Abstract
To investigate the ratio of mitochondrial DNA to genomic DNA (mt/gDNA) in embryo culture medium as a possible predictor for embryonic development and pregnancy outcome, we collected a total of 93 embryo biopsy specimens from 52 women at the corresponding Day 3 (D3) and Day 5 (D5) embryo culture medium of in vitro fertilization. With the multiple annealing and looping-based amplification cycles method of next-generation sequencing for whole genome amplification, we examined the karyotype of the biopsy samples and the mt/gDNA ratio in the culture medium. Results showed that the ratio of mt/gDNA had an upward trend with decreasing trophectoderm levels with no significant difference. At the same time, from D3 to D5, the mt/gDNA ratio in the medium of embryos that failed to become blastocysts showed an upward trend, and the mt/gDNA ratio of medium from embryos that reached blastulation with successful pregnancy showed a decreasing trend, but the differences were not statistically significant. We conclude that there is a certain correlation between mt/gDNA ratio and early embryonic development, but it does not reach a level that can be used as a clinical predictor.
Introduction
In vitro fertilization technology has become an important choice for many infertile couples as an effective treatment1. Assisted reproductive techniques (ART) account for the birth of more than 3.5 millions babies worldwide, and the number of IVF cycles performed increases every year1–5. However, the current average success rate is only about 20–30%1,2,5. Therefore, improving the success rate of embryo implantation has became a huge challenge for assisted reproductive technology.
Previous studies have shown that as women age (especially >35 years), the probability of aneuploidy appears to increase significantly6,7, contributing to infertility. The widely used embryo preimplantation genetic screening (PGS) technology can be broadly divided into nucleic acid detection and embryo imaging technique. The former is a traumatic procedure requiring cell biopsy that increases the risk of embryonic damage8. The latter is difficult to identify certain genetic defects that may not affect initial embryonic morphology but may seriously affect embryonic growth and development at later stages9. Therefore, there is an urgent need for a technology capable of non-invasively and rapidly assessing the genetic quality of embryos in order to improve the success rate of IVF.
Mitochondria are organelles that have an independent cell membrane within the eukaryotic cells and play an important role in cell survival10,11. Unlike other organelles, it has an independent DNA structure (mtDNA) that cords genetic information related to energy production and cellular metabolism12,13. Mitochondria and mtDNA also have a crucial impact on the early development of embryos14–16. Recent studies have found that when embryos entered the blastocyst stage, intracellular mitochondria multiplied significantly, and there were millions of mtDNA copies in a single trophoblast cell17,18. From blastocyst fluid and embryonic body DNA was released into the culture medium that can be extracted for measurement19–21. Ravichandran et al. suggested that the mtDNA/gDNA ratio in the embryo culture medium was related to embryonic development, and it could be used, instead of biopsied embryo cells, to predict the quality and the outcome of embryo for implantation22. Many attempts have been made to address this issue, Stigliani et al. and Hashimoto et al. believed that the mt/gDNA ratio of embryo culture medium was related to blastulation and was expected to have a predictive value, while Victor et al. believed that mtDNA content had no correlation with blastocyst ploidy, age or viability23–25. This controversy has not been solved up to now. Therefore we performed a study to address this controversy.
Results
The experimental group and the control group
We designed two kinds of controls, one was named NC1 that was culture medium underwent the corresponding culturing process but did not contain embryos, and the other was named NC2 that was not used for culturing. The average numbers of mt/gDNA ratios of the D3 group and the D5 group were 158.38 (SEM = 18.39) and 206.24 (SEM = 30.24), respectively, and there was no statistical difference (P = 0.09). The average numbers of mt/gDNA ratios of the D3-NC1 group, the D5-NC1 group and the NC2 group were 30.3 (SEM = 12.38), 34.2 (SEM = 12.27) and 31.10 (SEM = 7.64), respectively. The experimental results showed that the mt/gDNA ratio in the experimental groups (D3 and D5) are much higher than that of the corresponding control groups (D3-NC1 and D5-NC1) (P < 0.001). At the same time, there was also a significant difference between the experimental group and NC2 (P < 0.001), while there was no significant difference between NC2 and NC1 (D3-NC1 and D5-NC1) (Fig. 1).
Figure 1.
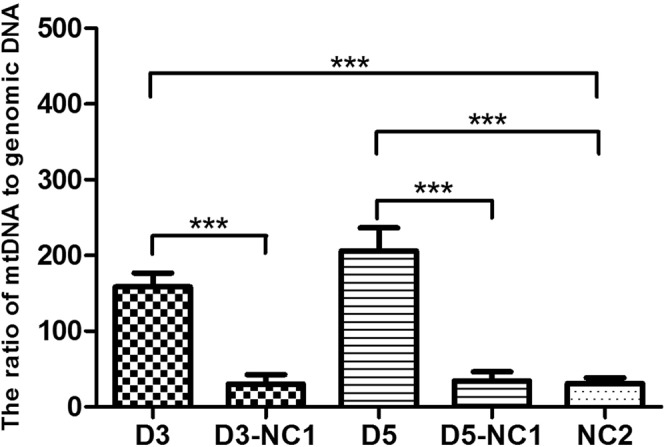
The ratio of mtDNA/gDNA in culture medium (n = 93) and negative control medium (n = 30). D3 and D5 were experimental groups, and D3-NC1 (n = 10) and D5-NC1 (n = 10) were the control groups that identically processed but did not contain embryo, NC2 (n = 10) is the unused culture medium. Data are means + SEM. ***P < 0.005.
Embryonic chromosome euploidy and mt/gDNA ratio
To investigate the relationship between mt/gDNA ratio and embryo chromosomal ploidy in culture medium, we compared the mt/gDNA ratio of the D3 and D5 embryos in 93 embryos and chromosome ploidy of the corresponding biopsy samples. The embryo is usually biopsied at D5 if the embryo was developed into the Grade 4 blastocyst or later stage. If on D5 the embryo had not matured to the Grade 4 blastocyst, we continued to culture it to D6 and then performed the biopsy. The embryos after biopsy were vitrified. The number of eggs biopsied can be found in Supplement Table 1. The results showed that 30 (32.2%) embryonic biopsy samples had a normal chromosomal euploidy, and 63 cases (67.7%) had uneuploidy. In D3 culture medium, the average numbers of mt/gDNA ratios of the euploid group and the aneuploid group were 78.95 (SEM = 35.19) and 124.9 (SEM = 20.57), respectively, and there was no statistical difference (P = 0.087). In D5 culture medium, the average numbers of mt/gDNA ratios of the aneuploid group and the abnormal group were 37.52 (SEM = 33.40) and 107 (SEM = 25.45), respectively, and there was no statistical difference neither (P = 0.21) (Fig. 2).
Figure 2.
