Abstract
Background:
Epigenetic mechanisms may alter the airway epithelial barrier and ultimately lead to atopic diseases such as asthma. Here we aim to identify DNA methylation profiles that are associated with -and accurately classify- atopy and atopic asthma in school-aged children.
Methods:
We conducted a genome-wide study of DNA methylation in nasal epithelium and atopy or atopic asthma in 483 Puerto Rican children ages 9–20 years, recruited from October 2013 through August 2017 using multistage probability sampling. Atopy was defined as as ≥1 positive IgE (≥0·35 IU/mL) to common aeroallergens, and asthma was defined as a physician’s diagnosis plus wheeze in the previous year. Significant methylation signals were correlated with gene expression, and top CpGs were validated by pyrosequencing. We then replicated our top methylation findings in a cohort including 72 predominantly African American children, and in 432 children from a European birth cohort. Next, we tested nasal methylation-based classification models of atopy or atopic asthma in all cohorts.
Findings:
DNA methylation profiles were markedly different between subjects with (n=312) and without (n=171) atopy in the Puerto Rico discovery cohort. After adjustment for covariates and multiple testing, we found 8,664 differentially methylated CpGs by atopy, with FDR-adjusted P-values ranging from 9·58×10−17 to 2·18×10−22 for the top 30 CpGs. These CpGs are in or near genes relevant to epithelial barrier function, including CDHR3 (cadherin-related family member 3) and CDH26 (cadherin 26), and in other biologically plausible genes like FBXL7 (F-box and leucine-rich repeat protein 7), NTRK1 (neurotrophic receptor tyrosine kinase 1), and SLC9A3 (solute carrier family 9 member A3). Moreover, 28 of the top 30 CpGs replicated in the same direction in both independent cohorts. Nasal methylation-based classification models of atopy performed well in the Puerto Rico cohort (area under the curve=0·93–0·94 and accuracy=85%−88%) and in both replication cohorts (AUC=0·74–0·92, accuracy=68%−82%). The models also performed well for atopic asthma in the Puerto Rico cohort (AUC=0·95–1.00, accuracy=88%) and the replication cohorts (AUC=0·82–0·88, accuracy=86%).
Interpretation:
We identified specific methylation profiles in airway epithelium that are associated with atopy and atopic asthma in children, and a nasal methylation panel that classifies children by atopy or atopic asthma. Our findings support the feasibility of using the nasal methylome for future clinical applications, such as predicting the development of asthma among wheezing infants.
INTRODUCTION
Sensitization to allergens (atopy) is a key component of atopic diseases such as asthma and atopic dermatitis. Over the last few decades, allergic diseases have increased in industrialized nations, likely due to environmental changes. Such changes could alter epigenetic regulation and expression of atopy susceptibility genes, ultimately leading to atopic diseases.(1) Indeed, a recent meta-analysis of genome-wide association studies (GWAS) identified 18 genetic loci associated with asthma, but such loci explained only 3.5% of disease risk, emphasizing the need to identify non-genetic, environmentally-mediated (i.e. epigenetic) causal mechanisms for atopy and asthma.(2)
By modulating activating and inhibitory signals, the airway epithelium regulates immune responses to environmental challenges and airway inflammation.(3) Tobacco smoke, allergens, or pollutants could penetrate defective epithelial barriers, interact with dendritic cells, alter immune responses, and cause atopy in children. Several susceptibility genes for asthma are expressed in airway epithelium (e.g., cadherin-related family member 3 [CDHR3], protocadherin-1 [PCDH1],(4) and orosomucoid-like 3 [ORMDL3/GSDMB]).(5) CDHR3 is a receptor for rhinovirus C (implicated in asthma and severe asthma exacerbations),(6) and bronchial epithelial expression of ORMDL3 suffices for induction of Alternaria-related allergic airway disease in murine models.(7) Similarly, polymorphisms of filaggrin, a filament-associated protein that binds to keratin in skin epithelium, confer susceptibility to atopic dermatitis.(8)
In the U.S., asthma affects ~7 million children and ~10–12% of adolescents report respiratory or skin allergies. Puerto Ricans are disproportionately affected by asthma and atopy,(9, 10) and are often exposed to environmental agents linked to both DNA methylation and atopic diseases. We therefore hypothesized that DNA methylation of airway epithelium is linked to atopy and atopic asthma in Puerto Ricans. To test this hypothesis, we conducted genome-wide studies of DNA methylation in nasal epithelium (a surrogate marker for bronchial epithelium(11, 12)) and atopy and atopic asthma in Puerto Rican children and adolescents, a high-risk group. We then conducted replication studies for the top methylation signals in two cohorts.(5, 13) Moreover, we tested nasal methylation-based predictive modeling to classify children according to whether they had atopy or atopic asthma, as a necessary step to provide “proof of concept” and show the feasibility of future clinical applications using the nasal methylome.
METHODS
Please see the Methods in the Online Supplement for additional details.
Study population and procedures
The Epigenetic Variation and Childhood Asthma in Puerto Ricans (EVA-PR) is a case-control study of asthma in subjects aged 9–20 years, recruited using a similar approach to that used in a prior study.(14, 15) Participants with and without asthma were recruited from households in San Juan (PR) from October 2013 through August 2017, using multistage probability sampling; 638 households had ≥1 eligible child, and 543 (85·1%) children (one per household) agreed to participate. There were no significant differences in age or sex between eligible children who did and did not participate. The study was approved by the institutional review boards of the University of Puerto Rico (San Juan, PR) and the University of Pittsburgh (Pittsburgh, PA). Written parental consent and assent were obtained from participants <18 years old, and consent was obtained from participants ≥18 years old. The study protocol included questionnaires on respiratory health, measurement of serum allergen-specific IgE, and nasal samples for DNA and RNA extraction. Atopy was defined as ≥1 positive IgE (≥0·35 IU/mL) to five common aeroallergens in Puerto Rico: house dust mite, cockroach, cat dander, dog dander, and mouse urinary protein. Asthma was defined as a physician’s diagnosis plus at least one episode of wheeze in the previous year.
Nasal genome-wide methylation (GWM) and RNA sequencing (RNA-Seq)
DNA and RNA were extracted from nasal specimens collected from the inferior turbinate. To account for potential effects of different cell types, we implemented a protocol in a subset of nasal samples (n=31) to select CD326(+) nasal epithelial cells before DNA and RNA extraction. Whole-genome methylation assays were performed using HumanMethylation450 BeadChips (Illumina, San Diego, CA). After QC, 227,836 CpG probes remained. Methylation β-values were calculated as a percentage: β=M/(M+U+α), where M and U represent methylated and unmethylated signal intensities, respectively, and α is an arbitrary offset to stabilize β-values where fluorescent intensities are low. β-values were then transformed to M-values as log2(β/(1-β)), and M-values were used in all downstream analyses. RNA-Seq was performed with the Illumina NextSeq 500 platform, paired-end reads at 75 cycles, and 80M reads/sample; reads were aligned to reference human genome (hg19) and TPM (Transcripts Per Kilobase Million) were used as proxy for gene expression level. After QC, 16,487 genes were retained; our analysis focused on genes in cis (<10kbp) with significant CpGs from the methylation analysis (see below). For both GWM and RNA-Seq, known batch effects (e.g. plates) were removed using an empirical Bayes framework, and sva was used to estimate latent factors (LFs) that capture unknown data heterogeneity.
