The Sydney Heart Bank (SHB) primarily contains tissues from hearts that were either derived from patients undergoing an isotopic heart transplantation or were from non-failing donor hearts. The latter had no apparent disease but were not required or not suitable for transplantation. Several review articles have been written (dos Remedios et al. 2017, 2018; Lal et al. 2015; Li et al. 2013), but none has attempted to summarise all causes of the failing hearts in the SHB. This review gathers the relevant information about the heart tissue from 450 patients with cardiomyopathies and 120 healthy donor hearts.
Our aim is to give readers the ability to assess whether the SHB contains tissue that might be useful for proposed experiments, particularly where prospective users have preliminary data based on animal models of human heart failure. We are often contacted by researchers who have data from failing or diseased human heart tissue but have no access to healthy donors. Few realise that the SHB samples are individually snap frozen in the heart transplant theatres. The left ventricles are all sampled and snap frozen with liquid nitrogen in cryotubes within two-10 min of cross-clamp (when the coronary artery flow ceases). We then snap-freeze samples from the right ventricles, interventricular septa, and finally, the left and right atria. No samples are collected later than 40 min. Donor hearts are collected by the surgeons at sites often up to 4-h travel time from St Vincent’s Hospital, and are perfused with ice-cold cardioplegia. Any ischemic damage that occurs within this 40-min period appears to be minimal. For example, there is no evidence for degradation RNA in samples used for RNA sequencing (Kong et al. 2010; Huang et al. 2016; Lin et al. 2016), probably because intrinsic myoglobin in the cardiomyocytes is still capable of slowly releasing oxygen as the pO2 falls.
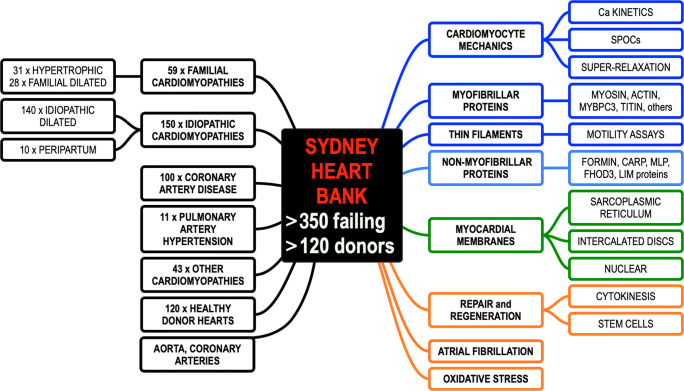
Figure 1 summarises the areas of published articles arising from tissue supplied by the SHB (see Supplementary Material). The figure is complex and requires explanation. It is intended to provide a quick visual guide to the range of patient cardiomyopathies (black boxes). A large portion of the failing hearts in the SHB were from patients diagnosed with idiopathic dilated cardiomyopathy (IDCM). We have ten hearts from an interesting subgroup of IDCM women who developed peripartum cardiomyopathy (PPCM) that was unexplained and where the patient failed to respond to medications. The hearts categorised as familial cardiomyopathies include patients with extensive hypertrophy (usually of the left ventricle) who were diagnosed with hypertrophic cardiomyopathy (HCM). We also have commenced collecting samples from HCM patients who underwent resection of the interventricular septum. Apart from HCM, there are increasing numbers (29 so far) of IDCM patients that have subsequently been shown to be caused by mutations in the sarcomere protein genes, particularly mutation in the giant TTN gene (Marston et al. 2015). This is discussed in more detail below.
Fig. 1.
This summarises the content of more than 130 publications from the Sydney Heart Bank (SHB) and its collaborators. Boxes on the left-hand side indicate the number and types of failing human hearts and non-failing healthy donor hearts that act as “controls” for the diseased hearts (black boxes). The right-hand side refers to experimental aspects of heart failure that have been published using SHB tissue. These include contractile mechanics of the cardiomyocytes and its sarcomeric proteins (dark blue boxes), the expression profile of cytoskeletal proteins (light blue boxes), membrane proteins (green boxes), and myocardial regeneration after injury (orange boxes). The entire publications arising from the SHB are provided in the supplementary information for this article. References to all of these studies can be found in the Supplementary Material
The SHB is currently undertaking whole genome DNA sequencing, RNA sequencing, and mass spectrometry to identify protein changes including post-translational modifications. All patient heart samples have associated deidentified patient clinical data. Permeabilized mechanical measurements have also been reported that involve a relatively new physiological state called Super-Relaxation (Fig. 1). This has opened a new field of experiments using failing hearts and donor controls.
