Abstract
Background
It is well known that hepatocellular carcinoma (HCC) has been one of the most life-threatening diseases all over the world. Plenty of internal and extrinsic factors have been proven to be related to HCC, such as gene mutation, viral hepatitis, and Nitrosamines. Though previous studies demonstrated that glutathione S-transferase (GST) genotypes are associated with HCC, the conclusions are inconsistent. Therefore, we carried on a renewed meta-analysis to expound the connection between the null GSTM1, GSTT1 polymorphisms and the risk of HCC.
Methods
We searched PubMed, Web of Science, Embase, and CNKI databases to select qualified researches which satisfied the inclusion criteria up to July 31, 2018. Finally, we selected 41 articles with 6124 cases and 9781 controls in this meta-analysis. We use ORs and 95% confidence interval (CI) to evaluate the correlation intension between the GSTM1 and GSTT1 null genes and the risk of HCC. All the statistical processes were executed by Stata (version 12.0).
Results
The pooled analysis showed that both GSTM1 null genotypes (OR = 1.37, 95% CI = 1.18–1.59) and GSTT1 null genotypes (OR = 1.43, 95% CI = 1.23–1.66) increased the risk of HCC. And GSTM1–GSTT1 dual-null genotypes also increased the risk of HCC (OR = 1.58, 95% CI = 1.22–2.05). In the subgroup analysis, we obtained significant results among Asians when stratified by race, and the results are GSTM1 null OR = 1.44, 95% CI = (1.22–1.71), GSTT1 null OR = 1.48, 95% CI = (1.25–1.77), GSTM1–GSTT1 null OR = 1.58, 95% CI = (1.19–2.09), while we didn’t obtain significant results among Caucasians or Africans. Stratified analyses on the type of control indicated a higher risk of HCC associated with GSTM1, GSTT1 single null genotypes and GSTM1–GSTT1 dual-null genotypes in healthy people. No evidence of significant connection was discovered in chronic liver disease (CLD) except in GSTT1 single null.
Conclusions
Our study indicated that an individual who carries the GSTM1, GSTT1 single null genotypes and GSTT1–GSTM1 dual-null genotypes is more likely to develop HCC.
Electronic supplementary material
The online version of this article (10.1186/s12935-019-0792-3) contains supplementary material, which is available to authorized users.
Keywords: Glutathione S-transferase, Hepatocellular carcinoma, GSTM1, GSTT1, Meta-analysis
Background
Hepatocellular carcinoma (HCC) has been one of the most common and terrifying cancer on account of a high mortality, which is turning into the third major cancer all over the world and is the second important factor of cancer-related deaths in China [1, 2]. It is evaluated that 70% to 90% liver cancers in the world are hepatocellular carcinoma. The pathogenesis of HCC is extremely complicated which including the internal and external factors, in other words, environmental conditions and individual genetic background both matters. Among those multiple risk factors, chronic infection with hepatitis B virus (HBV) has a strong relationship with the HCC prevalence. The high incidence rate of hepatocellular carcinoma in a certain district of Asia and Africa is prevailingly because of the high infection rate of chronic HBV, which follows the development of the path from hepatitis to liver cirrhosis and turns into HCC at last [3]. Besides, heavy consumption of alcohol and tobacco and family history of related tumor have been certified to have an intimate connection with a higher risk of HCC by numerous epidemiological and statistical researches [4–7].
When we talk about the internal factors, it is inevitable to study the effect of changes in genetic expression of certain genes or cytokines. The change of many signaling pathways and its molecules such as extracellular regulated protein kinases, antichymotrypsin, growth factor receptor-bound protein 2 (Grb2)—associated binding protein 2 have been found to be related to the blossom of HCC [8]. Among those expected genes, the GSTs have been a hot topic for its tremendous impact [9]. The GSTs play a crucial role in phase II isoenzymes which have tremendous defensive function because of its detoxication and regulating effect. Therefore, the decline of content of GSTs is highly possible to bring out carcinogenesis.
Among all the GSTs, the GSTM1 and GSTT1 have been extensively found related to various types of cancers [10]. Though former studies recognized the great value of GSTM1 and GSTT1 null genotypes, they did not meet a widely-agreed conclusion which could be generated by the limitation of sample size, statistical methods and other unsuspected reasons. For instance, certain studies included less than a few hundred HCC cases could lead to a conclusion which lacks credibility. To eliminate this interference factors in a better way, we therefore carried on a renewed meta-analysis to comprehensively evaluate the total harm of GSTM1 and GSTT1 null genotypes on the progress of HCC.
Methods
Identification and selection of relevant studies
We sought several databases including but not limited to PubMed, Web of Science, Embase, and CNKI for correlative researches up to July 31, 2018 for using various combination of those keywords: GSTM1, GSTT1, hepatocellular carcinoma, polymorphism and HCC.
Inclusion and exclusion criteria
We adopted the publications which assess the correlation of the GSTM1 and GSTT1 polymorphisms with HCC. Qualified studies must satisfy each requirement which was listed below: (1) must be a case–control study or a cohort study (2) have primitive and firsthand data and be independent from other researches, (3) have complete data to conduct statistical analysis.
Exclusion criteria are: (1) primitive and firsthand data are not available; (2) we excluded the studies which were based upon the same subjects and published by the same authors and selected the biggest samples or the latest data; (3) Additionally, meeting abstract, case reports, and other meta-analysis were not adopted.
Data extraction
Two participants independently extracted the data that we need cautiously from all qualified publications, and the related information included the first author, race of the samples, source of control, year of publication, respective number of each case and control, genotyping method, and type of control. Any potential inconsistency of the results was discussed and settled by consensus.
Statistical analysis
For GSTT1 and GSTM1, we respectively compared null genotype and normal genotype frequencies between cases and controls. And we calculated its ORs and 95% confidence interval (CI) to evaluate the correlation intension between the GSTM1 and GSTT1 null genes and the risk of HCC. Cochran’s Q statistic was used to assess heterogeneity and the I2 statistic to measure the inconsistency [11–13]. Subgroup analyses were executed by ethnicity (Asians, Africans, Caucasians), and type of control (healthy and chronic liver disease). We made Begg’s funnel plot to check whether there exists the potential publication bias. In order to measure the steadiness and reliability of our results, we carried out the sensitivity analyses by removing each single study and reanalyzing the rest to check the potential biases [14, 15]. All the statistical processes were executed by Stata (version 12.0). The P values were two-sided, and the P < 0.05 was taken for statistically significant. In addition, for GSTT1–GSTM1 both null, we use the same method to analyze the datum.
