Abstract
The objective of this study was to evaluate the frequency of different extended-spectrum β-lactamases (ESBL) as well as to associate these ESBL with antimicrobial (ATM) resistance in Escherichia coli and Klebsiella spp. isolates from outpatients and inpatients with urinary tract infections. The study included 435 consecutive nonduplicate clinical isolates, including 362 E. coli isolates, 62 Klebsiella pneumoniae isolates, and 11 K. oxytoca isolates. Isolates were obtained from patients who were treated in a University Hospital between August 2012 and July 2013. Three multiplex PCR were performed to identify the ESBL groups. A total of 48 (11%) ESBL-producing isolates were found. The risk for the ESBL presence was significantly higher in males (26.4%) than females (8%), from hospital-acquired infections (29.1%) than community-acquired infections (7.0%) and in Klebsiella spp. (27.4%) than in E. coli (7.7%). ESBL-producing isolates presented a significantly higher percentage of resistance in 21 of the 23 ATMs analyzed. The CTX-M-1 group was the most predominant ESBL identified. The blaCTX-M-1-group gene was found in 56% of the total ESBL producers from community and in 42.4% from hospital origins; it was followed in frequency by the blaCTX-M-8/25-group, also found in both environments. Klebsiella spp. presented the largest variety of β-lactamase enzyme combinations and a higher level of resistance to cefotaxime. These findings contribute to better knowledge of the epidemiology of ESBL enzymes and are alarming for the reduced therapeutic options available for the risk groups identified in the studied populations.
Keywords: ESBL, antimicrobial resistance, Escherichia coli, multidrug resistance, Klebsiella pneumoniae
Introduction
Multidrug-resistant bacteria and the subsequent limitation of effective antimicrobial (ATM) therapies are a worldwide major health concern. The overuse and misuse of ATMs is identified as a significant driver in the emergence of ATM resistance.1 Escherichia coli and Klebsiella pneumoniae are the most common causes of urinary tract infections (UTIs) for both community-acquired and nosocomial infections.2 The World Health Organization has classified these extended-spectrum β-lactamase (ESBL)-producing species as a group of bacteria that has a major impact on public health around the world.3
ESBL are enzymes capable of, through hydrolysis, conferring bacterial resistance to penicillins, cephalosporins (first, second, and third generation), and aztreonam, but not cephamycins or carbapenems, and which can be inhibited by clavulanic acid and other β-lactamase inhibitors.4 The major ESBL types are TEM, SHV, CTX-M, and OXA. SHV and TEM ESBL types are derived from SHV-1 and TEM-1 β-lactamases, respectively.5 The CTX-M family is formed by five main groups differing by ≥10% in amino acid sequence identity: CTX-M-1-group, CTX-M-2-group, CTX-M-8-group, CTX-M-9-group, and CTX-M-25-group.6 The OXA-ESBL type has been found more frequently in Pseudomonas aeruginosa, but with its genes distributed on plasmids; OXA are currently found in several members of the Enterobacteriaceae family too.7,8
Despite worldwide distribution of ESBL, their epidemiology varies according to the geographic region. The enzymes of the CTX-M group currently overlap with other ESBL and have expanded beyond hospital borders.9 Due to plasmid dissemination, it is now a reality in outpatients10,11 and is strongly associated with other classes of ATM resistance.12 Besides, the presence of different β-lactamases has influenced the minimum inhibitory concentration of cephalosporins and consequent breakpoints to determine susceptibility profile by microdilution methods.13 The objective of this study was to evaluate the frequency of different ESBL as well as to associate the presence of ESBL with resistance to ATMs in E. coli and Klebsiella spp. isolates from outpatients and inpatients with UTI in a hospital in Southern Brazil.
Methodology
Study setting
This is a retrospective, observational and analytical study that included 435 consecutive nonduplicate clinical isolates, with 362 isolates being identified as E. coli and 73 as Klebsiella spp. (62 K. pneumoniae and 11 K. oxytoca), obtained from patients with UTI treated at the University Hospital of the Universidade Federal do Rio Grande (HU-FURG), Southern Brazil, from August 2012 to July 2013.
To identify patient features that were associated with ESBL-producing strains, medical records were reviewed, including age, gender, and origin of infection (community or hospital acquired). Hospital-acquired infection was defined as a UTI occurring within the first 72 hours after hospital admission or up to 72 hours after the patient was discharged from the hospital.14 Bacterial-related data were obtained with the EpiCenter software, version 6.20A (Phoenix System; BD™), of the clinical laboratory register and included bacteria species, ESBL production, and the isolates' susceptibility profiles to ATMs.
The study was conducted based on Resolution 466/12 of the National Health Council and was approved by the Research Ethics Committee of the Health Area of the HU-FURG under number 177/2013.
Bacterial collection
Ten microliters of urine samples was seeded on cystine lactose electrolyte deficient and MacConkey agar and incubated aerobically at 35°C for 18–24 hours. After Enterobacteriaceae isolation on MacConkey agar, the BD Phoenix™ automated microbiology system (Becton Dickinson Diagnostic Systems, Sparks, MD) was used for identification of bacterial species, ATM susceptibility profile, and phenotypic identification of ESBL producers according to the manufacturer's instructions. The clinical isolates were stored in Luria Bertani broth with 15% glycerol at −70°C in the Medical Microbiology Research Center, FURG (NUPEMM-FURG) bacterial collection.
Determination of minimal inhibitory concentration to cefotaxime
The minimal inhibitory concentration (MIC) of cefotaxime (CTX) of ESBL producers was determined by Broth Microdilution Method according to the CLSI M07-A9.15 Concentrations tested ranged between 0.5 and 512 mg/L. E. coli 25922 was used as control in the susceptibility test.
