Abstract
Genome-wide association studies (GWAS) have identified genetic variants at 34 loci contributing to age-related macular degeneration (AMD)1–3. We generated transcriptional profiles of postmortem retina from 453 controls and cases at distinct stages of AMD and integrated retinal transcriptomes, covering 13,662 protein-coding and 1,462 non-coding genes, with genotypes at over 9 million common single nucleotide polymorphisms (SNPs) for expression quantitative trait loci (eQTL) analysis of a tissue not included in Genotype-Tissue Expression (GTEx) and other large datasets4, 5. Cis-eQTL analysis identified 10,474 genes under genetic regulation, including 4,541 eQTLs detected only in the retina. Integrated analysis of AMD-GWAS with eQTLs ascertained likely target genes at six reported loci. Using transcriptome-wide association analysis (TWAS), we identified three additional genes, RLBP1, HIC1 and PARP12, after Bonferroni correction. Our studies expand the genetic landscape of AMD and establish the Eye Genotype Expression (EyeGEx) database as a resource for post-GWAS interpretation of multifactorial ocular traits.
AMD is a leading cause of incurable vision impairment, resulting in progressive loss of photoreceptors particularly in the macular region of the retina1. AMD-GWAS have identified strong and highly replicated association of 52 independent SNPs at 34 genetic loci accounting for over 50% of the genetic heritability3. To derive mechanistic insights and further advance AMD genetics, we initiated the EyeGEx project to elucidate genetic regulation of gene expression in the human retina. We characterized 523 post-mortem donor retina using the Minnesota Grading System (MGS)6, with criteria similar to the Age-related Eye Disease Study (AREDS)7 (Supplementary Fig. 1, Supplementary Data 1). MGS1 donor retinas demonstrated no AMD features and serve as control, whereas MGS2 to MGS4 samples represent progressively more severe disease stages.
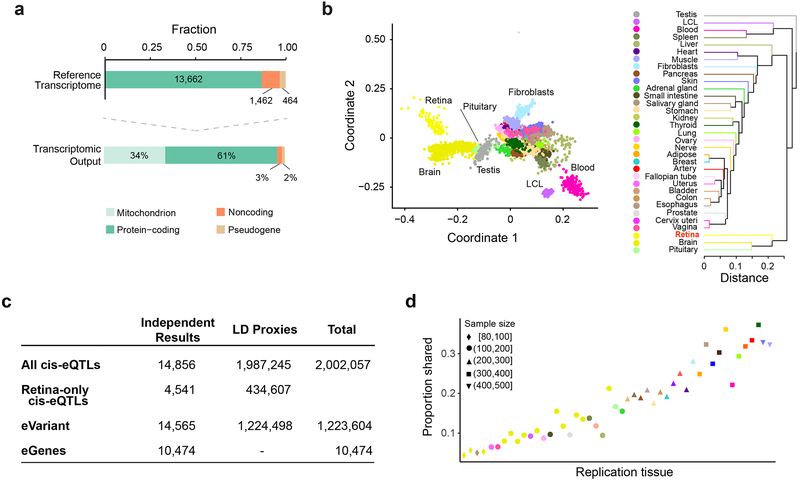
RNA-seq of donor retinas provided 32.5 million (median) uniquely mapped paired-end reads per sample with 94% mapping rate to Ensembl release GRCh38.p7 (Supplementary Fig. 2). After RNA-seq quality control (see Supplementary Notes), 105 MGS1, 175 MGS2, 112 MGS3, and 61 MGS4 samples were selected for further analyses. The reference transcriptome profile was generated from MGS1 control retinas (Fig. 1a; Supplementary Data 2) and included 67% of the protein-coding genes (13,662) and 6.7% of the non-coding genes (1,462) in Ensembl, consistent with a previous study8. High-abundance genes (186 genes showing ≥100 Fragments Per Kilobase of transcript per Million mapped reads; FPKM) accounted for half of the Ensembl-annotated transcripts in our RNA-seq data and were enriched for visual perception, metabolic processes, and energy homeostasis (Supplementary Fig. 3a; Supplementary Data 2). Overall, 34% of the retinal transcripts were of mitochondrial origin (Fig. 1a, Supplementary Fig. 3b), reflecting the high concentration of mitochondria in photoreceptors9, which are the predominant cell type in human retina10.
Figure 1.
EyeGEx: retinal transcriptome and eQTL analyses.
(a) Reference transcriptome output from 105 MGS1 control donor retinas. Top: Fraction of expressed genes in Ensembl gene biotypes. Below: Percentage of gene expression in distinct gene subtypes.
