Abstract
Background & Aims:
Neutrophils are among the most prevalent immune cells in the microenvironment of colon tumors, are believed to promote growth of colon tumors, and their numbers correlate with outcomes of patients with colon cancer. Trials of inhibitors of neutrophil trafficking are under way in patients with cancer, but it is not clear how neutrophils contribute to colon tumorigenesis.
Methods:
Colitis-associated colon cancer was induced mice with conditional deletion of neutrophils (LysMCre;Mcl1fl/fl) and wild-type littermates (LysMCre;Mcl1+/+, control mice) by administration of azoxythmethane and/or dextran sulfate sodium. Sporadic colon tumorigenesis was assessed in neutrophil-deficient and neutrophil-replete mice with conditional deletion of colon epithelial Apc (Cdx2-CreERT2;Apcfl/fl). Primary colon tumor tissues from these mice were assessed by histology, RNA-sequencing, quantitative PCR, and fluorescence in situ hybridization analyses. Fecal and tumor-associated microbiota were assessed by 16s rRNA sequencing.
Results:
In mice with inflammation-induced and sporadic colon tumors, depletion of neutrophils increased the growth, proliferation, and invasiveness of the tumors. RNA sequencing analysis identified genes that regulate anti-microbial and inflammatory processes that were dysregulated in neutrophil-deficient colon tumors, compared to colon tumors from control mice. Neutrophil depletion correlated with increased numbers of bacteria in tumors and proliferation of tumor cells, tumor cell DNA damage, and an inflammatory response mediated by interleukin 17 (IL17). 16s rRNA sequencing identified significant differences in the composition of the microbiota between colon tumors from neutrophil-deficient vs control mice. Administration of antibiotics or a neutralizing antibody against IL17 to neutrophil-deficient mice resulted in development of less-invasive tumors compared to mice given vehicle. We found bacteria in tumors to induce production of IL17, which promotes influx of intra-tumor B cells that promote tumor growth and progression.
Conclusions:
In comparisons of mice with vs without neutrophils, we found neutrophils to slow colon tumor growth and progression by restricting numbers of bacteria and tumor-associated inflammatory responses.
Keywords: PMNs, anti-tumor immune response, genetic, cytotoxic
Introduction
The tumor immune response plays a critical role in the neoplastic progression of colon cancer1. Anti-tumor immune cells, such as T-cells and natural killer cells inhibit tumorigenesis and tumor progression. Pro-tumorigenic immune cells such as T-regulatory cells and tumor associated macrophages foster tumor growth and immune evasion 2. Polymorphonuclear neutrophils (PMNs) are myeloid cells of the innate immune system known for their role in the acute host response to infection. PMN effector functions consist of bacterial phagocytosis, generation and release of cytotoxic granule proteins, and production of superoxide radicals 3. Evidence has emerged that PMNs are among the most prevalent immune cells type in several human cancers, including colon cancer, pancreatic cancer, and lung cancer 4. The functional role for PMNs in the progression of cancer has largely been suggested to be pro-tumorigenic through suppression of anti-tumor immunity and direct activation of tumor cell growth 5, 6. Recent meta-analysis demonstrates PMNs have the highest correlation with adverse outcomes across all cancers 7. Despite these observations, the primary contribution of PMNs to initiation of colon tumorigenesis is not clear. Therefore, a more thorough investigation of PMNs in cancer is necessary to identify their potential for therapeutic targeting in colon cancer.
The majority of studies have relied upon antibody mediated PMN depletion and PMN trafficking receptor inhibitors which may have off-target effects and may not completely eradicate intra-tumoral PMNs in colon cancer 8, 9. In the current study, a murine model of genetic PMN depletion was assessed in mouse models of colitis and colon cancer. Genetic depletion of PMNs robustly enhanced tumor progression and invasion in a colitis-associated tumor model as well as a sporadic colon tumor model. Genetic PMN depletion promoted a dramatic expansion of tumor-associated bacteria and Il17 expression that was critical for the increased tumor progression. Tumor-associated bacteria through IL17 led to accumulation of intra-tumoral B-cells which played an important role in increased tumor growth and invasion. Therapeutic targeting of granulocytic myeloid cells and PMNs is currently being evaluated for clinical utility in several cancer types 10. Our data delineates an important role for neutrophils in blunting colon tumor progression and invasion and provides caution against anti-PMN therapy in colon cancer, particularly in the context of preceding inflammation and in the earliest stages of neoplastic disease.