Results
Study characteristics
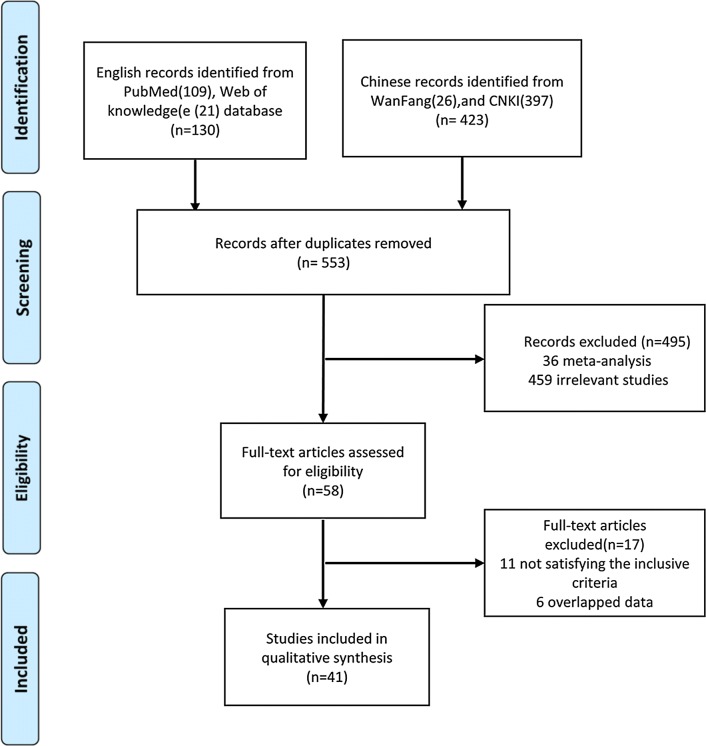
We have selected 553 correlative papers in term of the keywords from the databases, and 512 articles were removed for not meeting all the inclusion criteria. A total of 41 qualified papers were adopted in the end and all the studies obtained high-quality score according to the Newcastle–Ottawa Scale (NOS) (Table 1, Additional file 1: Table S1). Some articles included two groups in terms of a different type of control or source of control. We divided them into two studies to analyze them conveniently [16–20]. As a result, we got 46 studies and all the studies with 15905 objects for GSTM1 and 34 of them with 12589 objects for GSTT1, respectively [16–56]. The process of the selection of the literature was shown in Fig. 1. Thirty nine was involved in Asian, 4 was involved in Caucasian, 3 was involved in African among these studies. We exhibited the main characteristics of the studies and the main results in Tables 1 and 2, severally (more detailed information was showed in Additional file 1: Table S2).
Table 1.
Characteristics of the studies included in the meta-analysis
| Study | Year | Ethnicity | Source of control | No. of cases/controls | Genotyping method | Type of control | Study quality |
|---|---|---|---|---|---|---|---|
| Boccia [21] | 2015 | Caucasian | Hospital | 221/290 | RFLP-PCR | Healthy | 6 |
| Chen [22] | 2012 | Asian | Population | 76/68 | Multiplex PCR | Healthy | 8 |
| Tang [23] | 2012 | Asian | Population | 150/150 | RFLP-PCR | Healthy | 7 |
| Li [24] | 2012 | Asian | Population | 476/481 | PCR-CTPP | Healthy | 7 |
| Sarma [25] | 2012 | Asian | Hospital | 68/123 | PCR | CLD | 8 |
| Xiao [26] | 2011 | Asian | Hospital | 210/75 | PCR | Healthy | 8 |
| Wei [27] | 2010 | Asian | Population | 181/641 | PCR | Healthy | 7 |
| Kao [28] | 2010 | Asian | Population | 102/386 | PCR | Healthy | 8 |
| Asim [29] | 2010 | Asian | Hospital | 254/525 | Multiplex PCR | CLD | 8 |
| Yang [30] | 2009 | Asian | Population | 100/60 | Multiplex PCR | Healthy | 6 |
| Takeshi [16] | 2009 | Asian | Hospital | 209/256 | RFLP-PCR | Healthy | 8 |
| Takeshi [16] | 2009 | Asian | Hospital | 209/381 | RFLP-PCR | CLD | 8 |
| He [31] | 2008 | Asian | Population | 105/151 | Multiplex PCR | Healthy | 6 |
| Kiran [17] | 2008 | Asian | Hospital | 63/169 | RFLP-PCR | Healthy | 6 |
| Kiran [17] | 2008 | Asian | Hospital | 63/174 | RFLP-PCR | CLD | 6 |
| Elhamy [32] | 2008 | African | Hospital | 60/50 | PCR | CLD | 7 |
| Borentain [33] | 2007 | Caucasian | Population | 56/89 | ARMS-PCR | Healthy | 7 |
| Deng [34] | 2006 | Asian | Population | 88/115 | Multiplex PCR | Healthy | 6 |
| Long [35] | 2006 | Asian | Hospital | 257/649 | RFLP-PCR | CLD | 7 |
| Ladero [36] | 2006 | Caucasian | Population | 184/329 | Multiplex PCR | Healthy | 7 |
| Zhang [18] | 2005 | Asian | Population | 60/73 | Multiplex PCR | Healthy | 6 |
| Zhang [18] | 2005 | Asian | Hospital | 60/66 | Multiplex PCR | CLD | 6 |
| Guo [37] | 2005 | Asian | Population | 95/103 | Multiplex PCR | Healthy | 8 |
| Long [38] | 2005 | Asian | Population | 140/536 | PCR | Healthy | 8 |
| Deng [19] | 2005 | Asian | Population | 181/360 | PCR | Healthy | 7 |
| Deng [19] | 2005 | Asian | Population | 110/135 | PCR | Healthy | 7 |
| He [39] | 2005 | Asian | Population | 105/151 | Multiplex PCR | Healthy | 8 |
| Gelatti [40] | 2005 | Caucasian | Hospital | 200/400 | RFLP-PCR | Healthy | 7 |
| Kirk [41] | 2005 | African | Hospital | 149/294 | Multiplex PCR | Healthy | 7 |
| Ma [42] | 2005 | Asian | Population | 62/73 | PCR | CLD | 6 |
| Zhu [20] | 2005 | Asian | Population | 91/130 | Multiplex PCR | Healthy | 7 |
| Zhu [20] | 2005 | Asian | Hospital | 91/118 | Multiplex PCR | CLD | 7 |
| Li [43] | 2004 | Asian | Population | 207/207 | Multiplex PCR | Healthy | 8 |
| Yu [44] | 2003 | Asian | Hospital | 577/389 | RFLP-PCR | CLD | 7 |
| Liu [45] | 2003 | Asian | Population | 51/53 | RFLP-PCR | Healthy | 7 |
| Munaka [46] | 2003 | Asian | Hospital | 78/138 | Multiplex PCR | Healthy | 6 |
| Chen [47] | 2002 | Asian | Hospital | 93/35 | PCR | Healthy | 6 |
| Edine [48] | 2001 | African | Population | 112/194 | Multiplex PCR | Healthy | 6 |
| Sun [49] | 2001 | Asian | Hospital | 79/149 | PCR | CLD | 8 |
| Zhu [50] | 2001 | Asian | Population | 52/100 | Multiplex PCR | Healthy | 8 |
| Bian [51] | 2000 | Asian | Hospital | 63/88 | Multiplex PCR | Healthy | 8 |
| Wu [52] | 2000 | Asian | Population | 54/136 | Multiplex PCR | Healthy | 7 |
| Yu [53] | 1999 | Asian | Population | 88/375 | PCR | CLD | 7 |
| Dong [54] | 1997 | Asian | Population | 54/54 | PCR | Healthy | 8 |
| Dong [55] | 1997 | Asian | Population | 110/112 | PCR | Healthy | 8 |
| Yu [56] | 1995 | Asian | Hospital | 30/150 | PCR | CLD | 6 |
PCR polymerase chain reaction, RFLP restriction fragment length polymorphism, ARMS the amplification refractory mutation system, PCR-CTPP polymerase chain reaction with confronting two-pair primers, CLD chronic liver disease
Fig. 1.