Molecular detection of ESBL
Total DNA extraction of all ESBL-producing E. coli and Klebsiella spp. isolates was obtained by heat treatment.16 The products were subjected to three distinct multiplex PCR as described previously.17 Reaction 1 is for the detection of ESBL of blaTEM-group, blaSHV-group, and blaOXA-group, reaction 2 is for the detection of blaCTX-M from groups 1, 2, and 9, and reaction 3 is for the identification of blaCTX-M-8/25-group. The oligonucleotides used in all reactions are in Table 1. HotStarTaq® (Qiagen) was used for reaction 1 and Platinum® Taq DNA Polymerase (Invitrogen, Life Technologies Brazil) for reactions 2 and 3. The positive controls used were blaTEM-1, blaSHV-2, and blaOXA-1 for reaction 1, blaCTX-M-16, blaCTX-M-2, and blaCTX-M-9 for reaction 2, and blaCTX-M-8 for reaction 3, all kindly provided by Dr. Jorge Luiz Mello Sampaio. E. coli ATCC 25922 DNA was used as negative control.
Table 1.
Oligonucleotide Sequences Used to Detect blaESBL Genes
| Multiplex reaction PCR | Name—prime | Sequence (5′–3′) |
|---|---|---|
| TEM—F | CATTTCCGTGTCGCCCTTATTC | |
| TEM—R | CGTTCATCCATAGTTGCCTGAC | |
| Reaction 1 | SHV—F | AGCCGCTTGAGCAAATTAAAC |
| SHV—R | ATCCCGCAGATAAATCACCAC | |
| OXA—F | GGCACCAGATTCAACTTTCAAG | |
| OXA—R | GACCCCAAGTTTCCTGTAAGTG | |
| CTX-M-1—F | TTAGGAARTGTGCCGCTGYAa | |
| CTX-M-1—R | CGATATCGTTGGTGGTRCCATa | |
| Reaction 2 | CTX-M-2—F | CGTTAACGGCACGATGAC |
| CTX-M-2—R | CGATATCGTTGGTGGTRCCATa | |
| CTX-M-9—F | TCAAGCCTGCCGATCTGGT | |
| CTX-M-9—R | TGATTCTCGCCGCTGAAG | |
| Reaction 3 | CTX-M-8/25—F | AACRCRCAGACGCTCTACa |
| CTX-M-8/25—R | TCGAGCCGGAASGTGTYATa |
Y = T or C; R = A or G; S = G or C.
ESBL, extended-spectrum β-lactamases; F, forward; R, reverse.
DNA viability of samples that did not present any bla gene amplification was confirmed by amplification of the chromosomal gene gyrA.18
Sequencing of the blaSHV and blaTEM genes
The differentiation of ESBL from penicillinases was carried out by the sequencing of blaSHV and blaTEM genes. After purification of the amplicons,19 the sequencing was accomplished using the ABI 3500 Genetic Analyzer (Life Technologies Technology—Applied Biosystems). Nucleotide sequences were aligned with TEM-1 (KT314169.1 K. pneumoniae subsp. pneumoniae strain GF4K3 beta-lactamase TEM-1 gene) and SHV-1 (JX268740.1 K. pneumoniae strain AKH11/46 beta-lactamase SHV-1) sequences, using the program “Mega” software version 5.2.2. SHV and TEM types were defined by comparing the amino acid sequences by BLAST on the website https://blast.ncbi.nlm.nih.gov/Blast.cgi. The SHV β-lactamases found were also classified according to a functional classification scheme by Bush et al.20
Three blaOXA-like genes, three blaCTX-M-1-group genes, three blaCTX-M-2-group genes, two blaCTX-M-8/25-group genes, and one blaCTX-M-9-group gene were sequenced to confirm that they were β-lactamases of their respective groups. The sequences of those genes were analyzed using “Blast” in https://blast.ncbi.nlm.nih.gov/Blast.cgi
Data analysis
Consistency analysis was performed through creating and categorizing variables and verifying frequencies in SPSS 17.0. A dependent variable was used as the presence of an ESBL producer, and the following independent variables were considered: demographic data (age and gender), infection source (hospital or community acquired), and bacterial species (E. coli or Klebsiella spp.). The χ2 test at a 95% significance level was used to compare proportions. After obtaining the prevalence ratio and their respective confidence intervals (CIs, 95% CI), the variables with a p-value ≤ 0.20 were included in the multivariate analysis. Poisson regression with robust adjustment of variance was used in the following hierarchical levels: age and gender (first level), and infection source and bacterial species (second level). Variables with p ≤ 0.05 were considered significant. The χ2 test of linear association was used to evaluate the association of ATM resistance with the presence of ESBL, a p ≤ 0.05 was considered statistically significant.