Methods
Animals and treatments
For all experiments, male and female mice aged 6 to 8-weeks were used. All mice are C57BL/6 background. Mcl1fl/fl were crossed to LysMCre expressing mice as previously described 11. For azoxythmethane (AOM) / dextran sulfate sodium (DSS) experiments, animals were injected I.P. with 10mg/kg AOM then cycled on and off 2% (weight/volume) DSS in their drinking water. For acute DSS experiments, animals were treated with 2.5% DSS in drinking water for seven days then changed back to regular drinking water for two days. Cdx2-CreERT2;Apcfl/fl mice have been previously described 12. Mrp8-Cre expressing mice were previously described and crossed to Mcl1fl/fl mice 13. Bone marrow transplantation was performed by isolation of single cell suspension of bone marrow cells from femur and tibia of 6- to 8-week old mice LysMCre;Mcl1fl/fl, Mrp8Cre;Mcl1fl/f, and LysMCre;Mcl1wt/wt mice. 1×10^6 bone marrow cells were injected into the tail vein of recipient mice lethally irradiated with two split doses of 6gy radiation four-hours apart. Mice were supplemented with Neomycin (1mg/ml) in their drinking water for two-weeks. Four-weeks post transplantation, recipient mice were injected I.P. with tamoxifen (100mg/kg) in corn oil and sacrificed 14-days after administration. For antibiotics treatment, AOM/DSS was induced in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice with 2% DSS. 14-days after administration of AOM, mice were placed on antibiotics drinking water (neomycin 1mg/ml, ampicillin 1mg/ml, and streptomycin 0.5mg/ml). Every other day, mice were orally gavaged with antibiotics (Abx) solution (neomycin 2.5mg/ml, ampicillin 2.5mg/ml, streptomycin 1.25mg/ml, and metronidazole 1.25mg/ml) and sacrificed 72-days after AOM administration. For B-cell inhibition experiments, colon tumorigenesis was initiated in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice using AOM and 2% DSS. One-week following the third cycle of DSS, LysMCre;Mcl1fl/fl mice were randomized into Ctrl and anti-B220 treatment groups (BioXcell). Mice were treated every fourth day with 400ug I.P. anti-B220 diluted in sterile 1x PBS pH 7.0. For IL17 inhibition experiments, mice were injected with AOM and DSS (1.5%) was administered with 3 cycles of 3-5 days on DSS and 10 days of recovery. IL17 injections began 14 days following the last cycle of DSS. Mice were injected I.P. 3×/week for 3 weeks with 50μg of IL17 (anti-mouse IL17A BioXCell) diluted in sterile 1× PBS pH 7.0. Tumor burden was defined as summation of total tumor volume and tumor size was defined as average tumor diameter.
Flow cytometry
Collagenase Type II digested tissues were passed through a 40μm cell strainer. Peripheral blood was prepared by lysing one drop of tail blood for twenty minutes with RBC lysis buffer. Single-cell suspensions were stained with eFluor780- anti-CD45 (eBioscience), PE-anti-Ly6g (BD), APC-anti-Cd11b (eBioscience), FITC-anti-B220 (eBioscience) and eFluor450 anti-F4/80 (eBioscience). Flow cytometry was performed using an LSRFortessa (BD). Flow cytometry data was analyzed using FlowJo software (BD Biosciences).
Histology and immunofluorescence
Histological and tumor analysis was scored by a blinded gastroenterologist as previously described 14. Antibodies for immunofluorescence were as follows: Ki67 (1:100, Vector Labs), p-H2AX (1:200, Cell Signaling Technologies), and Alexa Fluor® 488 goat anti-rabbit IgG (1: 500, Molecular Probes Inc). B220 staining was performed following antigen retrieval of 5 μm paraffin slides using anti-B220 FITC antibody (eBioscience). Bacteria labeling was performed with Cy3 labeled EUB338 probe (5’-Cy3 GCTGCCTCCCGTAGGAGT). Paraffin tissue sections were deparaffinized in xylenes (2× for 5 minutes) followed by 100% ethanol (2× for 5 minutes) then rinsed in D.I. water. Slides were incubated with 100ul of 5ng/ul EUB338 for two hours at 46°C in hybridization buffer (35% formamide, 20mM Tris-HCl pH7.2, 0.9M Nacl, 0.01% SDS) then washed for 40 minutes at 48°C and mounted with ProLong™ Gold reagent with Dapi (Invitrogen). cCasp3 and p-H2AX were quantified using ImageJ software as number of positive cells per high powered field (HPF). EUB338 was quantified as percent of EUB338 positive area per Dapi positive area.