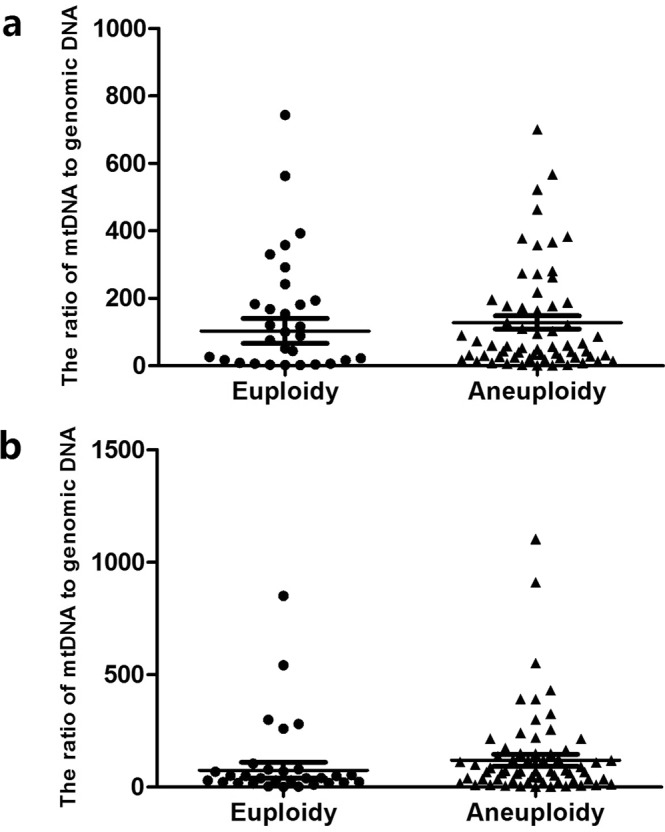
Corresponding mt/gDNA ratio in embryo culture medium when embryonic euploidy is different. At D3, the mt/gDNA ratio corresponding to different euploidy in the embryo culture medium (a). Mt/gDNA ratio corresponding to different euploidy in D5 embryo culture medium (b). We divided the samples into euploidy and aneuploidy based on chromosome ploidy of the embryo biopsy samples, as shown on the abscissa. The ordinate represents the ratio of mt/gDNA in the culture medium. One black dot or black triangle represents one sample. Data are means ± SEM.
Blastocyst quality and mt/gDNA ratio
-
We classified embryos based on inner cell mass (ICM) and TE morphology. There were 31 ICM Class A blastocysts (22 on day 5 and 9 on day 6), 38 ICM class B blastocysts (18 on day 5 and 20 on day 6) and 24 ICM class C (5 on day 5 and 19 on day 6).
The results showed that the average numbers of mt/gDNA in the D3 medium, with the ICM scores of A, B and C, were 115.24 (SEM = 32.75), 137 (SEM = 22.13), and 122.81 (SEM = 36.75), respectively. There was no significant difference among them (PAB = 0.64, PBC = 0.93, PAC = 0.70). At the same time, with the ICM scores A, B and C, the average numbers of mt/gDNA in D5 medium were 77.76 (SEM = 22.76), 84.9 (SEM = 33.28) and 93.7 (SEM = 68.54), respectively with no significant difference among them (PAB = 0.36, PBC = 0.19, PAC = 0.053) (Fig. 3).
There were 33 TE Class A blastocysts (22 on Day 5 and 11 on Day 6), 38 TE Class B blastocysts (18 on Day 5 and 20 on Day 6) and 22 TE C-grade blastocysts (5 on day 5 and 17 on day 6). The results showed that the average numbers of mt/gDNA in the D3 medium, with the TE scores of A, B and C, were 62 (SEM = 16.62), 144.25 (SEM = 24.35) and 159.56 (SEM = 42.24), respectively, and there was no significant difference (PAB = 0.45, PBC = 0.48, PAC = 0.42). At the same time, with the TE scores A, B and C, the average numbers of mt/gDNA in D5 medium were 29.25 (SEM = 14.19), 74.61 (SEM = 29.97) and 86.2 (SEM = 55.31), respectively with no significant difference (PAB = 0.36, PBC = 0.57, PAC = 0.35). Interestingly, although there was no significant difference between the two on D3 and D5, they all increased with the decrease of TE series (Fig. 4).
According to the proportion of fragments, the embryos were divided into three categories: Mild (0%), moderate (1–10%) and severe (>11%). The results showed that with the increase of the proportion of embryonic fragments from mild to moderate to severe, the average numbers of mt/gDNA in D3 medium were 134.48 (SEM = 21.57), 145.39 (SEM = 32.35) and 196.6 (SEM = 62.39), respectively, and there was no significant difference among them (PAB = 0.36, PBC = 0.65, PAC = 0.49). At the same time, with the increase of the proportion of embryonic fragments from mild to moderate to severe, the average numbers of mt/gDNA in D5 medium were 85.81 (SEM = 20.95), 85.28 (SEM = 30.63) and 117.84 (SEM = 87.02), respectively, and there was no significant difference (PAB = 0.90, PBC = 0.34, PAC = 0.40) (Fig. 5).
Blastulation: The average numbers of mt/gDNA ratio between embryos developing to blastocyst stage (85/93) and embryos developing to blastocyst stage (Grade 4) failed (8/93) on D3 were 141.43 (SEM = 17.76) and 89.43 (SEM = 56.24) respectively without significant difference (P = 0.054), and 83.39 (SEM = 19.63) and 101.57 (SEM = 80.67) on D5 respectively without significant difference (P = 0.61) (Fig. 6).
Blastulation time (D5/D6): A total of 44 (51.7%) embryos developed to blastocyst stage on D5, and 41 (48.3%) embryos developed to blastocyst stage on D6. In D3 culture medium, the average numbers of mt/gDNA ratio of the embryos developed to blastocyst stage on the D5 and D6 were 148.5 (SEM = 26.61) and 112.13 (SEM = 23.04) respectively with no significant difference (P = 0.27). In D5 culture medium, the average numbers of mt/gDNA ratios on D5 and D6 were 81.43 (SEM = 25.46) and 82.63 (SEM = 29.37) respectively with no significant difference (P = 0.61) (Fig. 7).
Figure 3.
