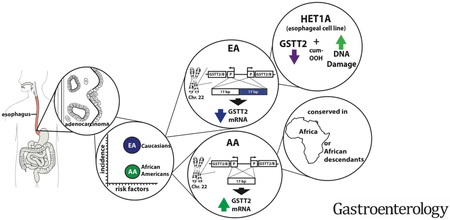
Abstract
Background & Aims:
African Americans and European Americans have a similar prevalence of gastroesophageal reflux disease (GERD) yet esophageal adenocarcinoma (EAC) disproportionately affects European Americans. We investigated whether the esophageal squamous mucosa of African Americans has features that protect against GERD-induced damage, compared with European Americans.
Methods:
We performed transcriptional profile analysis of esophageal squamous mucosa tissues from 20 African American and 20 European Americans (24 with no disease and 16 with Barrett’s esophagus and/or EAC). We confirmed our findings in a cohort of 56 patients and analyzed DNA samples from patients to identify associated variants. Observations were validated using matched genomic sequence and expression data from lymphoblasts from the 1000 Genomes Project. A panel of esophageal samples from African American and European American subjects were used to confirm allele-related differences in protein levels. The esophageal squamous-derived cell line Het-1A and a rat esophagogastroduodenal anastomosis model for reflux-generated esophageal damage were used to investigate the effects of the DNA-damaging agent cumene-hydroperoxide (cum-OOH) and a chemopreventive cranberry proanthocyanidin (C-PAC) extract, respectively, on levels of protein and mRNA.
Results:
We found significantly higher levels of glutathione S-transferase theta 2 (GSTT2) mRNA in squamous mucosa from African Americans compared with European Americans and associated these with variants within the GSTT2 locus in African Americans. We confirmed that 2 previously identified genomic variants at the GSTT2 locus, a 37-kb deletion and a 17-bp promoter duplication, reduce expression of GSTT2 in tissues from European Americans. The non-duplicated 17-bp promoter was more common in tissue samples from African descendant populations. GSTT2 protected Het-1A esophageal squamous cells from cum-OOH–induced DNA damage. Addition of C-PAC increased GSTT2 expression in Het-1A cells incubated with cum-OOH and in rats with reflux-induced esophageal damage. C-PAC also reduced levels of DNA damage in reflux-exposed rat esophagi, as observed by reduced levels of phospho-H2A histone family member X.
Conclusions:
We found GSTT2 to protect esophageal squamous cells against DNA damage from genotoxic stress and that GSTT2 expression can be induced by C-PAC. Increased levels of GSTT2 in esophageal tissues of African Americans might protect them from GERD-induced damage and contribute to the low incidence of EAC in this population.
Keywords: racial disparities, BE, glutathione transferase, cranberry proanthocyanidins
Graphical Abstract:
INTRODUCTION
EAC often arises from metaplastic mucosa called Barrett’s esophagus (BE), a predisposing condition in which the normal squamous epithelium of the esophagus (NE) is replaced by columnar intestinal-type epithelium. There are substantial but not uniform data to suggest that continued esophageal reflux leads to chronic esophagitis, which then acts as a trigger for the formation of BE1. To date, the characteristics that trigger the formation of BE and its exact cell of origin remain unclear. It has been extensively observed, however, that both BE and EAC primarily affect European Americans (EA) but not African Americans (AA), despite both populations having similar risk factors including obesity, gastroesophageal reflux disease (GERD), and smoking2, 3. More importantly, it is known that the primary risk factors including GERD and obesity4 do not differ between the two racial groups5-7. Nevertheless, AA demonstrate half the incidence of erosive esophagitis when compared to EA among subjects reporting weekly heartburn or reflux (24% vs 50%, P =0.03)5 and about a quarter the incidence of EAC8. To address this question, we sought to investigate the transcriptional profile of the normal squamous mucosa of individuals of both populations (EA vs. AA). Further, we sampled the squamous mucosa from both disease-free (no history of either BE or EAC) individuals and those with a disease history (presence of BE and/or development of EAC) from both racial groups. Our goal was to identify differentially-expressed genes and molecular pathways that might better protect the esophageal mucosa and thus reduce the risk of disease progression.
Here we report that a detoxifying enzyme responsible for inactivating reactive oxygen species and reducing DNA damage9, glutathione-s-transferase theta 2 (GSTT2, GSTT2), showed significantly higher average expression in esophageal mucosa of AA relative to EA. Moreover, we report on two underlying genomic events that negatively affect GSTT2 mRNA expression (a 37kb deletion and a 17bp promoter duplication in the GSTT2 locus) which are highly over-represented among individuals of European descent. Conversely, promoter non-duplication is associated with higher levels of GSTT2 mRNA and is proportionately more conserved in African and African descendant populations. In addition, we show that experimental reduction of the GSTT2 protein in an esophageal squamous cell line increases its susceptibility to DNA damage under genotoxic stress. Together, these observations suggest that increased GSTT2 expression may protect against esophageal mucosal damage caused by GERD and may be one factor that underlies the low incidence of BE and hence EAC in AA. Agents that increase GSTT2 in the esophagus could provide a chemopreventative strategy for patients at risk for BE and EAC.
MATERIALS AND METHODS
Biopsies of the squamous esophagus
Histologically-normal biopsies of esophageal squamous epithelium were collected from consented individuals without a history of Barrett’s esophagus or erosive esophagitis (population controls) of either AA or EA ethnicity who underwent a research upper endoscopy between 2008 and 2016 at the time of a scheduled, screening colonoscopy performed at the University of Michigan Health System, the University of North Carolina, or the Ann Arbor Veterans Affairs Medical Center. Samples from the normal squamous epithelium (NE) of AA and EA individuals who developed BE and/or EAC were collected from Case Western University, Johns Hopkins University, and the University of Michigan Health System between 1991 and 2004 using protocols approved by their respective institutional review boards (IRB). Racial group determination (AA or EA), age, smoking status, BMI (>25 classified as overweight), GERD status and BE/EAC status, as summarized in Supplemental Table S1, were based on self-reporting questionnaires completed by each recruited participant. None of the EAC patients from whom normal (NE) biopsies were obtained had received preoperative radiation or chemotherapy. All specimens were collected fresh and frozen in liquid nitrogen and stored at −80°C until use.
mRNA extraction and Affymetrix expression analysis
RNA was isolated from the normal esophageal squamous mucosa from individuals self-identified as either EA or AA, as previously described10. mRNA extraction and Human Gene ST 2.1 array (Affymetrix, Santa Clara, CA) hybridizations were performed as previously described10. Expression values for each gene were determined using the robust multi-array average (RMA) method11 in the Bioconductor package12 of the R statistical platform. Analyses were restricted to the 24,909 coding and non-coding genes for which annotation details were available. We fitted ANOVA models with terms for the four groups: AA without Barrett’s (AA-NE; n=12), AA with Barrett’s (AA-NE:BE; n=8), EA without Barrett’s (EA-NE; n=12), and EA with Barrett’s (EA-NE:BE; n=8). P values were determined and calculated for four pairwise comparisons as shown in Supplemental Table S2. We estimated false discovery rates by analyzing 10,000 data sets in which the sample labels were randomly permuted, and averaged the number of qualifying probe-sets across the 10,000 data sets.
Data availability
Both normalized and raw expression data for this experiment were deposited into the Gene Expression Omnibus (GEO series GSE77563).