RNA isolation and qPCR analysis
qPCR analysis was performed using Radiant Green qPCR mix (Alkali Scientific Inc.). Primers are listed in Table S1. For cytokine analysis PCR arrays from Qiagen were used.
Protein isolation and Western blotting
Proteins were separated and using SDS-PAGE and transferred to nitrocellulose membrane. Antibodies used are as follows: phosphor-P65 (1:1000, Cell Signaling Technology), total-P65 (1:1000 Cell Signaling Technology), phospho-STAT3 (1:1000, Cell Signaling Technology), total-STAT3 (1:1000 Cell Signaling Technology), GAPDH (1:1000, Santa Cruz Biotechnology), PCNA (1:1000, Cells Signaling Technology).
ELISA
Fecal IgA was measured using Mouse IgA ELISA Quantitation Kit according to manufacturer’s protocol (Bethyl Laboratories, Inc). IL17 was measured in the colon lysates or serum using mouse IL17 ELISA kit following manufacturers recommendations (Lifespan Biosciences Inc, Seattle, WA)
High throughput RNA sequencing (RNA-seq) and RNA-Seq data analysis
TruSeq RNA library prep kit v2 (Illumina) was used to prepare RNA sequencing libraries. Libraries were sequenced using single-end 50-cycle reads on an Illumina HiSeq 2500 sequencer. RNA-seq read quality was assessed utilizing FastQC. Reads were aligned to a splice junction aware build of the mouse genome (mm10) using STAR15 with the options “outFilterMultimapNmax 10” and “sjdbScore 2”. Differential expression testing between LysMCre;Mcl1wt/wt, LysMCre;Mcl1fl/wt, and LysMCre;Mcl1fl/fl mice colon samples was conducted with CuffDiff v 2.1.1 with the parameter settings “-compatible-hits-norm,” and “–frag-bias-correct”. UCSC mm10.fa was used and the GENCODE mouse M12 primary assembly annotation GTF as the reference genome and reference transcriptome, respectively. Genes were considered differentially expressed at a false-discovery rate-adjusted (FDR) P value of <0.05.
16S rRNA gene sequencing and bacterial community analysis
Feces were collected at 65 and 75-days after AOM/DSS induced colon tumorigenesis in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice. At day 75, 2 mm tumors located in the distal colon were collected and snap frozen. Bacterial DNAs were extracted from mouse intestines as described. The V4 region of the 16S rRNA gene (252 bp) was sequenced using an Illumina MiSeq sequencer and ~22K reads were analyzed by Mothur 16, 17. Operational taxonomic units (OTUs) were classified into taxonomic phylotypes at >97 % identity using Mothur 17.
Statistical analysis
P-values were calculated by student’s t-test, one-way, two-way ANOVA followed by Tukey’s multiple comparisons test. Error bars represent standard error of the mean. Statistical analysis for tumor invasion was assessed by assigning a score for each graded tumor and preforming a Mann-Whitney U test.
Results
Neutrophil deficiency enhances the acute inflammatory response in colitis
PMN infiltration in IBD is a marker of disease severity and progression 18. In a mouse model of acute DSS-induced colitis, neutrophils infiltrated inflamed mucosal tissues 4-days after initiation of colitis and were maintained through active inflammation at 7-days and were quickly lost during resolution (Fig. 1A & B). Macrophage influx was milder and less robust (Fig. S1A & B). To dissect the role for PMNs in the inflammatory progression of colitis, we utilized mice with a genetic neutrophil deficiency. Mcl-1 is an anti-apoptotic member of the Bcl-2 family that is selectively required for PMN survival11. Mice with myeloid deficiency of Mcl-1 using LysM-cre (Fig. 1C) had greater than 50% reduction of circulating PMNs in heterozygous floxed animals (LysMCre;Mcl1fl/wt) and greater than 90% reduction in PMNs in homozygous floxed animals (LysMCre;Mcl1fl/fl) (Fig. 1D & E). This was accompanied by no significant change in the number of circulating monocytes as previously reported (Fig. S2A) 19. Furthermore, no changes in basal histological colon architecture, cell proliferation and cytokine profiles were observed (Fig. S2B-D).
