Abstract
Background
The molecular aetiology of inflammatory bowel disease [IBD] and its two subtypes, ulcerative colitis [UC] and Crohn’s disease [CD], have been carefully investigated at genome and transcriptome levels. Recent advances in high-throughput proteome quantification has enabled comprehensive large-scale plasma proteomics studies of IBD.
Methods
The study used two cohorts: [1] The CERTIFI-cohort: 42 samples from the CERTIFI trial of anti-TNFα–refractory CD patients; [2] the PROgECT-UNITI-HCs cohort: 46 UC samples of the PROgECT study, 84 CD samples of the UNITI I and UNITI II studies, and 72 healthy controls recruited in Mount Sinai Hospital, New York, USA. The plasma proteome for these two cohorts was quantified using high-throughput platforms.
Results
For the PROgECT-UNITI-HCs cohort, we measured a total of 1310 proteins. Of these, 493 proteins showed different plasma levels in IBD patients to the plasma levels in controls at 10% false discovery rate [FDR], among which 11 proteins had a fold change greater than 2. The proteins upregulated in IBD were associated with immunity functionality, whereas the proteins downregulated in IBD were associated with nutrition and metabolism. The proteomic profiles were very similar between UC and CD. In the CERTIFI cohort, 1014 proteins were measured, and it was found that the plasma protein level had little correlation with the blood or intestine transcriptomes.
Conclusions
We report the largest proteomics study to date on IBD and controls. A large proportion of plasma proteins are altered in IBD, which provides insights into the disease aetiology and indicates a potential for biomarker discovery.
Keywords: Proteomics, inflammatory bowel disease, differential expression analysis, proteomic quantitative trait loci
1. Introduction
It is estimated that 568 per 100000 people in the USA have been diagnosed with inflammatory bowel disease [IBD], among which 249 have ulcerative colitis [UC] and 319 have Crohn’s disease [CD].1 The investigation of molecular phenotypes [e.g. transcriptome and proteome] and how they relate to disease susceptibility and progression can help close the gap in understanding between variation in the human genome that is associated with disease and the biological processes that lead to disease. The integration of genetic studies [e.g. genome-wide association studies, GWAS] and transcriptomic studies has proven particularly fruitful. Transcriptome profiling [e.g. microarray or RNA sequencing] has identified changes in the mRNA of many genes in the blood and intestine that have been associated with IBD2,3; however, the transcriptomic differences between colonic or blood UC and CD have been reported as minimal.3 Genome-wide association studies and genome sequencing have been used to identify genetic loci, including eQTLs and pQTLs [expressional and proteomics quantitative trait loci, respectively], that have proven to be powerful intermediate phenotypes in revealing the molecular aetiology underlying IBD genetic association loci.4–8
Circulating protein levels are critical measures of disease presence and activity. Only recently have researchers begun employing high-throughput screening technologies to measure circulating plasma protein levels in large human populations.9–13 Although extensive study of the transcriptional signatures of IBD in relevant tissues has been conducted, research on proteomic alterations in CD and UC is still in its infancy,14–18 and to date, there has been no comprehensive plasma proteomics study of IBD using a large sample size [e.g. N > 200]. Herein, we have attempted to close this gap. In this study, we employed high-throughput proteomics technology [SOMAscan panel, Materials and Methods] to assess variation in the levels of plasma proteins of three groups of individuals of well-characterized UC and CD, and non-IBD controls,19,20 and the controls were matched to IBD patients in terms of age, sex, and ethnicity. We examined the correlation between plasma protein levels and mRNA measured in blood and intestine. Further, we systematically explored the distinct pair-wise plasma proteomic profiles for UC vs control, CD vs control and UC vs CD.
2. Materials and methods
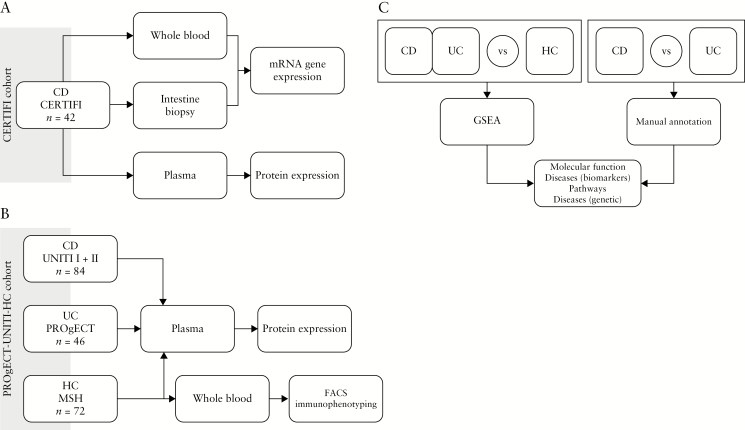
Figure 1 summarizes the work flow of data collection and downstream analysis. Details are reported in the following sections.
Figure 1.
Study design and workflow. [A] Data generation from the CERTIFI cohort; [B] data generation from the PROgECT-UNITI-HCs cohort; [C] differential protein analysis workflow.
2.1. Study cohorts
All subjects used in this study were of Caucasian ancestry.
The CERTIFI cohort [Figure 1] assessed were CD subjects from the CERTIFI study [ClinicalTrials.gov number, NCT00771667]. The range of disease duration was 6 months – 41 years, and the median was 11.1 years; the range of CD activity index was 223–449, with median = 331.5 [Table S1]. The transcriptome data of this cohort is publicly available at GEO [Accession number GSE100833].21
In the PROgECT-UNITI-HCs cohort [Figure 1], samples were selected from three different studies. [1] A total of 46 UC samples were collected as part of the PROgECT study [ClinitalTrials.gov number, NCT01988961].22 The UC subjects had a Mayo score of between 7 and 11 [median = 9], had a disease duration of between 0 and 11 years [median = 2.4 months], and were receiving corticosteroids [n = 16] and immunomodulators [n = 7], Azathioprine and 6-MP [n = 7], and 5-ASA [n = 38]. [2] A total of 84 CD samples were collected as part of UNITI I and UNITI II [ClinicalTrials.gov numbers NCT01369329 and NCT01369342, respectively].20 The CD subjects had a CD activity index of between 208 and 460 [median = 286], had involvement of the colon only [n = 11], colon and ileum [n = 50], and ileum only [n = 23], had a disease duration of between 5 months and 40 years [median = 7.3 years], and were under standard of care. [3] The 72 HC samples were recruited at Mount Sinai Medical Center in New York City, NY, USA. The CD, UC, and HCs in the ‘matched IBD and HC cohort’ were matched in terms of age and sex. Most importantly, plasma collections were conducted using identical protocols for the UC, CD, and control subjects, and proteome profiling was performed at the same time and under the same laboratory conditions.
Importantly, the CERTIFI cohort and the PROgECT-UNITI-HCs cohort were profiled using different versions of SOMAscan panels.
