Abstract
Tick microbiomes may play an important role in pathogen transmission. However, the drivers of microbiome variation are poorly understood, and this limitation has impeded mechanistic understanding of the functions of microbial communities for pathogen acquisition. The goal of this research was to characterize the role of the blood meal host in structuring the microbiome of Ixodes scapularis, the primary vector of Lyme disease in the eastern United States, and to determine if ticks that fed from different host species harbor distinct bacterial communities. We performed high-throughput 16S rDNA amplicon sequencing on I. scapularis nymphs that fed as larvae from known wildlife hosts: raccoon, Virginia opossum, striped skunk, red squirrel or gray squirrel. Using Analysis of Similarity, we found significant differences in the abundance-weighted Unifrac distance matrix among ticks fed from different host species (p = 0.048) and a highly significant difference in the weighted and unweighted Unifrac matrices for individuals within species (p < 0.01). This finding of associations between the blood meal host and I. scapularis microbiome demonstrates that the blood meal host may be a driver of microbiome variation that should be accounted for in studies of pathogen acquisition by ticks.
Keywords: Microbiome, Ixodes scapularis, Vector-host interactions, Lyme disease
1. Introduction
Research into the drivers of microbiome variation among ticks may improve mechanistic understanding of pathogen transmission (Clay et al., 2008; Narasimhan and Fikrig, 2015). Sources of microbiome variation may include species, life stage, sex, degree of engorgement and geographic location (Moreno et al., 2006; Van Treuren et al., 2015; Zolnik et al., 2016). However, the role of the host blood meal in structuring tick microbiomes remains uncertain. Ixodes scapularis, the principal vector of Lyme disease in the eastern United States (U.S.), makes direct contact with host fur, skin and blood (Barbour et al., 2009) that may influence its microbiome composition. Additionally, host immune cells may lead to alteration of the existing tick microbiome (Kuo et al., 2000; Kurtenbach et al., 2002). The importance of the host blood meal to microbiome variation of ticks is further supported by Swei and Kwan (2017) and by research into reservoir competancy (e.g. LoGiudice et al., 2003).
Lyme borreliosis is a tick-borne disease affecting an estimated 300,000 people per year in the United States (U.S.; CDC, 2013; Mead et al., 2013). The disease-causing agent of Lyme disease is the spirochetal bacterium Borrelia burgdorferi sensu stricto, transmitted to humans in the eastern U.S. through the bite of nymph and adult life-stage I. scapularis. Here, we seek to understand the influence of host identity on microbiome composition of I. scapularis nymphs, the life stage responsible for the majority of Lyme disease cases (Barbour and Fish, 1993).
2. Materials and Methods
Ticks analyzed in this study are known-host nymphs collected in 2001 for LoGiudice et al. (2003) and stored unprocessed for 16 years. Available ticks fed on gray squirrels (Sciurus carolinensis), raccoons (Procyon lotor), red squirrels (Tamiasciurus hudsonicus), striped skunks (Mephitis mephitis), and Virginia opossums (Didelphis virginiana; Table S1). Hosts were held in cages suspended over pans of water and engorged larvae were collected twice daily and held in vials until molting. Molted nymphs were then stored at 4°C for approximately three months and then maintained at −80°C. For the current study, whole and unfed nymphs were rinsed with 70% ethanol and homogenized on a tissue lyser with a sterile 5mm steel bead. DNA was extracted using Qiagen’s QIAamp DNA Micro Kit (Qiagen, Alameda, CA).
We used quantitative real-time PCR to determine tick infection status (presence/absence) and loading (number of B. burgdorferi organisms per tick). We used forward and reverse primers (5’-GCTGTAAACGATGCACACTTGGT-3’ and 5’-GGCGGCACACTTAACACGTTAG-3’ respectively) which target a 69 nt region of the B. burgdorferi and B. miyamotoi 16S gene. We simultaneously used a TaqMan™ probe (5’-TTCGGTACTAACTTTTAGTTAA-3’, including the FAM dye and MGBNFQ quencher) to detect only B. burgdorferi with this primer pair (Barbour et al., 2009). Each PCR run included amplicon standards of known concentration from which standard curves were generated. The PCR cycle included a two-minute activation step at 95°C, denaturation at 95°C for 5 s and a combined annealing/extension step of 51°C for 5 s. qPCR was performed on a Qiagen Rotor Gene Q PCR thermal cycler. We utilized “bead blanks”, tubes that contained a sterile bead but no tick, to test for laboratory contamination. DNA was quantified with the Qubit® dsDNA assay kit and Qubit® 2.0 fluorometer (Life Technologies, Carlsbad, CA).