(b) Within-tissue sample similarity and transcriptome comparison across the retina (n = 105 MGS1 retinas) and the GTEx tissues (v7) (n = 6,421 samples across all body sites) based on normalized gene expression levels. Each color represents a distinct tissue. Left: multidimensional scaling. Right: tissue hierarchical clustering.
(c) A summary of retinal cis-eQTLs, eGenes and eVariants. 1.8% of the top eVariants (14,565) regulate more than one eGene. Variants in LD with the most significant eVariant are indicated as LD proxies. LD, linkage disequilibrium.
(d) The proportion of cis-eQTLs in the retina (y-axis) that are detected in GTEx (x-axis), ordered by the sample size of each tissue. Color and shape of each point represent the tissue and sample size, respectively.
Genome-guided transcript assembly supplemented 410 putative novel lincRNA and 2,861 protein-coding isoforms of genes expressed in the retina (Supplementary Fig. 3c; Supplementary Data 2). Putative lincRNA isoforms were not enriched for any biological pathway. In contrast, predicted gene function and classification of novel protein-coding isoforms showed enrichment in Gene Ontology (GO) biological processes involving synapse structure or activity (adjusted P value = 1.37 × 10−2), sensory perception (adjusted P value = 1.64 × 10−2), regulation of membrane potential (adjusted P value = 3.45 × 10−2), and photoreceptor maintenance (adjusted P value = 3.45 × 10−2). The multidimensional scaling plot of the retina reference transcriptome against the GTEx v7 data distinguished tissue-specific clusters consistent with the defined biological replicates, whereas tissue hierarchical clustering on the mean gene expression levels revealed a high degree of similarity between brain and retina (Fig. 1b; Supplementary Fig. 3d, Supplementary Fig. 4). We identified 247 genes with 10-fold or higher expression in the retina relative to at least 42 of the 53 GTEx (v7) tissues (Supplementary Data 2).
Mapping of cis-eQTLs [as defined by SNP-gene combination within ± 1 Mb of the transcriptional start site (TSS) of each gene] (see Methods)identified 14,565 genetic variants (eVariants) that control expression of 10,474 genes (eGenes) at false-discovery rate (FDR) ≤ 0.05; these included 8,529 known protein-coding and 1,358 non-coding genes (Fig. 1c; Supplementary Data 3). The strength of association was contingent upon the eVariant’s distance from the TSS of its corresponding eGene (Supplementary Fig. 5). A majority of the retinal cis-eQTLs were present in at least one GTEx tissue, with more retinal eQTLs replicated with increase in GTEx tissue sample size (Fig. 1d). The proportion of GTEx cis-eQTLs replicated in the retina was larger for GTEx tissues with smaller sample sizes5 (see Supplementary Fig. 5f). Almost one-third of re tina-only eQTLs observed in our study, compared to those reported by GTEx for other tissues, can be attributed to the relatively larger sample size (Supplementary Fig. 6a,b).
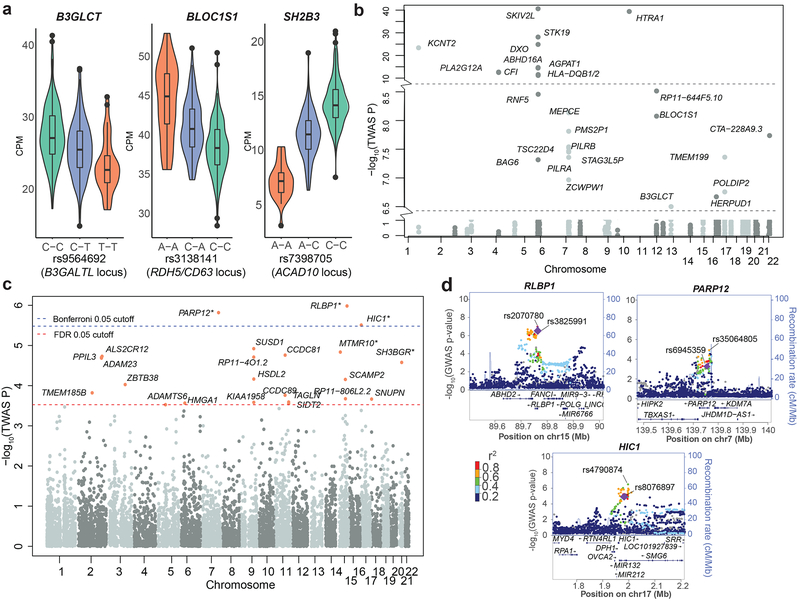
We examined the global role of eQTLs in the genetics of AMD. Q-Q plots identified cis-eQTL SNPs to be enriched for AMD associations with more pronounced enrichment for eVariants shared across several tissues11, 12, and this relationship remained relatively consistent across all other complex disease phenotypes examined (see Supplementary Fig. 5g). We then integrated retina eQTL results with associations reported across loci identified by AMD-GWAS (Supplementary Table 1). Nine lead SNPs at the GWAS loci were significant eQTLs in the retina for 19 SNP-gene associations. Similar analysis showed a comparable number of lead SNPs as eQTLs in several GTEx tissues (see Supplementary Data 3). To ascertain the most likely causal variants, we applied eCAVIAR, which calculates the colocalization posterior probability (CLPP) to identify the variant responsible for both AMD-GWAS and retina-eQTL signals, after accounting for local linkage disequilibrium (LD) patterns. At the recommended threshold of 1% CLPP13, we discovered likely causal SNPs and underlying target genes at six AMD loci (Supplementary Table 1, Fig. 2a). The lead GWAS signal at two loci (B3GALTL and RDH5/CD63) was identified as the most likely causal SNP, whereas the likely causal variant was distinct from the lead SNP at four other loci; SLC16A8 (rs5756908), ACAD10 (rs7398705), TMEM/VTN (rs241777), and APOE (rs157580) (Supplementary Table 1).
