Abstract
Introduction
Long noncoding RNAs (lncRNAs) play an important role in the origination and progression of hepatocellular carcinoma (HCC). However, the biological function of the long intergenic non-protein-coding RNA, LINC01296, in HCC remains unknown.
Methods
Here, we observed an increase in the expression levels of LINC01296 in HCC tissues and cell lines using reverse transcription quantitative PCR; these data were consistent with that obtained from The Cancer Genome Atlas database.
Results
A higher expression level was correlated with higher alpha fetoprotein levels, a larger tumor size, an advanced TNM stage, and a poorer overall survival rate. Upregulation of LINC01296 promoted the proliferation, migration, and invasion of HCC cells. Improvement of cell migration and invasion attributable to the overexpression of LINC01296 was related to an increase in epithelial–mesenchymal transition (EMT). Mechanistically, miR-122-5P can bind to LINC01296 and decrease its oncogenic effect.
Conclusion
Collectively, the results of this study revealed that LINC01296 is a tumor promoter that can promote the migration and invasion of HCC cells through EMT, while miR-122-5P is involved in the underlying mechanisms.
Keywords: hepatocellular carcinoma, LINC01296, miR-122-5P, EMT
Introduction
Hepatocellular carcinoma (HCC) is a major health problem worldwide.1,2 It affects more than 500,000 people worldwide every year,3 with a 5-year survival rate of 30%.4 The poor prognosis is mainly attributed to a high rate of recurrence and metastasis.5
Epithelial–mesenchymal transition (EMT) plays an important role in cancer metastasis.6,7 EMT regulators such as VEGF-R are potential therapeutic targets;7,8 therefore, the identification of biomarkers related to EMT is important.
Long noncoding RNAs (lncRNAs) comprise more than 200 nucleotides that do not code for proteins and are widely expressed in various organs.5,9 They are involved in diverse biological/pathological processes such as epigenetic transcription/post-transcriptional regulation,10–12 proliferation, and metastasis of cancer cells.13,14 Recently, lncRNAs have been verified to play a critical role in EMT.15 LINC01296 is located at 14q11.2 and is upregulated to promote the cellular proliferation and invasion of cancers of stomach,16 colon,17 bile duct,18 and prostate.19 Yang et al demonstrated a higher than normal level of LINC01296 in primary HCC;20 however, the biological implications of this remain unclear.
miRNAs and lncRNAs can interact with each other21,22 and affect cellular behavior;22 recently, this has become an interesting field of research. miRNA-triggered lncRNA decay is the primary form of interaction between miRNAs and lncRNAs;23 an example of this is the reduction in MALAT1 abundance by miR-9 directly binding to two sites in the sequence.24 Meanwhile, lncRNAs can act as a sponge to reduce the regulatory effect of miRNAs.25,26 miR-122 has been extensively studied for its association with HCC27 as it triggers EMT and suppresses cellular motility and invasion.28 However, the interaction between LINC01296 and miR-122 in HCC is not yet thoroughly understood.
The aim of this study was to investigate the role of LINC01296 in the progression and metastasis of HCC and to further explore the negative regulation of the biological effect of LINC01296 by miR-122-5P.
Materials and methods
Clinical samples and cell lines
Fifty pairs of HCC tissues and pericancerous tissues (PTs) were collected from 37 male and 13 female patients, who received treatment in The First Affiliated Hospital of Chongqing Medical University (Chongqing, China). The study used human tissues and it was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board. Signed informed consent was obtained from all the patients.
Human HCC cell lines (SMMC-7721, Hep3B, HepG2, and Huh7) and a normal hepatocyte cell line (HL-7702) were obtained from the China Center for Type Culture Collection (Wuhan, China). Cells were cultured in DMEM (HyClone, Logan, UT, USA) enriched with 10% FBS (Hyclone) and incubated in 5% CO2 at 37°C.
Microarray data
Differentially expressed lncRNAs with high abundance were screened from a microarray database supplied by Yang et al.20 In the database, 323 upregulated and 525 downregu-lated RNAs were summarized as recurrently dysregulated lncRNAs in primary HCC samples from 20 HCC patients sequenced by total RNA-seq.
Bioinformatics database
Differentially expressed lncRNAs were compared between HCC and normal liver tissues using data from Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/). Furthermore, RNA-seq analysis was used to identify the expression of LINC01296 in HCC cohorts from The Cancer Genome Atlas (TCGA, https://tcga-data.nci.Nih. gov/tcga/). In total, 424 samples (50 normal liver tissues and 374 HCC tissues) were available for analysis of LINC01296 expression. In addition, LINC01296 expression levels and 10-year survival time data were collected from 370 HCC patients in the TCGA database to perform a survival analysis.