Epigenome-wide (EWAS) and transcriptome-wide (TWAS) association studies
We conducted an EWAS using multivariable logistic regression with the general model: Atopy (or atopic asthma) = CpG M-value + age + sex + LFs + top five principal components from genotypic data + (for the analysis of atopy only) asthma status. Adjustment for multiple testing was performed using the false discovery rate (FDR); significance was defined as FDR-corrected P<0·01. Differentially methylated regions (DMRs) were analyzed using Python tool comb-p, with a seed p-value of 0·01 and a maximum extendable distance of 300bp. The regression output included the effect size and p-value for each CpG. For ease of interpretation, tables show the difference in mean betas between participants with atopy and those without atopy and expressed as a percent: Methylation Δ (%) = mean betaatopy – mean betano atopy.
To evaluate whether our significant methylation signals are associated with gene expression, we first conducted a TWAS (following a similar model but using log2-scaled TPM instead of M-values) and then performed an expression quantitative trait DNA methylation (eQTM) analysis of significant CpGs and genes in cis (<10 kbp) with those CpGs. We then performed a mediation analysis to evaluate whether any proportion of the association between methylation and atopy is mediated by differences in gene expression. In this mediation analysis, we assessed the contributions of the direct pathway between methylation and atopy (methylation → atopy) and the indirect (or mediated) pathway through changes in gene expression (methylation → expression → atopy). Finally, we conducted an Ingenuity Pathway Analysis (IPA) including all genes with FDR P-value <0·01 in the TWAS, EWAS, and eQTM analysis.
Classification models
To investigate whether GWM data can be used to classify atopy, we used three classic machine learning models to select the best performing panel for each method and their optimal probability cut-offs (see Online Supplement for details). For each method, we trained the model with the top 500 EWAS CpGs on all samples, and sorted all CpGs according to variable importance, defined as each CpG’s contribution to the model (Suplemental Figure 6). We then sequentially selected the top 10 through 100 CpGs to train and test the models (Supplementary Figures 6, 7) and to optimize the parameters (Supplementary Figure 8). The performance of different methods using different numbers of features were compared based on the testing AUCs (area under the ROC curve), with the largest average AUC among 10 cross-validations indicating the best performance. On this basis, we selected a 30-CpG panel and evaluated its diagnostic test performance (Supplementary Table 10). In order to further compare the performance of the CpG panel, we used two “negative controls”: 1) a model using 30 randomly selected CpGs; and 2) a model using only demographic/questionnaire variables: age, sex, household income, parental history of asthma, tobacco smoke exposure in early life, obesity, and allergic rhinitis.
Validation and external replications
We performed an internal validation of top CpGs using pyrosequencing in a random sub-sample of 40 participants with atopic asthma and 40 non-atopic non-asthmatic controls. We also performed external replication in two independent cohorts: a public dataset (GSE65205) from a case-control study of atopic asthma and nasal epithelial DNA methylation in 72 predominantly African American children(5); and data from 432 16-year-old participants in the Prevention and Incidence of Asthma and Mite Allergy (PIAMA) cohort of Dutch children born in 1996–1997.(16)
RESULTS
A total of 523 participants had complete data for atopy, atopic asthma, and all covariates; 483 had nasal genome-wide methylation (GWM) data and 456 had nasal RNA-Seq data (Supplementary Figure 1). The main characteristics of study participants are shown in Table 1. Atopic subjects had higher mean total IgE (790 IU/mL vs. 110 IU/mL) and were more likely to have asthma (57% vs. 39%) than those without atopy, without significant differences in age or sex.
Table 1 –
Characteristics of study participants in the discovery and replication cohorts
| Puerto Rico (discovery cohort) | Yang et al. | PIAMA | ||||||
|---|---|---|---|---|---|---|---|---|
| Atopy | No atopy | Atopic asthma | Non-atopic controls | Atopic asthma | Non-atopic controls | Atopy | No atopy | |
| N (%) | 312 (64·6%) | 171 (35·4%) | 169 (61·9%) | 104 (38·1%) | 36 (50%) | 36 (50%) | 207 (47·9%) | 225 (52·1%) |
| Age (years) | 15 (3) | 15 (3) | 15 (3) | 16 (3) | 11·1 (0·8) | 10·9 (0·9) | 16·4 (0·2) | 16·3 (0·2) |
| Female sex, n (%) | 140 (44·9%) | 93 (54·4%) | 66 (39·1%)* | 61 (58·7%) | 17 (47·2%) | 19 (52·8%) | 92 (44·4%) | 127 (56·4%) |
| Race/ethnicity | ||||||||
| • Hispanic/Latino | 100% | 13·9%a | 0% | |||||
| • African American | 0 | 91·7% | 0% | |||||
| • Non-Hispanic White | 0 | 6·9%a | 97.1% | |||||
| • Other/missing | 0 | 4·2% | 2.9% | |||||
| Asthma, n (%) | 169 (54·2%)* | 67 (39·2%) | 169 (100%)* | 0 (0%) | 36 (100%)* | 0 (0%) | 27 (13·0%)* | 6 (2·7%) |
| Total IgE (IU/mL) | 409 [207–816]* | 43 [22–93] | 386 [214–806]* | 42 [21–78] | 366 [185–785] | 29 [16·5–49·5] | 140 [55–140] | 20 [10–55] |
| Number of positive allergen-specific IgEs+ | 2 [1–3]* | 0 | 2 [1–3]* | 0 | n/a | 0 | 2 [1–3]* | 0 |
The Puerto Rico cohort (EVA-PR) is a case-control study of asthma. Yang et al. is a case-control study of atopic asthma. PIAMA is a birth cohort, unselected for either atopy or asthma. Numbers represent number of participants (%) for categorical variables and mean (SD) or median[interquartile range] for continuous variables.
P<0·05 for atopy vs. no atopy, or asthma vs. no asthma within each cohort. n/a: not available in public dataset.
Does not add up to 100% because participants could report more than one race/ethnicity.
Nasal DNA methylation and atopy
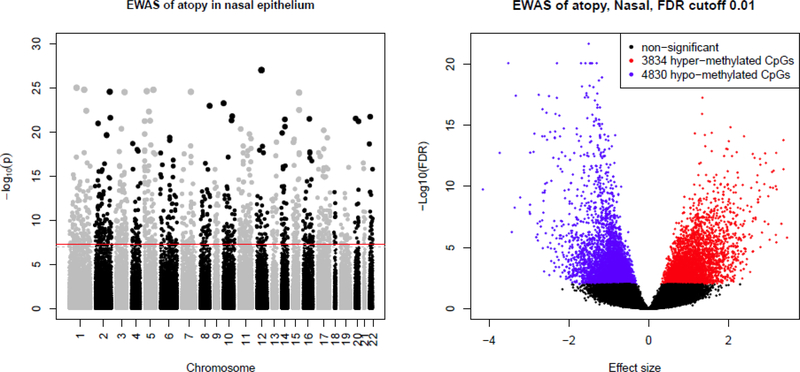
DNA methylation profiles were remarkably different between subjects with and without atopy (Figure 1). After adjustment for covariates and multiple testing, we found 8,664 differentially methylated CpGs by atopy (FDR P-value <0·01): 3,834 with higher methylation in atopic subjects and 4,830 with lower methylation in atopic subjects. Table 2 shows the top 30 differentially methylated CpGs, with FDR-adjusted P-values ranging from 9·58×10−17 to 2·18×10-22. Similarly, we found differentially methylated regions (DMRs) associated with those top genes (Supplementary Table 1). These CpGs and DMRs are in or near biologically plausible genes for atopy, such as FBXL7 (F-box and leucine-rich repeat protein 7, FDR-P=8·5×10−21), NTRK1 (neurotrophic receptor tyrosine kinase 1, FDR-P=8·5×10−21), CDHR3 (FDR-P=8·5×10−21), CDH26 (cadherin 26, FDR-P=5·8×10−18), CAPN14 (calpain 14, FDR-P=9·5×10−18), GRK5 (G protein-coupled receptor kinase 5, FDR-P=5·2×10−18), and SLC9A3 (solute carrier family 9 member A3, FDR-P=5·6×10−18). There was no residual clustering by sex, batch group, or processing protocol (i.e. whole nasal sample vs. CD326+ epithelial cells) (Supplementary Figure 2). Moreover, we obtained similar results in analyses restricted to the 31 cell-sorted samples despite small sample size (Supplementary Table 2) or after removing samples from 12 subjects using nasal steroids.
