Abstract
Microbiome-wide association studies on large population cohorts have highlighted associations between the gut microbiome and complex traits, including type 2 diabetes (T2D) and obesity1. However, the causal relationships remain largely unresolved. We leveraged information from 952 normo-glycemic individuals for whom genome-wide genotyping, gut metagenomic sequence and fecal short chain fatty acid (SCFA) levels were available2, and combined these with genome-wide association summary statistics for 17 metabolic and anthropometric traits. Using bidirectional Mendelian Randomization (MR) analyses to assess causality3, we found that host genetic-driven increase in gut production of the SCFA butyrate is associated with improved insulin response following an oral glucose test (P = 9.8 × 10−5), while abnormalities in production or absorption of another SCFA, propionate, are causally related to increased risk of T2D (P = 0.004). These data provide evidence of a causal effect of the gut microbiome on metabolic traits, and support the use of MR as a route to elucidate causal relationships from microbiome-wide association findings.
There is increasing evidence that the human gut microbiome plays a role in immune function and metabolic disease1,4,5. Manipulation of the gut microbiome offers an alternative to pharmacological interventions provided it can be demonstrated that altering microbiota composition and/or function (e.g. through personalized nutrition) has clinical benefit. To demonstrate this, it is essential to discriminate between microbiome features that are causal for disease, from those that are a consequence of disease or its treatment, and those that show statistical correlation due to confounding or pleiotropy.
Animal studies support a causal role for the gut microbiome in the development of type 2 diabetes (T2D), insulin resistance and obesity6,7, but translating these findings to humans and identifying the specific bacterial species responsible has proven challenging8. Cross-sectional studies have confirmed that gut microbiota composition is altered in subjects with pre-diabetes or T2D compared to controls, while fecal transplantation studies have shown that insulin sensitivity increases in obese subjects with metabolic syndrome after the transfer of gut microbiota from lean donors4,5,9,10. Whilst the specific microbiome features identified as responsible for these effects have differed between studies, one consistent finding in T2D subjects is a shift in microbiome composition away from species able to produce butyrate. Butyrate and other short-chain fatty acids (SCFAs), such as acetate and propionate, are produced by gut bacterial fermentation of undigested food components. Following absorption by the colonocytes, these SCFAs are either used locally as fuel for colonic mucosal epithelial cells or they enter the portal bloodstream11. While the bulk of evidence suggests that increased SCFA production benefits the host by exerting anti-obesity and anti-diabetic effects4,10,12–14, some in vitro and in vivo studies have indicated that over-production or accumulation of SCFAs in the bowel may also lead to obesity due to increased energy accumulation15,16. Resolution of these conflicting data requires a detailed understanding of the causal relationships between gut microbiome composition, SCFA abundance and host energy metabolism.
Using a Mendelian Randomization (MR) approach3, we set out to identify if any bacterial species or pathways, i.e. sets of species grouped according to the specific functions they play in the gut, have a causal effect on metabolic traits. We and others have recently shown that it is possible to detect variants in the host genome that influence the composition of the gut microbiota2,17,18. This allowed us to deploy a MR approach to infer causal relationships by asking whether genetic predictors of microbiome content influence metabolic traits—or the reverse. This formulation holds even though the quantitative contribution of host genetics to variation in microbiome composition may be limited19.
We assembled genome-wide genetic data, gut metagenomic-sequencing, measurements of fecal SCFAs, and clinical phenotypes for 952 normo-glycemic individuals from the LifeLines-DEEP (LL-DEEP) cohort. From consortium websites (GIANT, MAGIC and DIAGRAM, see URLs), we also gathered publically-available genome-wide association (GWA) summary statistics for 17 anthropometric and glycemic traits20–27 (Supplementary Table 1). We focused our analyses on 245 microbiome features (2 fecal SCFA levels, 57 unique taxa, 186 pathways) that were, in LL-DEEP, correlated (false discovery rate (FDR) < 0.1) with at least one of the measured anthropometric and metabolic traits (Methods, Supplementary Table 2 and 3).
For each of these features, we sought genetic predictors -- independent genetic variants (r2 ≤ 0.1), associated (P < 1 × 10−5) with the respective features – using GWAS data from LL-DEEP, reprocessed from our previous study2 (Methods). The threshold P < 1 × 10−5 for variant inclusion was identified by maximizing the amount of genetic variance explained by the genetic predictors in 445 independent normo-glycemic individuals (the 500FG cohort)28(Methods, Supplementary Figure 1), and designed to capture sets of variants likely to be enriched for association. On average, in LL-DEEP the identified genetic predictors explained 13% (range 2%−30%) of variance in their respective microbiome features. The average F-statistic, another measure of the strength of these genetic predictors, was 21.7 (range 15.3 – 25.5); an F-statistic >10 is considered sufficiently informative for MR analyses29.