2.2. Proteomics assays
Plasma protein levels were measured using a SOMAmer-based capture array, ‘SOMAscan’10,23 [web site: http://www.somalogic.com/Products-Services/SOMAscan]. On the CERTIFI cohort, 1014 proteins were measured, and on the PROgECT-UNITI-HCs cohort, a total of 1310 proteins were measured. The SOMAscan technology uses chemically modified nucleotides to convert a protein signal to a nucleotide signal that is measured as relative fluorescence units using a custom DNA microarray. Quality Control was based on assessment of batch effect, unsupervised clustering of the expression profiles, and Principal Components Analysis [PCA]. No sample or probe failed the Quality Control. Further, we fully randomized the sample across the Somalogics assay plates to eliminate possible confounding by batch effects. In data analyses, we adjusted for plate as a covariate.
2.3. Plasma CRP level
In the clinical lab [independent of the Somalogic technology], we measured plasma CRP levels in the majority of the PROgECT-UNITI-HCs cohort: 84 CD subjects, 42 UC subjects and 71 normal HCs. The clinical lab measurements were compared with the Somalogic values, where Pearson’s correlation was 0.817 [Figure S5].
2.4. Differential protein analysis
Differential proteins [DPs] refer to the proteins that showed different plasma levels in two groups of subjects. The analysis was performed on the PROgECT-UNITI-HCs cohort with linear regression models, using the log-2 transformed protein level as the outcome variable [y] and disease status plus other covariates as regressors. Specifically, we analyzed one protein at a time. The following regression model was fitted: y ~ Age + Sex + PlateID + disease_CD + disease_UC. Significance of the ‘CD+UC’ contrast (average of the CD vs controls and UC vs controls effects) and the ‘CD-UC’ contrast (difference between the CD vs controls and the UC vs controls effects) was assessed with a t test, using the ‘glht’ function from the ‘multcomp’ R package. Since 1310 proteins were examined, we accounted for multiple testing using the false discovery rate [FDR]. The FDR was estimated by repeating the analysis for all probes 1000 times, each time randomly permuting the subject labels. The FDR for an observed nominal p value ‘p0’ was then computed as:
2.5. Gene set enrichment analysis
Differential proteins [e.g identified comparing UC vs control] entered gene sets enrichment analysis to explore their common functions. Three tools for gene sets enrichment analysis were used. [1] The Molecular Signatures Database [MSigDB]. Differential proteins were compared with MSigDB24 using the GSEA software v2.2.0 and the MSigDB v5.1 [http://software.broadinstitute.org/gsea]. Because multiple gene sets were evaluated, the FDR was quantified separately for each contrast by running 1000 permutations. [2] MetaCore Suite [portal.genego.com]. The 1310 proteins measured by SOMAscan were used as the background for the enrichment analysis. The FDR was also calculated by the MetaCore software. [3] Enrichment for disease-related genes were conducted usingthe NHGRI-EBI GWAS catalog.25 Specifically, for each disease we tested for the enrichment of proteins upregulated in IBD, and proteins downregulated in IBD separately.
2.6. Genomic co-localization of protein level and IBD risk
Co-localization analysis is conducted to determine whether the plasma protein level and IBD risk are controlled by the same genetic variant[s], using COLOC version 2.3–6 in R.26 Our analysis focused on the IBD proteomic signature proteins that were also influenced by pQTL in the CD-only cohort.13 The aim of co-localization analysis is to find molecules that sit on the causal pathway of IBD, and the strategy is to identify the protein[s] that is controlled by the same genetic variant[s] that leads[lead] to IBD. Co-localization analysis uses two types of information [1] IBD GWAS, which use a large sample size [n ~ 60000] to convincingly link genetic variants to IBD incidence27; [2] pQTLs, which quantify the association between genetic variants and proteomic traits. Herein these two types of information come from [1] UKBB IBD GWAS,27 for which the results are publicly available; and [2] pQTLs quantified for the CERTIFI cohort.13 The analysis was conducted using the COLOC package version 2.3–6 in R,26 in which default priors of the software were used. COLOC analyzed one protein’s level and IBD risk at a time. In each analysis, we denoted Trait 1 as the level of the given protein; and Trait 2 as the IBD risk. In total, five hypotheses were evaluated. H0: No association with either Trait 1 or Trait 2; H1: Association with Trait 1, not with Trait 2; H2: Association with Trait 2, not with Trait 1; H3: Association with Trait 1 and Trait 2, with multiple single-nucleotide polymorphisms [SNPs]; H4: Association with Trait 1 and Trait 2, one shared SNP. When we observed a large posterior probability of H4 [i.e.. PP.H4 > 80%], we concluded a single genetic variant controls both IBD risk and the protein’s level in the plasma. When we observed high posterior probability of Hypothesis 3 and 4 [i.e. PP.H3+PP.H4 > 80%], the data multiple genetic variants may control both IBD risk and the protein’s level in the plasma.
2.7. FACS immunophenotyping profiles and FACS-protein association testing
FACS data was collected from the HCs of the PROgECT-UNITI-HCs cohort [Supplementary Information]. Association between 12 blood immunophenotyping traits levels and 1310 plasma protein levels were tested using a linear regression, including the inverse-normal transformed protein level as the outcome, and the immunophenotyping trait as a covariate. Additional model covariates were age, sex, proteomics plate, and FACS batch.
3. Results
3.1. The plasma proteome was only weakly correlated with the whole blood or intestine transcriptome
We first assessed whether plasma proteomics provided non-redundant biological insights compared with blood and intestinal transcriptomes. Whole blood transcriptomes, intestine tissue transcriptomes, and plasma proteomes were profiled for the CERTIFI cohort [42 CD subjects of the anti-TNF–refractory CERTIFI trial, Table S1; GEO accession number: GSE100833]. In total, 1014 plasma proteins were assessed by SOMAscan panel. The whole transcriptome [52810 probesets] was assessed in blood and intestinal tissue using an Affymetrix HU133 microarray. The plasma proteomics profile was only weakly correlated with the whole blood [median ρ = 0.03] or the intestinal transcriptome [median ρ = 0.01] [Figure S1A and B]. Although the median correlation [ρ] was still positive [Figure S1A, Figure S1B, and Table S2], such a weak correlation indicated that the plasma proteomics extracted information orthogonal to the transcriptomes of disease-relevant tissues in CD patients, and indicated further protein signature studies were necessary. We also conducted Co-inertia analysis [CIA],28 which only indicated a detectable correlation between plasma protein and whole blood mRNA [p = 0.054], but not between plasma protein and intestinal mRNA [Table S3]. The limited sample size for the intestinal data made it difficult to interpret the CIA test statistics. We conducted permutation [by randomizing the sample IDs in the SOMAscan dataset] to derive the empirical p value for the CIA test. The empirical p value was 0.054 for the correlation between the plasma proteome and the whole blood mRNA. For the intestine, CIA empirical p values were not significant [e.g. p value = 0.262 for the sigmoid colon; p value = 0.817 for the transverse colon]. Since the plasma proteome is secreted from multiple organs and cell types, and the blood and intestinal transcriptome are derived from intracellular sources, perhaps it is not surprising that the plasma protein – mRNA correlation is weak. Moreover, the correlation between the blood and intestinal mRNAs was also very weak [median ρ = 0.02, Figure S1C].
