SUMMARY
Acinetobacter baumannii is an important nosocomial pathogen capable of causing wound infections, pneumonia, and bacteremia. During infection, A. baumannii must acquire Zn to survive and colonize the host. Vertebrates have evolved mechanisms to sequester Zn from invading pathogens by a process termed nutritional immunity. One of the most upregulated genes during Zn starvation encodes a putative cell wall-modifying enzyme which we named ZrlA. We found that inactivation of zrlA diminished growth of A. baumannii during Zn starvation. Additionally, this mutant strain displays increased cell envelope permeability, decreased membrane barrier function, and aberrant peptidoglycan muropeptide abundances. This altered envelope increases antibiotic efficacy both in vitro and in an animal model of A. baumannii pneumonia. These results establish ZrlA as a crucial link between nutrient metal uptake and cell envelope homeostasis during A. baumannii pathogenesis, which could be targeted for therapeutic development.
In Brief
Acinetobacter baumannii must acquire zinc during infection. During zinc starvation, A. baumannii expresses a peptidase named ZrlA. Lonergan et al. discovered ZrlA is required for bacterial cell envelope integrity and overcoming zinc limitation. Inactivation of zrlA increases bacterial membrane permeability, which improves antibiotic efficacy in vitro and during infection.
Graphical Abstract:
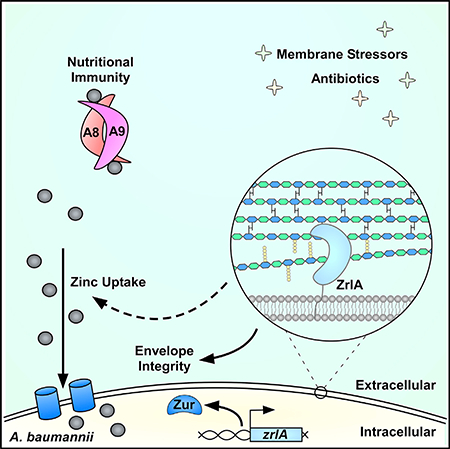
INTRODUCTION
In Gram-negative bacteria, the cell envelope comprises two membranes and a peptidoglycan (PG) layer that together coordinate to allow growth in diverse niches. The cell envelope is necessary for the maintenance and storage of essential molecules and provides a protective barrier against harsh environments. The genus Acinetobacter represents a diverse group of Gram-negative bacteria that inhabit several environmental niches (Baumann, 1968). Members of the genus are important opportunistic pathogens. Specifically, A. baumannii is a leading cause of ventilator-associated pneumonia and can cause wound and burn infections, urinary tract infections, and sepsis (Gaynes et al., 2005; Trouillet et al., 1998). The prevalence of multidrug-resistant strains prompted the World Health Organization to list A. baumannii as its most critical pathogen for the development of new therapeutics (WHO, 2017). Despite the global burden of A. baumannii infections, mechanistic studies of A. baumannii virulence and basic physiology are limited (Antunes et al., 2014; Harding et al., 2018).
Like other pathogens, A. baumannii must acquire nutrient metals from the host to replicate (Hood et al., 2012; Juttukonda et al., 2016). Metals are required for life and serve as protein structural components and enzymatic cofactors. For bacteria, these metals are essential for cell envelope maintenance because key enzymatic steps are metal dependent (Gattis et al., 2010; Rayman and MacLeod, 1975; Whittington et al., 2003). Vertebrates sequester metals from invading pathogens through a process termed nutritional immunity (Palmer and Skaar, 2016; Weinberg, 1975). One facet of nutritional immunity involves zinc (Zn) sequestration. Vertebrates withhold Zn from pathogens through the deployment of calprotectin (CP), also known as calgranulin A/B or myeloid-related protein 8/14. CP is the heterodimer of S100A8 and S100A9 (Hunter and Chazin, 1998). Two transition metal binding sites are formed at the dimer interface of CP that bind Zn and other nutrient metals (Baker et al., 2017; Corbin et al., 2008; Damo et al., 2013; Kehl-Fie et al., 2011; Nakashige et al., 2017). CP inhibits bacterial growth in vitro, and this inhibition is dependent on the metal-binding properties of the protein (Corbin et al., 2008; Hood et al., 2012; Kehl-Fie et al., 2011; Zackular et al., 2016). Furthermore, CP accumulates at infectious foci, underscoring the importance of CP and Zn withholding at the host-pathogen interface (Corbin et al., 2008; Hood et al., 2012; Juttukonda et al., 2017; Zackular et al., 2016).
Despite the evolution of host metal-sequestering strategies, A. baumannii and other bacteria have developed mechanisms to overcome Zn limitation (Ammendola et al., 2007; Desrosiers et al., 2010; Gaballa and Helmann, 1998; Hood et al., 2012; Liu et al., 2012; Patzer and Hantke, 1998; Stork et al., 2010). The response to Zn starvation in A. baumannii is primarily controlled by the Zn uptake-repressor Zur (Hood et al., 2012; Mortensen et al., 2014). This response includes accessing a labile histidine-Zn pool within the cell and elaboration of high-affinity Zn acquisition systems (Hood et al., 2012; Mortensen et al., 2014; Nairn et al., 2016). However, the effects of Zn starvation on other aspects of A. baumannii physiology are unknown.
We previously identified genes differentially expressed in a strain lacking zur through the application of a transcriptomics-based approach (Mortensen et al., 2014). From this, we discovered a putative PG-modifying enzyme; based on sequence prediction and experimental evidence, we herein name the gene zrlA (Zur-regulated lipoprotein A). The gene encoding ZrlA is regulated by Zur and is significantly upregulated in A. baumannii following exposure to CP (Mortensen et al., 2014). We hypothesized that ZrlA serves as an integral link between cell envelope and nutrient Zn homeostasis. ZrlA localizes to the inner membrane as a Zn-binding peptidase and is critical for the response of A. baumannii to Zn starvation. ZrlA also has a pivotal role in maintaining robust cell envelope barrier function, and a strain lacking zrlA (ΔzrlA) is sensitive to envelope stresses. This barrier defect sensitizes the ΔzrlA strain to many classes of antibiotics in vitro and in vivo, establishing ZrlA as a viable target for small-molecule drug development to combat A. baumannii infections.
RESULTS
ZrlA Is a Zn-Binding Peptidase Critical for A. baumannii Cellular Envelope Maintenance and Morphology
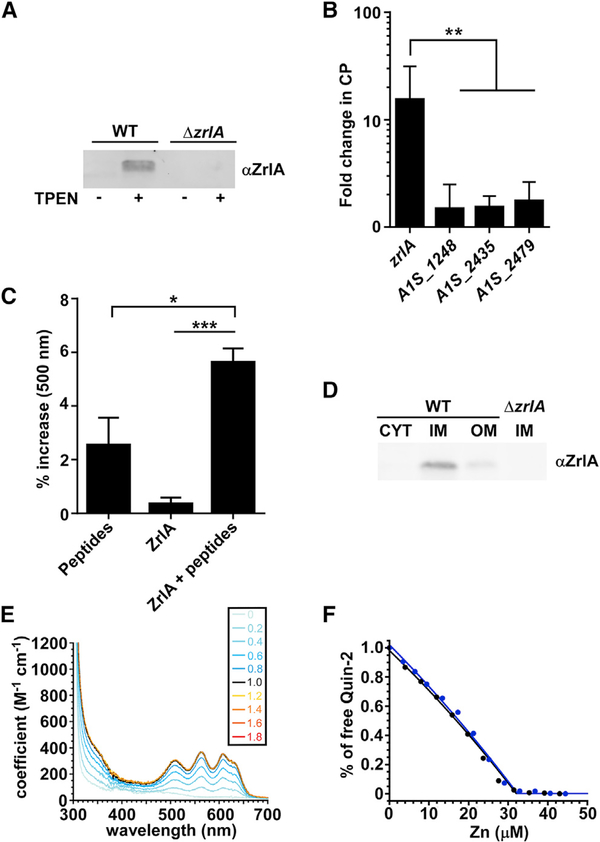
To assess whether ZrlA contributes to the response of A. baumannii to Zn starvation, we monitored the transcriptional induction of zrlA during Zn limitation and generated a polyclonal antibody against ZrlA to monitor protein abundance in those conditions. zrlA and its gene product are both detectable in Zn-replete conditions but are induced during Zn starvation by the small-molecule Zn chelator TPEN (N,N,N’,N’-tetrakis(2-pyr-idinylmethyl)-1,2-ethanediamine) and the Zn-chelating host protein CP (Figures 1A, 1B, and S1A-S1C). Sequence predictions suggest that ZrlA has d-alanine-d-alanine carboxypeptidase (d,d-CPase) activity and is a member of the M15 metallopeptidase family (Rawlings and Barrett, 1993). Peptidase activity was confirmed using a modified cadmium ninhydrin assay (Wu et al., 1995) to detect release of free amino acids from a peptide substrate (Figure 1C). Additionally, subcellular fractionation experiments revealed that ZrlA localizes to the inner membrane (Figures 1D and S1D-S1F). Finally, recombinant ZrlA coordinates Zn, and the binding of Zn is preferential over other divalent cations, which was validated by quantitative metal-binding experiments with ZnII, and CoII (Figures 1E, 1F, S1G, and S1H). Soluble ZrlA lacking the N-terminal 41 residue membrane-anchoring domain (Figure S1F), denoted ZrlA42C, binds a single molar equivalent of CoII or ZnII in a tetrahedral coordination geometry (Figure 1E) and is characterized by a high affinity for Zn, KZn of 3.6 (±0.5) × 1011 M−1 (Figure 1F). These results demonstrate that ZrlA is a Zn-binding lipoprotein that is maximally induced during nutrient Zn starvation and exhibits peptidase activity.