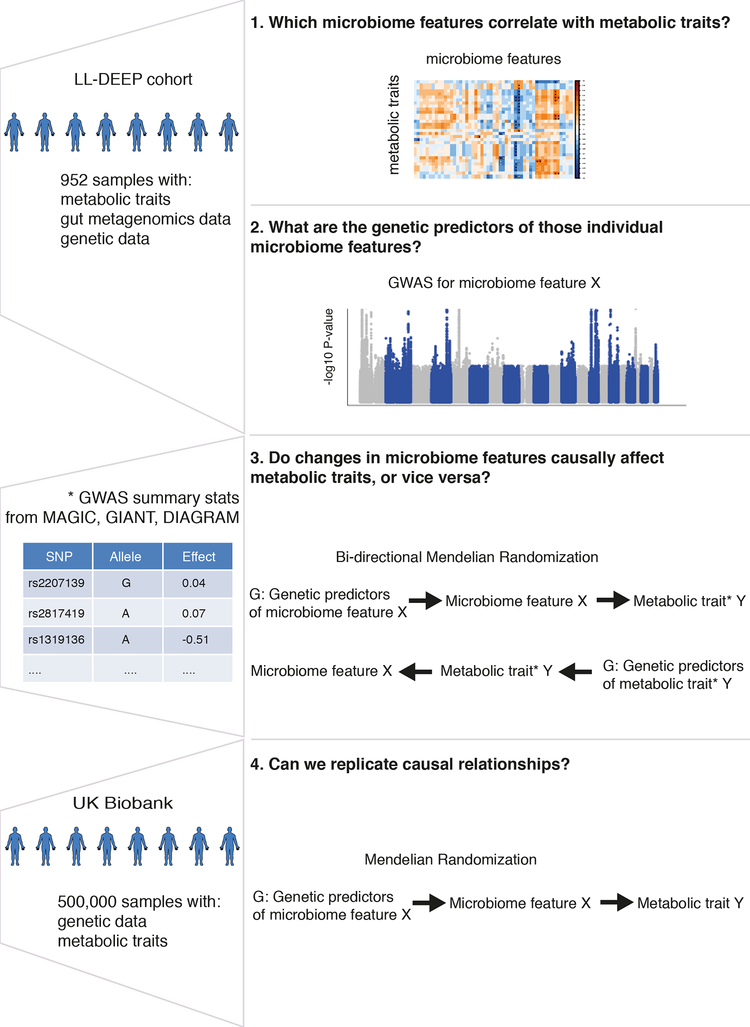
We used the inverse-variance weighted (IVW) test to identify causal relationships between the 245 microbiome features and the 17 traits of interest in a two-sample bidirectional MR analysis using pairs of GWAS summary statistics (one from a microbiome feature and one from a metabolic/anthropometric trait)29. Based on principal component analysis (PCA) and cluster analyses conducted on the microbiome and metabolic and anthropometric traits (Methods, Supplementary Figure 2), we adopted a conservative multiple testing adjusted threshold of P < 1.3 × 10−4 to declare a causal relationship significant. Because the presence of horizontal pleiotropy (where a genetic predictor has independent effects on the diseases through multiple traits) could bias the MR estimates, we investigated the robustness of significant findings to pleiotropy by using three additional MR tests: the MR-PRESSO30, the weighted median test31, and the MR-Egger32. We formally examined the presence of horizontal pleiotropy using the MR-PRESSO Global test30 and the modified Rücker’s Q’ test33,34. Finally, we sought to validate these causal relationships in an independent cohort (UK Biobank)35 (Figure 1).
Figure 1. Schematic representation of the study.
Figure 1 is a schematic representation of our study, highlighting for each step the research question we want to answer, the analysis workflow, and the data used. We first aimed to identify which microbiome feature (taxa, microbiome pathway or short-chain fatty acid (SCFA)) correlated with metabolic traits in the LifeLines-DEEP (LL-DEEP) cohort (Step 1). We then performed genome-wide association (GWA) analysis in LL-DEEP to identify genetic predictors of those microbiome features (Step 2), and used the genetic predictors to estimate causal relationships through bidirectional Mendelian Randomization analysis and effect sizes for metabolic traits extracted from summary statistics of large GWA studies (Step 3). Finally, we validated our causality results using the UK Biobank (Step 4).
