Abstract
Background:
Exploring the associations of air pollution and weather variables with blood leukocyte distribution is critical to understand the impacts of environmental exposures on the human immune system.
Objectives:
As previous analyses have been mainly based on data from cell counters, which might not be feasible in epidemiologic studies including large populations of long-stored blood samples, we aimed to expand the understanding of this topic by employing the leukocyte distribution estimated by DNA methylation profiles.
Methods:
We measured DNA methylation profiles in blood samples using Illumina HumanMethylation450 BeadChip from 1,519 visits of 774 Caucasian males participating in the Normative Aging Study. Leukocyte distribution was estimated using Houseman’s and Horvath’s algorithms. Data on air pollution exposure, temperature, and relative humidity within 28 days before each blood draw was obtained.
Results:
After fully adjusting for potential covariates, PM2.5, black carbon, particle number, carbon monoxide, nitrogen dioxide, sulfur dioxide, temperature, and relative humidity were associated with the proportions of at least one subtype of leukocytes. Particularly, an interquartile range-higher 28-day average exposure of PM2.5 was associated with 0.147-, 0.054- and 0.101-unit lower proportions (z-scored) of plasma cells, naïve CD8+ T cells, and natural killers, respectively, and 0.059- and 0.161-unit higher proportions (z-scored) of naïve CD4+ T cells and CD8+ T cells, respectively.
Conclusions:
Our study suggests that short-term air pollution exposure, temperature, and relative humidity are associated with leukocyte distribution. Our study further provides a successful attempt to use epigenetic patterns to assess the influences of environmental exposures on human immune profiles.
Keywords: air pollution, weather variations, DNA methylation, leukocyte distribution, epigenetic epidemiology, environmental health
1. Introduction
Air pollution, especially fine particulate matter [PM <2.5 μm (PM2.5)], and weather variations, such as changes in temperature and humidity, are critical factors for the increased risks of respiratory and cardiovascular diseases in varied populations (Brook et al. 2010; Koken et al. 2003; Schwartz et al. 2004). To date, one of the most widely accepted underlying mechanisms for these health effects is activation of systemic inflammatory responses (Brook et al. 2010; Halonen et al. 2010). Given that systemic inflammation activates/mobilizes inflammatory cells and may change the distribution of leukocytes (van Eeden and Hogg 2002), exploring the associations of air pollution and weather with leukocyte distribution is important to understand the impacts of environmental exposures on the immune system.
However, previous epidemiologic studies have mostly evaluated associations of environmental exposures with the distribution of leukocytes in freshly drawn blood samples using automated hematology cell counters, which mostly assess five types of blood leukocytes: neutrophils, basophils, eosinophils, lymphocytes, and monocytes (Bruske et al. 2010; Chuang et al. 2011; Halonen et al. 2010; Herr et al. 2010; Hertz-Picciotto et al. 2005; Ma et al. 2017; Pope et al. 2004; Rich et al. 2012; Salvi et al. 1999; Schwartz 2001; Steenhof et al. 2014; Viehmann et al. 2015; Zuurbier et al. 2011). Estimation accuracy of cell counters is limited by several technical aspects, including the need for large volumes of fresh cells, high requirements for fresh cell processing and protection, and time-consuming cell analyses (Cembrowski and Clarke 2015). Therefore, to explore the impacts of environmental exposures at the population level, simpler yet comprehensive methods to assess leukocyte composition are warranted.
DNA methylation, a major form of epigenetic changes with a heritable but dynamic nature (Bollati and Baccarelli 2010; Goldberg et al. 2007), can be used to distinguish cell lineages of blood leukocytes and describe differences in cell types with high sensitivity and specificity (Khavari et al. 2010). As epidemiologic studies often collect and archive blood DNA extracted from leukocytes, DNA methylation offers a new approach to estimate leukocyte distribution in large populations. This is especially useful when direct measurement of cell counts in fresh samples is not feasible, such as for long-stored blood samples, or was not performed at the time of sample collection with good reliability (Dugue et al. 2016). In 2012, Houseman et al. developed a reference-based algorithm to estimate cell-types (Houseman et al. 2012), which used cell-type-specific differentially methylated regions to infer the proportions of six subtypes of blood leukocytes: cytotoxic (CD8+) T cells, helper (CD4+) T cells, natural killers (NK), B cells, monocytes, and granulocytes. This method has now been widely accepted and used in epigenome analyses to control for confounding introduced by cell-type heterogeneity, especially in whole blood samples (Teschendorff and Zheng 2017; Titus et al. 2017). Horvath optimized this method by adding another four cell subtypes to his DNA methylation age predictors (Horvath 2013): plasma cells, exhausted CD8 T cells (defined as CD28-CD45RA- T cells), naïve CD8+ T cells, and naïve CD4+ T cells. Leukocyte distributions estimated by both methods are highly correlated with corresponding measures from automated hematology cell counters and may be minimally affected by external exposures (Horvath et al. 2016).
To expand our knowledge of the associations of leukocyte distribution with air pollutants and weather variations, we analyzed DNA methylation data from the Normative Aging Study (NAS), a cohort of older males in the greater Boston area. We estimated leukocyte distribution using algorithms from Houseman et al. and Horvath and investigated their associations with short-term exposures to ambient air pollution, temperature, and relative humidity.
2. Methods
2.1. Study design and population
The NAS is an ongoing longitudinal study on aging established by the U.S. Department of Veterans Affairs in 1963. Details of the study have been published previously (Bell et al. 1972). Briefly, the NAS is a closed cohort of 2,280 male veterans living in the greater Boston area. Participants were enrolled after an initial health screening to determine whether they were free of known chronic medical conditions. Most participants were examined up to four times during 1999–2013. They have been reevaluated every 3–5 years on a continuous rolling basis using detailed on-site physical examinations and questionnaires. Eligible participants for this study were those who had continued participation as of 2000, when data on air pollution, temperature, and relative humidity started to be collected. To control for race heterogeneity, a total of 1,519 medical visits from 774 Caucasian participants aged 55–85 years at initial visit were used in the analysis (visited in January 2003–December 2011). The NAS was approved by the Department of Veterans Affairs Boston Healthcare System, and written informed consent was obtained from each subject before participation.
2.2. Data collection
As previously described (Mordukhovich et al. 2015), participants were asked to provide detailed information about their lifestyles, dietary habits, activity levels, and demographic factors at each visit. Height and weight were used to calculate body mass index (BMI, in kg/m2). Blood samples were collected at each medical visit to assess blood-based biomarkers, such as total cholesterol (mg/dL), serum triglycerides (mg/dL), and high-density lipoprotein (HDL, mg/dL), and were stored for future analysis. Systolic and diastolic blood pressures (SBP and DBP, respectively) were measured once on each arm using a standard cuff, while the subject was seated. Major diseases were assessed based on participants’ medical histories and prior diagnoses (Nyhan et al. 2018).
2.3. DNA methylation data
As previously described (Gao et al. 2018), we used the QIAamp DNA Blood Kit (Qiagen, CA, USA) to extract DNA from stored buffy coats and performed bisulfite conversion with the EZ-96 DNA Methylation Kit (Zymo Research, CA, USA). To minimize batch effects, we randomized chips across plates and randomized samples based on a two-stage age-stratified algorithm so that age distributed similarly across chips and plates. We measured DNA methylation of CpG probes using the Illumina HumanMethylation450 BeadChip. After quality control, remaining samples were preprocessed using Illumina-type background correction, dye-bias adjustment, and BMIQ normalization (Teschendorff et al. 2013) to generate methylation status. Methylation status of a specific CpG site was quantified as a β-value ranging from 0 (no methylation) to 1 (full methylation).
