Abstract
Drug traversal across the blood-brain barrier has come under increasing scrutiny recently, particularly concerning the treatment of sicknesses, such as brain cancer and Alzheimer’s disease. Most therapies and medicines are limited due to their inability to cross this barrier, reducing treatment options for maladies affecting the brain. Carbon dots show promise as drug carriers, but they experience the same limitations regarding crossing the blood-brain barrier as many small molecules do. If carbon dots can be prepared from a precursor that can cross the blood-brain barrier, there is a chance that the remaining original precursor molecule can attach to the carbon dot surface and lead the system into the brain. Herein, tryptophan carbon dots were synthesized with the strategy of using tryptophan as an amino acid for crossing the blood-brain barrier via LAT1 transporter-mediated endocytosis. Two types of carbon dots were synthesized using tryptophan and two different nitrogen dopants, urea and 1,2-ethylenediamine. Carbon dots made using these precursors show excitation wavelength-dependent emission, low toxicity, and have been observed inside the central nervous system of zebrafish (Danio rerio). The proposed mechanism for these carbon dots abilities to cross the blood-brain barrier concerns residual tryptophan molecules which have attached to the carbon dots surface, enabling them to be recognized by the LAT1 transporter. The role of carbon dots for transport open promising avenues for drug delivery and imaging in the brain.
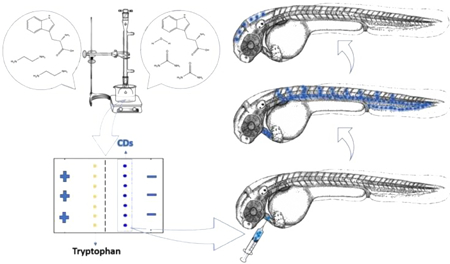
Graphical Abstract:
Summary of tryptophan carbon dots (CDs) hydrothermal preparation, purification through electrophoresis, and injection into zebrafish to assess the ability of tryptophan CDs to cross the blood-brain barrier.
Introduction
Since the discovery of carbon dots (CDs) in 2004, much research has been done to investigate their optical properties. They have been examined because they show promise as nanocarriers and bio-imaging materials due to their low toxicity, high quantum yield, and easily functionalized surface.1–3 Although CDs are usually non-toxic they are often made from toxic precursors, so the purification of CDs from the remaining precursor is an important consideration. Many different synthetic mechanisms have been studied attempting to optimize the optical properties and produce more useful CDs that can be used in bio-imaging and bio-sensing.3–5 A thermal preparation has generally been shown to produce a higher quantum yield than other common methods.6, 7 A great deal of work has also been done to compare which precursors will provide a higher quantum yield, but results in that area have been less conclusive.
1,2-ethylenediamine (EDA) is commonly used in the preparation of CDs as a nitrogen dopant because it has been shown to increase the quantum yield of photoluminescence.8–10 While it is toxic on its own, it has been shown that CDs prepared with EDA are not toxic at low and moderate concentrations.11, 12 Urea is used as a nitrogen dopant which is a less toxic alternative to EDA.7, 13 Since it is a solid, unlike EDA, a solvent is necessary to carry out the reaction with a solid carbon precursor.
EDA is commonly used with citric acid to synthesize CDs.8, 10, 11, 14 Replacing citric acid with a molecule that is already fluorescent should increase the quantum yield of the CDs formed under these conditions. The carbon precursor used in this study was tryptophan which is one of 20 essential amino acids and essential for many biological processes including the production of melatonin and seratonin.15 Because it is required in the brain, there is a specialized transporter to mediate in crossing the blood-brain barrier referred to as LAT1, which is composed of the glycoprotein CD98.16, 17 Because of this property, it would be advantageous to produce CDs which has tryptophan present on the surface so that it is possible for the CDs to cross the blood-brain barrier through the LAT1 transporter. This would remove the need for conjugations to receptors, which often possess low-yield reactions, and enable CDs to deliver drugs to the brain.
Herein, is reported CDs prepared from tryptophan and EDA (or urea) which have been characterized to have excitation wavelength-dependent emission, size less than 10 nm, and low toxicity. Surface and optical characterization shows tryptophan remaining on the surface. Additionally, the CDs prepared from EDA (CD-EDA) are shown to cross into the central nervous system of zebrafish, bypassing the blood-brain barrier. For the first time, tryptophan has been used as a precursor to develop self-targeting CDs. These CDs open the possibility to deliver drugs into the brain without tedious conjugation to receptors.