Reverse transcription quantitative PCR (RT-qPCR)
mRNAs and miRNAs were extracted from cells or tissues using the Hipure Universal mRNA/miRNA kit (Magen, Guangzhou, China). RT-qPCR was performed using the all-in-one mRNA qPCR or miRNA qPCR Primer (Gene-Copoeia, Inc., Rockville, MD, USA). The protocol was as follows: 95°C for 10 minutes, 40 cycles at 95°C for 10 seconds, 60°C for 20 seconds, and 72°C for 15 seconds. The 2–⊗⊗Cq method29 was used to calculate the relative gene expression normalized by GAPDH or U6. The primers used were: 5′-CAAAGAGGGAGTAGTTGAGCAAGGTATG-3′ and 5′-CTGTGGCGACGGTGGAGGGATGT GAGTG-3′ for LINC01296; 5′-TGGCAGAGTTGCACAG AAGA-3′ and 5′-ACCACTCACACTTTCATCGCT-3′ for ZFPM2-AS1; 5′-TGAAGTCATCACGAACCG CA-3′ and 5′-GGAGCCCCAAGTTAATGCGA-3′ for ELF3-AS1; 5′-AATTTGCCACCACCCTGTGA-3′ and 5′-ATGCCGTTTTAGGGGGACAG-3′ for LINC00978; 5′-CCAGGGCTGCTTTTAACTCT-3′ and 5′-GGACTC CACGACGTACTCA-3′ for GAPDH; 5′-GGGGTGGAGT GTGACAATG-3′ and 5′-CAGTGCGTGTCGTGGAGT-3′ for miR-122-5P; and 5′-FCTCGCTTCGGCAGCCACA-3′ and 5′-AACGCTTCACGAATTTGCGT-3′ for U6.
Cell transfection
Three shRNAs targeting LINC01296 were designed (Hanbio Biotechnology Co. Ltd., Shanghai, China), with sequences of 5′-GCCCTGCAGCAGCAGCGGCGTG GTCA-3′ (shRNA1), 5′-GAGCAGCGGCGTGGTCA GAGCGAGCT-3′ (shRNA2), and 5′-GCACACACACAC AAGCTGGCTCACAG-3′ (shRNA3). The shRNAs were inserted into the lentiviral vector HBLV-H-Scramble-GFP-PURO (Hanbio, Shanghai, China), that is, LINC01296 shRNA1/2/3; negative control was named control shRNA with a sequence of 5′-TTCTCCGAACGTGTCACGTAA-3′. Full-length cDNA of LINC01296 was inserted into the len-tiviral vector (ie, LINC01296), and the empty vector was used as the control. All the shRNAs were transfected into Huh7 cells, and LINC01296 and the corresponding control vector were transfected into SMMC-7721 cells using poly-brene (Hanbio, Shanghai, China). miR-122-5P mimics and negative control, and miR-122-5P inhibitor (anti-miR-122; 5′-CAAACACCAUUGUCACACUCCA-3′) and negative control (GeneCopeia, Guangzhou, China) were transfected into cells using Lipofectamine2000 (Thermo Fisher Scientific, Waltham, MA, USA). Puromycin was added into the medium to remove uninfected cells. Cells were incubated for 24 hours before the assays.
Cell viability
Cells were seeded into a 96-well plate (2,500 cells/well) and cell viability was determined using a CCK8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) at 0, 24, 48, 72, and 96 hours.
Colony formation assay
In a six-well plate, 500 cells/well were seeded. After incubation for 14 days, the cells were washed twice with PBS, fixed with methanol, and stained with crystal violet. The number of colonies containing >5 cells was counted under a microscope. This experiment was performed three times.
Cell migration and invasion
For analysis of cell migration, 4·104 cells were seeded in a transwell chamber (8-∝m pore size; EMD Millipore, Billerica, MA, USA) with 200 ∝L serum-free DMEM. A volume of 500 ∝L of the medium containing 10% FBS was added to the lower chamber. For invasion assay, following the above process, the Matrigel (diluted 1:9; Corning Incorporated, Corning, NY, USA) was spread on the upper surface of the membrane. After incubating for 24 hours (cell migration) or 36 hours (cell invasion), the cells on the lower surface of the membrane were fixed in 4% paraformaldehyde for 30 minutes and stained with 0.5% crystal violet. Cells in five predetermined fields were counted under a microscope (Ci-L; Nikon Corporation, Tokyo, Japan; magnification ·200). This experiment was performed five times.
Luciferase reporter assay
LINC01296 containing the predicted miR-122-5P binding site was cloned into pGL3-LINC01296-Wt (wild-type) and pGL3-LINC01296-Mut (mutant reporter) vectors (Pro-mega Corporation, Fitchburg, WI, USA). HEK-293T cells were co-transfected with miR-122-5P mimics or negative control, and pGL3-LINC01296-WT or pGL3-LINC01296-Mut using Lipofectamine2000 (Thermo Fisher Scientific). The luciferase activity was measured after 36 hours using the Dual-Luciferase Reporter Assay System (Promega Corporation).
Western blot
Proteins were extracted with the RIPA buffer (Beyotime Institute of Biotechnology, Haimen, China). About 20 ∝g of proteins was separated by 10% SDS-PAGE and transferred onto a nitrocellulose membrane (EMD Millipore). The membrane was blocked with 8% skimmed milk diluted with Tris-buffered saline with Tween-20 (Tris-HCl 24.23 g/L, NaCl 80.06 g/L [pH 7.5], and 0.1% Tween [1 mL]) at room temperature for 1 hour and then incubated with the primary antibody (1:1,000) overnight at 4°C. Horseradish peroxidase-conjugated secondary antibody (1:2,000) was added and incubated for 1 hour. The targeted proteins were visualized with the ECL kit (EMD Millipore). The primary antibodies were rabbit polyclonal antibodies: anti-E-cadherin, anti-N-cadherin (Wanlei Life Science, Shenyang, China, 1:2,000), anti-vimentin, and anti-GAPDH (Bioss, Beijing, China, 1:4,000). The secondary antibody was a goat anti-rabbit IgG antibody. The relative band density was determined using ImageJ software (NIH, Bethesda, ML, USA).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA). The results were expressed as mean ± SD. Student’s t-test was used to compare two groups. Differences among groups were analyzed using analysis of variance followed by the least significant difference post hoc analysis. The associations between LINC01296 expression and clinical pathological factors were analyzed using chi-squared test. A value of P<0.05 was set as the critical value.