3.2. Comparison of plasma proteomic profiles among UC, CD, and control subjects
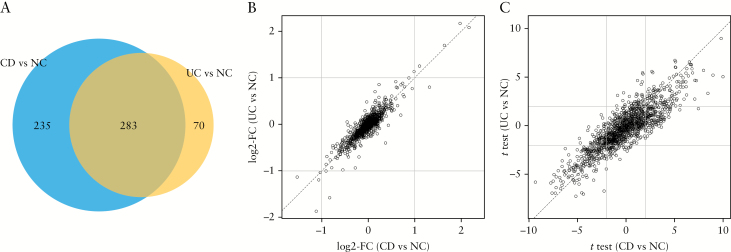
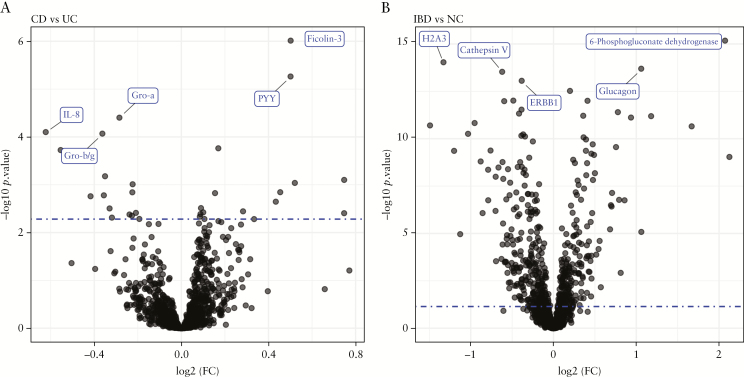
We tested 1310 probes for differential protein expression in the PROgECT-UNITI-HCs cohort: UC patients of the PROgECT trial [n = 46], CD patients of UNITI I and UNITI trials [n = 84], and HCs [HCs, n = 72]. The HC samples were matched to UC and CD patients in terms of age and sex [Table S1]. At 10% FDR, N = 353 proteins were differentially expressed between UC and HCs, among which N = 12 proteins had a fold change of ≥2 [Table S4]. These 353 proteins were termed ‘UC differential proteins’ [UC-DPs]. Comparing CD vs controls, N = 518 proteins were differentially expressed at 10% FDR, among which N = 9 proteins had a fold change of ≥2 [Table S4]. These 518 proteins were termed ‘CD-DPs’. Importantly, the UC- and CD-DPs were very similar, with 283 proteins in common [Figure 2A, overlap odds ratio = 12.4, Fisher test p value = 1.5E-75]. Furthermore, the UC- and CD-DPs had very similar fold changes ([Figure 2B] and significance level [Figure 2C], indicating the two IBD subtypes [i.e. UC and CD] were similar in term of alterations in plasma proteomics from healthy states). Next, we compared CD [UNITI I and II] with UC [PROgECT] patients. At 10% FDR, 16 proteins were upregulated and 15 downregulated, respectively [Figure 3 and Table S4]. Further, we pooled UNITI I and II and PROgECT as a single group [henceforth, simply ‘IBD’] and compared it with HCs [Table 1]. At 10% FDR, we found 219 probes upregulated and 274 downregulated, respectively, comparing IBD vs controls [Table 1]. Eleven proteins had a fold change of ≥2, including CRP, 6-Phosphogluconate dehydrogenase, Ferritin, Glucagon, Haptoglobin, PCI, SAA, Haemoglobin, LEAP-1, Cyclophilin F, and H2A3, for which the FDRs were all below 0.1%. These combined 493 up- and downregulated proteins [37.6% of all proteins measured by SOMAscan] were termed ‘IBD-DPs’.
Figure 2.
Comparison of CD and UC proteomic signatures. [A] Venn Diagram of the overlap of the 10% false discovery rate [FDR] signature probes lists. [B] Scatterplot of the estimated log2-fold change [FC] of CD vs HCs [horizontal axis] and UC vs HCs [vertical axis]; dotted lines mark the ±2 effect sizes thresholds. Each circle represents a somalogic probe. All tested probes are reported. [C] Scatterplot of the estimated t- test of the CD vs HCs [horizontal axis] and UC vs HCs [vertical axis]. Dotted lines mark the ±2 significance thresholds. Each circle represents somalogic probe. All tested probes are reported.
Figure 3.
Volcano plots of differential protein expression analysis. [A] CD vs UC signature. [B] IBD vs HCs signature; dotted blue line marks the empirical 10% false discovery rate [FDR] significance threshold, obtained by permuting the input dataset and repeating the analysis 1000 times. The top five significant proteins are labelled.
Table 1.
Plasma proteomic signatures of IBD and subtypes.
| Signature | Sample size | # significant probes [10% FDR] | Effect size: |log2[FC]| | ||
|---|---|---|---|---|---|
| Up | Down | Median | 95th percentile | ||
| IBD vs HCs | 130 vs 72 | 219 | 274 | 0.092 | 0.487 |
| CD vs UC | 84 vs 46 | 16 | 15 | 0.054 | 0.223 |
IBD: inflammatory bowel disease; HCs: healthy controls; UC: ulcerative colitis; CD: Crohns’ disease; FDR: false discovery rate; FC: fold change.
3.3. Functional annotation of differential proteins
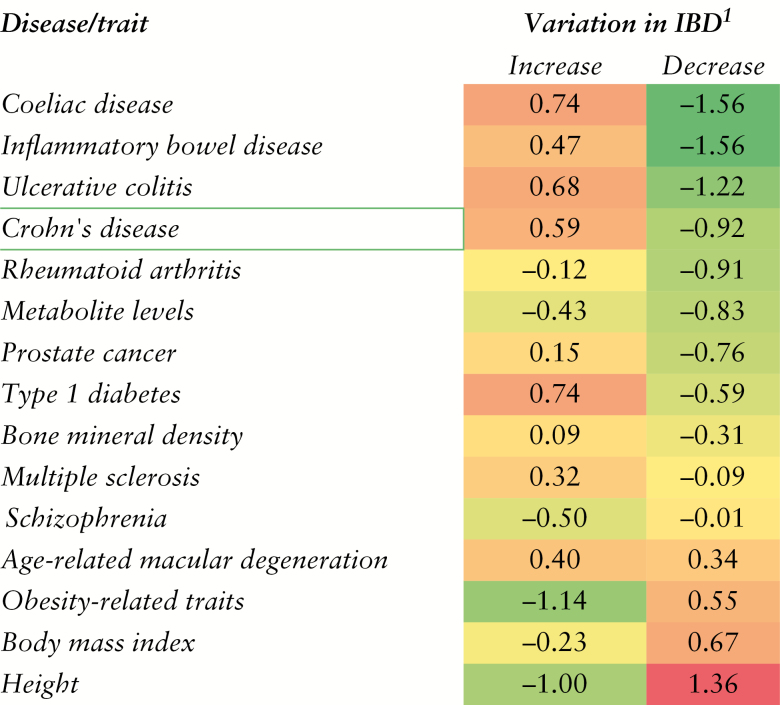
We characterized the annotation of the IBD-DPs by testing for enrichment of gene sets from three databases: MSigDB,24 MetaCore [portal.genego.com], and the NHGRI-EBI catalog.25 The full results are reported in Table S5 [MSigDB], Table S6 [MetaCore], and Table S7 [NHGRI-EBI GWAS catalog]. At 5% FDR, the ‘chemokine and cytokine activities’ gene set was enriched in the proteins upregulated in IBD vs control [Table S5], as were ‘biomarkers of CD’, ‘other immune disorders’, and ‘myocardial infarction and ischemia’ [Table S6]. The upregulated signature also enriched for IBD, CD, UC, and Coeliac disease GWAS loci and for IL-6-induced signalling and Th-17–derived cytokines [Table S6], potentially highlighting the anti-TNF–related pathways in disease. The upregulated signature was under enriched for GWAS loci of height, BMI, and obesity [Figure 4 and Table S7]. In contrast, the plasma proteins downregulated in IBD subjects were enriched for hyperlipidemia biomarkers [Table S5], as well as Obesity-, Height- and BMI–associated genetic loci. Proteins downregulated in IBD patients were further enriched for ‘c-Kit ligand signalling pathway during haemopoiesis’and ‘complement pathway disruption in thrombotic microangiopathy’.