In Fig. 1, the black box refers to the 120 non-failing donor hearts that were collected by the heart transplant team but were then not used or required. Often these hearts could not be used for transplantation because there was no tissue-type match for any eligible patient waiting for a heart transplant procedure. None of the SHB samples were collected post-mortem. A minority of donors were rejected by the transplant team because of concerns that the donors may have had coronary artery pathology (later not supported by pathology). These donor hearts are much sought after by research teams that have access to failing transplanted hearts but whom cannot obtain donor left ventricles that have the corresponding age and sex to their failing hearts. Publications that used these non-diseased, healthy, or normal donors as “controls” is arguably the best way to deal with sceptical reviewers who are concerned that these samples were obtained post-mortem. Of course, the donors are not “controls” in the strict sense.
The SHB also contains blood vessels (Fig. 1). The coronary arteries were the very first samples to be snap frozen and the samples of ascending aorta were harvested last.
All transplant patients were consented by consulting i.e cardiologist or a member of their team. Tissue was only collected when the SHB team had sighted the patient approval. Human Research Ethics Committee 5-year approvals were obtained from both the St. Vincent’s Hospital and The University of Sydney. All investigations conformed to the principles outlined in Declaration of Helsinki. The numbers in each box indicate how many transplant patients were involved in each category.
The orange boxes highlight a group of papers published by two groups with opposing explanations about whether adult human hearts can undergo repair and regeneration based on either cell division of cardiomyocytes (Mollova et al. 2013; Polizzotti et al. 2015) and/or repair by cells that are resident in the heart, and which can proliferate (Bergmann et al. 2015). This question remains under active investigation by both groups, but the disagreement has not yet been resolved. The orange box also refers to a group who investigated oxidative stress as a cause of human heart failure.
The right-hand side of Fig. 1 also provides details of the nature of the experiments. Many reports have used isolated and permeablized small bundles of cardiomyocytes or single cardiomyocytes for mechanical experiments that monitor stiffness and/or contraction, particularly involving Ca2+ activation in association with phosphorylation (references to these experiments are provided in the Supplementary Material). (Marston et al. 2015; Messer et al. 2016) Another group used motility assays based on the velocity of thin filaments moving on a bed of modified muscle myosin. Both groups are discussed in more detail below.
An entire special issue of Biophysical Reviews was devoted to articles that deal with heart failure caused by defective non-myofibrillar proteins (Gehmlich and Ehler 2018). The cardiomyocytes have specialised membrane structures called intercalated discs that make tight electrical and chemical connections between cardiomyocytes enabling them to contract in a concerted and synchronous way (Bennett 2018). Nuclear membranes have also been implicated in heart failure (Stroud 2018). Finally, there are internal sarcoplasmic reticulum membranes that are known to be defective in certain forms of human heart failure. All three are reported in the above special issue of this journal.
Below, we discuss an example where tissue provided by the SHB has recently played a crucial role in the development of unexpected information on the molecular and cellular basis of human heart failure.
How a homozygous mutation in cardiac troponin T provided insight into the mechanism of hypertrophic cardiomyopathy
Only a small number of human patients have been identified as carrying a homozygous mutation of a gene expressed in the heart. A recent report in 2017 investigated 2871 probands from patients with congenital heart disease but found no homozygous mutants (Jin et al. 2017). However, Klauke et al. reported a homozygous mutation in the PKP2 gene that encodes the protein plakophilin-2 (Klauke et al. 2017) which they located in the intercalated discs. They investigated both mRNA and protein levels but found neither were different in the patient compared to controls, so they concluded that the missense mutation probably plays no role in the heart failure.
A sample of left ventricle wall from a 26-year-old heart transplant recipient was sent to the van der Velden group in Amsterdam at the VU Medical Center. It was a part of a group of HCM patient samples that carried mutation(s) for several sarcomeric genes. The patient had earlier undergone cardiomyectomy to relieve an obstructive outflow tract of his left ventricle. Sequencing for myofibrillar proteins revealed a single mutation at K280N in the gene cardiac troponin T (TNNT2) but there seemed to be no haplotype. When everything was re-sequenced, it became clear that he was actually homozygous for cardiac TNNT2 (Sequeira et al. 2013). This patient presented a rare and perhaps unique opportunity to examine the effects of a thin filament mutation in the absence of any confounding response from the normal wild-type protein.
