Abstract
Polythene is the most widely used plastic around the globe. Among the total plastic waste generated, polythene contributes the maximum share (64%). Various strategies/methods are being utilized to deal with the increasing rate of plastic waste, but among all the methods, bioremediation is regarded as the ecofriendly and widely accepted method. In the current investigation, we have attempted to discover the elite polythene deteriorating fungi (isolated from the rhizosphere soil of Avicennia marina). From 12 different eco-geographical locations along the West Coast of India, total 109 fungal isolates were recorded. The polythene deteriorating fungi were screened at varied pH (3.5, 7 and 9.5) based on changes in weight and tensile strength of the treated polythene at ambient temperature with continuous shaking for 60 days. BAYF5 isolate (pH 7) results in maximum reduction in weight (58.51 ± 8.14) whereas PNPF15 (pH 3.5) recorded highest reduction in tensile strength (94.44 ± 2.40). Surprisingly, we have also reported weight gain, with highest percent weight gain (28.41 ± 6.99) with MANGF13 at pH 9.5. To test the reproducibility of the results, the elite polythene degrading fungal isolates based on weight loss and reduction in tensile strength were only used for repetition experiment and the results based on the reduction in tensile strength were found only reproducible. Polythene biodegradation was further confirmed using Scanning Electron Microscopy (SEM) and Fourier Transform Infrared Spectroscopy (FTIR) analysis. The most efficient polythene deteriorating fungal isolates were identified as Aspergillus terreus strain MANGF1/WL and Aspergillus sydowii strain PNPF15/TS using both morphological keys and molecular tools.
Introduction
The word plastic is originated from the root word ‘plastikos’ (‘grow’ or ‘form’: able to be molded into different shapes) of the Greek language1. It is a polymer, made up of high molecular weight (petrochemicals), long chain of hydrocarbons2. Plastic in various forms tender services in our day-to-day life from our kitchen to industry level3–5 and thus increase its demand around the globe. The production of plastic is doubled annually, and was estimated as 250 million tons in 20085. As per the report, the highest plastic consumer in the world is Asia (35%) followed by North America (26%), Western Europe (23%), Japan (6%) and India (5%)5. Due to the various beneficial properties of the plastic viz. stability, durability (mechanical and thermal property), the utilization of the plastic is at its peak and its demand is continuously increasing6–8. The synthetic plastic is non-biodegradable9 and/or having very slow or least rate of degradation, e.g. polythene needs about 1000 years to degrade under natural environment5,10. Mueller11 reveled that micro-organisms are unable to degrade plastic due to their short term presence in the environment and therefore during evolution microorganisms failed to design elite enzymes to degrade plastic completely. Due to slow degradation rate and increased utilization of plastic5,12, annually 25 million tons plastic waste gets accumulated in the environment13–15. Among the total accumulated plastic waste, polythene (PE) alone contributes about 64%16 and is considered as most problematic17,18. At dumping sites, terrestrial animals usually consume discarded plastic bags along with foodstuff and experiences severe health issues, which finally lead to their death12,19. All types of plastic wastes, finally enters into the marine environment through various routes and represents the maximum share (60–80%) of the marine waste by mass20. In the oceans, polythene waste emerged as a potential threat to the marine animals, leads to hamper their digestive tract and results in death of millions of marine animals8,21–23.
To minimize the production of plastic waste, different guidelines were adopted by various commissions and pollution control boards across the world. In India, Central Pollution Control Board working under the Ministry of Environment and Forests banned the production, dumping and marketing of the virgin/recycled carry bags with less than 20 micron thick24. Similarly Govt. of Maharashtra also banned the manufacturing and usage of the carrier bags below the thickness range of 50 micron25. Despite the ban imposed, various small grocery shops, fruit and vegetable stalls still uses these thin single use polythene bags of 20-micron thick illegally. Plastic wastes which forms an estimated quantity of 5–10% of total municipal solid waste, is being generated at the rate of about 1.2 lakh tons per day (TPD), of which 6000 TPD is plastic wastes26. The lack of provision for the proper disposal of post-consumer plastic wastes, results in littering on road which often chokes open drainage systems, and leads to flood like condition during rainy seasons. At dumping sites, it mixes with the soil and release of the toxic compounds makes fertile land, infertile. So in order to tackle the menace of 20 micron thick plastic, it is necessary to find out way for its degradation. Despite being imposing restrictions on the usage of the plastic, still the plastic waste is generating at an alarming rate, so the disposal of the plastic waste emerged as a major challenge to deal with, throughout the world. Since, the discovery of the polythene, people tried to dispose polythene (plastic) waste using various strategies viz. landfilling (65%)27–30, incineration (25%)27,31,32, recycling (10%)27,31,33, producing biodegradable plastic34–36, construction of roads37–39, production of fuel40–42, degradation3,12,43, and biodegradation44. Each of the method is having, either deteriorating effects on the environment or economic exploitation and among all the methods, biodegradation is considered as the most accepted and ecofriendly method3,12. The degradation of the synthetic plastic mediated by the microbes is known as biodegradation15,45,46. Biodegradation of natural and synthetic plastics is carried out by microbes like Bacteria, Fungi and Actinomycetes47 under optimal growth conditions of the respective microbes in soil10. Rate of biodegradation is directly proportional to the molecular weight of the plastic targeted8,47, and can be enhanced by various factors viz. abiotic hydrolysis, photo–oxidation, and physical disintegration10. Various microorganism are reported to produce some special enzymes viz. intracellular and extracellular, which enabled the microbes to disintegrate the polymer into several monomers and dimers, which are being used by the microbes as a carbon source48–50 and results in the conversion of the polythene waste into water, CO2 or methane3. Efficiency of biodegradation can be increased by making polythene susceptible for microbial attack by using starch and pro-oxidant as additives of the plastic51. By the addition of starch during the preparation of polythene, hydrophilic nature of the polythene gets improved and enables some microbes to get attached on the surface of the polythene and results in de-polymerization with ease due to release of amylase enzyme27. As per Muthukumar and Veerappapilli8 growth of fungi can penetrate into the polymer and leads to its degradation. In past, polythene deteriorating fungi were reported from plastic waste dumping sites52, mangrove rhizosphere soil4,53 and marine water45.
So, in the current investigation, efforts were made to select those plastic waste dumping sites with growing mangroves surrounded by marine water along the West Coast of India for collection of the rhizosphere soil of Avicennia marina (Forsk.) Vierh., to isolate, screen, and characterize the potential polythene degrading fungi.
Results
Collection of the soil samples
The rhizosphere soil samples of Avicennia marina (Forsk.) Vierh. (Fig. 1) were collected from 12 different eco-geographical locations along the West Coast of India (Supplementary Fig. S1). The longitude, latitude and altitude (from mean sea level) of each locality was also recorded (Supplementary Table S1).
Figure 1.
Representative collection sites of A. marina along the West Coast of India.
Isolation of Fungi
Total 109 fungal isolates were recorded from the collected rhizosphere soil. Sabouraud Dextrose Agar medium was found to be the best for the cultivation of the fungi (Supplementary Table S1). Maximum (31) fungal isolates were recorded from Mangalore locality whereas minimum (4) fungal isolates were reported in Surat, Mirya Bandar and Pudponnani.
Investigation of polythene (PE) biodegradation using the fungal isolates
The potential polythene degrading fungi were screened on the basis of reduction in weight (percent weight loss: %WL) and tensile strength (percent loss in Tensile strength: % loss in TS) of the polythene after 60 days of incubation at ambient temperature with continuous shaking.