2.4. Estimation of leukocyte distribution
Leukocyte distribution was estimated using two well-established methods based on background-corrected DNA methylation profiles. The proportions of CD8+ T cells (CD3+CD8+ T-lymphocytes), CD4+ T cells (CD3+CD4+ T-lymphocytes), NK cells (CD56+ NK cells), B cells (CD19+ B-lymphocytes), monocytes (CD14+ monocytes), and granulocytes (CD15+ granulocytes) were estimated with Houseman et al.’s algorithm (Houseman et al. 2012) embedded in R package ‘minfi’ with the default option (quantile normalization for blood samples). The proportion of exhausted CD8 T cells (CD28-CD45RA- T cells), naïve CD8+ T cells, naïve CD4+ T cells, and plasma cells (effector B cells) were estimated with the Horvath’s algorithm embedded in the ‘Advanced Blood Analysis’ of the online DNA methylation age predictor with internal normalization (Horvath 2013).
2.5. Assessments of air pollution exposure and weather variations
As previous studies demonstrated robust inflammatory responses after short-term exposures to air pollution and weather variations (Dauchet et al. 2018; Halonen et al. 2010; Zuurbier et al. 2011), we focused on short-term air pollution exposures, temperature, and relative humidity measured on the same day of the visit and mean values at 7, 14, 21, and 28 days before visits for each blood draw. The air pollutants were PM2.5, black carbon (BC), particle number (PN), carbon monoxide (CO), nitrogen dioxide (NO2), ozone (O3), and sulfur dioxide (SO2).
As previously described (Bind et al. 2014a; Bind et al. 2014b; Mehta et al. 2015), particulate concentrations were measured at the Harvard University supersite located near downtown Boston and approximately 1 km from the examination center. Because study participants lived in the greater Boston area with a median distance of 20 km from center, we assumed that the ambient air pollutant concentrations could serve as surrogates of participant exposures. Concentrations of PM2.5 (μg/m3) and BC (μg/m3) were measured hourly using a tapered element oscillation microbalance (Model 1400A, Rupprecht and Pastashnick) and aethalometer (Magee Scientific Co., Model AE-16). We measured hourly PN, which were fine and ultrafine particles with a diameter of 0.007–3 μm, using a condensation particle counter (TSI Inc., Model 3022A). Hourly CO, NO2, O3, and SO2 concentrations [parts per million (ppm)] were measured by local state monitors within the greater Boston area and were averaged based on data from all available sites.
We also obtained temperature and relative humidity data from the National Weather Service Station at Logan Airport (Boston, MA, USA), located approximately 12 km from the examination site. Because study participants lived throughout the metropolitan area, we assumed that the monitored temperature and humidity could serve as surrogates of their exposures.
2.6. Statistical analysis
Descriptive statistics were used to summarize socio-demographics and lifestyle factors for the first visit and all visits of the participants.
We examined whether the same-day and up to 28-day (7, 14, and 28 days prior to visit) average air pollution exposures, temperature, and relative humidity were associated with the distribution of 10 types of leukocytes estimated by DNA methylation data. We z-scored the distributions of 10 leukocytes for all visits to unify the scale of estimates from Houseman et al.’s and Horvath’s algorithms. We used time-varying linear mixed-effect regression models with random participant-specific intercepts (via PROC MIXED), accounting for the correlation of repeated measures. We treated the z-score of blood cells as outcomes and the exposures at certain exposure windows as predictors as shown in Formula 1:
| (1) |
where Yij is the z-scored proportion of a certain cell subtype of subject i at visit j, Eij is average levels of a certain air pollutant/weather variation before visit j, Covariate 1ij is subject i’s first covariate at visit j (this notation is relevant to all other potential covariates, because we used values assessed at each visit in our analyses), and ui is the random intercept that accounts for correlation within subjects. We adjusted for the following potential covariates: age (years), BMI (underweight or normal weight/overweight/obese), smoking status (current/former/never smoker), alcohol intake (<2 drinks per day or ≥2 drinks per day), total cholesterol, triglycerides, HDL, SBP, hypertension, stroke, coronary heart disease (CHD), diabetes and cancer diagnosed by physician (yes/no), and season of medical visits (spring/summer/fall/winter). Specifically, models of air pollution additionally adjusted for corresponding temperature and relative humidity in different exposure windows. Models for weather variations mutually adjusted for relative humidity and temperature, given that both factors were highly correlated, to understand their independent associations with leukocyte distributions. Effect estimates were reported as change in the z-score of blood cells per interquartile range (IQR) increase in exposure. Exposure IQRs were calculated separately for exposures on the same day of the visit and mean exposures computed up to 28 days before each visit. IQR reflects distribution (25th–75th percentile) in the observed data while also enabling comparison of effects of different exposure types measured with different units.
As one of the sensitivity analyses, PM2.5 was additionally added in the model for other pollutants to explore whether there was a confounding effect of PM2.5 on the associations between other pollutants and leukocyte distribution. Further, as with all longitudinal studies, healthier study participants may be more likely to participate in subsequent clinical examinations over time (Seaman and White 2013). To evaluate the validity of the missing at random assumption and assess the impact of potential selection bias caused by non-random unavailability for follow-up, we used inverse probability weighting (IPW) to correct for this potential survival bias as another sensitivity analysis. We estimated the inverse probability of coming to a subsequent clinical visit using logistic regressions given all relevant covariates at the previous visit among eligible participants: age, BMI, smoking status and pack-years, cholesterol level, hypertension, and medication use (diuretics and beta blockers). Rather than a constant of ‘1’, the inverse probability of being included in the analysis was assigned to the baseline visit. A weighted model simultaneously adjusted for the inverse probability and the aforementioned potential covariates of the main analysis.
Additionally, given the compositional nature of leukocytes, we speculated that this may influence the patterns we identified in the main analysis. Since we were unable to unify the proportions estimated by the two different methods using isometric log-ratio transformation (Egozcue et al. 2003), we conducted a sensitivity analysis using the cell counts of six subtypes calculated from the total number of leukocytes measured by cell counters (1000/mm3) and Houseman et al.’s cell proportion (counts = total number of leukocytes estimated by cell counters * proportions estimated by Houseman et al.’s algorithm) to understand to what extent the compositional nature could influence the identified patterns. We analyzed associations (z-scored) of the cell counts with environmental exposures using the same analysis models and compared outcomes with the main findings. Finally, we inspected the linearity of the associations between temperature and cell proportions using a generalized additive mixed model with the ‘mgcv’ R package. We fitted penalized splines for temperature to detect potential deviations from linearity.
SAS version 9.4 (SAS Institute Inc., Cary, NC, USA) was used to perform data cleaning and all analyses. A two-sided p-value of <0.05 was considered statistically significant.
3. Results
3.1. Characteristics of participants
Table 1 shows characteristics of the 774 study participants. Overall, average age at the initial visit was ~72 years. More than 60% of participants were former smokers, and <5% were current smokers. Most the participants were overweight or obese and consumed <2 drinks per day. Participants were healthier at baseline than at subsequent visits in terms of the prevalence of major diseases. Most visits (>80%) were in March–November. Average proportions of cell types in first-visit samples estimated by Houseman et al.’s algorithm were: ~5.2% for CD8+ T cells, ~10.1% for CD4+ T cells, ~4.7% for NK cells, ~3.1% for B cells, ~9.5% for monocytes (CD14+ monocytes), and ~67.0% for granulocytes. Proportions were similar in all visit samples. Most proportions of leukocyte subtypes were significantly correlated (p < 0.05, Table 2). However, correlation coefficients were relatively low, which might be explained by different immunological functions of those subtypes.
Table 1.