Figure 1 – Epigenome-wide association study (EWAS) of atopy in nasal epithelium.
Top panel shows the Manhattan plot, with –log10(P) on the y-axis and chromosome position in the x-axis; red line indicates FDP-P<0·01. Bottom panel shows the volcano plot, with –log10(FDR P-value) in the y-axis and effect size in the x-axis. Hypermethylated CpGs (i.e. higher methylation level in participants with atopy compared to those with no atopy) are shown in red; hypomethylated CpGs in blue; non-significant (FDR P>0·01) in black.
Table 2 –
Top 30 Epigenome-wide Association Study (EWAS) results for atopy
| Puerto Rican (Discovery) Cohort | Yang et al. | PIAMA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG | Gene | Chr | Position | Methylation Δ(%)a | P-value | FDR P-value | Methylation Δ(%)a | P-value | Methylation Δ(%)a | P-value | Combined P-valueb | |
| 1 | cg20372759 | METTL1 | 12 | 58162287 | −23·8% | 9·59×10−28 | 2·18×10−22 | −22·4% | 1·15×10−12 | −7·4% | 6·01×10−18 | 1·05×10−47 |
| 2 | cg15006973 | GJA4 | 1 | 35258933 | −16·0% | 9·56×10−26 | 8·53×10−21 | −15·4% | 3·61×10−10 | −4·4% | 2·72×10−14 | 4·20×10−40 |
| 3 | cg24707200 | NTRK1 | 1 | 156833163 | −10·3% | 1·62×10−25 | 8·53×10−21 | −8·0% | 1·21×10−07 | −3·8% | 1·21×10−13 | 5·39×10−37 |
| 4 | cg08844313 | PDE6A | 5 | 149240529 | −15·8% | 1·63×10−25 | 8·53×10−21 | −12·7% | 2·97×10−07 | −8·1% | 2·16×10−09 | 1·85×10−32 |
| 5 | cg00664723 | FBXL7 | 5 | 15927184 | −19·2% | 2·33×10−25 | 8·53×10−21 | −23·3% | 3·77×10−10 | −6·9% | 9·23×10−12 | 1·32×10−37 |
| 6 | cg10549071 | SPP2 | 2 | 235160451 | −18·3% | 2·74×10−25 | 8·53×10−21 | −20·4% | 1·40×10−09 | −5·2% | 4·52×10−07 | 1·82×10−32 |
| 7 | cg16027132 | CDHR3 | 7 | 105521116 | −23·1% | 2·78×10−25 | 8·53×10−21 | −23·6% | 2·68×10−13 | −7·6% | 2·23×10−14 | 2·95×10−43 |
| 8 | cg20790648 | SUCNR1 | 3 | 151619923 | −27·1% | 3·00×10−25 | 8·53×10−21 | −20·8% | 1·41×10−08 | −11·7% | 2·37×10−11 | 1·15×10−35 |
| 9 | cg01870976 | PCSK6 | 15 | 101887154 | −18·8% | 3·41×10−25 | 8·63×10−21 | −16·0% | 4·80×10−11 | −5·3% | 4·62×10−17 | 1·13×10−43 |
| 10 | cg03875819 | LINC00704 | 10 | 4386802 | −24·1% | 5·41×10−24 | 1·23×10−19 | −25·9% | 8·61×10−11 | −5·8% | 4·44×10−07 | 1·59×10−32 |
| 11 | cg01859321 | PLEC | 8 | 144970195 | −22·6% | 1·06×10−23 | 2·20×10−19 | −21·2% | 4·65×10−08 | −5·9% | 1·04×10−10 | 3·74×10−33 |
| 12 | cg18749617 | PCSK6 | 15 | 102028637 | −15·4% | 3·12×10−23 | 5·93×10−19 | −9·0% | 3·08×10−06 | −6·0% | 2·87×10−09 | 1·49×10−29 |
| 13 | cg09472600 | NCF2 | 1 | 183537770 | −19·0% | 3·84×10−23 | 6·73×10−19 | −14·4% | 4·29×10−05 | −7·2% | 3·51×10−04 | 1·88×10−23 |
| 14 | cg21158502 | ANKRD31 | 5 | 74348187 | −12·3% | 4·75×10−23 | 7·72×10−19 | −7·1% | 5·65×10−04 | −4·6% | 1·14×10−09 | 1·25×10−27 |
| 15 | cg15428140 | TEX36-AS1 | 10 | 127220354 | −15·0% | 1·58×10−22 | 2·40×10−18 | −6·5% | 1·48×10−04 | −2·4% | 2·25×10−04 | 1·38×10−22 |
| 16 | cg13586696 | C22orf31 | 22 | 29458723 | −15·9% | 1·81×10−22 | 2·58×10−18 | −16·3% | 1·91×10−10 | −4·2% | 1·74×10−09 | 3·03×10−33 |
| 17 | cg19497511 | LRRFIP1 | 2 | 238609807 | −11·3% | 2·40×10−22 | 3·21×10−18 | −7·6% | 2·52×10−04 | −2·6% | 2·91×10−07 | 4·92×10−25 |
| 18 | cg03387497 | BANF2 | 20 | 17680945 | −16·9% | 2·91×10−22 | 3·68×10−18 | −15·2% | 2·76×10−07 | −4·2% | 3·20×10−11 | 1·05×10−31 |
| 19 | cg07239613 | CES4A | 16 | 67051005 | −8·0% | 3·19×10−22 | 3·83×10−18 | −12·0% | 4·30×10−08 | −1·6% | 0·036 | 1·12×10−23 |
| 20 | cg19610615 | ADCK1 | 14 | 78446340 | −13·8% | 3·65×10−22 | 4·16×10−18 | −9·1% | 6·30×10−07 | −5·6% | 9·20×10−07 | 5·77×10−27 |
| 21 | cg00406211 | GRK5 | 10 | 121077022 | −17·4% | 4·82×10−22 | 5·23×10−18 | −13·6% | 7·13×10−04 | −3·2% | 5·16×10−04 | 2·98×10−21 |
| 22 | cg22862094 | EFNA5 | 5 | 106859865 | −14·9% | 5·16×10−22 | 5·35×10−18 | −11·1% | 7·61×10−09 | −4·7% | 8·04×10−14 | 1·31×10−35 |
| 23 | cg19107578 | SLC9A3 | 5 | 493262 | +11·8% | 5·64×10−22 | 5·59×10−18 | +0·4% | 0·51 | +4·3% | 2·17×10−06 | 9·17×10−21 |
| 24 | cg16518142 | CDH26 | 20 | 58533713 | −10·6% | 6·08×10−22 | 5·77×10−18 | −10·1% | 4·42×10−04 | −2·1% | 0·051 | 1·72×10−19 |
| 25 | cg04132353 | CAPN14 | 2 | 31440349 | −16·1% | 1·04×10−21 | 9·48×10−18 | −9·7% | 7·08×10−06 | −4·0% | 9·33×10−11 | 1·77×10−29 |
| 26 | cg22855021 | TSHR | 14 | 81610812 | −18·7% | 2·44×10−21 | 2·13×10−17 | −17·1% | 1·14×10−07 | −3·4% | 1·30×10−06 | 7·50×10−27 |
| 27 | cg08175352 | ZPLD1 | 3 | 101894206 | −18·7% | 4·20×10−21 | 3·55×10−17 | −17·3% | 1·03×10−09 | −4·0% | 2·53×10−07 | 2·59×10−29 |
| 28 | cg10830021 | NUP98 | 11 | 3815589 | −17·7% | 4·45×10−21 | 3·62×10−17 | −15·2% | 3·15×10−06 | −4·3% | 6·92×10−06 | 1·59×10−24 |
| 29 | cg20864568 | MAP3K14 | 17 | 43391599 | −11·1% | 6·27×10−21 | 4·93×10−17 | −3·9% | 9·10×10−05 | −3·8% | 1·14×10−04 | 8·29×10−22 |
| 30 | cg15388974 | PRKD1 | 14 | 30291201 | −9·5% | 1·26×10−20 | 9·58×10−17 | −7·1% | 0·082 | −3·1% | 7·59×10−06 | 8·12×10−20 |
Discovery cohort analysis adjusted for age, sex, asthma status, methylation latent factors, and genotypic principal components. Yang et al. adjusted for age, sex, and latent factors. PIAMA replication adjusted for age, sex, asthma status, batch, and study center. CpG position from hg19 reference genome. A list of the middle 1,000 significant CpGs in the Puerto Rico cohort is found in Supplementary Table 12, at the end of the Online Supplement; Supplementary Table 12 also contains the Bonferroni-corrected P-values for those CpGs. All significant CpGs in Puerto Rico are found in the Supplemental Excel file.