We performed amplicon sequencing of the V4 region of 16S rDNA gene to characterize the bacterial microbiome. For PCR amplification, equal concentrations of DNA isolates, along with primers targeting the V4 region of the 16S rDNA gene (515f/806r; Bergmann et al., 2011) were added to two 48-well Fluidigm Access Array plates (South San Francisco, CA) at the Roy J. Carver Center for Biotechnology (University of Illinois, Urbana, IL). The amplified gene products were then sequenced on a MiSeq PE v3 machine (lllumina Inc., San Diego CA), generating 2 × 300 nt reads. Sequence reads were processed using QIIME v 1.9 (Caporaso et al., 2010). Quality filtering followed the recommendations of Bokulich et al. (2013) and sequences were de-replicated with Open Reference OTU picking (Rideout et al., 2014) at the 97% identity threshold. Chimeras were identified with USearch61 (Edgar, 2010) and removed, as were OTUs that matched to chloroplasts and mitochondria. We removed singleton and doubleton sequences and sequences that were detected in 10 or more copies on the bead blanks.
For beta diversity we rarefied the dataset to 7,420 sequences per sample and calculated the abundance-weighted and -unweighted Unifrac distance matrices (Lozupone et al., 2011), using the OTU table and phylogenetic tree (Price et al., 2009). To test for an effect of host species on tick bacterial community assemblage, we used a nested Analysis of Similarity (ANOSIM; Clarke, 1993), with host species as a fixed factor and individual host as a nested random factor. By doing so, the average Unifrac distances between hosts are based on estimated centroids for each individual animal. We used principal coordinates (PCO) analysis to visualize the relationship between host species and bacterial community similarity among samples. The ANOSIM and PCO analyses were performed on both the weighted and unweighted Unifrac distance matrices.
We further refined the visualization of principal coordinate plots by performing a canonical analysis of principal coordinates (CAP; Anderson and Willis, 2003). CAP provides a constrained ordination of the distance matrix based on the hypothesis that host species is a significant factor and using a subset of the principal coordinates (PCOs); the ordination is selected to maximize the between-group distance and minimize the within-group distance. In order to assess which bacterial orders were driving differences between ticks fed from different host species, the CAPs were correlated with the most abundant bacterial orders (>1% relative abundance). These orders were then ranked according to the sum of the squared correlation coefficients (SSCC) for the CAPs, providing insight into the taxa making the greatest contribution to variation in bacterial community composition. Statistical analyses were performed in Primer (v 6) and R (v 3.4.1).