Figure 1. ZrlA Is a Peptidase Induced during Zn Starvation.
(A) ZrlA expression was assessed in wild-type (WT) and ΔzrlA (15 μg protein/lane) by immunoblot after growth in lysogeny broth (LB) ± 40 μM TPEN.
(B) Transcriptional changes for zrlA and other predicted d,d-CPases A1S_1248, A1S_2435, and A1S_2479 were assessed by qRT-PCR + 250 μg/ml CP. **p < 0.01 as determined by one-way ANOVA with Tukey multiple comparisons test on three independent experiments, means ± SD.
(C) Modified cadmium-ninhydrin assay was performed on recombinant MBP-ZrlA in the presence of a peptide substrate. *p < 0.05, ***p < 0.001 as determined by one-way ANOVA with Tukey multiple comparisons test on three independent experiments, means ± SD.
(D) ZrlA protein localization was assessed in WT and ΔzrlA (7.5 μg protein/lane) by immunoblot of membrane fractions following growth in LB ± 40 μM TPEN.
(E) CoII titration, where a tetrahedral or distorted tetrahedral coordination geometry is suggested with a d-d transition of 400 M−1 • cm−1 at 600 nm (Corwin and Koch, 1987). Inset, number of CoII:ZrlA42C mol • equivalents corresponding to each spectrum shown; note that all spectra acquired at CoII:ZrlA42C overlap, revealing a 1:1 metal binding stoichiometry.
(F) Normalized binding titration of a mixture of ZrlA42C and competitor quin-2 with Zn in two independent experiments: 17.0 μM ZrlA42C with 15.2 μM quin-2 (blue filled circles) and 15.1 μM ZrlA42C with 12.3 μM quin-2 (black filled circles). The continuous lines represent the results of a nonlinear, least-squares fit to a Zn:ZrlA42C = 1:1 binding model. The global fitting results in KZn = 3.6 (±0.4) × 1011 M− 1.
See also Figure S1.
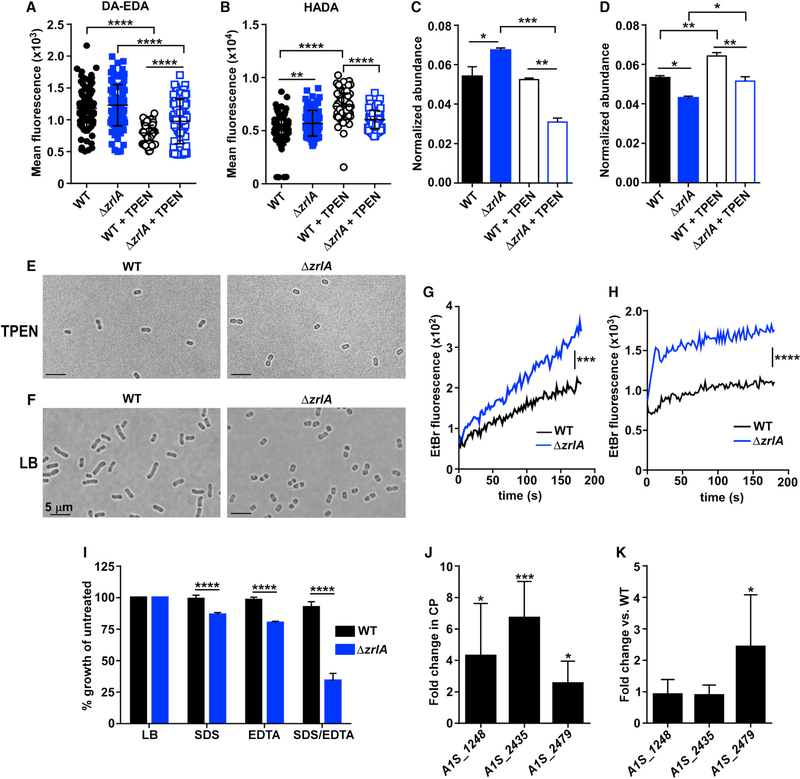
We hypothesized that the absence of ZrlA may change PG architecture. To test this, we labeled PG with fluorescent d-amino acids and their dipeptide derivatives to monitor changes in PG during Zn starvation. For these experiments, we used conjugated fluorescent molecules designed to label either the fourth or fifth position of PG stem peptides using HADA and DA-EDA, respectively, and then monitored changes in fluorescence intensity during Zn limitation (Kuru et al., 2012; Liechti et al., 2014). Wild-type (WT) A. baumannii had increased fourth-position labeling that corresponded with decreased fifth-position labeling during Zn starvation, which was diminished in the ΔzrlA strain (Figures 2A and 2B). This finding is consistent with ZrlA induction resulting in cleavage of terminal d-alanine residues and increased PG crosslinking. We next performed ultra performance liquid chromatography-mass spectrometry (UPLC-MS) on PG purified from WT or ΔzrlA A. baumannii after growth in Zn-replete or Zn-depleted conditions and quantified the normalized abundances of major monomeric and dimeric (crosslinked) muropeptide species (Figure S2A; Table S1). Tripeptide monomers were more abundant in the ΔzrlA strain in Zn-replete conditions but were significantly reduced during Zn starvation (Figure 2C). In contrast, tetrapeptide monomer species, although comparable in the WT and ΔzrlA strains in Zn-replete conditions, were increased in the ΔzrlA strain in Zn-starved conditions, relative to WT (Figure S2B). Conversely, dimeric tripeptide-tetrapeptide crosslinks were significantly reduced in the ΔzrlA strain in both Zn-replete and -depleted conditions (Figure 2D), whereas dimeric tetrapeptides were slightly elevated in both strains in Zn-depleted conditions (Figure S2C). Taken collectively, these data demonstrate that A. baumannii increases major cell-wall crosslinks during Zn starvation, and zrlA affects both homeostatic muropeptide abundance and specific PG alterations that occur during Zn limitation.
Figure 2. ZrlA Contributes to Cellular Morphology and Envelope Integrity.
(A) Mean fluorescence of WT or ΔzrlA ± 10 μΜ TPEN after DA-EDA labeling of the fifth position of PG pentapeptides. ****p < 0.0001 as determined by one-way ANOVA with Tukey multiple comparisons test (n = 100 cells/group).
(B) Mean fluorescence of WT or ΔzrlA ± 10 μM TPEN after HADA labeling of the fourth position of PG pentapeptides **p < 0.01, ****p < 0.0001 as determined by one-way ANOVA with Tukey multiple comparisons test (n = 100 cells/group).
(C) Normalized abundance of PG muropeptide tripeptide monomers (sum M3, M3(−N-acetylglucosamine [GlcNAc]; see Figure S2 for a representative liquid chromatography-tandem mass spectrometry (LC-MS/MS) chromatogram and Table S1 for muropeptide designations and expected and experimental masses) in WT or ΔzrlA ± 40 μM TPEN.
(D) Normalized abundance of muropeptide tetrapeptide monomers (sum M4,M4[–GlcNAc]) (Figure S2; Table S1) in WT or ΔzrlA ± 40 μM TPEN. *p < 0.05, **p < 0.01, ***p < 0.001 as determined by one-way ANOVA with Tukey multiple comparisons test on biological duplicates, means ± SD.
(E) Microscopy of WT or ΔzrlA A. baumannii grown to mid-log in LB + 40 μM TPEN (100×).
(F) Microscopy of WT or ΔzrlA A. baumannii grown to mid-log in LB (100×). Scale bar, 5 μm.
(G) Ethidium bromide uptake following growth in LB. ***p < 0.001 as determined by Student’s t test of mean line slopes from three independent experiments.
(H) Ethidium bromide uptake following growth in 40 μM TPEN, ****p < 0.0001 as determined by Student’s t test of mean line slopes from three independent experiments.
(I) Percentage of growth as determined by optical density 600 (OD600) at 8 h of growth in LB, 0.01% SDS, or 0.01 mM EDTA for WT and ΔzrlA compared with untreated samples. ****p < 0.05 as determined by Student’s t test from three independent experiments, means ± SD.
(J) ΔzrlA was subjected to qRT-PCR analysis of putative d,d-CPases in 250 μg/ml CP. *p < 0.05, ***p < 0.001 as determined by Student’s t test from three independent experiments, means ± SD.
(K) ΔzrlA was subjected to qRT-PCR analysis of putative d,d-CPases in LB compared with expression in the WT strain. *p < 0.05 as determined by Student’s t test from three independent experiments, means ± SD.
See also Table S1 and Figure S2.
To test whether the absence of zrlA leads to morphological changes, we performed live cell imaging on WT A. baumannii and on a ΔzrlA strain after growth in Zn-replete and -depleted conditions. WT A. baumannii and the ΔzrlA strain have a rounded morphology when grown in Zn-starved conditions (Figure 2E). When cultured in Zn-replete conditions, the WT strain displays morphological heterogeneity, with rounded cells and short and long rods detected, whereas the ΔzrlA strain remains rounded; this phenotype can be complemented by providing zrlA in trans (Figures 2F and S2D). These results suggest that ZrlA contributes to morphological plasticity in A. baumannii and the ability of this organism to adapt to changing environmental conditions.
Given the altered PG structure and shape in the ΔzrlA mutant, we hypothesized that ZrlA may contribute to the maintenance of overall cellular envelope integrity. To test that, the rate of ethidium bromide diffusion across the WT or ΔzrlA mutant envelope was assessed by monitoring fluorescence that occurs upon ethidium bromide-DNA interaction (Dalebroux et al., 2015). This assay revealed the ΔzrlA strain has increased fluorescence compared with the WT strain, indicative of increased envelope permeability, and the rate of uptake was increased in the ΔzrlA, strain during Zn starvation (Figures 2G and 2H). Furthermore, the ΔzrlA strain was more sensitive to the detergent SDS as well as to EDTA, which chelates metals associated with the outer membrane (Malinverni and Silhavy, 2009) (Figure 2I). These envelope defects are complemented by expressing zrlA in trans (Figures S2E and S2F).