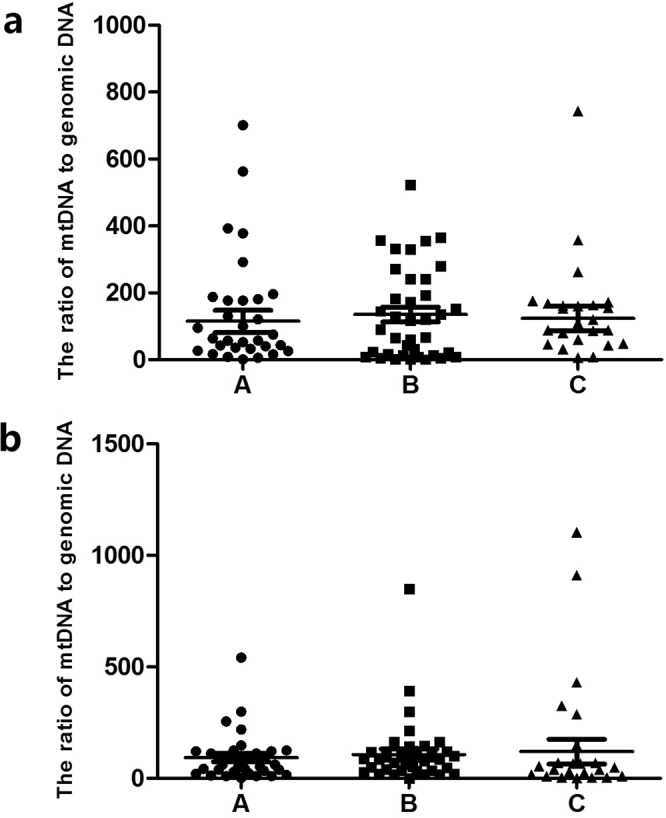
Corresponding mt/gDNA ratio in embryo culture medium when the score of ICM is different. At D3, the mt/gDNA ratio corresponding to different score of ICM in the embryo culture medium (a). Mt/gDNA ratio corresponding to different score of ICM in D5 embryo culture medium (b). According to the scores of ICM in each blastocyst (A–C), the samples were divided into three groups, A, B, and C, such as the abscissa. The ordinate represents the ratio of mt/gDNA in the culture medium. One black dot, black triangle or black square represents one sample. Data are means ± SEM.
Figure 4.
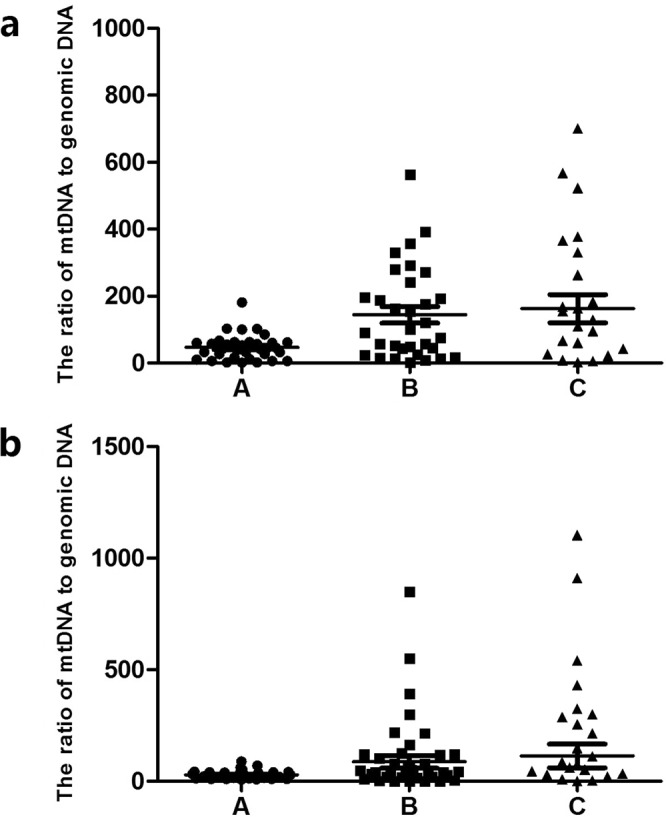
Corresponding mt/gDNA ratio in embryo culture medium when the score of TE is different. At D3, the mt/gDNA ratio corresponding to different score of TE in the embryo culture medium (a). Mt/gDNA ratio corresponding to different score of TE in D5 embryo culture medium (b). According to the scores of TE of each blastocyst (A–C), samples were divided into three groups, A, B, and C, such as the abscissa. The ordinate of the ordinate represents the ratio of mt/gDNA in the culture medium. One black dot, black triangle or black square represents one sample. Data are means ± SEM.
Figure 5.
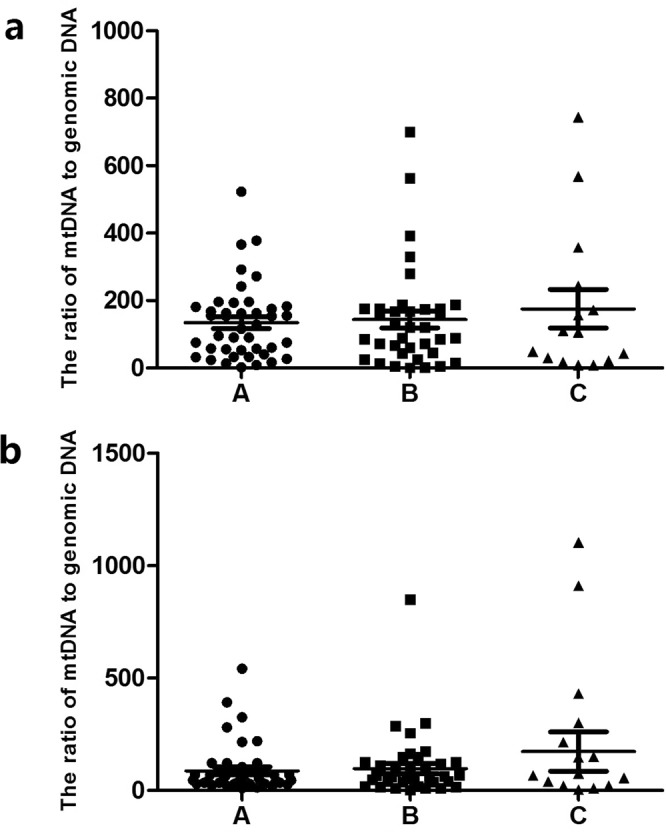
Corresponding mt/gDNA ratio in embryo culture medium when the proportion of embryo fragments is different. At D3, the mt/gDNA ratio corresponding to different proportion of embryo fragments in the embryo culture medium (a). Mt/gDNA ratio corresponding to different score of proportion of embryo fragments in D5 embryo culture medium (b). According to the scores of TE of each blastocyst (A–C), the samples were divided into three groups, A, B, and C, such as the abscissa. The ordinate of the ordinate represents the ratio of mt/gDNA in the culture solution. One black dot or black triangle or black square represents one sample. Data are means ± SEM.
Figure 6.