Methods
A new synthetic pathway was used in this study to generate CDs. Tryptophan and EDA were combined in a 1:14 molar ratio (1.0923 g and 5 mL respectively) and heated, with vigorous stirring, to 180 ° C for four hours. The reaction was carried out under argon gas to prevent the oxidation of the EDA. The product was transferred to a rotavapor machine to remove excess EDA. A yellow gel-like liquid remained after the removal of EDA. To remove excess, monomeric tryptophan, gel electrophoresis was performed. A 10% agarose gel was prepared in a 0.1 M Bis-Tris solution to buffer the gel (pH=5.90). The tryptophan control was stained with Coomassie blue and could be seen moving only slightly forward (figure S1), since the pH of 5.90 is just above the isoelectric point of tryptophan (5.89). Under the light of a UV lamp (365 nm), the blue photoluminescence (PL) of the CDs could be seen moving backward, toward the negative electrode. After 15 minutes, when the PL had completely separated from the wells (figures S1,S2), the gel was cut and dispersed in deionized water, and then centrifuged at 3000 rpm for 20 minutes. The supernatant was taken and filtered through a 0.2 μm syringe filter and lyophilized to obtain a gel-like substance (CD-EDA) with a very pale yellow color similar to previous reactions using EDA.10 This same process was repeated for a reaction with tryptophan and urea. The same molar ratio of 1:14 was used (0.5462 g and 2.2486 g respectively) and combined with 6 mL of deionized water with resistivity of 18.2 MΩ·cm. The evaporation step was omitted because there was no liquid precursor. In addition, the pH used for electrophoresis was 6.20 (this showed better separation for CD-Urea). This also yielded a gel-like substance (CD-Urea) which was less yellow than the corresponding product with EDA. The procedures used for toxicity assays and in vivo tests are detailed in supplementary information.
Results and Discussion
The electrophoresis separation provided some insight into the nature of the two different CDs, CD-EDA and CD-Urea. The luminescent band in both products moves backward (toward the negative electrode), in the opposite direction of tryptophan alone, suggesting that the surface of the CDs is positively charged, possibly due to protonated amine groups. The zeta potential for CD-EDA was −9.02 ± 0.17 and for CD-Urea was −10.8 ± 0.54. These measurements were recorded in deionized water of pH 7, so at the acidic pH of the electrophoresis separation, the surface of CDs was protonated, resulting in the backwards movement (toward the negative electrode). In addition, from electrophoresis, CD-EDA can be seen in a very narrow luminescent band while CD-Urea is much more widely distributed (figures S1d,S2d). Two possible explanations, based on electrophoresis separation, for this difference in the distribution of CDs are: the size distributions of the respective samples and differences in uniformity of the surfaces of the CDs in the two samples. The surface state could be affected by tryptophan and the amine groups of the precursor, but also by the carbonyl of urea which is not present in EDA. The optical properties and size measurements give insight into these options.
The UV/vis absorption spectrum of the product before purification shared similar peaks with tryptophan. After electrophoresis, the tryptophan absorption peak at 280 nm could be observed in both CDs, but there was also strong absorption between 200 and 280 nm (figures 1a,c). The peak at 280 nm can be attributed to the n→π* transition for C=O, as in tryptophan. The higher energy peak at 210 nm can be attributed to the π →π* transition in the aromatic sp2 carbons provided by tryptophan. The absorption moiety at 280 nm indicates that some tryptophan remains attached to the surface of the CDs. Based on the relative heights of the two peaks (210 and 280 nm) for each sample, it would appear that there is more tryptophan in CD-EDA than CD-Urea. This could potentially create a more homogenous surface state on CD-EDA. When the PL spectra are analyzed, greater differences can be seen between CD-EDA and CD-Urea (figures 1b,d). The photoluminescence spectrum for CD-EDA shows little shifting of the emission with increasing excitation wavelength, whereas CD-Urea shows a more excitation-dependent behavior. As mentioned previously, the absorption spectrum shows there is more tryptophan left on the surface of CD-EDA (cf. figures S3 and S4) and this may create a more homogenous surface which results in less shifting of the emission. Additionally, the maximum emission in CD-EDA was obtained from 400 nm excitation as opposed to 350 nm in CD-Urea. These values are an important consideration for imaging and enable CD-EDA to be more easily observed due to the agreement between the maximum excitation for CD-EDA and the blue excitation channel in the confocal microscope used (405 nm). The quantum yield was calculated with an excitation wavelength of 350 nm using quinine sulfate as a reference (Φ=54.6%). The result was 48.4 ± 3.6% for CD-EDA and 21.5 ± 3.2% for CD-Urea.