GSTT2 and GSTT2B probesets
Affymetrix probesets do not adequately distinguish between GSTT2 and GSTT2B transcripts, therefore we present mRNA expression as GSTT2/2B and only report the more complete 16933088 probeset which covers all 5 exons, rather than 16928115 which only has probes on exons 2 and 4. The regulatory and coding sequences of RefSeq genes GSTT2 (hg38:chr22:23980123-23983911, RefSeq: NM_000854) and GSTT2B (hg38: chr22:23957414-23961186, RefSeq: NM_001080843) are indistinguishable.
qRT-PCR validation
An extended normal squamous tissue cohort, consisting of the 12 AA and 12 EA examined by ST 2.1 array, plus an additional 9 AA, 9 EA, and one AA:BE were for quantitative real-time polymerase chain reaction (qRT-PCR) validation of selected gene transcripts. Total RNA was converted to cDNA and used for qRT-PCR as previously described10, with details of qRT-PCR primers and analysis provided in Supplementary Methods.
GSTT2B deletion genotype analysis
Genomic DNA was extracted from normal squamous biopsy material as described previously13 and GSTT2B genotypes were resolved using three-primer PCR as previously described14. The PCR reaction and electrophoretic distinction of alleles are detailed in Supplementary Methods.
GSTT2/2B promoter duplication analysis
The primers and PCR protocol from Marotta et al 15 and 2% agarose gels were used to genotype the 17bp promotor duplication status. Two examples of each promoter genotype were confirmed by Sanger sequencing as detailed in Supplementary Methods.
1000 Genomes and HapMap data analysis for GSTT2B and GSTT2 promoter genotypes
The 17bp promoter duplication is not present in the 1000 Genomes (1000G) set, primarily due to the duplicative nature of the variant, but exists in the raw alignment data. After manual alignment, the duplication appears as a deletion relative to the reference sequence (GRCh37). Subsequently, we could not use the 1000G variant calls and therefore counted raw reads containing either the duplicated allele or non-duplicated allele sequence:
Duplicated:
GTGCACGAAGTGGGAGCTCCCGCTGTCTGGCAGCTCCCGCTGTCTGGCAG
Non-duplicated:
GTGCACGAAGTGGGAGCTCCCGCTGTCTGGCAGCAGCTGCTCTGCAGGGG
We enumerated sequences that mapped around both paralogs, to take into account alignments that might align to one or another. We extracted RKPM expression data for 110 lymphoblast cell lines from the tabulated GEUVADIS RNAseq data (Express Array ID: E-GEUV-1)16. These samples (39 AFR and 77 EUR) match those for whom raw copy number variation data were available. We then used the non-parametric Kruskal-Wallis statistic to investigate the frequency of GSTT2 copy number variations and expression levels within these matched (RNA/DNA) samples.
Phospho-H2A histone family member X (γ-H2AX) detection in cumene-hydroperoxide (cum-OOH) treated of Het-1A and HeLa cell lines
Detailed methods for siRNA transfection and Western blotting are provided in Supplementary Methods. Briefly, cultured Het-1A (immortalized normal esophageal squamous mucosa) and HeLa (squamous cervical cancer) were exposed to mock control (RNAimax; Thermo Fisher), scramble non-target (NT) siRNA or four commercial siRNAs targeting GSTT2 (#LQ-011181-00-0005, Dharmacon, Lafayette, CO) at a concentration of 10nm for 48hrs before harvesting. Western blot lysates were prepared at 20μg total, resolved using commercial SDS 4-12% gradient gels then transferred to pre-activated nitrocellulose. Following blocking, membranes were incubated overnight at 4°C with primary GSTT2 (#514667; Santa Cruz, Dallas, TX; 1:500) or γ-H2AX (#05-636; Millipore, Burlington, MA; 1/1500) antibodies. Membranes were then washed and exposed to secondary antibodies for 1hr incubations at room temperature (RT) prior to imaging with ECL and X-ray film. Duplicate plates were run each day and the experiment was repeated twice.
Immunofluorescence analysis of γ-H2AX
After methanol fixation, PBS was removed from the coverslips containing fixed Het-1A or HeLa cells and protein-DNA cross-links were formed with 10% phosphate-buffered formalin for 20min at room temperature (RT). Cells attached to these coverslips were washed, permeabilized with 100% cold methanol (−20°C for 5min), incubated for 1hr in blocking buffer before overnight exposure to primary antibody (γ-H2AX; 1/1500 or GSTT2; 1:500) at 4°C in a humidified chamber. Coverslips were rinsed before 1hr incubation with secondary antibody at RT, washed and mounted onto slides using DAPI mounting solution (#P36935, Thermo Fisher). The ratio of γ-H2AX positive cells verse total viable (DAPI stained) nuclei was averaged across 2 replicate plates for each experiment (n=2), with both cell lines (n=2), siRNA treatments (control, siRNA05, siRNA06) and the presence/absence of cum-OOH (n=2) compared in a four-way ANOVA model.
Evaluation of GSTT2 levels in the Het-1A esophageal cell line treated with cranberry proanthocyanidins (C-PAC)
Het-1A (150,000) cells were seeded in triplicate 35mm tissue culture treated dishes (Corning; ThermoFisher) and adhered overnight prior to treatment with C-PAC [50 and 75μg/mL] as previously described17 or vehicle control (0.1% ethanol) dissolved in DMEM with 10% fetal bovine serum (Thermo Fisher). Cell lysates were harvested at 0, 24 and 48hr post-treatment using lysis buffer (1% Triton X-100, 50mM HEPES, pH 7.4, 150mM NaCl, 1.5mM MgCl2, 1mM EGTA, 100mM NaF, 10mM sodium pyrophosphate, 1mM sodium orthovanadate, 10% glycerol) with cOmplete™ EDTA-free protease and PhosSTOP phosphatase inhibitors (Sigma-Aldrich) following published protocols18. Immunoblotting was performed using commercially available antibodies from Cell Signaling Technology (Danvers, MA): GAPDH (#2118; 1:40,000) and Santa Cruz Biotechnology: GSTT2 (#514667; 1:500) and HSP60 (#13115; 1:1000). Images were captured via the ChemiDoc Molecular Imager and band quantification with ImageLab analysis software (both Bio-Rad). Expression values normalized to loading controls were determined by chemiluminescent immunodetection with fold-change from vehicle reported.
Evaluation of GSTT2 and γ-H2AX levels in rat esophageal tissue
Sprague Dawley rats were purchased from Charles River Laboratories (Wilmington, MA) at 5-6 weeks of age and an esophagogastroduodenal anastomosis (EGDA) was performed as previously reported19 to create chronic reflux of bile salts and stomach acid which leads to esophageal adenocarcinoma and precursor lesions. Animals were group-housed in a dedicated animal facility under a 12-hr light/dark cycle and fed AIN93M (Dyets Inc, Bethlehem, PA) ad libitum. Rats were provided water ad libitum or water with C-PAC [690μg/rat/day] and euthanized at 40 weeks of study. Rat lower esophageal lysates (n=3/experimental group) were prepared by homogenization (PRO Scientific Inc., Oxford, CT) in T-PER® Tissue Protein Extraction Reagent (Thermo Fisher) with cOmplete™ EDTA-free protease inhibitor cocktail and PhosSTOP phosphatase inhibitors (Sigma-Aldrich) according to the manufacturer’s instructions. Equivalent protein amounts (30μg/lane) were loaded into precast 4-20% Mini-Protean TGX gels (Bio-Rad). Immunoblotting was performed as indicated above and fold-change from water treated animals reported for each treatment group. Phospho-H2AXSer139 protein levels were detected using the commercially available Cell Signaling Technologies (Danvers, MA) rabbit antibody #2577 at a 1:1000 dilution.