Results
LINC01296 was upregulated in HCC tissues
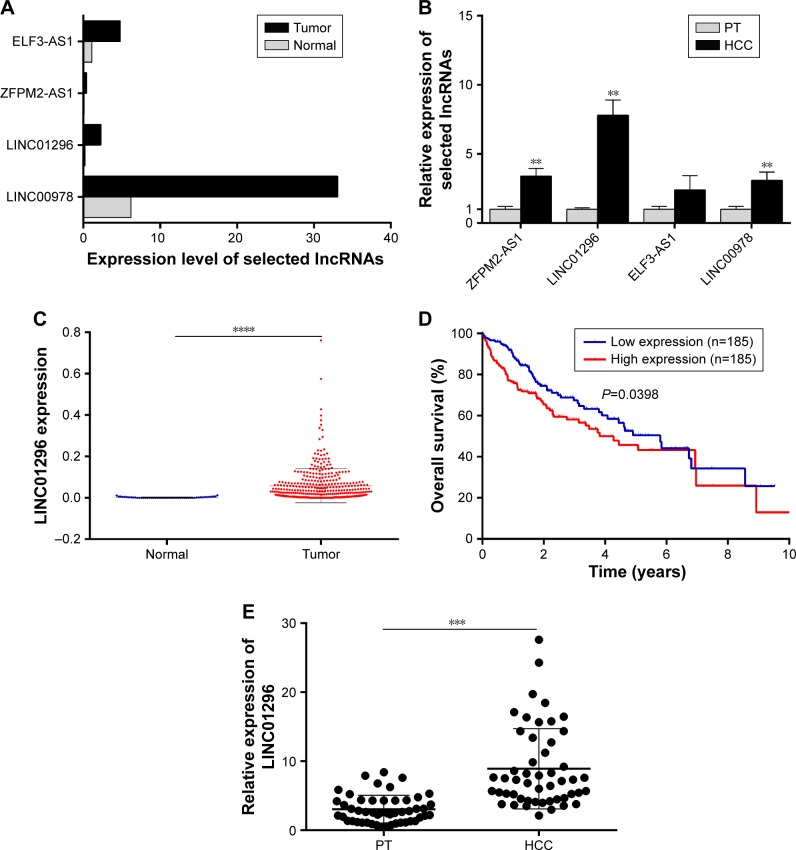
Microarray data indicated that there were four upregulated lncRNAs with higher abundance (average fragments per kilobase of transcript per million fragments mapped [FPKM] [normal] >0.5, average FPKM [tumor] >5) and differential expression (log2 fold change [tumor/normal] >2) in primary HCC tissues (Table 1). GEPIA was performed to compare the expression of these four lncRNAs between HCC and paired normal liver tissues, and the data demonstrated that they were more highly expressed in HCC (Figure 1A). To further verify the difference, RT-qPCR was performed by using tissues from five patients, and the results indicated that three lncRNAs (ZFPM2-AS1, LINC01296, and LINC00978) were upregulated in HCC tissues compared with the PT (Figure 1B).
Table 1.
Four selected lncRNAs from microarray database
| Gene ID | Gene | Average FPKM (normal) | Average FPKM (tumor) | Average FPKM Log2 fold change (tumor/normal) | P-value |
|---|---|---|---|---|---|
| ENSG00000251003 | ZFPM2-AS1 | 0.60374475 | 8.51625825 | 3.818206357 | 0.000850677 |
| ENSG00000244306 | LINC01296 | 0.7626481 | 9.6995527 | 3.66883394 | 1.91E–5 |
| ENSG00000234678 | ELF3-AS1 | 0.98327255 | 5.493271 | 2.482001516 | 1.91E–6 |
| ENSG00000172965 | LINC00978 | 6.9771975 | 35.876685 | 2.362326804 | 5.72E–6 |
Abbreviation: FPKM, fragments per kilobase of transcript per million fragments mapped.
Figure 1.
LINC01296 is upregulated in HCC. (A) Expression levels of LINC00978, LINC01296, ZFPM2-AS1, and ELF3-AS1 in HCC and paired normal liver tissues were analyzed from GEPIA database. (B) Expression levels of four selected lncRNAs were qualified with RT-qPCR in five HCC and paired PTs (**P<0.01). (C) Expression levels of LINC01296 in 374 HCC tissues and 50 normal liver tissues were analyzed from TCGA database (****P<0.0001). (D) Overall survival was analyzed in 370 HCC tissues from TCGA database (P=0.0398). (E) Expression levels of LINC01296 were detected by performing RT-qPCR in 50 HCC and paired PTs (***P<0.001).
Abbreviations: HCC, hepatocellular carcinoma; GEPIA, Gene Expression Profiling Interactive Analysis; RT-qPCR, reverse transcription quantitative PCR; TCGA, The Cancer Genome Atlas; PT, pericancerous tissue.
LINC01296 presented the highest difference and was therefore selected as the focus of the research. In the TCGA data set, a higher expression level of LINC01296 was confirmed in HCC tissues (n=374) compared with that in normal tissues (n=50) (Figure 1C), and a survival analysis was performed using the median expression level of LINC01296 as the cutoff value (0.030756) in 370 HCC patients. Patients with higher LINC01296 levels in tumor tissues had a lower overall survival rate (Figure 1D). These data manifested that LINC01296 was highly expressed in HCC and that a higher expression level predicted a poor prognosis.