Assessment of the PE deteriorating fungi based on percent reduction in weight
Screening of the PE degrading fungi at pH 3.5
After 60 days of incubation with continuous shaking at ambient temperature, among 109 fungal isolates, maximum percent reduction in weight or percent loss in weight (% WL) of the pretreated PE strips was recorded with BAYF6 (23.31 ± 1.88) at pH3.5 whereas least %WL was recorded with VASF8 (0.98 ± 0.02) (Supplementary Fig. S2). In addition to % WL, we also recorded weight gain with various isolates. Maximum percent weight gain (%WG) of the polythene was recorded with PODPF2 (13.37 ± 4.72).
Screening of PE degrading fungi at pH 7
Among the 109 fungal isolates maximum % WL was recorded with MANGF1 (58.51 ± 8.14) followed by ERNF1 with 37.94 ± 3.06%WL whereas minimum % WL (1.38 ± 0.54) was recorded with SURF3 at pH 7 (Supplementary Fig. S3) after 60 days of continuous shaking at ambient temperature. Similar to pH 3.5 percent weight gain (%WG) was also recorded at pH 7. Maximum weight gain (7.43 ± 1.98) was recorded with OLDGF2.
Screening of PE degrading fungi at pH 9.5
Maximum % WL (41.82 ± 5.47) was recorded with MANGF1, among the total 109 fungal isolates after 2 months of regular shaking at room temperature at pH 9.5 (Supplementary Fig. S4). With various isolates no loss or gain of the percent weight was recorded. Least % WL (0.64 ± 0.22) was recorded with SURF3. Similar to pH 3.5 and pH7, weight gain was also reported at pH 9.5. At pH 9.5 highest percent weight gain (28.41 ± 6.99) was recorded with MANGF13 compared to pH 3.5 and pH 7.
Among the 109 fungi, maximum %WL (58.51 ± 8.14) was recorded at pH7 with BAYF5 (Supplementary Table S2) followed by MANGF1 (41.82 ± 5.47) at pH 9.5.
Reproducibility of the results based on %WL
Reproducibility of the data is the most important component of a successful experiment. To validate the replicability the results, three most efficient polythene degrading fungi were used for repetition experiment. It was noticed that the results based on the repetition experiment were different than the previous results. In screening BAYF5 leads maximum % WL at pH 7 but in repetition experiment we observed maximum reduction in weight (≈50%WL) with MANGF1 (Supplementary Fig. S5).
Assessment of the PE deteriorating fungi based on reduction in tensile strength (TS)
Screening of PE degrading fungi at pH 3.5
Among the 109 fungi, maximum percent reduction in TS or % loss in TS (94.44 ± 2.41) was reported with PNPF15 at pH 3.5 (Supplementary Fig. 6) and the least % loss in TS (2.5 ± 0.42) was reported with the isolate BAYF8 respectively.
Screening of PE degrading fungi at pH 7
Based on % loss in TS, the most efficient polythene degrading fungal isolate at pH 7 was VASF1 with potential of 76.04 ± 5.21% loss in TS in 60 days of incubation period (Supplementary Fig. S7). Fungal isolate JAMNF5 was found to have least activity and leads only 1.67 ± 0.42% loss in TS in the same period.
Screening of PE degrading fungi at pH 9.5
At pH 9.5 the fungal isolate VASF6 recorded 62.50 ± 4.17% loss in TS of polythene which is the maximum among the total fungi (Supplementary Fig. S8) whereas the least % loss in TS (1.25 ± 0.42) was documented with BAYF6 at the same pH after 60 days of incubation period.
Among the 109 fungal isolates at three different pH, the top 5 elite potential polythene deteriorating fungi based % loss in TS are enlisted in Supplementary Table S3.
Reproducibility of the results based on percent reduction in TS
During the repetition experiment the results were found similar to that of screening (Supplementary Fig. 9) and proved reproducible compared to the %WL after 60 days of continuous shaking at ambient temperature.
Confirmation of PE degradation with fungi
Scanning electron microscopic (SEM) analysis
The results of degradation of the polythene strips by the fungal isolates were confirmed by the formation of the cracks/holes/scions and were visualized in Scanning electron microscopic photographs (Fig. 2).
Figure 2.
Scanning Electron Microscopic image of the polythene strips: (a–d) SEM of the PE with most %weight gain (24.4%) by JAMNF at pH9.5; (e–h) SEM of PE strip with maximum (94) % loss in TS by PNPF-15 at pH 3.5; (i–l) SEM of PE strips of maximum %WL (41%) by MANGF1.
Fourier-transform infrared spectroscopy (FTIR) analysis
The degradation of the polythene was also confirmed with FTIR analysis (Fig. 3; Supplementary Table S4) in terms of changes in Carbonyl Index. Maximum change in Carbonyl Index was recorded in untreated polythene (4.36 and 4.34) with both the fungi (MANGF1/WL and PNP15/TS respectively) compared to pre-treated PE strips (2.56 and 2.63).
Figure 3.
FTIR spectra: A. pretreated PE strip. a: control, b: MANGF1/WL, c: PNP15/TS. B. untreated PE strip. a: control, b: MANGF1/WL, c: PNP15/TS.
Characterization of the most efficient polythene degrading fungal Isolates
Identification of fungi based at morphological keys
Based on the morphological keys, the fungal isolates were identified up to genus level. Among the ten elite polythene degrading fungi, 8 isolates (BAYF5/WL; MANGF1/WL; ERNF1/WL; BAYF7/WL; MANGF2/WL; PNPF15/TS; VASF1/TS and VASF6/TS) were reported to belong the genus Aspergillus, whereas, one isolate (ERNF3/WL) was not identified at morphological level and the isolate MIRF3/TS represent the genus Penicillium sp. (Supplementary Fig. S10; Supplementary Fig. S11).
Molecular characterization of the potential PE degrading fungi
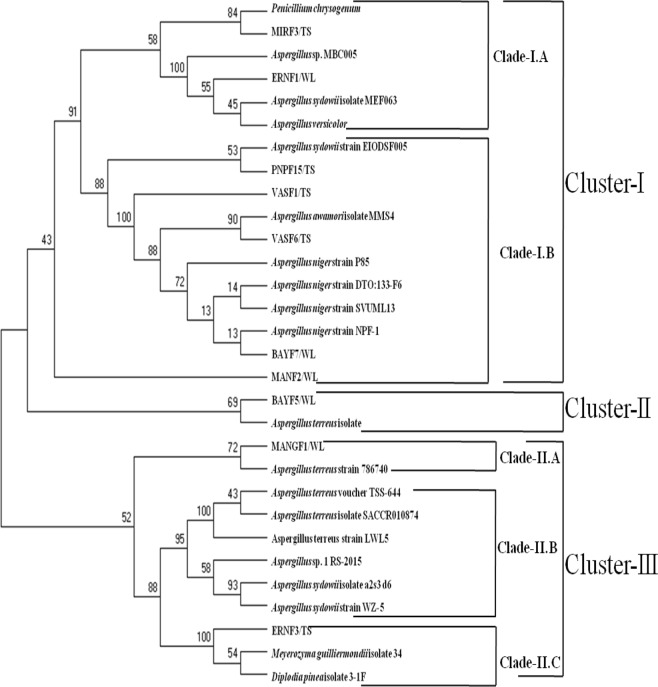
The amplified ITS genes of the polythene degrading fungi were separated on 1.2% Agarose gel against 100 base DNA ladder along with the negative control (Supplementary Fig. S12). All the sequences were accessioned by the gene bank (NCBI) (Supplementary Table S5). The polythene degrading fungal isolates were characterized based on sequence homology of internal transcribed spacer (ITS) gene. All the ITS sequences of the top 10 fungal isolates along with the ITS gene homology sequences retrieved from the gene bank were clustered into three main groups (Fig. 4). Group one is the largest cluster and further sub-clustered into two clades (clade I.A and clade I.B). In clade-I.A only two fungal isolates, MIRF3/TS and ERNF1/TS were grouped with other homologous sequences obtained from the gene bank and were identified as Penicillium chrysogenum strain MIRF3/TS and Aspergillus sydowii strain ERNF1/TS. Four fungal isolates, BAYF7/WL, VASF6/TS, VASF1/TS and PNPF15/TS were grouped with clade I.B and were identified as Aspergillus niger strain BAYF7/WL, Aspergillus awamori strain VASF6/TS, Aspergillus awamori strain VASF1/TS and Aspergillus sydowii strain PNPF15/TS. In cluster II only BAYF5/WL was grouped, and was identified as Aspergillus terreus strain BAYF5/WL. Cluster-III was grouped into three main clades (clade-II.A, clade-II.B and clade-II.C). In clade-II.A only one fungal isolate was clustered and identified as Aspergillus terreus strain MANGF1/WL. Similar to clade-II.A, only one fungal isolate was grouped with clade-II.C and was identified as Meyerozyma guilliermondii strain ERNF3/TS.