Figure 4.
Enrichment for genetic risk loci. Log-2 odds ratios of overlap between IBD signature (10% false discovery rate [FDR]) proteins and genes/diseases from the NHGRI-EBI catalog of genome-wide association studies. The IBD signature was further split into a list of IBD-increased and IBD-decreased proteins, respectively.
3.4. Weighted gene correlation network analysis
Weighted gene correlationi network analysis [WGCNA] was used as a systems biology method to describe the correlation patterns among the proteins measured in the PROgECT-UNITI-HCs cohort.29,30 Networks were built in an unsupervised manner for PROgECT, UNITI, HCs and combined cohorts, resulting in three disease group–specific co-expression networks and one combined co-expression network [Table S8]. For example, in the combined network, we found seven modules of highly correlated proteins [Table S8, the modules are named by colors], with 485 proteins not fitting into a module therefore being placed into the ‘Grey’ module. The functional annotations of each module, using the Kyoto Encyclopedia of Genes and Genomes [KEGG] database,31 are summarized in Table S9. The first principle components [i.e. eigen-peptide]30 of the combined co-expression network, Pooled_blue, Pooled_brown and Pooled_green, were significantly associated with IBD GWAS, with a Wilcoxon test p value of 7.86E-09, 1.03E-03, and 1.03E-5, respectively [Table S10]. We explored the function annotations of these three modules using the Kyoto Encyclopedia of Genes and Genomes [KEGG] database.31 All three modules were closely related to the immune functions [Table S9], and they were significantly enriched for differential proteins between CD vs CL and UC vs CL [Table S11], indicating that these modules were disease associated. Further, the differential expression–tagged modules were also identified for PROgECT, UNITI, and HCs co-expression networks as they were also significantly enriched for CD vs CL and UC vs CL differential proteins [Table S11].
3.5. Genetic co-localization of plasma protein level and IBD risk
Fifteen proteins in the IBD-DPs were previously reported that were influenced by nearby genetic variants [i.e. proteomic quantitative trait loci, pQTL].13 The genetic variants in association with the protein levels were termed pSNPs. Co-localization analysis examined whether the IBD, UC, and CD risk was controlled by the same genetic variants that influence protein levels. We evaluated the causality of how genetic variants modulate disease variance in IBD through protein level alteration. Integrating the pQTLs that we previously reported on the CERTIFI trial of anti-TNFα–refractory CD patients13 and the most recent GWAS on IBD,27 we found two proteins [LYZ and MSP] co-localized with IBD risk [Table S11]. Macrophage-stimulating protein [MSP, aka MST1] showed a large posterior probability of H4 [i.e. >0.99, Table S12], indicating that the same genetic variant controls both IBD risk and plasma MSP levels. Genetic co-localization analysis on LYZ [lysozyme] resulted in a large posterior of H3+H4 probability [i.e. >0.8, Table S12], suggesting that multiple variants at the LYZ locus control IBD susceptibility and the plasma LYZ levels.
3.6. Contrast of CD vs UC signature
Comparing CD [UNITI I & II] vs UC [PROgECT] subjects, 31 proteins showed significantly different plasma levels [FDR≤10%, Table 2]. We manually annotated the signature proteins to broad biological categories. Of the 31 proteins, 17 were related to immune response. Further, by querying the STRING database,32 we found 10 out of the 31 proteins formed a tight network of interacting proteins [Figure S2]: CXCL1, CXCL2/3, GCC, GZMB, IFNA7, IL23R, IL8, PYY, RPS3, and TYK2, known for their roles in immune response. Although several of the proteins [TYK2, CXCL1, EGF, REG4, GZMB, and IL8] had different plasma levels in UC vs CD subjects, they were also in CD- and/or UC-DPs [Table S4].
Table 2.
Differentially expressed proteins: UC vs CD [FDR≤10%]*.