RESULTS
Transcriptional Profile of Normal Squamous Mucosa in AA vs. EA
To determine whether the NE of AA demonstrates a different transcription profile from that of EA, we used Affymetrix GeneChip 2.1 ST analysis. Total RNA was isolated from the NE of four subject groups: healthy control individuals (no history of BE or EAC) from both racial groups (AA-NE n=12; Cau-NE n=12) and patients histologically positive for BE and/or EAC (AA-NE:BE n=8; Cau-NE:BE n=8). We summarize key demographic data available for these patient groups in Supplementary Table S1. We fitted a one-way ANOVA model with terms for the means of four groups to log-transformed gene expression data, for gene probesets annotated with both gene symbol and Entrez Gene ID (n=24,909), and tested differences in means for specific pairs of groups as follows: between control populations (AA-NE vs EA-NE); between disease groups (AA-NE:BE vs. EA-NE:BE) and control vs. disease comparisons within each population (AA-NE vs. AA-NE:BE and EA-NE vs. EA-NE:BE). Similar to previous reports20, 21, we found strong differences between the squamous of EAC patients compared to both BE and non-disease patients (detailed in Supplementary Array Analysis) consistent with the idea of an etiologic field effect22. This result directed our initial comparisons towards expression differences between nondisease groups, within esophageal squamous tissue.
When comparing NE profiles of healthy AA and EA groups 5 probe-sets gave fold-changes greater than 3 and P<0.01 in an ANOVA model with means for 4 groups (GSTT2, GSTT2B, HLA-DBP1, IGHD and IGHA1; Supplementary Table S2), whereas 10,000 data-sets with random permutations of the sample labels gave just 2.28 probe-sets with these properties on average, so that nearly half (false discovery rate = 0.46) of the 5 selected probe-sets were expected to be false-positives. The FDR for a list satisfying "P < 0.01 and fold-change >3" can be much different than for a list merely satisfying "P < 0.01”. Previous studies also found few AA vs EA gene expression differences23-25. Nevertheless, the appearance of both GSTT2 and GSTT2B, and a review of their possible functions, lead us to investigate them further.
The GSTT2 mRNA demonstrated the largest FC difference (FC=5.15) between the two racial groups, while the paralog, GSTT2B, demonstrated the fourth largest FC (Figure 1A; Supplementary Table S2). Both genes produce the same protein and have nearly identical, but inverted, structure and sequence, accounting for their strongly correlated transcript abundance (Pearson coefficient of 0.95 across log2 array data for all 40 samples; data not shown). Since the Affymetrix probesets do not adequately distinguish between GSTT2 and GSTT2B transcripts, we present mRNA expression as GSTT2/2B (detailed in Methods). We observed a similar trend of increased GSTT2/2B expression (FC=2.17) when comparing the population-based disease groups, but without a supportive P value (P=0.14; AA-NE:BE to EA-NE:BE in Supplementary Table S2). Within the AA population there was a trend (P=0.048; FC=0.39) towards lower expression in the disease group, when compared to the control group (Supplementary Table S2). We then validated the array results using an extended cohort of non-diseased squamous mucosa samples (AA-NE n=20 vs. EA-NE n=21) with reverse transcription followed by qRT-PCR. This confirmed that GSTT2/2B was overexpressed in the NE of AA relative to EA (FC=6.3, P=0.0013; one-way ANOVA with Tukey’s post-hoc adjusted); Supplementary Figure S1A) and correlated with the array data across the arrayed samples (r=0.86; Supplementary Figure S1B). Other between-group comparisons were not significant.
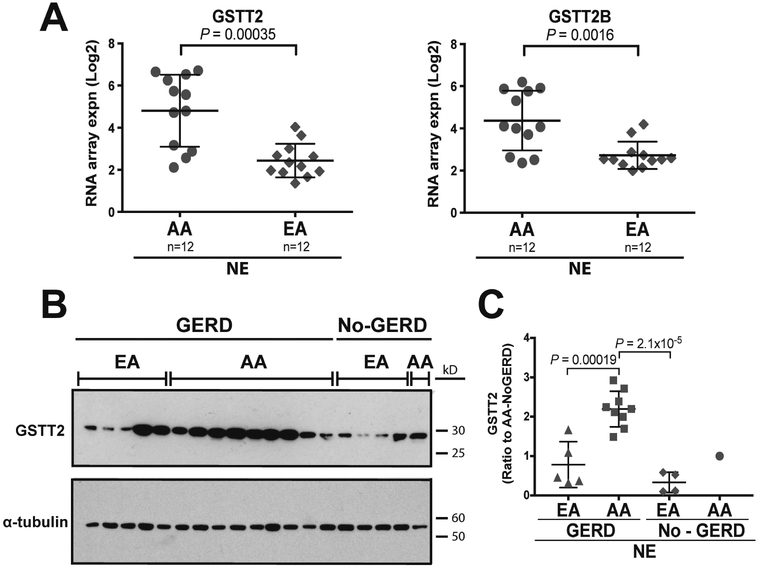
Figure 1. GSTT2 and GSTT2B expression in the NE of AA vs EA.
(A) Array analysis of normal squamous esophageal tissue (NE) from AA and EA reveals differential mRNA expression of GSTT2 and GSTT2B (right panel), respectively. One-way ANOVA P values are quoted, as shown in Supplementary Table S2. (B) Western blot depicting GSTT2 expression in an independent cohort of AA and EA normal esophagus with α-tubulin as the loading control. (C) Quantification of protein expression of GSTT2 in AA and EA normal esophagus biopsies (GSTT2 expression in each sample relative to α-tubulin). AA individuals with a history of GERD have significantly higher GSTT2 protein levels than either EA individuals with or without a history of GERD. One-way ANOVA with Bonferroni multiple comparison adjustment (allowing for three pairwise tests) was used to compare between groups with multiple samples. The EA-GERD vs. EA-nonGERD comparison was not significant (P=0.50).
Further, we investigated whether GSTT2 protein was differentially expressed in the NE of AA and EA individuals, with or without GERD. We obtained biopsies of NE from an independent cohort of individuals that self-identify as either AA or EA and had either a history of GERD or no history of GERD, BE or EAC. We measured GSTT2 protein levels in the NE from these individuals using Western blot analysis, normalized against α-tubulin. A history of GERD did not appear to change the level of GSTT2 expression among EA (P=0.50). We observed that GSTT2 is over-expressed in the mucosa of AA compared to EA (AA with GERD vs. EA with GERD P=0.00019; AA with GERD vs. EA without GERD P=2.1×10−5, each using one-way ANOVA with Bonferroni multiple comparison adjustment) (Figure 1B, C). Together the above observations suggest that GSTT2 expression is lower in the NE of a healthy EA population as well as in those who develop BE or EAC.