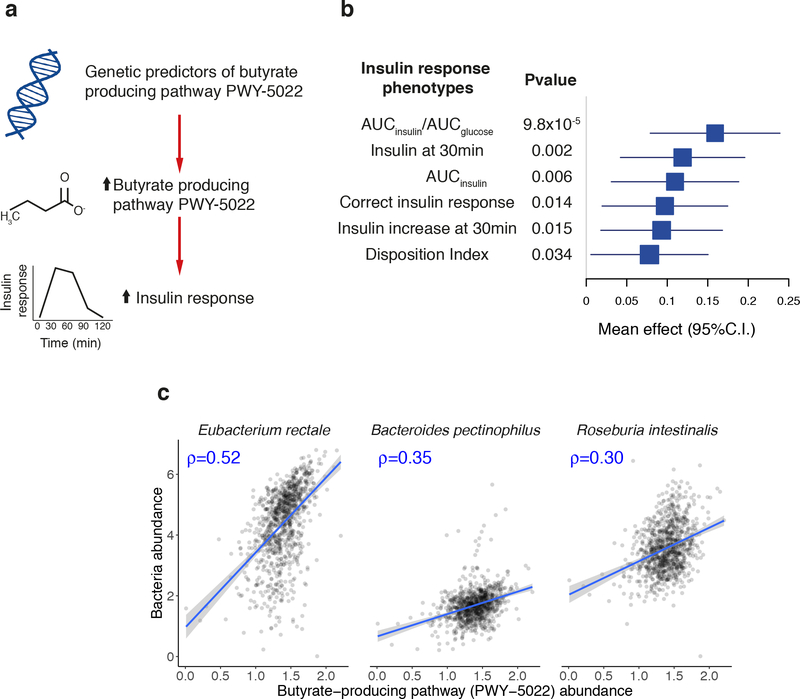
We observed a significant causal influence for one specific microbiome feature, a microbial pathway involved in 4-aminobutanoate (GABA) degradation (MetaCyc designation PWY-5022: 4-aminobutanoate degradation V) on increased insulin secretion, and specifically the ratio of the areas under the curve for insulin and glucose, AUCinsulin/AUCglucose, measured during an oral glucose tolerance test (oGTT) (Figure 2a). Using nine genetic predictors (variance explained = 16%; F-statistic = 21, Supplementary Table 4), we estimated that each standard deviation (SD) increase in the abundance of PWY-5022 generated a 0.16 mU/mmol increase in AUCinsulin/AUCglucose (P = 9.8 × 10−5) (Supplementary Table 5, Supplementary Figure 3). This causal relationship was robust when additional MR tests were performed (PMR-PRESSO = 0.02, PWeighted-Median = 0.02 and PMR-EGGER = 0.02), and there was no evidence for horizontal pleiotropy (PMR-PRESSOglobal = 0.18 and PRückerQ’(modified) = 0.77) (Supplementary Figure 4). The reverse MR analysis (testing the relationship between genetic predictors of AUCInsulin/AUCglucose and PWY-5022 abundance) was not significant (P > 0.1, Supplementary Table 6). There was no evidence of causality with seven metabolic and anthropometric traits (body-mass index (BMI), body fat %, waist-hip ratio (WHR), visceral adipose tissue, abdominal subcutaneous adipose tissue, obesity and T2D) in a MR analyses that used UK Biobank summary statistics (Supplementary Table 7); insulin secretion phenotypes after oGTT were not available. We also found supportive evidence (P < 0.05) for a causal impact of this pathway on other insulin response parameters (Figure 2b). Though other types of causal relationship are possible, these data are consistent with a model whereby host genetic variation that influences gut microbiome composition so as to modulate GABA degradation activity results in improvements in the capacity of the pancreatic islets to secrete insulin in response to a physiological glucose challenge.
Figure 2. Causal effect of butyrate-producing activity of the gut on glucose-stimulated insulin response.
a) Schematic representation of the Mendelian Randomization analysis results: genetic predisposition to higher abundance of butyrate-producing microbiome pathway PWY-5022 (4-aminobutanoate degradation V pathway) is associated with insulin response after glucose challenge. The causal effect of PWY-5022 was also seen on other insulin response parameters, and the forest plot in panel (b) represents the magnitude of the effect on each parameter per one-standard-deviation increase in pathway abundance, as estimated in the inverse-variance weighted Mendelian Randomization (MR) analysis. MR analysis was carried out using up to nine genetic predictors and their effect size from LL-DEEP (952 samples) and MAGIC summary statistics (trait specific sample sizes are: AUCinsulin/AUCglucose = 4213; insulin at 30 min = 4,409; AUCinsulin = 4,324; correct insulin response = 4,789; insulin increase at 30 min = 4,447; Disposition index = 5,130) (Methods, Supplementary Table 4 and 5). Corresponding two-sided P-values are given in the annexed table. (c) Correlation plots with PWY-5022 abundance and the bacteria correlating the most with it in 950 LL-DEEP samples (subset of the 952 normo-glycemic samples for which presence of those bacteria was detected). The Spearman correlation coefficient ρ is given in blue in each panel.
Butyrate and acetate are products of GABA degradation. In our taxonomic analyses, the bacterial species most correlated with the abundance of PWY-5022 were Eubacterium rectale and Roseburia intestinalis (Spearman ρ = 0.52 and 0.30, respectively, Figure 2c), both well-known butyrate-producing bacteria36,37. Plasma butyrate levels were not measured in our study; current assays are challenging to perform and provide unreliable estimates38. Whilst we consider the abundance of the PWY-5022 pathway to act as a proxy for butyrate production in the gut, we were unable to directly link PWY-5022 abundance to the amount of butyrate absorbed by the host. The abundance of PWY-5022 was poorly correlated with fecal butyrate levels (Spearman ρ = 0.1), and we did not detect any causal relationships between this SCFA and the 17 traits (P > 0.05), indicating that fecal butyrate is a poor proxy for butyrate production and absorption.
These results point towards a causal role for gut-produced butyrate that is focused on the dynamic insulin response to food ingestion, rather than the homeostatic mechanisms involved in the maintenance of glucose metabolism in the fasted state. Independent clinical studies support this hypothesis. For example, an intervention study evaluating the role of Bifidobacteria-increasing prebiotics (fructo-oligosaccharides) in 35 healthy individuals showed that prebiotics decrease levels of butyrate-producing bacteria and result in adverse effect on glucose metabolism following an oGTT39.
