaction stopped, the height of foam that remained constant for 15 minutes was measured in cm using a ruler. For each sample, three repeats of catalase activity measurements were performed. For each sample, height of foam was expressed as percentage change by 100 × [{height of foam(exposed cells) − height of foam(unexposed control)} ⁄ height of foam(unexposed control)].
Statistical analysis
Statistical analysis was performed with GraphPad Prism version 5.03. Data normality was verified by D’Agostino and Pearson omnibus test. Differences between groups were determined using two-tailed paired t-test. Data are presented as the mean ± standard error of mean (SEM). A p-value less than 0.05 was considered significant.
Supplementary information
Acknowledgements
We thank the management of the KCRH for facilitating sample collection. We also thank Godfrey Sande of Kitale County Referral Hospital PCR laboratory for technical assistance with qPCR experiments. Finally, we thank Prof. Samuel Kariuki of Centre for Microbiology Research, Kenya Medical Research Institute for donating the ATCC reference strain. This study was supported by a grant from the National Commission for Science, Technology and Innovation, Kenya. The funder had no role in conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript.
Author Contributions
C.O.O., P.O.A. and D.O.O. designed the study. C.O.O. conducted the experiments and analyzed the data. C.O.O., P.O.A. and D.O.O. interpreted the results, discussed the analyses, interpretation and presentation. C.O.O., P.O.A. and D.O.O. critically read and contributed to the final version of the manuscript. All authors read and approved the final manuscript.
Data Availability
All data generated or analyzed during this study are included within this article and its Supplementary Information files.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41797-8.
References
- 1.Fey P, Oslon M. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 2010;5:917–933. doi: 10.2217/fmb.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Otto M. Molecular basis of Staphylococcus epidermidis infections. Semin Immunopathol. 2012;34:201–214. doi: 10.1007/s00281-011-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammer A, Carson F, Rileya V. Effects of Melaleuca alternifolia (tea tree) essential oil and the major monoterpene component terpinen-4-ol on the development of single- and multistep antibiotic resistance and antimicrobial susceptibility. Antimicrob Agents Chemother. 2012;56:909–915. doi: 10.1128/AAC.05741-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peeters E, Nelis J, Coenye T. Evaluation of the efficacy of disinfection procedures against Burkholderia cenocepacia biofilms. J Hosp Infect. 2008;70:361–8. doi: 10.1016/j.jhin.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Francolini I, Donelli G. Prevention and control of biofilm-based medical-device-related infections. FEMS Immun. Med Microbiol. 2010;59:227–238. doi: 10.1111/j.1574-695X.2010.00665.x. [DOI] [PubMed] [Google Scholar]
- 6.Garrett T, Bhakoo M, Zhang Z. Bacterial adhesion and biofilms on surfaces. Prog Nat Sci. 2008;18:1049–1056. doi: 10.1016/j.pnsc.2008.04.001. [DOI] [Google Scholar]
- 7.Joo H, Otto M. Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem Biol. 2012;19:1503–13. doi: 10.1016/j.chembiol.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fux A, Costerton W, Stewart P, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Urish L, et al. Antibiotic-tolerant Staphylococcus aureus biofilm persists on arthroplasty materials. Clin Orthop Relat Res. 2016;474:154–156. doi: 10.1007/s11999-016-4720-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao Y, Sturdevant E, Otto M. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J Infect Dis. 2005;191:289–298. doi: 10.1086/426945. [DOI] [PubMed] [Google Scholar]
- 11.Hall C, Mah T. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 12.Paharik E, Horswill R. The staphyloccal biofilm: adhesins, regulation and host response. Microbiol Spectr. 2016;4:1–48. doi: 10.1128/microbiolspec.VMBF-0022-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cebráin G, et al. Role of the alternative sigma factor B on Staphylococcus aureus resistance to stresses of relevance to food preservation. J Appl Microbiol. 2009;107:187–196. doi: 10.1111/j.1365-2672.2009.04194.x. [DOI] [PubMed] [Google Scholar]