Furthermore, HCC tissues and paired PT were collected from 50 patients, and the relationship between LINC01296 expression and clinicopathological variables was analyzed. Higher expression levels of LINC01296 were observed in HCC tissues compared with the PT (Figure 1E), and the expression level of LINC01296 was positively correlated with TNM stage, tumor size, and the alpha fetoprotein (AFP) level (Table 2). These suggest that the expression level of LINC01296 may be a biomarker for HCC.
Table 2.
Correlation of LINC01296 expression with clinico-pathologic variables
| Clinicopathologic parameter | No of patients | LINC01296 expression | Chi-squared P-value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | 0.5287 | |||
| ≤60 | 15 | 5 | 10 | |
| >60 | 35 | 15 | 20 | |
| Gender | 0.4814 | |||
| Male | 37 | 13 | 24 | |
| Female | 13 | 6 | 7 | |
| HBsAg | 0.3295 | |||
| Yes | 38 | 13 | 25 | |
| No | 12 | 6 | 6 | |
| AFP (ng/mL) | 0.0105* | |||
| ≤400 | 17 | 11 | 6 | |
| >400 | 33 | 9 | 24 | |
| Tumor size (cm) | 0.0375* | |||
| ≤5 | 30 | 18 | 12 | |
| >5 | 20 | 6 | 14 | |
| Vascular invasion | 0.8634 | |||
| Yes | 34 | 14 | 20 | |
| No | 16 | 7 | 9 | |
| Tumor number | 0.3357 | |||
| Single | 19 | 10 | 9 | |
| Multiple | 31 | 12 | 19 | |
| TNM stage | 0.0003*** | |||
| I–II | 16 | 13 | 3 | |
| III–IV | 34 | 9 | 25 | |
Notes: The median expression level was used as the cutoff. Low expressions of LINC01296 in 50 patients was classified as values below the 50th percentile. High expressions of LINC01296 in 50 patients was classified as values above the 50th percentile.
P<0.05.
P<0.001.
Abbreviations: HBsAg, Australia antigen; AFP, alpha fetoprotein.
LINC01296 promoted cell proliferation, migration, and invasion
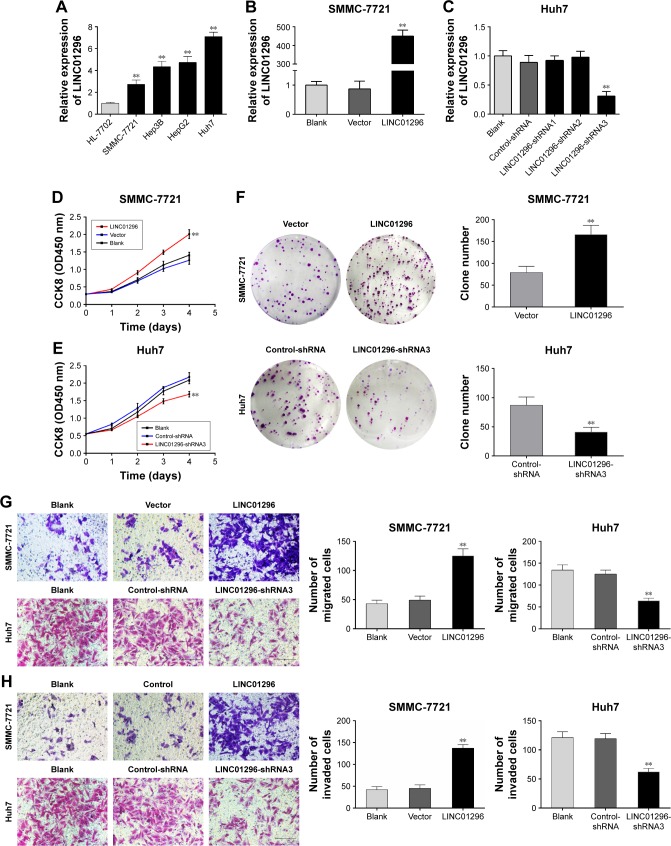
The mRNA levels of LINC01296 were increased in HCC cell lines (Hep-3B, SMMC-7721, HepG2, and Huh7) compared with that in a normal hepatocyte cell line (HL-7702), with the highest basal level observed in Huh7 cells and the lowest basal level observed in SMMC-7721 cells (Figure 2A). Therefore, to investigate its biological functions, LINC01296 was overexpressed in SMMC-7721 cells (Figure 2B) and conversely knocked down in Huh7 cells. (Figure 2C). Cell proliferation was increased in SMMC-7721 cells after LINC01296 overexpression, while it was inhibited in Huh7 cell with LINC01296 knockdown (Figure 2D–F). Moreover, LINC01296 overexpression promoted cell migration, while LINC01296 knockdown inhibited cell migration (Figure 2G); similar effects were also observed in the invasion behavior (Figure 2H).
Figure 2.
LINC01296 promotes proliferation, migration, and invasion of HCC cells. (A) Expression levels of LINC01296 in four HCC cell lines and HL-7702 cells. (B) Expression levels of LINC01296 in SMMC-7721 cells transfected with LINC01296. (C) Expression levels of LINC01296 in Huh7 cells transfected with three LINC01296 shRNAs. (D and E) CCK8 assays and (F) clone formation assays in SMMC-7721 cells with LINC01296 overexpression and Huh7 cells with LINC01296 knockdown. (G) Migration assays and (H) Invasion assays in SMMC-7721 cells with LINC01296 overexpression and Huh7 cell with LINC01296 knockdown. (**P<0.01 compared to blank/control group).