Characteristics of Normative Aging Study participants on the first visit and at all visitsa
| Characteristic | First visit (N=774) | All visits (N=1519) |
|---|---|---|
| Age (years) | 72.64 (6.82) | 74.60 (7.06) |
| Total cholesterol (mg/dL) | 192.28 (37.70) | 187.62 (38.42) |
| Serum triglyceride (mg/dL) | 141.18 (92.07) | 132.27 (81.69) |
| HDL cholesterol (mg/dL) | 49.21 (12.87) | 48.82 (12.87) |
| Systolic blood pressure (mm Hg) | 131.98 (17.12) | 128.91 (17.56) |
| Smoking status | ||
| Current smoker | 32 (4.1%) | 60 (3.9%) |
| Former smoker | 503 (65.0%) | 981 (64.6%) |
| Never smoker | 239 (30.9%) | 478 (31.5%) |
| Body mass index | ||
| Underweight or normal weight (<25.0) | 152 (19.6%) | 335 (22.1%) |
| Overweight (≥25 to <30) | 415 (53.6%) | 787 (51.8%) |
| Obese (≥30.0) | 207 (26.7%) | 397 (26.1%) |
| Alcohol consumption (≥2 drinks per day) | 147 (19.0%) | 278 (18.3%) |
| Major diseases | ||
| Hypertension | 550 (71.1%) | 1123 (73.9%) |
| Stroke | 56 (7.2%) | 121 (8.0%) |
| Coronary heart disease | 228 (29.5%) | 497 (32.7%) |
| Diabetes | 107 (13.8%) | 234 (15.4%) |
| Cancer | 383 (49.5%) | 833 (54.8%) |
| Season of visit | ||
| Spring (March-May) | 113 (23.2%) | 358 (23.6%) |
| Summer (June-August) | 120 (24.6%) | 412 (27.1%) |
| Fall (September-November) | 168 (34.4%) | 483 (31.8%) |
| Winter (December-February) | 87 (17.8%) | 266 (17.5%) |
Mean values (standard deviation) for continuous variables and n (%) for categorical variables;
Table 2.
Correlation matrix of proportions of leukocyte subtypes (Spearman coefficient)a
| Cell type | Plasma cells | Exhausted CD8 T cells | Naïve CD8+ T cells | Naive CD4+ T cells | CD8+ T cells | CD4+ T cells | NK cells | B cells | Monocytes | Granulocytes |
|---|---|---|---|---|---|---|---|---|---|---|
| Plasma cells | 1 | |||||||||
| Exhausted CD8 T cells | 0.192 | 1 | ||||||||
| Naïve CD8+ T cells | 0.080 | −0.484 | 1 | |||||||
| Naïve CD4+ T cells | −0.189 | −0.204 | 0.249 | 1 | ||||||
| CD8+ T cells | −0.395 | 0.119 | −0.124 | −0.045 | 1 | |||||
| CD4+ T cells | −0.399 | −0.331 | 0.139 | 0.438 | −0.185 | 1 | ||||
| NK cells | −0.136 | 0.276 | −0.179 | −0.285 | 0.207 | 0.014 | 1 | |||
| B cells | −0.142 | −0.041 | −0.107 | −0.184 | −0.067 | 0.341 | 0.250 | 1 | ||
| Monocytes | 0.096 | 0.092 | −0.165 | 0.019 | 0.050 | −0.206 | −0.078 | −0.230 | 1 | |
| Granulocytes | 0.577 | −0.022 | 0.198 | −0.051 | −0.455 | −0.469 | −0.557 | −0.463 | −0.068 | 1 |
Bolded coefficients have p < 0.05.
3.2. Distributions of air pollution, temperature, and relative humidity
Average values of environmental exposures over the follow-up period remained stable across the 28-day exposure window (Table 3). The 28-day average exposure was 10.00±2.77 μg/m3 (IQR = 3.23) for PM2.5, 0.74±0.18 μg/m3 (IQR = 0.28) for BC, 2.29±1.02 ×104 counts/cm3 (IQR = 1.39) for PN, 4.18±1.98 ×10−1 ppm (IQR = 2.65) for CO, 1.91±0.36 ×10−2 ppm (IQR = 0.56) for NO2, 2.45±0.72 ×10−2 ppm (IQR = 1.19) for O3, and 4.07±2.28 ×10−3 ppm (IQR = 3.03) for SO2. The average temperature was 12.58°C with an IQR of 13.58°C, and the relative humidity was 68.27% with an IQR of 8.11%. Table 4 lists correlations of 28-day average air pollution exposures, temperature, and relative humidity. Most mutual correlations were statistically significant (p < 0.05), except for correlations between PM2.5 and PN, and between CO and relative humidity. For a given exposure, its levels in different time windows were highly correlated (p < 0.01, Table S1).
Table 3.
Distributions of air pollution, temperature, and relative humidity for all NAS visits, January 2003 – December 2011
| Exposure (unit) | Exposure window (days) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7 | 14 | 21 | 28 | |||||||
| Mean (SD) | IQR | Mean (SD) | IQR | Mean (SD) | IQR | Mean (SD) | IQR | Mean (SD) | IQR | ||
| Air pollution | PM2.5 (μg/m3) | 10.16 (6.58) | 6.58 | 9.96 (3.62) | 4.18 | 9.94 (3.13) | 3.66 | 9.95 (3.13) | 3.66 | 10.00 (2.77) | 3.23 |
| Black carbon (μg/m3) | 0.75 (0.43) | 0.52 | 0.74 (0.23) | 0.33 | 0.73 (0.19) | 0.30 | 0.74 (0.18) | 0.26 | 0.74 (0.18) | 0.28 | |
| Particle number (counts/cm3, ×104) | 2.37 (1.27) | 1.82 | 2.31 (1.07) | 1.55 | 2.31 (1.05) | 1.49 | 2.31 (1.03) | 1.39 | 2.29 (1.02) | 1.39 | |
| CO (ppm, ×10−1) | 4.14 (2.49) | 2.88 | 4.19 (2.11) | 2.92 | 4.17 (2.03) | 2.76 | 4.17 (2.00) | 2.67 | 4.18 (1.98) | 2.65 | |
| NO2 (ppm, ×10−2) | 1.90 (0.65) | 0.81 | 1.91 (0.42) | 0.59 | 1.91 (0.39) | 0.58 | 1.91 (0.37) | 0.56 | 1.91 (0.36) | 0.56 | |
| O3 (ppm, ×10−2) | 2.38 (1.16) | 1.64 | 2.41 (0.85) | 1.31 | 2.42 (0.77) | 1.25 | 2.43 (0.74) | 1.22 | 2.45 (0.72) | 1.19 | |
| SO2 (ppm, ×10−3) | 4.13 (3.10) | 3.19 | 4.08 (2.39) | 2.97 | 4.08 (2.33) | 2.97 | 4.08 (2.14) | 2.87 | 4.07 (2.28) | 3.03 | |
| Temperature (°C) | 12.52 (8.63) | 13.55 | 12.60 (8.04) | 13.39 | 12.56 (7.94) | 13.30 | 11.89 (8.36) | 13.91 | 12.58 (7.91) | 13.58 | |
| Relative humidity (%) | 68.58 (15.60) | 23.98 | 68.41 (8.91) | 12.51 | 68.22 (6.49) | 9.24 | 67.26 (6.69) | 8.80 | 68.27 (5.94) | 8.11 | |
SD = standard deviation; IQR = interquartile range
Table 4.
Correlation matrix of 28-day average air pollution exposure, temperature, and relative humidity (Spearman coefficient)a
| Exposure | PM2.5 | Black carbon | Particle number | CO | NO2 | O3 | SO2 | Temperature | Relative humidity | |
|---|---|---|---|---|---|---|---|---|---|---|
| Air pollution | PM2.5 | 1 | ||||||||
| Black carbon | 0.716 | 1 | ||||||||
| Particle number | −0.017 | −0.154 | 1 | |||||||
| CO | −0.249 | 0.295 | 0.507 | 1 | ||||||
| NO2 | 0.371 | 0.304 | 0.687 | 0.736 | 1 | |||||
| O3 | 0.064 | −0.206 | −0.424 | −0.319 | −0.312 | 1 | ||||
| SO2 | 0.204 | 0.175 | 0.592 | 0.684 | 0.716 | −0.469 | 1 | |||
| Temperature | 0.323 | 0.376 | −0.738 | −0.362 | −0.448 | 0.533 | −0.638 | 1 | ||
| Relative humidity | 0.215 | 0.506 | −0.103 | −0.024 | −0.056 | −0.174 | −0.146 | 0.376 | 1 | |
Bolded coefficients have p < 0.05.