Figure 2.
Genes and variants associated with AMD using retina eQTL data (n = 406 retinas) and summary level AMD-GWAS data (based on z-scores of two-sided t-tests using 33,976 individuals)3.
(a) Violin plots of the relationship between the variant at a GWAS locus and the target gene identified by eCAVIAR. At three loci, the target gene shown was the only one significantly associated (FDR ≤ 0.05) by TWAS. The y-axis represents the distribution of expression levels (CPM) of each gene, whereas the x-axis shows the genotype (orange; homozygous minor allele, green; homozygous major allele, and blue; heterozygous) for a given SNP. Box plots depict the median (thick black horizontal bar), the interquartile range, and minimum and maximum CPM values.
(b) TWAS results (n = 406 retinas) for genes that pass Bonferroni-corrected significance identified within 1 Mb on either side of the lead SNP at previously-reported GWAS loci. PLEKHA1 (TWAS P value = 7.91 × 10−119) was omitted for appropriate scaling, and the horizontal lines indicate y-axis break.
(c) Manhattan plot of TWAS-identified genes outside the reported lead SNP (> 1 Mb on either side) at the GWAS loci. Of the genes with expression model R2 > 0.01, 23 genes met the FDR threshold of 0.05 (red line), and three of these passed Bonferroni-corrected significance (cutoff shown as blue line).
(d) LocusZoom plots showing empirical GWAS association for top three TWAS signals outside GWAS loci. The diamonds indicate top eVariants for independent eQTL signals. The coloration of the points is determined by their LD with respect to the eQTL in purple. The top GWAS variant in the region is also labeled. The recombination rate is shown as a blue line.
We leveraged retinal eQTLs and the most recent GWAS data3 to detect novel AMD risk genes in a transcriptome-wide association study (TWAS)14 using our retina transcriptome data. Gene expression was modeled using SNPs within a 1-Mb window using mixed models, Least Absolute Shrinkage and Selection Operator (LASSO), and elastic net. The TWAS identified 61 transcriptome-wide significant gene-AMD associations (FDR ≤ 0.05), which passed a gene expression model fit filter (R2 > 0.01) (Supplementary Data 4). We detected 38 genes within 1 Mb of 13 AMD-GWAS loci, and of these, 28 passed genome-wide Bonferroni correction (Fig. 2b). TWAS analysis also identified 23 genes outside the GWAS loci (Fig. 2c); these genes fell within 16 separate regions (± 1 Mb). Three of these – RLBP1, PARP12 and HIC1 – were the only significant genes in the region and remained so even after Bonferroni correction, thus representing the strongest new candidate AMD-associated genes (Fig. 2d). Conditional testing of the full 61 significant (FDR ≤ 0.05) candidates identified 47 independent signals (α = 0.05). A permutation test (see Methods) demonstrated two of the genes (MTMR10 and SH3BGR) at least 1 Mb outside of any GWAS region, with TWAS associations significantly informed by eQTL data after Bonferroni correction for the number of genes permuted (α = 0.05; Supplementary Data 4). However, we note that the test is overly conservative in the presence of LD.
We compared the data from eQTL, eCAVIAR, and TWAS to highlight the most plausible target genes; B3GLCT and BLOC1S1 were each identified as the only target gene at two AMD loci by all three methods, whereas SH2B3, PLA2G12A, PILRB and POLDIP2/TMEM199 were likely targets at four additional loci by two methods (Table 1, Supplementary Fig. 7). A comparison of these findings with those reported in GTEx5, 15 showed that the contribution of these SNPs to gene regulation varied across different tissues (Supplementary Data 3; Section 3.4 in Supplementary Notes). Specifically, no single non-retina tissue showed replication of retinal findings for all SNP-target gene combinations (see Supplementary Data 3).