Methylation Δ shows the difference between the mean methylation beta in participants with atopy and those without ayopy: Mean betaatopy – mean betano-atopy (e.g. −23·8% means that the mean beta for participants with amiddley was 0·238 lower than for participants without atopy).
Fisher combined P-value.
Epigenomic and transcriptomic integration
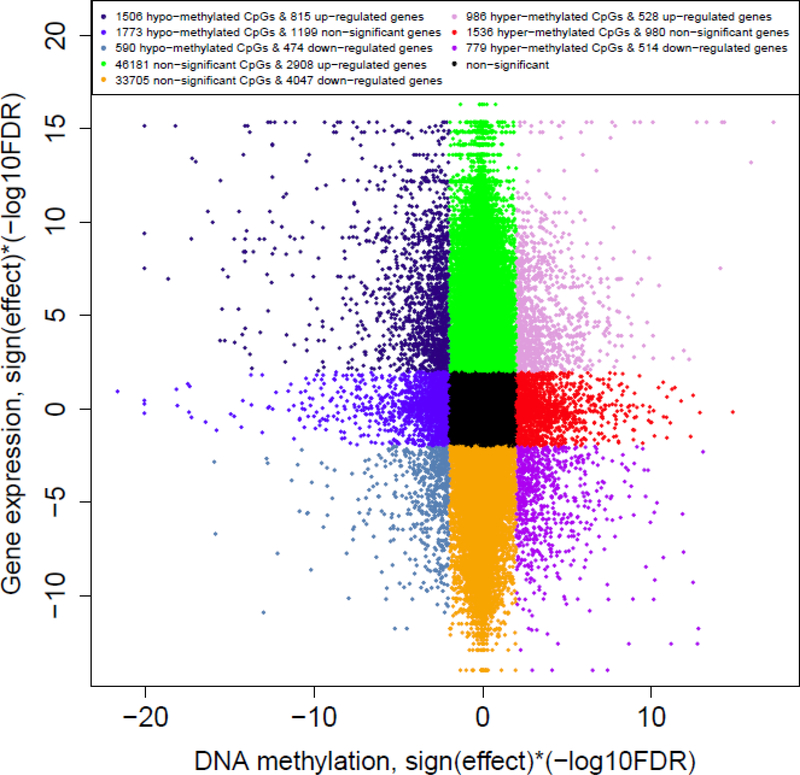
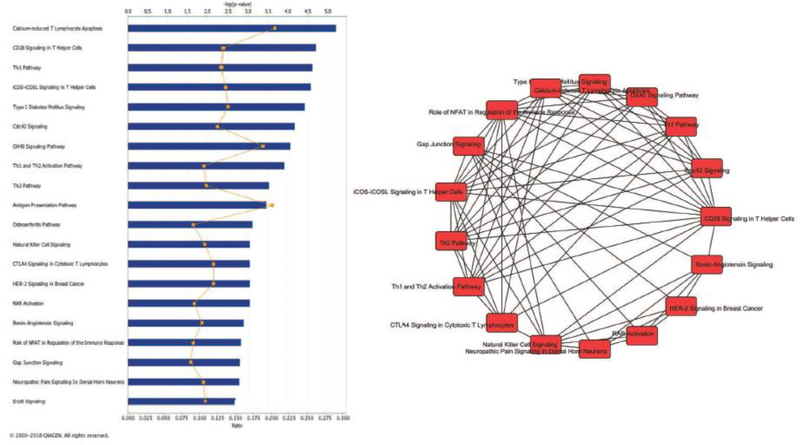
There was substantial overlap between differential methylation and expression (Figure 2), including 779 hyper-methylated CpGs corresponding to 514 down-regulated genes, and 1,506 hypomethylated CpGs corresponding to 815 up-regulated genes. Among genes at or near our top EWAS results, 13/30 (43·3%) showed differential expression by atopy, including SLC9A3, PCSK6, CDH26, FBXL7, and NTRK1 (Supplementary Table 3). The eQTM analysis revealed 1,570 CpG-gene expression pairs, including 11/30 (36·7%) of the top 30 EWAS results (Supplementary Table 4). Based on these findings, we performed a mediation analysis for each top 30 CpG/gene pair to evaluate direct (methylation → atopy) and indirect (methylation → expression → atopy) pathways: we found significant mediation for 12/30 (40%) genes (Supplementary Table 5), including CDH26, PCSK6, PRKD1, SLC9A3, and MAP3K14. Results from the pathway analysis of the 724 genes that were significant in the EWAS, TWAS, and eQTM analyses are shown in Supplementary Table 6 and Figure 3. The top 20 enriched pathways were related to immune regulation (Th1 and Th2 pathways; antigen presentation; T helper, Treg and NK cell signaling; etc), as well gap junction signaling and the ErbB or epidermal growth factor receptor pathway.
Figure 2 – Integration of Epigenome-wide Association Study (EWAS) and Transcriptome-wide Association Study (TWAS) results for atopy.
Figure shows the -log10(FDR P-value) for DNA methylation (EWAS) in the x-axis, and for gene expression (TWAS) in the y-axis; positive values indicate hypermethylation or over-expression, and negative values indicate hypomethylation or under-expression. Pairs where both EWAS and TWAS results are non-significant (FDR P>0·01) are shown in black. Significantly hypermethylated CpGs with under-expressed corresponding genes are shown in purple (right lower quadrant; 779 CpGs, 514 genes); hypomethylated and over-expressed genes shown in dark blue (left upper quadrant; 1506 CpGs, 815 genes).