To determine whether inactivation of zrlA imbalanced other cell wall-modifying enzymes, expression of predicted carboxypeptidases in A. baumannii was monitored in WT and ΔzrlA in Zn-replete and Zn-depleted conditions. Those carboxypeptidases were identified based on sequence comparisons with defined d,d-CPases in Escherichia coli, as well as through manual genome analysis (Ghosh et al., 2008). During Zn starvation, WT A. baumannii only induced expression of zrlA (Figure 1B). However, the ΔzrlA strain has aberrant d,d-CPase expression in Zn-starved conditions as well as in Zn-replete coniditions, relative to the WT strain (Figures 2J and 2K). Protein sequence alignment revealed that one of these putative d,d-CPases, A1S_1248, was more closely related to ZrlA but was not predicted to be regulated by Zur. This protein was purified as described for ZrlA and was found to possess a metal-coordination geometry (Figures S2G and S2H) and KZn of 2.2 (±0.1) × 1011 M−1 (Figures S2G and S2H) that is nearly indistinguishable from ZrlA. This finding effectively rules out a “zinc sparing” mechanism in which Zn-starved A. baumannii overexpresses a non-Zn requiring paralog (ZrlA) relative to constitutively expressed A1S_1248 in WT cells (Blaby-Haas et al., 2011; Nanamiya et al., 2004). Taken together, these data establish ZrlA as a critical factor for maintaining cellular morphology and envelope integrity in A. baumannii.
ZrlA Is Required for A. baumannii to Overcome Nutrient Zn Limitation
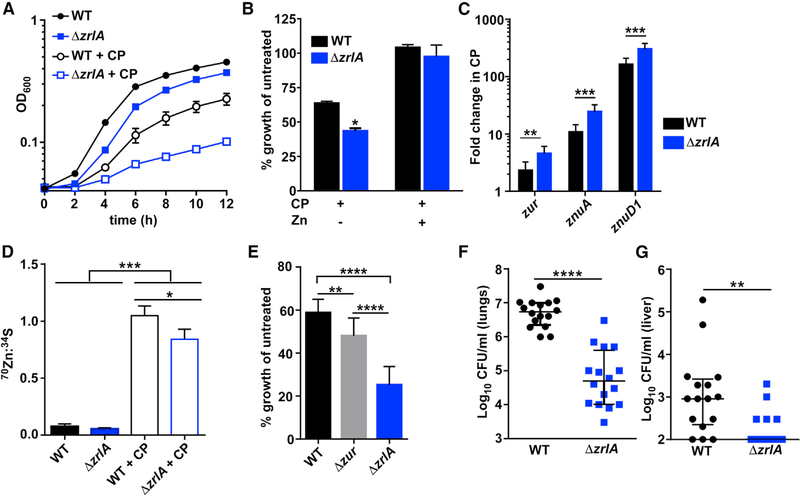
We next hypothesized that ZrlA has a role in the response of A. baumannii to Zn starvation. Upon growth of WT or ΔzrlA in the presence of CP or the Zn chelator TPEN, ΔzrlA exhibits a growth defect (Figures 3A, S3A, and S3B) that can be fully rescued by Zn supplementation (Figure 3B) or expression of zrlA in trans (Figure S3C). Additionally, the predicted metal-coordinating residues of ZrlA (Figure S1F) are required for this complementation because a mutant zrlA predicted to no longer bind Zn was unable to restore growth, despite expression at levels comparable to the WT ZrlA (Figures S3D and S3E). Furthermore, the requirement of zrlA for overcoming Zn limitation is conserved in a recently isolated multidrug-resistant strain of A. baumannii (Figure S3F). To determine whether that growth defect was due to alterations in cytosolic Zn buffering or acquisition of extracellular Zn, Zn levels were monitored via inductively coupled plasma mass spectrometry (ICP-MS). No differences were found in total cellular Zn in the ΔzrlA mutant relative to WT (Figure S3G). However, qRT-PCR analysis of Zur-regulated Zn uptake genes revealed that the ΔzrlA strain has increased expression of zur, znuA, and znuD1 relative to WT (Figure 3C), all of which are transcriptionally induced during Zn limitation (Mortensen et al., 2014). Because ZrlA affects cellular envelope integrity and homeostasis, we hypothesized that ZrlA specifically contributes to Zn uptake. Cultures were pulsed with a rare Zn isotope (70Zn) and analyzed by ICP-MS to quantify cellular 70Zn levels; that experiment revealed that WT A. baumannii is capable of competing with CP for Zn but that the ΔzrlA mutant was impaired in Zn uptake in conditions of Zn limitation (Figure 3D). To determine whether aberrant expression of Zn uptake genes was the primary contributor to the envelope defects observed in the ΔzrlA mutant, a strain lacking zur (Δzur), which overexpresses Zn-uptake genes (Mortensen et al., 2014), was used to assess outer-membrane envelope integrity after exposure to SDS/EDTA. This experiment revealed that the Δzur strain has a modest growth defect in these conditions but grew significantly better than the ΔzrlA mutant (Figure 3E). This suggests that overexpression of the Zn-uptake genes in the ΔzrlA mutant was insufficient to cause cell envelope defects and that the direct effect of ZrlA on cell-wall architecture is a major contributor to maintaining envelope integrity and overcoming Zn limitation.
Figure 3. ZrlA Is Critical for Full Growth in Low Zn Environments and During Infection.
(A) WT or ΔzrlA were grown ± 250 μg/ml CP with OD600 monitored over time.
(B) Percentage of growth as determined by OD600 at 8 h after growth in 250 μg/ml CP ± 50 μM ZnCl2 compared with untreated strains. *p < 0.05 as determined by Student’s t test from three independent experiments, means ± SD.
(C) qRT-PCR analysis was performed on WT or ΔzrlA after growth in 250 μg/ml CP. **p < 0.01, ***p < 0.001 as determined by Student’s t test from three independent experiments, means ± SD.
(D) Cellular 70Zn was quantified by ICP-MS and normalized to 34S for WT and ΔzrlA. *p < 0.05, ***p < 0.001 as determined by one-way ANOVA with Tukey multiple comparisons test from three independent experiments, means ± SD.
(E) Percentage of growth as determined by OD600 at 6 h for WT, Δzur, or ΔzrlA grown in 0.01% SDS/0.1 mM EDTA relative to untreated strains. **p < 0.01, ****p < 0.0001 as determined by one-way ANOVA with Tukey multiple comparisons test from three independent experiments, means ± SD.
(F) Mice were intranasally infected with a 1:1 mixture of WT and ΔzrlA, and bacterial burdens were assessed at 36 hpi in the lungs. ****p < 0.0001 as determined by Mann-Whitney U test.
(G) WT and ΔzrlA bacterial burdens recovered from the liver at 36 hpi. **p < 0.01 as determined by Mann-Whitney U test; error bars are median ± interquartile range (n = 16).
See also Figure S3
Given that the vertebrate host is a harsh environment with limited metal availability, we investigated the contribution of ZrlA to A. baumannii pathogenesis with a mouse model of A. baumannii pneumonia. This experiment revealed that ΔzrlA is significantly attenuated in lung colonization (Figure 3F) and is defective at disseminating to the liver (Figure 3G). Taken together, these results suggest that A. baumannii uses ZrlA to coordinate nutrient Zn uptake and cellular envelope maintenance to persist within the vertebrate host.
ZrlA Is Critical for Surviving Antibiotic Exposure
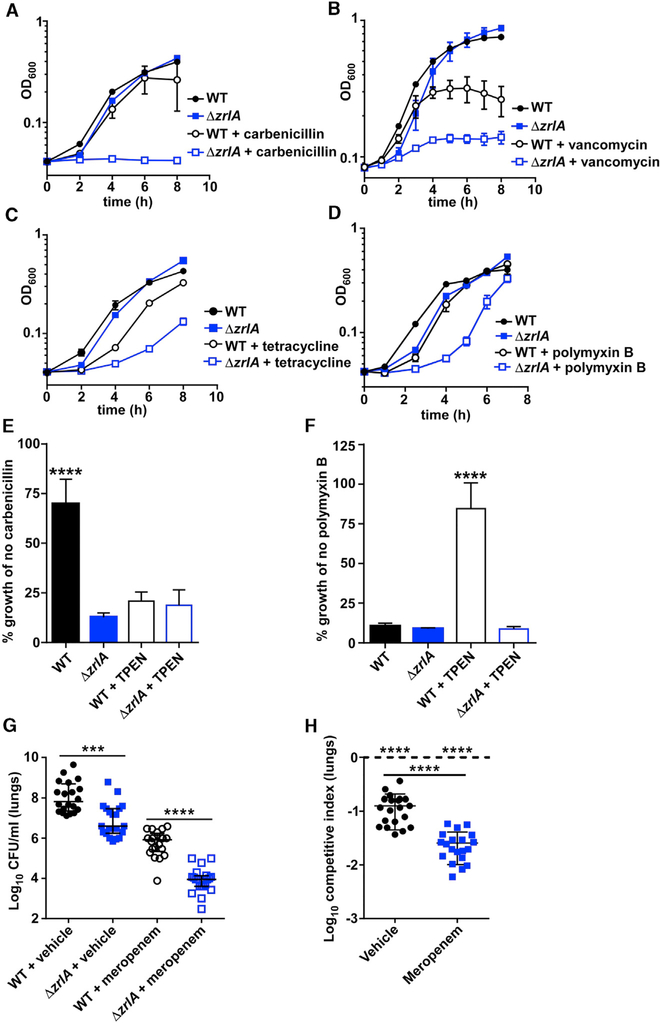
To test whether ZrlA provides a fitness advantage upon exposure to other cellular stressors, such as antibiotics, WT and Δzr/A A. baumannii were exposed to subinhibitory concentrations of several antibiotic classes. Incubation of those strains with antibiotics revealed that Δzr/A exhibits increased sensitivity to several different antimicrobial molecules, including β-lactams, tetracyclines, polymyxins, and vancomycin (Figures 4A-4D). WT and Δzr/A A. baumannii were then subjected to Zn starvation and exposed to antibiotics. We found that the β-lactam antibiotic carbenicillin synergized with Zn restriction to inhibit A. baumannii growth, and the Δzr/A strain remained more sensitive relative to the WT strain (Figures 4E and S4A). Surprisingly, Zn starvation coupled with exposure to polymyxin B decreased antibiotic efficacy and permitted more A. baumannii growth compared with polymyxin B alone (Figure 4F). The Δzur mutant was also sensitive to carbenicillin (Figure S4B), despite improved outer-membrane barrier function compared with the Δzr/A mutant (Figure 3E). These results illustrate that ZrlA contributes to antibiotic resistance in A. baumannii and that Zn availability alters antibiotic efficacy in a class-dependent manner.