| Probe ID | Protein | UniProt | Gene symbol | Log2-FC | SE | t-value | p value |
|---|---|---|---|---|---|---|---|
| SL009341 | BASI | P35613 | BSG | 0.09 | 0.028 | 3.12 | 4.10E-03 |
| SL008382 | CYTD | P28325 | CST5 | 0.45 | 0.132 | 3.44 | 1.43E-03 |
| SL016566 | DRAK2 | O94768 | STK17B | 0.08 | 0.027 | 3.08 | 4.65E-03 |
| SL000084 | EGF | P01133 | EGF | –0.32 | 0.104 | –3.07 | 4.87E-03 |
| SL000420 | Ferritin | P02794 P02792 | FTH1 FTL | 0.75 | 0.207 | 3.60 | 7.93E-04 |
| SL004337 | FGF-19 | O95750 | FGF19 | –0.42 | 0.123 | –3.38 | 1.74E-03 |
| SL002086 | Ficolin-3 | O75636 | FCN3 | 0.50 | 0.096 | 5.21 | 9.70E-07 |
| SL004271 | GFAP | P14136 | GFAP | –0.33 | 0.103 | –3.21 | 3.13E-03 |
| SL000433 | Glucagon | P01275 | GCG | 0.52 | 0.146 | 3.56 | 9.17E-04 |
| SL004068 | Granzyme B | P10144 | GZMB | –0.51 | 0.131 | –3.92 | 2.48E-04 |
| SL003173 | Gro-a | P09341 | CXCL1 | –0.28 | 0.065 | –4.37 | 3.96E-05 |
| SL017610 | Gro-b/g | P19876 P19875 | CXCL2/3 | –0.36 | 0.087 | –4.19 | 8.54E-05 |
| SL019978 | IFNA7 | P01567 | IFNA7 | –0.23 | 0.073 | –3.10 | 4.45E-03 |
| SL005177 | IL-1F6 | Q9UHA7 | IL36A | 0.43 | 0.131 | 3.30 | 2.26E-03 |
| SL005185 | IL-23 R | Q5VWK5 | IL23R | 0.28 | 0.089 | 3.16 | 3.60E-03 |
| SL000039 | IL-8 | P10145 | CXCL8 | –0.62 | 0.148 | –4.21 | 7.93E-05 |
| SL004536 | LEAP-1 | P81172 | HAMP | 0.75 | 0.238 | 3.14 | 3.94E-03 |
| SL013872 | LRP1B | Q9NZR2 | LRP1B | –0.21 | 0.066 | –3.14 | 3.87E-03 |
| SL000645 | MMP-10 | P09238 | MMP10 | –0.36 | 0.105 | –3.39 | 1.66E-03 |
| SL000522 | MMP-12 | P39900 | MMP12 | –0.55 | 0.139 | –3.99 | 1.90E-04 |
| SL007237 | MP2K4 | P45985 | MAP2K4 | 0.33 | 0.110 | 3.05 | 5.23E-03 |
| SL008933 | PARK7 | Q99497 | PARK7 | –0.24 | 0.076 | –3.12 | 4.19E-03 |
| SL016555 | PDE11 | Q9HCR9 | PDE11A | 0.09 | 0.028 | 3.21 | 3.08E-03 |
| SL011404 | PDE4D | Q08499 | PDE4D | 0.10 | 0.032 | 3.15 | 3.78E-03 |
| SL011405 | PDE5A | O76074 | PDE5A | 0.11 | 0.035 | 3.05 | 5.20E-03 |
| SL011509 | PYY | P10082 | PYY | 0.50 | 0.104 | 4.83 | 5.45E-06 |
| SL012561 | REG4 | Q9BYZ8 | REG4 | –0.35 | 0.096 | –3.65 | 6.65E-04 |
| SL008059 | RS3 | P23396 | RPS3 | –0.23 | 0.066 | –3.44 | 1.44E-03 |
| SL010384 | Testican-1 | Q08629 | SPOCK1 | –0.22 | 0.063 | –3.55 | 9.78E-04 |
| SL007181 | TYK2 | P29597 | TYK2 | 0.17 | 0.042 | 4.01 | 1.73E-04 |
| SL018946 | UB2G2 | P60604 | UBE2G2 | 0.15 | 0.045 | 3.42 | 1.50E-03 |
UC: ulcerative colitis; CD: Crohns’ disease; SE: standard error; FDR: false discovery rate; FC: fold change. *The sign of the t-value represents the direction, and the positive value indicates the protein level is higher in CD than inUC; the negative value indicates vice versa.
Other downregulated proteins in CD vs UC include RS3 [40S ribosomal protein S3], involved in apoptosis in response to oxidative stress33,34; PARK7, involved in NADPH function,35 GFAP36,37 and MMP12. MMP12 was linked to dendritic cells as part of a tissue remodelling network that contributes to granuloma formation in a gene network that programs phagocytosis in leprosy.38–40 Proteins upregulated in CD vs UC, included FCN3,41–43 involved in the complement pathway; PYY,44 involved in serotonin receptor signalling, IL36A45–47; a potent regulator of dendritic cells and T cells and highly correlated with IL1 and IL17; MAP2K4,48 involved in IL1 signalling to P38; UBE262,49,50 which has a role in ER stress recovery; PDE5A with a putative role in anti-oxidant status, and BASI, a protein that associates with NOD2.51,52
4. Discussion
High-throughput technology for protein quantitation in large sample sizes has only recently become tractable. We conducted a systematic plasma proteomics study on UC subjects [from the PROgECT trial], CD subjects [from UNITI I and II], and matched HCs, as well as paired measures of protein and mRNA levels in a CD cohort [from the CERTIFI trial]. Two key observations were made. [1] CD and UC plasma proteomics profiles were significantly different from those of the HCs, and the up- and downregulated proteins revealed distinct biological functions, whereas the contrast between CD and UC proteomics was minimal. [2] The blood and intestine transcriptomes and plasma proteomics had very modest correlation and reflected different biological insights.
Up- and downregulated proteins in IBD subjects belonged to different functional categories. Proteins upregulated in IBD versus controls were significantly enriched in chemokine and cytokine activity, immune disease biomarkers, and autoimmune disease GWAS signals [Tables S5–S7, Figure 4]. In contrast, the proteins downregulated in IBD were enriched for nutrition and metabolism [e.g. height, BMI, and obesity GWAS gene], potentially related to impaired gastrointestinal function in IBD.53–55 Comparing IBD [UC subjects from the PROgECT trial and CD subjects from the UNITI I and II trials] vs HCs, we identified proteins of large fold changes [i.e. FC≥2]. Not unexpected, CRP, an inflammatory marker upregulated in IBD, was among the most upregulated in IBD compared with in the HCs [Table 3]. We further observed a large increase in PGD concentration (6-Phosphogluconate dehydrogenase; log2 fold change [FC] = 2.07), an enzyme in the pentose pathway that produces NADPH and is increased with the oxidative stress that is associated with CD.56,57 The substantial fold changes enabled us to classify IBD vs HCs using select blood protein markers. As a proof of concept, in Figure S3, we display the joint distribution of PGD and CRP in our cohort. PGD and CRP levels were moderately correlated [Pearson correlation = 0.211, 95% CI: 0.075–0.339]. Together, PGD and CRP separated IBD patients from controls, validating a known blood biomarker for IBD.
Table 3.
Top IBD differentially expressed proteins [FDR≤10% and |log2FC| > 1].
| Probe ID | Target | Gene symbol | Log2-FC | SE | p value | CD vs UC |
|---|---|---|---|---|---|---|
| SL000247 | 6-Phosphogluconate dehydrogenase | PGD | 2.07 | 0.193 | 6.66E-16 | NS |
| SL000051 | CRP | CRP | 1.18 | 0.159 | 6.45E-12 | NS |
| SL005793 | Cyclophilin F | PPIF | –1.03 | 0.146 | 5.51E-11 | NS |
| SL000420 | Ferritin | FTH1 FTL | –1.2 | 0.179 | 4.37E-10 | higher in CD |
| SL000433 | Glucagon | GCG | 1.06 | 0.126 | 2.02E-14 | higher in CD |
| SL019979 | H2A3 | HIST3H2A | –1.33 | 0.156 | 9.21E-15 | NS |
| SL000437 | Haptoglobin, Mixed Type | HP | 1.06 | 0.224 | 8.28E-06 | NS |
| SL000836 | Haemoglobin | HBA1 HBB | –1.13 | 0.241 | 1.09E-05 | NS |
| SL004536 | LEAP-1 | HAMP | –1.49 | 0.206 | 1.99E-11 | higher in CD |
| SL000550 | PCI | SERPINA5 | 1.67 | 0.231 | 2.22E-11 | NS |
| SL000572 | SAA | SAA1 | 2.12 | 0.324 | 9.15E-10 | NS |
IBD: inflammatory bowel disease; HCs: healthy controls; UC: ulcerative colitis; CD: Crohns’ disease; FDR: false discovery rate; FC: fold change.; NS: non-significant.