Figure 3 – Pathway analysis of nasal epigenomics and atopy.
Left panel shows the top 20 enriched canonical pathways for our analyses of atopy. Blue bars depict -log(P-value) and orange symbols depict enrichment ratios. Right panel shows the overlap between these top 20 pathways. Only connections between pathways with ≥5 genes in common are shown. Pathway analysis performed including all genes that were significant (FDR P<0·01) in the EWAS, TWAS and eQTM analyses (n=724 genes). See supplementary table 6 for further details.
Atopic asthma
The results of the analysis of atopic asthma are shown in the Online Supplement. DNA methylation profiles were markedly different between children with atopic asthma and those without atopy or asthma: FDR-adjusted P-values for the top 30 CpGs in atopic asthma ranged from 2·2×10−13 to 1·3×10−16 (Supplementary Figure 3, Supplementary Table 7). There was substantial overlap between the results for atopy and those for atopic asthma (Supplementary Figure 4), and the top EWAS results for atopic asthma were similar to our results for atopy.
Validation and external replication
Pyrosequencing of 15 top CpGs (nine for atopy, six for atopic asthma) showed strong correlation with microarray results (Supplementary Figure 5; correlation coefficients ~0·84–0·96, P-values 1·4×10−22 to 3·2×10−42). Furthermore, methylation of all CpGs measured by pyrosequencing were significantly associated with atopy in the same direction as the microarray EWAS (Supplementary Table 8).
Results from the replication analysis of our EWAS of atopy are shown in Table 2: using data from Yang et al.,(5) 28/30 CpGs replicated in the same direction with P<0·01. Likewise, in PIAMA, 29/30 CpGs replicated in the same direction with P<0·01. Fisher combined P-values across the three cohorts ranged from 1·72×−19 to 1·05×10−47 (Table 2). In addition, we replicated most of the top results reported by Yang et al. (Supplementary Table 9).
Atopy classification panels
Based on our analysis using three models to determine the best “predictive” CpGs, optimal number of markers, and probability cut-offs (Supplementary Figures 6–8), we selected a 30-CpG panel (Supplementary Table 10). Results from applying this panel to classify participants by atopy are shown in Figure 4: depending on the model, in the Puerto Rico cohort the AUC ranged from 0·93 to 0·94; accuracy was 85%−88%, sensitivity 0·85–0·87, specificity 0·85–0·89, positive predictive value (PPV) 0·91–0·94, and negative predictive value (NPV) 0·76–0·80. Similar results were obtained after applying the same panel and model to data from Yang et al. for atopic asthma: AUC 0·91–0·92, accuracy 81%−82%, sensitivity 0·67–0·92, specificity 0·75–0·97, PPV 0·77–0·96, and NPV 0·74–0·90. In PIAMA, we obtained AUC 0·74–0·79, accuracy 68%−73%, sensitivity 0·51–0·62, specificity 0·74–0·87, PPV 0·69–0·80, and NPV 0·65–0·69 for atopy. In contrast, our “negative control” models yielded AUCs of 0·57–0·63 (for a panel of 30 randomly selected CpGs) and 0·65–0·69 (for the model including only demographic and questionnaire data) among Puerto Rican children in EVA-PR (Supplementary Figure 9).
Figure 4 – DNA methylation panel and classification/prediction of atopy in study cohorts.
Top panels show the receiver operating characteristic (ROC) curves for Puerto Rico (left), Yang (middle), and PIAMA (right) using three different statistical approaches (GLMNET, GBM, RF; see Methods for details). Middle row shows heat maps for the three cohorts using the 30 CpG in the GLMNET model. Bottom row shows classification tables for atopy in the three cohorts. Results shown are from using the same 30-CpG panel, coefficients, and probability cut-offs trained from the Puerto Rico cohort data, then applied to both independent cohorts. Sens: sensitivity. Spec: specificity. PPV: positive predictive value. NPV: negative predictive value. Accuracy: (True positives + true negatives) / (total N).
Next, we applied the same CpG panel to classify atopic asthma (Supplementary Figure 10). In Puerto Rico, the panel yielded AUC 0·95–0·99 and accuracy of 88%. In PIAMA, the analysis of atopic asthma yielded AUC 0·82–0·88 and accuracy of 86%. Because Yang et al. compared atopic asthmatics vs non-atopic controls, the analysis of atopy and atopic asthma yielded the same results as that of atopy vs. no atopy.
DISCUSSION
Nasal epithelium is a non-invasive surrogate marker for bronchial epithelium in children.(11, 12) We show that epigenomic profiles in nasal epithelium are significantly associated with atopy or atopic asthma in three cohorts of children and adolescents. Moreover, many top DNA methylation signals are associated with expression of their corresponding genes and, in several instances, the association between methylation and atopy is mediated by gene expression. Importantly, the 30-CpG panel we selected accurately identified atopy or atopic asthma. To our knowledge, this is the first genome-wide study of DNA methylation and atopy, and the largest such study of atopic asthma. Our results strongly suggest a key role for a dysfunctional airway epithelium in atopy and asthma.
Our enrichment analysis revealed that the methylation signals linked to atopy or atopic asthma are in pathways related to gap junction signaling and immune regulation, including antigen presentation and Th1/Th2 signaling. Indeed, several genes identified in this study are implicated in epithelial barrier processes and immune regulation, including CDH26, a cadherin family protein that is involved in allergic responses, modulates CD4+ T cells and IL-2 production,(17) and regulates airway epithelial cell structure and polarity(18). CDHR3, in the same family, has been strongly implicated in interactions between rhinovirus infection and asthma exacerbations, and in bronchiolitis.(19, 20) GJA4 (gap-junction protein alpha-4) expression in bronchiolar epithelium is markedly decreased in murine asthma.(21) CAPN14, a susceptibility locus for eosinophilic esophagitis (EoE), is induced by IL-13, and together they alter epithelial function and repair.(22–24) MTRNL (meteorin-like), part of our classification panel and a cytokine present in mucosal barrier and skin, is over-expressed in atopic dermatitis.(25) Our findings suggest that DNA methylation may alter airway epithelial integrity and function, leading to antigen penetration of the epithelial barrier, antigen presentation to dendritic cells, altered Th1/Th2 immune responses, and –ultimately– atopy or atopic asthma.