Results
Characterization of the patients and clinical isolates
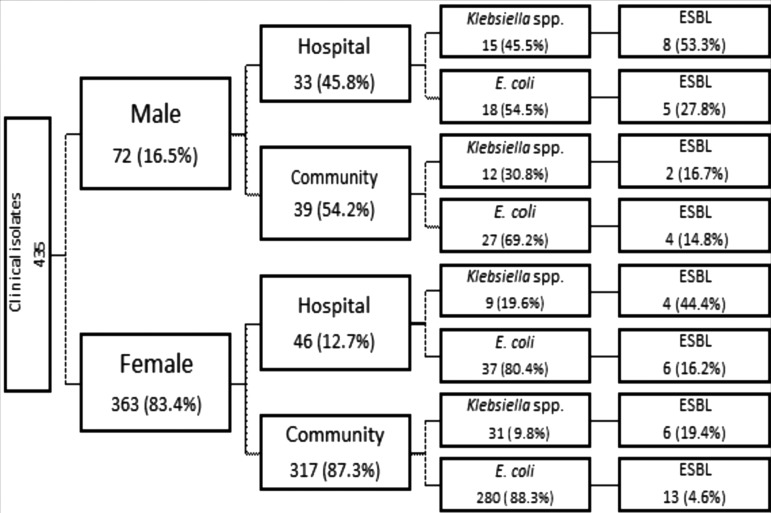
Of the 435 clinical isolates, 72 (16.5%) were obtained from male and 363 (83.5%) from female patients with UTI (Fig. 1). Of the 72 isolates from males, 33 (45.8%) were hospital acquired and 39 (54.2%) were community acquired; of the 363 isolates from females, 46 (12.7%) were hospital acquired and 317 (87.3%) were community acquired. Figure 1 shows the distribution of bacteria according to infection source and gender. In males, Klebsiella spp. represented 45.5% of the hospital-acquired isolates and 30.8% of the community-acquired isolates, while in females, 19.6% were hospital acquired and 9.8% were community acquired.
FIG. 1.
Distribution of the clinical isolates according to gender, infection source, type of Enterobacteria, and frequency of ESBL. ESBL, extended-spectrum β-lactamases.
Frequency of ESBL
A total of 48 (11%) isolates were identified as ESBL producers, 26 from outpatients and 22 from inpatients (Fig. 1). The risk for the ESBL presence was significantly higher in males (26.4%) than females (8%) (p = 0.047), from hospital-acquired infections (29.1%) than community-acquired infections (7.0%) (p = 0.004), and in Klebsiella spp. (27.4%) than in E. coli (7.7%) (p = 0.002) (Table 2). In relation to age of the patients, the risk of ESBL was progressive according to population age, but there was no significant association between the three age ranges examined (Table 2).
Table 2.
Frequency and Adjusted Analysis of Associated Factors to Extended-Spectrum β-Lactamases Producers
| Categories | Total number | ESBL n (%) | Adjusted analysis PR (CI95%) | p-Value |
|---|---|---|---|---|
| Age | ||||
| ≥61 | 173 | 25 (14.5) | 1.0 | |
| 41–60 | 120 | 15 (12.5) | 0.83 (0.39–1.74) | 0.628 |
| 21–40 | 96 | 6 (6.2) | 0.54 (0.20–1.46) | 0.230 |
| Gender | ||||
| Female | 363 | 29 (8.0) | 1.0 | |
| Male | 72 | 19 (26.4) | 2.15 (1.01–4.58) | 0.047* |
| Origin | ||||
| Community | 356 | 25 (7.0) | 1.0 | |
| Hospital | 79 | 23 (29.1) | 2.93 (1.42–6.05) | 0.004* |
| Bacteria | ||||
| Escherichia coli | 362 | 28 (7.7) | 1.0 | |
| Klebsiella spp. | 73 | 20 (27.4) | 3.07 (1.50–6.25) | 0.002* |
Statistically significant.
PR, prevalence ratio; CI, confidence interval.
Antimicrobial resistance associated with the presence of ESBL
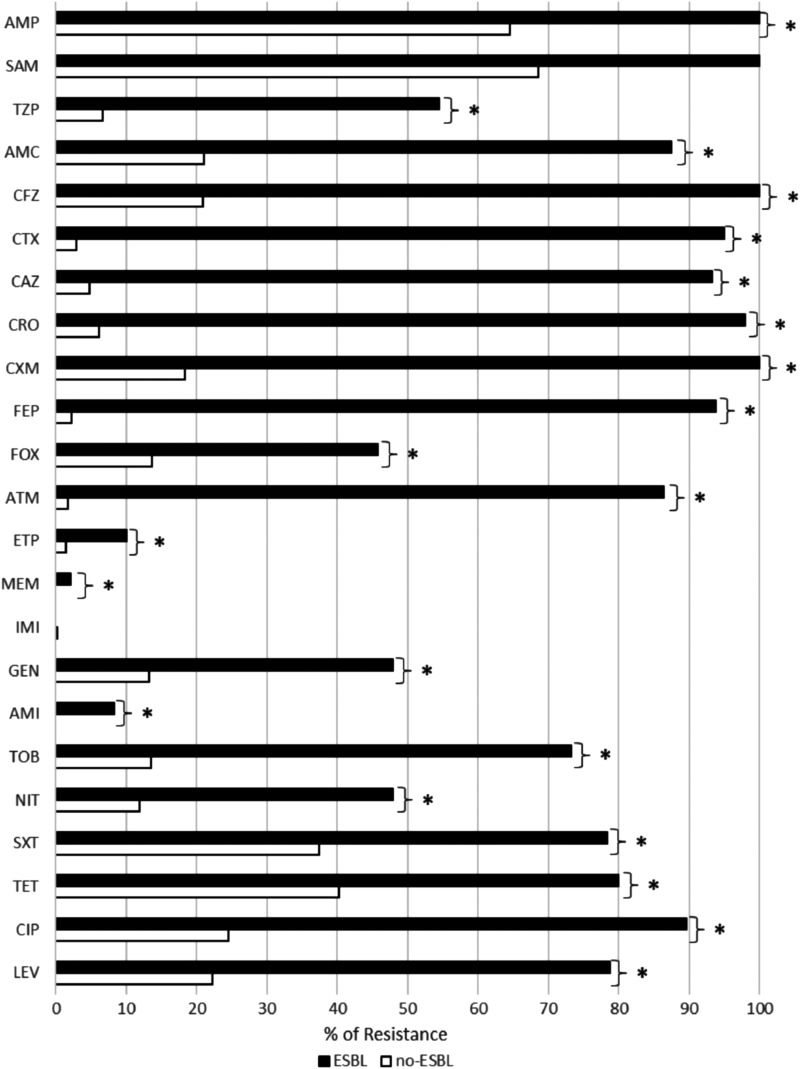
ESBL-producing isolates presented a significantly higher percentage of resistance to 21 of the 23 agents tested (Fig. 2), including resistance to penicillins (AMP, TZP, and AMC), cephalosporins (CFZ, CXM, CAZ, CRO, and CTX e FEP), ATM, fluoroquinolones (CIP and LEV), aminoglycosides (AMI, GEN, and TOB), and carbapenems (ETP and MEM), FOX, NIT, SXT, and TET. However, the resistance rate to imipenem and ampicillin with sulbactam was not significantly in relation to non-ESBL-producing isolates (Fig. 2).