We found downregulation of REG4 [SOMAscan probeID: SL012561] in CD compared with UC. REG4 has been shown to be involved in the regeneration of damaged gastrointestinal mucosa in UC patients, where it has higher expression compared with HC subjects.58 REG4 has been further suggested as a biomarker for the discrimination of CD from UC patients, as it showed differential expression in the inflamed and uninflamed biopsies of six CD patients when compared with six UC patients.59,60 This study also detected higher REG4 plasma levels in CD vs control and UC vs control, supporting the strategy of applying high-throughput proteomics technology to larger patient populations to identify plasma protein biomarkers for diagnostics.
Although UC and CD are two IBD subtypes with key clinical differences,2,61 they displayed similar plasma proteomic profiles, comparable with those of previous findings at the mRNA level.3 At 10% FDR, 31 proteins [2.4% of the proteins measured in this study] showed significantly different levels in CD vs UC subjects [Table 2], and of these 17 were related to the immune response. Our observation supports the clinical description that UC and CD are two subsets of the same disease. The common aetiology explained the highly similar proteomics profile. The UC and CD subjects we used in the study represented heterogeneous phenotypes within each cohort, in terms of disease duration and disease severity [Table S1], which is a limitation. We tested the association between plasma protein levels and disease duration or disease severity [Figure S4]. The Spearman correlation coefficient was centered at zero [Figure S4A and C], indicating disease duration or severity did not systematically shift the protein levels upward or downward. The p-values of Spearman correlation largely follow a diagonal line in the quantile–quantile plots [Figure S4B and D], indicating that the association between the plasma proteome and disease duration or disease severity was not more significant than random chance. In other words, the differential proteins identified in this study were not likely attributable to heterogeneity in the samples’ clinical characters.
There are previous proteomics studies on IBD,14–18 summarized in Table 4. However, these studies used small sample sizes and reported limited proteins or mass spectrometry peaks as different in IBD vs control, or as different between subtypes of IBD. Herein, we report a proteomics study on IBD of the largest sample size to date. The improved statistical power enabled us to identify 493 proteins with significantly different plasma levels in IBD vs controls, substantially more than the number of proteins reported by previous studies [Table 4]. Our findings showed certain degrees of consistency with previous reports, even though the assay platform and clinical states of the study subjects were different.
Table 4.
Comparison of differential proteins of present study with published IBD proteome studies.
| Comparison groups and reference* | Bio-sample | Sample size by group | Published differential proteins | Published differential proteins measured in the current study |
Published differential proteins validated in the current study |
|---|---|---|---|---|---|
| IBD vs HCs62 | serum | CD: 30 UC: 30 Inflammatory controls: 30 HCs: 30 | PF4, MRP8, FIBA, HPA2 | PF4 | – |
| IBD vs HCs63 | blood | CD: 22 UC: 48 Colorectal Cancer: 5 Infectious colitis: 6; HCs: 13 | HNP1, NHP2, HNP3 | – | – |
| CD vs UC64 | blood | CD: 13 UC: 17 HCs: 17 | ABHD14B, ACTB, ANXA6, CA2, PPIA, PRDX2, S100A9 | ANXA6, CA2, PPIA, S100A9 | CA2, PPIA |
| CD vs HCs65 | blood | HCs: 48 CD: 15 | APOA4, APOE, C3, C3F, FETUA, FIBA, FPA | APOE | – |
| IBD vs HCs66 | serum | CD: 48 UC: 62 | APOC1, ITIH4, PF4 | ITIH4, PF4 | ITIH4 |
| IBD vs HCs67 | intestine | CD: 3 UC: 4 Inflammatory polyps: 2 Normal colon: 3 | TTBK2, SYNE2, SUCLG2, POSTN, ANXA2, EPX, LAP3, RDX, S100A8, MPO, DEFA1B, PRG2, LCP1, PSME1 | POSTN, ANXA2, PSME1, MPO | POSTN, ANXA2, PSME1 |
| IBD vs HCs68 | serum | UC: 27 CD: 56 HCs: 14 RA controls: 12 | SPP24, alpha-1-microglobulin | – | – |
| CD w/wo stricturing69 | serum | CD: 18 UC: 9 | A2M, LDHB, CAPNS1, MBL2, APOB, CD5L, ALB, FLNA, SCGB1A1, BRDT, CTSD, PSMA7, CP, OIT3, F2, TNXA | A2M, APOB, CAPNS1, CD5L, CTSD, F2, LDHB, MBL2 | A2M, LDHB, APOB |
| IBD vs HCs70 § |
serum | HCs: 48 CD: 15 UC: 26 | – | – | – |
| CD vs UC71 § |
colon | CD: 27 UC: 24 | – | – | – |
| CD vs UC72 § |
colon | CD: 26 UC: 36 | – | – | – |
* IBD: inflammatory bowel disease; HCs: healthy controls; UC: ulcerative colitis; CD: Crohns’ disease.
§Study only reported mass spectrometry peaks but not proteins.
We showed that the plasma protein levels in IBD subjects were weakly positively correlated with the mRNA level in two IBD relevant tissues [blood and intestine]. The explanations are two-fold. [1] The plasma proteome is secreted from multiple organs and cell types, and the liver is a dominant source.13 In 72 HCs of the PROgECT-UNITI-HCs cohort, we quantified immune cell compositions [FACS, Materials and Methods]. The plasma proteome was not associated with immune cell compositions [Table S13]. [2] It has been recognized that levels of mRNAs and the levels of the proteins they encode only correlate modestly.73,74 For many genes, the functional products are the proteins rather than transcripts. Complex regulatory mechanisms are used by biological systems to modulate protein levels in response to environmental exposure or disease conditions. The regulatory mechanisms include translational regulation [e.g. small RNAs and regulatory proteins], ribosome density, and protein half-life.73 Nevertheless, our results indicate that research into the IBD transcriptome and proteome reveal different aspects of the disease and are complementary.
Conflict of Interest
The authors have no conflict of interest to disclose.
Funding
The work was partially supported by NIH grants U24DK062429 and 1R01DK092235-02. Dr Ke Hao is partially supported by the National Natural Science Foundation of China [Grant Nos 21477087 and 91643201] and by the Ministry of Science and Technology of China [Grant No. 2016YFC0206507].
Supplementary Material
Glossary
Abbreviations:
- CD
Crohn’s disease
- UC
ulcerative colitis
- HCs
healthy controls
- GWAS
genome-wide association study
- pQTL
proteomic quantitative trait loci
- eQTL
expression quantitative trait loci.
Author Contributions
Conception and design: AFDN, ST, CB, JC, EES, AK, RD, and KH; analysis and interpretation: AFDN, LAP, CA, KL, ST, CB, BK, and KH; drafting the manuscript for important intellectual content: AFDN, ST, CB, LAP, CA, AK, JD, JC, KL, ST, RD, and KH.