Figure 4.
Molecular phylogenetic analysis of polythene degrading fungi by maximum likelihood method along with the homologous ITS sequences retrieved from the gene bank (NCBI).
Discussion
Polythene waste alone shares 64% of the total plastic waste produced annually across the globe12,16. Among all the methods available to deal with plastic waste disposal, bioremediation technology succeeded with wide range of acceptance throughout the globe3,12. In literature, polythene deteriorating fungi were reported from various sources viz. marine water, plastic dumping sites and mangrove rhizosphere soil3,12,16. In the current investigation, we selected only those sites, for collecting of the rhizosphere soil samples to isolate polythene degrading fungi which represent all these sources. There are two reports4,54 from East Coast of India and one report4 from South East Coast of India and each report exhibits either utilization of mangrove rhizosphere soil or marine water for isolation of polythene degrading fungi. To the best of our knowledge, from the West coast of India, we have reported for the first time, polythene degrading fungi from all the available polythene degrading sources (dumping site, mangrove rhizosphere, marine water). Polythene degradation using fungal isolates had been assessed by determining the changes in some of the key characteristics of the polythene before and after the treatment of the fungal isolates viz. reduction in weight, reduction in tensile strength, reduction in percent elongation, reduction in viscosity, reduction in crystallinity, formation of cracks/scars/holes on the surface of the polythene, changes in the carbonyl index, estimation of CO2 released etc3,12. The rate of polythene degradation is highly influenced with various other factors viz. incubation time, pH, temperature, treatment of the polythene with some acids e.g. nitric acid (to remove the plasticizers) etc3,12. The incubation time period (period from the date of treatment to harvesting of the polythene strips for assessing the level of polythene deterioration) of the fungal isolates used to determine the rate of polythene degradation reports to be varied from 10 days to 32 years.
Previously after 30 days of incubation maximum 28.80 ± 2.40 percent weight loss (%WL) of the polythene was recorded with Aspergillus glaucus4, 23.11% WL (pre-treated UV and nitric acid) with Aspergillus niger55, 12.25%WL with Aspergillus niger56, 11.11% WL (LDPE) with Aspergillus japonicas and 5.8% with Aspergillus niger57. Even after increasing the incubation duration to 60 days, only 28–40% WL the polythene with Aspergillus niger was recorded but our results are more promising and efficient comparatively, we have reported ≈50% WL of the pretreated polythene strips with Aspergillus terreus MANGF1/WL at pH 9.5 during the same incubation period (60 days). Various workers used 3 months incubation time to determine the level of polythene degradation using fungi and recorded maximum 58.45% WL in pretreated polythene (2 days old chemically treated polythene followed by UV irradiation for 50 minutes before treatment) with Aspergillus oryzae, whereas in case of untreated polythene, only 6.3%WL with the same fungi during the same test period was recorded58 followed by 5.95% WL (with Aspergillus niger)59, 1.2% WL (with Curvularia lunata), 0.8% WL (with Alternaria alternate), 7.7% WL (Penicellium simplicissimum) and 0.7% WL (with Fusarium sp.) but the consortia of all these fungi (Curvularia lunata + Alternaria alternate + Penicellium simplicissimum + Fusarium sp.) results in 27% WL during the same test period60. After 6 months of incubation, only 26.17% WL (with Aspergillus niger)61 and 20.63%WL (with Aspergillus flavus)62 of polythene was documented. Aswale63 reported maximum 50%WL (with Phanerochaete chrysososporium) of the polythene at pH 4 after 8 months of incubation period. Abdullahi et al.64 recorded polythene degradation after 9 months of test period in terms of percent weight loss in two different types of degradation sets (polythene seeding in cow dung mixed fadama soil (PECDS) and poultry dropping mixed fadama soil (PEPDS) and reported 18.1% WL in the set of PECDS whereas only 6.0% WL with set PEPDS. Finally they concluded Aspergillus niger, A. fumigatues, and A. flavus mixed with PECDS and PEPDS leads to highest weight loss of the polythene compared to fadama soil mixed with inorganic fertilizer (NPK) and control. The possible reason for weight reduction in polythene in all the studies is due to breakdown of carbon backbone (enzymatic degradation)65 and utilizing the resulting monomers and oligomers as a carbon source by the fungi61,66. Otake et al.67 buried different kinds of plastic (including polythene) in the soil and assessed the level of degradation after 32 years and reported only whitening patches on the surface of the polythene due to the microbes (both fungi and bacteria) and recorded no evidence of degradation of other types of plastics. Among the various reports68–72 published during 2017–2018 on fungal based plastic degradation, Penicillium sp. was recorded as the most efficient fungi with percent weight reduction 43.4%70 in just 30 days.
Besides reduction in weight due to the degradation potential of the fungi, reduction of the tensile strength is also one of the widely studied parameter by different research groups around the globe. In the current investigation, we reported highest percent reduction in TS (94.44 ± 2.40%) with A. sydowii strain PNPF15/TS at pH 3.5 after 60 days of continuous shaking at ambient temperature. Our results are in agreement with the previous studies, previously, after 10 days of incubation maximum reduction (60%) in tensile strength (TS) of the heat treated polythene was reported with Mucor rouxii [NRRL 1835]73. After three months of testing period maximum reduction in TS (63%) of the polythene was reported with A. oryzae58 followed by 51% reduction in PE (Mangnease sterate treated LDPE exposed to UV irradiation) with the same fungi (A. oryzae)74. Vijaya and Reddy52 followed the ASTM standard and assessed the degradation (by compositing) of polythene (HDPE) along with municipality solid waste and recorded highest 20% reduction in tensile of HDPE after 1 year of testing duration. Vijaya and Reddy52 studied correlation coefficient among WL and TS and suggested strong correlation coefficient; if one factor is affected by microbial attack other factor also gets affected at the same time.