- 14.Chan P, Foster S, Ingham E, Clements M. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J Bacteriol. 1998;180:6082–6089. doi: 10.1128/jb.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaik W, Tempelaars M, Wouters J, de Vos W, Abee T. The alternative sigma factor σB of Bacillus cereus: response to stress and role in heat adaptation. J Bacteriol. 2004;186:316–325. doi: 10.1128/JB.186.2.316-325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voelker U, Maul B, Hecker M. Expression of the sigma B dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J Bacteriol. 1999;181:3942–3948. doi: 10.1128/jb.181.13.3942-3948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abram F, et al. Identification of components of the sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl Environ Microbiol. 2008;74:6848–6858. doi: 10.1128/AEM.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker L, Çetin M, Hutkins R, Benson A. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J Bacteriol. 1998;180:4547–4554. doi: 10.1128/jb.180.17.4547-4554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, et al. Vancomycin activates σB in vancomycin-resistant Staphylococcus aureus resulting in the enhancement of cytotoxicity. PLoS ONE. 2011;6:1–8. doi: 10.1371/journal.pone.0024472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poole K. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother. 2012;67:2069–2089. doi: 10.1093/jac/dks196. [DOI] [PubMed] [Google Scholar]
- 21.Pintens V, et al. The role of σB in persistence of Staphylococcus epidermidis foreign body infection. Microbiology. 2008;154:2827–2836. doi: 10.1099/mic.0.2007/015768-0. [DOI] [PubMed] [Google Scholar]
- 22.Knobloch J, Jäger S, Horstkotte M, Rohde H, Mack D. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor B by repression of the negative regulator gene icaR. Infect Immun. 2004;72:3838–3848. doi: 10.1128/IAI.72.7.3838-3848.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knobloch J, Sabottke A, Rohde H, Feucht H, Mack D. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J Bacteriol. 2001;183:2624–2633. doi: 10.1128/JB.183.8.2624-2633.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotter J, O’Gara J, Mack D, Casey E. Oxygen-mediated regulation of biofilm development is controlled by the alternative sigma factor σB in Staphylococcus epidermidis. Appl Environ Microbiol. 2009;75:261–264. doi: 10.1128/AEM.00261-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho Y, Lee J, Roe J. A developmentally regulated catalase required for proper differentiation and osmoprotection of Streptomyces coelicolor. Mol Microbiol. 2000;35:150–160. doi: 10.1046/j.1365-2958.2000.01685.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Heo Y, Lee K, Cho Y. KatA, the major catalase, is critical for osmoprotection and virulence in Pseudomonas aeruginosa PA14. Infect Immun. 2005;73:4399–4403. doi: 10.1128/IAI.73.7.4399-4403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu W, et al. Comparative analysis of the responses of related pathogenic and environmental bacteria to oxidative stress. Microbiology. 2012;158:636–647. doi: 10.1099/mic.0.057000-0. [DOI] [PubMed] [Google Scholar]
- 28.Su S, et al. Catalase (katA) plays a role in protection against anaerobic nitric oxide in Pseudomonas aeruginosa. PLoS ONE. 2014;9:1–15. doi: 10.1371/journal.pone.0091813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks, G., Carroll, K., Butel, J. & Morse, S. Jawetz, Melnick, & Adelberg’s Medical Microbiology. (The McGraw-Hill Companies, Inc 2007).
- 30.Cowan, K. Microbiology: a systems approach. (The McGraw-Hill Companies, Inc 2012).
- 31.O’Byrne P, Karatzas A. The role of sigma B (sigma B) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Adv Appl Microbiol. 2008;65:115–140. doi: 10.1016/S0065-2164(08)00605-9. [DOI] [PubMed] [Google Scholar]
- 32.Gaio V, Lima C, Oliveira F, França Â, Cerca N. Carvacrol is highly disruptive against coagulase-negative staphylococci in in vitro biofilms. Future Microbiol. 2017;12:1487–1496. doi: 10.2217/fmb-2017-0122. [DOI] [PubMed] [Google Scholar]
- 33.Sihto H, Tasara T, Stephan R, Johler S. Temporal expression of the staphylococcal enterotoxin D gene under NaCl stress conditions. FEMS Microbiol Lett. 2015;362:1–7. doi: 10.1093/femsle/fnv024. [DOI] [PubMed] [Google Scholar]
- 34.Rutala, W., Weber, D. & HICPAC. Guideline for disinfection and sterilization in healthcare facilities, 2008 (2008).
- 35.Perdew, G., Vanden, J. & Peters, J. Regulation of gene expression. (Humana press, 2007).