Figure 1:
(a) Absorption and normalized photoluminescence spectra of CD-EDA. (b) Photoluminescence spectrum of CD-EDA. (c) Absorption and normalized photoluminescence spectra of CD-Urea. (d) Photoluminescence spectrum of CD-Urea. Optical pathlength, 1 cm. Solvent, water.
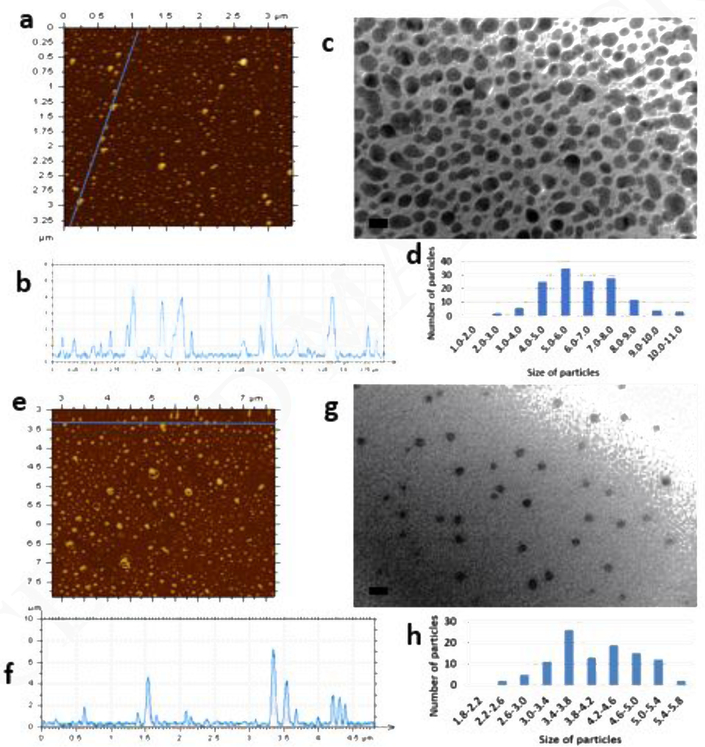
The AFM and TEM images in figure 2 show the sizes of the obtained CDs. The mean sizes from TEM measurement for CD-EDA and CD-Urea were 6.2 and 4.1 nm, respectively. The sizes obtained from AFM images are in general agreement with this, which means both CDs are roughly spherical in shape. The size distribution for CD-EDA is much wider than for CD-Urea, which was not expected based on the electrophoresis images. Therefore, it confirms that the particle distribution observed in electrophoresis resulted from the surface electronic state rather than the size-effect.
Figure 2:
(a) AFM image of CD-EDA. (b) AFM height profile of CD-EDA. (c) TEM image and histogram of CD-EDA. Scale bar is 10 nm. (d) AFM image of CD-Urea. (e) AFM height profile of CD-Urea. (f) TEM image and histogram of CD-Urea. Scale bar in TEM images is 10 nm.
To examine the surface functionalization of CDs, the two methods used were attenuated total reflection Fourier transform infrared spectroscopy (ATR-FTIR) and x-ray photoelectron spectroscopy (XPS). The peaks and assigned functional groups are summarized in Table 1a and the spectra in figures S5,S6,S7. The FTIR spectra are similar and indicate similar functional groups, which are similar to other CDs except for the lack of carboxylic group. The main difference between the two spectra can be observed in the two peaks (1665 and 1643 cm−1) above 1600 cm-1. The higher energy peak indicates either C=O of an amide group or C=N (imine group) and the slightly lower energy peak is assigned to the N-H stretch of amines or amides. The lower intensity of the higher energy peak in CD-EDA indicates a lower presence of imine or C=O groups. From the elemental analysis given by XPS (Table 1b), it can be seen that there is 10% less imine groups in CD-EDA which would explain the differences in the FTIR spectra.