Zhang et al.25 noted differential HLA-DPB1 expression while comparing non-disease AA and EU groups using the Affymetrix ST array platform and were able to show that this was due to differential binding of one ST array probeset as a result of a frequent, population-variable SNP causing difference in response of one probe.
Genomic events associated with mRNA levels of GSTT2
To further address the basis for the differential mRNA expression of GSTT2/2B in AA vs EA, we examined the chromosomal region surrounding the GSTT2 locus. Resulting from an inverted chromosomal segmentation duplication, both genes (GSTT2 and GSTT2B) are located in chromosome band 22q11.23, with DDT and its inverted homolog DDTL, located between them26 (Supplementary Figure S2C). GSTT1, the only other Theta class GST present in humans, shares 55% protein homology and is located ~50kb telomeric to both GSTT2 and GSTT2B26 (Supplementary Figure S2C). Greater expression of GSTT2 and GSTT2B (FC>3, P<0.002; as discussed above and Supplementary Figure S2A-B, Supplementary Table S2), but not neighboring genes DDT (FC=0.85, P=0.013), GSTTP1 (FC=0.80, P=0.16), or GSTT1 (FC=0.70, P=0.23), was observed between AA-NE vs. EA-NE (Supplementary Figure S2D-F). This suggests the increased expression of GSTT2/2B in AA is not due to regional DNA amplification and/or chromosomal translocation, but is GSTT2 and GSTT2B specific. We further assessed other genomic events within the chromosome 22q11.23 loci that may affect the regulation of GSTT2/2B expression. Zhao et al.14 characterized a 37kb deletion of GSTT2B (Figure 2A; Supplementary Figure S3A), which they associated with lower levels of GSTT2 expression. We genotyped GSTT2B copy number for both AA and EA NE cohort samples using the method of Zhao et al.4 and observed a higher frequency (Fisher Exact P=0.018) of the GSTT2B deletion in EA-NE (80.6%) relative to AA-NE (54.8%; Supplementary Figure S3B). However, the GSTT2B deletion genotype alone did not explain the expression differences observed between NE from AA and EA groups (Supplementary Figure S3C-D).
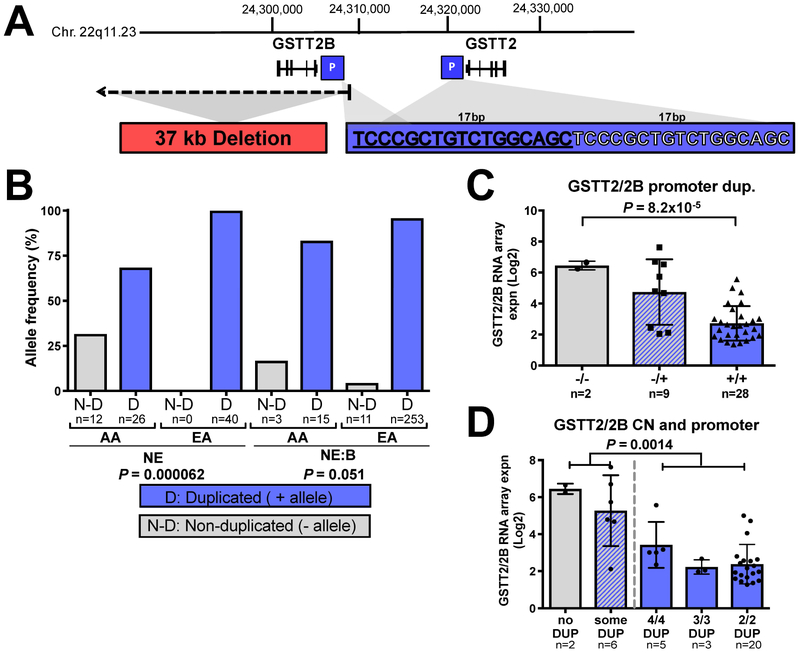
Figure 2. GSTT2/2B 17bp promoter duplication is associated with reduced mRNA expression.
(A) Two genomic events influence GSTT2/2B mRNA levels: a 37kb deletion that removes the GSTT2B gene, and the 17bp tandem GSTT2/2B promoter duplication. (B) The 17bp GSTT2/2B promoter duplication frequency is significantly lower in AA vs. EA NE (non-disease) populations, with a similar trend between NE:BE disease groups by Fisher Exact test. Allele numbers are shown below each bar. (C) When all squamous samples are combined and analyzed (one-way ANOVA, with a post-test for linear trend P value of 0.00078) we observed that the 17bp promoter duplication shows a dose-dependent association with GSTT2/2B mRNA expression. (D) The combination of the 17bp GSTT2/2B promoter duplication with the gene-dose effect of the GSTT2B whole gene deletion shows that individuals with only (homozygous) the 17bp promotor duplication had lower expression as compared to individuals having at least one copy of the non-duplicated promoter, irrespective of how many GSTT2 copies they have. We used a student T-test and Mann-Whitney tests to compare genotypes that included at least one non-duplicated 17bp allele (first 2 bars) to those with only alleles containing 17bp duplicated GSTT2 promotors (3 bars on the right). We quote the MWU P value as it was more conservative than the two-sample T-test (P=0.00004).
In a study examining tandem duplications across the genome, Marotta et al.15 identified a 17bp tandem duplication within the promoter of GSTT2 and GSTT2B (Figure 2A), and using a luciferase-linked assay to assess promoter function, showed the presence of the 17bp tandem duplication associates with lower GSTT2 and GSTT2B promoter activity. Utilizing a PCR-based methodology, and Sanger sequencing confirmation (Supplementary Figure S4), to differentiate the 17bp promoter duplication, we genotyped gDNA from the EA and AA samples. We observed a significantly higher frequency (EA-NE vs. AA-NE: 97.5% to 67.5%, P=0.000062 by two-sided Fisher Exact Test) of the promoter duplication in EA relative to AA (Figure 2B). The promoter duplication frequency between AA-NE and AA-NE:BE was not significantly different (P=0.34), possibly due to there being only 9 AA-NE:BE samples. We then examined matched lymphoblast genotype and mRNA expression data available for a subset of the 1000 Genomes Project samples27 and confirmed individuals of African descent tended to have higher GSTT2 mRNA levels than those of European ancestry (P=0.030) (Figure 3A). Furthermore, we confirmed homozygosity for the GSTT2 promoter duplication correlated with lower levels of GSTT2 expression in both local esophageal (one-way ANOVA P=8.2×10−5 and P=0.00078 with a post-hoc linear trend test; Figure 2C) and 1000G (ANOVA P=3.9×10−6; 4.7×10−6, with linearity test; Figure 3B) cohorts. The GSTT2B deletion also strongly correlated with GSTT2 expression in the 1000G data (Figure 3C), however, the most important contribution to low GSTT2/2B expression occurs when the 17bp GSTT2/2B promoter duplication is homozygous (Figure 3D) and thus similar to the results seen in the esophagus (Figure 2D). These results suggest the presence of the promoter duplication is more frequent in EA and may underlie reduced esophageal GSTT2/2B expression in this population. In terms of potential confounding effects, we did not observe significant differences in GSTT2/2B mRNA levels between males and females (Supplementary Figure S5A-B) nor in relation to GERD history or smoking status (Supplementary Figure S5C-D). We did note a trend (Welsh test P=0.056) towards an older age among AA relative to EA controls (Supplementary Table S1), but no evidence for interaction between age and GSTT2/2B mRNA expression, nor the 17bp promoter or GSTT2B deletion variant genotypes (2-way ANOVA P values of 0.27, 0.84, 0.95 respectively).