The degradation of the polythene was further authenticated using SEM and FTIR analysis. The SEM analysis revealed the degradation level on the surface of the polythene in the form craks/scions/holes (Fig. 2). Our observations are similar with the previous reports. Due to use of SEM analysis, structural changes and erosions on the surface of the polythene in the form of porosity, cavities, holes/scions/cracks were reported with fungal consortia60, Mucor circinilloides and Aspergillus flavus45, Aspergillus and Penicillium75, Chaetomium globosum76, Aspergillus niger and Aspergillus japonicas57,77. After SEM analysis, the level of polythene degradation was further authenticated by FTIR analysis. In the present investigation, the FTIR data confirmed the level of structural changes in the polythene. Abiotically treated sample (HNO3 treatment, 20 min UV treatment) shows generation of carbonyl peak, carboxylic acid and its derivatives. We observed the peak of carboxylic acid in the range of 1633.73–1812.08 cm−1 and reported the reduction of this peak up to 1629.53 cm−1 and no peak was observed at 1812.08 cm−1 on the PE strips degraded by A. terreus strain MANGF1/WL. A. sydowii strain PNPF15/TS based degraded polythene strip depicts the reduction of peak from 1633.73–1812.08 cm−1 to 1628.60 cm−1 and similarly no peak was recorded at 1812.08. Similarly in past, Konduri et al.74 also reported carboxylic acid peak in the range of 1630–1840 cm−1.
Chatterjee et al.78 reported the formation of C-H stress group at peak 2915 and we also reported similar peak at 2912.10 cm−1 in abiotic treated PE (control) and documented the reduction in the peak with both the fungal strains (A. terreus strain MANGF1/WL and A. sydowii strain PNPF15/TS) to 2912.09 cm−1 and 2912.92 cm−1 respectively. Chatterjee et al.78 reported occurrence of CH2 peak at 718, same functional group was observed in our study in the control PE strip at peak 721.52 cm−1 and compared to control we reported reduction in the peak with A. terreus strain MANGF1/WL (720.88 cm−1) and A. sydowii strain PNPF15/TS (721.05 cm−1). Balasubramanian et al.79 reported Keto carbonyl band at 1715, we reported the similar peak (1716.66 cm−1) in the control PE strips, but due to action of both the fungal strains this Keto carbonyl peak was not recorded on the degraded PE strips. Konduri et al.74 reported the peak of C=O stretching in between 1710–1740 cm−1, we got nearly same but smallar peak in between 1716–1766 cm−1 in the control PE strips and similar to Keto carbonyl band, C=O stretching was also not observed in degraded PE strips with both the fungal strains. In case of untreated PE strips (control), carboxylic group peak was not recorded, whereas CH2 was recorded at peak 721.28 cm−1 and reduction in CH2 peak was reported in the untreated PE strips degraded by both the fungi (718.45 cm−1 by A. terreus strain MANGF1/WL and 720.51 cm−1 by A. sydowii strain PNPF15/TS). CH stress peak was observed in the control PE (untreated) strips at 2913.03 cm−1 and only A. sydowii strain PNPF15/TS was reported to lead reduction of CH peak to 2912.02. In agreement with the Konduri et al.74 we also recorded C=C stretching at two peaks 1739.35 cm−1 and 1792.57 cm-1 only in case of PE strips degraded by A. terreus strain MANGF1/WL. Microbes are also reported to be responsible for decreasing the carbonyl index58 which in turn depicts the level of degradation. Manzur et al.80 reported maximum (40%) reduction in Carbonyl Index (CI) after 3 months of incubation. Yamada et al.81 studied the effect of Penicillium simplicissimum (soil fungi) on the degradation of low density polythene and after 3 months of incubation in liquid culture, Penicillium simplicissimum was reported to utilize polythene as a carbon source before irradiating with UV and nitric acid treatment. They also suggested that time required for degradation of polythene is depend on the time needed for the growth phase pure culture and they also reported that degradation is directly proportional to the addition of functional groups. Konduri et al.58,74 observed reduction in carbonyl group after three and six months of incubation with the fungi A. oryzae and A. flavus. Similarly in the current investigation there was a change in carbonyl groups, carboxylic groups after incubation with A. terreus strain MANGF/WL and A. sydowii strain PNPF15/TS for 60 days of continuous shaking at ambient temperature.
The previous literature depicts that polythene deteriorating fungi were mostly characterized based on morphological keys82–84. There are only few reports of identification of polythene degrading fungi at biochemical level82,85 and at molecular level5,58,69. As per the literature the traditional methods of fungal identification are time consuming, labour extensive and needs the utility of wide range of culture media with experienced personnel to characterize commonly occurring fungal strain variants86–88. The traditional methods are usually based on morphological keys and biochemical tests such as the identification of yeast based on biochemical test such as carbohydrate assimilation and fermentation tests which are unmanageable in non-specialized laboratory of microbiology89. There are various kits available in the market which leads to the rapid identification of the fungi but these kits are also not reliable and may takes few weeks to get the final results90,91. So, it is needed to have fast and accurate method of fungi identification. In the present scenario, the strategy utilized to identify many important fungi is the combined usage of morphological keys and biochemical tests with molecular diagnostics92. Presently, molecular tools are employed to aid the traditional method of fungi identification at greater pace93–95. Analysis of the variation in the internal transcribed spacer (ITS) regions of the rDNA is widely used for accurate identification of fungi96. Identification species and strain are more accurate based on variation in the ITSl/ITS2 domains than the 18S region (small subunit), the 5.8S region and the 28S region (large subunit)96,97. As per reports98,99 sequence based method is most rapid and authentic. Furthermore, molecular tools are the authentic and more reliable than the morphological and biochemical analysis and are considered as gold standard for the identification of any micro-organism. In the current investigation, based on morphological and molecular level (ITS gene sequence variation analysis), Aspergillus, Penicillium and Meyerozyma were reported as three main polythene degrading fungal genera. In case of genus Aspergillus, only four species in agreement with the previous reports such as Aspergillus awamori100; A. niger52,83,101, A. terreus57,102 and A. versicolor102 were recorded with polythene degradation potential. Besides the above species of the Aspergillus 7 more species such as A. candidus52, A. cremeus52, A. glaucus4,101, A. japonicus57, A. nidulans82,62, A. flavus52,82, A. oryzae103, A. ornatus52 were reported to have polythene degradation potential. The fungus, Aspergillus sydowii from the genus Aspergillus was reported for the first time with the polythene degradation potential, however, it was reported to degrade PVC plastic5. In genus Penicillium only Penicillium chrysogenum was recorded in the current investigation to degrade polythene. Sowmya et al.60 studied the degradation of rubber due to Penicillium chrysogenum, however, in literature Penicillium simlicimmum81, Penicilliumsp.82, P. pinophilum83, P. frequentans82, P. funiculosum104, also reported to have polythene degradation capacity. In the literature there is no report of Meyerozyma guilliermondii with polythene deteriorating potential, instead gasoline was reported to be degraded with same fungi105. Further, the polythene degradation-products (PE-DP) produced with the elite polythene degrading fungi (Aspergillus terreus strain MANGF1/WL and Aspergillus sydowii strain PNPF15/TS) were subjected to Gas Chromatography and Mass Spectra analysis followed by followed by their deleterious potential effect on Sorghum seeds and tiger shark fish were assessed, fungi based by-products of the polythene were found least toxic to both the plants and animal system106.
Methods
Collection and transportation of mangrove rhizosphere soil
Soil samples were collected from rhizosphere of A. marina from 12 different locations along the West Coast of India through individual and group visits (Supplementary Fig. S1 and Supplementary Table S1) and were transported to the laboratory as per standard method stated in our previous study107.
Isolation of the fungi using serial dilution
From the collected rhizosphere soil samples, fungal isolates were obtained by following serial dilution method, the fungal isolates were grown on Sabouraud’s Agar media (SA media), and the axenic cultures were also maintained as per the standard protocols108.
Screening of the polythene degrading fungi
From each fungal isolate (from six days old pure culture grown on Sabouraud’s broth) 1 ml (fungal) culture was used as an inoculum (average inoculum size 9.52 × 102 CFU) were screened based on their potential to degrade polythene at varied pH with regular shaking at room temperature. After 60 days of continuous shaking, polythene degradation was assessed using percent reduction or loss in weight (%WL) and percent reduction or loss in tensile strength (% loss in TS).