The PWY-5022 finding led us to consider the role of other SCFAs in metabolic and anthropometric traits. In our cross-sectional analysis within LL-DEEP, we had detected associations between fecal propionate levels and BMI (FDR < 0.1). Propionate is produced by different bacteria than those producing butyrate40, and its three genetic predictors (variance explained = 6.3%, F-statistic = 21) were independent of those implicated in PWY-5022 abundance (Supplementary Table 4, 8). In MR analyses for the 17 traits of interest, we found that each SD increase in fecal propionate levels was causally associated with a 0.03 SD increase in BMI (P = 0.0068) and an odd’s ratio (OR) = 1.15 for T2D (P = 0.004) (Supplementary Table 9), although these did not pass our significance threshold. No associations were evident in the reverse MR analysis testing the effect of T2D and BMI on fecal propionate levels (P > 0.1, Supplementary Table 10).
Of the two observed effects of fecal propionate, on BMI and T2D, the latter was more robust. The causal relationship on increased T2D risk was robust when other MR tests were performed (PMR-PRESSO = 0.03, PWeighted-Median = 0.03) and there was no evidence for pleiotropy (PMR-PRESSOglobal 0.75, PRückerQ’(modified) = 0.50) (Supplementary Figure 5). By contrast, the effect of propionate on increased BMI was not significant when using other MR tests and there was also evidence for pleiotropy (PMR-PRESSOglobal = 2.0 × 10−3, PRückerQ’(modified) = 9.2 × 10−4)(Supplementary Table 9, Supplementary Figure 6). The pleiotropy in the BMI effect could be accounted for by SNP rs7142308 (NC_000014.8:g.79482379A>G) (PMR-PRESSOOutlierTest = 0.01), located within a BMI-associated locus20 but independent of the lead variant (rs7141420 (NC_000014.8:g.79899454C>T), r2 = 0.01 with rs7142308 in 1000 Genomes Europeans).
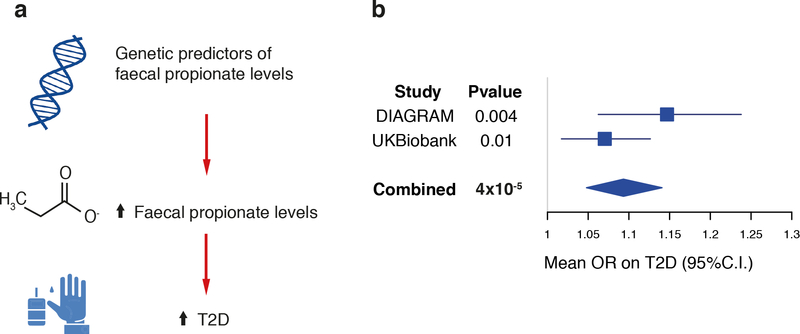
Applying MR analyses to UK Biobank summary statistics, we replicated the relationship between fecal propionate levels and increased risk of T2D (PIVW = 0.01, PMR-PRESSO = 0.007, PWeighted-Median = 0.04; PIVWcombined = 4 × 10−5, Figure 3), and there was no evidence of pleiotropy (PMR-PRESSOglobal = 0.97, PRückerQ’(modified) = 0.99). The relationship between fecal propionate and BMI was again not robust to pleiotropy, highlighting the need for caution in interpreting this effect as causal (Supplementary Table 11).
Figure 3. Causal effect of fecal propionate on type 2 diabetes (T2D).
a) Schematic representation of the Mendelian Randomization analysis results: genetic predisposition to higher fecal propionate levels is associated with increased risk of T2D. b) A forest plot depicts the magnitude of the causal effect on T2D per each one-standard-deviation increase in fecal butyrate levels, as estimated by the inverse-variance weighted Mendelian Randomization (MR) analysis. MR analysis was carried out using the three genetic predictors derived in LifeLines-DEEP (LL-DEEP) and their effects in the discovery data set (DIAGRAM; 26,676 T2D cases and 132,532 controls) and in the replication cohort (UK Biobank; 19,119 T2D cases and 423,698 controls). The effect derived combining the two causal effects (from discovery and replication) with an inverse-variance weighted meta-analysis approach is also given. Corresponding two-sided P-values are listed in the annexed table. OR, odd’s ratio.
Over 95% of gut-produced SCFAs are absorbed by the host41, such that increases in fecal propionate levels could be the consequence of either increased production or reduced absorption. The latter (which would link increased fecal propionate to reduced circulating levels) would be more consistent with the preponderance of evidence that indicates that SCFAs have a largely beneficial effect on energy balance and metabolic homeostasis4,10,12–14. As with plasma butyrate, plasma propionate levels were not measured in our cohorts. Further studies are warranted to explore the mechanisms underlying this relationship between fecal propionate levels and T2D.
In summary, these data are consistent with a causal role for gut-produced SCFAs, specifically butyrate and propionate, with respect to energy balance and glucose homeostasis in man. We have shown that a genetically-influenced shift in the gut microbiome towards increased production of butyrate has beneficial effects on beta-cell function, though no impact on T2D risk could be detected. We have also demonstrated that host genetic variation that results in increased fecal propionate levels (reflecting some combination of increased production or impaired absorption) has impact on T2D-risk.