Figure 4. ZrlA Is Required for Overcoming Antibiotic Exposure In Vitro and In Vivo.
(A-D) WT and Δzr/A were grown (A) ± carbenicillin, (B) ± vancomycin, (C) ± polymyxin B, and (D) ± tetracycline with OD600 monitored overtime.
(E) WT and Δzr/A were grown in the presence of carbenicillin ± TPEN with OD600 monitored over time; data depict growth at 8 h relative to growth with no carbenicillin. ****p < 0.0001 as determined by one-way ANOVA with Tukey multiple comparison’s test from three independent experiments, means ± SD.
(F) WT and Δzr/A were grown in the presence of polymyxin B ± TPEN with OD600 monitored over time; data depict growth at 24 h relative to growth with no polymyxin B. ****p < 0.0001 as determined by one-way ANOVA with Tukey multiple comparisons test from three independent experiments, means ± SD.
(G) WT or Δzr/A bacterial burdens recovered from the lungs at 36 hpi. ***p < 0.001, ****p < 0.0001 as determined by Mann-Whitney U test; error bars are median ± interquartile range (n = 20).
(H) Competitive index ([input Δzr/A/WT])/[output Δzr/A/WT]) in the lungs at 36 hpi. ****p < 0.0001 as determined by Student’s t test with an arbitrary value of 1 or between treatments; error bars are median ± interquartile range (n = 20).
See also Figure S4.
We next evaluated whether inactivation of zr/A improves antibiotic efficacy during A. baumannii pneumonia. To test that, mice were intranasally inoculated with WT and Δzr/A A. baumannii and administered either meropenem, a β-lactam antibiotic frequently used to treat A. baumannii infections, or a vehicle control at 0, 12, and 24 h postinfection (hpi). At 36 hpi, mice were humanely euthanized, and bacterial colony-forming units (CFUs) were enumerated. Meropenem significantly reduced WT A. baumannii bacterial burden in the lungs (Figure 4G). Strikingly, Δzr/A burden was further reduced with meropenem treatment (Figures 4G and 4H), and that reduction in burden was not due to the presence of the antibiotic marker in the mutant strain (Figure S4C). These results demonstrate that targeting ZrlA has the potential to increase the efficacy of existing antimicrobials against A. baumannii.
DISCUSSION
Overcoming nutrient metal limitation is a requirement for bacterial pathogens to survive and cause disease. In A. baumannii, Zn starvation promotes upregulation of the znuABCD Zn-uptake system and the predicted Zn metallochaperone zigA (Hood et al., 2012; Mortensen et al., 2014; Nairn et al., 2016). However, the capacity of A. baumannii to elaborate the Zn acquisition systems and continue cellular division requires the maintenance of the cell envelope. A. baumannii coordinates its response to Zn starvation with resistance to extracellular stressors through the action of ZrlA. ZrlA is required to maintain proper membrane barrier function and contributes to nutrient Zn acquisition. Antibiotics are more efficacious against a strain lacking zrlA both in vitro and in an animal model of A. baumannii pneumonia, suggesting that ZrlA inhibitors may improve therapeutic treatments during infection with A. baumannii.
Bacterial cell envelope maintenance is a dynamic process, involving many enzymes and pathways and the ability to modify the envelope promotes infection (Guo et al., 1997; Sycuro et al., 2010). Given the essentiality of the envelope for viability and pathogenesis, functional redundancy is common among envelope-modifying proteins, particularly among PG-modifying enzymes. Although in-depth studies investigating PG biosynthesis and maintenance in A. baumannii are lacking, functional redundancy among d,d-CPases in E. coli is well-described (Peters et al., 2016). This redundancy may allow bacteria to use different subsets of d,d-CPases to adapt to unique environments (Ghosh et al., 2008). We identified four putative d,d-CPases in the A. baumannii genome; however, a large portion of A. baumannii genes remains unannotated, and therefore, the existence of others cannot be excluded. Of these four putative d,d-CPases, zrlA is the only one that is induced during Zn starvation, and ZrlA is a Zn-binding peptidase. Some M15 family peptidases also have multiple enzymatic activities, including dipeptidase, transpeptidase, and endopeptidase activity, which suggests ZrlA may have additional enzymatic functions that contribute to muropeptide abundances detected in our analyses (Podmore and Reynolds, 2002; Stefanova et al., 2004). We predict that the other d,d-CPases in A. baumannii are induced in unique niches and that this likely contributes to the notable ability of pathogenic and nonpathogenic Acinetobacter species to persist in diverse environments.
Our observation that the ΔzrlA strain accumulates ethidium bromide at a faster rate than WT A. baumannii suggests that the ΔzrlA mutant has increased cellular permeability, although the contribution of bacterial efflux pumps to this phenotype was unexplored. We also discovered that the ΔzrlA strain is defective in Zn uptake despite overexpression of Zn-acquisition genes. We suggest that the activity of ZrlA promotes more efficient Zn uptake through PG modification. Although we have not shown a direct effect on membrane localization of the Zn-acquisition machinery, other large membrane-anchored molecular structures, such as secretion systems, require PG modification for effective elaboration; that, in turn, suggests similar cell-wall changes may be required to elaborate transition metal uptake systems (Dijkstra and Keck, 1996; Scheurwater and Burrows, 2011; Wang et al., 2015).
Zn availability modulates antibiotic efficacy, and the β-lactam antibiotic carbenicillin is more efficacious in low-Zn environments. However, polymyxin B is less effective in Zn-limited conditions, which is consistent with previous observations that Pseudomonas aeruginosa is less susceptible to polymyxin B when other cations are restricted and also suggests Zn may be an important metal associated with the A. baumannii cell envelope (Nicas and Hancock, 1983) (Brown and Melling, 1969). These observations also suggest that changes in antibiotic efficacy may in part be driven by nutrient Zn availability and the capacity of pathogenic bacteria to alter local metal levels. We previously observed that, during A. baumannii sepsis, the host actively restricts ferrous iron, which is consistent with numerous reports that the host limits iron availability during infection (Aron et al., 2017; Schade and Caroline, 1946; Weinberg, 1974; Wright et al., 1981). Zn is also restricted from pathogens; only 0.1% of total body Zn is located in the plasma, and that percentage is reduced during inflammation through sequestration by metal-binding proteins and through Zn transporters promoting tissue metal accumulation (Aydemir et al., 2012; Haase and Rink, 2014). However, that metal sequestration is not uniform, and bacteria may experience different metal levels within the host (Cassat et al., 2018). Therefore, microenvironments within the host can alter bacterial metabolic processes in a way that may change drug efficacy, so multifaceted approaches to target A. baumannii infections may be a long-term solution to combat multidrug resistance.
This work demonstrates that ZrlA is induced during metal starvation, contributes to cell envelope maintenance, and is critical for A. baumannii to survive exposure to antibiotics. Moreover, ZrlA contributes to the ability of A. baumannii to overcome Zn starvation and survive within the metal-restricted environment of a vertebrate host. Finally, we identified that antibiotics are more efficacious at treating A. baumannii infections in the absence of zrlA. These data highlight an exciting intersection between bacterial cell physiology, PG remodeling, and metal-acquisition strategies, and they underscore the therapeutic potential of targeting bacterial metal-acquisition systems to treat infections.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eric Skaar (eric.skaar@vumc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Strains
Experiments were performed using Acinetobacter baumannii strain ATCC 17978 and its derivatives unless otherwise noted. Transposon mutants in the A. baumannii ABUW5075 background were purchased from the University of Washington A. baumannii mutant library, and transposon insertion was confirmed by PCR (Gallagher et al., 2015). Cloning was performed in E. coli DH5α, and protein expression was performed in E. coli BL21 (DE3). All experiments involving liquid cultures were performed in LB at 37°C with aeration unless otherwise stated. Carbenicillin for plasmid maintenance was used at 50 μg/ml for E. coli or 75 μg/ml for A. baumannii. Kanamycin (Km) was used at 40 μg/ml. Relevant primers are listed in Table S2.
Animal Models
Eight to ten-week-old male C57BL/6J mice were purchased from Jackson Laboratories. Mice were housed on standard VUMC facility chow and bedding with a 12 hour light-dark cycle. All animal experiments were approved by the Vanderbilt University Medical Center (VUMC) Institutional Care and Use Committee and conform to policies and guidelines established by VUMC, the Animal Welfare Act, the National Institutes of Health, and the American Veterinary Medical Association.