3.3. Associations of leukocyte distribution with air pollution, temperature, and relative humidity
After adjusting for all potential covariates, PM2.5, PN, CO, NO2, and SO2 showed strong associations with the proportions of at least two subtypes of leukocytes (Figures 1–5), while BC, temperature, and relative humidity only showed relatively weak associations with one cell type (Figures S1, S3, S4). O3 showed no associations with any leukocyte subtype (Figure S2). Corresponding estimations by IQR for 28-day average environmental exposures are summarized in Table 5. Tables S2–S5 show the estimates for 1-day, 7-day, 14-day, and 21-day average exposures, respectively.
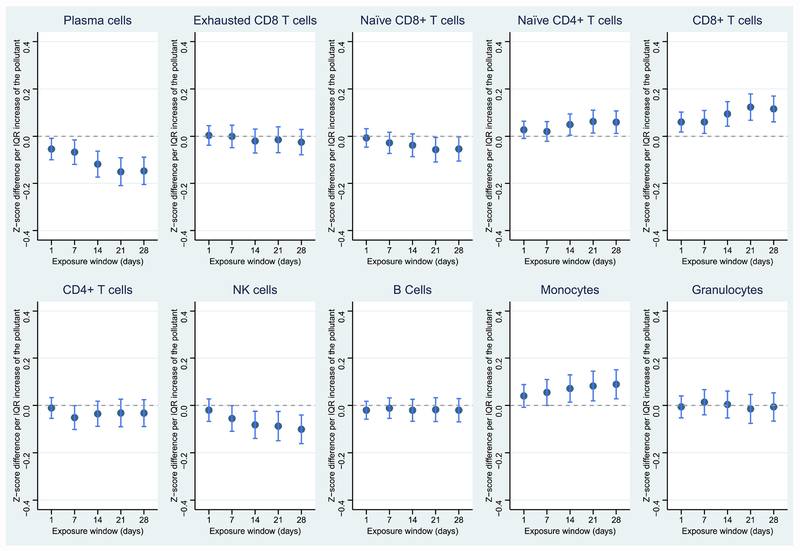
Figure 1.
Associations between PM2.5 and leukocyte distribution in the 28-day exposure window. Dark blue dots represent point estimates; light blue lines represent 95% confidence levels. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Figure 5.
Associations between SO2 and leukocyte distribution in the 28-day exposure window. Dark blue dots represent point estimates; light blue lines represent 95% confidence levels. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Table 5.
Associations of 28-day average air pollution exposure, temperature, and relative humidity with leukocyte distribution (z-score)a
| Exposure (unit) | Distribution of leukocytes [z-score, change per IQR (SE)] | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma cells | Exhausted CD8 T cells | Naïve CD8+ T cells | Naïve CD4+ T cells | CD8+ T cells | CD4+ T cells | NK cells | B cells | Monocytes | Granulocytes | ||
| Air pollution | PM2.5 (Mg/m3) | −0.147 (0.030)**** | −0.025 (0.028) | −0.054 (0.026)* | 0.059 (0.024)* | 0.115 (0.028)**** | −0.032 (0.029) | −0.101 (0.031)** | −0.020 (0.025) | 0.089 (0.031)** | −0.006 (0.030) |
| Black carbon (μg/m3) | −0.138 (0.047)** | 0.051 (0.044) | −0.001 (0.042) | 0.006 (0.039) | 0.085 (0.045) | −0.035 (0.046) | −0.073 (0.049) | −0.008 (0.040) | 0.088 (0.050) | −0.010 (0.048) | |
| Particle number (counts/cm3) | −0.257 (0.052)**** | 0.031 (0.049) | −0.166 (0.047)*** | −0.016 (0.044) | 0.145 (0.049)** | 0.051 (0.052) | −0.081 (0.054) | 0.053 (0.041) | 0.136 (0.058)* | −0.124 (0.054)* | |
| CO (ppm) | −0.031 (0.035) | 0.044 (0.032) | −0.011 (0.031) | −0.073 (0.029)* | −0.016 (0.033) | 0.047 (0.034) | 0.013 (0.035) | 0.094 (0.030)** | −0.018 (0.036) | −0.080 (0.035)* | |
| NO2 (ppm) | −0.149 (0.043)*** | 0.030 (0.039) | −0.093 (0.038)* | −0.056 (0.036) | 0.044 (0.040) | 0.024 (0.042) | −0.035 (0.044) | 0.064 (0.037) | 0.084 (0.044) | −0.090 (0.043)* | |
| O3 (ppm) | −0.063 (0.082) | −0.097 (0.075) | −0.105 (0.070) | 0.048 (0.065) | 0.061 (0.076) | 0.012 (0.079) | −0.017 (0.086) | 0.061 (0.067) | −0.149 (0.087) | −0.017 (0.084) | |
| SO2 (ppm) | −0.200 (0.044)**** | 0.057 (0.041) | −0.040 (0.039) | 0.00001 (0.037) | 0.185 (0.041)**** | 0.020 (0.043) | 0.047 (0.045) | 0.049 (0.038) | 0.044 (0.046) | −0.154 (0.045)*** | |
| Temperature (°C) | 0.073 (0.068) | −0.061 (0.063) | −0.055 (0.059) | 0.002 (0.055) | −0.150 (0.064)* | −0.019 (0.067) | −0.047 (0.071) | 0.056 (0.056) | 0.079 (0.073) | 0.028 (0.070) | |
| Relative humidity (%) | −0.004 (0.032) | 0.052 (0.029) | 0.032 (0.027) | 0.046 (0.025) | 0.078 (0.030)** | −0.033 (0.031) | −0.009 (0.033) | −0.049 (0.026) | 0.016 (0.034) | 0.012 (0.033) | |
p < 0.05;
p < 0.01;
p < 0.001;
p < 0.0001
Model adjusted for age (years), body mass index (BMI; underweight or normal weight/overweight/obese), smoking status (current/former/never smoker), alcohol intake (<2 drinks per day or ≥2 drinks per day), total cholesterol, triglycerides, high-density lipoprotein (HDL), systolic blood pressure (SBP), hypertension, stroke, coronary heart disease, diabetes and cancer diagnosed by physician (yes/no), and season of medical visit (spring/summer/fall/winter). For air pollution exposures, models also adjusted for corresponding temperature and relative humidity. Weather models mutually adjusted for humidity and temperature.
PM2.5 was negatively associated with plasma cells, naïve CD8+ T cells, and NK cells, and positively associated with naïve CD4+ T cells and CD8+ T cells (Figure 1, p < 0.05). An IQR-higher 28-day average exposure to PM2.5 was associated with 0.147-, 0.054- and 0.101-unit lower proportions (z-scored) of plasma cells, naïve CD8+ T cells, and NK cells, respectively, and 0.059- and 0.161-unit higher proportions (z-scored) of naïve CD4+ T cells and CD8+ T cells, respectively (Table 5). As an important component of PM2.5, BC showed similar but weaker associations with leukocyte distribution than PM2.5 and was only significantly associated with the reduced proportion of plasma cells (Figure S1). An IQR-higher 28-day average BC exposure was associated with a 0.138-unit lower proportion (z-scored) of plasma cells (Table 5).