Reproducibility of the fungal based polythene degradation results
After screening, most efficient polythene degrading fungi based on percent reduction or loss in weight (Top 2 fungal isolates) and reduction or loss in tensile strength (Top 2 fungal isolates) along with control, were subjected to repetition under the same conditions as applied during screening, to infer the reproducibility of the polythene degradation results. At this stage, two types of polythene strips viz. pre-treated and untreated strips were used.
Confirmation of the polythene degrading potential of the fungi
After screening, the most efficient polythene degrading fungi were subjected to Scanning Electron Microscopy analysis and Fourier-transform infrared spectroscopy (FTIR) analysis to confirm the level of polythene degradation as per the methods stated in our previous study107.
Characterization of the most efficient polythene degrading fungal isolates
The most efficient fungal isolates with potential to degrade polythene (5 based on %WL and 5 based on % loss in TS) were characterized based on morphological keys109 and molecular tools. At morphological level, microphotographs of the selected fungal isolates were captured using trinocular microscope (Leica DM3000, Germany) equipped with cooled CCD camera (Leica DFC450, Germany). The captured photographs were processed by Leica Application Suite (Version 4.5.0). Cetyl Trimethyl Ammonium Bromide (CTAB) method110 was used to isolate genomic DNA from the 6 days old fungal cultures (100 mg of fungal mycelium) to identify the elite polythene degrading fungi at molecular level. The PCR reaction (25 μl) was carried out using 1.5 mM MgCl2, 0.25 mM of each dNTPs, 1X Taq buffer, 1 U/μl Taq DNA polymerase, 10 pmole of each primer of ITS gene93 (Supplementary Table S6) and 50 ng DNA template. The PCR (Veriti, gradient thermocyler cycler, Applied Biosystem, USA) was programmed at initial denaturation 95 °C 5 min, 40 cycles with denaturation at 95 °C 1 min, annealing 59 °C 1 min, extension 72 °C 1 min followed by final extension at 72 °C 10 min. The PCR products were separated on 1.2% Agarose gel (Invitrogen) prepared using 1X TAE buffer against negative control and 100 bp DNA ladder (Invitrogen). All the amplified bands were eluted from the Agarose gel using Qiagen gel extraction kit (Cat. No. 28115) as per the instruction manual and were given to commercial lab along with primers for purification and sequencing. The sequences obtained from the commercial lab were viewed using chromas lite and were curated to make contig using MEGA 6 software. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura 3-parameter model111. The bootstrap consensus tree inferred from 1000 replicates111 was taken to represent the evolutionary history of the taxa analyzed111. Branches corresponding to partitions reproduced in less than 100% bootstrap replicates were collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was shown above the branches111. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pair wise distances estimated using the Maximum Composite Likelihood (MCL) approach. The analysis involved 30 nucleotide sequences. There were a total of 1088 positions in the final dataset. Evolutionary analyses were conducted in MEGA6112.
All the ITS gene sequences of the top 10 polythene degrading fungi were submitted to gene bank (NCBI) and were accessioned (KU551273–KU551282). The pure fungal cultures of two most efficient polythene deteriorating fungal isolates in slants (duplicate) were submitted at Col. Sir R. N. Chopra, Microbial Resource Center Jammu (MRCJ), CSIR-Indian Institute of Integrative Medicine, Jammu, India for general deposition and were also accessioned (MRCJ-791 and MRCJ-792).
Conclusions
Among the 109 fungal isolates, Aspergillus terreus strain MANGF1/WL (more than 50.00 ± 4% WL, pH 9.5) and Aspergillus sydowii strain PNPF15/TS (94.44 ± 2.40% loss in TS, pH 3.5) are the most efficient and elite polythene deteriorating fungi based on reduction in weight, reduction in tensile strength, SEM and FTIR analysis. SEM analysis of the surface of the degraded polythene showed disturbances such as cracks, scions, fissures and holes which confirms corrosion. FTIR analysis shows formation of carbonyl group (1710–1740 cm−1), carboxylic group (1630–1840 cm−1), CH stress (2915 cm−1) and CH2 group (720 cm−1) after the UV and chemical treatment in control. These peaks were found to be reduced after fungal treatment. These decreasing peaks are due to the consumption of carbonyl and carboxylic acid derivatives by fungi indicating the de-polymerization of the polythene chain.
Supplementary information
Supplementary Information for Potential of fungi isolated from the dumping sites mangrove rhizosphere soil to degrade polythene
Acknowledgements
This work was a part of the project funded by Board of College and University Development (BCUD-2012–14), Savitribai Phule Pune University, Pune. MKS is thankful to UGC-BSR (UGC232 (004)) for providing the research fellowship to carry out the work. MS is also indebted to University Grants Commission-Maulana Azad National Fellowship (UGC-MANF-2013-14-MUS-JAM-22369) and Department of Science and Technology-Science Engineering and Research Board, National Post-doctoral Fellowship (DST-SERB N.PDF) (PDF/2017/000178) for financial assistance.
Author Contributions
M.K.S.: designed the problem, collected the samples, executed the experiments, data analysis, proof reading of the manuscript. M.S.: co-designed the problem, helped MKS in sample collection to experiment execution, data analysis, statistical analysis, written and communicated the manuscript. A.B.A.: Designed the problem, helped in data analysis, proof reading of the manuscript.
Data Availability
Data would be available on request to corresponding author.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Manisha K. Sangale and Mohd. Shahnawaz contributed equally.
Contributor Information
Mohd. Shahnawaz, Email: mskhakii@unipune.ac.in
Avinash B. Ade, Email: avinashade@unipune.ac.in
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41448-y.
References
- 1.Seymour RB. Polymer science before and after 1899: notable developments during the lifetime of Maurits Dekker. Journal of Macromolecular Science-Chemistry. 1989;26:1023–1032. [Google Scholar]
- 2.Shimao M. Biodegradation of plastics. Current Opinion in Biotechnology. 2001;12:242–247. doi: 10.1016/s0958-1669(00)00206-8. [DOI] [PubMed] [Google Scholar]
- 3.Shah AA, Hasan F, Hameed A, Ahmed S. Biological degradation of plastics: a comprehensive review. Biotechnology Advances. 2008;26:246–265. doi: 10.1016/j.biotechadv.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Kathiresan K. Polythene and plastics-degrading microbes from the mangrove soil. Revista de Biologia Tropical. 2003;51:629–633. [PubMed] [Google Scholar]
- 5.Ali MI, et al. Isolation and molecular characterization of polyvinyl chloride (PVC) plastic degrading fungal isolates. Journal of Basic Microbiology. 2014;54:18–27. doi: 10.1002/jobm.201200496. [DOI] [PubMed] [Google Scholar]
- 6.Rivard C, Moens L, Roberts K, Brigham J, Kelley S. Starch esters as biodegradable plastics: Effects of ester group chain length and degree of substitution on anaerobic biodegradation. Enzyme and Microbial Technology. 1995;17:848–852. [Google Scholar]
- 7.Ibrahim IN, Maraqa A, Hameed KM, Saadoun IM, Maswadeh HM. Assessment of potential plasticdegrading fungi in Jordanian habitats. Turkish Journal of Biology. 2011;35:551–557. [Google Scholar]
- 8.Muthukumar A, Veerappapillai S. Biodegradation of Plastics: A Brief Review. International Journal of Pharmaceutical Sciences Review and Research. 2015;31:204–209. [Google Scholar]
- 9.Corcoran PL, Biesinger MC, Grifi M. Plastics and beaches: a degrading relationship. Marine Pollution Bulletin. 2009;58:80–84. doi: 10.1016/j.marpolbul.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 10.Mukherjee S, Chatterjee S. A comparative study of commercially available plastic carry bag biodegradation by microorganisms isolated from hydrocarbon effluent enriched soil. International Journal of Current Microbiology and Applied Sciences. 2014;3:318–325. [Google Scholar]
- 11.Mueller R-J. Biological degradation of synthetic polyesters— enzymes as potential catalysts for polyester recycling. Process Biochemistry. 2006;41:2124–2128. [Google Scholar]
- 12.Sangale MK, Shahnawaz M, Ade AB. A review on biodegradation of polythene: the microbial approach. Journal of Bioremediation and Biodegradation. 2012;3:1–9. [Google Scholar]
- 13.Gu, J. D., Ford, T. E., Mitton, D. B. & Mitchell, R. Microbial corrosion of metals. The Uhlig Corrosion Handbook. 2nd Edition. New York: Wiley, 915–927 (2000).