Table 1:
Significant target genes and variants for AMD susceptibility at GWAS loci after eQTL, eCAVIAR and TWAS analyses
| AMD Locus | Lead GWAS SNP |
Chr:Position | GWAS_pval | eQTL_pval | Target gene(s) | % Variability Explained |
Significant TWAS gene at the locus (FDR) |
|---|---|---|---|---|---|---|---|
| B3GALTL | rs9564692 | 13:31821240 | 3.31 × 10−10 | 2.36 × 10−11* | B3GLCT† | 10.47 | B3GLCT (1.34 × 10−4) |
| RDH5/CD63 | rs3138141 | 12:56115778 | 4.3 × 10−9 | 5.69 × 10−19* | BLOC1S1† | 17.8 | BLOC1S1 (7.06 × 10−6) |
| ACAD10 | rs61941274 | 12:112132610 | 1.07 × 10−9 | 8.95 × 10−2 | SH2B3† | 0.71 | SH2B3 (0.0217) |
| CFI | rs10033900 | 4:110659067 | 5.35 × 10−17 | 3.98 × 10−7* | PLA2G12A | 6.17 | CFI (3.01 × 10−10), PLA2G12A (4.30 × 10−10) |
| PILRB/PILRA | rs7803454 | 7:99991548 | 4.76 × 10−9 | 3.57 × 10−77* | PILRB, PILRA, ZCWPW1, TSC22D4 | 57.51 | MEPCE (6.51 × 10−6), PILRB (2.06 × 10−5) |
| TMEM97/VTN | rs11080055 | 17:26649724 | 1.04 × 10−8 | 8.37 × 10−19* | POLDIP2, SLC13A2**, TMEM199† | 17.65 | TMEM199 (2.55 × 10−5), POLDIP2 (8.60 × 10−5) |
eQTL is significant after correction for multiple testing.
Target of causal variant identified by eCAVIAR.
Retina-specific eQTL.
Only protein-coding genes are shown here. B3GLCT is the new gene symbol for B3GALTL. SH2B3 was identified by GWAS co-localization (eCaviar) and TWAS, two of the three criteria used to identify target genes in our study. Despite its high eQTL P value, SH2B3 is an excellent biological candidate for AMD because of its association with inflammation. eQTL analysis was based on 406 post-mortem donor retina samples.
Differential expression (DE) analysis of retinal transcriptomes identified 14 genes with and 161 genes without age correction in advanced AMD (FDR ≤ 0.20) (Supplementary Data 5; Supplementary Fig. 8a). Thus, like other complex diseases16, 17, our DE analysis did not detect many gene expression changes, probably because of heterogeneity caused by aging, polygenic inheritance, and environmental factors. We then examined biological pathways by gene set enrichment analysis (GSEA). Immune regulation and cholesterol metabolism pathways, previously implicated in GWAS3, were upregulated in early and advanced AMD, whereas pathways associated with synapse development and function were largely and exclusively downregulated in intermediate AMD (Supplementary Data 5). We note that a majority of the genes within susceptibility loci for advanced AMD do not appear to be associated with intermediate AMD despite having sufficient power3. Thus, intermediate AMD may not be a transitional stage between early and advanced AMD, but a separate entity with unique and distinct genetic underpinning(s) that require further exploration. Furthermore, Weighted Gene-Co-expression Network Analysis (WGCNA) of all samples suggested that several of the pathways implicated in AMD operate through closely connected networks in the retina (Supplementary Fig. 8b,c; Supplementary Data 6).
GWAS have successfully identified variants at hundreds of loci that contribute to healthy and disease traits, thereby defining their broad genetic architecture18, 19. Interpretation of GWAS findings, however, remains a major challenge since a large proportion of associated variants is not in the protein-coding genomic regions and their impact on specific phenotypes often individually appears to be small20, 21. eQTL analysis in disease-relevant tissues appears to be a prominent tool for biological interpretation of GWAS loci11, 22. Owing to the large sample size, we were able to identify 14,856 eQTLs that modulate retinal gene expression, and a significant proportion is not reported in GTEx v7 data. More significantly, we could connect the lead GWAS signal to specific target genes at six known AMD loci by at least two lines of evidence. Two of the target genes were validated by three independent methods: B3GLCT encodes a glucosyltransferase23, and its loss of function leads to Peters Plus syndrome24; BLOC1S1 encodes a subunit of a multiprotein complex associated with the biogenesis of an organelle of endosome-lysosome system25, and its altered function can affect synaptic function26. Thus, altered expression of B3GLCT and BLOC1S1 might impact extracellular matrix stability or signaling and degradation of unwanted/recycled proteins, respectively, thereby contributing to AMD pathogenesis. We attribute the lack of obvious target genes at remaining AMD-GWAS loci to multiple factors, including LD structure, variants affecting expression in trans or in other AMD-relevant tissues (such as retinal pigment epithelium and choroid) and power of this study. Interpretation of eVariants that regulate multiple genes at a particular locus requires further biological validation.