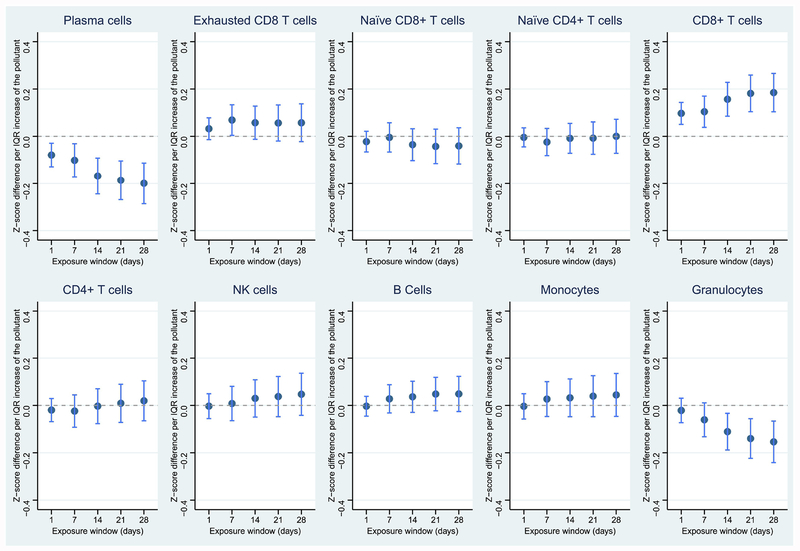
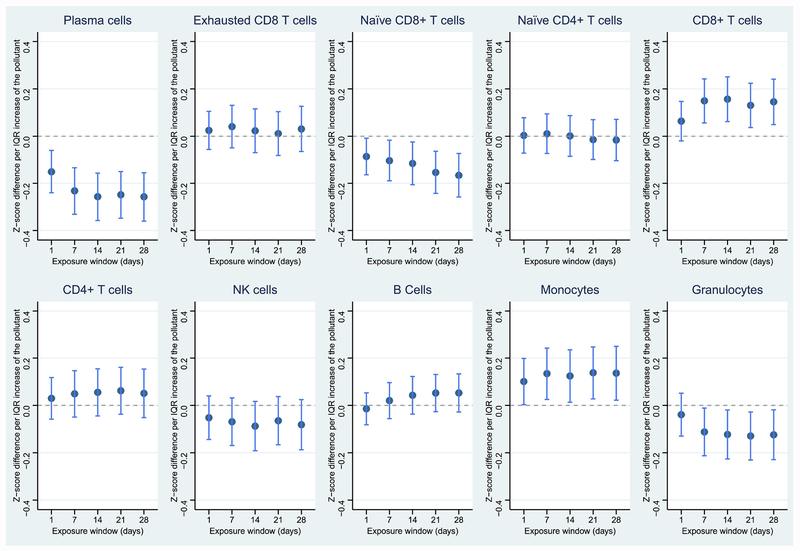
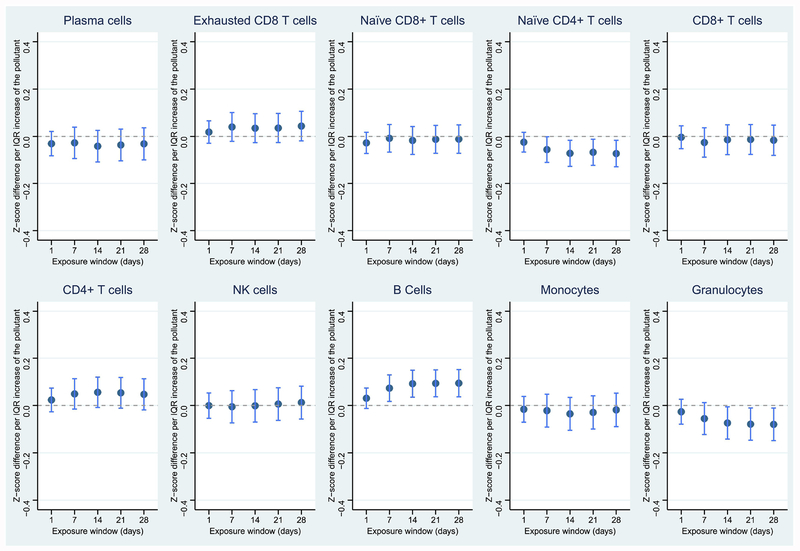
Similar to PM2.5, PN was also associated with reduced proportions of plasma cells and naïve CD8+ T cells, in addition to reduced total granulocytes. However, the proportions of CD8+ T cells and monocytes slightly increased in response to PN (Figure 2). Specifically, an IQR-higher 28-day average exposure to PN was associated with a 0.257-unit decreased proportion (z-scored) of plasma cells (Table 4). CO, NO2, and SO2 were also highly associated with reduced total granulocytes, and exposures to NO2, and SO2 were also associated with decreased proportions of plasma cells (Figures 3–5). CO was additionally associated with a decreased proportion of naïve CD4+ T cells and increased proportion of B cells. NO2 was associated with a decreased proportion of naïve CD8+ T cells, while SO2 was associated with an increased proportion of CD8 T cells. Temperature and relative humidity were only associated with CD8+ T cells, but in opposite directions (Figures S3 & S4). The effect of environmental exposures on leukocyte distribution became progressively stronger with increasing exposure length for several cell types and exposures (e.g., PM2.5 & plasma cells, PM2.5 & CD8+ T cells, PN & plasma cells).
Figure 2.
Associations between PN and leukocyte distribution in the 28-day exposure window. Dark blue dots represent point estimates; light blue lines represent 95% confidence levels. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Figure 3.
Associations between CO and leukocyte distribution in the 28-day exposure window. Dark blue dots represent point estimates; light blue lines represent 95% confidence levels. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Sensitivity analyses separately controlling for PM2.5 and IPW yielded essentially unchanged estimates for each environmental exposure from the main analysis models (data not shown), suggesting that results were not biased by loss to follow-up. There was no confounding effect of PM2.5 on the associations between other air pollutants and leukocyte distribution. In another sensitivity analysis with cell counts, most directions of effects of environmental exposures on cell counts were essentially unchanged or slightly attenuated compared to main results, and trends of the effects across 28-day windows were also similar (Figures S5–S13, detailed data not shown). This further suggests that the compositional nature of leukocyte distribution may influence our main findings to a limited extent. Finally, regarding linearity tests for temperature–cell proportions relationships, penalized splines estimated 1 degree of freedom for each exposure window, suggesting that the effect of temperature on cell proportions was linear.
4. Discussion
In the present study, we investigated the associations of air pollution, temperature, and relative humidity with leukocyte distribution estimated by DNA methylation profiles from 774 older males within 28 days before the blood draw. After adjusting for potential covariates, most air pollutants (except O3), temperature, and relative humidity were associated with the proportion of at least one subtype of leukocytes. Particularly, an IQR increase in the 28-day average PM2.5 level was associated with lower proportions of plasma cells, naïve CD8+ T cells, and NK cells and higher proportions of naïve CD4+ T cells and CD8+ T cells. We successfully demonstrated differences in leukocyte proportions in response to short-term air pollution exposures, temperature, and relative humidity using DNA methylation-based approaches.
To the best of our knowledge, this is the first study to use leukocyte distribution estimated by DNA methylation profiles to predict the impacts of air pollution and weather variations on human immune profiles. Since DNA methylation profiles can also be retrieved from different blood samples (e.g., fresh, frozen, and dry), our study further implies the possibility of assessing immune modulations in virtually any blood sample including archived samples previously precluded from such analysis. Further, our results suggest that the effect of environmental exposures on leukocyte distribution may accumulate over time with sustained exposure. This implies that medium-term (>one month) to long-term (up to one year) effects of those exposures on cell subtypes warrant further investigation. Additionally, we note the compositional nature of leukocytes warrants caution when interpreting causal associations of exposures with certain cell subtypes. Although we demonstrated that this had limited impact on our main findings, optimized DNA methylation-based methods to describe the full landscape of leukocytes are still needed to address this concern.