Figure 1. Neutrophil depletion exacerbates acute colitis.
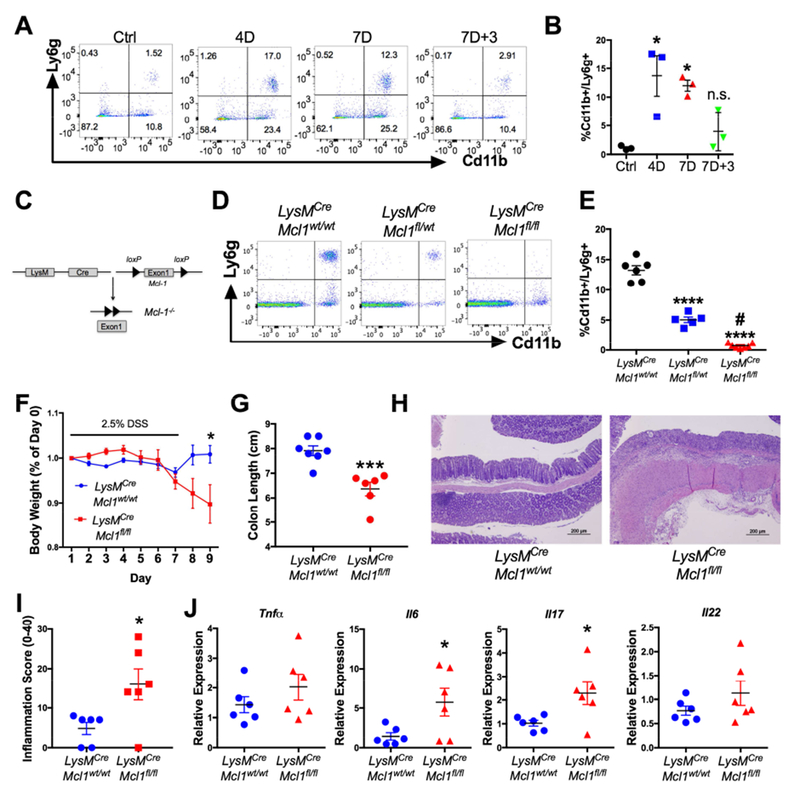
(A) Flow cytometry dot plots (D=days of DSS treatment) and (B) quantification of peripheral blood neutrophils in WT mice treated with 2.5% DSS then changed back to regular drinking water (n=3). (C) Schematic of Mcl-1 deletion in myeloid cells. (D) Representative dot plots and (E) quantification of flow cytometric analysis of peripheral blood neutrophils in LysMCre;Mcl1wt/wt, LysMCre;Mcl1fl/wt, and LysMCre;Mcl1fl/fl mice. ***p<0.001 and ****p<0.0001 relative to LysMCre;Mcl1wt/wt, #p<0.05 compared to LysMCre;Mcl1fl/wt, (n=5-6). (F) Body weight, (G) colon length, (H) hematoxylin & eosin staining, and (I) histopathologic inflammation score of LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl (n=6-7) mice treated with 2.5% DSS. ***p<0.001, *p<0.05. (J) qPCR analysis of indicated genes animals treated with 2.5% DSS. *p<0.05 relative to LysMCre;Mcl1wt/wt. Statistical analysis was performed with student’s t-test or one-way ANOVA followed by Tukey’s multiple comparisons test.
LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice were treated with 2.5% DSS in their drinking water for 7-days then changed back to regular drinking water for 2-days. Compared to LysMCre;Mcl1wt/wt mice, LysMCre;Mcl1fl/fl mice failed to recover body weight by 9-days and had significant colon length shortening (Fig. 1F & G). Histologic analysis showed significant destruction of colonic architecture and higher histopathologic inflammation in LysMCre;Mcl1fl/fl colon tissue compared to LysMCre;Mcl1wt/wt (Fig. 1H & I). PMN-deficiency did not change expression of Tnfa and Il22 but correlated with increased expression of Il17 and Il6 (Fig. 1J).
Neutrophils inhibit colitis-associated colon tumorigenesis.
PMNs are among the most prevalent immune cell type in the colon tumor microenvironment in the AOM/DSS model (Fig. S3A & B). The function for PMNs in the progression of inflammation-induced colon cancer has largely been suggested to be pro-tumorigenic 9. However, genetic PMN ablation prior to initiation of colon tumorigenesis has not been assessed. To address this, LysMCre;Mcl1wt/wt, LysMCre;Mcl1fl/wt, and LysMCre;Mcl1fl/fl mice were subjected to an AOM/DSS colon tumor model (Fig. 2A). The AOM/DSS model recapitulates many aspects observed in initiation of human CAC such as Wnt signaling activation, heightened inflammatory responses, and elevated c-myc 20. By the end of the third cycle of DSS, LysMCre;Mcl1fl/fl mice failed to recover body weight (Fig. 2B). Compared to LysMCre;Mcl1wt/wt animals, LysMCre;Mcl1fl/fl mice developed significantly larger tumors (Fig. 2C & D). No change in the total tumor number was observed between LysMCre;Mcl1wt/wt, LysMCre;Mcl1fl/wt, and LysMCre;Mcl1fl/fl mice (Fig. 2E). However, the tumor burden and average tumor size were significantly increased in LysMCre;Mcl1fl/fl mice (Fig. 2F & G). Flow cytometric analysis of primary tumor tissue conformed the depletion of intra-tumoral neutrophils (Fig. 2H and I). No change in the percentage of tumor-associated macrophages was observed (Fig. S4A & B). This suggests that PMNs play a critical role in inhibiting colitis-associated colon tumorigenesis by restricting tumor size and growth.
Figure 2. Neutrophils restrict colitis-associated colon tumor progression.
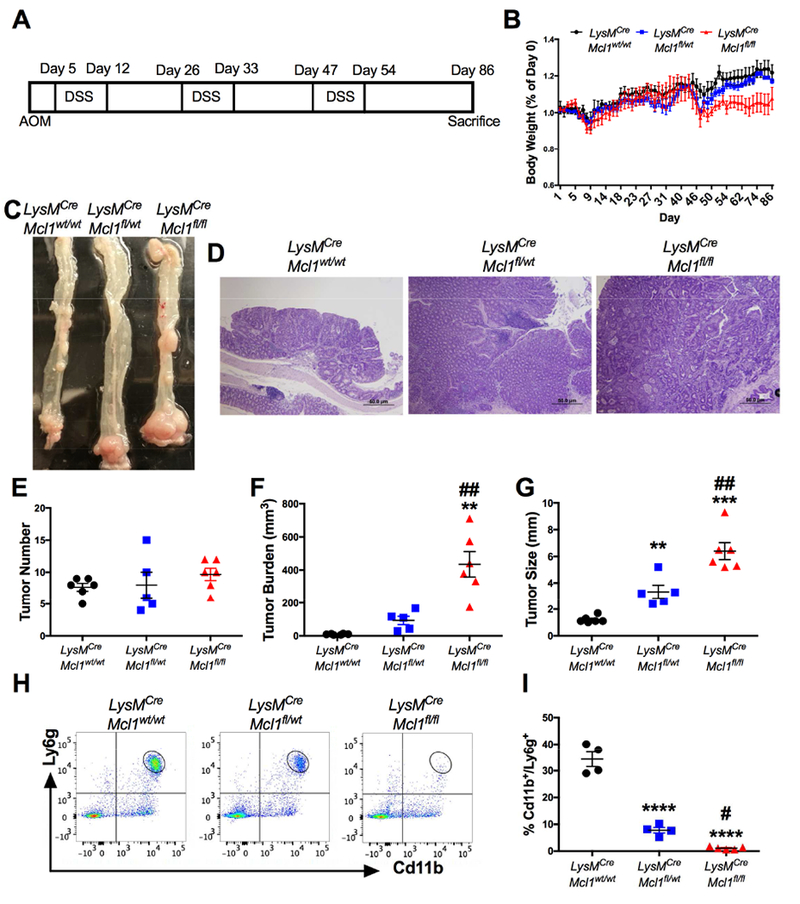