AMD is fairly unique among complex traits because of its high heritability and large effect sizes for individual GWAS SNPs3. We show that variants associated with gene expression across many tissues as eQTLs, as opposed to those with tissue-specific associations only, are enriched for AMD associations despite high tissue specificity (see Supplementary Data 3). We hypothesize that, at least in part, such associations reflect larger, more robust effects among the shared eQTLs. Not surprisingly, retina is the only tissue for which we detected regulation consistently across all six identified SNPs (Supplementary Data 3). In addition, 36 of the 61 retina-identified TWAS candidates were significant (FDR ≤ 0.05) in at least one GTEx tissue. The remaining candidates could not be analyzed because of either no expression or heritability in the GTEx tissues or were not replicated in any other tissue. Our results corroborate recent studies12, 27 and suggest that the best way to increase power for discovery of genes using TWAS and similar approaches is to increase the diversity of tissues for greater resolution of the impact of regulatory variants. We emphasize, however, that eQTL effects detected only in a non-biologically relevant tissue, but not in a relevant one, would be difficult to interpret for disease-specific phenotypes. Although other tissues may contribute to AMD, retinal effects of eQTLs are more likely to be directly relevant. We suggest that eQTL analyses of retinal pigment epithelium and choroidal endothelial cells would further contribute to understanding of genes involved in AMD pathobiology. AMD-associated genes uncovered by TWAS provide additional insights into the relevance of gene regulation on phenotypic consequences in this complex disease.
EyeGEx complements the GTEx project and provides a reference for biological interpretation of genetic variants associated with common ocular traits, including glaucoma and diabetic retinopathy. Comparative analysis of retinal transcriptomes and eQTLs with the GTEx data should assist in exploring biological questions relating to visual function in syndromic and multifactorial traits.
ONLINE METHODS
Study subjects.
Post-mortem human donor eyes were procured by the Minnesota Lions Eye Bank after informed consent from the donor or next of kin and in accordance with the tenets of the Declaration of Helsinki. Exclusion criteria for donors included a history of diabetes or glaucoma. Donors were also excluded from this study if, upon examination of donor macular images, there were clinical symptoms of diabetic retinopathy, advanced glaucoma, myopic degeneration, or the presence of atypical debris in the eyes. Donor eyes were enucleated within four hours of death and stored in a moist chamber at 4°C until retinal dissection was performed. Dissection and classification of donor retinas for AMD were carried out according to the four-step Minnesota Grading System (MGS) as previously described6, 28. Tissue sections were flash frozen in liquid nitrogen and stored in −80°C until further processing. Samples with ambiguous or no MGS levels were excluded from downstream analysis. Details of donor characteristics are described in the Supplementary Notes.
GTEx data.
RNA-seq and genotyping data from the Genotype-Tissue Expression (GTEx) release v7 were downloaded from the Database of Genotypes and Phenotypes (dbGaP) under accession phs000424.v7.p2 and from the GTEx portal (see URLs), respectively.
RNA-seq, genotyping, and QC.
Details of RNA-seq, genotyping, and quality control are provided in the Supplementary Notes.
Batch correction.
Surrogate variables were identified and estimated for known batch effects as well as latent factors using the supervised SVA (SSVA) (version 3.28.0/3.24.4) method29–31 based on the model;
Negative control genes for SSVA were selected from a reported list of 3,804 housekeeping genes that are uniformly expressed across 16 human tissues32. The Pearson method was used to observe correlations between all significant surrogate variables identified by SSVA and possible sources of variation, including biological and technical factors. Known batch effects were assessed using Principal Variance Component Analysis (PVCA) (version 1.23.0) before and after batch correction33. All surrogate variables identified by SSVA were used for batch correction. Additional details are described in the Supplementary Notes.
Reference transcriptome.