Abbreviations: HCC, hepatocellular carcinoma; CCK8, cell counting kit 8.
LINC01296 promoted EMT
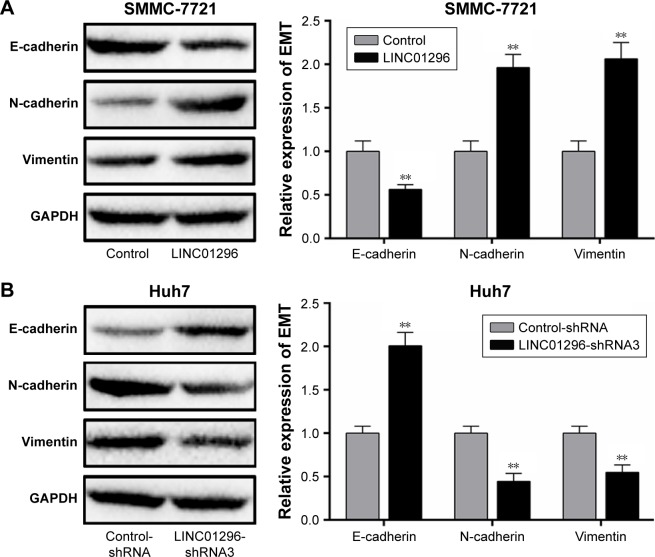
E-cadherin, N-cadherin, and vimentin are EMT markers involved in cancer metastasis. It has been reported that LINC01296 could promote metastasis by affecting EMT in colon carcinoma and pancreatic ductal adenocarcinoma.17,30 In order to verify the role of LINC01296 on HCC metastasis, the expression of the EMT markers was measured using Western blot. LINC01296 overexpression decreased the E-cadherin level and increased the levels of N-cadherin and Vimentin in SMMC-7721 cells (Figure 3A). LINC01296 knockdown resulted in an increase in E-cadherin and a decrease in N-cadherin and Vimentin in Huh7 cells (Figure 3B).
Figure 3.
LINC01296 promotes EMT. (A) Western blot analysis of EMT-related markers in SMMC-7721 cells transfected with LIN01296. (B) Western blot analysis of EMT-related markers in Huh7 cell transfected with LIN01296-shRNA3. **P<0.01 compared to control group.
Abbreviation: EMT, epithelial–mesenchymal transition.
LINC01296 interacted with miR-122-5P
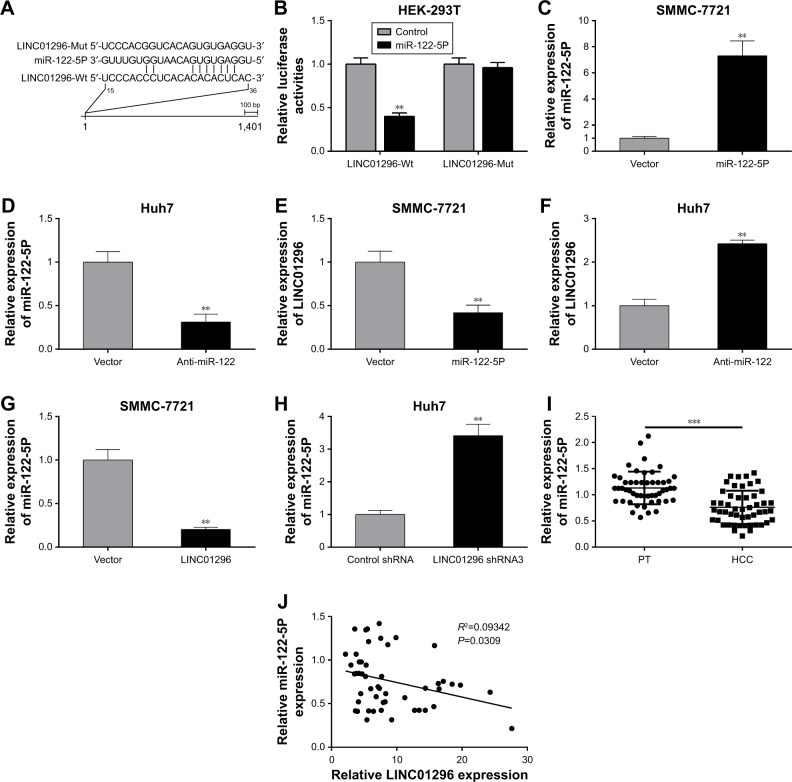
Bioinformatics analysis showed that miR-122-5P has the most binding sites closely targeting the 3′-UTR of LINC01296 (Figure 4A). The binding within LINC01296-Wt and miR-122-5P was also confirmed in HEK-293T cells by luciferase reporter assay (Figure 4B). In order to confirm that miR-122-5P could induce the decay of LINC01296, miR-122-5P was overexpressed in SMMC-7721 cells (Figure 4C) and was knocked down in Huh7 cells (Figure 4D). The expression levels of LINC01296 were reduced in SMMC-7721 cells after miR-122-5P overexpression (Figure 4E) and were elevated in Huh7 cells after miR-122-5P knockdown (Figure 4F).
Figure 4.