3. Results
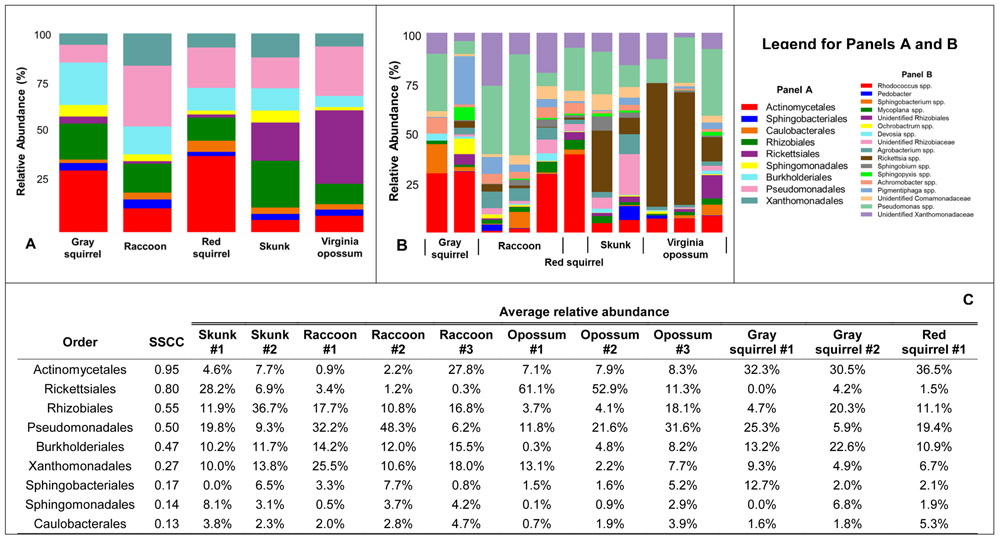
We obtained 5.5 million paired end reads from 92 tick samples, with one sample returning no sequences. After quality filtering and rarefaction, 88 samples were retained for analysis. There were a total of 2,232 OTUs at the 97% identity threshold. Orders with an average relative abundance among samples of >1% were Actinomycetales (6.0% - 36.5% among host species), Rickettsiales (1.2% - 36.3%), Rhizobiales (10.1 % - 22.9%), Pseudomonadales (8.9% - 29.8%), Burkholderiales (5.5% - 21.2%), Xanthomonadales (5.6% - 15.8%), Sphingomonadales (1.7% - 5.9%), Sphingobacteriales (2.1% - 4.4%) and Caulobacterales (1.7% - 5.3%, Fig. 1A). The most abundant genera (>5% abundance) were Pseudomonas spp. (8.4% - 29.1% among host species), Rhodococcus spp. (4.7% - 34.6%), Rickettsia spp. (0.9% - 36.3%), an unidentified Xanthomonadaceae (4.3% - 15.4%) and Pigmentiphaga spp. (0.9 - 17.1%). Other genera with a relative abundance >1% were Agrobacterium spp., Achromobacter spp., Mycoplana spp., Sphingobacterium spp., Sphingopyxis spp., Ochrobactrum spp., Devosia spp. as well as single unidentified genera within Rhizobiaceae, Comamonadaceae, Rhizobiales, and Phyllobacteriaceae (Fig. 1B). One of the 88 ticks was positive for B. burgdorferi, with an estimated load of 62 spirochetes. No Borrelia spp. or other human bacterial pathogens (e.g. Anaplasma spp.) were detected in the 16S rDNA amplicon sequencing data.
Figure 1.
Graphs of mean relative abundance of bacterial taxa associated with each host species for bacterial families (Panel A) and genera (Panel B). Genera and families individually comprising <1% of total community are not included. Panel C shows values of relative abundance at the order level. Orders are ranked by the sum of squares of their correlation coefficients (SSCC) with canonical analysis of principal coordinates (CAPs) 1 and 2 (see Figure S1 and the text).
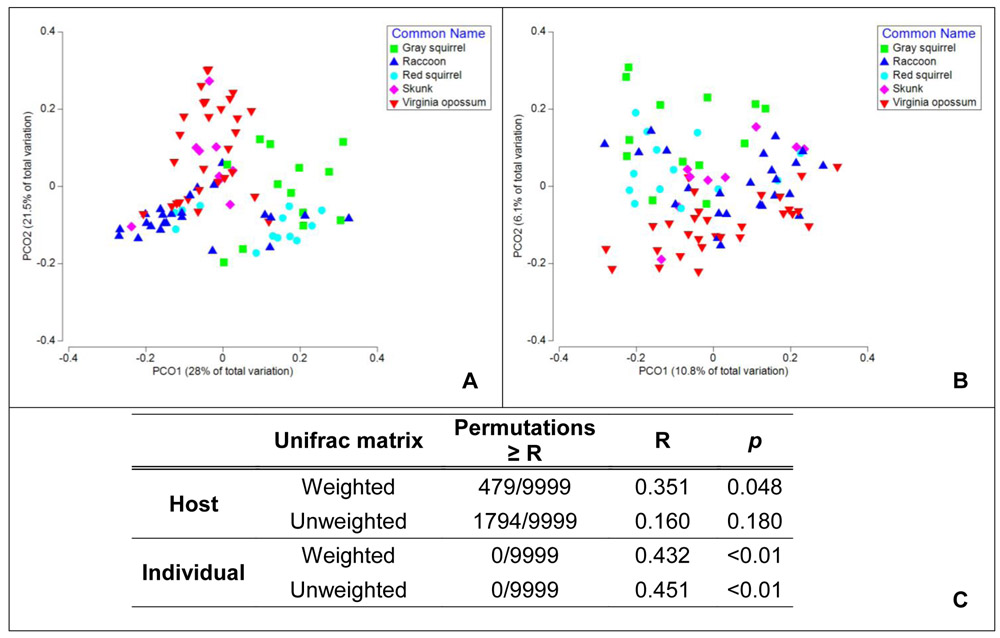
We found a significant effect of blood meal host species identity on bacterial community similarity from the weighted Unifrac distance matrix (Spearman’s R = 0.351; p = 0.048; Fig. 2) and a highly significant effect of individual hosts (R = 0.432; p < 0.001). For the unweighted Unifrac distance matrix there were no significant effects for host species (R = 0.16; p = 0.18) but a highly significant effect of individual hosts (R = 0.451, p < 0.001).