Our findings suggest that short-term environmental exposures may contribute to systemic inflammation and may partly explain the established connection between ambient PM exposure and the risks of cardiovascular disease and hypertension (Chi et al. 2016; Miller et al. 2007). The change in leukocytes may mechanistically linked atherosclerosis to inflammatory responses to PM (Adar et al. 2013; Brook and Rajagopalan 2010; Diez Roux et al. 2008; Kunzli et al. 2005; Perez et al. 2015) and may be related to the activation of adhesion and coagulation molecules after the PM inhalation, which could lead to increased leukocyte content (Baccarelli et al. 2007; Bind et al. 2012; O’Neill et al. 2007; CA Pope et al. 2016; Rückerl et al. 2006; Tsai et al. 2012). Our results share some similarities with previous studies of the impact of air pollution on blood cells using data assessed by automatic cell counters. We observed positive associations between levels of most air pollutants (except O3) and the proportion of monocytes. This is in line with Steenhof et al.’s report of increased monocytes numbers in response to PM10 and PM2.5 exposures among 31 healthy adults (Steenhof et al. 2014). This positive association can be explained by higher production of monocytes from bone marrow in response to the acute inflammation triggered by air pollution. As granulocytes are a major type of leukocytes, a large decrease in granulocytes may also drive a decrease in total leukocytes (Monie 2017). In our study, PN, NO2, and SO2 showed robust negative relations with the proportion of total granulocytes. These results are consistent with a study showing negative associations of neutrophils and total leukocytes with PN among 34 healthy adults (Zuurbier et al. 2011) and another study showing significantly decreased eosinophils after exposure to NO2 (Steenhof et al. 2014). However, other previous studies have yielded different results. For instance, we did not find an association between total proportion of granulocytes and PM2.5, whereas Frampton et al. (Frampton et al. 2004) found a decreased proportion of basophils and increased proportion of neutrophils among 28 people in response to PM2.5 exposure. Riediker et al. (Riediker et al. 2004) also found an increased proportion of neutrophils in response to PM2.5 among 9 healthy men in four successive days.
Unlike most previous investigations, which mostly focused on total blood lymphocytes measured only by blood cell counters, our study examined associations of environmental exposures with each subtype of lymphocytes and plasma cells. We found that PM2.5, BC, PN, NO2, and SO2 showed strong negative associations with the proportion of plasma cells, and most pollutants (except NO2) were associated with an increased proportion of CD8+ T cells. Also, the five air pollutants showed much weaker associations with the proportion of B cells. Therefore, we speculate that these air pollutants might act in the lung and then elicit a systemic inflammatory response by triggering upregulation of T-cell-mediated immunity, rather than humoral immunity mediated by macromolecules (e.g., antibodies released by plasma cells).
Increased T cells induced by air pollution have been observed in different populations and tissues. For instance, Salvi et al. (Salvi et al. 1999) found increased CD8+ T cells in bronchial biopsies of healthy volunteers obtained six hours after the exposure to diesel exhaust, a major contributor to PM pollution. Ma et al. (Ma et al. 2017) also observed elevated cytotoxicity in CD8+ T cells in response to PM2.5 exposure in peripheral blood cells of healthy individuals. Similar patterns also appeared in cord blood samples collected from pregnant women at early gestation, in which Herr et al. found significant associations of PM2.5 exposure with lower proportions of CD19+ B cell and NK cells (Herr et al. 2010). However, all these reports showed higher CD4+ cells associated with PM2.5, which we did not find in our study. Further, our study was the first to describe an association of temperature and humidity with CD8+ T cells, although previous studies have found that outdoor temperature and relative humidity are associated with global DNA methylation and gene-specific methylation in blood cells (Bind et al. 2014b; Bind et al. 2016; Lim et al. 2017).
Because leukocyte distribution was estimated by reference-based methods using DNA methylation data, a critical question is whether and to what extent the epigenetic patterns used to estimate leukocyte distribution mediate the associations between environmental exposures and change in blood cells. Even though previous epigenome-wide association studies (EWASs) have identified several CpG sites associated with air pollution, including PM2.5 (Panni et al. 2016) and NO2 (Gruzieva et al. 2017), and with leukocyte proportions, such as CD4+ T cells (Martino et al. 2014), no CpG sites associated with both air pollution and leukocytes have been identified. Since leukocyte distribution is indeed a critical factor that affects the results of EWAS based on whole blood samples — as it may reflect indirect effects of any external exposures on measured DNA methylation profiles—it is also important to understand the biological connections of the CpG sites used in Houseman et al.’s and Horvath’s algorithms with leukocytes. Such CpG sites may indicate genomic areas that are not only related to hematopoiesis and immune cell development (Teschendorff and Zheng 2017) but also may be sensitive to environmental exposures (Bauer et al. 2012; CA Pope, 3rd et al. 2016). Future multidisciplinary studies are required to establish the roles of CpG sites used to estimate leukocyte distributions in the biological connections of air pollution and weather variations with systemic inflammatory responses.
Strengths of the present study include measurements of DNA methylation profiles, multiple air pollutants, and weather variations in a longitudinal setting. Several limitations should also be noted when interpreting these results. First, the DNA methylation-based methods we used remain at the population level and cannot provide more detail about the change in immune cells at the individual level or account for cell-cell interactions, which might come from minor leukocytes comprising <5% of total leukocytes (Teschendorff and Zheng 2017; Titus et al. 2017). Second, each participant’s real exposure to some air pollutants might differ from the average city level that we used in our analyses, as exposure also depends on the time spent at home, rates of penetration of ambient particles into the house, and presence of indoor sources of particles. However, we noted that it is more likely that NAS participants spent a large part of their days at home. Exposure misclassification is likely to be non-differential and to bias results toward the null, rather than causing the observed associations (Kioumourtzoglou et al. 2014). Additionally, multiple comparisons between cell types and exposures may also potentially induce findings by chance, but we observed consistencies of the significant associations between certain exposures and cell subtypes across each time window. Together with similar findings from previous reports based on data from cell counters, these results indicate that our findings were not incidental. Lastly, the selected participants of this study were Caucasians and older males, which limits generalizability of our results to other racial/ethnic groups and to women.
5. Conclusions
In conclusion, our study represent a successful attempt to use DNA methylation data to assess the impact of air pollution on human immune profiles. Our findings confirm that short-term ambient exposure to air pollutants, temperature, and relative humidity may be associated with subclinical, but epidemiologically relevant, inflammatory responses across the population and that these associations may be captured using DNA methylation data. Our study also demonstrates that DNA methylation markers are highly likely to be implemented to describe ambient exposure impacts. Future interdisciplinary studies with population-based mediation analyses and explorations of biological functions will provide novel insights into epigenetic mechanisms (e.g., DNA methylation) underlying the impact of environmental exposures on the human immune system and provide new hints for large-scale immunological studies of environmental health.
Supplementary Material
Figure 4.
Associations between NO2 and leukocyte distribution in the 28-day exposure window. Dark blue dots represent point estimates; light blue lines represent 95% confidence levels. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Highlights.
We estimated leukocyte distribution using DNA methylation data.
Environmental exposures are found to be associated with the change of estimated leukocyte distribution.
Changes in leukocyte distribution varied by the exposures.
Epigenetic patterns can assess the influences of environmental exposures on human immune profiles.
Acknowledgments:
The authors would like to thank all Normative Aging Study participants and Dr. Sarah Rasmussen for sharing the codes adapted for inverse probability weighting.
Sources of funding: This work was supported by the National Institute of Environmental Health Sciences (grants P30ES009089, R01ES021733, R01ES025225, and R01ES027747). The VA Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the U.S. Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center in Boston, MA. JCN is supported by an NIH/NIA Ruth L. Kirschstein National Research Service Award (1 F31AG056124–01A1).
Abbreviations
- PM2.5
fine particulate matter with diameter <2.5 μm
- NK
natural killers
- BMI
body mass index
- HDL
high-density lipoprotein
- SBP
systolic blood pressure
- DBP
diastolic blood pressure
- CpG
cytosine-phosphate-guanine
- IPW
inverse probability weighting
- NAS
Normative Aging Study
- BC
black carbon
- PN
particle number
- PPM
parts per million
- IQR
interquartile range
- SD
standard deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have no conflict of interest to disclose.