METHOD DETAILS
Bacterial mutant generation
To generate the zrlA::km strain, approximately 1000 base pairs of DNA in both the 5ˊ and 3ˊ flanking regions surrounding the open reading frame were amplified from A. baumannii genomic DNA, and the kanamycin resistance gene aph was amplified from pUC18K1 (Ménard et al., 1993). The three PCR products were joined together using overlap extension PCR, and the construct was cloned into pFLP2 and sequence verified (Hoang et al., 1998). pFLP2 was then electroporated into A. baumannii, plated onto LB Km40 agar, and grown overnight at 37°C. Transformants were patched to LB Km40 or LB agar with 10% sucrose to isolate Km-resistant (KmR) and sucrose-sensitive merodiploids. Merodiploid strains were grown in LB overnight at 37°C to resolve the plasmid. Cultures were serially diluted, plated on LB agar with 10% sucrose, and incubated at 37°C overnight. The resulting KmR sucrose-resistant strains were screened for loss of zrlA and replacement with aph by multiple PCR reactions and Southern blot using gene-specific probes. To generate the zrlA unmarked strain, recombineering was utilized as described previously (Tucker et al., 2014). Briefly, the pUC18K1 aph gene was amplified using primers that contained 120 bp regions of homology flanking the zrlA open reading frame. The resulting PCR product was purified, concentrated to > 1 μg/μl, and transformed via electroporation into WT A. baumannii containing pAT02 (Tucker et al., 2014). RecAb recombinase expression was induced with 2 mM isopropyl β-D-1-thiogalactopyranoxide (IPTG). Cultures were grown for four hours and plated to LB agar supplemented with Km. Resulting colonies were screened using primers external to zrlA and restreaked to LB agar containing Km or carbenicillin to screen for loss of pAT02. Resulting KmR carbenicillinS strains were transformed with PAT03 and plated to carbenicillin. From here, colonies were streaked to LB agar containing 1 mM IPTG to induce FLP recombinase expression and excision of the Km resistance cassette. Colonies were then screened for KmS and carbencillinS and verified by PCR. Relevant primers are listed in Table S2.
Construction of zrlA expression and complementation vectors
To generate the zrlA complementation vector, the zrlA gene and its native promoter were amplified with a C-terminal cMyc tag and cloned into the digested pWH1266 vector using the BamHI/SalI restriction sites (Hunger et al., 1990). To generate the non-metal binding variant, a gene block with a C-terminal cMyc tag containing the following open reading frame nucleotide substitutions was synthesized: C448G, A449C, A470C, A617C (Integrated DNA Technologies, San Jose, CA) and cloned into pWH1266 under the native zrlA promoter. The integrity of the promoter and gene sequences were confirmed by sequencing. The empty vector and expression vectors were each transformed into A. baumannii by electroporation. All relevant primers are listed in Table S2.
Polyclonal antibody generation
ZrlA was purified as described below and submitted to the Vanderbilt Antibody and Protein Resource core for generation of two rabbit polyclonal antibodies against ZrlA through Cocalico Biologicals (Stevens, PA). The antisera were affinity purified for increased ZrlA specificity. All antibodies, reactive sera, and pre-immune sera collected from the rabbits were tested for reactivity and specificity in enzyme-linked immunosorbent assays with purified ZrlA protein as well as immunoblot analysis of WT and ΔzrlA whole cell lysates.
Quantitative RT-PCR
Overnight cultures of A. baumannii were diluted 1:50 in LB and grown for 1 h at 37°C. Cultures were then diluted 1:100 into LB plus or minus 200 μg/ml calprotectin or 40 μM TPEN and grown to mid-log. Cultures were pelleted at 4°C and resuspended in 1:1 acetone:ethanol prior to storage at −80°C until processing. For RNA extraction, cells were pelleted and resuspended in LETS buffer (0.1 M LiCl, 0.01 M Na2EDTA, 0.01 M Tris-HCl pH 7.4, 0.2% SDS) and lysed using Lysis Matrix B tubes (MP Biologicals) and a FastPrep-24 (MP) bead beater. Samples were heated to 55°C for 5 min and pelleted at 15,000 rpm for 10 min. The top phase was combined with TRIzol and incubated at room temperature for 5 min. Chloroform was mixed with each sample, incubated for 3 min, and centrifuged for 15 min at 4°C at 15,000 rpm. Following centrifugation, the upper aqueous phase was transferred to a new tube and incubated with isopropyl alcohol for 10 min at room temperature to precipitate the RNA. Samples were centrifuged at 4°C for 10 min at 15,000 rpm. Supernatant was removed, and the pellet was washed twice with 70% ethanol and dissolved in water. DNA contamination was removed by adding RQ1 and RQ1 buffer (Promega), and RNase inhibitor (Thermo Fisher Scientific), and the samples were incubated at 37°C for 2 h. Following DNase treatment, RNA was purified using RNease mini kit (QIAGEN) following manufacturer’s recommendation. RNA was quantified, and 2 μg of RNA was used for cDNA synthesis using M-MLV reverse transcriptase (Thermo Fisher Scientific). cDNA synthesis and qRT-PCR was performed as previously described using the ΔΔCT method with the iQ SYBR Green Supermix (BioRad) (Mortensen et al., 2014; Nairn et al., 2016). Data are combined from at least 3 biological replicates. Relevant primers were published previously or are listed in Table S2 (Hood et al., 2012; Mortensen et al., 2014).
Membrane fractionation
Overnight cultures of A. baumannii were diluted 1:50 in LB and grown for 1 h at 37°C. Cultures were then diluted 1:100 into LB plus or minus 200 μg/ml calprotectin or 40 μM TPEN and grown to mid log. Cells were pelleted, and whole cell lysates were generated by resuspending pellets in lysis buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5), transferring samples to Lysis Matrix B tubes (MP Biologicals), and lysing on a FastPrep-24 (MP) bead beater. Crude membrane fractions were separated from cytoplasmic fractions by ultracentrifugation at 100,000 × g for 90 min, with the cytoplasmic fraction being in the supernatant. The crude membrane fraction pellet was solubilized in lysis buffer containing 0.5% sarkosyl and incubated at room temperature with shaking for 30 min. Insoluble outer membrane fractions were isolated from the cytoplasmic membrane fraction by centrifugation at 100,000 × g for 60 min. Protein concentrations for each fraction were determined by BCA assay (Thermo Scientific) using bovine serum albumin as a standard. Samples were analyzed by immunoblot using mouse anti-cMyc 9E10 or rabbit anti-ZrlA, along with goat anti-mouse or anti-rabbit Alexa Fluor 680. Presented immunoblots are representative of at least 3 biological replicates.
ZrlA Purification
ZrlA was cloned into pMALc5× to produce recombinant ZrlA tagged with a maltose binding protein (MBP). E. coli BL21 (DE3) pREL containing the ZrlA expression vector was grown at 37°C shaking to an OD600 of 0.5. Protein expression was induced with 0.5 mM IPTG and grown for an additional 6 h at 37°C. Cells were harvested by centrifugation at 6000 × g and suspended in lysis buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5) supplemented with lysozyme (1 mg/ml) and bovine deoxyribonuclease (3 mg). Cells were lysed by five passes through an Emulsiflex homogenizer (Aventin, Inc.) at 20,000 psi. Lysates were centrifuged at 8,000 × g to remove intact cells, and insoluble debris was removed by ultracentrifugation at 100,000 × g for 1 h at 4°C. Supernatants were applied to amylose resin pre-equilibrated with lysis buffer. The column was washed with 12 column volumes of wash buffer (500 mM NaCl, 20 mM Tris-HCl pH 7.5, 1 mM EDTA), and ZrlA was eluted in a single elution with 10 mM maltose. In some cases, the MBP tag was cleaved by incubation with Factor Xa (New England Biolabs) during an overnight dialysis at 4°C into dialysis buffer (150 mM NaCl, 20 mM Tris-HCl pH 7.5, 2.5 mM CaCl2) with a buffer change after 2 h. To separate the MBP tag from ZrlA, the cleaved proteins were applied to Ni-NTA resin pre-equilibrated with dialysis buffer, where ZrlA was retained on the resin and eluted with an imidazole gradient (50–250 mM). ZrlA-containing fractions and sample purity were assessed by SDS-PAGE, and protein concentrations were determined by BCA assay (Thermo Scientific). To assess metals bound to ZrlA-MBP, purified protein was diluted to 1 mg/ml into lysis buffer, and Millipore water was added to a final volume of 10 mL in 15 mL metal-free conical tubes. Element quantification was assessed by ICP-MS. ZrlA (42-end) (ZrlA42C) was cloned into a pET22b(+) plasmid between NcoI and EcoRI sites and was expressed without the His-tag. This resulting plasmid was transformed into BL21 (DE3) for expression in LB with ampicillin (100 μg/ml) and grown to OD600 of 0.6, at which time 0.75 mM IPTG was added, and the cells allowed to grow at 16°C overnight. For each 1 L cell pellet, cells were suspended in 40 mL lysis buffer A (25 mM MES, 50 mM NaCl, pH 5.5) and sonicated at 60% power (3 s on; 9 s off, on ice), for total 60 min/L cells. This solution was then subjected to low-speed centrifugation at 10,000 × g (20 min, 4°C). The supernatant was filtered with 0.22 μm and loaded onto a self-packed SP-Sepharose™ FastFlow (GE Healthcare) column. The SP-column was run at 2 mL/min, and ZrlA42C was eluted with a 0%−50% buffer B (25 mM MES, 1 M NaCl, pH 5.5) gradient over 90 min. ZrlA42C-containing fractions were pooled. EDTA to a final concentration of 2 mM was added. The protein sample were concentrated and dialyzed again G75 (Hi Load™ 16/60 Superdex™ 75 prep grade) running buffer (25 mM HEPES, 150 mM NaCl, pH 7.5). ZrlA42C eluted at a flow-rate of 1 ml/min. ZrlA42C-containing fractions were flash-frozen in liquid nitrogen and stored at −80°C, and buffer exchanged prior to use. Protein was judged to be 90% pure by overloaded SDS-PAGE gels. The ZrlA42C concentration was measured using λ280 = 36,690 M−1 ⋅ cm−1. An N-terminally deleted version of A1S_1248 lacking the 55 N-terminal residues, termed A1S_124856C, was purified essentially the same way, and concentration measured using λ280 = 40,910 M−1 ⋅ cm−1. Relevant primers are listed in Table S2.