LINC01296 interacts with miR-122-5P. (A) miR-122-5P and its predicted binding sites in the LINC01296 sequence. (B) Luciferase activities in HEK-293T cells co-transfected with miR-122-5P mimics/control mimics and LINC01296-Wt/LINC01296-Mut. (C and D) Expression levels of miR-122-5P in SMMC-7721 cells transfected with miR-122-5P mimics and Huh7 cells transfected with a miR-122-5P inhibitor. (E and F) Expression levels of LINC01296 in SMMC-7721 cells transfected with miR-122-5P mimics and Huh7 cells transfected with a miR-122-5P inhibitor. (G and H) Expression levels of miR-122-5P in SMMC-7721 cells transfected with LINC01296 and Huh7 cells transfected with LINC01296-shRNA3. (I) Expression levels of miR-122-5P in 50 HCC and paired PTs. (J) Correlation between LINC01296 and miR-122-5P in 50 HCC tissues. **P<0.01, ***P<0.001 compared to control group.
Abbreviations: HCC, hepatocellular carcinoma; PT, pericancerous tissue.
Meanwhile, LINC01296 as a “sponge” of miR-122-5P could suppress the expression of miR-122-5P. Indeed, LINC01296 overexpression decreased the expression levels of miR-122-5P in SMMC-7721 cells (Figure 4G), while its knockdown increased the levels of miR-122-5P in Huh7 cells (Figure 4H). In 50 patients with HCC, miR-122-5P was found to be downregulated relative to that in the PT (Figure 4I), and a negative correlation between LINC01296 and miR-122-5P was noted in the 50 HCC tissues (Figure 4J). These demonstrated that LINC01296 and miR-122-5P could suppress each other’s expression by mutual binding.
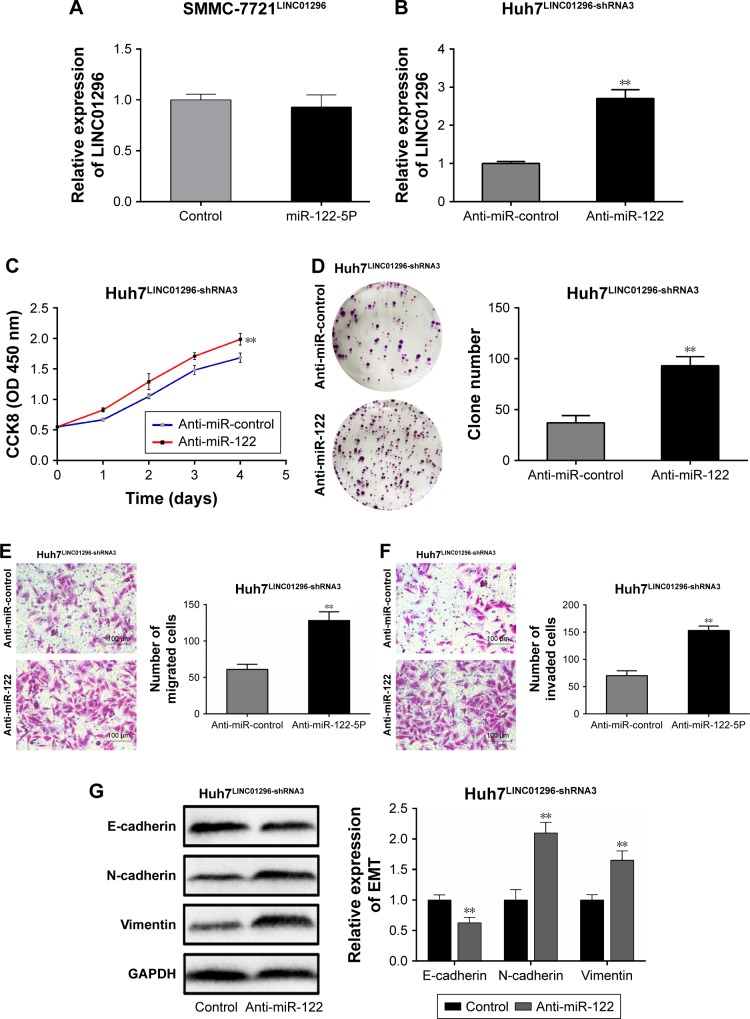
miR-122-5P reversed the function of LINC01296
In order to further investigate the role of miR-122-5P in the biological response of HCC induced by LINC01296, SMMC-7721 cells were transfected with LINC01296 with/ without miR-122-5P mimics (Figure 5A), and Huh7 cells were transfected with LINC01296-shRNA3 with/without a miR-122-5P inhibitor (Figure 5B). There were no differences in the LINC01296 expression levels between miR-122-5P and control mimics in SMMC-7721 cells transfected with LINC01296, which may be ascribed to the overexpression of LINC01296 induced by lentivirus. Therefore, Huh7 cells transfected with LINC01296-shRNA3 with/without a miR-122-5P inhibitor were used for rescue experiments. The cellular proliferation, migration, and invasion induced by LINC01296 knockdown were reversed by a miR-122-5P inhibitor (Figure 5C–F). Similar reversal effects were also observed in EMT (Figure 5G). These indicated that the effect of LINC01296 on HCC could be attributed to the inhibition of miR-122-5P.
Figure 5.
miR-122-5P reverses the biological function of LINC01296 in HCC cells. (A) Expression levels of LINC01296 in SMMC-7721 cells transfected with LINC01296 and/or miR-122-5P. (B) Expression levels of LINC01296 in Huh7 cells transfected with LINC01296-shRNA3 and/or miR-122-5p inhibitor. (C) CCK8 assays, (D) Clone formation assays, (E) Migration assays, and (F) Invasion assays in Huh7 cells transfected with LINC01296-shRNA3 and/or miR-122-5p inhibitor. (G) Western blot analysis of EMT-related markers in Huh7 cells transfected with LINC01296-shRNA3 and/or miR-122-5p inhibitor. **P<0.01 compared to control group.