Although the LL-DEEP cohort represents the largest population study on the genetics of microbiome2,17,18, it is still underpowered to capture the limited genetic component that has been estimated for microbiome features19. The results from this and other microbiome GWAS2,17,18 showed only limited direct overlap, highlighting the need for standardized protocols for data analyses and larger sample size42. This will be crucial also in the context of MR analyses, as expanded GWAS will deliver more robust genetic predictors43. A better understanding of the complex interplay between gut microbiome and host metabolism will require expansion of current analyses and the ability to fold in measures of circulating SCFAs. Nevertheless, this study demonstrates that microbiome GWAS provide a route to causal inference that can guide and complement more direct experimental approaches, such as those based around fecal transplantation and animal models. We envisage that with expanded microbiome-genetic studies, for example the MiBioGen consortium44, MR will become a standard tool for systematically screening a large number of hypotheses generated in current and future microbiome-wide association studies.
Online Methods
Study samples
The discovery cohort of this study is LifeLines-DEEP (LL-DEEP), a population-based cohort of 1,539 individuals from Northern Netherlands (age range 18–84 years) that is a subset of the largest Lifelines biobank (N = 167,000). For all LL-DEEP volunteers, an extensive dataset of measured and self-reported phenotypic information has been collected, as well as blood and stool specimens, all as described previously45,46. Measurement of SCFAs in stool was carried out by gas chromatography-mass spectrometry following the method of Garciá-Villalba et al47.
To identify the appropriate threshold for the selection of genetic predictors of microbiome features we used the 500 Functional Genomics (500FG) cohort28, an independent cohort of 534 healthy individuals from the Netherlands (age-range 18–75 years). Protocols for stool collection and metagenomic sequencing were similar to those used in LL-DEEP, as previously described48.
All participants from both studies signed an informed consent form. The LL-DEEP study was approved by the institutional ethics review boards of the UMCG (ClinicalTrials.gov NCT00775060). The 500FG study was approved by the Ethical Committee of Radboud University Nijmegen (NL42561.091.12, 2012/550).
To replicate our findings we used genotype and phenotype data from the UK Biobank, a study of 500,000 subjects from the United Kingdom aged 45–65 years of age35. Each participant provided a blood sample for DNA extraction and completed a detailed questionnaire providing baseline data. Individuals are also linked to electronic medical records on a number of traits including BMI and T2D.
Data generation and pre-processing
Genotyping
Genotype data was available for 1,268 LL-DEEP volunteers, as previously described2,45. In brief, genotyping was carried out using two Illumina arrays, HumanCytoSNP-12 BeadChip and ImmunoChip. After standard per-sample and per-SNP quality control filters, data from the two arrays were merged and additional markers were imputed using the HRC reference panel v1.149 on the Michigan server (see URLs). For our analyses, we focused on 15,001,957 variants with imputation accuracy RSQR > 0.3. In the 500FG cohort, 516 samples were genotyped using the Illumina Human OmniExpress Exome-8 v1.0 SNP chip and, after standard quality controls checks28, were imputed using the same procedure and reference panel used with LL-DEEP. The UK Biobank samples were genotyped using the Affymetrix UK BiLEVE Axiom array on an initial 50,000 participants. The remaining 450,000 participants were genotyped using the Affymetrix UK Biobank Axiom® array35. Quality control on samples and genotypes were performed centrally and subsequent imputation was performed using the HRC reference panel at the Wellcome Centre Human Genetics.
Metagenomic sequencing
Metagenomic sequencing of the gut microbiome was performed using the Illumina HiSeq platform on 1,179 LL-DEEP samples. After applying per-sample and per-read quality filters2, the profile of microbial composition was determined using Bracken pipeline (see URLs). In total, 903 taxonomies were identified and normalized using a log transformation; normalized non-zero values were then adjusted for age, sex and read depth using linear regression.
Functional profiling was performed using HUMAnN2 (v 0.4.0), which maps reads to a customized database of functionally annotated pan-genomes50. This analysis identified 742 pathways from the MetaCyc metabolic pathway database51. Similar to the taxonomy data, we normalized pathway abundances using log transformation and corrected the normalized nonzero values for age, sex and read depth. We considered only non-zero values for analyses, and therefore restricted analyses to microbiome features (taxonomies and pathways) that had non-zero values in less than 50% of the samples and retained only one of pairs of pathways or bacteria showing > 0.99 Spearman correlation. This filtering resulted in a final set of 796 features (273 taxa and 523 pathways) that were used for analyses.
We further confined all statistical analyses to normo-glycemic samples with good quality genetic and microbiome data. Normo-glycemic status was assigned to samples from individuals not reported to have diabetes or to be taking oral anti-diabetes medications and who had fasting glucose levels < 7 mml/L. We also removed individuals who were taking antibiotics at the time of the stool collection. This filtering resulted in a final set of 952 samples available for analyses. In the 500FG cohort, we used the same filters and selected 445 normo-glycemic samples with both genetic and microbiome data for analyses.