- 14.Cooper DA, Corcoran PL. Effects of mechanical and chemical processes on the degradation of plastic beach debris on the island of Kauai, Hawaii. Marine Pollution Bulletin. 2010;60:650–654. doi: 10.1016/j.marpolbul.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 15.Bhatia M, Girdhar A, Tiwari A, Nayarisseri A. Implications of a novel Pseudomonas species on low density polyethylene biodegradation: an in vitro to in silico approach. SpringerPlus. 2014;3:497. doi: 10.1186/2193-1801-3-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee B, Pometto AL, Fratzke A, Bailey TB. Biodegradation of degradable plastic polyethylene by Phanerochaete and Streptomyces species. Applied and Environmental Microbiology. 1991;57:678–685. doi: 10.1128/aem.57.3.678-685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potts, J. E. & Jelink, H. H. G. Biodegredation aspect of biodegradation and stabilization of polymers. (Elsevier, New York, 1978).
- 18.Andrew, N. Analysis of polyethylene degrading potentials of microorganisms isolated from compost soil. International Journal of Pharmaceutical & Biological Archive3 (2012).
- 19.Singh B. Harmful effect of plastic in animals. The Indian Cow: The Scientific and Economic Journal. 2005;2:10–18. [Google Scholar]
- 20.Derraik JGB. The pollution of the marine environment by plastic debris: a review. Marine Pollution Bulletin. 2002;44:842–852. doi: 10.1016/s0025-326x(02)00220-5. [DOI] [PubMed] [Google Scholar]
- 21.Denuncio P, et al. Plastic ingestion in Franciscana dolphins, Pontoporia blainvillei (Gervais and d’Orbigny, 1844), from Argentina. Marine Pollution Bulletin. 2011;62:1836–1841. doi: 10.1016/j.marpolbul.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Secchi ER, Zarzur S. Plastic debris ingested by a Blainville’s beaked whale, Mesoplodon densirostris, washed ashore in Brazil. Aquatic Mammals. 1999;25:21–24. [Google Scholar]
- 23.Laist, D. W. Impacts of marine debris: entanglement of marine life in marine debris including a comprehensive list of species with entanglement and ingestion records, In Marine Debris 99–139 (Springer, 1997).
- 24.Anonymous. State of Environment - Plastic Waste Management. (Centre for Environmental Studies, Bhubaneswar, 2006).
- 25.Malik, F. In Hindustan Times, Maharashtra government set to ban plastic containers, banners, boards, not just bags, (Mumbai, 2018).
- 26.Anonymous. Overview of Plastic Waste Management. (Central Pollution Control Board, Delhi, 2013).
- 27.Zheng Y, Yanful EK, Bassi AS. A review of plastic waste biodegradation. Critical Reviews in Biotechnology. 2005;25:243–250. doi: 10.1080/07388550500346359. [DOI] [PubMed] [Google Scholar]
- 28.Tollner EW, Annis PA, Das KC. Evaluation of strength properties of polypropylene-based polymers in simulated landfill and oven conditions. Journal of Environmental Engineering. 2010;137:291–296. [Google Scholar]
- 29.Andrady AL. Microplastics in the marine environment. Marine Pollution Bulletin. 2011;62:1596–1605. doi: 10.1016/j.marpolbul.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 30.Tansel B, Yildiz BS. Goal-based waste management strategy to reduce persistence of contaminants in leachate at municipal solid waste landfills. Environment, Development and Sustainability. 2011;13:821–831. [Google Scholar]
- 31.Sinha V, Patel MR, Patel JV. PET waste management by chemical recycling: a review. Journal of Polymers and the Environment. 2010;18:8–25. [Google Scholar]
- 32.Zhang J, Wang X, Gong J, Gu Z. A study on the biodegradability of polyethylene terephthalate fiber and diethylene glycol terephthalate. Journal of Applied Polymer Science. 2004;93:1089–1096. [Google Scholar]
- 33.Awaja F, Pavel D. Recycling of PET. European Polymer Journal. 2005;41:1453–1477. [Google Scholar]
- 34.Gross RA, Kalra B. Biodegradable polymers for the environment. Science. 2002;297:803–807. doi: 10.1126/science.297.5582.803. [DOI] [PubMed] [Google Scholar]
- 35.Poirier Y, Nawrath C, Somerville C. Production of polyhydroxyalkanoates, a family of biodegradable plastics and elastomers, in bacteria and plants. Nature Biotechnology. 1995;13:142. doi: 10.1038/nbt0295-142. [DOI] [PubMed] [Google Scholar]
- 36.Flieger M, Kantorova M, Prell A, Rezanka T, Votruba J. Biodegradable plastics from renewable sources. Folia Microbiologica. 2003;48:27. doi: 10.1007/BF02931273. [DOI] [PubMed] [Google Scholar]
- 37.Batayneh M, Marie I, Asi I. Use of selected waste materials in concrete mixes. Waste Management. 2007;27:1870–1876. doi: 10.1016/j.wasman.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 38.Verma SS. Roads from plastic waste. The Indian Concrete. Journal. 2008;3:43–44. [Google Scholar]
- 39.Menon, A. Roads Made of Plastic Waste in India? Yes! Meet the Professor Who Pioneered the Technique, https://www.thebetterindia.com/43685/plastic-waste-in-road-construction-plastic-man-india-prof-vasudevan/ (2016).
- 40.Mani M, Subash C, Nagarajan G. Performance, emission and combustion characteristics of a DI diesel engine using waste plastic oil. Applied Thermal Engineering. 2009;29:2738–2744. [Google Scholar]
- 41.Panda AK, Singh RK, Mishra DK. Thermolysis of waste plastics to liquid fuel: A suitable method for plastic waste management and manufacture of value added products - A world prospective. Renewable and Sustainable Energy Reviews. 2010;14:233–248. [Google Scholar]
- 42.Sarker M. Converting waste plastic to hydrocarbon fuel materials. Energy Engineering. 2011;108:35–43. [Google Scholar]
- 43.Pospisil, J. & Nespurek, S. Highlights in chemistry and physics of polymer stabilization, In Macromolecular Symposia. 143–163 (1997).
- 44.Rutkowska M, Heimowska A, Krasowska K, Janik H. Biodegradability of polyethylene starch blends in sea water. Polish Journal of Environmental Studies. 2002;11:267–272. [Google Scholar]
- 45.Pramila R, Ramesh KV. Biodegradation of low density polyethylene (LDPE) by fungi isolated from marine water a SEM analysis. African. Journal of Microbiology Research. 2011;5:5013–5018. [Google Scholar]
- 46.Kyaw BM, Champakalakshmi R, Sakharkar MK, Lim CS, Sakharkar KR. Biodegradation of low density polythene (LDPE) by Pseudomonas species. Indian. Journal of Microbiology. 2012;52:411–419. doi: 10.1007/s12088-012-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu, J., Ford, T. E. & Mitchell, R. Microbiological corrosion of concrete, In Uhlig’s corrosion handbook (2nd edition) 477–491 (John Wiley & Sons, 2000).