The flow diagram of the meta-analysis, according to the PRISMA 2009. CNKI: China National Knowledge Infrastructure
Table 2.
Main results of the meta-analysis on the association between GSTM1 and GSTT1 null polymorphisms and HCC risk
| Null genotype | No. of studies | OR (95% CI) | P(Q) | I2 (%) | P(H) |
|---|---|---|---|---|---|
| GSTM1 | 46 | 1.37 (1.18–1.59) | < 0.001 | 77.2 | < 0.001 |
| Subgroup analysis | |||||
| Ethnicity | |||||
| Asian | 39 | 1.44 (1.22–1.71) | < 0.001 | 78.2 | < 0.001 |
| African | 3 | 1.31 (0.98–1.76) | 0.398 | 0 | 0.073 |
| Caucasian | 4 | 0.93 (0.77–1.13) | 0.312 | 16.0 | 0.476 |
| Type of control | |||||
| Healthy | 33 | 1.37 (1.17–1.59) | < 0.001 | 68.0 | < 0.001 |
| CLD | 13 | 1.36 (0.94–1.95) | < 0.001 | 87.3 | 0.099 |
| GSTT1 | 34 | 1.43 (1.23–1.66) | < 0.001 | 69.3 | < 0.001 |
| Subgroup analysis | |||||
| Ethnicity | |||||
| Asian | 28 | 1.48 (1.25–1.77) | < 0.001 | 71.9 | < 0.001 |
| African | 3 | 1.21 (0.77–1.90) | 0.122 | 52.4 | 0.404 |
| Caucasian | 3 | 1.19 (0.93–1.51) | 0.273 | 23.0 | 0.164 |
| Type of control | |||||
| Healthy | 24 | 1.42 (1.21–1.66) | < 0.001 | 58.2 | < 0.001 |
| CLD | 10 | 1.45 (1.02–2.08) | < 0.001 | 82.8 | 0.041 |
| GSTM1–GSTT1 | 19 | 1.58 (1.22–2.05) | < 0.001 | 46.1 | 0.001 |
| Subgroup analysis | |||||
| Ethnicity | |||||
| Asian | 17 | 1.58 (1.19–2.09) | < 0.001 | 64.1 | 0.002 |
| African | 1 | – | – | – | – |
| Caucasian | 1 | – | – | – | – |
| Type of control | |||||
| Healthy | 14 | 1.61 (1.17–2.22) | < 0.001 | 64.7 | 0.003 |
| CLD | 5 | 1.20 (0.94–1.53) | 0.179 | 36.3 | 0.153 |
GSTM1 glutathione S-transferase M1, GSTT1 glutathione S-transferase T1, CI confidence interval, OR odds ratio, CLD chronic liver disease, OR odds ratio, P(Q) P value of Q test; P(H) P value of heterogeneity
Correlation of GSTM1, GSTT1 single null genotype and GSTM1–GSTT1 double null polymorphism with HCC risk
In the conjoint analysis, we found an obvious association between the null genotype of GSTM1 and the HCC risk (OR = 1.37, 95% CI = 1.18–1.59, P < 0.001, Pheterogeneity < 10−3). When we conducted a subgroup analysis in term of race, we acquired the similar results in Asian (OR = 1.44, 95% CI = 1.22–1.71, P < 0.001, Pheterogeneity < 10−3) (Fig. 2a). However, we found no significant estimates on Caucasian (OR = 0.93, 95% CI = 0.77–1.13, P = 0.476, Pheterogeneity = 0.312) and African (OR = 1.31, 95% CI = 0.98–1.76, P = 0.073, Pheterogeneity = 0.398) populations (Fig. 2a, Table 2). Subgroup analyses in term of type of control showed a valuable result for healthy controls (OR = 1.37, 95% CI = 1.17–1.59, P < 0.001, Pheterogeneity < 10−3), but for CLD controls, we did not find any positive results (OR = 1.36, 95% CI = 0.94–1.95, P = 0.099, Pheterogeneity < 10−3) (Fig. 2b, Table 2).
Fig. 2.
Stratified analysis for the association between GSTM1 null genotype with hepatocellular carcinoma risk (a stratified analysis by ethnicity; b stratified analysis by type of control). CI confidence interval, OR odds ratio
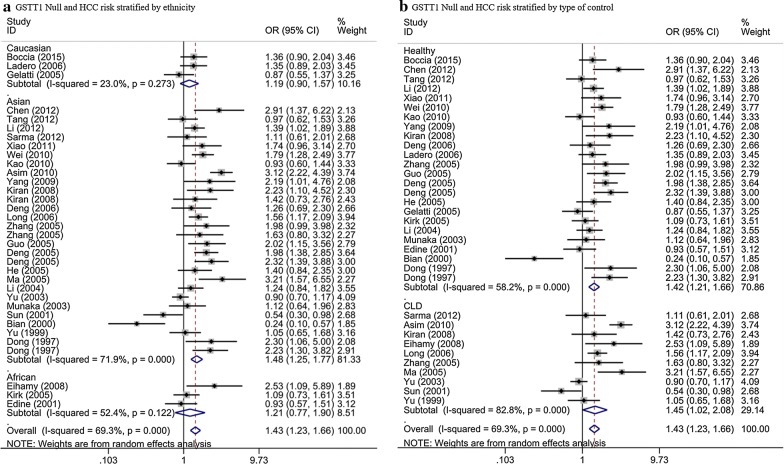
The results of 34 studies showed that GSTT1 null genotype increased the risk of HCC (OR = 1.43, 95% CI = 1.23–1.66, P < 0.001, Pheterogeneity < 10−3). When stratified by race, positive results were discovered among Asians (OR = 1.48, 95% CI = 1.25–1.77, P < 0.001, Pheterogeneity < 10−3) (Fig. 3a). Nevertheless, no connections were discovered among Caucasian (OR = 1.19, 95% CI = 0.93–1.51, P = 0.164, Pheterogeneity = 0.273) and African (OR = 1.21, 95% CI = 0.77–1.90, P = 0.404, Pheterogeneity = 0.122) (Fig. 3a). In the subgroup analysis based on type of control, we obtained positive results in healthy objects (OR = 1.42, 95% CI = 1.21–1.66, P < 0.001, Pheterogeneity < 10−3). In addition, we got similar conclusion in CLD controls (OR = 1.45, 95% CI = 1.02–2.08, P = 0.041, Pheterogeneity < 10−3) (Fig. 3b; Table 2).