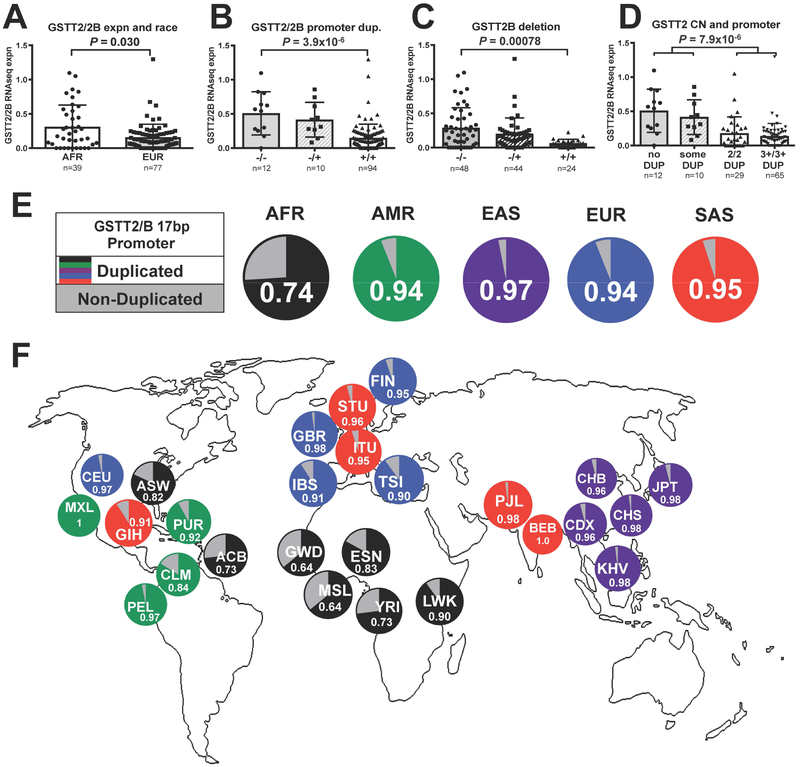
Fig. 3. 1000 Genomes Data analysis of GSTT2/2B genotype and mRNA.
We used publicly available, matched RNAseq and DNA copy number data from cultured lymphoblasts from a subset of 1000G population controls (n=116) from normal African (AFR) and European (EUR) individuals that show: (A) the same trend as our esophageal NE samples of higher average GSTT2/2B expression in individuals of African descent. We also confirmed that both the (B) GSTT2/2B promoter duplication and (C) GSTT2B deletion have gene dose-related effects upon expression such that (D) individuals homozygous for the promoter duplication have much lower GSTT2/2B expression than individuals with at least one non-duplicated copy. (E) Frequency of GSTT2/2B promoter duplication and non-duplicated alleles in super populations (1000G data) (P<0.0001 by Fisher Exact for each of the other 4 superpopulations against AFR). (F) Frequency of the non-duplicated GSTT2/2B promoter is highest among African and African descent populations. Subpopulation abbreviations are described in Supplementary Table S3.
To elucidate the promoter duplication frequency, we used 1000G data to examine the distribution of the 17bp GSTT2/2B promoter duplication allele across world populations and found it to be more prevalent in each of the non-African super-populations (AMR=92%, EUR=93%, SAS=96% and EAS=95%) when compared to AFR (AFR=71%) (P<0.0001 by Fisher Exact Test for each of the four vs AFR; Figure 3E). In particular West African populations showed the lowest frequency, with <70%, which is similar to the 64% we observed in our American population with African descent (Figure 3F; Supplementary Table S3).
GSTT2 functions to prevent DNA damage in cells undergoing genotoxic stress
To investigate the effect of GSTT2/2B gene dose (GSTT2B deletion) and GSTT2 promoter duplication in vitro we genotyped a cohort of cell lines including those derived from normal esophageal squamous mucosa (Het-1A), as well as cancers of squamous cell origins from head/neck and cervical cancer (HeLa) (Supplementary Figure S6A). We observed that Het-1A is homozygously deleted for the 37kb fragment, indicating that it only carries 2 copies of GSTT2/2B while HeLa cells are homozygously non-deleted, and therefore have 4 copies of GSTT2/2B (Supplementary Figure S6A, top panel). We further assessed whether these gene copies of GSTT2/2B harbor the promoter duplication. Het-1A is homozygous and HeLa is heterozygous for the promoter duplication, respectively (Supplementary Figure S6A bottom panel). We found that HeLa cells had about 4 times more endogenous GSTT2, at both the mRNA and protein level, with comparable amounts of housekeeping gene/protein levels (Supplementary Figure S6B-C). The relative mRNA difference between these two cell lines is consistent with their GSTT2 genotypes, based esophageal squamous cohort (Figure 2D) and the 1000 Genomes lymphoblast cell line panel (Figure 3D), comparing bars for “some Dup” sample average to the “2/2 DUP” average. Thus we chose to investigate the potential importance of GSTT2 in the DNA damage response of these two squamous cell lines with key differences in GSTT2 structural alleles.
GSTT2 has been previously shown to protect cells against DNA damage following exposed to the oxidative agent, cumene-hydroperoxide (cum-OOH)28. To confirm that GSTT2 plays a role in protecting squamous cells against genotoxic stress, we performed knockdown experiments in both Het-1A and HeLa cell lines, with and without genotoxic stress induced by cum-OOH treatment. We transfected Het-1A and HeLa cells using four commercial GSTT2 targeting siRNAs and confirmed successful GSTT2 mRNA and protein knockdown of >50-80% in both cell lines (Supplementary Figure S7A-G). While we observed knockdown of GSTT2 across all four siRNAs, 05 and 06 had the better knockdown performance overall, and were chosen for subsequent experiments.
We hypothesized that Het-1A cells may be more susceptible to DNA damage following cum-OOH exposure. DNA damage was measured using immunofluorescence staining for the DNA double strand break marker, gamma-H2AX (γ-H2AX) as a ratio against nuclear foci (DAPI stained, see Methods). Cum-OOH treated, GSTT2 knocked-down Het-1A cells showed dramatically more DNA damage compared to controls (control vs. siRNA-05, 06) (P=4.8×10−5, 1.0×10−5, respectively), with more than 50% of the cell population positive for DNA damage response (Figure 4A-B). We saw a similar trend in HeLa cells (Supplementary Figure S7I-J). These data confirm that GSTT2 protects cells against genotoxic stress, however, we saw no evidence that Het-1A had more γ-H2AX-positive nuclear foci than HeLa cells, either with or without genotoxic stress (Supplementary Figure S6D). Both cell lines showed a significant shift from about 1-2% of cells with active damage (>10 γ-H2AX nuclear foci) to around 20% following 100μM cum-OOH treatment, but the change was not different between the two cell lines. Thus, while the level of GSTT2 mRNA and protein in these cell lines may be different, we did not see this translate into a difference in DNA damage response under the genotoxic stress conditions (Supplementary Figure S6C-D).