CoII titration
These experiments were performed as described previously using a Hewlett-Packard model 8452A spectrophotometer (Ma et al., 2011). The experiments were carried out in chelated titration buffer (25 mM HEPES, 150 mM NaCl, pH 7.4) at ambient temperature. Apo ZrlA42C or Apo A1S_124856C (1 mL of 100 μM) was titrated with stock solution of CoCl2. The binding reaction was monitored using 1-cm path length. Five minutes of equilibration time was allowed before the UV-vis spectra were acquired. UV-vis spectra from 230–800 nm were recorded after each addition and baseline-corrected as previously described (Ma et al., 2011).
Quin-2 Zn competition assays
These chelator competition experiments were carried out as described in previous work using a Hewlett-Packard model 8452A spectrophotometer (Reyes-Caballero et al., 2010). The experiments were carried out in chelated titration buffer (25 mM HEPES, 150 mM NaCl, pH 7.4) at ambient temperature. Apo ZrlA42C or apo A1S_124856C (1 mLof 15~17 μM and 12~15 μM, respectively) in the presence of 12~15 μM (for ZrlA42C) or 20~27 μM (for A1S_124856C) Quin-2 was titrated with a solution of ZnSO4. Equilibrium time was 10 min between measurements. Peak intensities at 265 nm from two independent experiments were globally fit to a simple 1:1 binding competition model with KZnquin−2 = 2.7 × 1011 M− 1 using Dynafit (Kuzmic, 1996).
PAR Zn binding assay
4-(2-pyridylazo)-resorcinol (PAR) Zn binding assays were performed as previously described (Hood et al., 2012; Mortensen et al., 2014; Nairn et al., 2016). Briefly, free PAR exhibits a peak absorbance of 410 nm that shifts to 500 nm upon Zn binding. To determine if ZrlA binds Zn, increasing concentrations of recombinant ZrlA-MBP (0–12.5 μM) were added to solutions containing 20 μM PAR and 10 μM ZnCl2 in 20 mM Tris-HCl pH 7.5, 150 mM NaCl. Spectra were collected for free and Zn-bound PAR and compared to spectra obtained in the presence of ZrlA-MBP. Spectra are representative of at least 3 biological replicates.
70Zn uptake assay
Cells were grown for 7 h ± calprotectin, and optical densities were normalized across strains. 70ZnO was spiked into each sample for a final concentration of 25 μM, and samples were incubated at 37°C for 15 min. A 1:1 mixture of acetone:ethanol was added to the cells and pelleted for 10 min at 6,000 rpm. Cells were then washed twice in PBS. Following the second wash, samples were transferred to metal-free 15 mL conical tubes (VWR). Samples were digested overnight in 50% Optima-grade nitric acid (Fisher Scientific) at 50°C and diluted with Millipore water prior to inductively-coupled plasma mass spectrometry analysis. 70Zn values were normalized to total 34S.
Inductively-Coupled Plasma Mass Spectrometry
Element quantification analysis on acid-digested protein samples was performed using ELEMENT 2 high resolution inductively coupled plasma mass spectrometry (HR-ICPMS, Thermo Fisher Scientific, Bremen, Germany) coupled with ESI auto sampler (Elemental Scientific, Omaha, NE). The ICPMS is equipped with a PFA microflow nebulizer (Elemental Scientific, Omaha, NE), a Scott double-pass spray chamber (at room temperature), a magnetic sector followed by an electric sector, and a second electron multiplier. Liquid sample was up-taken by self-aspiration via 0.50 mm ID sample probe and sample capillary. Interested metals were measured at medium resolution (R = 4200). Elemental quantification on 70Zn-labeled samples was performed using an Agilent 7700 inductively coupled plasma mass spectrometer (Agilent, Santa Clara, CA). The following settings were fixed for the analysis Cell Entrance = −40 V, Cell Exit = −60 V, Plate Bias = −60 V, OctP Bias = −18 V, and collison cell Helium Flow = 4.5 ml/min. Optimal voltages for Extract 2, Omega Bias, Omega Lens, OctP RF, and Deflect were determined empirically before each sample set was analyzed. Element calibration curves were generated using ARISTAR ICP Standard Mix (VWR, Radnor, PA). Samples were introduced by peristaltic pump with 0.5 mm internal diameter tubing through a MicroMist borosilicate glass nebulizer (Agilent). Samples were initially up taken at 0.5 rps for 30 s followed by 30 s at 0.1 rps to stabilize the signal. Samples were analyzed in Spectrum mode at 0.1 rps collecting three points across each peak and performing three replicates of 100 sweeps for each element analyzed. Data were acquired and analyzed using the Agilent Mass Hunter Workstation Software version A.01.02.
Modified Cd-Ninhydrin Assay
The Cd-ninhydrin reagent was generated as previously described (Doi et al., 1981). Briefly, ninhydrin (Sigma) was dissolved in 99.5% EtOH and acetic acid, and CdCl2 was added to the mixture. Enzymatic assays were performed as described elsewhere (Wu et al., 1995). Briefly, recombinant MBP-ZrlA (1 mM) was combined with Nα,Nε-Diacetyl-Lys-D-Ala-D-Ala (10 mM; Sigma) in the reaction buffer (50 mM HEPES pH 8) and incubated for 15 min at 37°C. Samples were mixed at a ratio of 1:2 with the Cd-ninhydrin reagent and heated for 5 min at 85°C. Samples cooled briefly at room temperature, and the absorbance at 500 nm was determined using a Varian Cary 50 Bio UV-Visible Spectrophotometer. Data are combined from at least 3 independent experiments.
Fluorescent D-amino acid labeling
Overnight cultures of WT or ΔzrlA A. baumannii were diluted 1:50 into LB for 2 h and subsequently diluted 1:10 into M9 minimal media with 0.5% sodium succinate and Vishniac’s trace metal mix (Vishniac, 1955) ± 10 μM TPEN. HADA (7-hydroxycoumarin-3-carboxylic acid-amino-d-alanine) and DA-EDA (d-alanyl-ethynyl-d-alanine dipeptide) were synthesized as described previously and added at a final concentration of 1 mM to each culture (Kuru et al., 2012; Liechti et al., 2014). Cells were incubated with shaking at 37°C for 2 h and fixed in 70% ethanol for 1 h at 0°C. Cells were pelleted and washed twice with PBS. DA-EDA-labeled cells were conjugated with the Atto 488 azide label (Sigma) using the Click-iT cell reaction buffer kit (Thermo) to generate fluorescence signal. Cells were imaged using a Nikon Ti-E inverted microscope equipped with a 1.4NA Plan Apo 60X oil objective and Andor iXon ENCCD camera. NIS-Elements AR software was used for image acquisition. Quantitative analysis of fluorescence intensity was performed using the MicrobeJ plugin in FIJI (Ducret et al., 2016). Data are pooled from 100 cells per treatment.
Sacculi Purification and UPLC-MS Analysis
WT A. baumannii and ΔzrlA were grown for 12 h in LB ± 40 μM TPEN. Cells were collected, and sacculi were purified as described previously (Kuru et al., 2012; Litzinger et al., 2010). Briefly, cell pellets were resuspended in water, added dropwise to boiling 5% sodium dodecyl sulfate (SDS, w/v), and incubated with stirring for 30 min. The SDS insoluble material was collected by ultracentrifugation at 39,000 × g at 30°C. Insoluble material was resuspended in water and boiled for 30 min with stirring in 4% SDS. Samples were collected again by ultracentrifugation, washed repeatedly with water, resuspended in a digestion buffer (10 mM Tris-HCl pH 7.0, 10 mM NaCl, 0.32 M imidazole, 1 mM MgSO4) and digested with DNase I (Thermo Fisher Scientific) and α-amylase (Promega) for 2 h at 37°C. Samples were pelleted and resuspended in 0.05 M Tris-pH 7.8 and pronase (type XXV from Streptomyces griseus) and incubated at 60°C for 2 h. Following this digestion, samples were pelleted, resuspended in water, and added dropwise to boiling 1% SDS with stirring for 30 min. Samples were collected by centrifugation and washed repeatedly in water before storage at −80°C in a minimal amount of water. For UPLC-MS, the purified sacculi were thawed and digested with mutanolysin for 16 h at 37°C. Before applying the sample to the UPLC-MS analysis, digested PG samples were reduced with 10 mg/ml sodium borohydrate in 0.5 M borax in miliQ water at pH 9.0. The reduction was stopped with 98% phosphoric acid, with the resultant pH adjusted to 2~3. An Acquity UPLC coupled to a PDA detector and SynaptG2S mass spectrometer, using a Waters C18 CSH 130 Å, 1.7 μm, 2.1 mm × 100 mm column and associated guard column C18 CSH 130 Å, 1.7 mm, 2.1 mm × 35 mm were used for UPLC-MS analysis. Solvent A was 0.1% TFA and solvent B was 0.1% TFA in 30% methanol and a linear gradient to 100% B over 60 min at a flow rate of 0.176 ml/min was used at a column temperature of 52°C. In a typical analytical run, 10 μL samples were injected and analyzed. PDA detects 210 nm. Muropeptides were identified with theoretical m/z ± 0.02 Da. Integration of peak areas were performed with Masslynx. Peak area for each muropeptide was normalized to the total peak area of all identified muropeptides (muropeptide peaks 1~10 in Table S1 correspond to ≥ 95% of the total peak area), and biological duplicates were grouped for statistical analyses.