Figure 2.
First two principal coordinates for the abundance-weighted Unifrac distance matrix (Panel A) and the abundance-unweighted Unifrac matrix (Panel B). Panel C shows the results of the nested Analysis of Similarity (ANOSIM), with host species as a fixed factor and individual host as a nested random factor. There were 9,999 permutations, randomly sampled from 69,300 possible permutations.
The CAP analysis yielded a re-parameterization of the first five PCOs, which cumulatively explained 82% of the variation in the abundance-weighted Unifrac distance. In comparison to the PCO plot (Fig. 2), the first two CAP coordinates improved visual separation among hosts (Fig. S1A). The highest SSCC values between orders and the CAP coordinates were found for Actinomycetales (SSCC = 0.95), Rickettsiales (SSCC = 0.80), Rhizobiales (SSCC = 0.55), Pseudomonadales (SSCC = 0.50) and Burkholderiales (SSCC = 0.47), indicating that these groups made the greatest contribution to bacterial community variation across these coordinates (Figs. 1C and S1B). Additional orders with SSCC values > 0.1 were Xanthomonadales (SSCC = 0.27), Sphingobacteriales (0.17), Sphingomonadales (0.14) and Caulobacterales (0.13). To visualize which taxa contributed to variation in bacterial community composition by host, the most abundant (>1%) bacterial orders were plotted according to their correlations with CAPs 1 and 2, and in relation to the average CAPs per host species (Fig S1B).
4. Discussion
Here, we find evidence that the microbiome of I. scapularis is affected by both the individual and species identity of the blood meal host. We detected significant effects of host species identity on the composition of the I. scapularis microbiome despite limited replication (11 hosts) and the use of a conservative, non-parametric (rank-based) permutation test. Variability in microbial communities between individuals of the same host species may be due to differences in host genotype, diet, physiology and microbial interactions, as well as by stochastic processes (Adair and Douglas, 2017; Grabowski and Hill, 2017). Considering the preliminary nature of these findings, additional research is needed to determine how consistently host identity influences tick microbiomes, as well as possible mechanisms and importance to the transmission of pathogens.
Previous research has led to mixed conclusions about whether host identity influences the tick microbiome. Swei and Kwan (2017) found differences in bacterial diversity between lizard-(Sceloporus occidentalis) and mouse-fed (Peromyscus maniculatus) Ixodes pacificus, potentially due to antimicrobial properties of lizard blood (Lane and Quistad, 1998; Kuo et al., 2000). Additionally, host identity influences the likelihood that human pathogens will be acquired by feeding larval I. scapularis (e.g. LoGiudice et al., 2003; Keesing et al., 2012). On the other hand, Hawlena et al. (2013) found no differences in microbiome composition between I. scapularis collected from two different rodent species and Rynkiewicz et al. (2015) found that I. scapularis and D. variabilis harbored distinct bacterial communities, despite having fed from the same host. These opposing conclusions are not directly comparable as the studies showing no host effect utilized different tick species.
Several of the dominant genera we detected have been identified as inhabitants of internal structures of I. scapularis. For example, Pseudomonas, one of the most abundant genera in our dataset (Fig. 1), is believed to reside in the midgut (Narasimhan and Fikrig, 2015). In our dataset it was the primary representative from the Pseudomonadales, an order that was a strong driver of microbiome variation among host species (SSCC = 0.50; Fig. 1 and S1B). Three other abundant taxa - Ochrobactrum spp., Agrobacterium spp. (Order: Rhizobiales) and the family Xanthomonadaceae (Order: Xanthomonadales, Fig. S1) were detected in rodent blood (Zhang et al., 2014; Zolnik et al., 2018), suggesting they may be acquired during the blood meal. Taxa present in lower abundances (<1%) in our samples that may reside internally include Stenotrophomonas (Order: Xanthomonadales), Brevundimonas (Order: Caulobacterales) and Acinetobacter (Order: Pseudomonadales; Narasimhan and Fikrig, 2015). These purported internal inhabitants of I. scapularis may interact with B. burgdorferi and could influence infection status. In particular, Pseudomonas spp. may have a negative interaction with B. burgdorferi (Ross et al., 2018).