Genome-wide association scans of anthropometric and glycemic traits
We downloaded full GWAS summary statistics from 9 studies that represented 17 GWAS for different anthropometric and glycemic traits. These traits were BMI and waist-hip-ratio (WHR), fasting glucose, insulin and pro-insulin, 2hr glucose, HOMA-derived measurements of insulin resistance (HOMA-IR) and sensitivity (HOMA-B)), glycated hemoglobin (HbA1c), T2D, and 7 insulin response parameters measured during an oral glucose tolerance test (oGTT) (Supplementary Table 1 and URLs). SNP names and genomic positions were aligned to the genomic build GRCh37/hg19.
Statistical Analysis
Correlation of short chain fatty acids and microbiome features with anthropometric and glycemic traits
We correlated 5 short chain fatty acids (Acetate, Butyrate, Propionate, Calproate and Valerate) and 796 other microbiome features (taxa or pathways) with measured anthropometric (BMI and WHR) and glycemic traits (fasting glucose, insulin, HbA1c, HOMA-IR and HOMA-B) in the LL-DEEP cohort. Anthropometric and glycemic traits were adjusted for age, sex, and BMI (except for BMI phenotype). We used the non-parametric Spearman correlation test (cor.test(method=“Spearman”) function in R (v3.3)) and considered results significant when the multiple-testing-adjusted two-sided P-value was < 0.1. The multiple-testing-adjusted P-value, FDR P, was calculated using the Benjamini-Hochberg procedure in the p.adjust() function in R (v3.3) (see URLs).
Genome-wide association analyses of short-chain fatty acids and microbiome features
For each microbiome feature and short-chain fatty acid, we performed a genome-wide association scan in LL-DEEP samples by re-processing data from our previous study in a different manner2. In particular: a) we re-mapped metagenomic reads to a more recent database, b) we restricted analyses to only normo-glycemic samples and those who were no under antibiotics, and c) we performed genetic analyses using a linear mixed model that accounts for population structure instead of the Spearman correlation method. In particular for genetic analyses, we used EPACTS (v3.2.6)52, a software program that performs a linear mixed model adjusted with a genomic-based kinship matrix that is calculated using all quality-checked genotyped autosomal SNPs with minor allele frequency > 1%. The advantage of this model is that the kinship matrix encodes a wide range of sample structures, including both cryptic relatedness and population stratification; this produces more robust results than standard linear regression. All traits were inverse quantile normalized before genetic analysis. Specifically for SFCAs, age, sex, Chromogranin A, stool type according to Bristol scale, and BMI were added as covariates.
The variance explained (adjusted R squared) and the F-statistic for each microbiome feature were extracted from a linear model that fitted all the selected genetic predictors on the normalized, covariates-adjusted microbiome feature.
Mendelian Randomization analyses with 17 GWAS traits
The Mendelian Randomization procedure consists of two steps: i) identification of proper instrumental variables or genetic predictors, e.g. variants independently associated with the exposure factor, and ii) calculation of causal estimates. For each GWAS summary statistic, we first selected independent SNPs using the clumping procedure in PLINK v1.9 (see URLs) and setting a linkage disequilibrium threshold of r2 < 0.1 in a 500-Kb window. Linkage disequilibrium was calculated using the LL-DEEP cohort when running the clumping procedure on the GWAS of microbiome features and short chain fatty acids, whereas for GWAS of anthropometric and glycemic traits we used the linkage disequilibrium estimates from the 1000 Genomes phase 3 European samples.
Furthermore, since the majority of the downloaded GWAS were based on the HapMap2 genetic map, for each independently associated variant, we identified the best HapMap2 proxy (r2 > 0.8) or discarded that variant if no proxy was available.
Finally, we selected only variants that showed association at P < 1 × 10−5. We identified this as the optimal P-value threshold to use for selection of genetic predictors associated with microbiome features because this threshold led to a larger variance explained, on average, of the same microbiome features in the 500FG cohort (Supplementary Figure 1). For consistency, we used the same threshold and procedure for selecting genetic predictors from the downloaded GWAS on anthropometric and glycemic traits.
To calculate causal estimates, we used the inverse-variance weighted (IVW) method32 as a two-sample MR analysis of summary association statistics of the exposure and the outcome. Specifically, we estimated the causal effect in a fixed-effect meta-analysis framework, i.e. as a sum of single-SNP causal effects (derived as a ratio of the SNP-effect on the outcome by the SNP-effect on the exposure) weighted by the inverse of their variance (derived as a squared ratio of the SNP-standard deviation on the outcome on SNP-effect on the exposure). The P-value was calculated as P = 2* (1- Φ(Z)), where Φ(Z) is the standard normal cumulative distribution function and Z is ratio of the combined (using inverse variance weights) causal effect and its standard error. Of note, the causal estimate is equivalent to that obtained as a weighted linear regression of the outcome SNP-effects on the exposure SNP-effects with a fixed intercept of 0 and with the inverse of the variance of the effect sizes on the outcome as weights. For analyses, we set the effect allele of the genetic predictors to be the allele with the positive direction. We also calculated causal estimates using additional MR methods: MR-PRESSO30, which removes pleiotropy by identifying and discarding influential outlier predictors from the IVW test and uses a t-test to calculate P-values; the weighted-median test31, which uses a statistical estimator that is robust to the presence of pleiotropy in a subset (< 50%) of the predictors; and the MR-Egger32, which adjusts for average horizontal pleiotropy and assumes that > 50% of the predictors have pleiotropy. Furthermore, we specifically evaluated presence of pleiotropy using MR-PRESSO Global test30 and the modified Rücker’s Q’ test33.