Live cell microscopy
WT and mutant cells were grown to mid to late-exponential phase in LB ± 40 μM TPEN and immediately spotted onto agarose-PBS pads (0.05% agarose). Cells were allowed to dry briefly before sealing and imaging using brightfield microscopy on an Olympus BX60 microscope at 100X magnification. Images are representative of at least 3 independent experiments.
Ethidium bromide uptake
WT and mutant cells were grown in LB ± 40 μM TPEN, normalized to OD600 of 0.3, and washed once with PBS. Cell resuspensions were placed into black 96-well plates and spiked with ethidium bromide (1 μg/ml, Sigma) before immediately reading fluorescence at 2 s intervals (Ex: 530; Em: 600) using a BioTek Cytation 5 imaging reader. Data are combined from at least 3 independent experiments.
Bacterial growth in experimental conditions
Overnight cultures of WT or ΔzrlA were subcultured 1:50 in LB for 1 h. Back-diluted cultures were then inoculated 1:100 into LB containing various concentrations of SDS, ethylenediamine tetraacetic acid (EDTA) (Sigma), tetrakis-(2-pyridylmethyl)ethylenedi-amine (TPEN), calprotectin, carbenicillin (6.25 μg/ml), tetracycline (AlfaAesar, 78 ng/ml), polymyxin B (Alexis Biochemicals, 250 ng/ml), or vancomycin (Fisher Scientific, 100 μg/ml) and growth was monitored over time by monitoring OD600 using a BioTek Synergy 2 or Epoch 2 microplate reader. For growth in antibiotics with TPEN, cells were grown in carbenicillin (6.25 μg/ml) ± 20 μM TPEN or polymyxin B (1 μg/ml) ± 10 μM TPEN. For growth in calprotectin, back-diluted cultures were inoculated 1:100 into LB containing 40% calprotectin buffer (20 mM Tris-HCl pH 7.5,100 mM NaCl, 5 mM β-mercaptoethanol, 3 mM CaCl2) supplemented with calprotectin. Recombinant human calprotectin was used for all in vitro experiments and was expressed and purified as described previously (Corbin et al., 2008; Kehl-Fie et al., 2011). Percent growth panels are combined from at least 3 biological replicates.
Mouse model of A. baumannii pneumonia
Eight to ten-week-old male mice were inoculated 1:1 with a mixture of A. baumannii (WT and Km-marked ΔzrlA, or unmarked ΔzrlA and Km-marked ΔzrlA) totaling 3 × 108 CFU in 40 μL PBS. For meropenem dosing experiments, mice were administered meropenem (Hospira, Inc.) dissolved in PBS (12.5 mg/kg) or vehicle via intraperitoneal injection at 0, 12, and 24 hours post-infection (hpi). At 36 hpi, mice were euthanized, and CFU were enumerated in the lungs and livers following tissue homogenization and dilution plating on LB and LB Km 40. The limit of detection of this assay is 100 CFU/ml.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using GraphPad Prism 7 and Microsoft Excel. Statistical significance was generally assessed using a two-tailed unpaired Student’s t test, one-way ANOVA with Tukey multiple comparisons test, or Mann Whitney U test. Significance was defined as p < 0.05, and data were only excluded on the basis of technical errors associated with the experiment. Exact statistical tests used, significance values, group sizes, and dispersion and precision of measurements are defined in the figure legends.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-ZrlA | This paper | N/A |
| Mouse anti-c-Myc | Vanderbilt Antibody & Protein Resource | VAPR9E10 |
| Goat anti-rabbit Alexa fluor 680 | Thermo Fisher Scientific | A-21076; RRID:AB_141386 |
| Goat anti-mouse Alex fluor 680 | Thermo Fisher Scientific | A-21057; RRID: AB_141436 |
| Bacterial and Virus Strains | ||
| Acinetobacter baumannii 17978 | ATCC | 5377 [NCDC KC 755] |
| A. baumannii zrlA::km | This paper | N/A |
| A. baumannii ΔzrlA (clean deletion) | This paper | N/A |
| A. baumannii zur::km | Mortensen et al., 2014 | N/A |
| A. baumanniizrlA::tn (ABUW_0072) | University of Washington | AB00192 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RQ1 Dnase | Thermo Fisher Scientific | M6106 |
| Ribolock RNase inhibitor | Thermo Fisher Scientific | EO0382 |
| iQ Sybr Green Supermix | Biorad | 1708880 |
| M-MLV reverse transcriptase | Thermo Fisher Scientific | M1705 |
| Factor Xa | New England Biolabs | P8010S |
| SP-Sepharose | GE Healthcare Life sciences | 17515701 |
| Hi Load 16/60 Superdex | GE Healthcare Life sciences | 28-9893-33 |
| 4–2(2-pyridylazo) resorcinol (PAR) | Fluka | 82970 |
| 70ZnO | Cambridge Isotope Laboratories | ZNLM-4275-PK |
| N,N,N′,N′-Tetrakis(2-pyridylmethyl)ethylenediamine | Sigma | P4413-50MG |
| Nα,Nε-Diacetyl-Lys-D-Ala-D-Ala | Sigma | D9904-25MG |
| Human calprotectin | Corbin et al., 2008; Kehl-Fie et al., 2011 | N/A |
| MBP-ZrlA | This paper | N/A |
| ZrlA42C | This paper | N/A |
| A1S_124856c | This paper | N/A |
| HADA | Kuru et al., 2012 | N/A |
| DA-EDA | Liechti et al., 2014 | N/A |
| Meropenem | Hospira | NDC 0409-3505-01 |
| Critical Commercial Assays | ||
| Rneasy Mini Kit | QIAGEN | 74104 |
| Click-iT reagents | Fisher Scientific | N/A |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J mice | The Jackson Laboratory | Cat #664 |
| Oligonucleotides | ||
| Primers for cloning and qRT-PCR: Table S2. | ||
| Software and Algorithms | ||
| Agilent Mass Hunter Workstation | Agilent | Version A.01.02 |
| Dynafit | BioKin, Ltd. | http://www.biokin.com/dynafit/license/index.html |
| MicrobeJ | Ducret et al., 2016 | http://www.microbej.com/ |
| GraphPad Prism | GraphPad | Version 7 |
| Canvas X16 | Canvas | Version 16 |
Highlights.
During zinc starvation, A. baumannii expresses a putative peptidase named ZrlA
ZrlA is critical for bacterial cell envelope integrity and overcoming Zn limitation
Inactivation of zrlA increases antibiotic efficacy in vitro and during infection
ACKNOWLEDGMENTS
The work presented in this manuscript was supported by grants R01 AI101171 (to W.J.C., D.P.G., and E.P.S.), R35 GM118157 (to D.P.G.), and R01 GM113172 (to M.S.V. and Y.P.H.) from the NIH. Z.R.L. was supported by NIH F31 AI136255 and T32 ES007028. B.L.N. was supported by NIH F32 AI108192 and by the Childhood Infection Research Program (ChIRP, NIH T32 AI095202). L.E.H. was supported by NIH T32 GM065086. W.N.B was supported by NIH T32 HL069765.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2019.01.089.