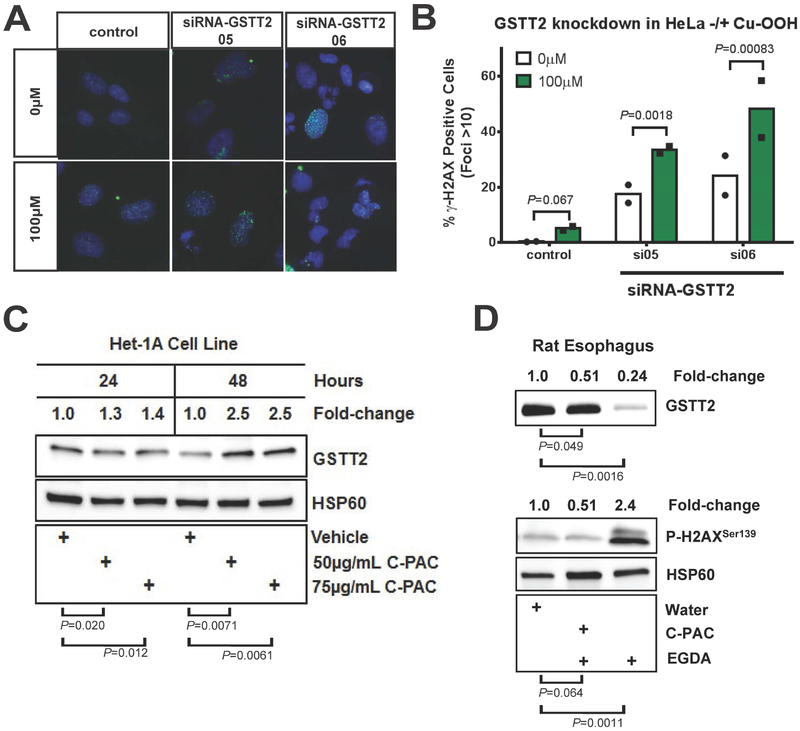
Fig. 4. Effect of GSTT2 knockdown, cum-OOH and cranberry proanthocyanidin extract (C-PAC) on GSTT2 expression and DNA damage response.
(A-B) Quantification of γ-H2AX positive foci in Het-1A, with and without GSTT2 specific siRNA knockdown. Nuclei with >10 γ-H2AX-stained foci considered as positive for DNA damage, for duplicate experiments. Analyses were modelled as a 4-way ANOVA with terms of cum-OOH (0 vs 100μM), cell type (Het-1A vs HeLa), treatment (control, siRNA05, siRNA06) and experiment day, as detailed in Supplementary Methods. In Het-1A both GSTT2 siRNA knockdowns dramatically increased the response to cum-OOH. Contrasts between 0μM Cum-OOH treatments were P=0.026 and P=0.14 for control to siRNA05 or siRNA6, respectively, and P=0.34 for siRNA05 to siRNA06, with 7-20% more cell showing damage. Both siRNAs gave significantly more foci than control (P=3.9E-05 and 2.9E-05, respectively) at 100μM Cum-OOH, but were not different from each other (P=0.83). (C) Western blot of Het-1A showing that treatment with C-PAC induces GSTT2 protein. Log2 ratios (GSTT2 vs HSP60) for 24 and 48hr time points were used in separate 2-way ANOVA models, each with terms for experiment (triplicate ratios) and C-PAC treatments. Unadjusted vehicle vs treatment group comparison P values are shown, while between treatment (50 vs 75 μg/mL C-PAC) comparisons were not significant (P=0.58 and P=0.84 for 24 and 48hr time points respectively). (D) Western blot showing that C-PAC mitigation of GSTT2 loss coincides with protection against the DNA damaging effects of gastroduodenal reflux in an esophagogastroduodenal anastomosis (EGDA) rat model, with relative esophageal protein levels of GSTT2 and γ-H2AX contrasted with and without C-PAC treatment. Log 2 ratios (HSP60 as reference) for GSTT2 and γ-H2AX were used in separate 2-way ANOVA models, each with terms for experiment (triplicate ratios) and treatment groups. Unadjusted water vs treatment group comparison P values are shown. EGDA rats with C-PAC had significantly more GSTT2 (P=0.00041) and less γ-H2AX foci (P=0.0088) than EGDA rats not given C-PAC.
C-PAC induces GSTT2 expression and reduces DNA damage in rats with esophagogastroduodenal anastomosis (EGDA)
Cranberries are rich in bioactive constituents, including proanthocyanidins (C-PAC), which are known to possess cancer inhibitory properties in preclinical models and thus an avenue for potential chemoprevention17. We observed that C-PAC significantly induces the expression of GSTT2 in Het-1A esophageal cells after 48hrs exposure (FC=2.5) (Figure 4C). In a rat esophagogastroduodenal anastomosis (EGDA) model for reflux-induced development of EAC, esophageal tissue showed significantly reduced GSTT2 and increased γ-H2AX levels relative to surgically naive rats, as measured at 40 weeks (Figure 4E). In contrast, the C-PAC treated EGDA rats had low esophageal γ-H2AX levels, similar to the non-surgical controls, yet maintained significantly high GSTT2 levels compared to EGDA positive rats without C-PAC treatment (Figure 4E). C-PAC treatment therefore reduced esophageal DNA damage associated with surgically-induced chronic acid reflux (EGDA) and concomitantly maintained protective levels of GSTT2.
DISCUSSION
When we compared the transcriptome profiles of normal esophageal squamous mucosa (NE), we observed very few group differences between mRNA from individuals that self-identified as AA or EA. The most striking difference was the relative over-expression of GSTT2/2B among the AA group (Figure 1; Supplementary Table S2). We observed a substantially higher frequency of the 17bp promoter duplication in EA as compared to AA that strongly correlated with lower levels of GSTT2/2B expression in both our NE cohort (Figure 2) and the 1000G cohort (Figure 3A). This promoter variant was a much stronger predictor of GSTT2/2B mRNA level than the whole gene duplication variation that leads to humans inheriting 2, 3 or 4 GSTT2/2B copies (Supplementary Figure S3). Our data confirm the original report15 that homozygosity for the tandem-duplicated GSTT2/2B form associates with lower expression (Figure 2D), but we now extend this to human tissues. With this in mind, we observed the homozygous frequency of the 17bp duplication as 100% in EA-NE (Figure 2B), compared to 88% for the EUR super population (homozygous carrier frequency estimate of Figure 3E value), vs. 46% of AA-NE (compared to 56% for the AFR super-population, with considerable variation (41% to 69%) seen between West African populations (homozygous carrier frequency estimate of Figure 3E, F values). Using esophageal tissue samples and the 1000G/HapMap data, we observed that the non-duplicated promoter is conserved in populations of African descent. Our findings suggest the change in the 17bp promoter duplication frequency arose in conjunction with the trans-global migration of ancient humans, perhaps in response to new, or diverse selection pressures. It is interesting to note that Denisovan sequencing tracks (USCS browser tracks for GSTT2B and GSTT2)29, 30 show that several individuals from this ancient population had the 17bp duplication variant, while, so far, none of the Neanderthal sequences do (JBrowse regions for GSTT2B and GSTT2)31. Thus, we speculate that the distribution of these variants accompanied ancient hominid migration/integration. Lower levels of the only other human theta-class GST family member, GSTT1, have been similarly associated with an increased prevalence of cancer of the esophageal squamous mucosa, but in this case restricted to Asian populations32.