(A) Schematic of AOM/DSS-induced colon tumorigenesis. (B) Body weight, (C) representative images of whole mount specimens, (D) hematoxylin and eosin (H&E) staining of indicated mice after AOM/DSS. Body weight shown as a percentage of Day 0. of AOM/DSS treated colon tissue from LysMCre;Mcl1wt/wt, LysMCre;Mcl1fl/wt, and LysMCre;Mcl1fl/fl (n=5-6) mice. (E) Tumor number, (F) tumor burden, and (G) average tumor size of colon tumors. *p<0.05, **p<0.01, ***p<0.001 relative to LysMCre;Mcl1wt/wt, ##p<0.01 relative to LysMCre;Mcl1fl/wt. (H) Flow cytometric analysis and (I) quantification of Cd11b and Ly6g staining from primary colon tumors. ***p<0.001 relative to LysMCre;Mcl1wt/wt. ##p<0.01 relative to LysMCre;Mcl1fl/wt. Statistical analysis was performed with one-way ANOVA followed by Tukey’s multiple comparison’s test.
Neutrophil depletion increases colon tumor invasion and proliferation.
In the AOM/DSS model, highly invasive disease is rare and in many cases requires additional genetic hits 14. Further histologic characterization showed that no colon tumors in LysMCre;Mcl1wt/wt mice progressed into invasive adenocarcinomas (Fig. 3A). Strikingly, 25% of tumors from LysMCre;Mcl1fl/wt mice were invasive adenocarcinomas and this was potentiated in the LysMCre;Mcl1fl/fl mice where greater than 60% of colon tumors were invasive (Fig. 3B). Invasion through the muscularis propria was seen in 16% of tumors from LysMCre;Mcl1fl/fl, and many tumors could be detected in perirectal fat (Fig. 3C & D). In addition to increased tumor invasion, PMN depletion dramatically increased colon tumor proliferation as measured by Ki67 incorporation and proliferating cell nuclear antigen (PCNA) expression (Fig. 3E & F). These data delineate an important function for intra-tumoral PMNs in restricting tumor invasion and proliferation.
Figure 3. Neutrophils limit tumor progression and invasion.
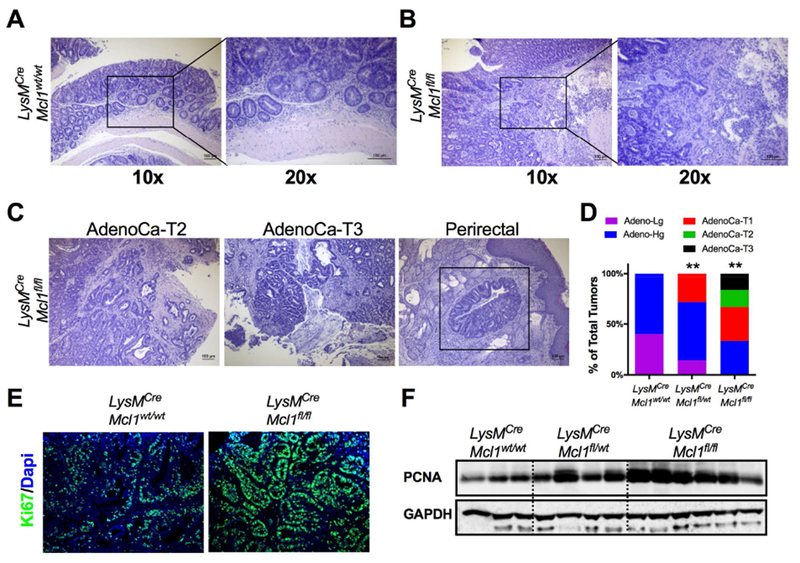
(A) Representative H&E of staining of adenoma from LysMCre;Mcl1wt/wt mice and (B) invasive adenocarcinoma from LysMCre;Mcl1fl/f mice at 10× and 20× magnification. (C) Representative image of H&E of invasive AdenoCa-T2, AdenoCa-T3, and colon tumor cells in perirectal fat in LysMCre;Mcl1fl/fl mice. (D) Quantification of percentage of low-grade adenomas (Adeno-lg), high-grade adenomas (Adeno-Hg), submucosal invasive adenocarcinomas (AdenoCa-T1), muscularis invasive adenocarcinoms (AdenoCa-T2), and invasive through muscularis (AdenoCa-T3) in at least 20 individual tumors from LysMCre;Mcl1wt/wt, LysMCre;Mcl1fl/wt, and LysMCre;Mcl1fl/fl mice. **p<0.01 relative to LysMCre;Mcl1wt/wt. Statistical analysis was performed with Mann-Whitney U test. (E) Representative image of Ki67 staining of colon tumors in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice. (F) Western blot analysis of proliferating cell nuclear antigen (PCNA) of colon tumors from LysMCre;Mcl1wt/wt, LysMCre;Mcl1fl/wt, and LysMCre;Mcl1fl/fl mice.