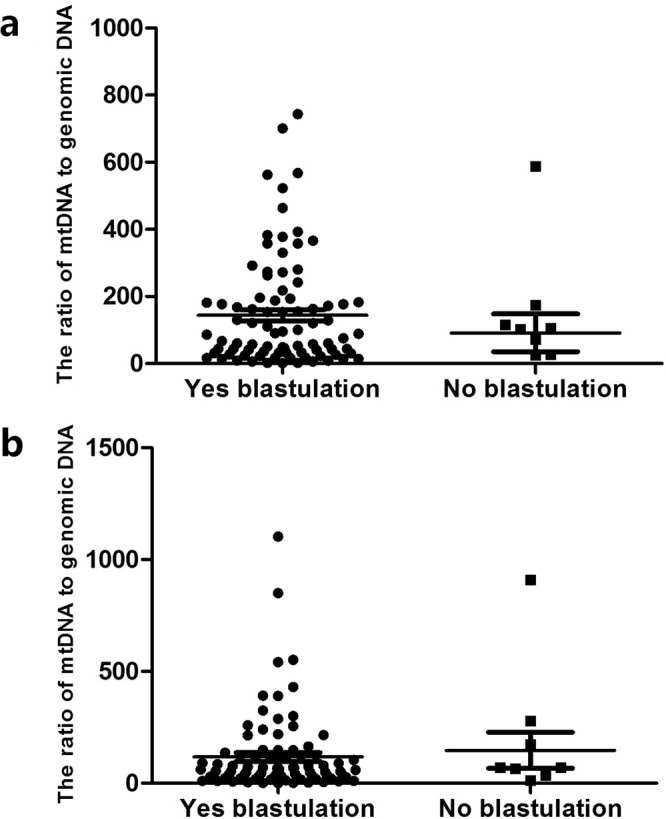
Mt/gDNA ratio in embryo culture medium corresponding to different outcome of blastulation. At D3, the mt/gDNA ratio corresponding to different outcome of blastulation in the embryo culture medium (a). Mt/gDNA ratio corresponding to different outcome of blastulation in D5 embryo culture medium (b). According to whether embryos grow to the blastocyst stage, the samples were divided into two groups, Yes and No, such as the abscissa. The ordinate of the ordinate represents the ratio of mt/gDNA in the culture solution. One black dot or black square represents one sample. Data are means ± SEM.
Figure 7.
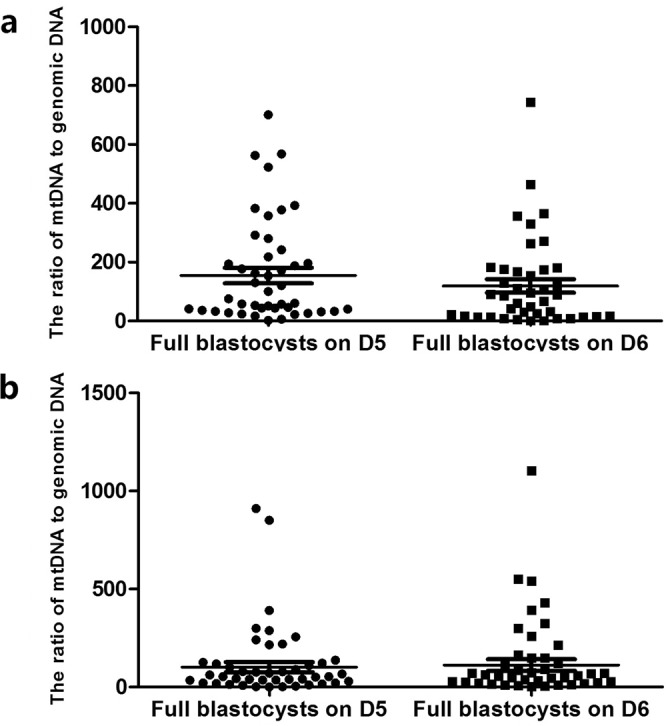
Mt/gDNA ratio in embryo culture medium corresponding to different time of blastulation. At D3, the mt/gDNA ratio corresponding to different time of blastulation in the embryo culture medium (a). Mt/gDNA ratio corresponding to different time of blastulation in D5 embryo culture medium (b). According to the time when embryos grow to the blastocyst stage, samples were divided into two groups, D5 and D6, such as the abscissa. The ordinate represents the ratio of mt/gDNA in the culture broth. One black dot or black square represents one sample. Data are means ± SEM.
Age of women and mt/gDNA ratio
According to age, the patients were divided into three groups of <=30 years old (A), 31–34 years old (B), and >=35 years old (C). There were 8 cases and 15 embryos in group A, 5 cases and 7 embryos in group B, 6 cases and 8 embryos in group C. When analyzing euploid embryos only, the average numbers of D3 mt/gDNA ratios were 136.86 (SEM = 26.48), 161.17 (SEM = 59.95) and 267.5 (SEM = 93.48), respectively with no significant difference (PAB = 0.82, PBC = 0.47, PAC = 0.40). The average numbers of D5 mt/gDNA were 53.21 (SEM = 18.21), 70.83 (SEM = 51.99) and 109.43 (SEM = 77.23), respectively with no significant difference (PAB = 0.60,PBC = 0.18,PAC = 0.24) (Fig. 8).
Figure 8.
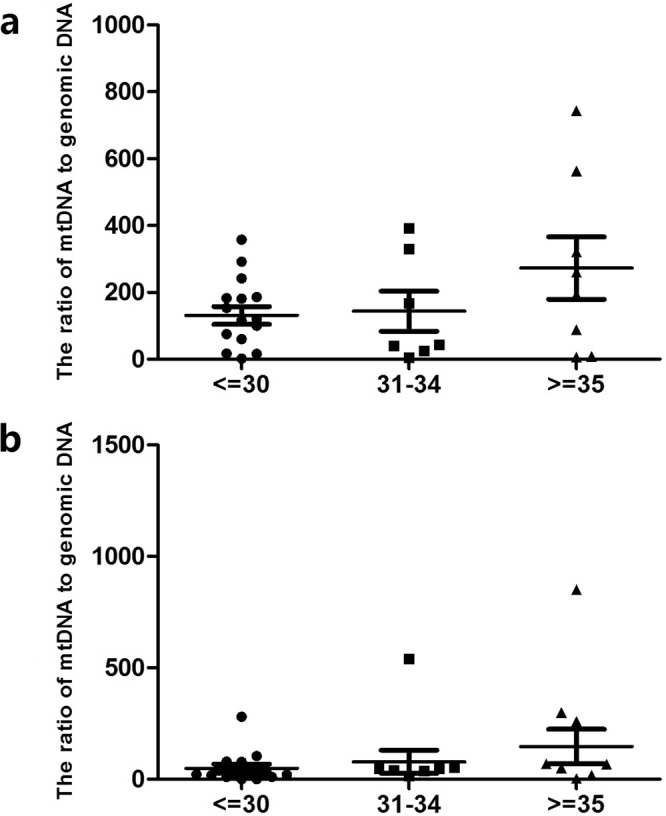
Mt/gDNA ratio in embryo culture medium corresponding to different age of woman (regardless of euploidy). At D3, the mt/gDNA ratio in the embryo culture medium corresponding to different age of woman (a). Mt/gDNA ratio corresponding to different time of blastulation in D5 embryo culture medium (b). According to the patient’s age, they were divided into three groups, which are less than or equal to 30 years old, greater than 31 years old and less than 34 years old and greater than or equal to 35 years old (<=30, 31–34 and >=35), such as the abscissa. The ordinate represents the ratio of mt/gDNA in the culture medium. Data are means ± SEM.