One of the most influential orders driving variation in bacterial community composition was the Rickettsiales (SSCC = 0.80), a group that was composed almost entirely of Rickettsia spp. (Fig. 1), with >96% of sequences from a single OTU. A nucleotide BLAST search revealed Rickettsia buchneri, a non-pathogenic, obligate intracellular bacterium that is transmitted transovarially and transstadially (Macaluso and Paddock, 2014; Kurtti et al., 2015; Gulia-Nuss et al., 2016), as the most likely match. Our observed differences in relative abundance of Rickettsia spp. by host species (Figs. 1 and S1B) may suggest that hosts vary in their capacity to support horizontal and/or transstadial transmission of this organism. As Rickettsia in I. scapularis may be nutritional endosymbionts (e.g. Rio et al., 2016), the potential role of host species to affect I. scapularis fitness should be studied further.
Due to variability in 16S gene copy number among taxa (Větrovský and Baldrian, 2013; Krehenwinkel et al., 2017), relative abundance may be over-estimated for some groups. Additionally, recent studies suggest that the bacterial diversity of ticks may be lower than reported here (e.g. Narasimhan et al., 2017; Ross et al., 2018). As our analysis was performed on whole ticks, our ability to distinguish between surface-colonizing and internal bacteria is limited and could account for the higher diversity. Several identified orders (e.g. Actinomycetales, Xanthomonadales, Burkholderiales, Rhizobiales, Sphingobacteriales, Fig. 1) contain species present on plants and in soil that may have colonized I. scapularis larvae from contact with their environment either prior to attachment (Estrada-Pena et al., 2013; Zolnik et al., 2016) or during contact with host hair and skin during feeding. At least one dominant genus (Rhodococcus spp.) is a potential inhabitant of mammalian fecal matter (Bell et al., 1998) and could have been acquired by engorged larvae from the water pan. However, we found very little Enterobacteriaceae, a group that is also expected to be present in fecal matter. Such surface-dwelling microbes, while likely reflective of a host signature, may have limited interactions with tick physiology and microbiome and thus not affect pathogen transmission. On the other hand, bacteria on the host surface and near the bite wound could have mixed with host blood and entered the tick during the blood meal (Hynes, 2014); this transmission route warrants further investigation.
Conclusions
This research suggests that despite high within species differences in the tick microbiome, the microbial community varies significantly between host species. Future studies should attempt to account for host identity as one of myriad factors that may influence the composition of the complex tick microbiome in order to better understand how host-microbiome-pathogen interactions may affect pathogen transmission.
Supplementary Material
Acknowledgements
We thank Bonnie Nightingale, Andrea Silvestri, Jada Lee and L. Page Fredericks for performing DNA extractions and laboratory assistance and Obadiah Mulder for help with data analysis. We thank Tim Hunter of the Advanced Genome Technologies Core Facilities at the University of Vermont and Sheryl White of the University of Vermont’s Neuroscience COBRE Molecular Core (Burlington, VT) for consultation on qPCR and for preparation of standards. We thank Dr. Robert Cluss of Middlebury College for his advice throughout the project.