In 2013, two reports were published using the homozygous K280N patient. One used an established motility assay (where filaments of F-actin were observed moving on a surface coated with the muscle motor protein, myosin (Bayliss et al. 2013). In healthy donor samples, the Ca2+-sensitivity is threefold higher when troponin I (TnI) is unphosphorylated. In the K280N TnT sample, the level of phosphorylation of TnI was similarly low but there was no increase in Ca2+-sensitivity. Thus, the TnT mutation causes an uncoupling of thin filament Ca2+-sensitivity to the level of phosphorylation. Attempts to increase the level of TnI phosphorylation using protein kinase A also failed, but when the K280N mutant protein was exchanged out with wild-type troponin (extracted from donor hearts) in the disease assay, Ca2+-sensitivity was restored to donor levels (Bayliss et al. 2013).
About the same time, Sequeira et al. (2013) reported the functional consequences of mutation in several sarcomeric genes including: four missense mutations in MYBPC3, six in MYH7, one in TPM1, and two in TNNI3 and the TNNT2 HCM patient with the homozygous missense mutation at K280N. Single isolated cardiomyocytes were examined for sarcomere-length-dependent Ca2+-sensitivity and protein kinase A (PKA) phosphorylation of troponins under the control of phenylephrine. The effect of these mutations was reversed in permeabilized cardiomyocytes by exchanging wild-type troponins for the mutated ones, but the exchange demonstrated that as little as 14% of mutant protein was sufficient to recapitulate the effect of homozygous protein. Does the HCM phenotype result from a “poison peptide” effect from the mutated gene which perhaps was initially corrected by the expression of wild-type protein, and that over time that correction failed? Or was there was another explanation? The contractile performance of single cardiomyocytes from the K280N patient contained no wild-type protein, but length-dependent Ca2+ activation was also fully reversed by exchange with human recombinant wild-type TnT protein. Unlike Bayless et al. whose studies measured unloaded thin filament velocity, Sequeira et al. examined isometric contractions. They reported that the increase in length-dependence maximum force seen in the controls was entirely absent in any of the HCM patient with mutations in thin filament protein genes.
Two years later, Messer et al. (2016) examined mutations in TnT from seven patients with HCM, again using motility assays to study Ca2+-activation, showed that the TnT K280N mutation uncoupled the connection between TnT and TnI phosphorylation. They then tested the effect of epigallocatechin-3-gallate which reversed the Ca2+-sensitivity and restored the lost coupling (Papadaki et al. 2015).
Perhaps the last word in this story was published last month (Piroddi et al. 2019). They simultaneously measured maximal isometric ATPase activity and Ca2+-activated tension in demembranated strips of myocardium and showed that the most probable underlying cause of the HCM is the higher energy cost of tension than in control hearts. Replacement of the mutant TnT with wild-type troponin in isolated single myofibrils restores the energy cost to control level. So, TNNT2 mutation directly affects cross-bridge kinetics, causing inefficient use of ATP that leads to pathogenesis (Witjas-Paalberends et al. 2013; Witjas-Paalberends et al. 2014).
If readers are stimulated by this article and wish to examine samples of human failing and non-failing hearts, we advise they refer to (Lal et al. 2015) or contact the corresponding author by email (sydneyheartbank@gmail.com).
Electronic supplementary material
(DOCX 79 kb)
Compliance with ethical standards
Conflict of interest
Amy Li declares that she has no conflict of interest. Sean Lal declares that he has no conflict of interest. Cristobal G. dos Remedios declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bayliss CR, Jacques AM, Leung MC, Ward DG, Redwood CS, et al. Myofibrillar Ca2+−sensitivity is uncoupled from troponin I phosphorylation in hypertrophic obstructive cardiomyopathy due to abnormal troponin T. Cardiovasc Res. 2013;97:500–508. doi: 10.1093/cvr/cvs322. [DOI] [PubMed] [Google Scholar]
- Bennett PM. Riding the waves of the intercalated disc of the heart. Biophys Rev. 2018;10:955–959. doi: 10.1007/s12551-018-0438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, et al. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- dos Remedios CG, Lal SP, Li A, McNamara J, Keogh A, et al. The Sydney Heart Bank: improving translational research while eliminating or reducing the use of animal models of human heart disease. Biophys Rev. 2017;9:431–441. doi: 10.1007/s12551-017-0305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Remedios CG, Li A, Lal S (2018) Non-sarcomeric causes of Heart failure: a Sydney Heart Bank perspective. Biophys Rev 10(4):949–954. 10.1007/s12551-018-0441-4 [DOI] [PMC free article] [PubMed]