- 36.Noventa-Jordão M, et al. Catalase activity is necessary for heat-shock recovery in Aspergillus nidulans germlings. Microbiology. 1999;145:3229–3234. doi: 10.1099/00221287-145-11-3229. [DOI] [PubMed] [Google Scholar]
- 37.Terzenbach D, Blaut M. Purification and characterization of a catalase from the nonsulfur phototrophic bacterium Rhodobacter sphaeroides ATH 2.4.1 and its role in the oxidative stress response. Arch Microbiol. 1998;169:503–508. doi: 10.1007/s002030050603. [DOI] [PubMed] [Google Scholar]
- 38.Stewart J, et al. Effect of antimicrobial and physical treatments on growth of multispecies staphylococcal biofilms. Appl Environ Microbiol. 2017;83:1–14. doi: 10.1128/AEM.03483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Small A, Chang W, Toghrol F, Bentley E. Toxicogenomic analysis of sodium hypochlorite antimicrobial mechanisms in Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2007;74:176–185. doi: 10.1007/s00253-006-0644-7. [DOI] [PubMed] [Google Scholar]
- 40.Hassett D, et al. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol. 1999;34:1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 41.Panmanee W, Hassett D. Differential role of OxyR controlled antioxidant enzymes alkyl hydroperoxide reductase (ahpCF) and catalase (katB) in the protection of Pseudomonas aeruginosa against hydrogen peroxide in biofilm vs. planktonic culture. FEMS Microbiol Lett. 2009;295:238–244. doi: 10.1111/j.1574-6968.2009.01605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olwal C, Ang’ienda P, Onyango D, Ochiel D. Susceptibility patterns and the role of extracellular DNA in Staphylococcus epidermidis biofilm resistance to physico-chemical stress exposure. BMC Microbiol. 2018;18:1–13. doi: 10.1186/s12866-018-1183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coraça-Huber D, Fille M, Hausdorfer J, Pfaller K, Nogler M. Staphylococcus aureus biofilm formation and antibiotic susceptibility tests on polystyrene and metal surfaces. J Appl Microbiol. 2012;112:1235–1243. doi: 10.1111/j.1365-2672.2012.05288.x. [DOI] [PubMed] [Google Scholar]
- 44.França Â, Freitas A, Henriques A, Cerca N. Optimizing a qPCR gene expression quantification assay for Staphylococcus epidermidis biofilms: a comparison between commercial kits and a customized protocol. PLOS ONE. 2012;7:1–9. doi: 10.1371/journal.pone.0037480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Divya G, Vyshnavi R. Incidence of biofilm producing Staphylococcus epidermidis isolated from clinical samples and skin of healthy individuals: a correlative study. International Journal of Multidisciplinary Research and Development. 2015;2:16–19. [Google Scholar]
- 46.Freitas A, Vaconcelos C, Vilanova M, Cerca N. Optimization of an automatic counting system for the quantification of Staphylococcus epidermidis cells in biofilms. J Basic Microbiol. 2014;54:750–757. doi: 10.1002/jobm.201200603. [DOI] [PubMed] [Google Scholar]
- 47.Oslon E, Garvin L, Fey D, Rupp E. Adherence of Staphylococcus epidermidis to biomaterials is augmented by PIA. Clin Orthop Relat Res. 2006;451:21–24. doi: 10.1097/01.blo.0000229320.45416.0c. [DOI] [PubMed] [Google Scholar]
- 48.Linnes J, Hongyan M, Bryers J. Giant extracellular matrix binding protein expression in Staphylococcus epidermidis is regulated by biofilm formation and osmotic pressure. Curr Microbiol. 2013;66:627–633. doi: 10.1007/s00284-013-0316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atshan S, et al. Improved method for the isolation of RNA from bacteria refractory to disruption, including S. aureus producing biofilm. Gene. 2012;494:219–224. doi: 10.1016/j.gene.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 50.França Â, Melo L, Cerca N. Comparison of RNA extraction methods from biofilm samples of Staphylococcus epidermidis. BMC Research Notes. 2011;4:1–5. doi: 10.1186/1756-0500-4-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gomes F, Teixeira P, Cerca N, Ceri H, Oliveira R. Virulence gene expression by Staphylococcus epidermidis biofilm cells exposed to antibiotics. Microbial Drug Resistance. 2011;17:191–196. doi: 10.1089/mdr.2010.0149. [DOI] [PubMed] [Google Scholar]
- 52.Iwase T, et al. A simple assay for measuring catalase activity: a visual approach. Sci Rep. 2013;3:1–4. doi: 10.1038/srep03081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell G, et al. Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2- heptylquinoline-N-oxide. BMC Microbiol. 2010;3:1–15. doi: 10.1186/1471-2180-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karmakar A, Dua P, Ghosh C. Biochemical and molecular analysis of Staphylococcus aureus clinical isolates from hospitalized patients. Canadian Journal of Infectious Diseases and Medical Microbiology. 2016;2016:1–7. doi: 10.1155/2016/9041636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included within this article and its Supplementary Information files.