Relationship between mt/gDNA ratio and pregnancy outcome
A total of 12 of the 52 cases were transplanted, and one embryo was transplanted in each case (Table S1). And the rest were frozen embryos that had were not transplanted. The 28-day B-ultrasound gestational sac and fetal heart rate were the criteria for success. Of the transplanted cases, 6 were successful and 6 failed. The results showed that on D3, the average numbers of mt/gDNA corresponding to pregnancy success and failure were 210 (SEM = 66.86) and 55.17 (SEM = 26.68) (P = 0.089). On D5, the average numbers of mt/gDNA corresponding to the success and failure of pregnancy were 46.33 (SEM = 18.65) and 239.17 (SEM = 175.7), (P = 0.37) (Fig. 9).
Figure 9.
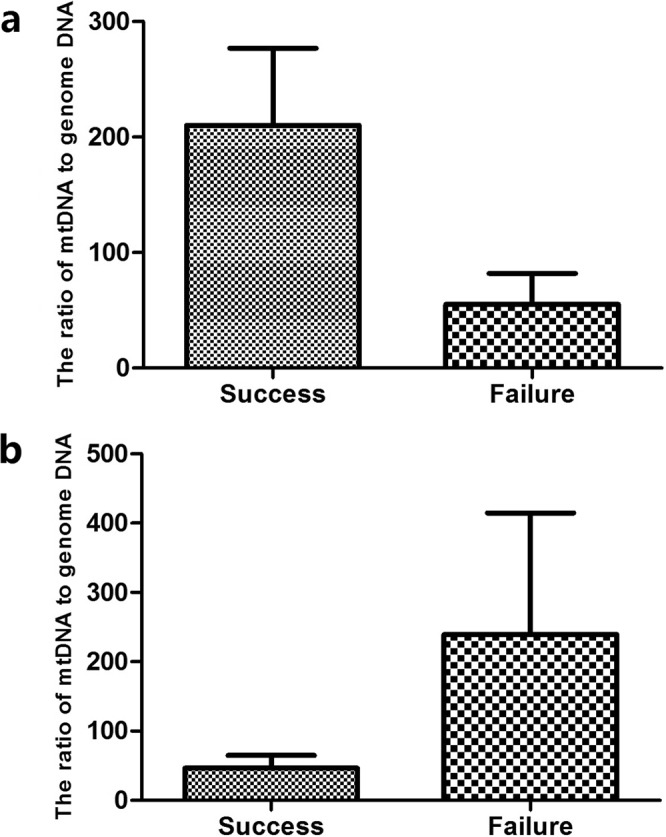
The mt/gDNA ratio for different pregnancy outcomes. At D3, the mt/gDNA ratio in the embryo culture medium corresponding to different pregnancy outcomes (a). Mt/gDNA ratio corresponding to different pregnancy outcomes in D5 embryo culture medium (b). Based on the pregnancy outcome of the embryo, the samples were divided into two groups, success and failure, such as the abscissa. The 28-day B-ultrasound can monitor the gestational sac and fetal heartbeat as the success criteria. The ordinate of the ordinate represents the ratio of mt/gDNA in the culture medium. Data are means + SEM.
Discussion
In this study, culture medium of the embryo on D3 and D5 before freezing (transplantation) was collected, and the mt/gDNA in the culture medium was detected using NGS to examine changes of released DNA during the culture process. We believe that the observed low baseline levels of DNA were derived from protein supplements originally contained in the culture medium. The main protein supplement was human serum albumin, which had a primary binding affinity of DNA26,27. When the commercial medium and protein supplements were analyzed separately, the highest level of DNA was found in 100% serum replacement supplement28. In our study, there was no significant difference in mt/gDNA ratio between NC1 and NC2, indicating that human operation and culture environment did not significantly affect the DNA content in the culture medium. Results showed that there was no significant correlation between euploidy and mt/gDNA of the embryo, but the average number of D3 and D5 in the normal group was lower than that in the abnormal group. There was a certain association between the two, but mt/gDNA could not be used as a clinical indicator to predict the ploidy of embryonic chromosomes as the changes were not statistically significant.
Based on our results, the ratio of mt/gDNA in D3 and D5 was increased slightly with the decrease of embryonic ICM score, but there was no statistical difference. Similarly, as the TE score decreased, the mt/gDNA in D3 and D5 also showed a small increase, and there was no statistical difference either. In addition, the proportion of fragments contained in the blastocyst did not significantly affect the mt/gDNA ratio in the medium (D3 and D5). Therefore, it appeared that mt/gDNA was related to the quality of embryos to a certain extent, but it could not be used as a clinical indicator in predicting the formation of capsules and superior embryos.
From D3 to D5, the mt/gDNA of embryos with successful pregnancy showed a downward trend, while the trend of non-pregnancy showed an upward trend, which was consistent with the trend of forming blastocysts. However, the changes did not reach the level of statistical difference. It could be speculated that mt/gDNA might have a certian correlation with embryo quality, but it could not be used as a clinical indicator to predict pregnancy outcome.
In recent years, the role of mitochondria in embryonic growth and development has attracted wide attention. There are different options on whether mitochondria can be used as a predictor of embryo quality. Some believed that mitochondrial DNA copy number in culture medium could be used as a non-invasive predictive index23,24. However, others believed that mtDNA content had no correlation with blastocyst ploidy, age or viability25. The results of our study, however, are inclined to show a relationship between mitochondrial DNA and embryo quality and development, but the relationship was not significant enough to be used as an indicator to judge the quality of the embryos.
The conclusion of our study is different from those of the previous reports for several reasons: (1) Most of the studies that support mtDNA as a clinical predictor were based on the recognition of the “silent embryo hypothesis”. This hypothesis was derived from early research on cattles and believed that there was a “mitochondrial genetic bottleneck”29–31. The copy number of mtDNA is significantly increased during oogenesis12,32 and is actively reduced during pre-implant development29. The mtDNA sequence variants segregate between generations despite the high mtDNA copy number in the oocyte23. This led to the concept of “mitochondrial genetic bottleneck”, which limits the process by which mtDNA molecules travel from the mother to the fetus and the offspring. Subsequent studies suggested that the copy number of mtDNA was significantly increased during egg production in large mammals12,32–36 and then rapidly decreased during the pre-implantation developed phase31,37. However, a similar study on mice reached a different conclusion33,38,39. It appeared that similar studies of different species yielded different results. (2) At the same time, the mechanism of the mitochondrial genetic bottleneck mentioned above is still controversial. Some studies suggested that mitosis of mtDNA occured before primordial germ cell expansion to gonadal colonization40. On the other hand, other studies suggested that mtDNA bottlenecks occurred in the reduction of mtDNA copy number during development before implantation39. We collected the culture medium on D3 and D5 of early embryo development, and the results only reflected the changes of this stage. (3) Concerning target genes and detection technology, previous studies selected D-loop gene or other short fragments in mitochondrial DNA as the detection target to represent the copy number of mitochondrial DNA35,41. Studies showed that environmental stress might affect the transient synthesis of mitochondria after fertilization42. The D-loop region is more susceptible to variation due to environmental stress than other regions of mitochondrial DNA43. At the same time, previous studies used qPCR combined with specific probes to detect the D-loop region of mitochondrial DNA35, which increased the possibility of unstable results. Another study chose Alu, a multiple copies gene, as a detection fragment44, which was not technically superior to that of a single copy gene, and the frequency and composition of the Alu sequence are significantly different in population45. multiple annealing and looping-based amplification cycles (MALBAC) and NGS has been successfully used in various studies of DNA at single cell level, such as detecting single nucleotide variation (SNV) and copy number variation (CNV), analyzing meiotic recombination46,47, and performing PGS of in-vitro fertilized embryos47,48. Studies have also shown that NGS is sensitive enough to detect small differences between subculture populations from the same cell line and that MALBAC-NGS is also suitable to study mtDNA copy number in single cells with good reproducibility49. Most of the samples used in previous studies were from multiple clinics22–24,28,50, and the samples used in this study were from a single clinic. Factors such as media, culture temperature, biopsy techniques, and equipment used during sample collection would affect the outcome. (4) There were other contradictions among previous reports. In some studies samples were collected from both the cleavage and/or the blastocyst stages28,50, while other studies collected samples from the cleavage stage only23. The above may provide some but not all explanations for the discrepancy.