FIG. 2.
Resistance to antimicrobials in ESBL (black bar)- and non-ESBL (white bar)-producing isolates. *Indicates a significant difference between ESBL and non-ESBL with p ≤ 0.05. AMP, ampicillin; SAM, ampicillin with sulbactam; TZP, piperacillin with tazobactam; AMC, amoxicillin with clavulanic acid; CFZ, cefazolin; CTX, cefotaxime; CAZ, ceftazidime; CRO, ceftriaxone; CXM, cefuroxime; FEP, cefepime; FOX, cefoxitin; ATM, aztreonam; ETP, ertapenem; MEM, meropenem; IMI, imipenem; GEN, gentamicin; AMI, amikacin; TOB, tobramycin; NIT, nitrofurantoin; SXT, sulfamethoxazole/trimethoprim; TET, tetracycline; CIP, ciprofloxacin; LEV, levofloxacin.
Molecular characterization of ESBL
Table 3 shows the frequency of ESBL genes found in the 48 ESBL producers, of which the blaCTX-M-1-group was the most predominant in E. coli as well as in Klebsiella spp. This gene was found in 56% of the total ESBL producers from community origin and in 42.4% from hospital origin; it was followed in frequency by the blaCTX-M-8/25-group, also found in both environments.
Table 3.
Frequency of Extended-Spectrum β-Lactamases Gene Families Found in Escherichia coli and Klebsiella spp. Clinical Isolates
| E. coli n (%) | Klebsiella spp. n (%) | Total n (%) | |||
|---|---|---|---|---|---|
| Genes | Community17 (100) | Hospital11 (100) | Community8 (100) | Hospital12 (100) | 48 (100) |
| blaCTX-M-1-group | 8 (47.1) | 6 (54.5) | 6 (75.0) | 8 (66.7) | 28 (58.3) |
| blaCTX-M-2-group | 1 (5.9) | 1 (9.1) | 0 (0.0) | 0 (0.0) | 2 (4.2) |
| blaCTX-M-8/25-group | 5 (29.4) | 2 (18.2) | 1 (12.5) | 5 (41.6) | 13 (27.1) |
| blaCTX-M-9-group | 1 (5.9) | 0 (0.0) | 1 (12.5) | 0 (0.0) | 2 (4.2) |
| blaSHV-2a | 1 (5.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (2.1) |
| blaSHV-11 | 0 (0.0) | 0 (0.0) | 1 (12.5) | 1 (8.3) | 2 (4.2) |
| blaSHV-26 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (8.3) | 1 (2.1) |
| blaESBL no detected | 1 (5.9) | 2 (18.2) | 0(0.0) | 0 (0.0) | 3 (6.3) |
Among the genes of the blaCTX-M family, at lower frequency, two E. coli isolates presented the blaCTX-M-2-group and one E. coli and one Klebsiella spp. isolate presented the blaCTX-M-9-group. In relation to the blaSHV family, after sequencing of the 14 blaSHV genes, the blaSHV-2a genotype was found in one E. coli isolate, the blaSHV-11 in two Klebsiella spp. isolates, and the blaSHV-26 in one Klebsiella spp. isolate. In two isolates of ESBL-producing E. coli, none of studied genes was found.
Among isolates that presented the blaSHV gene, there were four with SHV enzymes presenting phenotype (2br or 2be) consisting of one SHV-2a, two SHV-11, and one SHV-26. Sample A27 showed a single glutamine to histidine change at amino acid 150, which did not allow for characterization of the SHV enzyme type (Table 4). The blaTEM gene was detected in 13 isolates, being identified as encoding the TEM-1 penicillinase, and none was characterized as encoding ESBL.
Table 4.
Characterization of Isolates Presenting the blaSHV Gene with Mutation(s) in Relation to the Penicillinase blaSHV-1 Gene
| Sample | Bacteria | ESBL by Phoenix | blaSHV type | Mutation (aa) | Phenotypea | Other genes (bla) found |
|---|---|---|---|---|---|---|
| A3 | Klebsiella spp. | Yes | SHV-26 | A183T | 2br | CTX-M-8/25-group and TEM-1 |
| A27 | Klebsiella spp. | Yes | SHV | Q150H | — | CTX-M-9-group |
| A29 | Klebsiella spp. | Yes | SHV-11 | L31Q | 2be | CTX-M-8/25-group and TEM-1 |
| A43 | Klebsiella spp. | Yes | SHV-11 | L31Q | 2be | CTX-M1-group and TEM-1 |
| A109 | E. coli | Yes | SHV-2a | L31Q and G234S | 2be | TEM-1 |
2br = β-lactamase resistant to inhibitors and 2be = ESBL.
Mutation (aa): A, alanine; T, threonine; Q, glutamine; H, histidine; L, leucine; G, glycine; S, serine.