- 48.Fuhs* GW. Der mikrobielle Abbau von Kohlenwasserstoffen. Archiv fur Mikrobiologie. 1961;39:374–422. [PubMed] [Google Scholar]
- 49.Iiyoshi Y, Tsutsumi Y, Nishida T. Polyethylene degradation by lignin-degrading fungi and manganese peroxidase. Journal of Wood Science. 1998;44:222. [Google Scholar]
- 50.Jen-hou* L. Zum Verhalten von bakteriengemischen gegenuber polyathylen verschiedenen mittleren Molekulargewichts. Kunststoffe. 1961;51:317–320. [Google Scholar]
- 51.Muthukumar T, Aravinthan A, Mukesh D. Effect of environment on the degradation of starch and prooxidant blended polyolefins. Polymer Degradation and Stability. 2010;95:1988–1993. [Google Scholar]
- 52.Vijaya C, Reddy RM. Impact of soil composting using municipal solid waste on biodegradation of plastics. Indian Journal of Biotechnology. 2008;7:235–239. [Google Scholar]
- 53.Mehdi, F. S. & Saifullah, S. M. Occurrence of fungi on mangroves of Karachi. Status of Plant Pathology in Pakistan, (Ghaffar, A. & Shehzad, S. eds) pp, 177–182 (1992).
- 54.Kumar S, Hatha AAM, Christi KS. Diversity and effectiveness of tropical mangrove soil microflora on the degradation of polythene carry bags. Revista de Biologia Tropical. 2007;55:777–786. doi: 10.15517/rbt.v55i3-4.5954. [DOI] [PubMed] [Google Scholar]
- 55.Sagar K, Namrata P, Shruti K, Priya VGS, Sastry DN. Bio-degradation of 40 micron plastic bags by Aspergillus niger and optimization of pre-treatment methods. Environment Conservation. Journal. 2013;14:61–68. [Google Scholar]
- 56.Priyanka N, Archana T. Biodegradability of polythene and plastic by the help of microorganism: a way for brighter future. Journal of Environmental & Analytical Toxicology. 2011;1:2161–0525. [Google Scholar]
- 57.Raaman N, Rajitha N, Jayshree A, Jegadeesh R. Biodegradation of plastic by Aspergillus spp. isolated from polythene polluted sites around Chennai. Journal of Academia and Industrial Research. 2012;1:313–316. [Google Scholar]
- 58.Konduri MKR, Anupam KS, Vivek JS, Kumar DBR, Narasu ML. Synergistic effect of chemical and photo treatment on the rate of biodegradation of high density polyethylene by indigenous fungal isolates. International Journal of Biotechnology and Biochemistry. 2010;6:157–174. [Google Scholar]
- 59.Constantin M, Iuliana R, Vasilescu G, Arsene ML, Luiza J. Colonization and degradation of polyethylene composites by fungal strains isolated. Scientific Bulletin. Series F. Biotechnologies. 2012;16:109–112. [Google Scholar]
- 60.Sowmya HV, Ramalingappa B, Nayanashree G, Thippeswamy B, Krishnappa M. Polyethylene degradation by fungal consortium. International Journal of Environmental Research. 2015;9:823–830. [Google Scholar]
- 61.Deepika S, Jaya MR. Biodegradation of low density polyethylene by microorganisms from garbage soil. Journal of Experimental Biology and Agricultural Sciences. 2015;3:15–21. [Google Scholar]
- 62.Usha R, Sangeetha T, Palaniswamy M. Screening of polyethylene degrading microorganisms from garbage soil. Libyan Agriculture Research Center Journal International. 2011;2:200–204. [Google Scholar]
- 63.Aswale, P. Studies on bio-degradation of polythene Ph. D. thesis, Dr. Babasaheb Ambedkar Marathwada University (2010).
- 64.Abdullahi M, Mohammed AS, Racheal ST. Biodegradation of polythene and plastic using fadama soil amended with organic and inorganic fertilizer. Indian Journal of Scientific Research. 2013;4(1):17–24. [Google Scholar]
- 65.Raut S, et al. Enhancing degradation of low density polyethylene films by Curvularia lunata SG1 using particle swarm optimization strategy. Indian Journal of Microbiology. 2015;55:258–268. doi: 10.1007/s12088-015-0522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vasile, C. Colonization and degradation of polyethylene composites by fungal strains isolated, In Handbook of polyolefins synthesis and properties Vol. 148 (eds Vasile, C. & Seymour, R. B.) 479–506 (Marcel Dekker Inc, New York, 1993).
- 67.Otake Y, Kobayashi T, Asabe H, Murakami N, Ono K. Biodegradation of low-density polyethylene, polystyrene, polyvinyl chloride, and urea formaldehyde resin buried under soil for over 32 years. Journal of Applied Polymer Science. 1995;56:1789–1796. [Google Scholar]
- 68.El-Morsy EM, Hassan HM, Ahmed E. Biodegradative activities of fungal isolates from plastic contaminated soils. Mycosphere. 2017;8:1071–1087. [Google Scholar]
- 69.Ojha N, et al. Evaluation of HDPE and LDPE degradation by fungus, implemented by statistical optimization. Scientific Reports. 2017;7:39515. doi: 10.1038/srep39515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alshehrei F. Biodegradation of low density polyethylene by fungi isolated from Red sea water. International Journal of Current Microbiology and Applied Sciences. 2017;6:1703–1709. [Google Scholar]
- 71.Munir, E., Harefa, R. S. M., Priyani, N. & Suryanto, D. Plastic degrading fungi Trichoderma viride and Aspergillus nomius isolated from local landfill soil in Medan, In IOP Conference Series: Earth and Environmental Science, IOP Publishing 126, 012145 (2018).
- 72.Brunner I, Fischer M, Ruthi J, Stierli B, Frey B. Ability of fungi isolated from plastic debris floating in the shoreline of a lake to degrade plastics. PloS One. 2018;13:e0202047. doi: 10.1371/journal.pone.0202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.El-Shafei HA, El-Nasser NHA, Kansoh AL, Ali AM. Biodegradation of disposable polyethylene by fungi and Streptomyces species. Polymer Degradation and Stability. 1998;62:361–365. [Google Scholar]
- 74.Konduri MKR, Koteswarareddy G, Rohini Kumar DB, Venkata Reddy B, Lakshmi Narasu M. Effect of pro-oxidants on biodegradation of polyethylene (LDPE) by indigenous fungal isolate, Aspergillus oryzae. Journal of Applied Polymer Science. 2011;120:3536–3545. [Google Scholar]
- 75.Mahalakshmi, V., Siddiq, A. & Andrew, N. Analysis of polyethylene degrading potentials of microorganisms isolated from compost soil. International Journal of Pharmaceutical & Biological Archive3 (2012).