Despite different substrate specificities, GSTs function to protect cells against genotoxic stress, and in colon mucosa it has been shown than having high or low levels of GSTT2 contributes to a cell’s susceptibility to DNA damage9. Here we report that GSTT2 functions in the same manner in the Het-1A esophageal squamous mucosa cell line. We observed that after knockdown of GSTT2, cells are more susceptible to DNA damage following genotoxic stress (Figure 4A-B). While we did see relative differences in mRNA and protein levels of GSTT2 between Het-1A and HeLa consistent with the particular GSTT2 genotypes they carry, this did not translate into a differential DNA damage response, either endogenously, or when we applied genotoxic stress via cum-OOH treatment. While background differences between these cell lines may mask a differential GSTT2 response, or alternatively DNA damage induction treatment may be concentration dependent, it is also possible that phenotypically differential GSTT2 responses are subtle. Even though the incidence of esophageal cancer has risen dramatically over the past few decades, the average age of onset has not changed, based on SEER data from the 1970s, 80s, 90s and into the 2000s33. So while risk factor exposure appears to have increased, the average time for cancer detection has not decreased, suggesting that time may be a key factor in disease etiology, and perhaps reflux-induced genotypic expression differences are subtle.
While EAC continues to be more prevalent among EA, its incidence has continued to increase among other racial groups within developed countries. El-Serag et al.5 and others have reported no difference in the frequency of GERD symptoms (weekly heartburn and/or regurgitation: AA 29%, EA 28%, other 25%, P=0.80) as well as other risk factors including obesity, smoking, or age between ethnic groups present in the USA5-7. However, AA participants had a much lower risk of esophagitis (adjusted OR 0.22–0.46, P<0.001)5, which translates into reduced tissue damage for comparable reflux exposure. AA continue to have a substantially lower EAC incidence than EA. Our findings provide the first evidence for a genetic and a molecular basis to explain part of the disparity between these two populations.
The prevalence of GERD in East Asia is estimated to be 2.5-7.8% while populations in the US and Europe, with higher EAC risk, have more frequent GERD (18-28%)34. Global obesity levels follow a similar pattern, with higher rates among high-risk EAC populations (15-25%) when compared to lower risk populations in Asia (5-15%)35. Thus, the potential intersection between lower GSTT2/2B levels and obesity should be evaluated to determine the combined influence an individual’s risk of developing BE and EAC. Among non-African populations, where the 17bp promoter duplication is the prominent allele, obesity rates may offer a partial explanation for the difference in EAC incidences. If true, then the current trend of increasing global obesity levels should be viewed with concern in terms of esophageal adenocarcinoma cancer risk.
Non-steroidal anti-inflammatory drugs (NSAIDs) have been shown to reduce the risk of EAC development by >40-50%36. In addition, NSAIDs and apple polyphenols have been observed to induce the levels of GSTT29, 28, 37. Although it has been suggested that the effect of NSAIDs in reducing the risk of EAC is through inhibition of inflammation, we propose that NSAIDs could also lower an individual’s risk for GERD-related damage through increasing esophageal GSTT2 expression. Specific studies looking at the effect of NSAIDs on esophageal GSTT2 levels in subjects at various stages of disease ‘progression’ would be needed to test this hypothesis. Potentially, treatment with these or other compounds in high-risk populations with GERD may increase protective GSTT2 protein levels and help reduce or delay the risk of progression towards EAC. We show that a cranberry extract enriched for proanthocyanidins, known to have inhibitory effects on multiple cancer types38 including EAC cell lines17, 18 mitigates GSTT2 protein loss in a rat surgical model for reflux-induced EAC, potentially reducing oxidative damage caused by bile salts, thus offering a possible alternative, or adjunct, to prophylactic NSAID treatment.
Strengths and Limitations
We report a significant difference in the levels of a key-detoxifying enzyme, GSTT2, which is differentially-expressed between EA and AA populations. Two genetic variants (37kb deletion and 17bp promoter duplication) negatively associate with the levels of GSTT2 and are highly frequent in the EA population. We also examine a large cohort of patients who develop EAC and confirm these associations. In addition, we report that esophageal squamous cells with low levels of GSTT2 are more susceptible to DNA damage following treatments that induce genotoxic stress. We provide evidence that GSTT2 can be induced via cranberry proanthocyanidin exposure in human and rat esophageal cells and tissue, suggesting a novel preventive approach for individuals at greatest risk for the disease. Altogether, these observations suggest that increased GSTT2 expression may be protective against esophageal mucosal damage caused by GERD and might underlie the low incidence of EAC in AA populations. We have limited the scope of our analysis to the normal squamous epithelium of patients and controls and have not measured the expression of GSTT2 or variants in BE or EAC since the 1000G data indicate that the allelic differences in GSTT2 are constitutive with genotype. We also have not yet examined methods to induce GSTT2 in BE patients and are at present limited to model systems but these studies are currently being planned.
Supplementary Material
Editor’s Notes.
BACKGROUND AND CONTEXT
Esophageal adenocarcinoma (EAC) primarily affects European Americans and is rare in African Americans. The basis for this racial difference is unknown and insights into its underlying mechanism may have significant impact on strategies to reduce the incidence of EAC.
NEW FINDINGS
We demonstrate that the protective detoxifying enzyme, glutathione Stransferase theta 2 (GSTT2), is more abundantly expressed in the normal esophageal mucosa of African Americans relative to European Americans and find that a 17bp GSTT2/2B promoter duplication associates strongly with expression levels. Further, we demonstrate that GSTT2 functions to protect esophageal cells from DNA damage under genotoxic stress, and in a preclinical model of EAC, chemopreventative cranberry proanthocyanidins (C-PAC) increased expression of GSTT2 while reducing esophageal DNA damage.
LIMITATIONS
We have limited the scope of this paper to understanding the differential expression of GSTT2 in the normal squamous tissue of patients and controls, since we hypothesized the presence of a chemopreventative factor. Thus, we have not presented the expression level, nor differential variant level in BE or EAC tissue. However, the 1000G data offers the expression in transformed (environmental influence normalized) white cells, suggesting that the allelic differences in GSTT2 are constitutive with genotype.
IMPACT
These results have important implications for understanding EAC risk and offer the potential for chemopreventative treatment of at-risk patients.
Acknowledgments:
Funding: Supported by NCI Grants: CA163059, CA967622, CA200113, CA009676, CA046592, CA158319, and the John and Carla Klein family research fund.
Grant support: Supported by NCI Grants: CA163059, CA967622, CA200113, CA009676, CA046592, CA158319, and the John and Carla Klein family research fund
Abbreviations:
- EAC
[esophageal adenocarcinoma]
- GSTT2
[glutathione S-transferase theta 2]
- AA
[African American]
- EA
[European American]
- GERD
[gastroesophageal reflux disease]
- BE
[Barrett’s esophagus]
- NE
[normal esophageal squamous mucosa]
- 1000G
[1000 Genome]
- EGDA
[esophagogastroduodenal anastomosis]
- cum-OOH
[cumene-hydroperoxide]
- C-PAC
[cranberry proanthocyanidin extract]
- γ-H2AX
[phospho-H2A histone family member X]
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: The authors report no conflict of interest
Data and materials availability: Both normalized and raw expression data for “Expression profile comparison of African American and American Caucasian esophageal squamous epithelium from subjects with and without Barrett's Esophagus” were deposited into the Gene Expression Omnibus (GEO series GSE77563).