Several clinical isolates presented multiple genes encoding ESBL. Table 5 shows the combination of genes found in the 48 ESBL-producing strains, including the non-ESBL genes blaOXA, blaTEM-1, and blaSHV-1, which are penicillinases. The blaOXA was always found in presence of the blaCTX-M-1-group (Table 5). In E. coli, this combination was found in all 14 clinical isolates. For Klebsiella spp., the combination of blaCTX-M-1-group and blaOXA was also found, as well as the combination with blaSHV-1 and/or blaTEM-1. Table 5 shows that the majority of cases of blaSHV-1 β-lactamase found also presented the gene blaCTX-M-1-group, except for one isolate that presented only the gene responsible for the TEM-1 β-lactamase.
Table 5.
Profile of bla Genes Found in ESBL-Producing Escherichia coli and Klebsiella spp. and the MIC to CTX
| Combination of genes (bla) | E. coli n (%) | Range MIC (mg/L) | Klebsiella spp. n (%) | Range MIC (mg/L) |
|---|---|---|---|---|
| CTX-M-1-group | — | 1 (5%) | 256 | |
| CTX-M-1-group + OXA-1-like | 14 (50%) | 8–32; 256a | 4 (20%) | 256–512 |
| CTX-M-1-group + OXA-1-like + SHV-1 | — | 2 (10%) | 256–512 | |
| CTX-M-1-group + OXA-1-like + SHV-1 + TEM-1 | — | 4 (20%) | 256–512 | |
| CTX-M-1-group + SHV-11 + TEM-1 | — | 1 (5%) | NP | |
| CTX-M-1-group + SHV-1 | — | 1 (5%) | 256 | |
| CTX-M-2-group | 2 (7.1%) | >512 | — | |
| CTX-M-8/25-group | 6 (21.4%) | 4–32 | — | |
| CTX-M-8/25-group + TEM-1 | 1 (3.6%) | 32 | 2 (10%) | 256 |
| CTX-M-8/25-group + SHV-1 + TEM-1 | — | 1 (5%) | NP | |
| CTX-M-8/25-group + CTX-M-1-group + OXA-1-like + SHV-1 | — | 1 (5%) | 64 | |
| CTX-M-8/25-group + SHV-11 + TEM-1 | — | 1 (5%) | 32 | |
| CTX-M-8/25-group + SHV-26 + TEM-1 | — | 1 (5%) | 256 | |
| CTX-M-9-group | 1 (3.6%) | 64 | — | |
| CTX-M-9-group + SHVb | — | 1 (5%) | NP | |
| TEM-1 | 1 (3.6%) | 8 | — | |
| SHV-2a + TEM-1 | 1 (3.6%) | NP | — | |
| Not detected | 2 (7.2%) | 64 | — | |
| Total | 28 (100%) | 20 (100%) |
Value of only one isolate.
Indicates the protein contained a change of glutamine to histidine at position 150 (mutation Q150H). This was not enough to identify the β-lactamase type.
MIC, minimal inhibitory concentration; NP, not performed.
Of the 48 ESBL-producing isolates, the MIC for CTX was determined in 40 (23 E. coli and 17 Klebsiella spp.), while 8 isolates presented a loss of viability during the study. In E. coli, for most of the isolates presenting the blaCTX-M-1-group, blaCTX-M-8/25-group, or blaCTX-M-9-group genes, as well as the three samples in which blaESBL genes were not detected (including blaTEM-1 alone), the MIC ranged from 4 to 64 mg/L. The two exceptions were the blaCTX-M-1-group isolate that had an MIC of 256 mg/L and the blaCTX-M-2-group isolate that had an MIC ≥512 mg/L (Table 5). Klebsiella spp. presented MIC ranging from 256 to 512 mg/L in isolates with the blaCTX-M-1-group and 32 to 256 mg/L in isolates with the blaCTX-M-8/25-group (Table 5). Figure 3 shows that Klebsiella spp. presented higher MIC to CTX than E. coli.
FIG. 3.
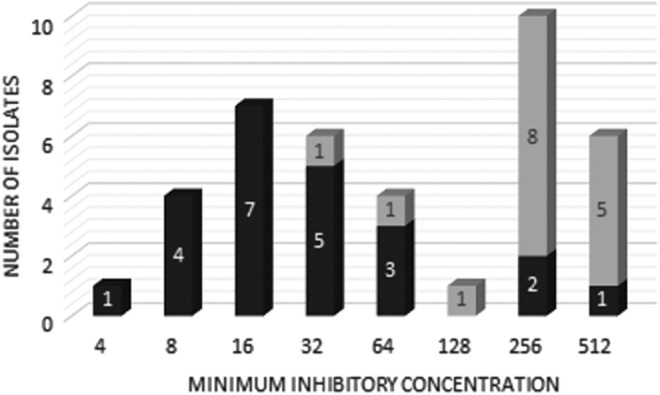
Minimal inhibitory concentration (mg/L) to cefotaxime of ESBL-producing Escherichia coli and Klebsiella spp. Dark gray bar: E. coli; light gray bar: Klebsiella spp.
Discussion
In this study, the frequency of E. coli and Klebsiella spp. producing ESBL was 11%; however, 7.0% of community- and 29.1% of hospital-acquired infections were ESBL producers. were hospital-acquired infections. Regarding the community-acquired origin, the frequency in this study was similar to other studies in different regions of Brazil, with percentages ranging from 7.1% to 7.6%,21,22 and elevated in comparison with developed countries such as France (2.4%) and Sweden (2.8%).23 Nevertheless, ESBL was more associated with nosocomial infections (2.9-fold higher risk than in community-acquired infections), male gender, and Klebsiella spp. infections. Previous studies that compared the frequency of ESBL from inpatients and outpatients treated in the same hospital also showed a predominance of ESBL producers in hospital-acquired infections.24 This environment is where ESBL originally emerged and where more third-generation cephalosporins are consumed, which are often used to treat complicated UTI.25 In addition, men were more prone to develop hospital-acquired UTI and consequently more at risk for acquisition of ESBL producers than women. In our study, the relationship can be supported since 45.8% of UTI in males were hospital acquired, while in females, the percentage was only 12.7%.