Other genes present in both our top CpGs and the classification panel are involved in atopy, lung disease, or inflammation. We found decreased methylation (and increased expression) of NTRK1 in atopy. NTRK1 has been linked to circulating nerve growth factor (NRF) in children with asthma, with epistasis between both genes;(26) NTRK1 is induced by IL-13, enhances allergic inflammation, and is increased in EoE.(27) SLC9A3 codes for an Na+/H+ exchanger and has been linked to lower lung function in cystic fibrosis;(28, 29) to our knowledge, this is the first report linking SLC9A3 to atopy or asthma. Among genes significant at multiple levels (EWAS, eQTM, and mediation analyses), PCSK6 was among the top findings from Yang and colleagues; we also found decreased methylation of PCSK6 in atopy, and further report a corresponding increase in gene expression. PCSK6 activates NF-κB, IL-1, and IL-6,(30) and may have a paracrine role in activating matrix metalloproteases.(31) We also found hypomethylation (and up-regulation) of FBXL7 in atopic subjects; FBXL7 expression has been associated with decreased inhaled corticosteroid response in asthma(32), and FBXL7 has been implicated in aspirin-induced urticaria and angioedema.(33) GRK5 (also hypo-methylated and up-regulated among atopic children in our study) may mediate β2-adrenergic receptor desensitization,(34) and PRKD1 polymorphisms could interact with cleaning products on adult-onset asthma.(35) Finally, among genes that were not among the most significant by p-values but included in the classification panel, DUOX1 (dual oxidase-1) mediates inflammation, mucous cell metaplasia, and airway remodeling in asthma;(36) IL-4 signaling can activate IRS2 (insulin receptor substrate 2) leading to M2 polarization of lung macrophages, which has been linked to poor lung function in atopic asthma;(37) and LRP1 (LDL receptor-related protein 1) modulates dendritic cell responses and attenuates dust mite-induced eosinophilic airway inflammation.(38)
Our methylation results are biologically relevant. Beyond plausibility from existing literature, our TWAS and eQTM analyses examined whether methylation is associated with gene expression, and our mediation analysis further evaluated whether the methylation-atopy relationship is explained by transcriptomic differences. Several CpG/gene pairs showed significant mediation, including SLC9A3, CDH26, PCSK6, and MAP3K14; others, like FBXL7 or NTRK1, showed little or non-significant mediation, suggesting that the link between methylation and atopy occurs through other mechanisms. Moreover, we replicated >93% of our top EWAS results in two cohorts. Of note, two of our top genes (METTL1 and PCSK6) were also among the genes reported by Yang et al,(5) as were two genes in our CpG classification panel. We also provide the first replication of the top results in that study, which linked the nasal methylome to atopic asthma but had limited statistical power and lacked external replication.
With regard to the clinical significance of our findings, developing predictors of asthma or atopy in early life is a major need in pediatrics, as only ~41% of children who report any wheeze by age 3 years will go on to have “true asthma” by age 6 years, while the remaining 59% of young children with “early wheeze” will report no wheeze at age 6 years. Using data from four clinical parameters and peripheral blood eosinophil count, Castro-Rodriguez et al developed an Asthma Predictive Index (API) to help differentiate young children with transient wheeze from those who go on to develop asthma(39). However, this API has limited predictive accuracy. For asthma at age 11 years, the “stringent API” has a sensitivity of 15%, a PPV of 42%, and a NPV of 85.6%. At present, neither a child’s parents nor a child’s physician knows with certainty whether an infant with wheeze will go on to develop atopy or asthma at school age. Such knowledge could help guide treatment or reduce anxiety in the child’s family.
Our study is cross-sectional, and thus we cannot determine whether specific methylation marks preceded or were a consequence of atopy or atopic asthma. Moreover, we are unable to examine non-atopic asthma because of limited or no data on this outcome. However, our findings demonstrate the feasibility of using the nasal methylome to develop clinical tools in future longitudinal studies, as highlighted by the ability of the 30-CpG panel to identify atopic subjects in three studies despite racial/ethnic differences: our cohort included Puerto Ricans with ~24% African ancestry, the study by Yang predominantly (~92%) included African Americans, and PIAMA included mosty (~97%) Western Europeans. Furthermore, PIAMA participants were not selected for atopy or asthma and thus had lower total IgE (Table 1); this likely explains the somewhat smaller effect sizes in PIAMA, though a small contribution of European ancestry effects is also possible. Of note, the same CpG panel was able to reliably classify atopic asthma, with accuracy of 88% in Puerto Rico, 82% in the data from Yang, and 86% in PIAMA. While the PPV in PIAMA was lower at 0·42, this is expected from an unselected birth cohort with ~10% prevalence of the outcome (atopic asthma). These very promising results are far superior to predictive models using genetic variants, and also in clear contrast to those of our “negative control” analyses, in which panels composed of either 30 random CpGs (AUC ~0·57–0·63) or demographic and questionnaire data (AUCs of ~0·65–0·69) performed markedly worse than our selected CpG panel. Although nasal transcriptomics offers an alternative approach for asthma or atopy classification, DNA is more stable than RNA, and methylation marks offer a more direct assessment of environmental exposures. Moreover, preliminary (nonreplicated) findings from a small study in adults suggest that a panel of 90 genes (considerably larger than our 30-CpG panel) may be needed for asthma diagnosis using transcriptomics(40). Our results suggest that nasal methylation patterns can reliably classify atopy or atopic asthma in children differing in age, race/ethnicity, and degree of atopy. Furthermore, novel genes in our predictive panel (e.g., MAP2K6, EPHA4, ACOX2, BBOX1, or AXIN2) are worthy of further study.
Our findings were robust to nasal sample collection and processing protocols. Indeed, our results were unchanged when excluding CD326+ sorted samples, and were consistent when analyzing only the CD326+ subset. Notably, the study by Yang used histology and gene expression to ensure high proportion of ciliated epithelial cells, and PIAMA included whole nasal samples collected using a protocol similar to ours. Of interest, none of our top results overlapped with the 14 CpGs reported in a recent EWAS of asthma in whole blood(13) (Supplementary Table 11), further highlighting the novelty of our findings in airway epithelium and the critical importance of tissue specificity in epigenomic studies.
In summary, we have identified novel and biologically plausible methylation markers of atopy and atopic asthma in nasal epithelium, which are located in or near genes related to immune regulation and airway epithelial integrity. Most of these genes were not identified in GWAS, and thus our results support a key role of epigenetic changes in the nasal epithelium in the causality of atopy and atopic asthma in children. Moreover, we report a nasal methylation profile that accurately classifies subjects according to atopy or atopic asthma, thus supporting future longitudinal studies to develop predictive nasal methylation panels. Such panels could be used for early prediction of asthma, identifying early-life environmental risk factors for asthma, and assessing response to asthma therapies in children.
Supplementary Material
RESEARCH IN CONTEXT.
Evidence before this study:
A large meta-analysis of genome-wide association studies (GWAS) identified 18 loci associated with asthma but explaining only 3.5% of disease risk. Given that genes associated with childhood asthma are often expressed in airway epithelium, environmentally-influenced epigenetic regulation of that tissue could play a role in the pathogenesis of atopic asthma. We searched PubMed for articles in English and found prior reports of DNA methylation in blood or white blood cells and asthma, but minimal evidence of a link between airway epithelium DNA methylation and atopy or asthma.
Added value of this study:
To our knowledge, this is the first genome-wide study of the nasal epithelium methylome and atopy, and the first such study of atopic asthma in a large sample of children. We identified 8,664 CpG sites that were significantly associated with atopy and atopic asthma in Puerto Rican children; FDR P-values ranged from 9·58×10−17 to 2·18×10−22 for the top 30 CpGs. Several top CpGs were located near genes associated with immune responses or epithelial barrier function, and a high proportion of significant CpGs were associated with changes in gene expression. We replicated 28 of the top 30 CpGs in two independent cohorts from different ethnic backgrounds; combined P-values for the three cohorts ranged from 1·72×−19 to 1·05×10−47. Moreover, we designed a 30-CpG panel that accurately classified children according to atopy (area under the curve [AUC] 0·93–0·94 for the original cohort and 0·74–0·92 for the replication cohorts) or atopic asthma (AUC of 0·95–0·99 in the original cohort, and 0·85–0·88 in the replication cohorts).