References
- Adar SD, Sheppard L, Vedal S, Polak JF, Sampson PD, Diez Roux AV, et al. 2013. Fine particulate air pollution and the progression of carotid intima-medial thickness: A prospective cohort study from the multi-ethnic study of atherosclerosis and air pollution. PLOS Medicine 10:e1001430. doi: 10.1371/journal.pmed.1001430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Zanobetti A, Martinelli I, Grillo P, Hou L, Giacomini S, et al. 2007. Effects of exposure to air pollution on blood coagulation. J Thromb Haemost 5:252–260. doi: 10.1111/j.1538-7836.2007.02300.x [DOI] [PubMed] [Google Scholar]
- Bauer RN, Diaz-Sanchez D, Jaspers I. 2012. Effects of air pollutants on innate immunity: The role of toll-like receptors and nucleotide-binding oligomerization domain-like receptors. J Allergy Clin Immunol 129:14–24; quiz 25–16. doi: 10.1016/j.jaci.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell B, Rose CL, Damon A. 1972. The Normative Aging Study: An interdisciplinary and longitudinal study of health and aging. Int J Aging Hum Dev 3:5–17. [Google Scholar]
- Bind M-A, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, et al. 2012. Air pollution and markers of coagulation, inflammation, and endothelial function: Associations and epigene-environment interactions in an elderly cohort. Epidemiology 23:332–340. doi: 10.1097/EDE.0b013e31824523f0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bind MA, Lepeule J, Zanobetti A, Gasparrini A, Baccarelli A, Coull BA, et al. 2014a. Air pollution and gene-specific methylation in the normative aging study: Association, effect modification, and mediation analysis. Epigenetics 9:448–458. doi: 10.4161/epi.27584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bind MA, Zanobetti A, Gasparrini A, Peters A, Coull B, Baccarelli A, et al. 2014b. Effects of temperature and relative humidity on DNA methylation. Epidemiology 25:561–569. doi: 10.1097/EDE.0000000000000120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bind MA, Coull BA, Baccarelli A, Tarantini L, Cantone L, Vokonas P, et al. 2016. Distributional changes in gene-specific methylation associated with temperature. Environ Res 150:38–46. doi: 10.1016/j.envres.2016.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A. 2010. Environmental epigenetics. Heredity (Edinb) 105:105–112. doi: 10.1038/hdy.2010.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook RD, Rajagopalan S. 2010. Particulate matter air pollution and atherosclerosis. Current Atheroscler Rep 12:291–300. doi: 10.1007/s11883-010-0122-7 [DOI] [PubMed] [Google Scholar]
- Brook RD, Rajagopalan S, Pope CA 3rd, Brook JR, Bhatnagar A, Diez-Roux AV, et al. 2010. Particulate matter air pollution and cardiovascular disease: An update to the scientific statement from the American Heart Association. Circulation 121:2331–2378. doi: 10.1161/CIR.0b013e3181dbece1 [DOI] [PubMed] [Google Scholar]
- Bruske I, Hampel R, Socher MM, Ruckerl R, Schneider A, Heinrich J, et al. 2010. Impact of ambient air pollution on the differential white blood cell count in patients with chronic pulmonary disease. Inhal Toxicol 22:245–252. doi: 10.3109/08958370903207274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cembrowski GS, Clarke G. 2015. Quality control of automated cell counters. Clin Lab Med 35:59–71. doi: 10.1016/j.cll.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Chi GC, Hajat A, Bird CE, Cullen MR, Griffin BA, Miller KA, et al. 2016. Individual and neighborhood socioeconomic status and the association between air pollution and cardiovascular disease. Environ Health Perspect 124:1840–1847. doi: 10.1289/EHP199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang KJ, Yan YH, Chiu SY, Cheng TJ. 2011. Long-term air pollution exposure and risk factors for cardiovascular diseases among the elderly in taiwan. Occup Environ Med 68:64–68. doi: 10.1136/oem.2009.052704 [DOI] [PubMed] [Google Scholar]
- Dauchet L, Hulo S, Cherot-Kornobis N, Matran R, Amouyel P, Edme JL, et al. 2018. Short-term exposure to air pollution: Associations with lung function and inflammatory markers in non-smoking, healthy adults. Environ Int 121:610–619. doi: 10.1016/j.envint.2018.09.036 [DOI] [PubMed] [Google Scholar]
- Diez Roux AV, Auchincloss AH, Franklin TG, Raghunathan T, Barr RG, Kaufman J, et al. 2008. Long-term exposure to ambient particulate matter and prevalence of subclinical atherosclerosis in the multi-ethnic study of atherosclerosis. Am J Epidemiol 167:667–675. doi: 10.1093/aje/kwm359 [DOI] [PubMed] [Google Scholar]
- Dugue PA, English DR, MacInnis RJ, Jung CH, Bassett JK, FitzGerald LM, et al. 2016. Reliability of DNA methylation measures from dried blood spots and mononuclear cells using the humanmethylation450k beadarray. Sci Rep 6:30317. doi: 10.1038/srep30317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egozcue JJ, Pawlowsky-Glahn V, Mateu-Figueras G, Barcelo-Vidal C. 2003. Isometric logratio transformations for compositional data analysis. Math Geol 35:279–300. doi:Doi 10.1023/A:1023818214614 [DOI] [Google Scholar]
- Frampton MW, Utell MJ, Zareba W, Oberdorster G, Cox C, Huang LS, et al. 2004. Effects of exposure to ultrafine carbon particles in healthy subjects and subjects with asthma. Res Rep Health Eff Inst:1–47; discussion 49–63. [PubMed] [Google Scholar]
- Gao X, Colicino E, Shen J, Just AC, Nwanaji-Enwerem JC, Coull B, et al. 2018. Accelerated DNA methylation age and the use of antihypertensive medication among older adults. Aging (Albany NY) 10:3210–3228. doi: 10.18632/aging.101626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. 2007. Epigenetics: A landscape takes shape. Cell 128:635–638. doi: 10.1016/j.cell.2007.02.006 [DOI] [PubMed] [Google Scholar]
- Gruzieva O, Xu CJ, Breton CV, Annesi-Maesano I, Anto JM, Auffray C, et al. 2017. Epigenome-wide meta-analysis of methylation in children related to prenatal no2 air pollution exposure. Environ Health Perspect 125:104–110. doi: 10.1289/EHP36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halonen JI, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. 2010. Associations between outdoor temperature and markers of inflammation: A cohort study. Environ Health 9:42. doi: 10.1186/1476-069X-9-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr CE, Dostal M, Ghosh R, Ashwood P, Lipsett M, Pinkerton KE, et al. 2010. Air pollution exposure during critical time periods in gestation and alterations in cord blood lymphocyte distribution: A cohort of livebirths. Environ Health 9:46. doi: 10.1186/1476-069X-9-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz-Picciotto I, Herr CE, Yap PS, Dostal M, Shumway RH, Ashwood P, et al. 2005. Air pollution and lymphocyte phenotype proportions in cord blood. Environ Health Perspect 113:1391–1398. doi: 10.1289/ehp.7610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S 2013. DNA methylation age of human tissues and cell types. Genome Biol 14:R115. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, et al. 2016. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol 17:171. doi: 10.1186/s13059-016-1030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86. doi: 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari DA, Sen GL, Rinn JL. 2010. DNA methylation and epigenetic control of cellular differentiation. Cell Cycle 9:3880–3883. doi: 10.4161/cc.9.19.13385 [DOI] [PubMed] [Google Scholar]