With ESBL producers presenting a significantly higher frequency of resistance to 21 of 23 ATMs tested (except to imipenem and ampicillin with sulbactam), appropriate treatment is limited, and the identification of risk groups for ESBL is critical. Aside from fourth-generation cephalosporins, aztreonam and ampicillin, ESBL presented resistance rates in over 50% of cases to fluoroquinolones (LEV and CIP), tetracycline (TET), sulfa-trimethoprim (SXT), aminoglycoside (TOB), and beta-lactams associated with beta-lactamase inhibitor (AMC, TZP, and SAM). The associated resistance to these ATMs has been shown to be related to the presence of a conjugative plasmid that has accumulated genes that confer resistance. These genes can include rmtA to rmtD-, armA-, and npmA-encoded 16S rRNA methylases associated with aminoglycoside resistance,26 the blaampC gene encoding the AmpC enzyme, resulting in resistance to the β-lactamase inhibitor-β-lactam combination27; and qnr, qepA, and aac(6′)-Ib-cr genes associated with resistance to some fluoroquinolones.25 Recently, our group showed that the presence of ESBL producers in UTI isolates increased the risk to fluoroquinolone resistance 11-fold,28 and we found an elevated presence of the aac(6′)-Ib-cr gene associated with chromosomal mutations in gyrA and parC, which encode the main targets of fluoroquinolones in E. coli and Klebsiella spp.29 Fortunately, in our study, carbapenems and amikacin displayed activity over 90% in ESBL producers, allowing these ATMs to remain as options for the treatment of complicated UTI. However, it is important to ration their use, since the resistance rates are increasing in Enterobacteriaceae members, especially in K. pneumoniae. This emergence is mainly associated with the spread of plasmids containing genes encoding carbapenemases and has resulted in outbreaks in several countries, including the United States and China.30
The profile of multidrug resistance associated with ESBL has previously been shown in E. coli and K. pneumoniae to involve the presence of acquired blaCTX-M genes, which are carried on conjugative plasmids.31 Actually, in our study, CTX-M was the major ESBL family, identified in 85.6% of E. coli and in 100% of Klebsiella spp. Among the CTX-M family, the predominant group was CTX-M-1, followed by CTX-M-8/25, and in lower frequency, the groups CTX-M-2 and CTX-M-9. Worldwide, the CTX-M-1 is the most widely disseminated ESBL group in the clinical settings, mainly for pandemic E. coli clones bearing the CTX-M-15 type.12 In relation to the CTX-M-2-group, our results differ from a systematic review of the spread of CTX-M in Brazil that determined the prevalent groups are CTX-M-1 and CTX-M-2.32 While we found a rate of 4.1% of CTX-M-2-group, the rate of CTX-M-2-group reached nearly 60% in some studies from southern and southeastern Brazil.33,34 Previous reports from Brazil and Spain determined that CTX-M-9 was more prevalent in Spain,35,36 as well as the CTX-M-2-group; we found a low frequency of the CTX-M-9 group (4.1%), but outside of Brazil, its frequency ranges from 11.0% to 24.2%.34,37 Finally, in relation to CTX-M-8/25 groups, the primer pair can amplify the bla gene of both groups, but 2 of the 13 isolates detected were sequenced and showed similarity with the CTX-M-8 type, which belongs to CTX-M-8-group. The first report of CTX-M-8 was in Brazil,38 and it has subsequently been found at a frequency ∼12% in E. coli ESBL producers from a different region of Brazil,33,39 which is lower than the 25% rate that was found in our study. In addition, in Klebsiella spp., the frequency of this group was 30%, demonstrating its successful spread in both Enterobacteria species.
The detection of the blaCTX-M-group was successfully performed by multiplex PCR.17 This tool could be useful for rapid epidemiological studies screening for blaCTX-M groups. However, the multiplex PCR to detect blaSHV, blaTEM, and blaOXA had limited efficacy, since the presence of amplified PCR product did not differentiate between ESBL or simply penicillinases. For blaSHV and blaTEM, it was necessary to sequence the amplified product to differentiate ESBL from penicillinases (e.g., TEM-1 or 2 and SHV-1). The blaSHV presented at frequency of 8.3% of the ESBL in the studied population, and this lower frequency of SHV is in agreement with a previous study.40 This result demonstrates an inversion in the prevalence of CTX-M enzymes on TEM and SHV. Although two samples did not show any gene that was screened for and one showed just penicillinase TEM-1, these bacteria should contain another ESBL family that was not studied in this work. Some possible ESBL would be the PER, GES, and BES enzymes, which have already been detected in Brazil.33,38,41
Due to variation in the combination of genes encoding β-lactamase enzymes, no statistical analysis was able to be performed to associate MICs with ESBL enzymes. Nevertheless, it was possible to observe that Klebsiella spp. presented higher levels of resistance to CTX than E. coli. Klebsiella spp. also contained more combinations of genes, such as the presence of penicillinases TEM-1, SHV-1, SHV-derived-ESBL, and CTX-M groups. The variation in MIC may indirectly represent the levels of expression of the genes involved,42 but they may also represent the involvement of other resistance mechanisms to CTX, such as efflux pumps, lower permeability due to the reduction of porins, or the production of AmpC.43–45
This study showed a relevant frequency of ESBL principally in Klebsiella spp. isolated from male gender with UTI of hospital origin. CTX-M was the major family of ESBL identified, with a higher frequency of the CTX-M-1-group, but the CTX-M-8/25 groups were also present. Klebsiella spp. also presented more combinations of β-lactamase enzymes and higher levels of resistance to CTX. The strong association with different ATM classes, but low resistance to carbapenems, shows that despite having therapeutic options for ESBL producers, it is necessary to ration ATM use to control the spread of ESBL producers in the studied populations.