Table 1:
(a) Comparison and classification XPS and FTIR characteristic peaks. (b) Elemental analysis from XPS data.
| a | |||||
|---|---|---|---|---|---|
| XPS | FTIR | ||||
| C1s | 284.9 eV | alkenyl C=C | C=C | 1690–1640 cm−1 (1665 cm−1) (shared with amide C=O and C=N) | |
| C-O bond not present in XPS analysis | -O-H | 3500 – 3200 cm−1 (3303 cm−1) | |||
| C-O | 1320–1000 cm−1 (1030 cm−1) | ||||
| O1s | 531.8 eV | amide C=O | C=ONH2 | 1690 – 1630 cm−1 (1665 cm−1) | |
| N1s | 398.0 eV | -C=N- | C=N | 1690–1640 cm−1 (1665 cm−1) (shared with amide C=O and C=C) | |
| 399.2 eV | C-N (amide or amine) | N-H stretch | 3400 – 3250 cm−1 (3303 cm−1) (obscured by –OH peak) | ||
| N-H bend | 1650–1580 cm−1 (1643 cm−1) | ||||
| C-N | 1250–1020 cm−1 (1120 cm−1) | ||||
| b | |||
|---|---|---|---|
| Element | Percent of CD-EDA | Percent of CD-Urea | |
| Carbon | 55.0% | 60.6% | |
| Oxygen | 44.7% | 39.2% | |
| Nitrogen | 0.3% (49.1% imine and 50.9% amine/amide) | 0.2% (59.4% imine and 40.6% amine/amide) |
All of the functional groups shown in the XPS data are able to be correlated with the FTIR peaks. The FTIR peaks corresponding to the C-OH functional group cannot be correlated with XPS due to the lack of a C-O peak. This can be explained by the fact that ATR-FTIR can probe samples at depths of up to 2 microns while XPS samples the surface at a depth of less than 100 Å.18, 19 Based on this, the alcohol groups shown by FTIR are further from the surface than the functional groups seen by XPS. The absence of carboxylic groups from both spectra is interesting because of the presence of the characteristic absorption of tryptophan. However, this can be explained by the formation of peptide bonds. There is a ratio between carboxylic groups and amine groups of 1:28 in the starting materials, so the probability of peptide bond formation between the two functional groups is high and would explain the disappearance of the carboxylic groups and the appearance of amide groups. The imine group could result from the isomerization of the indole group in tryptophan to an indolenine group.20, 21 This is normally not the most favorable isomer, but the carbon dot core and surface could stabilize the structure and provide the C=N functional group observed in the XPS spectra. The oxygen in the samples results from the oxidation of the carbon dot surface and not the oxidation of the amine groups during the reaction (due to the protected atmosphere by argon). This oxidation results in a very strong alcohol peak (ca. 3300 cm−1) in both CDs samples.
Because of the low percentage of nitrogen present in CD-EDA and CD-Urea, 0.3% and 0.2% respectively as seen from XPS, the only nitrogen present on the surface of CDs is likely the result of the tryptophan moieties still present. CD-EDA has a higher percentage of heteroatoms in general than CD-Urea (Table 1b). The greater presence of functional groups on the surface resulting from the greater amount of tryptophan (cf. absorption, figures 1a,c) creates the higher uniformity seen in the electrophoresis process and the fluorescence spectra for CD-EDA when compared to CD-Urea. Also, the presence of amine groups confirms the analysis of electrophoresis and zeta potential. The amine groups are easily protonated in acidic media, which makes the surface of the CDs more positive. Since the zeta potential value for both CDs is only slightly negative, the CDs displayed the positively charged characteristics in the acidic buffer that was used for electrophoresis.
Toxicity analysis show both CD samples to be non-toxic as shown in figure S8. CD-EDA showed minimal toxicity in two cancer cell lines, SJGBM2 and CHLA266, but the assay was performed at 1 mM, which is a higher concentration than is needed for most applications. These results show that the CD-EDA and CD-Urea samples are prime candidates for in vivo testing to examine their ability to cross the blood-brain barrier.