We conclude that the number of mitochondrial DNA copies released from embryos into the culture medium is related to embryo quality and development during in vitro culture, but it would not be a reliable clinical indicator to evaluate or forecast embryo development.
Methods
Patients selection
We recruited 52 couples (median maternal age 32.7 years) from Jinjiang Women and Children’s Hospital, Chengdu, China., who gave written informed consent. All couples underwent intracytoplasmic sperm injection (ICSI). This prevented paternal contamination from sperm and limited the maternal contamination from cumulus cells. Ethical approval was obtained from the Scientific and Ethical Committee of Jinjiang Women and Children Hospital for the experimental protocol (2018 KY-001). All clinical procedures including ova collection, in vitro fertilization, embryo culture and implantation were performed according to the standard hospital protocol (CDXN/QD-EMBYO-02-17) that has been approved by the Ethical Committee of Jinjiang Hospital which was accredited by ISO9001 and JCI (Joint Commission on Accreditation of Healthcare Organizations). Patient consent was obtained for each embryo sample used in this study.
Gamete preparation and ICSI procedure
Oocyte preparation
After the follicle became mature, human chorionic gonadotropin (HCG) was injected. Thirty-four to 36 hours later transvaginal ultrasound-mediated oocyte retrieval was performed. The ova were repeatedly pipetted in GMOPs (Ref. No. 10130, Vitrolife, Sweden) containing hyaluronidase (Ref. No. ART-4007-A, SAGE, US) for 10–30 s before moving to GMOPS (Ref. No. 10129, Vitrolife, Sweden) without HSA. When the granulosa cells were removed completely, the ova was transferred into culture medium G-IVF PLUS (Ref. No. 10136, Vitrolife, Sweden) covered with OVOIL (Ref. No. 10029 Vitrolife, Sweden). The culture dish was incubated in an incubator containing 6% CO2 in 37 °C for 1–1.5 hours.
Sperm preparation
We use the upstream method for semen optimization. Detailed steps are as follows:
The sample cup containing the specimen was placed in a 30 ± 0.5 °C electrothermal incubator for liquefaction. Specimens were processed within 60 minutes of sperm extraction.
Add 1.5 ML Sperm Rinse (Ref. No. 10101, Vitrolife, Sweden) solution to the centrifuge tube (Ref. No. 352099, BD Falcon, United States).
Mix the semen sample thoroughly and carefully add the semen to the 1.5 ML Sperm Rinse, taking care to maintain the liquid interface. The tube was tilted at 45° and incubated at 37 °C for 60 minutes.
Gently return the tube to erect and transfer the top 1 mL culture to the centrifuge tube (Ref. No. 352003, BD Falcon, United States). The semen containing the semen was placed in a 30 °C, 5% CO2 incubator for later use.
Then sperms were magnified 200 to 400 times under an inverted microscope. According to the protocol recommended by World Health Organization (WHO) criteria (2010), sperm morphology was assessed. Sperms of good morphology were selected and immobilized with the injection pipette.
ICSI
We used natural cycle ICSIs but did not use FSH for in vitro ovum maturation. We only selected naturally occurring MII (second metaphase) cells for intracytoplasmic sperm injection. A total of 585 eggs were taken from 52 cases, of which 512 oocytes were in MII. The number of eggs taken in each case and the number of oocytes reached MII in each case can be found in Supplement Table 2. With polar body at the 12 o’clock position, oocytes were held by the holding pipette and the injection pipette was inserted into the oocyte at the 3 o’clock position.
Embryo culture
Each ova was cultured in a micro-culture well Embryoscope Culture Dish (Vitrolife, Sweden). There were 25 µl G-1 PLUS (Ref. No. 10128, Vitrolife, Sweden) culture medium containing 5% HSA (Ref. No. 10064, Vitrolife, Sweden) in each well covered with OVOIL (Ref. No. 10029 Vitrolife, Sweden) and. Embryo culture was under an oxygen concentration of 5%. In order to allow the embryo culture to reach a temperature suitable for embryonic growth and development, and to bring the pH of the medium to the optimal pH range required by the instructions, we usually incubate the embryo culture media for 10 h before use. On the third day, the embryos that were developed to 6–8 cells were transferred into 25 ul of G-2 PLUS (Ref. No. 10132, Vitrolife, Sweden) containing 5% HSA and cultured until the 5–6th day. The blastocysts were scored according to the method of Gardner et al.51. The blastocysts with good scores were selected for transplantation.
Sample collection
Embryo culture medium
The culture fluid was replaced on the third day of embryo culture. At this time, 20 ul of the original culture fluid was collected into a RNase-DNase free PCR tube as a D3 medium sample. At the fifth or sixth day, 20 μl of blastocyst culture fluid corresponding to each blastocyst was collected as the D5 medium sample.
TE
The blastocysts were then placed individually in a biopsy dish containing 20 µL of G-MOPS PLUS (Ref. No. 10130, Vitrolife, Sweden) under oil for biopsy. Trophectoderm cells were encouraged to herniate from the zona by applying gentle suction with the biopsy pipette.
Negative control (NC)
The culture medium identically processed but did not contain embryo was used as a negative control (NC1) and unused culture medium was also used as a negative control (NC2).
WGA and NGS
The MALBAC method was applied to WGA according to manufacture’s protocols (catalog no. YK001B; YIKON Genomics). Briefly, amplification begins by annealing DNA to a random primer pool, each with a common 27 nucleotide sequence and 8 variable nucleotides. Prior to amplifying the DNA index to ~2 μg for NGS, a quasi-linear pre-amplification step was performed. Using a Covaris S2 system (Covaris, USA), approximately 400 ng of purified DNA was sonicated into a fragment of approximately 150–250 bp and purified using the Qiagen PCR purification kit (Qiagen, Germany). A sequencing library DNA library preparation was then constructed using NEB Next Ultra (New England Biolabs, UK). The constructed library was sequenced on an Illumina HiSeq2500 platform (Illumina, USA).