Implications of all the available evidence:
We demonstrate that DNA methylation profiles in airway epithelium are significantly associated with atopy and atopic asthma, particularly near genes related to epithelial barrier integrity or function and other immune regulatory processes. Our findings are robust to differences in ethnic/racial background, geographic location, and environmental exposures across cohorts. None of the top 30 genes in our study overlaps with the 18 asthma-susceptibility genes identified in a recent GWAS meta-analysis, emphasizing the novelty and importance of our results. Moreover, we identified a CpG panel that accurately classifies atopy and atopic asthma across three diverse populations. Our findings suggest a key role of epigenetic regulation of the airway epithelium in the pathogenesis of atopy and asthma, and support the feasibility of using the nasal methylome to develop much-needed research and clinical tools in future longitudinal studies. Such studies should help identify methylation marks that precede disease development or treatment response, and thus lead to clinical applications for the prediction of atopy or asthma in infants, and therapeutic responses in children with asthma.
Acknowledgments:
We thank all the participating children and families. This study was supported by grants HL079966, HL117191, and MD011764 (PI: Celedón JC) from the National Heart, Lung, and Blood Institute (NHLBI) of the U.S. National Institutes of Health (NIH). Dr. Forno’s contribution was supported by NIH grant HL125666. Dr. Celedón’s contribution was additionally supported by The Heinz Endowments. Research operations at the University of Puerto Rico were additionally supported by grant U54MD007587 from the National Institute on Minority Health and Health Disparities (NIMHD) and the National Institute of Allergy and Infectious Diseases (NIAID) of the U.S. NIH. PIAMA was supported by The Netherlands Organization for Health Research and Development; The Netherlands Organization for Scientific Research; The Netherlands Lung Foundation (with methylation studies supported by AF 4.1.14.001); The Netherlands Ministry of Spatial Planning, Housing, and the Environment; and The Netherlands Ministry of Health, Welfare, and Sport. Dr. Qi is supported by a grant from the China Scholarship Council.
Role of the funding source: Dr. Celedón had full access to all of the study data and takes responsibility for the integrity of the data and the accuracy of the data analysis. The study sponsors had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Footnotes
Conflict of interest statement: Dr. Koppelman has received grants from Lung Foundation of the Netherlands, during the conduct of the study; and grants from TEVA the Netherlands, GSK, Vertex, Ubbo Emmius Foundation, and TETRI foundation, outside the submitted work. Dr. Celedón has received research materials from GSK and Merck (inhaled steroids) and Pharmavite (vitamin D and placebo capsules) to provide medications free of cost to participants in NIH-funded studies, unrelated to the current work. The rest of authors reported no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Davidson EJ, Yang IV. Role of epigenetics in the development of childhood asthma. Curr Opin Allergy Clin Immunol. 2018. April;18(2):132–8. PubMed PMID: Epub 2018/02/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WOC, Altmuller J, Ang W, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018. January;50(1):42–53. PubMed PMID: Pubmed Central PMCID: PMC5901974. Epub 2017/12/24. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loxham M, Davies DE. Phenotypic and genetic aspects of epithelial barrier function in asthmatic patients. J Allergy Clin Immunol. 2017. June;139(6):1736–51. PubMed PMID: Pubmed Central PMCID: PMC5457128. Epub 2017/06/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koning H, Sayers I, Stewart CE, de Jong D, Ten Hacken NH, Postma DS, et al. Characterization of protocadherin-1 expression in primary bronchial epithelial cells: association with epithelial cell differentiation. FASEB J. 2012. January;26(1):439–48. PubMed PMID: Epub 2011/10/11. [DOI] [PubMed] [Google Scholar]
- 5.Yang IV, Pedersen BS, Liu AH, O’Connor GT, Pillai D, Kattan M, et al. The nasal methylome and childhood atopic asthma. J Allergy Clin Immunol. 2017. May;139(5):1478–88. PubMed PMID: Pubmed Central PMCID: PMC5391298. Epub 2016/10/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci U S A. 2015. April 28;112(17):5485–90. PubMed PMID: Pubmed Central PMCID: 4418890. Epub 2015/04/08. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loser S, Gregory LG, Zhang Y, Schaefer K, Walker SA, Buckley J, et al. Pulmonary ORMDL3 is critical for induction of Alternaria-induced allergic airways disease. J Allergy Clin Immunol. 2017. May;139(5):1496–507 e3 PubMed PMID: Pubmed Central PMCID: PMC5415707. Epub 2016/09/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers AJ, Celedon JC, Lasky-Su JA, Weiss ST, Raby BA. Filaggrin mutations confer susceptibility to atopic dermatitis but not to asthma. J Allergy Clin Immunol. 2007. December;120(6):1332–7. PubMed PMID: Epub 2007/12/13. eng. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs TS, Forno E, Brehm JM, Acosta-Perez E, Han YY, Blatter J, et al. Underdiagnosis of allergic rhinitis in underserved children. J Allergy Clin Immunol. 2014. September;134(3):737–9 e6. PubMed PMID: Epub 2014/05/07. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szentpetery SS, Gruzieva O, Forno E, Han YY, Bergstrom A, Kull I, et al. Combined effects of multiple risk factors on asthma in school-aged children. Respir Med. 2017. December;133:16–21. PubMed PMID: Pubmed Central PMCID: PMC5728683. Epub 2017/11/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O’Connor BP, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol. 2014. March;133(3):670–8 e12. PubMed PMID: Pubmed Central PMCID: 4043390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baccarelli A, Rusconi F, Bollati V, Catelan D, Accetta G, Hou L, et al. Nasal cell DNA methylation, inflammation, lung function and wheezing in children with asthma. Epigenomics. 2012. February;4(1):91–100. PubMed PMID: Pubmed Central PMCID: 3297414. Epub 2012/02/16. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu CJ, Soderhall C, Bustamante M, Baiz N, Gruzieva O, Gehring U, et al. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. The lancet Respiratory medicine. 2018. February 26 PubMed PMID: Epub 2018/03/03. [DOI] [PubMed] [Google Scholar]