References
- 1. Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012;142:46–54.e42; quiz e30. [DOI] [PubMed] [Google Scholar]
- 2. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011;474:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Granlund Av, Flatberg A, Østvik AE, et al. Whole genome gene expression meta-analysis of inflammatory bowel disease colon mucosa demonstrates lack of major differences between Crohn’s disease and ulcerative colitis. PLoS One 2013;8:e56818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cleynen I, Boucher G, Jostins L, et al. ; International Inflammatory Bowel Disease Genetics Consortium Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016;387:156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium [IIBDGC] Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu JZ, van Sommeren S, Huang H, et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luo Y, de Lange KM, Jostins L, et al. Exploring the genetic architecture of inflammatory bowel disease by whole-genome sequencing identifies association at ADCY7. Nat Genet 2017;49:186–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu WKK, Sun R, Zuo T, et al. A novel susceptibility locus in MST1 and gene–gene interaction network for Crohn’s disease in the Chinese population. J Cell Mol Med 2018;22:2368–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gold L, Walker JJ, Wilcox SK, Williams S. Advances in human proteomics at high scale with the SOMAscan proteomics platform. N Biotechnol 2012;29:543–9. [DOI] [PubMed] [Google Scholar]
- 10. Hensley P. SOMAmers and SOMAscan—a protein biomarker discovery platform for rapid analysis of sample collections from bench top to the clinic. J Biomol Tech 2013;24:S5. [Google Scholar]
- 11. Sattlecker M, Kiddle SJ, Newhouse S, et al. ; AddNeuroMed Consortium Alzheimer’s disease biomarker discovery using SOMAscan multiplexed protein technology. Alzheimers Dement 2014;10:724–34. [DOI] [PubMed] [Google Scholar]
- 12. Menni C, Kiddle SJ, Mangino M, et al. Circulating proteomic signatures of chronological age. J Gerontol A Biol Sci Med Sci 2015;70:809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Di Narzo AF, Telesco SE, Brodmerkel C, et al. High-throughput characterization of blood serum proteomics of IBD patients with respect to aging and genetic factors. PLoS Genet 2017;13:e1006565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chan PP, Wasinger VC, Leong RW. Current application of proteomics in biomarker discovery for inflammatory bowel disease. World J Gastrointest Pathophysiol 2016;7:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang F, Xu C, Ning L, et al. Exploration of serum proteomic profiling and diagnostic model that differentiate Crohn’s disease and intestinal tuberculosis. PLoS One 2016;11:e0167109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vaiopoulou A, Gazouli M, Papadopoulou A, et al. Serum protein profiling of adults and children with Crohn disease. J Pediatr Gastroenterol Nutr 2015;60:42–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Viennois E, Zhao Y, Han MK, et al. Serum miRNA signature diagnoses and discriminates murine colitis subtypes and predicts ulcerative colitis in humans. Sci Rep 2017;7:2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen CS, Sullivan S, Anderson T, et al. Identification of novel serological biomarkers for inflammatory bowel disease using Escherichia coli proteome chip. Mol Cell Proteomics 2009;8:1765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Telesco SE, Brodmerkel C, Zhang H, et al. Gene expression signature for prediction of golimumab response in a phase 2a open-label trial of patients with ulcerative colitis. Gastroenterology 2018;155:1008–1011.e8. [DOI] [PubMed] [Google Scholar]
- 20. Sandborn WJ, Gasink C, Gao LL, et al. ; CERTIFI Study Group Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N Engl J Med 2012;367:1519–28. [DOI] [PubMed] [Google Scholar]
- 21. Peters LA, Perrigoue J, Mortha A, et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet 2017;49:1437–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Telesco S, Greenbaum L, Zhang H, et al. Prediction of non-response to anti-TNF therapy in ulcerative colitis: Implications for patient stratification for IBD trials and novel treatment paradigms. Gastroenterology 2017;152:S984–S5. [Google Scholar]
- 23. Gold L, Ayers D, Bertino J, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 2010;5:e15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014;42:D1001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 2014;10:e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Lange KM, Moutsianas L, Lee JC, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 2017;49:256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Culhane AC, Perrière G, Higgins DG. Cross-platform comparison and visualisation of gene expression data using co-inertia analysis. BMC Bioinformatics 2003;4:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gibbs DL, Baratt A, Baric RS, et al. Protein co-expression network analysis [ProCoNA]. J Clin Bioinforma 2013;3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patil AV, Hsieh TS. Ribosomal protein S3 negatively regulates unwinding activity of RecQ-like helicase 4 through their physical interaction. J Biol Chem 2017;292:4313–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wier EM, Fu K, Hodgson A, Sun X, Wan F. Caspase-3 cleaved p65 fragment dampens NF-κB–mediated anti-apoptotic transcription by interfering with the p65/RPS3 interaction. FEBS Lett 2015;589:3581–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu W, Wu H, Chen L, Wen Y, Kong X, Gao WQ. Park7 interacts with p47[phox] to direct NADPH oxidase–dependent ROS production and protect against sepsis. Cell Res 2015;25:691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. von Boyen GB, Steinkamp M, Geerling I, et al. Proinflammatory cytokines induce neurotrophic factor expression in enteric glia: A key to the regulation of epithelial apoptosis in Crohn’s disease. Inflamm Bowel Dis 2006;12:346–54. [DOI] [PubMed] [Google Scholar]
- 37. von Boyen GB, Steinkamp M, Reinshagen M, Schäfer KH, Adler G, Kirsch J. Proinflammatory cytokines increase glial fibrillary acidic protein expression in enteric glia. Gut 2004;53:222–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Inkeles MS, Teles RM, Pouldar D, et al. Cell-type deconvolution with immune pathways identifies gene networks of host defense and immunopathology in leprosy. JCI Insight 2016;1:e88843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Biancheri P, Brezski RJ, Di Sabatino A, et al. Proteolytic cleavage and loss of function of biologic agents that neutralize tumor necrosis factor in the mucosa of patients with inflammatory bowel disease. Gastroenterology 2015;149:1564–74.e3. [DOI] [PubMed] [Google Scholar]
- 40. Pender SL, Li CK, Di Sabatino A, Sabatino AD, MacDonald TT, Buckley MG. Role of macrophage metalloelastase in gut inflammation. Ann N Y Acad Sci 2006;1072:386–8. [DOI] [PubMed] [Google Scholar]