Data analysis
The MALBAC amplified genome of each sample was sequenced using the Illumina HiSeq. 2500 platform for a total of approximately 1.5 million reads. The NGS data was analyzed and the chromosomal copy number variation was investigated as described in previous reports48,52,53. The high quality read numbers were counted along the whole genome with a bin size of 1.5~2 Mb and normalized by the GC content and a reference dataset. An increase of 2 to 3 copies of the copy number results in a 50% increase in the reading of the particular genomic fragment, while a decrease in the copy number from 2 to 1 results in a 50% reduction in the genomic fragment reading. Mitochondrial content is quantified in the form of copy number/nuclear genome. Sequencing reads were mapped to the mitochondrial genome and were counted and normalized to autosomes. Mitochondrial copy numbers per nuclear genome (MCN) were calculated with the following equation (1). Independent sample t test was used to assess the relationship between mt/gDNA and different embryo quality indicators.
| 1 |
Statistical analyses
The Mann-Whitney test was used to compare the mtDNA/gDNA ratio in spent embryo media and media controls (GraphPad; GraphPad Software). Additionally, it was used to compare differences in mtDNA/gDNA between groups when chromosome euploidy, cystic outcome (means blastulation outcome), and capsular time were different. Independent sample t-test was used to compare differences in mtDNA/gDNA between groups when ICM score, TE score, fragment ratio, and female age were different (SPSS). The independent sample t-test was also used to compare differences in mtDNA/gDNA between the two groups (n = 12) when pregnancy outcomes were different (SPSS). The results were considered statistically significant at P < 0.05.
Supplementary information
Acknowledgements
We thank the support of the Applied Basic Research Programs of Sichuan Province (No. 2017JY0096).
Author Contributions
X.Z., X.D. and T.H.H. designed the project; J.G. and X.Z. analyzed data; X.Z., Y.S., J.Z., F.S., T.T.H., P.L., R.M., X.G., P.L. J.H., Q.W., T.Q. and L.L. performed experiments and collected samples; J.G. Y.Z. and X.Z. wrote the paper; Y.S. contributed new reagents and analytic tools.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jianming Zhou, Fubo Sun, Tingting Han, Ping Lei, Rurong Mao, Xuzhou Guo and Qi Wang contributed equally
Contributor Information
Ying Zhong, Email: zhongy@scivf.com.
Jiang Gu, Email: 2523381625@qq.com.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41801-1.
References
- 1.Qiao J, Feng HL. Assisted reproductive technology in China: compliance and non-compliance. Translational pediatrics. 2014;3:91–97. doi: 10.3978/j.issn.2224-4336.2014.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kupka MS, et al. Assisted reproductive technology in Europe, 2011: results generated from European registers by ESHRE. Human reproduction. 2016;31:233–248. doi: 10.1093/humrep/dev319. [DOI] [PubMed] [Google Scholar]
- 3.De Mouzon J, et al. Assisted reproductive technology in Europe, 2006: results generated from European registers by ESHRE. Human reproduction. 2010;25:1851–62. doi: 10.1093/humrep/deq124. [DOI] [PubMed] [Google Scholar]
- 4.Kovalevsky G, Patrizio P. High rates of embryo wastage with use of assisted reproductive technology: a look at the trends between 1995 and 2001 in the United States. Fertility and sterility. 2005;84:325–330. doi: 10.1016/j.fertnstert.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 5.Ghazal S, Patrizio P. Embryo wastage rates remain high in assisted reproductive technology (ART): a look at the trends from 2004-2013 in the USA. Journal of assisted reproduction and genetics. 2017;34:159–166. doi: 10.1007/s10815-016-0858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munne S, Lee A, Rosenwaks Z, Grifo J, Cohen J. Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Human reproduction. 1993;8:2185–2191. doi: 10.1093/oxfordjournals.humrep.a138001. [DOI] [PubMed] [Google Scholar]
- 7.Munne S, Alikani M, Tomkin G, Grifo J, Cohen J. Embryo morphology, developmental rates, and maternal age are correlated with chromosome abnormalities. Fertility and sterility. 1995;64:382–391. doi: 10.1016/S0015-0282(16)57739-5. [DOI] [PubMed] [Google Scholar]
- 8.LaBonte ML, et al. An analysis of US fertility centre educational materials suggests that informed consent for preimplantation genetic diagnosis may be inadequate. Journal of medical ethics. 2012;38:479–484. doi: 10.1136/medethics-2011-100154. [DOI] [PubMed] [Google Scholar]
- 9.Kang HJ, Melnick AP, Stewart JD, Xu K, Rosenwaks Z. Preimplantation genetic screening: who benefits? Fertility and sterility. 2016;106:597–602. doi: 10.1016/j.fertnstert.2016.04.027. [DOI] [PubMed] [Google Scholar]
- 10.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 11.Mercer TR, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.May-Panloup P, et al. Low oocyte mitochondrial DNA content in ovarian insufficiency. Human reproduction. 2005;20:593–597. doi: 10.1093/humrep/deh667. [DOI] [PubMed] [Google Scholar]
- 13.Cummins JM. The role of mitochondria in the establishment of oocyte functional competence. European journal of obstetrics, gynecology, and reproductive biology. 2004;115(Suppl 1):S23–29. doi: 10.1016/j.ejogrb.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Houghton FD. Energy metabolism of the inner cell mass and trophectoderm of the mouse blastocyst. Differentiation. 2006;74:11–18. doi: 10.1111/j.1432-0436.2006.00052.x. [DOI] [PubMed] [Google Scholar]
- 15.Hewitson LC, Leese HJ. Energy metabolism of the trophectoderm and inner cell mass of the mouse blastocyst. The Journal of experimental zoology. 1993;267:337–343. doi: 10.1002/jez.1402670310. [DOI] [PubMed] [Google Scholar]
- 16.Dumollard R, Duchen M, Carroll J. The role of mitochondrial function in the oocyte and embryo. Current topics in developmental biology. 2007;77:21–49. doi: 10.1016/S0070-2153(06)77002-8. [DOI] [PubMed] [Google Scholar]
- 17.St John JC, Facucho-Oliveira J, Jiang Y, Kelly R, Salah R. Mitochondrial DNA transmission, replication and inheritance: a journey from the gamete through the embryo and into offspring and embryonic stem cells. Human reproduction update. 2010;16:488–509. doi: 10.1093/humupd/dmq002. [DOI] [PubMed] [Google Scholar]
- 18.Eichenlaub-Ritter U, Wieczorek M, Luke S, Seidel T. Age related changes in mitochondrial function and new approaches to study redox regulation in mammalian oocytes in response to age or maturation conditions. Mitochondrion. 2011;11:783–796. doi: 10.1016/j.mito.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Palini S, et al. Genomic DNA in human blastocoele fluid. Reproductive biomedicine online. 2013;26:603–610. doi: 10.1016/j.rbmo.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Gianaroli L, et al. Blastocentesis: a source of DNA for preimplantation genetic testing. Results from a pilot study. Fertility and sterility. 2014;102:1692–1699 e1696. doi: 10.1016/j.fertnstert.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 21.Tobler KJ, et al. Blastocoel fluid from differentiated blastocysts harbors embryonic genomic material capable of a whole-genome deoxyribonucleic acid amplification and comprehensive chromosome microarray analysis. Fertility and sterility. 2015;104:418–425. doi: 10.1016/j.fertnstert.2015.04.028. [DOI] [PubMed] [Google Scholar]