Funding
This research was funded by an Institutional Development Award (IDeA) to W.J.L. from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103449, an award by the Nathan Cummings Foundation to R.S.O. and National Institutes of Health Grant AI40076 to R.S.O. and F.K. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
Declarations of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adair KL and Douglas AE, 2017. Making a microbiome: the many determinants of host-associated microbial community composition. Curr. Opin. Microbiol 35, 23–29. [DOI] [PubMed] [Google Scholar]
- Anderson MJ and Willis TJ, 2003. Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84, 511–525. [Google Scholar]
- Barbour AG and Fish D, 1993. The Biological and Social Phenomenon of Lyme Disease. Science 260, 1610–1616. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Bunikis J, Travinsky B, Hoen AG, Diuk-Wasser MA, Fish D and Tsao JI, 2009. Niche partitioning of Borrelia burgdorferi and Borrelia miyamotoi in the same tick vector and mammalian reservoir species. Am. J. Trop. Med. Hyg 81, 1120–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell K, Philp J, Aw D and Christofi N, 1998. The genus Rhodococcus. J. Appl. Microbiol 85, 195–210. [DOI] [PubMed] [Google Scholar]
- Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, Knight R and Fierer N, 2011. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol. Biochem 43, 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA and Caporaso JG, 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J and Knight R, 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC, 2013. Reported cases of Lyme disease by state or locality, 2002-2011. [Google Scholar]
- Clarke KR, 1993. Non-parametric multivariate analyses of changes in community structure. Austral Ecol. 18, 117–143. [Google Scholar]
- Clay K, Klyachko O, Grindle N, Civitello D, Oleske D and Fuqua C, 2008. Microbial communities and interactions in the lone star tick, Amblyomma americanum. Mol. Ecol 17, 4371–4381. [DOI] [PubMed] [Google Scholar]
- Edgar RC, 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- Estrada-Pena A, Gray JS, Kahl O, Lane RS and Nijhof AM, 2013. Research on the ecology of ticks and tick-borne pathogens -- methodological principles and caveats. Front. Cell Infect. Microbiol 3, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski JM and Hill CA, 2017. A Roadmap for Tick-Borne Flavivirus Research in the "Omics" Era. Front Cell Infect Microbiol 7, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulia-Nuss M, Nuss AB, Meyer JM, Sonenshine DE, Roe RM, Waterhouse RM, Sattelle DB, de la Fuente J, Ribeiro JM, Megy K, Thimmapuram J, Miller JR, Walenz BP, Koren S, Hostetler JB, Thiagarajan M, Joardar VS, Hannick LI, Bidwell S, Hammond MP, Young S, Zeng Q, Abrudan JL, Almeida FC, Ayllon N, Bhide K, Bissinger BW, Bonzon-Kulichenko E, Buckingham SD, Caffrey DR, Caimano MJ, Croset V, Driscoll T, Gilbert D, Gillespie JJ, Giraldo-Calderon GI, Grabowski JM, Jiang D, Khalil SM, Kim D, Kocan KM, Koci J, Kuhn RJ, Kurtti TJ, Lees K, Lang EG, Kennedy RC, Kwon H, Perera R, Qi Y, Radolf JD, Sakamoto JM, Sanchez-Gracia A, Severo MS, Silverman N, Simo L, Tojo M, Tornador C, Van Zee JP, Vazquez J, Vieira FG, Villar M, Wespiser AR, Yang Y, Zhu J, Arensburger P, Pietrantonio PV, Barker SC, Shao R, Zdobnov EM, Hauser F, Grimmelikhuijzen CJ, Park Y, Rozas J, Benton R, Pedra JH, Nelson DR, Unger MF, Tubio JM, Tu Z, Robertson HM, Shumway M, Sutton G, Wortman JR, Lawson D, Wikel SK, Nene VM, Fraser CM, Collins FH, Birren B, Nelson KE, Caler E and Hill CA, 2016. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun 7, 10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawlena H, Rynkiewicz E, Toh E, Alfred A, Durden LA, Hastriter MW, Nelson DE, Rong R, Munro D, Dong Q, Fuqua C and Clay K, 2013. The arthropod, but not the vertebrate host or its environment, dictates bacterial community composition of fleas and ticks. ISME J. 7, 221–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes WL, 2014. How ticks control microbes: innate immune responses, in: Sonenshine DE and Roe RM, (Eds.), Biology of Ticks. Oxford University Press, New York, pp. 129–146. [Google Scholar]
- Keesing F, Hersh MH, Tibbetts M, McHenry DJ, Duerr S, Brunner J, Killilea M, LoGiudice K, Schmidt KA and Ostfeld RS, 2012. Reservoir competence of vertebrate hosts for Anaplasma phagocytophilum. Emerg. Infect. Dis 18, 2013–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krehenwinkel H, Wolf M, Lim JY, Rominger AJ, Simison WB and Gillespie RG, 2017. Estimating and mitigating amplification bias in qualitative and quantitative arthropod metabarcoding. Sci. Rep 7, 17668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MM, Lane RS and Giclas PC, 2000. A comparative study of mammalian and reptilian alternative pathway of complement-mediated killing of the Lyme disease spirochete (Borrelia burgdorferi). J. Parasitol 86, 1223–1228. [DOI] [PubMed] [Google Scholar]