The transcriptome profile of control human retina was generated from 105 MGS1 control retinas by applying two criteria for gene expression, the first to remove lowly expressed genes across all MGS stages [i.e., ≥ 1 Counts Per Million (CPM) in ≥ 10% of all 453 samples], and the second to describe the transcriptomic landscape in retina with greater confidence (i.e., ≥ 2 CPM in ≥ 50% of all 105 MGS1 samples). We calculated the cumulative transcriptional output as previously defined 34 by converting CPM into fragments per kilobase of transcript per million mapped reads (FPKM) values to take gene length into account. Similarities in transcriptomes between the retina and 53 GTEx tissues were observed with a gene filter of ≥ 1 CPM in ≥ 10% of all samples across all tissues whereas a different gene filter, namely ≥ 1 CPM in ≥ 10% of samples within each tissue, was applied to identify genes that were expressed at least 10-fold higher in retina compared to other tissues. Pathway enrichment analysis was performed using Gene Ontology (GO) biological process terms35, 36 within clusterProfiler version 3.4.437 using a Benjamini-Hochberg adjusted P value ≤ 0.05 as the significance threshold. The analysis and classification of potentially novel isoforms of known genes and unknown, intergenic transcripts were performed using the Cufflinks suite version 2.2138, 39, and further details are provided in the Supplementary Notes.
Comparison of transcriptomes across retina and GTEx tissues.
Raw GTEx v7 RNA-seq data were analyzed through our bioinformatics pipeline as aforementioned for retina. Effects due to differences in bioinformatics pipelines between our analysis and that of GTEx were compared as noted in the Supplementary Notes.
cis-eQTL mapping.
The analysis included 406 individuals for whom genotype and retina gene expression data were available, 17,389 genes that were expressed at ≥ 1 CPM in at least 10% of the retina samples, and 8,924,684 genotyped and imputed common variants. cis-eQTL analysis was conducted with QTLtools version 1.040, using a linear model to adjust for disease status (MGS level), age, sex, population stratification (10 principle components), and batch effects (21 surrogate variables). In the first step of the analysis, the variant most associated with each gene was selected, and then permutation was used to determine the distribution of its test statistic under the null. This was subsequently used to obtain the P value for each gene. These P values were adjusted for multiple testing using the q-value approach 41 at the desired Type I error level. The second step of the analysis involved the identification of all eVariants with independent effects on a given eGene (significant gene from the first stage). This was done by using the gene-level thresholds derived from the first stage, and then identifying which variants exhibit nominal P values below these thresholds based on the forward-backward stepwise regression algorithm.
GTEx comparison.
To calculate π1 we compared our cis-eQTL discoveries using the following definition:
π1 = P(cis-eQTL in discovery tissue is significant in replication tissue ∣ cis-eQTL in discovery tissue was also analyzed in the replication tissue)
Thus, for each cis-eQTL (gene-variant combination) we required that the combination was analyzed in both tissues being compared.
GWAS Lead Variant analysis.
Forty-one lead variants from AMD-GWAS3 were analyzed. Those not found were either not in the reference dataset used for imputation (6 variants) or did not pass our MAF threshold of < 1% (5 variants). Matrix eQTL version 2.1.142 was then used to obtain the marginal associations using the same cis criteria, which were then corrected for multiple testing only for the number of variants tested using the Bonferroni method with a Type I error rate of 5%.
Enrichment.
Q-Q plots for each GWAS dataset were processed in general by removing all SNPs within ± 1 Mb of the known GWAS signals, sub-setting to variants with MAF of at least 5%, and after removing variants in the major histocompatibility region. The remaining variants were then grouped based on eQTL characteristics. See Supplementary Notes for details.
Colocalization.
Likely colocalizing variants between the eQTL and the GWAS data were identified using eCAVIAR version 2.013 (see Supplementary Notes) based on marginal statistics from the cis-eQTL analysis and from AMD GWAS3.
TWAS.
To perform the transcript-wide association study, the log-transformed, SSVA-corrected expression data from the 406 samples in our dataset that both passed RNA-seq and genotyping quality control were inverse-normal transformed (rank offset = 3/8)43 to moderate the influence of potential outliers. Expression was then controlled for gender, age, and the ten population structure variables determined by Eigenstrat version 7.2.144, 45. For each gene, we took the subset of SNPs within 1 Mb of its start or end site that had GWAS statistics3 using VCFtools version 0.1.1546. We used Gusev et al.’s TWAS implementation14; heritability was calculated using GCTA version 1.2147, and genetic control of expression was modeled with either mixed models, LASSO, or elastic net (α = 0.5), depending on which of the three methods produced the highest five-fold cross-validation R2.
The effect sizes from these models acted as weights. Weighted z-scores were summed for each gene, and this gene-trait association statistic was divided by its standard deviation while accounting for LD between GWAS statistics. Standardized gene-level scores were tested against the standard normal distribution on both sides. The FDR was calculated to account for multiple testing across genes with calculated P values; genes that had an FDR < 0.05 were considered significant. We also determined whether genes passed a 0.05 significance threshold after Bonferroni correction. Genes were then filtered by their model expression fit; genes which had a genetic model R2 < 0.01 were discarded.