- 76.Sowmya HV, Krishnappa M, Thippeswamy B. Low density polyethylene degrading fungi isolated from local dumpsite of shivamogga district. International Journal of Biological Research. 2014;2:39–43. [Google Scholar]
- 77.Annamalai J, Nallamuthu T. Assessment of potential plastic degrading fungi isolated from soil buried plastic pieces at Coovam river bank. Research in Environment and Life Sciences. 2011;4:125–128. [Google Scholar]
- 78.Chatterjee S, Roy B, Roy D, Banerjee R. Enzyme-mediated biodegradation of heat treated commercial polyethylene by Staphylococcal species. Polymer Degradation and Stability. 2010;95:195–200. [Google Scholar]
- 79.Balasubramanian V, et al. High-density polyethylene (HDPE)-degrading potential bacteria from marine ecosystem of Gulf of Mannar, India. Letters in Applied Microbiology. 2010;51:205–211. doi: 10.1111/j.1472-765X.2010.02883.x. [DOI] [PubMed] [Google Scholar]
- 80.Manzur A, Limon-Gonzalez M, Favela-Torres E. Biodegradation of physicochemically treated LDPE by a consortium of filamentous fungi. Journal of Applied Polymer Science. 2004;92:265–271. [Google Scholar]
- 81.Yamada-Onodera K, Mukumoto H, Katsuyaya Y, Saiganji A, Tani Y. Degradation of polyethylene by a fungus, Penicillium simplicissimum YK. Polymer Degradation and Stability. 2001;72:323–327. [Google Scholar]
- 82.Priyanka N, Archana T. Biodegradability of polythene and plastic by the help of microorganism: a way for brighter future. Journal of Environmental and Analytical Toxicology. 2012;1:1–4. doi: 10.4172/2161-0525.1000111. [DOI] [Google Scholar]
- 83.Volke-Sepúlveda T, Saucedo-Castañeda G, Gutiérrez-Rojas M, Manzur A, Favela-Torres E. Thermally treated low density polyethylene biodegradation by Penicillium pinophilum and Aspergillus niger. Journal of Applied Polymer Science. 2002;83:305–314. [Google Scholar]
- 84.Nwachukwu S, Obidi O, Odocha C. Occurrence and recalcitrance of polyethylene bag waste in Nigerian soils. African Journal of Biotechnology. 2010;9:6096–6104. [Google Scholar]
- 85.Seneviratne G, Tennakoon NS, Nandasena KA, Weerasekara M. Polyethylene biodegradation by a developed Penicillium-Bacillus biofilm. Current Science. 2006;90:20–21. [Google Scholar]
- 86.Summerbell, R. C. Physiological and other special tests for identifying dermaophytes. Laboratory Handbook of Dermatophytes, 45–79 (1997).
- 87.Dixon, D. M., Rhodes, J. C. & Fromtling, R. A. Taxonomy, classification, and morphology of the fungi. Murray, P. R., Baron, E. J. & Pfaller, M. A. et al. Manual of Clinical Microbiology. 7th Ed. Washington DC: American Society for Microbiology, 1161–1196 (1999).
- 88.Howell, S. A. & Hazen, K. C. Candida, Cryptococcus, and other yeasts of medical importance, In Manual of Clinical Microbiology, 11th Edition (eds Jorgensen, J. et al.) Ch. 11, 1984–2014 (ASM Press, Washington, DC, 2011).
- 89.Freydiere AM, Guinet R, Boiron P. Yeast identification in the clinical microbiology laboratory: phenotypical methods. Sabouraudia. 2001;39:9–33. doi: 10.1080/mmy.39.1.9.33. [DOI] [PubMed] [Google Scholar]
- 90.Dooley DP, Beckius ML, Jeffrey BS. Misidentification of clinical yeast isolates by using the updated Vitek Yeast Biochemical Card. Journal of Clinical Microbiology. 1994;32:2889–2892. doi: 10.1128/jcm.32.12.2889-2892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kellogg JA, Bankert DA, Chaturvedi V. Limitations of the current microbial identification system for identification of clinical yeast isolates. Journal of Clinical Microbiology. 1998;36:1197–1200. doi: 10.1128/jcm.36.5.1197-1200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.de Hoog, G. S. & Guarro, J. Atlas of clinical fungi. (Centraalbureau voor Schimmelcultures, 1995).
- 93.White TJ, Bruns T, Lee S, Taylor JL. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. 1990;18:315–322. [Google Scholar]
- 94.Guarro J, Gene J, Stchigel AM. Developments in fungal taxonomy. Clinical Microbiology Reviews. 1999;12:454–500. doi: 10.1128/cmr.12.3.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jackson CJ, Barton RC, Evans EGV. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions. Journal of Clinical Microbiology. 1999;37:931–936. doi: 10.1128/jcm.37.4.931-936.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iwen PC, Hinrichs SH, Rupp ME. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Medical Mycology. 2002;40:87–109. doi: 10.1080/mmy.40.1.87.109. [DOI] [PubMed] [Google Scholar]
- 97.Lott TJ, Burns BM, Zancope-Oliveira R, Elie CM, Reiss E. Sequence analysis of the internal transcribed spacer 2 (ITS2) from yeast species within the genus Candida. Current Microbiology. 1998;36:63–69. doi: 10.1007/s002849900280. [DOI] [PubMed] [Google Scholar]
- 98.Henry T, Iwen PC, Hinrichs SH. Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. Journal of Clinical Microbiology. 2000;38:1510–1515. doi: 10.1128/jcm.38.4.1510-1515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hennequin C, et al. Identification of Fusarium species involved in human infections by 28S rRNA gene sequencing. Journal of Clinical Microbiology. 1999;37:3586–3589. doi: 10.1128/jcm.37.11.3586-3589.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nowak B, Pajak J, Drozd-Bratkowicz M, Rymarz G. Microorganisms participating in the biodegradation of modified polyethylene films in different soils under laboratory conditions. International Biodeterioration & Biodegradation. 2011;65:757–767. [Google Scholar]
- 101.Aswale PN, Ade AB. Effect of pH on biodegradation of polythene by Serretia marscence. The Ecotech. 2009;1:152–153. [Google Scholar]
- 102.Singh V, Dubey M, Bhadauria S. Biodeterioration of polyethylene high density by Aspergillus versicolor and Aspergillus terreus. Journal of Advanced Laboratory Research in Biology. 2012;3:47–49. [Google Scholar]
- 103.Nanda, S., Sahu, S. & Abraham, J. Studies on the biodegradation of natural and synthetic polyethylene by Pseudomonas spp. Journal of Applied Sciences and Environmental Management 14 (2012).
- 104.Weiland M, Daro A, David C. Biodegradation of thermally oxidized polyethylene. Polymer Degradation and Stability. 1995;48:275–289. [Google Scholar]
- 105.Goulart GG, Coutinho J, Monteiro AS, Siqueira EP, Santos VL. Isolation and characterization of gasoline-degrading yeasts from refined oil-contaminated residues. Journal of Bioremediation & Biodegredation. 2014;5:1. [Google Scholar]
- 106.Sangale MK, Shahnawaz M, Ade AB. Gas chromatography-Mass Spectra analysis and deleterious potential of fungal based polythene-degradation products. Scientific Reports. 2019;9:1–6. doi: 10.1038/s41598-018-37738-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shahnawaz M, Sangale MK, Ade AB. Rhizosphere of Avicennia marina (Forsk.) Vierh. as a landmark for polythene degrading bacteria. Environmental Science and Pollution Research. 2016;23:14621–14635. doi: 10.1007/s11356-016-6542-3. [DOI] [PubMed] [Google Scholar]
- 108.Aneja, K. R. Experiments in microbiology, plant pathology and biotechnology. (New Age International, 2003).
- 109.Gilman, J. C. & Joseph, C. A manual of soil fungi. (Daya Books, 1998).
- 110.Doyle JJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- 111.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information for Potential of fungi isolated from the dumping sites mangrove rhizosphere soil to degrade polythene
Data Availability Statement
Data would be available on request to corresponding author.