Fig. 3.
Stratified analysis for the association between GSTT1 null genotype with hepatocellular carcinoma risk (a stratified analysis by ethnicity; b stratified analysis by type of control). CI confidence interval, OR odds ratio
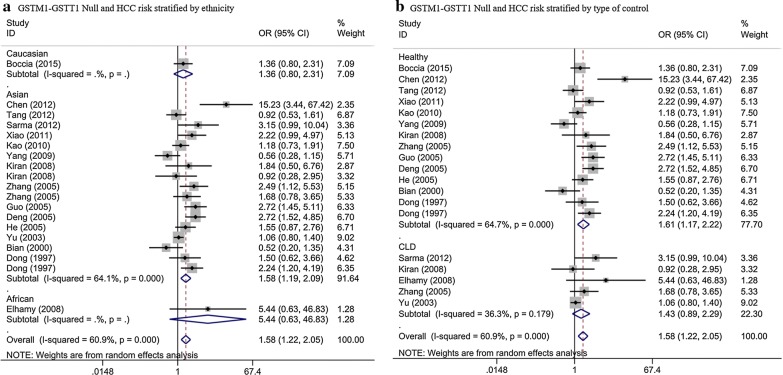
19 studies with 5063 subjects exhibited the connection between GSTM1 and GSTT1 double null and the risk of HCC. The result showed that GSTM1–GSTT1 double null genotype was related with enhanced risk of HCC (OR = 1.58, 95% CI = 1.22–2.05, P = 0.001, Pheterogeneity < 10−3). When stratified by race, positive results were found among Asians (OR = 1.58, 95% CI = 1.19–2.09, P = 0.002, Pheterogeneity < 10−3) (Fig. 4a) while no associations were found among African and Caucasian (Fig. 4a). When stratified by type of control, significant associations were found in healthy objects (OR = 1.61, 95% CI = 1.17–2.22, P = 0.003, Pheterogeneity < 10−3) (Fig. 4b). But for CLD controls, we did not find positive results (OR = 1.20, 95% CI = 0.94–1.53, P = 0.153, Pheterogeneity = 0.179) (Fig. 4b, Table 2).
Fig. 4.
Stratified analysis for the association between GSTM1 and GSTT1 double null genotype with hepatocellular carcinoma risk (a stratified analysis by ethnicity; b stratified analysis by type of control). CI confidence interval, OR odds ratio
Publication bias and sensitivity analysis
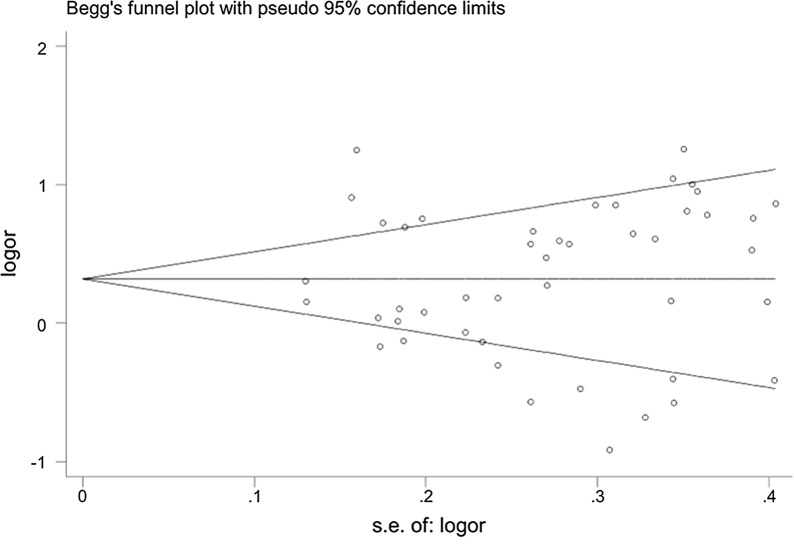
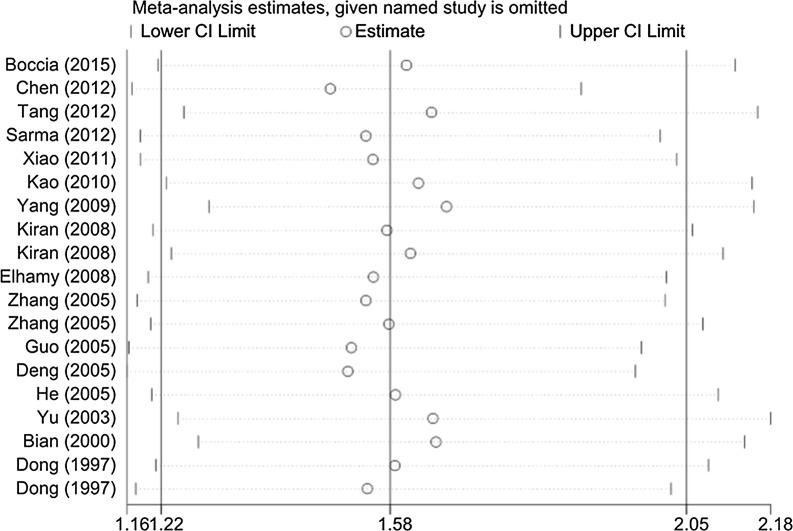
We utilized Begg’s and Egger’s test to detect the possible sources of bias. The funnel plots were symmetrical as showed in Fig. 5, indicating that there were no publication bias (P > 0.05). Sensitivity analysis was performed to measure the stability of the conclusive results, and we tell that there was no substantial transformation by removing any data set according to sensitivity analysis (Fig. 6). There were no small study effects among the adopted studies showed by Egger’s test.