Neutrophil depletion enhances tumor progression in genetic colon cancer models.
The vast majority of colon tumors are not preceded by chronic inflammation but develop due to mutations in the Adenomatous polyposis coli (APC) gene 21. In mice, truncation of a single Apc allele is sufficient to cause spontaneous intestinal tumorigenesis, albeit the vast majority of these tumors are localized to the small intestine 22. Mice with a colon-specific disruption of Apc using the colon-specific homeobox 2 (Cdx2) Cre (Cdx2-CreERT2; Apcfl/fl) develop colon tumors after administration of tamoxifen 12. To understand the role of PMN depletion in sporadic tumorigenesis, lethally-irradiated Cdx2-CreERT2; Apcfl/fl mice were transplanted with bone marrow from either LysMCre,Mcl1wt/wt or PMN-deficient LysMCre;Mcl1fl/fl mice (Fig. 4A). Chimeras transplanted with LysMCre;Mcl1fl/fl bone marrow had depleted circulating PMNs (Fig. 4B and C). Cdx2-CreERT2;Apcfl/fl mice were administered a single injection of tamoxifen (100mg/kg, I.P.) and were sacrificed and analyzed for early neoplastic changes. 14-days after tamoxifen injection. In LysMCre;Mcl1wt/wt chimeras, only low-grade adenomas were observed, whereas 3 out of 7 LysMCre;Mcl1fl/fl chimeras developed submucosal invasive adenocarcinoma (AdenoCa-T1) (Fig. 4D & E). This suggests that PMNs restrict tumor progression in colon cancer independent of acute exacerbations of colitis in the AOM/DSS model.
Figure 4. Neutrophils restrict progression of sporadic colon tumorigenesis.
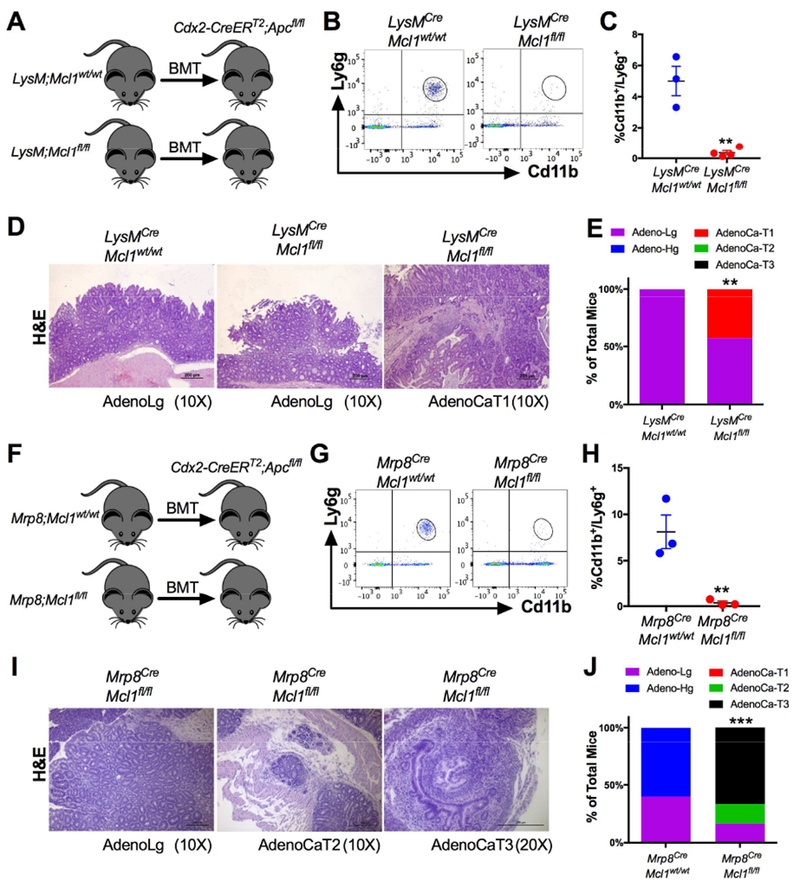
(A) Schematic of bone marrow transplantation from LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl (n=6-7) mouse donors to Cdx2-CreERT2;Apcfl/fl mice. (B) Flow cytometric dot plot analysis and (C) quantification of Cd11b and Ly6g staining of peripheral blood. **p<0.01 relative to LysMCre;Mcl1wt/wt. (D) Representative H&E images of low-grade adenomas (AdenoLg) in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl and invasive adenocarcinoma T1 (AdenoCaT1) in LysMCre;Mcl1fl/fl mice. (E) Quantification of colon tumor stage in indicated mice was assessed in at least 20 individual tumors. **p<0.01 relative to LysMCre;Mcl1wt/wt. Statistical analysis was performed