- Kioumourtzoglou MA, Spiegelman D, Szpiro AA, Sheppard L, Kaufman JD, Yanosky JD, et al. 2014. Exposure measurement error in PM2.5 health effects studies: A pooled analysis of eight personal exposure validation studies. Environ Health 13:2. doi: 10.1186/1476-069X-13-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koken PJ, Piver WT, Ye F, Elixhauser A, Olsen LM, Portier CJ. 2003. Temperature, air pollution, and hospitalization for cardiovascular diseases among elderly people in Denver. Environ Health Perspect 111:1312–1317. doi: 10.1289/ehp.5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzli N, Jerrett M, Mack WJ, Beckerman B, LaBree L, Gilliland F, et al. 2005. Ambient air pollution and atherosclerosis in Los Angeles. Environ Health Perspect 113. doi: 10.1289/ehp.7523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YH, Han C, Bae S, Hong YC. 2017. Modulation of blood pressure in response to low ambient temperature: The role of DNA methylation of zinc finger genes. Environ Res 153:106–111. doi: 10.1016/j.envres.2016.11.019 [DOI] [PubMed] [Google Scholar]
- Ma QY, Huang DY, Zhang HJ, Wang S, Chen XF. 2017. Exposure to particulate matter 2.5 (PM2.5) induced macrophage-dependent inflammation, characterized by increased Th1/Th17 cytokine secretion and cytotoxicity. Int Immunopharmacol 50:139–145. doi: 10.1016/j.intimp.2017.06.019 [DOI] [PubMed] [Google Scholar]
- Martino D, Joo JE, Sexton-Oates A, Dang T, Allen K, Saffery R, et al. 2014. Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with ige-mediated food allergy. Epigenetics 9:998–1006. doi: 10.4161/epi.28945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AJ, Kubzansky LD, Coull BA, Kloog I, Koutrakis P, Sparrow D, et al. 2015. Associations between air pollution and perceived stress: The Veterans Administration Normative Aging Study. Environ Health 14:10. doi: 10.1186/1476-069X-14-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KA, Siscovick DS, Sheppard L, Shepherd K, Sullivan JH, Anderson GL, et al. 2007. Long-term exposure to air pollution and incidence of cardiovascular events in women. New Engl J Med 356:447–458. doi: 10.1056/NEJMoa054409 [DOI] [PubMed] [Google Scholar]
- Monie TP. 2017. Section 1 - a snapshot of the innate immune system In: The innate immune system, (Monie TP, ed):Academic Press, 1–40. [Google Scholar]
- Mordukhovich I, Coull B, Kloog I, Koutrakis P, Vokonas P, Schwartz J. 2015. Exposure to sub-chronic and long-term particulate air pollution and heart rate variability in an elderly cohort: The Normative Aging Study. Environ Health 14:87. doi: 10.1186/s12940-015-0074-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyhan MM, Coull BA, Blomberg AJ, Vieira CLZ, Garshick E, Aba A, et al. 2018. Associations between ambient particle radioactivity and blood pressure: The NAS (Normative Aging Study). J Am Heart Assoc 7. doi: 10.1161/JAHA.117.008245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill MS, Veves A, Sarnat JA, Zanobetti A, Gold DR, Economides PA, et al. 2007. Air pollution and inflammation in type 2 diabetes: A mechanism for susceptibility. Occup Environ Med 64:373–379. doi: 10.1136/oem.2006.030023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panni T, Mehta AJ, Schwartz JD, Baccarelli AA, Just AC, Wolf K, et al. 2016. Genome-wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the Normative Aging Study. Environ Health Perspect 124:983–990. doi: 10.1289/ehp.1509966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez L, Wolf K, Hennig F, Penell J, Basagaña X, Foraster M, et al. 2015. Air pollution and atherosclerosis: A cross-sectional analysis of four european cohort studies in the escape study. Environ Health Perspect 123:597–605. doi: 10.1289/ehp.1307711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CA, Bhatnagar A, McCracken JP, Abplanalp W, Conklin DJ, O’Toole T. 2016. Exposure to fine particulate air pollution is associated with endothelial injury and systemic inflammation. Circ Res 119:1204–1214. doi: 10.1161/circresaha.116.309279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CA 3rd, Hansen ML, Long RW, Nielsen KR, Eatough NL, Wilson WE, et al. 2004. Ambient particulate air pollution, heart rate variability, and blood markers of inflammation in a panel of elderly subjects. Environ Health Perspect 112:339–345. doi: 10.1289/ehp.6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich DQ, Kipen HM, Huang W, Wang G, Wang Y, Zhu P, et al. 2012. Association between changes in air pollution levels during the beijing olympics and biomarkers of inflammation and thrombosis in healthy young adults. JAMA 307:2068–2078. doi: 10.1001/jama.2012.3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riediker M, Cascio WE, Griggs TR, Herbst MC, Bromberg PA, Neas L, et al. 2004. Particulate matter exposure in cars is associated with cardiovascular effects in healthy young men. Am J Respir Crit Care Med 169:934–940. doi: 10.1164/rccm.200310-1463OC [DOI] [PubMed] [Google Scholar]
- Rückerl R, Ibald-Mulli A, Koenig W, Schneider A, Woelke G, Cyrys J, et al. 2006. Air pollution and markers of inflammation and coagulation in patients with coronary heart disease. Am J Respir Crit Care Med 173:432–441. doi: 10.1164/rccm.200507-1123OC [DOI] [PubMed] [Google Scholar]
- Salvi S, Blomberg A, Rudell B, Kelly F, Sandstrom T, Holgate ST, et al. 1999. Acute inflammatory responses in the airways and peripheral blood after short-term exposure to diesel exhaust in healthy human volunteers. Am J Respir Crit Care Med 159:702–709. doi: 10.1164/ajrccm.159.3.9709083 [DOI] [PubMed] [Google Scholar]
- Schwartz J 2001. Air pollution and blood markers of cardiovascular risk. Environ Health Perspect 109 Suppl 3:405–409. doi: 10.1289/ehp.01109s3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J, Samet JM, Patz JA. 2004. Hospital admissions for heart disease: The effects of temperature and humidity. Epidemiology 15:755–761. [DOI] [PubMed] [Google Scholar]
- Seaman SR, White IR. 2013. Review of inverse probability weighting for dealing with missing data. Stat Methods Med Res 22:278–295. doi: 10.1177/0962280210395740 [DOI] [PubMed] [Google Scholar]
- Steenhof M, Janssen NA, Strak M, Hoek G, Gosens I, Mudway IS, et al. 2014. Air pollution exposure affects circulating white blood cell counts in healthy subjects: The role of particle composition, oxidative potential and gaseous pollutants - the RAPTES project. Inhal Toxicol 26:141–165. doi: 10.3109/08958378.2013.861884 [DOI] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. 2013. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29:189–196. doi: 10.1093/bioinformatics/bts680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Zheng SC. 2017. Cell-type deconvolution in epigenome-wide association studies: A review and recommendations. Epigenomics 9:757–768. doi: 10.2217/epi-2016-0153 [DOI] [PubMed] [Google Scholar]
- Titus AJ, Gallimore RM, Salas LA, Christensen BC. 2017. Cell-type deconvolution from DNA methylation: A review of recent applications. Hum Mol Genet 26:R216–R224. doi: 10.1093/hmg/ddx275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai D-H, Amyai N, Marques-Vidal P, Wang J-L, Riediker M, Mooser V, et al. 2012. Effects of particulate matter on inflammatory markers in the general adult population. Part Fibre Toxicol 9:24–24. doi: 10.1186/1743-8977-9-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden SF, Hogg JC. 2002. Systemic inflammatory response induced by particulate matter air pollution: The importance of bone-marrow stimulation. J Toxicol Environ Health A 65:1597–1613. doi: 10.1080/00984100290071685 [DOI] [PubMed] [Google Scholar]
- Viehmann A, Hertel S, Fuks K, Eisele L, Moebus S, Mohlenkamp S, et al. 2015. Long-term residential exposure to urban air pollution, and repeated measures of systemic blood markers of inflammation and coagulation. Occup Environ Med 72:656–663. doi: 10.1136/oemed-2014-102800 [DOI] [PubMed] [Google Scholar]
- Zuurbier M, Hoek G, Oldenwening M, Meliefste K, Krop E, van den Hazel P, et al. 2011. In-traffic air pollution exposure and CC16, blood coagulation, and inflammation markers in healthy adults. Environ Health Perspect 119:1384–1389. doi: 10.1289/ehp.1003151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.