REFERENCES
- Ammendola S, Pasquali P, Pistoia C, Petrucci P, Petrarca P, Rotilio G, and Battistoni A (2007). High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect. Immun 75, 5867–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes LC, Visca P, and Towner KJ (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis 71, 292–301. [DOI] [PubMed] [Google Scholar]
- Aron AT, Heffern MC, Lonergan ZR, Vander Wal MN, Blank BR, Spangler B, Zhang Y, Park HM, Stahl A, Renslo AR, et al. (2017). In vivo bioluminescence imaging of labile iron accumulation in a murine model of Acinetobacter baumannii infection. Proc. Natl. Acad. Sci. USA 114, 12669–12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydemir TB, Chang SM, Guthrie GJ, Maki AB, Ryu MS, Karabiyik A, and Cousins RJ (2012). Zinc transporter ZIP14 functions in hepatic zinc, iron and glucose homeostasis during the innate immune response (endotoxemia). PLoS ONE 7, e48679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TM, Nakashige TG, Nolan EM, and Neidig ML (2017). Magnetic circular dichroism studies of iron(ii) binding to human calprotectin. Chem. Sci. (Camb.) 8, 1369–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P (1968). Isolation of Acinetobacter from soil and water. J. Bacteriol 96, 39–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby-Haas CE, Furman R, Rodionov DA, Artsimovitch I, and de Crécy-Lagard V (2011). Role of a Zn-independent DksA in Zn homeostasis and stringent response. Mol. Microbiol 79, 700–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MR, and Melling J (1969). Role of divalent cations in the action of polymyxin B and EDTA on Pseudomonas aeruginosa. J. Gen. Microbiol 59, 263–274. [DOI] [PubMed] [Google Scholar]
- Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, and Brun YV (2012). Polar growth in the Alphaproteobacterial order Rhizobiales. Proc. Natl. Acad. Sci. USA 109, 1697–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat JE, Moore JL, Wilson KJ, Stark Z, Prentice BM, Van de Plas R, Perry WJ, Zhang Y, Virostko J, Colvin DC, et al. (2018). Integrated molecular imaging reveals tissue heterogeneity driving host-pathogen interactions. Sci. Transl. Med 10, eaan6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, et al. (2008). Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319, 962–965. [DOI] [PubMed] [Google Scholar]
- Corwin DFR, and Koch S (1987). Four- and five-coordinate cobalt(II) thiolate complexes: models for the catalytic site of alcohol dehydrogenase. Inorg. Chem 26, 3079–3080. [Google Scholar]
- Dalebroux ZD, Edrozo MB, Pfuetzner RA, Ressl S, Kulasekara BR, Blanc MP, and Miller SI (2015). Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe 17, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, Zhang Y, Betz C, Hench L, Fritz G, et al. (2013). Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc. Natl. Acad. Sci. USA 110, 3841–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers DC, Bearden SW, Mier I Jr., Abney J, Paulley JT, Fetherston JD, Salazar JC, Radolf JD, and Perry RD (2010). Znu is the pre-dominant zinc importer in Yersinia pestis during in vitro growth but is not essential for virulence. Infect. Immun 78, 5163–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra AJ, and Keck W (1996). Peptidoglycan as a barrier to transenvelope transport. J. Bacteriol 178, 5555–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi E, Shibata D, and Matoba T (1981). Modified colorimetric ninhydrin methods for peptidase assay. Anal. Biochem 118, 173–184. [DOI] [PubMed] [Google Scholar]
- Ducret A, Quardokus EM, and Brun YV (2016). MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol 1, 16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, and Helmann JD (1998). Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol 180, 5815–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, and Manoil C (2015). Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J. Bacteriol 197, 2027–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattis SG, Hernick M, and Fierke CA (2010). Active site metal ion in UDP-3-O-((R)-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase (LpxC) switches between Fe(II) and Zn(II) depending on cellular conditions. J. Biol. Chem 285, 33788–33796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynes R, and Edwards JR; National Nosocomial Infections Surveillance System (2005). Overview of nosocomial infections caused by Gram-negative bacilli. Clin. Infect. Dis 41, 848–854. [DOI] [PubMed] [Google Scholar]
- Ghosh AS, Chowdhury C, and Nelson DE (2008). Physiological functions of d-alanine carboxypeptidases in Escherichia coli. Trends Microbiol. 16, 309–317. [DOI] [PubMed] [Google Scholar]
- Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, and Miller SI (1997). Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276, 250–253. [DOI] [PubMed] [Google Scholar]
- Haase H, and Rink L (2014). Multiple impacts of zinc on immune function. Metallomics 6, 1175–1180. [DOI] [PubMed] [Google Scholar]
- Harding CM, Hennon SW, and Feldman MF (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol 16, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, and Schweizer HP (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. [DOI] [PubMed] [Google Scholar]
- Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, Chazin WJ, Caprioli RM, and Skaar EP (2012). Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog. 8, e1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger M, Schmucker R, Kishan V, and Hillen W (1990). Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87, 45–51. [DOI] [PubMed] [Google Scholar]
- Hunter MJ, and Chazin WJ (1998). High level expression and dimer characterization of the S100 EF-hand proteins, migration inhibitory factor-related proteins 8 and 14. J. Biol. Chem 273, 12427–12435. [DOI] [PubMed] [Google Scholar]
- Juttukonda LJ, Chazin WJ, and Skaar EP (2016). Acinetobacter baumannii coordinates urea metabolism with metal import to resist host-mediated metal limitation. MBio 7, e01475–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juttukonda LJ, Berends ETM, Zackular JP, Moore JL, Stier MT, Zhang Y, Schmitz JE, Beavers WN, Wijers CD, Gilston BA, et al. (2017). Dietary manganese promotes staphylococcal infection of the heart. Cell Host Microbe 22, 531–542 e538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, and Skaar EP (2011). Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe 10, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, and VanNieuwenhze MS (2012). In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew. Chem. Int. Ed. Engl 51, 12519–12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmic P (1996). Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem 237, 260–273. [DOI] [PubMed] [Google Scholar]
- Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, and Maurelli AT (2014). A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506, 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litzinger S, Duckworth A, Nitzsche K, Risinger C, Wittmann V, and Mayer C (2010). Muropeptide rescue in Bacillus subtilis involves sequential hydrolysis by β-N-acetylglucosaminidase and N-acetylmuramyl-L-alanine amidase. J. Bacteriol 192, 3132–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, et al. (2012). Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11, 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Gabriel SE, and Helmann JD (2011). Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res. 39, 9130–9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinverni JC, and Silhavy TJ (2009). An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc. Natl. Acad. Sci. USA 106, 8009–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard R, Sansonetti PJ, and Parsot C (1993). Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol 175, 5899–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen BL, Rathi S, Chazin WJ, and Skaar EP (2014). Acinetobacter baumannii response to host-mediated zinc limitation requires the transcriptional regulator Zur. J. Bacteriol 196, 2616–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairn BL, Lonergan ZR, Wang J, Braymer JJ, Zhang Y, Calcutt MW, Lisher JP, Gilston BA, Chazin WJ, de Crécy-Lagard V, et al. (2016). The response of Acinetobacter baumannii to zinc starvation. Cell Host Microbe 19, 826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashige TG, Zygiel EM, Drennan CL, and Nolan EM (2017). Nickel sequestration by the host-defense protein human calprotectin. J. Am. Chem. Soc 139, 8828–8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanamiya H, Akanuma G, Natori Y, Murayama R, Kosono S, Kudo T, Kobayashi K, Ogasawara N, Park SM, Ochi K, and Kawamura F (2004). Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol. Microbiol 52, 273–283. [DOI] [PubMed] [Google Scholar]
- Nicas TI, and Hancock RE (1983). Alteration of susceptibility to EDTA, polymyxin B and gentamicin in Pseudomonas aeruginosa by divalent cation regulation of outer membrane protein H1. J. Gen. Microbiol 129, 509–517. [DOI] [PubMed] [Google Scholar]
- Palmer LD, and Skaar EP (2016). Transition metals and virulence in bacteria. Annu. Rev. Genet 50, 67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzer SI, and Hantke K (1998). The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol 28, 1199–1210. [DOI] [PubMed] [Google Scholar]
- Peters K, Kannan S, Rao VA, Biboy J, Vollmer D, Erickson SW, Lewis RJ, Young KD, and Vollmer W (2016). The redundancy of peptidoglycan carboxypeptidases ensures robust cell shape maintenance in Escherichia coli. MBio 7, e00819–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podmore AH, and Reynolds PE (2002). Purification and characterization of VanXYC, a D,D-dipeptidase/D,D-carboxypeptidase in vancomycin-resistant Enterococcus gallinarum BM4174. Eur. J. Biochem 269, 2740–2746. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, and Barrett AJ (1993). Evolutionary families of peptidases. Biochem. J 290, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman MK, and MacLeod RA (1975). Interaction of Mg-2+ with peptidoglycan and its relation to the prevention of lysis of a marine pseudomonad. J. Bacteriol 122, 650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Caballero H, Guerra AJ, Jacobsen FE, Kazmierczak KM, Cowart D, Koppolu UM, Scott RA, Winkler ME, and Giedroc DP (2010). The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor. J. Mol. Biol 403, 197–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade AL, and Caroline L (1946). An iron-binding component in human blood plasma. Science 104, 340–341. [DOI] [PubMed] [Google Scholar]
- Scheurwater EM, and Burrows LL (2011). Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol. Lett 318, 1–9. [DOI] [PubMed] [Google Scholar]
- Stefanova ME, Tomberg J, Davies C, Nicholas RA, and Gutheil WG (2004). Overexpression and enzymatic characterization of Neisseria gonorrhoeae penicillin-binding protein 4. Eur. J. Biochem 271, 23–32. [DOI] [PubMed] [Google Scholar]
- Stork M, Bos MP, Jongerius I, de Kok N, Schilders I, Weynants VE, Poolman JT, and Tommassen J (2010). An outer membrane receptor of Neisseria meningitidis involved in zinc acquisition with vaccine potential. PLoS Pathog. 6, e1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, and Salama NR (2010). Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell 141, 822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouillet JL, Chastre J, Vuagnat A, Joly-Guillou ML, Combaux D, Dombret MC, and Gibert C (1998). Ventilator-associated pneumonia caused by potentially drug-resistant bacteria. Am. J. Respir. Crit. Care Med 157, 531–539. [DOI] [PubMed] [Google Scholar]
- Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, and Davies BW (2014). Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. MBio 5, e01313–e01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishniac HS (1955). The nutritional requirements of isolates of Labyrinthula spp. J. Gen. Microbiol. 12, 455–463. [DOI] [PubMed] [Google Scholar]
- Wang T, Si M, Song Y, Zhu W, Gao F, Wang Y, Zhang L, Zhang W, Wei G, Luo ZQ, and Shen X (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11, e1005020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg ED (1974). Iron and susceptibility to infectious disease. Science 184, 952–956. [DOI] [PubMed] [Google Scholar]
- Weinberg ED (1975). Nutritional immunity: host’s attempt to withold iron from microbial invaders. JAMA 231, 39–41. [DOI] [PubMed] [Google Scholar]
- Whittington DA, Rusche KM, Shin H, Fierke CA, and Christianson DW (2003). Crystal structure of LpxC, a zinc-dependent deacetylase essential for endotoxin biosynthesis. Proc. Natl. Acad. Sci. USA 100, 8146–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2017). Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in health care facilities (World Health Organization; ). [PubMed] [Google Scholar]
- Wright AC, Simpson LM, and Oliver JD (1981). Role of iron in the pathogenesis of Vibrio vulnificus infections. Infect. Immun 34, 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Wright GD, and Walsh CT (1995). Overexpression, purification, and characterization of VanX, a D-,D-dipeptidase which is essential for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 34, 2455–2463. [DOI] [PubMed] [Google Scholar]
- Zackular JP, Moore JL, Jordan AT, Juttukonda LJ, Noto MJ, Nicholson MR, Crews JD, Semler MW, Zhang Y, Ware LB, et al. (2016). Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat. Med 22, 1330–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.