- 14.Forno E, Wang T, Yan Q, Brehm J, Acosta-Perez E, Colon-Semidey A, et al. A Multiomics Approach to Identify Genes Associated with Childhood Asthma Risk and Morbidity. Am J Respir Cell Mol Biol. 2017. October;57(4):439–47. PubMed PMID: Pubmed Central PMCID: PMC5650086. Epub 2017/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forno E, Sordillo J, Brehm J, Chen W, Benos T, Yan Q, et al. Genome-wide interaction study of dust mite allergen on lung function in children with asthma. J Allergy Clin Immunol. 2017. October;140(4):996–1003 e7. PubMed PMID: Pubmed Central PMCID: PMC5544591. Epub 2017/02/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunekreef B, Smit J, de Jongste J, Neijens H, Gerritsen J, Postma D, et al. The prevention and incidence of asthma and mite allergy (PIAMA) birth cohort study: design and first results. Pediatr Allergy Immunol. 2002;13 Suppl 15:55–60. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 17.Caldwell JM, Collins MH, Kemme KA, Sherrill JD, Wen T, Rochman M, et al. Cadherin 26 is an alpha integrin-binding epithelial receptor regulated during allergic inflammation. Mucosal Immunol. 2017. September;10(5):1190–201. PubMed PMID: Pubmed Central PMCID: PMC5496811. Epub 2017/01/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lachowicz-Scroggins ME, Gordon ED, Wesolowska-Andersen A, Jackson ND, MacLeod HJ, Sharp LZ, et al. Cadherin-26 (CDH26) regulates airway epithelial cell cytoskeletal structure and polarity. Cell Discov. 2018;4:7 PubMed PMID: Pubmed Central PMCID: PMC5809386. Epub 2018/02/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kantor DB, Phipatanakul W, Hirschhorn JN. Gene-Environment Interactions Associated with the Severity of Acute Asthma Exacerbation in Children. Am J Respir Crit Care Med. 2018. March 1;197(5):545–7. PubMed PMID: Epub 2017/11/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonnelykke K, Coleman AT, Evans MD, Thorsen J, Waage J, Vissing NH, et al. Cadherin-related Family Member 3 Genetics and Rhinovirus C Respiratory Illnesses. Am J Respir Crit Care Med. 2018. March 1;197(5):589–94. PubMed PMID: Epub 2017/11/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park SJ, Lee KS, Kim SR, Min KH, Lee KY, Choe YH, et al. Change of connexin 37 in allergen-induced airway inflammation. Exp Mol Med. 2007. October 31;39(5):629–40. PubMed PMID: Epub 2007/12/07. [DOI] [PubMed] [Google Scholar]
- 22.Sleiman PM, Wang ML, Cianferoni A, Aceves S, Gonsalves N, Nadeau K, et al. GWAS identifies four novel eosinophilic esophagitis loci. Nat Commun. 2014. November 19;5:5593 PubMed PMID: Pubmed Central PMCID: PMC4238044. Epub 2014/11/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014. August;46(8):895–900. PubMed PMID: Pubmed Central PMCID: PMC4121957. Epub 2014/07/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Litosh VA, Rochman M, Rymer JK, Porollo A, Kottyan LC, Rothenberg ME. Calpain-14 and its association with eosinophilic esophagitis. J Allergy Clin Immunol. 2017. June;139(6):1762–71 e7. PubMed PMID: Pubmed Central PMCID: PMC5461191. Epub 2017/01/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ushach I, Burkhardt AM, Martinez C, Hevezi PA, Gerber PA, Buhren BA, et al. METEORIN-LIKE is a cytokine associated with barrier tissues and alternatively activated macrophages. Clin Immunol. 2015. February;156(2):119–27. PubMed PMID: Pubmed Central PMCID: PMC4336607. Epub 2014/12/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szczepankiewicz A, Rachel M, Sobkowiak P, Kycler Z, Wojsyk-Banaszak I, Schoneich N, et al. Neurotrophin serum concentrations and polymorphisms of neurotrophins and their receptors in children with asthma. Respir Med. 2013. January;107(1):30–6. PubMed PMID: Epub 2012/12/01. [DOI] [PubMed] [Google Scholar]
- 27.Rochman M, Kartashov AV, Caldwell JM, Collins MH, Stucke EM, Kc K, et al. Neurotrophic tyrosine kinase receptor 1 is a direct transcriptional and epigenetic target of IL-13 involved in allergic inflammation. Mucosal Immunol. 2015. July;8(4):785–98. PubMed PMID: Pubmed Central PMCID: PMC4429043. Epub 2014/11/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorfman R, Taylor C, Lin F, Sun L, Sandford A, Pare P, et al. Modulatory effect of the SLC9A3 gene on susceptibility to infections and pulmonary function in children with cystic fibrosis. Pediatr Pulmonol. 2011. April;46(4):385–92. PubMed PMID: Epub 2010/10/23. [DOI] [PubMed] [Google Scholar]
- 29.Corvol H, Blackman SM, Boelle PY, Gallins PJ, Pace RG, Stonebraker JR, et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat Commun. 2015. September 29;6:8382 PubMed PMID: Pubmed Central PMCID: PMC4589222. Epub 2015/09/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang H, Wang L, Wang F, Pan J. Proprotein convertase subtilisin/kexin type 6 promotes in vitro proliferation, migration and inflammatory cytokine secretion of synovial fibroblastlike cells from rheumatoid arthritis via nuclearkappaB, signal transducer and activator of transcription 3 and extracellular signal regulated 1/2 pathways. Mol Med Rep. 2017. December;16(6):8477–84. PubMed PMID: Epub 2017/09/26. [DOI] [PubMed] [Google Scholar]
- 31.Brant KA, Leikauf GD. Dysregulation of FURIN by prostaglandin-endoperoxide synthase 2 in lung epithelial NCI-H292 cells. Mol Carcinog. 2014. March;53(3):192–200. PubMed PMID: Epub 2012/10/16. [DOI] [PubMed] [Google Scholar]
- 32.Park HW, Dahlin A, Tse S, Duan QL, Schuemann B, Martinez FD, et al. Genetic predictors associated with improvement of asthma symptoms in response to inhaled corticosteroids. J Allergy Clin Immunol. 2014. March;133(3):664–9 e5. PubMed PMID: Epub 2014/02/04. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornejo-Garcia JA, Liou LB, Blanca-Lopez N, Dona I, Chen CH, Chou YC, et al. Genome-wide association study in NSAID-induced acute urticaria/angioedema in Spanish and Han Chinese populations. Pharmacogenomics. 2013. November;14(15):1857–69. PubMed PMID: Epub 2013/11/19. [DOI] [PubMed] [Google Scholar]
- 34.Wang WC, Mihlbachler KA, Bleecker ER, Weiss ST, Liggett SB. A polymorphism of G-protein coupled receptor kinase5 alters agonist-promoted desensitization of beta2-adrenergic receptors. Pharmacogenet Genomics. 2008. August;18(8):729–32. PubMed PMID: Pubmed Central PMCID: PMC2699179. Epub 2008/07/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rava M, Ahmed I, Kogevinas M, Le Moual N, Bouzigon E, Curjuric I, et al. Genes Interacting with Occupational Exposures to Low Molecular Weight Agents and Irritants on Adult-Onset Asthma in Three European Studies. Environ Health Perspect. 2017. February;125(2):207–14. PubMed PMID: Pubmed Central PMCID: PMC5289825 the local studies (Bergen and Paris, respectively) in ECHRS II. The authors declare they have no other actual or potential competing financial interests. Epub 2016/08/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Habibovic A, Hristova M, Heppner DE, Danyal K, Ather JL, Janssen-Heininger YM, et al. DUOX1 mediates persistent epithelial EGFR activation, mucous cell metaplasia, and airway remodeling during allergic asthma. JCI Insight. 2016. November 3;1(18):e88811 PubMed PMID: Pubmed Central PMCID: PMC5085603. Epub 2016/11/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warren KJ, Fang X, Gowda NM, Thompson JJ, Heller NM. The TORC1-activated Proteins, p70S6K and GRB10, Regulate IL-4 Signaling and M2 Macrophage Polarization by Modulating Phosphorylation of Insulin Receptor Substrate-2. The Journal of biological chemistry. 2016. November 25;291(48):24922–30. PubMed PMID: Pubmed Central PMCID: PMC5122764. Epub 2016/10/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mishra A, Yao X, Saxena A, Gordon EM, Kaler M, Cuento RA, et al. Low-density lipoprotein receptor-related protein 1 attenuates house dust mite-induced eosinophilic airway inflammation by suppressing dendritic cell-mediated adaptive immune responses. J Allergy Clin Immunol. 2017. December 21 PubMed PMID: Epub 2017/12/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castro-Rodriguez JA, Holberg CJ, Wright AL, Martinez FD. A clinical index to define risk of asthma in young children with recurrent wheezing. Am J Respir Crit Care Med. 2000. October;162(4 Pt 1):1403–6. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 40.Pandey G, Pandey OP, Rogers AJ, Ahsen ME, Hoffman GE, Raby BA, et al. A Nasal Brush-based Classifier of Asthma Identified by Machine Learning Analysis of Nasal RNA Sequence Data. Sci Rep. 2018. June 11;8(1):8826 PubMed PMID: Pubmed Central PMCID: PMC5995932. Epub 2018/06/13. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.