Acknowledgments
We would like to thank Dr. Jorge Luiz Mello Sampaio for kindly providing strains as positive controls and our colleague Lourdes Helena Rodrigues Martins for the support with the bench work. E. coli INCQS 00033—(ATCC 25922) was courtesy of Coleção de Microrganismos de Referência em Vigilância Sanitária-CMRVS, FIOCRUZ-INCQS, Rio de Janeiro, RJ.
Disclosure Statement
No competing financial interests exist.
References
- 1. Holmes A.H., Moore L.S.P., Sundsfjord A., Sundsfjord A., Steinbakk M., Regmi S., Karkey A., Guerin P.J, and Piddock L.J. 2016. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387:176–187 [DOI] [PubMed] [Google Scholar]
- 2. Foxman B. 2014. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect. Dis. Clin. North Am. 28:1–13 [DOI] [PubMed] [Google Scholar]
- 3. World Health Organization (WHO). 2014 Antimicrobial resistance: global report on surveillance. Available at www.who.int/drugresistance/documents/surveillancereport/en/ (accessed September4, 2017).
- 4. Paterson D.L., and Bonomo R.A. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradford P.A. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shaikh S., Fatima J., Shakil S., Rizvi S.M.D., and Kamal M.A. 2015. Antibiotic resistance and extended spectrum beta-lactamases: types, epidemiology and treatment. Saudi J. Biol. Sci. 22:90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans B.A., and Sebastian G.B. 2014. OXA β-lactamases. Clin. Microbiol. Rev. 27: 241–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Torres E., l. López-Cerero, Rodríguez-Martínez J.M., and Pascual A. 2016. Reduced susceptibility to cefepime in clinical isolates of Enterobacteriaceae producing OXA-1 beta-lactamase. Microb. Drug Resist. 22:141–146 [DOI] [PubMed] [Google Scholar]
- 9. Brolund A. 2014. Overview of ESBL-producing Enterobacteriaceae from a Nordic perspective. Infect. Ecol. Epidemiol. 4:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reuland E.A., Naiemi N.A., Kaiser A.M., Heck M., J.Kluytmans A.J.W., Savelkoul P.H.M., Elders P.J.M., and C.M.J.E. Vandenbroucke-Grauls. 2016. Prevalence and risk factors for carriage of ESBL-producing Enterobacteriaceae in Amsterdam. J. Antimicrob. Chemother. 71:1076–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shodavaram U.V.R., Ramanamma M.V., Mukherjee A.L., and Jacob A.S. 2016. Prevalence of ESBL-mediated resistance among hospital and community isolates of Klebsiella pneumoniae in Warangal. Int. J. Med. Res. Rev. 4:1005–1009 [Google Scholar]
- 12. Rogers B.A., Sidjabat H.E., and Paterson D.L. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14 [DOI] [PubMed] [Google Scholar]
- 13. Dudley M.N., Ambrose P.G., Bhavnani S.M., Craig W.A., Ferraro M.J., Jones R.N., for the Antimicrobial Susceptibility Testing Subcommittee of the Clinical and Laboratory Standards Institute. 2013. Background and rationale for revised clinical and laboratory standards institute interpretive criteria (breakpoints) for Enterobacteriaceae and Pseudomonas aeruginosa: I. cephalosporins and aztreonam, Clin. Infect. Dis. 56:1301–1309 [DOI] [PubMed] [Google Scholar]
- 14. Ministry of Health, Brazil. 1998. Ordinance 2616/98, of May 12, 1998. Regulates the actions to control hospital infection in the country [In Portuguese]. Available at http://bvsms.saude.gov.br/bvs/saudelegis/gm/1998/prt2616_12_05_1998.html (accessed October17, 2017).