Fig. 5.
Funnel plots of publication bias for GSTM1 null genotype
Fig. 6.
Sensitivity analysis of the associations between GSTM1 and GSTT1 double null genotype with hepatocellular carcinoma risk
Discussion
Previous studies already showed that the null GSTM1 and GSTT1 genotypes were likely to have an association with higher risk of a different type of cancers. The study by Wang zhao-yang et al. reported the null GSTT1 and null GSTM1 might have connection with higher death risk in gastric cancer [57]. Vesna M. Coric’s research showed that the carriers with GSTM1 null genotype is more likely to get renal cell carcinoma [58]. Neha Sharmaa et al. reported that the drop content of GST genotypes seemed to have close relationship with lung cancer and its histological subtypes [59]. There are also many studies focused on the relationship of GSTM1 and GSTT1 null genotypes and HCC risk, however, they did not meet a widely-agreed conclusion. Donna L. White’s study showed that the GSTM1 null and GSTT1 null genotype nearly had no connection with HCC [60]. Kang Song’s research showed that GSTM1 and GSTT1 null genotype may slightly increase the risk of HCC [61]. Ying-Hao Shen’s research found that the GSTM1 and GSTT1 null genotype was associated with an increased risk of HCC [62]. We therefore carried on this renewed meta-analysis to comprehensively evaluate the total risk of HCC when there exists GSTM1 and GSTT1 null genotypes. Our study includes 46 studies and 15905 objects in total compared to previous studies. Furthermore, we evaluated the risk of GSTM1–GSTT1 double null in HCC. Our results showed that the deficient of GSTM1 and GSTT1 genotype may be connected with a higher risk of HCC for its results is statistically significant. Compared to single null of GSTM1 and GSTT1 genotype, we furthermore discovered that individuals with dual genotypes of GSTs genes may be easier to develop HCC, but it still needs further exploration. The number of patients with hepatocellular carcinoma is large, and the diagnosis is late, also the prognosis is poor. These significant results may prompt us to conduct genetic testing on high-risk patients with hepatocellular carcinoma as early as possible, which plays an important role in early diagnosis, improvement of prognosis and reduction of the economic burden.
In the subgroup analysis in term of ethnicity, HCC risk was increased in Asians for single null of GSTM1 and GSTT1 and GSTM1–GSTT1 double-null genotype. However, we could not get the similar conclusion in Caucasians or Africans. This may be affected by diverse living surroundings and different inherited backgrounds. There may exist many other polymorphisms with combined effects that affect cancer susceptibility. As a consequence, the effect of certain individual polymorphism on the risk of cancer could be weakened. Our data also showed that the respectively null genotypes of GSTM1 and GSTT1 genotypes stand a good chance of having a duplicate effect on the HCC. When stratified by ethnicity, significant connection was found in Asian for GSTM1, but not in African and Caucasian. This could be explained by the potential possibility that the African and Caucasian may have certain unknown repair mechanism to avoid the damage caused by deficiency of the GSTM1 gene. It is said that the infection of HBV can easily evolve to liver cirrhosis and finally to HCC [63]. In addition, foodstuffs polluted with aflatoxin B1 (AFB1) are also a threat to livers. AFB1 is a kind of hepatotoxic mycotoxin. It could be produced by fungi of the Aspergillus species when the grains were stored in moist conditions. Once ingested, AFB1 was turned into an active intermediate, then produce certain a substance which may damage DNA [64]. Evidence from literature implies that the GSTs play an important role in the second metabolism phase of AFB1. Kimet al. reported that subjects which carried the null genotype of GSTT1 seem to increase the possibility of occurrence of chromosomal aberrations [65].
Despite our great efforts to check the association between the GSTs genes and the risk of HCC, we must pay attention to some limitations. Firstly, our analysis is a kind of cursory estimate since we had no chance to analyze each individual data. Secondly, the selected trials differ in certain aspects, including differences in trial populations and other potential diseases, and it is possible that methodological differences might have confused the conclusions of subgroups of trials. Thirdly, by including studies published a long time may affect the authenticity of the result.
Conclusions
All in all, our study showed that the single null of GSTM1, GSTT1 and GSTM1–GSTT1 double null genotypes are all connected with a higher risk of HCC. It is noteworthy that our data revealed that individuals with double-null genotypes of GSTs genes may be more likely to develop into HCC which worth further exploration. Last but not least, we need larger sample epidemiological studies of high quality to make current conclusions more reliable and stable.
Additional files
Additional file 1: Table S1. The evaluation of the data quality in this meta-analysis.
Additional file 2: Table S2. The detailed characteristics of the included studies in this meta-analysis.
Authors’ contributions
SL L, FX and SC conceived and designed the study. SL L, YZ, PT Y, SL, YJ D, PX, and LH Z collected and processed data. SL L, YZ, QH and ZZ analyzed data. SL L, YZ, YW and SL prepared tables and figures. SL L and YZ drafted the manuscript. ZJD and SC revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All the original data presented in additional files. Please contact authors for other data request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81471670), Science and Technology Plan of Innovation Project, Shaanxi Province, People’s Republic of China (No. 2015KTCL03-06), and the Key research and development plan, Shaanxi Province, China (2017ZDXM-SF-066).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- GSTM1
glutathione S-transferase M1
- GSTT1
glutathione S-transferase T1
- CI
confidence interval
- OR
odds ratio
- CLD
chronic liver disease
- PCR
polymerase chain reaction
- RFLP
restriction fragment length polymorphism
- ARMS
the amplification refractory mutation system
- PCR-CTPP
polymerase chain reaction with confronting two-pair primers
Contributor Information
Shanli Li, Email: 532469265@qq.com.
Feng Xue, Email: xue863386@126.com.
Yi Zheng, Email: 1079000475@qq.com.
Pengtao Yang, Email: 351249142@qq.com.
Shuai Lin, Email: voyage420@163.com.
Yujiao Deng, Email: 939477445@qq.com.
Peng Xu, Email: 164911802@qq.com.
Linghui Zhou, Email: 2544928065@qq.com.
Qian Hao, Email: 369182730@qq.com.
Zhen Zhai, Email: 907571680@qq.com.
Ying Wu, Email: 1468345892@qq.com.
Zhijun Dai, Email: dzj0911@126.com.
Shu Chen, Email: drchenshu@163.com.