Abbreviations: HCC, hepatocellular carcinoma; EMT, epithelial–mesenchymal transition; CCK8, cell counting kit 8.
Discussion
lncRNAs are a large subgroup of noncoding transcripts that have emerged as crucial regulators and prognostic markers in multiple cancers including HCC.31 Currently, lncRNAs are considered as the key in the development of novel cancer treatments because targeting oncogenic lncRNAs is a way to more directly induce an anticancer effect,32 such as EGFR and VEGFR as potential target for biological therapies in HCC cells.8 In the present study, a novel lncRNA, LINC01296, was identified using microarray data to be a recurrently dysregulated lncRNA in human primary HCC. Its expression was increased in HCC cell lines and tissues, which is consistent with the results obtained through the GEPIA and TCGA databases. In addition, LINC01296 expression was increased in patients with larger tumor sizes (>5 cm), higher AFP levels (>400 ng/mL), higher TNM grades, and a poorer 10-year survival. These suggested that LINC01296 is a potential diagnostic and prognostic biomarker. Furthermore, LINC01296 was involved in oncogenesis and promoted cellular proliferation, migration, and invasion in HCC cells. In previous studies, LINC01296 was shown to be an oncogene in some cancers, including gastric cancer, colon carcinoma, cholangiocarcinoma, and prostate cancer,16–19 indicating that LINC01296 is an extensively carcinogenic gene.
HCC is a serious malignant tumor characterized by metastasis.33 Currently, evidence that shows a crucial role of EMT in tumor metastasis is accumulating.34 Due to the lncRNA-miRNA-mRNA ceRNA network, lncRNAs open new opportunities in understanding the role of EMT in metastasis; moreover, lncRNAs are more known as ceRNAs in the regulation of EMT.35 Further efforts toward elucidating the underlying function of lncRNAs are needed. In previous studies, LINC01296 was confirmed as the ceRNA of miR-21a in colon carcinoma17 and miR-5095 in cholangiocarcinoma.18 LINC01296 silencing inhibited the proliferation and invasion of cells due to EMT in colon carcinoma.17 In the present study, LINC01296 promoted EMT and positively correlated with HCC metastasis. In addition, miR-122-5P was verified to be a binding target of LINC01296 and it could reverse the proliferation, migration, invasion, and EMT of HCC cells induced by LINC01296. Though it is known that miR-122-5P negatively regulates the EMT induced by LINC01296, the pathway downstream of LINC01296 that lead to EMT is still not clear.
lncRNAs can act as a sponge for miRNAs to reduce miRNA regulatory effects;25,26 the concentration of miRNAs can be titrated by the abundant lncRNAs that harbor similar microRNA target sequences and sequester miRNAs away from their target mRNAs.23 Another interesting finding in the study is that LINC01296 could decrease the expression of miR-122-5P, demonstrating a negative correlation between LINC01296 and miR-122-5P in HCC tissues. These results suggest that LINC01296 could act as a sponge for miR-122-5P to regulate its biological effects. miR-122 is the most abundant miRNA in the adult liver, and it is a central player in liver biology and disease.36 A previous study showed that miR-122 is a tumor inhibitor that is downregulated in HCC,37 which is consistent with our results. Furthermore, miR-122 has been shown to be an essential host factor for hepatitis C virus infection and is an antiviral target that is complementary to the standard of care using direct-acting antivirals or interferon-based treatments.36 Based on the above discussion, we can speculate that LINC01296, as a direct binding molecule of miR-122, could play a key role in the pathophysiology and treatment of HCC. Further study is needed to confirm the regulatory effect of LINC01296 on HCC. In addition, some signaling pathways of miR-122 in EMT have been demonstrated. For example, miR-122 can suppress EMT by targeting Snail1, Snail2,38 and RhoA,28 as well as suppress the WNT/®-cadherin signaling pathway.38 A systematic study concerning the mechanism of EMT mediated by the interaction of LINC01296 and miR-122-5P is essential.
Conclusion
In summary, here we report for the first time that LINC01296 could promote progression and metastasis of HCC cells by promoting EMT, while miR-122-5P could interact with LINC01296 and reverse its biological effects. Further research on LINC01296 is expected to provide new diagnostic and therapeutic targets for the treatment of HCC in the clinic.
Acknowledgments
We express our sincere thanks to Professor Tinghe Yu, at the Key Medical Laboratory of Obstetrics and Gynecology, The Second Affiliated Hospital, Chongqing Medical University, for reviewing the manuscript. This work was supported by grants from the Chongqing Science and Technology Commission of China (numbers cstc2015shmszx0496 and cstc2015jcsf10001-03).