We also performed a permutation test to determine the role the eQTL data played in the associations: for genes with a TWAS P value of less than 0.001, weights were randomly assigned to SNPs and the gene-level z-scores was recomputed for an adaptive number of iterations to generate a null distribution against which the original TWAS statistic was tested14. See Supplementary Notes for details on the methods used for the conditional TWAS test.
Differential expression.
Differential expression was assessed using the limma package in R version 3.34.248 with a significance threshold of FDR ≤ 0.20. MGS was treated as an ordinal variable in pairwise comparisons between controls and each AMD stage. Differential expression was performed with adjustments for sex and batch effects (22 surrogate variables), and with and without age as a covariate. Age is the most significant non-genetic risk factor for AMD, and age-related gene expression changes would likely be relevant to AMD. We therefore also performed differential expression analysis without correcting for age to generate a comprehensive list of candidate genes that require further investigation to ascertain their contribution to AMD pathogenesis. Additional differential expression analyses, performed after removing samples with conditions such as hypertension, high cholesterol and cardiovascular disease, were consistent across all comparisons made (data not shown).
Gene Set Enrichment Analysis and Leading-Edge Analysis.
Gene set enrichment analysis (GSEA) was performed by pre-ranking genes by significance and direction of fold change from differential expression analysis, and then testing for association with the Gene Ontology biological process gene set deposited in the GSEA MSigDB resource version 2.2.449. Leading edge analysis was performed on gene sets reaching a significance threshold of FDR ≤ 0.25 and absolute normalized enrichment score of ≥ 2.0. Significant gene sets were further classified into common functional categories by visualizing the gene ontology structure as described in Supplementary Notes (see URLs).
Weighted Gene-correlation Network Analysis.
WGCNA50 was performed on all 453 samples that passed RNA-seq QC in order to group genes by expression profile, using its associated software version 1.51. log-transformed expression values were corrected for age, sex, and batch effects (determined by SSVA29–31). Adjacency was calculated using Spearman correlation, and the power with which to raise the absolute values of the correlation to obtain the adjacency matrix was k = 3. Using hypergeometric testing at a significance threshold of 0.05 alpha-level after Bonferroni correction for multiple testing, modules were assessed for the enrichment of the following types of genes: (1) genes deemed relevant to macular degeneration pathogenesis in the literature, (2) genes within 500 kb of the 34 AMD loci identified through GWAS3, and (3) genes identified as leading edge by GSEA49. A list of genes that were relevant to AMD was obtained from one of the previous published studies51 and was updated through extensive PubMed search (through December 2017) using one of several search terms (See Supplementary Notes). Pathway analysis was performed on each module using Gene Ontology biological process terms35, 36 through clusterProfiler version 3.4.437. The connections between genes in modules were visualized using Cytoscape version 3.5.152.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Bernhard Weber for providing liver eQTL data. We thank members of the Swaroop Laboratory, especially Hyunjin Yang, John Bryan, Ash Police Reddy, and Felipe Giuste for assistance, and the Lions Gift of Sight members for procuring human retina. This work was supported by the Intramural Research Program of the National Eye Institute EY000450 and EY000474 (to A.S.), NIH grants EY028554 and EY026012, The Lindsay Family Foundation, an anonymous benefactor, and the Minnesota Lions Vision Foundation (to D.A.F.), and Johns Hopkins Bloomberg Distinguished Professorship Endowment (to N.C.). This study utilized the high-performance computational capabilities of the Biowulf Linux cluster (see URLs).
Footnotes
Accession codes
These studies were approved by respective Institutional Review Boards. The sequencing data are available at Gene Expression Omnibus (GEO) accession GSE115828 and NEI Commons (see URLs). The GTEx data used here were obtained from the GTEx Portal on 03/26/18 and/or dbGaP accession number phs000424.v7.p2.
Competing Financial Interests
The authors declare no competing financial interests, except that G.R.A. is now employed by Regeneron Pharmaceuticals.