References
- 1.Zheng R, Zeng H, Zhang S, Chen W. Estimates of cancer incidence and mortality in China, 2013. Chin J Cancer. 2017;36(1):66. doi: 10.1186/s40880-017-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei K, Peng X, Liang Z, Cen H. Liver cancer epidemiology world. China Cancer. 2015;24(08):621–630. [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: A Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Abdel-Rahman O, Helbling D, Schob O, Eltobgy M, Mohamed H, Schmidt J, Giryes A, Mehrabi A, Iype S, John H, et al. Cigarette smoking as a risk factor for the development of and mortality from hepatocellular carcinoma: an updated systematic review of 81 epidemiological studies. J Evid-Based Med. 2017;10(4):245–254. doi: 10.1111/jebm.12270. [DOI] [PubMed] [Google Scholar]
- 5.Yan G, Wang X, Sun C, Zheng X, Wei H, Tian Z, Sun R. Chronic alcohol consumption promotes diethylnitrosamine-induced hepatocarcinogenesis via immune disturbances. Sci Rep. 2017;7(1):2567. doi: 10.1038/s41598-017-02887-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassard AM, Gerard P, Perlemuter G. Microbiota, liver diseases, and alcohol. Microbiol Spect. 2017 doi: 10.1128/microbiolspec.bad-0007-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turati F, Edefonti V, Talamini R, Ferraroni M, Malvezzi M, Bravi F, Franceschi S, Montella M, Polesel J, Zucchetto A, et al. Family history of liver cancer and hepatocellular carcinoma. Hepatology. 2012;55(5):1416–1425. doi: 10.1002/hep.24794. [DOI] [PubMed] [Google Scholar]
- 8.Cheng J, Zhong Y, Chen S, Sun Y, Huang L, Kang Y, Chen B, Chen G, Wang F, Tian Y, et al. Gab2 mediates hepatocellular carcinogenesis by integrating multiple signaling pathways. FASEB J. 2017;31(12):5530–5542. doi: 10.1096/fj.201700120RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li R, Boerwinkle E, Olshan AF, Chambless LE, Pankow JS, Tyroler HA, Bray M, Pittman GS, Bell DA, Heiss G. Glutathione S-transferase genotype as a susceptibility factor in smoking-related coronary heart disease. Atherosclerosis. 2000;149(2):451–462. doi: 10.1016/s0021-9150(99)00483-9. [DOI] [PubMed] [Google Scholar]
- 10.Hayes JD, Strange RC. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology. 2000;61(3):154–166. doi: 10.1159/000028396. [DOI] [PubMed] [Google Scholar]
- 11.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai Z, Feng C, Zhang W, Liu J, Cao X, Zhang H, Liu Y, Wang M, Liu X, Dai Z. Lack of association between cytotoxic T-lymphocyte antigen-4 gene polymorphisms and lymphoid malignancy risk: evidence from a meta-analysis. Ann Hematol. 2016;95(10):1685–1694. doi: 10.1007/s00277-016-2753-4. [DOI] [PubMed] [Google Scholar]
- 13.Zhou L, Zheng Y, Tian T, Liu K, Wang M, Lin S, Deng Y, Dai C, Xu P, Hao Q, et al. Associations of interleukin-6 gene polymorphisms with cancer risk: evidence based on 49,408 cancer cases and 61,790 controls. Gene. 2018;670:136–147. doi: 10.1016/j.gene.2018.05.104. [DOI] [PubMed] [Google Scholar]
- 14.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 16.Imaizumi T, Higaki Y, Hara M, Sakamoto T, Horita M, Mizuta T, Eguchi Y, Yasutake T, Ozaki I, Yamamoto K, et al. Interaction between cytochrome P450 1A2 genetic polymorphism and cigarette smoking on the risk of hepatocellular carcinoma in a Japanese population. Carcinogenesis. 2009;30(10):1729–1734. doi: 10.1093/carcin/bgp191. [DOI] [PubMed] [Google Scholar]
- 17.Kiran M, Chawla YK, Kaur J. Glutathione-S-transferase and microsomal epoxide hydrolase polymorphism and viral-related hepatocellular carcinoma risk in India. DNA Cell Biol. 2008;27(12):687–694. doi: 10.1089/dna.2008.0805. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Deng C, Zhu Y. Study on genetic polymorphisms of xenobiotic metabolizing enzymes in hepatitis B virus-associated hepatic diseases. J Wenzhou Med Coll. 2005;35(06):464–467. [Google Scholar]
- 19.Deng Z, Wei Y, Ma Y. Genetic deletion of GSTM1 and GSTT1 detoxicated enzymes in relation to hepatocellular carcinoma in Guangxi. Guangxi Sciences. 2005;12(01):55–57. [Google Scholar]
- 20.Zhu M, Chen X, Zhou L. Association of genetic polymorphisms in glutathione S-transferases M1 with hepatitis B-related hepatocellular carcinoma. J Zhejiang Univ. 2005;34(02):35–39. doi: 10.3785/j.issn.1008-9292.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Boccia S, Miele L, Panic N. The effect of CYP GST, and SULT polymorphisms and their interaction with smoking on the risk of hepatocellular carcinoma. Biomed Res Int. 2015;2015:179867. doi: 10.1155/2015/179867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Ding F, Xie Y-A. Study on gene polymorphisms of glutathione S-transferase GSTM1 and GSTT1 in high-risk families for liver cancer in Fusui county, Guangxi. Chin J of Oncol Prev and Treat. 2012;4(02):140–144. [Google Scholar]
- 23.Tang Y, Li X, Liu T. A study of genetic polymorphisms of glutathione S-transferase in patients with hepatocellular carcinoma. Chin J Lab Diagn. 2012;16(04):660–662. [Google Scholar]
- 24.Li CG, Zhao ZM, Hu MG, Liu R. Predictive role of glutathione-S-transferase gene polymorphisms in risk and prognosis of hepatocellular carcinoma. Asian Pac J Cancer Prev APJCP. 2012;13(7):3247–3252. doi: 10.7314/apjcp.2012.13.7.3247. [DOI] [PubMed] [Google Scholar]
- 25.Sarma MP, Asim M, Medhi S, Bharathi T, Kar P. Hepatitis C virus related hepatocellular carcinoma: a case control study from India. J Med Virol. 2012;84(7):1009–1017. doi: 10.1002/jmv.23290. [DOI] [PubMed] [Google Scholar]
- 26.Xiao K, Li L, Peng M, Qin X, Peng T, Guo Y. Gene polymorphisms of GSTM1 and GSTT1 in the clustering families of hepatocellular carcinoma. Chin J Oncol Prev Treat. 2011;3(04):287–290. [Google Scholar]