Footnotes
Author contributions
Yafeng Wan is the senior author and writer of this manuscript. He participated in every step of this study. Molin Li participated in clinical studies and statistical analysis. Ping Huang gave the overall idea of the study and controlled the quality of all the work throughout the entire process. All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 3.Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22(33):5093–5107. doi: 10.1038/sj.onc.1206557. [DOI] [PubMed] [Google Scholar]
- 4.Si T, Chen Y, Ma D, et al. Transarterial chemoembolization prior to liver transplantation for patients with hepatocellular carcinoma: A meta-analysis. J Gastroenterol Hepatol. 2017;32(7):1286–1294. doi: 10.1111/jgh.13727. [DOI] [PubMed] [Google Scholar]
- 5.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27(20):2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65(4):798–808. doi: 10.1016/j.jhep.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Giannelli G, Sgarra C, Porcelli L, Azzariti A, Antonaci S, Paradiso A. EGFR and VEGFR as potential target for biological therapies in HCC cells. Cancer Lett. 2008;262(2):257–264. doi: 10.1016/j.canlet.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durzynska J, Lesniewicz K, Poreba E. Human papillomaviruses in epigenetic regulations. Mutat Res Rev Mutat Res. 2017;772:36–50. doi: 10.1016/j.mrrev.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Shi L, Peng F, Tao Y, Fan X, Li N. Roles of long noncoding RNAs in hepatocellular carcinoma. Virus Res. 2016;223:131–139. doi: 10.1016/j.virusres.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Van Roosbroeck K, Pollet J, Calin GA. miRNAs and long noncoding RNAs as biomarkers in human diseases. Expert Rev Mol Diagn. 2013;13(2):183–204. doi: 10.1586/erm.12.134. [DOI] [PubMed] [Google Scholar]
- 13.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 15.Liao JY, Wu J, Wang YJ, et al. Deep sequencing reveals a global reprogramming of lncRNA transcriptome during EMT. Biochim Biophys Acta. 2017;1864(10):1703–1713. doi: 10.1016/j.bbamcr.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Qin QH, Yin ZQ, Li Y, Wang BG, Zhang MF. Long intergenic noncoding RNA 01296 aggravates gastric cancer cells progress through miR-122/MMP-9. Biomed Pharmacother. 2018;97:450–457. doi: 10.1016/j.biopha.2017.10.066. [DOI] [PubMed] [Google Scholar]
- 17.Wang K, Zhang M, Wang C, Ning X. Long noncoding RNA LINC01296 harbors miR-21a to regulate colon carcinoma proliferation and invasion. Oncol Res. 2018 Apr;:19. doi: 10.3727/096504018X15234931503876. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D, Li H, Xie J, et al. Long noncoding RNA LINC01296 promotes tumor growth and progression by sponging miR-5095 in human cholangiocarcinoma. Int J Oncol. 2018;52:1777–1786. doi: 10.3892/ijo.2018.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Cheng G, Zhang C, et al. Long noncoding RNA LINC01296 is associated with poor prognosis in prostate cancer and promotes cancer-cell proliferation and metastasis. Onco Targets Ther. 2017;10:1843–1852. doi: 10.2147/OTT.S129928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Chen L, Gu J, et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun. 2017;8:14421. doi: 10.1038/ncomms14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CW, Lin PY, Yang PC. Noncoding RNAs in tumor epithelial-to-mesenchymal transition. Stem Cells Int. 2016;2016:2732705. doi: 10.1155/2016/1243659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulitsky I. Interactions between short and long noncoding RNAs. FEBS Lett. 2018;592:2874–2883. doi: 10.1002/feb2.2018.592.issue-17. [DOI] [PubMed] [Google Scholar]
- 23.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leucci E, Patella F, Waage J, et al. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep. 2013;3:2535. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin N, Tong GF, Sun LW, Xu XL. Long noncoding RNA MEG3 suppresses glioma cell proliferation, migration, and invasion by acting as a competing endogenous RNA of miR-19a. Oncol Res. 2017;25(9):1471–1478. doi: 10.3727/096504017X14886689179993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 27.Jopling C. Liver-specific microRNA-122: biogenesis and function. RNA Biol. 2012;9(2):137–142. doi: 10.4161/rna.18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang SC, Lin XL, Li J, et al. MicroRNA-122 triggers mesenchymal-epithelial transition and suppresses hepatocellular carcinoma cell motility and invasion by targeting RhoA. PLoS One. 2014;9(7):e101330. doi: 10.1371/journal.pone.0101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Yuan Q, Zhang Y, Feng L, Jiang Y. Upregulated long noncoding RNA LINC01296 indicates a dismal prognosis for pancreatic ductal adeno-carcinoma and promotes cell metastatic properties by affecting EMT. J Cell Biochem. 2019;120(1):552–561. doi: 10.1002/jcb.27411. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Lu C, Xiao M, Jiang F, Qu L, Ni R. Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. Biomed Pharmacother. 2017;89:857–863. doi: 10.1016/j.biopha.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45(8):1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 33.Huang C, Zhuang W, Feng H, et al. Analysis of therapeutic effectiveness and prognostic factor on argon-helium cryoablation combined with transcatheter arterial chemoembolization for the treatment of advanced hepatocellular carcinoma. J Cancer Res Ther. 2016;12(Supplement):C148–C152. doi: 10.4103/0973-1482.200605. [DOI] [PubMed] [Google Scholar]
- 34.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39(3):305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Lu L, Feng B, et al. Non-coding RNAs as emerging regulators of epithelial to mesenchymal transition in non-small cell lung cancer. Oncotarget. 2017;8(22):36787–36799. doi: 10.18632/oncotarget.16375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122–a key factor and therapeutic target in liver disease. J Hepatol. 2015;62(2):448–457. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Wang N, Wang Q, Shen D, Sun X, Cao X, Wu D. Downregulation of microRNA-122 promotes proliferation, migration, and invasion of human hepatocellular carcinoma cells by activating epithelial-mesenchymal transition. Onco Targets Ther. 2016;9:2035–2047. doi: 10.2147/OTT.S92378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin Y, Wang J, Han J, Luo D, Sun Z. MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing WNT/beta-cadherin signaling pathway. Exp Cell Res. 2017;360(2):210–217. doi: 10.1016/j.yexcr.2017.09.010. [DOI] [PubMed] [Google Scholar]