Calculation of significance threshold
To define our significance threshold for the IVW-based MR analyses, we first run a principal component analysis of the 245 microbiome features, and observed that the total variability could be explained by the first 57 principal component axes. To derive the number of independent anthropometric and metabolic traits out of the 17 of interest, we used pairwise genetic correlation calculated using LDScore regression (LDscore v1.0.0). Variants were restricted to those from HapMap3 and pre-computed LD Scores estimated in subjects of European descent were used as recommended by the authors53. Traits were hierarchically clustered based on genetic correlation values ρg, with dissimilarity metric (1-ρg)/2 (Supplementary Figure 2). The number of resulting clusters was used to define the number of independent traits. Genetic correlation could not be calculated with four insulin secretion traits so we counted these as fully independent traits. We set our multiple testing significance threshold at 1.3 × 10−4 (0.05/(57*7)).
Mendelian randomization analyses in UK Biobank
We first calculated association of the 12 genetic predictors (9 for PWY-5022 and 3 for fecal propionate) with 7 metabolic and anthropometric traits (BMI, body Fat %, WHR, visceral adipose tissue (VAT), subcutaneous adipose tissue (SAT), obesity and T2D) using a linear mixed model as implemented in BOLT-LMM (v2.3.2)54. T2D status was defined according to the definition used by Eastwood et al.55; BMI was defined according to that used by the GIANT consortium20 and obesity was defined by ICD code 278. Analyses were restricted to 442,817 unrelated individuals of European descent and were adjusted for age, sex, genotyping array and 6 genetic principal components; WHR was also adjusted for BMI. We then used the summary statistics at these 12 variants to estimate causal relationships and investigate presence of pleiotropy by applying the same statistical tests that were used with the GWAS summary statistics and described in the previous paragraph.
Supplementary Material
Acknowledgements
We thank the participants and staff of the LL-DEEP cohort for their collaboration, the UMCG Genomics Coordination center, the UG Center for Information Technology and their sponsors BBMRI-NL & TarGet for storage and compute infrastructure. We are also grateful to Marc Jan Bonder for help in formatting summary statistics, to Rinse K. Weersma and Yang Li for discussions and to Kate Mc Intyre for editing the manuscript. Part of this work was conducted using the UK Biobank resource under application number 9161.
This project was funded by: IN-CONTROL CVON grant CVON2012–03 to M.G.N., A.Z., L.J. and J.F.; Top Institute Food and Nutrition (TiFN, Wageningen, the Netherlands) grant TiFN GH001 to C.W.; the Netherlands Organization for Scientific Research (NWO) grants NWO-VENI 016.176.006 to M.O., NWO-VIDI 864.13.013 to J.F., and NWO-VIDI 016.Vidi.178.056 to A.Z; NWO Spinoza Prizes SPI 92–266 to C.W. and SPI 94–212 to M.G.N.; the European Research Council (ERC)-Starting grant ERC #715772 to A.Z.; the FP7/2007–2013/ERC Advanced Grant (agreement 2012–322698) to C.W.; the ERC Consolidator Grant ERC#310372 to M.G.N.; Tripartite Immunometabolism consortium (TrIC) – Novo Nordisk Foundation Grant#NNF15CC0018486 to M.M.; and Wellcome grants 090532, 098381, 106130, and 203141 to M.M.. A.Z. also holds a Rosalind Franklin Fellowship from the University of Groningen. M.M. is a Wellcome Senior Investigator, and a National Institute of Health Research Senior Investigator. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health.
Footnotes
URLs
MAGIC: https://www.magicinvestigators.org/
GIANT: http://portals.broadinstitute.org/collaboration/giant/index.php/Main_Page
DIAGRAM: http://www.diagram-consortium.org/
UK Biobank: http://www.ukbiobank.ac.uk/
Human Functional Genomics Project: http://www.humanfunctionalgenomics.org/
Bracken: https://github.com/jenniferlu717/Bracken
MetaCyc metabolic pathway database: http://www.metacyc.org/
PLINK: www.cog-genomics.org/plink2
Michigan imputation server: https://imputationserver.sph.umich.edu/
LDScore: https://github.com/bulik/ldsc
MR-PRESSO: https://github.com/rondolab/MR-PRESSO
Competing Interests statement
M.M serves on advisory panels for Pfizer, NovoNordisk, Zoe Global; has received honoraria from Pfizer, NovoNordisk and Eli Lilly; has stock options in Zoe Global; has received research funding from Abbvie, Astra Zeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, NovoNordisk, Pfizer, Roche, Sanofi Aventis, Servier, Takeda. All other authors declare no competing financial interests.
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Data availability
The LifeLines-DEEP metagenomics sequencing data are available at the European Genome-phenome Archive (EGA), with access code EGAS00001001704. Genotype and phenotype data can be requested from the Lifelines Biobank https://www.lifelines.nl/researcher/biobank-lifelines/application-process.
Summary statistics for metabolic traits were downloaded from MAGIC, GIANT and DIAGRAM websites (see URLs).