- 27.Wei Y, Long X, Liu Z, Ma Y, Deng Z. Genetic polymorphism of glutathione-S-transferase M1 and T1 in hepatocellular carcinoma and nasopharyngeal carcinoma. Cancer Prev Res. 2010;37(10):1162–1165. [Google Scholar]
- 28.Kao CC, Chen MK, Kuo WH, Chen TY, Su SC, Hsieh YH, Liu CL, Chou MC, Tsai HT, Yang SF. Influence of glutathione-S-transferase theta (GSTT1) and micro (GSTM1) gene polymorphisms on the susceptibility of hepatocellular carcinoma in Taiwan. J Surg Oncol. 2010;102(4):301–307. doi: 10.1002/jso.21643. [DOI] [PubMed] [Google Scholar]
- 29.Asim M, Khan LA, Husain SA, Husain S, Sarma MP, Ahmad I, Sharma JK, Sharma A, Sharma S, Thayumanavan L, et al. Genetic polymorphism of glutathione S transferases M1 and T1 in Indian patients with hepatocellular carcinoma. Dis Markers. 2010;28(6):369–376. doi: 10.3233/DMA-2010-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z, Xie Y-A, Kuang Z, Luo X, Zhang W, Leng C. Relationship between genetic polymorphisms of glutathione-S-transferase M1, T1 genes and susceptibility to hepatocellular carcinoma in population of Fusui District of Guangxi Zhuang Autonomous Region. Chin J Caner Prev Treat. 2009;16(13):970–973. [Google Scholar]
- 31.He S, Gu Y, Liao Z. Relationship between GSTM1 gene polymorphism, tobacco and alcohol habits and susceptibility to primary liver cancer. J Guangxi Medi Univ. 2008;25(04):567–568. [Google Scholar]
- 32.Abd El-Moneim E, Younis FA, Allam N, Gameel K, Osman M. Gene deletion of glutathione S-transferase M1 and T1 and risk factors of hepatocellular carcinoma in Egyptian patients. Egypt J Immunol. 2008;15(2):125–134. [PubMed] [Google Scholar]
- 33.Borentain P, Gerolami V, Ananian P, Garcia S, Noundou A, Botta-Fridlund D, Le Treut YP, Berge-Lefranc JL, Gerolami R. DNA-repair and carcinogen-metabolising enzymes genetic polymorphisms as an independent risk factor for hepatocellular carcinoma in Caucasian liver-transplanted patients. Eur J Cancer. 2007;43(17):2479–2486. doi: 10.1016/j.ejca.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Deng W. The study of correlation between the polymorphism of GST genes and familial susceptibility to hepatocellular carcinoma in the high incidence area of Guangxi. Master: Guangxi Medical University; 2006. [Google Scholar]
- 35.Long XD, Ma Y, Wei YP, Deng ZL. The polymorphisms of GSTM1, GSTT1, HYL1*2, and XRCC1, and aflatoxin B1-related hepatocellular carcinoma in Guangxi population, China. Hepatol Res. 2006;36(1):48–55. doi: 10.1016/j.hepres.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Ladero JM, Martinez C, Garcia-Martin E, Ropero P, Briceno O, Villegas A, Diaz-Rubio M, Agundez JA. Glutathione S-transferase M1 and T1 genetic polymorphisms are not related to the risk of hepatocellular carcinoma: a study in the Spanish population. Eur J Cancer. 2006;42(1):73–77. doi: 10.1016/j.ejca.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 37.Guo H, Bian J, Jiang F, Wang Q, Zhang Z, Fan W, Wang Q, Zhu X, Tang B. The null genotypes ofGSTM1 and GSTT1 and the genetic susceptibility of prmiary liver cancer in Luoyang, China. Tumor. 2005;25(01):58–61. [Google Scholar]
- 38.Long X, Ma Y, Wei Y, Deng Z. A study about the association of detoxication gene GSTM1 polymorphism and the susceptibility to aflatoxin B1-related hepatocellular carcinoma. Chin J Hepatol. 2005;13(09):668–670. [PubMed] [Google Scholar]
- 39.He S, Qin J, Gu Y, Zhong W, Su S. Polymorphism analysis of GSTM1 and GSTT1 genes in patients with hepatocellular carcinoma. J Guangxi Med Univ. 2005;22(06):875–877. [Google Scholar]
- 40.Gelatti U, Covolo L, Talamini R, Tagger A, Barbone F, Martelli C, Cremaschini F, Franceschi S, Ribero ML, Garte S, et al. N-Acetyltransferase-2, glutathione S-transferase M1 and T1 genetic polymorphisms, cigarette smoking and hepatocellular carcinoma: a case-control study. Int J Cancer. 2005;115(2):301–306. doi: 10.1002/ijc.20895. [DOI] [PubMed] [Google Scholar]
- 41.Kirk GD, Turner PC, Gong Y, Lesi OA, Mendy M, Goedert JJ, Hall AJ, Whittle H, Hainaut P, Montesano R, et al. Hepatocellular carcinoma and polymorphisms in carcinogen-metabolizing and DNA repair enzymes in a population with aflatoxin exposure and hepatitis B virus endemicity. Cancer Epidemiol Biomarkers Prev. 2005;14(2):373–379. doi: 10.1158/1055-9965.EPI-04-0161. [DOI] [PubMed] [Google Scholar]
- 42.Ma D, Chen Y, Li Y, Zhao H, Xie X, Lu X. Glutathione-S-transferase M1 and T1 polymorphisms and susceptibility to liver cancer in hepatitis B surface antigen positive population. Med J Chin People’ s Armed Police Forces. 2005;16(09):660–662. [Google Scholar]
- 43.Li S, Wu J, Ding J, Gao C, Cao H, Zhou X. Impact of Genetic polymorphisms of glutathione S-transferase T1, M1 on the risk of primary hepatocellular carcinoma in alcohol drinkers. Pract J Cancer. 2004;19(03):229–232. [Google Scholar]
- 44.Yu MW, Yang SY, Pan IJ, Lin CL, Liu CJ, Liaw YF, Lin SM, Chen PJ, Lee SD, Chen CJ. Polymorphisms in XRCC1 and glutathione S-transferase genes and hepatitis B-related hepatocellular carcinoma. J Natl Cancer Inst. 2003;95(19):1485–1488. doi: 10.1093/jnci/djg051. [DOI] [PubMed] [Google Scholar]
- 45.Liu Z. Genetic polymorphism of GSTT1 and EPHX and the risk of Hepatocellular carcinoma from Guangxi district. Master: Guangxi Med Univ; 2003. [Google Scholar]
- 46.Munaka M, Kohshi K, Kawamoto T, Takasawa S, Nagata N, Itoh H, Oda S, Katoh T. Genetic polymorphisms of tobacco- and alcohol-related metabolizing enzymes and the risk of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2003;129(6):355–360. doi: 10.1007/s00432-003-0439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen SY, Wang LY, Lunn RM, Tsai WY, Lee PH, Lee CS, Ahsan H, Zhang YJ, Chen CJ, Santella RM. Polycyclic aromatic hydrocarbon-DNA adducts in liver tissues of hepatocellular carcinoma patients and controls. Int J Cancer. 2002;99(1):14–21. doi: 10.1002/ijc.10291. [DOI] [PubMed] [Google Scholar]