- Kurtenbach K, De Michelis S, Etti S, Schäfer SM, Sewell H-S, Brade V and Kraiczy P, 2002. Host association of Borrelia burgdorferi sensu lato – the key role of host complement. Trends Microbiol. 10, 74–79. [DOI] [PubMed] [Google Scholar]
- Kurtti TJ, Felsheim RF, Burkhardt NY, Oliver JD, Heu CC and Munderloh UG, 2015. Rickettsia buchneri sp. nov., a rickettsial endosymbiont of the blacklegged tick Ixodes scapularis. Int. J. Syst. Evol. Microbiol 65, 965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane RS and Quistad G, 1998. Borreliacidal factor in the blood of the western fence lizard (Sceloporus occidentalis). J. Parasitol 29–34. [PubMed] [Google Scholar]
- LoGiudice K, Ostfeld RS, Schmidt KA and Keesing F, 2003. The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proc. Natl. Acad. Sci. USA 100, 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D, Stombaugh J and Knight R, 2011. UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaluso K and Paddock CD, 2014. Tick-borne spotted fever group rickettsioses and Rickettsia species, in: Sonenshine DE and Roe RM, (Eds.), Biology of ticks. Oxford University Press, New York, NY, pp. 211–250. [Google Scholar]
- Mead PS, Nelson C, Hinckley A, Hook S, Kugeler K, Perea A and Beard B, 2013. Estimating the public health burden of Lyme disease in the United States. International Conference on Lyme Borreliosis and Other Tick-Borne Diseases [Google Scholar]
- Moreno CX, Moy F, Daniels TJ, Godfrey HP and Cabello FC, 2006. Molecular analysis of microbial communities identified in different developmental stages of Ixodes scapularis ticks from Westchester and Dutchess Counties, New York. Environ. Microbiol 8, 761–772. [DOI] [PubMed] [Google Scholar]
- Narasimhan S and Fikrig E, 2015. Tick microbiome: the force within. Trends Parasitol. 31, 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan S, Schuijt TJ, Abraham NM, Rajeevan N, Coumou J, Graham M, Robson A, Wu M-J, Daffre S and Hovius JW, 2017. Modulation of the tick gut milieu by a secreted tick protein favors Borrelia burgdorferi colonization. Nat. Commun 8, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS and Arkin AP, 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol 26, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout JR, He Y, Navas-Molina JA, Walters WA, Ursell LK, Gibbons SM, Chase J, McDonald D, Gonzalez A, Robbins-Pianka A, Clemente JC, Gilbert JA, Huse SM, Zhou HW, Knight R and Caporaso JG, 2014. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2, e545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio RVM, Attardo GM and Weiss BL, 2016. Grandeur Alliances: Symbiont Metabolic Integration and Obligate Arthropod Hematophagy. Trends Parasitol. 32, 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross BD, Hayes B, Radey MC, Lee X, Josek T, Bjork J, Neitzel D, Paskewitz S, Chou S and Mougous JD, 2018. Ixodes scapularis does not harbor a stable midgut microbiome. ISME J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rynkiewicz EC, Hemmerich C, Rusch DB, Fuqua C and Clay K, 2015. Concordance of bacterial communities of two tick species and blood of their shared rodent host. Mol. Ecol 24, 2566–2579. [DOI] [PubMed] [Google Scholar]
- Swei A and Kwan JY, 2017. Tick microbiome and pathogen acquisition altered by host blood meal. ISME J. 11, 813–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treuren W, Ponnusamy L, Brinkerhoff RJ, Gonzalez A, Parobek CM, Juliano JJ, Andreadis TG, Falco RC, Ziegler LB, Hathaway N, Keeler C, Emch M, Bailey JA, Roe RM, Apperson CS, Knight R and Meshnick SR, 2015. Variation in the Microbiota of Ixodes Ticks with Regard to Geography, Species, and Sex. Appl. Environ. Microbiol 81,6200–6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Větrovský T and Baldrian P, 2013. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8, e57923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-C, Yang Z-N, Lua B, Ma X-F, Zhang C-X and Xua H-J, 2014. The composition and transmission of microbiome in hard tick, Ixodes persulcatus, during blood meal. Ticks Tick Borne Dis. 5, 864–870. [DOI] [PubMed] [Google Scholar]
- Zolnik CP, Prill RJ, Falco RC, Daniels TJ and Kolokotronis SO, 2016. Microbiome changes through ontogeny of a tick pathogen vector. Mol. Ecol 25, 4963–4977. [DOI] [PubMed] [Google Scholar]
- Zolnik CP, Falco RC, Daniels TJ and Kolokotronis SO, 2018. Transient influence of blood meal and natural environment on blacklegged tick bacterial communities. Ticks Tick Borne Dis. 9, 563–572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.