References
- 1.Zhernakova A et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonder MJ et al. The effect of host genetics on the gut microbiome. Nat Genet 48, 1407–1412 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Evans DM & Davey Smith G Mendelian Randomization: New Applications in the Coming Age of Hypothesis-Free Causality. Annu Rev Genomics Hum Genet 16, 327–50 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Larsen N et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5, e9085(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlsson FH et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Ley RE et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102, 11070–5 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreznar JH et al. Host Genotype and Gut Microbiome Modulate Insulin Secretion and Diet-Induced Metabolic Phenotypes. Cell Rep 18, 1739–1750 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunkwall L & Orho-Melander M The gut microbiome as a target for prevention and treatment of hyperglycaemia in type 2 diabetes: from current human evidence to future possibilities. Diabetologia 60, 943–951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kootte RS et al. Improvement of Insulin Sensitivity after Lean Donor Feces in Metabolic Syndrome Is Driven by Baseline Intestinal Microbiota Composition. Cell Metab 26, 611–619 e6 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Zhang X et al. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One 8, e71108(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rios-Covian D et al. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front Microbiol 7, 185(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pingitore A et al. The diet-derived short chain fatty acid propionate improves beta-cell function in humans and stimulates insulin secretion from human islets in vitro. Diabetes Obes Metab 19, 257–265 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Chambers ES et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64, 1744–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao L et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Peng L, He Z, Chen W, Holzman IR & Lin J Effects of butyrate on intestinal barrier function in a Caco-2 cell monolayer model of intestinal barrier. Pediatr Res 61, 37–41 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Schwiertz A et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 18, 190–5 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Turpin W et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet 48, 1413–1417 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Goodrich JK et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 19, 731–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothschild D et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Locke AE et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shungin D et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 518, 187–196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manning AK et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 44, 659–69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strawbridge RJ et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes 60, 2624–34 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soranzo N et al. Common variants at 10 genomic loci influence hemoglobin A(1)(C) levels via glycemic and nonglycemic pathways. Diabetes 59, 3229–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prokopenko I et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet 10, e1004235(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saxena R et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet 42, 142–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott RA et al. An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes 66, 2888–2902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y et al. A Functional Genomics Approach to Understand Variation in Cytokine Production in Humans. Cell 167, 1099–1110 e14 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Burgess S, Butterworth A & Thompson SG Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37, 658–65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verbanck M, Chen CY, Neale B & Do R Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 50, 693–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowden J, Davey Smith G, Haycock PC & Burgess S Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol 40, 304–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowden J, Davey Smith G & Burgess S Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44, 512–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowden J et al. Improving the accuracy of two-sample summary data Mendelian randomization: moving beyond the NOME assumption. Preprint at https://www.biorxiv.org/content/early/2018/10/11/159442 (2018). [DOI] [PMC free article] [PubMed]
- 34.Rucker G, Schwarzer G, Carpenter JR, Binder H & Schumacher M Treatment-effect estimates adjusted for small-study effects via a limit meta-analysis. Biostatistics 12, 122–42 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Bycroft C et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duncan SH, Hold GL, Barcenilla A, Stewart CS & Flint HJ Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int J Syst Evol Microbiol 52, 1615–20 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Pryde SE, Duncan SH, Hold GL, Stewart CS & Flint HJ The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217, 133–9 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Jakobsdottir G, Bjerregaard JH, Skovbjerg H & Nyman M Fasting serum concentration of short-chain fatty acids in subjects with microscopic colitis and celiac disease: no difference compared with controls, but between genders. Scand J Gastroenterol 48, 696–701 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Liu F et al. Fructooligosaccharide (FOS) and Galactooligosaccharide (GOS) Increase Bifidobacterium but Reduce Butyrate Producing Bacteria with Adverse Glycemic Metabolism in healthy young population. Sci Rep 7, 11789(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Louis P & Flint HJ Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19, 29–41 (2017). [DOI] [PubMed] [Google Scholar]
- 41.den Besten G et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54, 2325–40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurilshikov A, Wijmenga C, Fu J & Zhernakova A Host Genetics and Gut Microbiome: Challenges and Perspectives. Trends Immunol 38, 633–647 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Taylor AE et al. Mendelian randomization in health research: using appropriate genetic variants and avoiding biased estimates. Econ Hum Biol 13, 99–106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J et al. Meta-analysis of human genome-microbiome association studies: the MiBioGen consortium initiative. Microbiome 6, 101(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-Only References
- 45.Tigchelaar EF et al. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open 5, e006772(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li N et al. Pleiotropic effects of lipid genes on plasma glucose, HbA1c, and HOMA-IR levels. Diabetes 63, 3149–58 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Garcia-Villalba R et al. Alternative method for gas chromatography-mass spectrometry analysis of short-chain fatty acids in faecal samples. J Sep Sci 35, 1906–13 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Schirmer M et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 167, 1125–1136 e8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCarthy S et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48, 1279–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franzosa EA et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 15, 962–968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vatanen T et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 165, 842–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang HM et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42, 348–54 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loh PR et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet 47, 284–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eastwood SV et al. Algorithms for the Capture and Adjudication of Prevalent and Incident Diabetes in UK Biobank. PLoS One 11, e0162388(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.