URLs
1000 Genomes Project reference panel: http://www.internationalgenome.org/
Retinal Information Network (RetNet): https://sph.uth.edu/retnet/
GTEx: https://www.gtexportal.org/home/https://www.gtexportal.org/home/
Gene ontology structure: http://www.informatics.jax.org/vocab/gene_ontology/
HMMER: http://hmmer.org/
FastQC: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Precomputed TWAS weights: http://gusevlab.org/projects/fusion/
NEI Commons: https://neicommons.nei.nih.gov/#/
Biowulf Linux cluster: http://biowulf.nih.gov
REFERENCES
- 1.Fritsche LG, et al. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet 15, 151–171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grassmann F, Ach T, Brandl C, Heid IM & Weber BHF What Does Genetics Tell Us About Age-Related Macular Degeneration? Annu Rev Vis Sci 1, 73–96 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Fritsche LG, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet 48, 134–143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Small KS, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet 43, 561–564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium, G.T., et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olsen TW & Feng X The Minnesota Grading System of eye bank eyes for age-related macular degeneration. Invest Ophthalmol Vis Sci 45, 4484–4490 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Ferris FL, et al. A simplified severity scale for age-related macular degeneration: AREDS Report No. 18. Arch Ophthalmol 123, 1570–1574 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinelli M, et al. An atlas of gene expression and gene co-regulation in the human retina. Nucleic Acids Res 44, 5773–5784 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoang QV, Linsenmeier RA, Chung CK & Curcio CA Photoreceptor inner segments in monkey and human retina: mitochondrial density, optics, and regional variation. Vis Neurosci 19, 395–407 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Curcio CA, Sloan KR, Kalina RE & Hendrickson AE Human photoreceptor topography. J Comp Neurol 292, 497–523 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Finucane HK, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamazon ER, et al. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat Genet 50, 956–967 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hormozdiari F, et al. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am J Hum Genet 99, 1245–1260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gusev A, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48, 245–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strunz T, et al. A mega-analysis of expression quantitative trait loci (eQTL) provides insight into the regulatory architecture of gene expression variation in liver. Sci Rep 8, 5865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 19, 1442–1453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gandal MJ, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359, 693–697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck T, Hastings RK, Gollapudi S, Free RC & Brookes AJ GWAS Central: a comprehensive resource for the comparison and interrogation of genome-wide association studies. Eur J Hum Genet 22, 949–952 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45, D896–D901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakravarti A, Clark AG & Mootha VK Distilling pathophysiology from complex disease genetics. Cell 155, 21–26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallagher MD & Chen-Plotkin AS The Post-GWAS Era: From Association to Function. Am J Hum Genet 102, 717–730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown CD, Mangravite LM & Engelhardt BE Integrative modeling of eQTLs and cis-regulatory elements suggests mechanisms underlying cell type specificity of eQTLs. PLoS Genet 9, e1003649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozma K, et al. Identification and characterization of abeta1,3-glucosyltransferase that synthesizes the Glc-beta1,3-Fuc disaccharide on thrombospondin type 1 repeats. J Biol Chem 281, 36742–36751 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Lesnik Oberstein SA, et al. Peters Plus syndrome is caused by mutations in B3GALTL, a putative glycosyltransferase. Am J Hum Genet 79, 562–566 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langemeyer L & Ungermann C BORC and BLOC-1: Shared subunits in trafficking complexes. Dev Cell 33, 121–122 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Mullin AP, et al. Gene dosage in the dysbindin schizophrenia susceptibility network differentially affect synaptic function and plasticity. J Neurosci 35, 325–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hormozdiari F, et al. Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits. Nat Genet 50, 1041–1047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Decanini A, Nordgaard CL, Feng X, Ferrington DA & Olsen TW Changes in select redox proteins of the retinal pigment epithelium in age-related macular degeneration. Am J Ophthalmol 143, 607–615 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gagnon-Bartsch JA & Speed TP Using control genes to correct for unwanted variation in microarray data. Biostatistics 13, 539–552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leek JT svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res 42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leek JT, Johnson WE, Parker HS, Jaffe AE & Storey JD The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eisenberg E & Levanon EY Human housekeeping genes, revisited. Trends Genet 29, 569–574 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Scherer A ed. Batch effects and noise in microarray experiments: Sources and solutions (John Wiley & Sons, Ltd, 2009). [Google Scholar]
- 34.Mele M, et al. Human genomics. The human transcriptome across tissues and individuals. Science 348, 660–665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The Gene Ontology, C. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 45, D331–D338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu G, Wang LG, Han Y & He QY clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts A, Pimentel H, Trapnell C & Pachter L Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325–2329 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Delaneau O, et al. A complete tool set for molecular QTL discovery and analysis. Nat Commun 8, 15452 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Storey JD & Tibshirani R Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100, 9440–9445 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shabalin AA Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beasley TM, Erickson S & Allison DB Rank-based inverse normal transformations are increasingly used, but are they merited? Behav Genet 39, 580–595 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patterson N, Price AL & Reich D Population structure and eigenanalysis. PLoS Genet 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Danecek P, et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Lee SH, Goddard ME & Visscher PM GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langfelder P & Horvath S WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newman AM, et al. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Medicine 4, 16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.