- 41. Man-Kupisinska A, Michalski M, Maciejewska A, et al. A new ligand-based method for purifying active human plasma–derived ficolin-3 complexes supports the phenomenon of crosstalk between pattern-recognition molecules and immunoglobulins. PLoS One 2016;11:e0156691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Honoré C, Hummelshoj T, Hansen BE, Madsen HO, Eggleton P, Garred P. The innate immune component ficolin 3 [Hakata antigen] mediates the clearance of late apoptotic cells. Arthritis Rheum 2007;56:1598–607. [DOI] [PubMed] [Google Scholar]
- 43. Schaffer T, Flogerzi B, Schoepfer AM, Seibold F, Müller S. Increased titers of anti–Saccharomyces cerevisiae antibodies in Crohn’s disease patients with reduced H-ficolin levels but normal MASP-2 activity. J Crohns Colitis 2013;7:e1–10. [DOI] [PubMed] [Google Scholar]
- 44. Kojima SI, Kojima K, Fujita T. Investigation of 5-HT3 receptor–triggered serotonin release from guinea-pig isolated colonic mucosa: A role of PYY-containing endocrine cell. Eur J Pharmacol 2017;799:196–200. [DOI] [PubMed] [Google Scholar]
- 45. Boutet MA, Bart G, Penhoat M, et al. Distinct expression of interleukin [IL]-36α, β and γ, their antagonist IL-36Ra and IL-38 in psoriasis, rheumatoid arthritis and Crohn’s disease. Clin Exp Immunol 2016;184:159–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nerlich A, Ruangkiattikul N, Laarmann K, et al. C/EBPβ is a transcriptional key regulator of IL-36α in murine macrophages. Biochim Biophys Acta 2015;1849:966–78. [DOI] [PubMed] [Google Scholar]
- 47. Carrier Y, Ma HL, Ramon HE, et al. Inter-regulation of Th17 cytokines and the IL-36 cytokines in vitro and in vivo: Implications in psoriasis pathogenesis. J Invest Dermatol 2011;131:2428–37. [DOI] [PubMed] [Google Scholar]
- 48. Banerjee S, McGee DW. ROCK activity affects IL-1–induced signaling possibly through MKK4 and p38 MAPK in Caco-2 cells. In Vitro Cell Dev Biol Anim 2016;52:878–84. [DOI] [PubMed] [Google Scholar]
- 49. Yan L, Liu W, Zhang H, et al. Ube2g2-gp78–mediated HERP polyubiquitylation is involved in ER stress recovery. J Cell Sci 2014;127:1417–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu W, Shang Y, Zeng Y, et al. Dimeric Ube2g2 simultaneously engages donor and acceptor ubiquitins to form Lys48-linked ubiquitin chains. EMBO J 2014;33:46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maukonen J, Aura AM, Niemi P, et al. Interactions of insoluble residue from enzymatic hydrolysis of Brewer’s spent grain with intestinal microbiota in mice. J Agric Food Chem 2017;65:3748–56. [DOI] [PubMed] [Google Scholar]
- 52. Peng C, Zhang S, Lei L, et al. Epidermal CD147 expression plays a key role in IL-22–induced psoriatic dermatitis. Sci Rep 2017;7:44172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mijac DD, Janković GL, Jorga J, Krstić MN. Nutritional status in patients with active inflammatory bowel disease: prevalence of malnutrition and methods for routine nutritional assessment. Eur J Intern Med 2010;21:315–9. [DOI] [PubMed] [Google Scholar]
- 54. Nguyen DL, Limketkai B, Medici V, Saire Mendoza M, Palmer L, Bechtold M. Nutritional strategies in the management of adult patients with inflammatory bowel disease: Dietary considerations from active disease to disease remission. Curr Gastroenterol Rep 2016;18:55. [DOI] [PubMed] [Google Scholar]
- 55. Forbes A, Escher J, Hébuterne X, et al. ESPEN guideline: Clinical nutrition in inflammatory bowel disease. Clin Nutr 2017;36:321–47. [DOI] [PubMed] [Google Scholar]
- 56. Campbell K, Vowinckel J, Keller MA, Ralser M. Methionine metabolism alters oxidative stress resistance via the pentose phosphate pathway. Antioxid Redox Signal 2016;24:543–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ceyhan D, Danişan A, Oğüş IH, Ozer N. Purification and kinetic properties of 6-phosphogluconate dehydrogenase from rat small intestine. Protein J 2005;24:293–301. [DOI] [PubMed] [Google Scholar]
- 58. Nanakin A, Fukui H, Fujii S, et al. Expression of the REG IV gene in ulcerative colitis. Lab Invest 2007;87:304–14. [DOI] [PubMed] [Google Scholar]
- 59. von Stein P, Lofberg R, Kuznetsov NV, et al. Multigene analysis can discriminate between ulcerative colitis, Crohn’s disease, and irritable bowel syndrome. Gastroenterology 2008;134:1869–81; quiz 2153–4. [DOI] [PubMed] [Google Scholar]
- 60. Heiskala K, Andersson LC. Reg IV is differently expressed in enteroendocrine cells of human small intestine and colon. Regul Pept 2013;183:27–34. [DOI] [PubMed] [Google Scholar]
- 61. Qin X. Etiology of inflammatory bowel disease: A unified hypothesis. World J Gastroenterol 2012;18:1708–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Meuwis MA, Fillet M, Geurts P, et al. Biomarker discovery for inflammatory bowel disease, using proteomic serum profiling. Biochem Pharmacol 2007;73:1422–33. [DOI] [PubMed] [Google Scholar]
- 63. Kanmura S, Uto H, Numata M, et al. Human neutrophil peptides 1–3 are useful biomarkers in patients with active ulcerative colitis. Inflamm Bowel Dis 2009;15:909–17. [DOI] [PubMed] [Google Scholar]
- 64. Hatsugai M, Kurokawa MS, Kouro T, et al. Protein profiles of peripheral blood mononuclear cells are useful for differential diagnosis of ulcerative colitis and Crohn’s disease. J Gastroenterol 2010;45:488–500. [DOI] [PubMed] [Google Scholar]
- 65. Nanni P, Levander F, Roda G, Caponi A, James P, Roda A. A label-free nano-liquid chromatography–mass spectrometry approach for quantitative serum peptidomics in Crohn’s disease patients. J Chromatogr B Analyt Technol Biomed Life Sci 2009;877:3127–36. [DOI] [PubMed] [Google Scholar]
- 66. Subramanian V, Subramanian D, Pollok RC. S1182 serum protein signatures determined by mass spectrometry [SELDI-ToF] accurately distinguishes Crohn’s disease [CD] from ulcerative colitis [UC]. Gastroenterology 2008;134:A–196. [Google Scholar]
- 67. Han NY, Choi W, Park JM, Kim EH, Lee H, Hahm KB. Label-free quantification for discovering novel biomarkers in the diagnosis and assessment of disease activity in inflammatory bowel disease. J Dig Dis 2013;14:166–74. [DOI] [PubMed] [Google Scholar]
- 68. Wasinger VC, Yau Y, Duo X, et al. Low mass blood peptides discriminative of IBD severity: A quantitative proteomic perspective. Mol Cell Proteomics 2016;15:256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Townsend P, Zhang Q, Shapiro J, et al. Serum proteome profiles in stricturing Crohn’s disease: A pilot study. Inflamm Bowel Dis 2015;21:1935–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nanni P, Parisi D, Roda G, et al. Serum protein profiling in patients with inflammatory bowel diseases using selective solid-phase bulk extraction, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and chemometric data analysis. Rapid Commun Mass Spectrom 2007;21:4142–8. [DOI] [PubMed] [Google Scholar]
- 71. M’Koma AE, Seeley EH, Washington MK, et al. Proteomic profiling of mucosal and submucosal colonic tissues yields protein signatures that differentiate the inflammatory colitides. Inflamm Bowel Dis 2011;17:875–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Seeley EH, Washington MK, Caprioli RM, M’Koma AE. Proteomic patterns of colonic mucosal tissues delineate Crohn’s colitis and ulcerative colitis. Proteomics Clin Appl 2013;7:541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett 2009;583:3966–73. [DOI] [PubMed] [Google Scholar]
- 74. Ghazalpour A, Bennett B, Petyuk VA, et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet 2011;7:e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.