- 15. Clinical and Laboratory Standards Institute Edition. Approved standard—ninth. 2012. CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 16. Xu L., Ensor V., Gossain S., Nye K., and Hawkey P. 2005. Rapid and simple detection of blaCTX–M genes by multiplex PCR assay. J. Med. Microbiol. 54:1183–1187 [DOI] [PubMed] [Google Scholar]
- 17. Dallenne C., Da Costa A., Decré D., Favier C., and Arlet G. 2010. Development of aset of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 [DOI] [PubMed] [Google Scholar]
- 18. Bansal S., and Tandon V. 2011. Contribution of mutations in DNA gyrase and topoisomerase IV genes to ciprofloxacin resistance in Escherichia coli clinical isolates. Int. J. Antimicrob. Agents 37:253–255 [DOI] [PubMed] [Google Scholar]
- 19. Wu L.T., Chang S.Y., Yen M.R., Yang T.C., and Tseng Y.H. 2007. Characterization of extended-host-range pseudo-T-even bacteriophage Kpp95 isolated on Klebsiella pneumoniae. Appl. Environ. Microbiol. 73:2532–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bush K., Jacoby G.A., and Medeiros A.A. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abreu A.G., Marques S.G., Monteiro-Neto V., and Gonçalves A.G. 2013. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in community-acquired urinary tract infections in São Luís, Brazil’. Braz. J. Microbiol. 44:469–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gonçalves L.F., de Oliveira Martins-Júnior P., de Mello A.B.F., R.da Silva C.R.M., de Paulo Martins V., Pitondo-Silva A., and de Campos T.A. 2016. Multidrug resistance dissemination by extended-spectrum beta-lactamase-producing Escherichia coli causing community-acquired urinary tract infection in the Central-Western Region, Brazil. J. Glob. Antimicrob. Resist. 6:1–4 [DOI] [PubMed] [Google Scholar]
- 23. Kahlmeter G., J. Åhman, and Matuschek E. 2015. Antimicrobial resistance of Escherichia coli causing uncomplicated urinary tract infections: a European update for 2014 and comparison with 2000 and 2008. Infect. Dis. Ther. 4:417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manyahi J., Moyo S.J., Tellevik M.G., Ndugulile F., Urassa W., Blomberg B., and Langeland N. 2017. Detection of CTX-M-15 beta-lactamases in Enterobacteriaceae causing hospital- and community-acquired urinary tract infections as early as 2004, in Dar es Salaam, Tanzania. BMC Infect. Dis. 17:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Melekos M.D., and Naber K.G. 2000. Complicated urinary tract infections. Int. J. Antimicrob. Agents 15:247–256 [DOI] [PubMed] [Google Scholar]
- 26. Doi Y., and Arakawa Y. 2007. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 45:88–94 [DOI] [PubMed] [Google Scholar]
- 27. Jacoby G.A. 2009. AmpC β-lactamases. Clin. Microbiol. Rev. 22:161–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Volcão L.M., Pereira J.L., Ramis I.B., Ramos D.F., Damati G.S., Gonçalves C.V., Silva P.E.A., and Groll A.V. 2016. Factors associated with resistance to ciprofloxacin and levofloxacin in Gram-negative isolates from urinary tract infections. Rev. Epidemiol. Control. Infecção. 6:18–23 [Google Scholar]
- 29. Volcão L.M., Pereira J.L., Gewehr M.F., Real V.L., Ramos D.F., Gonçalves C.V., Possuelo L.G., Minarini L.A.R., da Silva P.E.A., and von Groll A. 2018. High frequency of the aac (6′)-Ib-cr gene associated with double mutations in gyrA and parC in Escherichia coli isolates from patients with urinary tract infections. J. Glob. Antimicrob. Resist. 13:180–183 [DOI] [PubMed] [Google Scholar]
- 30. Nordmann P., Cuzon G., and Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet. Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 31. Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rocha F.R., Pinto V.T.P., and F.C.B. Barbosa. 2016. The spread of CTX-M-type extended-spectrum β-lactamases in Brazil: a systematic review. Microb. Drug Resist. 22:301–311 [DOI] [PubMed] [Google Scholar]
- 33. Nogueira K.S., Conte D., Maia F.V., and Dalla-Costa L.M. 2015. Distribution of extended-spectrum β-lactamase types in a Brazilian tertiary hospital. Rev. Soc. Bras. Med. Trop. 48:162–169 [DOI] [PubMed] [Google Scholar]
- 34. Chagas T.P.G., Alves R.M., Vallim D.C., Seki L.M., Campos L.C., and Asensi M.D. 2011. Diversity of genotypes in CTX-M-producing Klebsiella pneumoniae isolated in different hospitals in Brazil. Braz. J. Infect. Dis. 15:420–425 [DOI] [PubMed] [Google Scholar]
- 35. Sabaté M.R., Tarrago F., Navarro E., Miró E., Vergés C., Barbé J., and Prats G. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing beta-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bonnet R., Dutour C., Sampaio J.L.M., Chanal C., Sirot D., Labia R., De Champs C., and Sirot J. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240 → Gly. Antimicrob. Agents Chemother. 45:2269–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dexheimer G.M., Prediger J., Weidlich L., and Pozzobon A. 2015. Prevalence of resistance and molecular characterization of extended spectrum beta-lactamase (ESBL)-producing bacteria isolated in a hospital in Southern Brazil. Afr. J. Microbiol. Res. 9:294–300 [Google Scholar]
- 38. Bonnet R., Sampaio J.L.M., Labia R., De Champs C., Sirot D., Chanal C., and Sirot J. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peirano G., Asensi M.D., Pitondo-Silva A., and Pitout J.D.D. 2011. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from Rio de Janeiro, Brazil. Clin. Microbiol. Infect. 17:1039–1043 [DOI] [PubMed] [Google Scholar]
- 40. Cantón R., González-Alba J.M., and Galán J.C. 2012. CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Poirel L., Le Thomas I., Naas T., Karim A., and Nordmann P. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kjeldsen T.S., Overgaard M., Nielsen S.S., Bortolaia V., Jelsbak L., Sommer M., Guardabassi L., and Olsen J.E. 2015. CTX-M-1 β-lactamase expression in Escherichia coli is dependent on cefotaxime concentration, growth phase and gene location. J. Antimicrob. Chemother. 70:62–70 [DOI] [PubMed] [Google Scholar]
- 43. Nikaido H., Basina M., Nguyen V.Y., and Rosenberg E.Y. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side chains. J. Bacteriol. 180:4686–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Doménech-Sánchez A., Hernández-Allés S., Martínez-Martínez L., Benedí V.J., and Albertí S. 1999. Identification and characterization of a new porin gene of Klebsiella pneumoniae: its role in β-lactam antibiotic resistance. J. Bacteriol. 181:2726–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Winokur P.L., Brueggemann A., DeSalvo D.L., Hoffmann L., Apley M.D., Uhlenhopp E.K., Pfaller M.A., and Doern G.V. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC β-lactamase. Antimicrob. Agents Chemother. 44:2777–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]