Transcriptional Profiling: Gene Expression Omnibus (GEO series GSE77563)
REFERENCES
- 1.Reid BJ, Li X, Galipeau PC, et al. Barrett's oesophagus and oesophageal adenocarcinoma: time for a new synthesis. Nat Rev Cancer 2010;10:87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pickens A, Orringer MB. Geographical distribution and racial disparity in esophageal cancer. Ann Thorac Surg 2003;76:S1367–9. [DOI] [PubMed] [Google Scholar]
- 3.Thrift AP, Whiteman DC. The incidence of esophageal adenocarcinoma continues to rise: analysis of period and birth cohort effects on recent trends. Ann Oncol 2012;23:3155–62. [DOI] [PubMed] [Google Scholar]
- 4.Domper Arnal MJ, Ferrandez Arenas A, Lanas Arbeloa A. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol 2015;21:7933–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Serag HB, Petersen NJ, Carter J, et al. Gastroesophageal reflux among different racial groups in the United States. Gastroenterology 2004;126:1692–9. [DOI] [PubMed] [Google Scholar]
- 6.El-Serag HB, Sonnenberg A. Associations between different forms of gastro-oesophageal reflux disease. Gut 1997;41:594–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spechler SJ, Jain SK, Tendler DA, et al. Racial differences in the frequency of symptoms and complications of gastro-oesophageal reflux disease. Aliment Pharmacol Ther 2002;16:1795–800. [DOI] [PubMed] [Google Scholar]
- 8.Thrift AP, El-Serag HB. Sex and Racial Disparity in Incidence of Esophageal Adenocarcinoma: Observations and Explanations. Clin Gastroenterol Hepatol 2016;14:330–2. [DOI] [PubMed] [Google Scholar]
- 9.Petermann A, Miene C, Schulz-Raffelt G, et al. GSTT2, a phase II gene induced by apple polyphenols, protects colon epithelial cells against genotoxic damage. Mol Nutr Food Res 2009;53:1245–53. [DOI] [PubMed] [Google Scholar]
- 10.Ferrer-Torres D, Nancarrow DJ, Kuick R, et al. Genomic similarity between gastroesophageal junction and esophageal Barrett's adenocarcinomas. Oncotarget 2016;7:54867–54882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003;31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blin N, Stafford DW. General Method for Isolation of High Molecular-Weight DNA from Eukaryotes. Nucleic Acids Research 1976;3:2303–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Marotta M, Eichler EE, et al. Linkage disequilibrium between two high-frequency deletion polymorphisms: implications for association studies involving the glutathione-S transferase (GST) genes. PLoS Genet 2009;5:e1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marotta M, Piontkivska H, Tanaka H. Molecular trajectories leading to the alternative fates of duplicate genes. PLoS One 2012;7:e38958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lappalainen T, Sammeth M, Friedlander MR, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013;501:506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kresty LA, Weh KM, Zeyzus-Johns B, et al. Cranberry proanthocyanidins inhibit esophageal adenocarcinoma in vitro and in vivo through pleiotropic cell death induction and PI3K/AKT/mTOR inactivation. Oncotarget 2015;6:33438–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weh KM, Howell AB, Kresty LA. Expression, modulation, and clinical correlates of the autophagy protein Beclin-1 in esophageal adenocarcinoma. Mol Carcinog 2016;55:1876–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Yang G, Ding WY, et al. An esophagogastroduodenal anastomosis model for esophageal adenocarcinogenesis in rats and enhancement by iron overload. Carcinogenesis 1999;20:1801–8. [DOI] [PubMed] [Google Scholar]
- 20.Brabender J, Marjoram P, Lord RV, et al. The molecular signature of normal squamous esophageal epithelium identifies the presence of a field effect and can discriminate between patients with Barrett's esophagus and patients with Barrett's-associated adenocarcinoma. Cancer Epidemiol Biomarkers Prev 2005;14:2113–7. [DOI] [PubMed] [Google Scholar]
- 21.Saadi A, Shannon NB, Lao-Sirieix P, et al. Stromal genes discriminate preinvasive from invasive disease, predict outcome, and highlight inflammatory pathways in digestive cancers. Proc Natl Acad Sci U S A 2010;107:2177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lochhead P, Chan AT, Nishihara R, et al. Etiologic field effect: reappraisal of the field effect concept in cancer predisposition and progression. Mod Pathol 2015;28:14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dluzen DF, Noren Hooten N, Zhang Y, et al. Racial differences in microRNA and gene expression in hypertensive women. Sci Rep 2016;6:35815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Storey JD, Madeoy J, Strout JL, et al. Gene-expression variation within and among human populations. Am J Hum Genet 2007;80:502–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Duan S, Kistner EO, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet 2008;82:631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landi S Mammalian class theta GST and differential susceptibility to carcinogens: a review. Mutat Res 2000;463:247–83. [DOI] [PubMed] [Google Scholar]
- 27.Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miene C, Klenow S, Veeriah S, et al. Impact of apple polyphenols on GSTT2 gene expression, subsequent protection of DNA and modulation of proliferation using LT97 human colon adenoma cells. Mol Nutr Food Res 2009;53:1254–62. [DOI] [PubMed] [Google Scholar]
- 29.Green RE, Krause J, Briggs AW, et al. A draft sequence of the Neandertal genome. Science 2010;328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer M, Kircher M, Gansauge MT, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 2012;338:222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deviese T, Karavanic I, Comeskey D, et al. Direct dating of Neanderthal remains from the site of Vindija Cave and implications for the Middle to Upper Paleolithic transition. Proc Natl Acad Sci U S A 2017;114:10606–10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi SM, Li GY. Null genotype of GSTT1 contributes to esophageal cancer risk in Asian populations: evidence from a meta-analysis. Asian Pac J Cancer Prev 2012;13:4967–71. [DOI] [PubMed] [Google Scholar]
- 33.Njei B, McCarty TR, Birk JW. Trends in esophageal cancer survival in United States adults from 1973 to 2009: A SEER database analysis. J Gastroenterol Hepatol 2016;31:1141–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Serag HB, Sweet S, Winchester CC, et al. Update on the epidemiology of gastro-oesophageal reflux disease: a systematic review. Gut 2014;63:871–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.WHO. Global status report on noncommunicable diseases World Health. Volume 2014 Geneva, Switzerland: World Health Organization, 2014:176. [Google Scholar]
- 36.Liao LM, Vaughan TL, Corley DA, et al. Nonsteroidal anti-inflammatory drug use reduces risk of adenocarcinomas of the esophagus and esophagogastric junction in a pooled analysis. Gastroenterology 2012;142:442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Lieshout EM, Tiemessen DM, Roelofs HM, et al. Nonsteroidal anti-inflammatory drugs enhance glutathione S-transferase theta levels in rat colon. Biochim Biophys Acta 1998;1381:305–11. [DOI] [PubMed] [Google Scholar]
- 38.Weh KM, Clarke J, Kresty LA. Cranberries and cancer: An update of preclinical studies evaluating the cancer inhibitory potential of cranberry and cranberry derived constituents. Antioxidants (Basel) 2016;5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Both normalized and raw expression data for this experiment were deposited into the Gene Expression Omnibus (GEO series GSE77563).