- 48.Tiemersma EW, Omer RE, Bunschoten A, van’t Veer P, Kok FJ, Idris MO, Kadaru AM, Fedail SS, Kampman E. Role of genetic polymorphism of glutathione-S-transferase T1 and microsomal epoxide hydrolase in aflatoxin-associated hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2001;10(7):785–791. [PubMed] [Google Scholar]
- 49.Sun CA, Wang LY, Chen CJ, Lu SN, You SL, Wang LW, Wang Q, Wu DM, Santella RM. Genetic polymorphisms of glutathione S-transferases M1 and T1 associated with susceptibility to aflatoxin-related hepatocarcinogenesis among chronic hepatitis B carriers: a nested case–control study in Taiwan. Carcinogenesis. 2001;22(8):1289–1294. doi: 10.1093/carcin/22.8.1289. [DOI] [PubMed] [Google Scholar]
- 50.Zhu W, Chen Q, Luo C, Chu X, Wu M. Relationship study between gene polymorphism of CYP1A1, GSTM1 and genetic susceptibility of primary hepatocellar carcinoma. China J Cancer Prev Treat. 2001;8(06):572–574. [Google Scholar]
- 51.Bian JC, Shen FM, Shen L, Wang TR, Wang XH, Chen GC, Wang JB. Susceptibility to hepatocellular carcinoma associated with null genotypes of GSTM1 and GSTT1. World J Gastroenterol. 2000;6(2):228–230. doi: 10.3748/wjg.v6.i2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu H, Chen M, Liu P, Zhang R. Relationship between GSTM1 gene polymorphism and genet ic susceptibility to primary hepatocellular carcinoma. Pract J Cancer. 2000;15(05):463–465. [Google Scholar]
- 53.Yu MW, Chiu YH, Chiang YC, Chen CH, Lee TH, Santella RM, Chern HD, Liaw YF, Chen CJ. Plasma carotenoids, glutathione S-transferase M1 and T1 genetic polymorphisms, and risk of hepatocellular carcinoma: independent and interactive effects. Am J Epidemiol. 1999;149(7):621–629. doi: 10.1093/oxfordjournals.aje.a009862. [DOI] [PubMed] [Google Scholar]
- 54.Dong C, Zi X, Yu S, Han J. Relationship between deletion of slutathion S-trausferase gene and susceptibility to primary hepatocellular carcinoma. Chin J Public Health. 1997;16(03):15–16. [Google Scholar]
- 55.Dong C, Yu S, Chen G, Deming Z, Fu Y. Polymorphisms of GSTT1 and M1 genotypes and their effects on elevated aflatoxin exposure and increased risk of hepatocellular carcinoma. Cancer Prev Res. 1997;24(06):327–329. [Google Scholar]
- 56.Yu MW, Gladek-Yarborough A, Chiamprasert S, Santella RM, Liaw YF, Chen CJ. Cytochrome P450 2E1 and glutathione S-transferase M1 polymorphisms and susceptibility to hepatocellular carcinoma. Gastroenterology. 1995;109(4):1266–1273. doi: 10.1016/0016-5085(95)90587-1. [DOI] [PubMed] [Google Scholar]
- 57.Wang ZY, Zhou J, Luo L, Huang YL, Dong PD. Predictive role of glutathione-S-transferase gene polymorphisms in the survival of gastric cancer cases. Asian Pac J Cancer Prev: APJCP. 2012;13(4):1515–1518. doi: 10.7314/apjcp.2012.13.4.1515. [DOI] [PubMed] [Google Scholar]
- 58.Coric VM, Simic TP, Pekmezovic TD, Basta-Jovanovic GM, Savic-Radojevic AR, Radojevic-Skodric SM, Matic MG, Suvakov SR, Dragicevic DP, Radic TM, et al. GSTM1 genotype is an independent prognostic factor in clear cell renal cell carcinoma. Urol Oncol. 2017;35(6):409–417. doi: 10.1016/j.urolonc.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 59.Sharma N, Singh A, Singh N, Behera D, Sharma S. Genetic polymorphisms in GSTM1, GSTT1 and GSTP1 genes and risk of lung cancer in a North Indian population. Cancer Epidemiol. 2015;39(6):947–955. doi: 10.1016/j.canep.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 60.White DL, Li D, Nurgalieva Z, El-Serag HB. Genetic variants of glutathione S-transferase as possible risk factors for hepatocellular carcinoma: a HuGE systematic review and meta-analysis. Am J Epidemiol. 2008;167(4):377–389. doi: 10.1093/aje/kwm315. [DOI] [PubMed] [Google Scholar]
- 61.Song K, Yi J, Shen X, Cai Y. Genetic polymorphisms of glutathione S-transferase genes GSTM1, GSTT1 and risk of hepatocellular carcinoma. PLoS ONE. 2012;7(11):e48924. doi: 10.1371/journal.pone.0048924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen YH, Chen S, Peng YF, Shi YH, Huang XW, Yang GH, Ding ZB, Yi Y, Zhou J, Qiu SJ, et al. Quantitative assessment of the effect of glutathione S-transferase genes GSTM1 and GSTT1 on hepatocellular carcinoma risk. Tumour Biol. 2014;35(5):4007–4015. doi: 10.1007/s13277-013-1524-2. [DOI] [PubMed] [Google Scholar]
- 63.Yuan BH, Li RH, Yuan WP, Xiang BD, Zheng MH, Yang T, Zhong JH, Li LQ. Perioperative entecavir for patients with HBV-related hepatocellular carcinoma and low levels of viral DNA: analysis using propensity score matching. Oncotarget. 2017;8(31):51810–51816. doi: 10.18632/oncotarget.15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chu YJ, Yang HI, Wu HC, Liu J, Wang LY, Lu SN, Lee MH, Jen CL, You SL, Santella RM, et al. Aflatoxin B1 exposure increases the risk of cirrhosis and hepatocellular carcinoma in chronic hepatitis B virus carriers. Int J Cancer. 2017;141(4):711–720. doi: 10.1002/ijc.30782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao SS, Chen XY, Zhu RZ, Choi BM, Kim SJ, Kim BR. Dual effects of phloretin on aflatoxin B1 metabolism: activation and detoxification of aflatoxin B1. BioFactors. 2012;38(1):34–43. doi: 10.1002/biof.190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The evaluation of the data quality in this meta-analysis.
Additional file 2: Table S2. The detailed characteristics of the included studies in this meta-analysis.
Data Availability Statement
All the original data presented in additional files. Please contact authors for other data request.