The central nervous system (CNS) of zebrafish includes the brain and spinal cord which are connected by the cerebral spinal fluid that circulates through ventricles of the brain that are contiguous with the central canal of the spinal cord. Any fluorescent species that is able to penetrate the blood-brain barrier will be easily observed in the central canal of the spinal cord. Therefore, the ability of CDs injected into the vasculature to reach the spinal cord and especially the central canal is our main assay for crossing the blood-brain barrier in Figure S9.
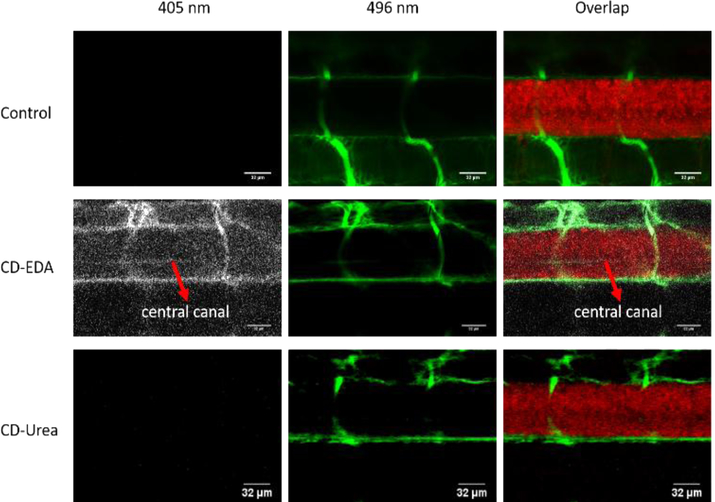
CD-EDA and CD-Urea were co-injected intravascularly with 10,000 molecular weight fluorescein dextran into the heart of transgenic zebrafish expressing mcherry (tg:HuC-mCherry) in all neurons to examine the ability of CDs to cross the blood-brain barrier (Figure 3). The 10,000 molecular weight of fluorescein dye is too large for the molecule to cross the blood-brain barrier. When the injection accurately targets the heart, fluorescein is restricted to the vasculature as can be seen in the control experiment. When CD-Urea was injected with the dye, similar images were obtained as the control experiment. These CDs cannot be observed excited at 405 nm, which was probably caused by the low quantum yield (21.5 ± 3.2%) and the discrepancy between the excitation of the channel (405 nm) and the maximum excitation wavelength of these CDs (350 nm). In contrast, upon the injection of CD-EDA mixed with the dye, we can observe the blue photoluminescence of the zebrafish vasculature in the CDs channel. Also, in the location of the central canal of spinal cord, blue photoluminescence was weak but also seen, as highlighted in Figure 3. (Notice: for convenience of display, blue photoluminescence was replaced by white color). This injection was repeated 3 times on batches of 6 fish (18 fish total) and an additional image is provided in Figure S10. This provides evidence of the CD-EDA penetrating the blood-brain barrier to reach the CNS. The reason for CD-EDA crossing the blood-brain barrier is hypothesized to be due to the greater amount of tryptophan molecules on the surface of CD-EDA as shown by UV-vis and XPS data. It is also easier to observe CD-EDA than CD-Urea due to the differences in quantum yield. Since, the LAT1 transporter is not specific for tryptophan, but recognizes several large amino acids, it is able to recognize tryptophan on the surface of CD-EDA and transports the whole system across the blood-brain barrier into the central nervous system.
Figure 3:
Confocal microscopic images of a six-day-old, transgenic zebrafish larva expressing mcherry (585 nm) in the central nervous system. The larvae were injected with either 10,000 MW fluorescein dextran dye (496 nm) alone (control, top row), or a combination of dye and CD-EDA (second row) or a combination of dye and CD-Urea (third row). Fluorescence from CDs (405 nm) that cross the blood brain barrier can be seen in the central canal that is highlighted with the red arrows.
Conclusion
The two CDs samples synthesized in this study showed the desired property of having the tryptophan moiety present on the surface of the CDs after purification. The CD-EDA sample shows the advantage of having more tryptophan on its surface and a higher quantum yield. This sample was tested with zebrafish to confirm the ability to cross the blood-brain barrier. The possibilities in bio-imaging or drug delivery to the brain without the need for receptor-conjugation dictates further investigation into these CDs samples, particularly CD-EDA because of its high quantum yield.
Supplementary Material
Highlights.
Carbon dots were prepared which appear to have tryptophan moieties on the surface.