- 22.Ravichandran K, et al. Mitochondrial DNA quantification as a tool for embryo viability assessment: retrospective analysis of data from single euploid blastocyst transfers. Human reproduction. 2017;32:1282–1292. doi: 10.1093/humrep/dex070. [DOI] [PubMed] [Google Scholar]
- 23.Stigliani S, et al. Mitochondrial DNA in Day 3 embryo culture medium is a novel, non-invasive biomarker of blastocyst potential and implantation outcome. Molecular human reproduction. 2014;20:1238–1246. doi: 10.1093/molehr/gau086. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto S, et al. Quantitative and qualitative changes of mitochondria in human preimplantation embryos. Journal of assisted reproduction and genetics. 2017;34:573–580. doi: 10.1007/s10815-017-0886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Victor AR, et al. Accurate quantitation of mitochondrial DNA reveals uniform levels in human blastocysts irrespective of ploidy, age, or implantation potential. Fertility and sterility. 2017;107:34–42 e33. doi: 10.1016/j.fertnstert.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 26.Malonga H, Neault JF, Arakawa H, Tajmir-Riahi HA. DNA Interaction with human serum albumin studied by affinity capillary electrophoresis and FTIR spectroscopy. DNA Cell Biol. 2006;25:63–68. doi: 10.1089/dna.2006.25.63. [DOI] [PubMed] [Google Scholar]
- 27.Morbeck DE, et al. Composition of protein supplements used for human embryo culture. J Assist Reprod Genet. 2014;31:1703–1711. doi: 10.1007/s10815-014-0349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond ER, et al. Characterizing nuclear and mitochondrial DNA in spent embryo culture media: genetic contamination identified. Fertility and sterility. 2017;107:220–228 e225. doi: 10.1016/j.fertnstert.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 29.Leese HJ. Quiet please, do not disturb: a hypothesis of embryo metabolism and viability. BioEssays: news and reviews in molecular, cellular and developmental biology. 2002;24:845–849. doi: 10.1002/bies.10137. [DOI] [PubMed] [Google Scholar]
- 30.Reynier P, et al. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol Hum Reprod. 2001;7:425–429. doi: 10.1093/molehr/7.5.425. [DOI] [PubMed] [Google Scholar]
- 31.May-Panloup P, et al. Increase of mitochondrial DNA content and transcripts in early bovine embryogenesis associated with upregulation of mtTFA and NRF1 transcription factors. Reprod Biol Endocrinol. 2005;3:65. doi: 10.1186/1477-7827-3-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olivo PD, Van de Walle MJ, Laipis PJ, Hauswirth WW. Nucleotide sequence evidence for rapid genotypic shifts in the bovine mitochondrial DNA D-loop. Nature. 1983;306:400–402. doi: 10.1038/306400a0. [DOI] [PubMed] [Google Scholar]
- 33.Thundathil J, Filion F, Smith LC. Molecular control of mitochondrial function in preimplantation mouse embryos. Mol Reprod Dev. 2005;71:405–413. doi: 10.1002/mrd.20260. [DOI] [PubMed] [Google Scholar]
- 34.El Shourbagy SH, Spikings EC, St. Freitas M, John JC. Mitochondria directly influence fertilisation outcome in the pig. Reproduction. 2006;131:233–245. doi: 10.1530/rep.1.00551. [DOI] [PubMed] [Google Scholar]
- 35.Santos TA, St. El Shourbagy S, John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril. 2006;85:584–591. doi: 10.1016/j.fertnstert.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 36.McConnell JM, Petrie L. Mitochondrial DNA turnover occurs during development and can be modulated by environmental factors. Reprod Biomed Online. 2004;9:418–424. doi: 10.1016/S1472-6483(10)61277-1. [DOI] [PubMed] [Google Scholar]
- 37.Smith LC, Thundathil J, Filion F. Role of the mitochondrial genome in preimplantation development and assisted reproductive technologies. Reprod Fertil Dev. 2005;17:15–22. doi: 10.1071/RD04084. [DOI] [PubMed] [Google Scholar]
- 38.Cao L, et al. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nature genetics. 2007;39:386–390. doi: 10.1038/ng1970. [DOI] [PubMed] [Google Scholar]
- 39.Cree LM, et al. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet. 2008;40:249–24. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- 40.Chiaratti MR, et al. Embryo mitochondrial DNA depletion is reversed during early embryogenesis in cattle. Biology of reproduction. 2010;82:76–85. doi: 10.1095/biolreprod.109.077776. [DOI] [PubMed] [Google Scholar]
- 41.McConnell JM, Petrie L. Mitochondrial DNA turnover occurs during preimplantation development and can be modulated by environmental factors. Reproductive biomedicine online. 2004;9:418–424. doi: 10.1016/S1472-6483(10)61277-1. [DOI] [PubMed] [Google Scholar]
- 42.Zhou J, et al. Sequence variations of mitochondrial DNA D-loop region in patients with acute myeloid leukemia. Oncology letters. 2017;14:6269–6276. doi: 10.3892/ol.2017.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmid CW. Alu: structure, origin, evolution, significance and function of one-tenth of human DNA. Progress in nucleic acid research and molecular biology. 1996;53:283–319. doi: 10.1016/S0079-6603(08)60148-8. [DOI] [PubMed] [Google Scholar]
- 44.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nature reviews Genetics. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 45.Lu S, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hou Y, et al. Genome analyses of single human oocytes. Cell. 2013;155:1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 47.Wells D, et al. Clinical utilisation of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J. Med. Genet. 2014;51:553–562. doi: 10.1136/jmedgenet-2014-102497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shang W, et al. Comprehensive chromosomal and mitochondrial copy number profiling in human IVF embryos. Reproductive biomedicine online. 2018;36:67–74. doi: 10.1016/j.rbmo.2017.10.110. [DOI] [PubMed] [Google Scholar]
- 49.Diez-Juan A, et al. Mitochondrial DNA content as a viability score in human euploid embryos:less is better. Fertil Steril. 2015;104:534–41.e1. doi: 10.1016/j.fertnstert.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 50.Fragouli E, et al. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS Genet. 2015;11:e1005241. doi: 10.1371/journal.pgen.1005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardner, D. K. & Schoolcraft, W. B. In virto culture of human blastocyst. In: Janson, R., Mortimer, D. (Eds), Towards Reproductive Certainty: Infertility and Genetics Beyond 1999. Parthenon Press, Carnforth (1999).
- 52.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juanjuan Xu, et al. Noninvasive chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilization. PNAS. 2016;42:11907–11912. doi: 10.1073/pnas.1613294113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