- Gehmlich L, Ehler E. Non-sarcomeric causes of heart failure. Biophys Rev. 2018;10:943–947. doi: 10.1007/s12551-018-0444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z-P, Ding Y, Chen J, Wu G, Kataoka M, Hu Y, Yang J-H, Liu J, Drakos SG, Selzman CH, Kyselovic J, Qu L-H, dos Remedios CG, Pu WT, Wang D-Z. Long non-coding RNAs link extracellular matrix gene expression to ischemic cardiomyopathy. Cardiovasc Res. 2016;112:543–554. doi: 10.1093/cvr/cvw201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49:1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauke B, Gaertner-Rommel A, Schulz U, Kassner A, Zu Knyphausen E, et al. High proportion of genetic cases in patients with advanced cardiomyopathy including a novel homozygous Plakophilin 2-gene mutation. PLoS One. 2017;12:e0189489. doi: 10.1371/journal.pone.0189489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong SW, Hu Y, Ho J, Ikeda S, Polster S, John R, Hall JL, Bisping E, Pieske B, dos Remedios CG, Pu WT. Heart failure associated changes in RNA splicing of sarcomere genes. Circ Cardiovasc Genet. 2010;3:138–146. doi: 10.1161/CIRCGENETICS.109.904698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal S, Li A, Allen D, Allen PD, Bannon P, et al. Best practice BioBanking of human heart tissue. Biophys Rev. 2015;7:399–406. doi: 10.1007/s12551-015-0182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Estigoy C, Raftery M, Cameron D, Odeberg J, et al. Heart research advances using database search engines, human protein atlas and the Sydney Heart Bank. Heart Lung Circ. 2013;22:819–826. doi: 10.1016/j.hlc.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Lin Z, Guo H, Cao Y, Zohrabian S, Zhou P, Ma Q, VanDusen N, Guo Y, Zhang J, Stevens SM, Liang F, Quan Q, van Gorp PR, Li A, dos Remedios C, He A, Bezzerides VJ, Pu WT. Acetylation of VGLL4 regulates hippo-YAP signaling and postnatal growth. Dev Cell. 2016;39:466–479. doi: 10.1016/j.devcel.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston S, Montgiraud C, Munster AB, Copeland O, Onjee C, et al. OBSCN mutations associated with dilated cardiomyopathy and haploinsufficiency. PLoS One. 2015;10:e0138568. doi: 10.1371/journal.pone.0138568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer AE, Bayliss CR, El-Mezgueldi M, Redwood CS, Ward DG, et al. Mutations in troponin T associated with hypertrophic cardiomyopathy increase Ca2+−sensitivity and suppress the modulation of Ca2+−sensitivity by troponin I phosphorylation. Arch Biochem Biophys. 2016;601:113–120. doi: 10.1016/j.abb.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollova M, Bersell K, Walsh S, Savla J, Das LT, et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci U S A. 2013;110:1446–1451. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadaki M, Vivhorev PG, Marston SB, Messer A. Uncoupling of myofilament Ca2+−sensitivity from troponin I phosphorylation by mutations can be reversed by epigallocatechin-3-gallate. Cardiovasc Res. 2015;108:99–110. doi: 10.1093/cvr/cvv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piroddi N, Witjas-Paalberends ER, Ferrara C, Ferrantini C, Vitale G, et al. The homozygous K280N troponin T mutation alters cross-bridge kinetics and energetics in human HCM. J Gen Physiol. 2019;151:18–29. doi: 10.1085/jgp.201812160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzotti BD, Ganapathy B, Walsh S, Choudhury S, Ammanamanchi N et al (2015) Stimulation of cardiomyocyte regeneration in neonatal mice and in human myocardium with neuregulin reveals a therapeutic window. Sci Translat Medic 7. 10.1126/scitranslmed.aaa5171 [DOI] [PMC free article] [PubMed]
- Sequeira V, Wijnker PJM, Nijenkamp LAM, Najafi A, Rosalie Witjas-Paalberends (2013) Perturbed length-dependent activation in human hypertrophic cardiomyopathy with sarcomere mutations in myosin and thin filament proteins. Circ Res 112:1491–1505 [DOI] [PMC free article] [PubMed]
- Stroud MJ. Linker of nucleoskeleton and cytoskeleton complex proteins in cardiomyopathy. Biophys Rev. 2018;10:1033–1051. doi: 10.1007/s1255-1-018-0431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witjas-Paalberends P, Tencate FJ, Michels M, Niessen JWM, Poggesi C, et al. Cellular dysfunction in hypertrophic cardiomyopathy. Cardiovasc Res. 2013;99:432–441. doi: 10.1093/cvr/cvt119. [DOI] [PubMed] [Google Scholar]
- Witjas-Paalberends ER, Güçlü A, Germans T, Knaapen P, Harms HJ, et al. Gene-specific increase in energetic cost of contraction in hypertrophic cardiomyopathy caused by thick filament mutations. Cardiovasc Res. 2014;103:248–257. doi: 10.1093/cvr/cvu127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 79 kb)