Tryptophan carbon dots showed bright photoluminescence (QY=48%) and low toxicity.
Confocal microscopy shows carbon dots in the central nervous system of zebrafish.
Acknowledgements
Funding sources include the National Science Foundation through grant 1809060 and the National Institute of Health through grant R21AR072226.
Footnotes
Conflicts of interest
There are no conflicts of interest to declare
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Xu X, Ray R, Gu Y, Ploehn HJ, Gearheart L, Raker K and Scrivens WA, J. Am. Chem. Soc, 2004, 126, 12736–12737. [DOI] [PubMed] [Google Scholar]
- 2.Luo PG, Yang F, Yang S-T, Sonkar SK, Yang L, Broglie JJ, Liu Y and Sun Y-P, RSC Adv, 2014, 4, 10791–10807. [Google Scholar]
- 3.Yang S-T, Wang X, Wang H, Lu F, Luo PG, Cao L, Meziani MJ, Liu J-H, Liu Y and Chen M, J. Phys. Chem. C, 2009, 113, 18110–18114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahu S, Behera B, Maiti TK and Mohapatra S, Chem. Comm, 2012, 48, 8835–8837. [DOI] [PubMed] [Google Scholar]
- 5.Zhu S, Meng Q, Wang L, Zhang J, Song Y, Jin H, Zhang K, Sun H, Wang H and Yang B, Angew. Chem, 2013, 125, 4045–4049. [DOI] [PubMed] [Google Scholar]
- 6.Shi L, Yang JH, Zeng HB, Chen YM, Yang SC, Wu C, Zeng H, Yoshihito O and Zhang Q, Nanoscale, 2016, 8, 14374–14378. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Gao P, Wang Y, Guo J, Zhang K-Q, Du D, Dai X and Zou G, APL Mater, 2015, 3, 0861021–0861027. [Google Scholar]
- 8.Wen Z-H and Yin X-B, RSC Adv, 2016, 6, 27829–27835. [Google Scholar]
- 9.Yu S, Chen K, Wang F, Zhu Y and Zhang X, Luminescence, 2017, 32, 970–977. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Desserre A, Sharma SK, Li S, Marksberry M, Chusuei C, Blackwelder P and Leblanc RM, ChemPhysChem, 2017, 18, 890–897. [DOI] [PubMed] [Google Scholar]
- 11.Parvin N and Mandal TK, Microchim. Acta, 2017, 184, 1117–1125. [Google Scholar]
- 12.Yang Y, Kong W, Li H, Liu J, Yang M, Huang H, Liu Y, Wang Z, Wang Z and Sham T-K, ACS Appl. Mater. Interfaces, 2015, 7, 27324–27330. [DOI] [PubMed] [Google Scholar]
- 13.Qu S, Liu X, Guo X, Chu M, Zhang L and Shen D, Adv. Funct. Mater, 2014, 24, 2689–2695. [Google Scholar]
- 14.Choi Y, Kang B, Lee J, Kim S, Kim GT, Kang H, Lee BR, Kim H, Shim S-H and Lee G, Chem. Mater, 2016, 28, 6840–6847. [Google Scholar]
- 15.Pereira JC, Hallinan MP and Alves RC, Med. Hypotheses, 2017, 98, 69–75. [DOI] [PubMed] [Google Scholar]
- 16.Boado RJ, Li JY, Nagaya M, Zhang C and Pardridge WM, Proc. Natl. Acad. Sci. U.S.A, 1999, 96, 12079–12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padbury JF, Diah SK, McGonnigal B, Miller C, Fugere C, Kuzniar M and Thompson NL, Biochem. Biophys. Res. Commun, 2004, 318, 529–534. [DOI] [PubMed] [Google Scholar]
- 18.Bhardwaj NK, Hoang V and Nguyen KL, Bioresour. Technol, 2007, 98, 962–966. [DOI] [PubMed] [Google Scholar]
- 19.Gardner SD, Singamsetty CS, Booth GL, He G-R and Pittman CU, Carbon, 1995, 33, 587–595. [Google Scholar]
- 20.Jackson A and Smith P, Tetrahedron, 1968, 24, 2227–2239. [Google Scholar]
- 21.Laskin A and Lifshitz A, J. Phys. Chem. A, 1997, 101, 7787–7801. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.