with Mann-Whitney U test. (F) Schematic of bone marrow transplantation from Mrp8Cre;Mcl1wt/wt and Mrp8Cre;Mcl1fl/fl (n=6-7) mouse donors to Cdx2-CreERT2;Apcfl/fl mice. (G) Flow cytometric dot plot analysis and (H) quantification of Cd11b and Ly6g staining of peripheral blood. **p<0.01 relative to Mrp8Cre;Mcl1wt/wt . (I) Representative H&E images of low grade adenomas (AdenoLg), invasive adenocarcinoma T2 (AdenoCa-T2), and adenocarcinoma T3 (AdenCa-T3) in Mrp8Cre;Mcl1fl/fl mice. (J) Quantification of colon cancer stage in indicated mice. Quantification of colon tumor stage in indicated mice was assessed in at least 20 individual tumors. ***p<0.001 relative to Mrp8Cre;Mcl1wt/wt. Statistical analysis was performed with Mann-Whitney U test.
LysM-Cre is expressed in multiple myeloid cell types including monocytes, mature macrophages, and granulocytes 23. To address the dependence of PMN in enhanced tumor progression, we specifically deleted Mcll in PMNs by crossing Mcll floxed animals with the PMN-specific Mrp8-Cre transgenic mice 13. Mrp8Cre;Mcl1fl/fl mice had no significant difference in circulating monocytic cells (Fig. S5A & B). Cdx2-CreERT2;Apcfl/fl mice that were transplanted with Mrp8Cre;Mcl1fl/fl bone marrow had significant reduction of circulating PMNs (Fig. 4F-H). Strikingly, 14- days after tamoxifen administration, Mrp8Cre;Mcl1fl/fl bone marrow recapitulated the increased invasiveness observed in LysM;Mcl1fl/fl mice with areas of invasion into and through the muscularis propria (Fig. 4I & J). These data indicate that PMN depletion in sporadic colon tumorigenesis dramatically increased progression and invasion at early time points.
Neutrophils restrict colon tumor-associated bacteria expansion
To identify mechanisms by which PMN-depletion enhanced colon tumor progression, RNA-seq analysis was performed on tumor tissue from LysMCre;Mcl1wt/wt, LysMCre;Mcl1fl/wt, and LysMCre;Mcl1fl/fl mice. Several genes with known roles in anti-microbial defense were significantly reduced (Cfd, Lyz2, Defa27, Mcpt1) and several B-cell associated genes (Ighd, Ighg1, Fcmr) were highly upregulated (Fig. 5A, full gene list in Table S2). Neutrophils are well described for their anti-microbial and phagocytic functions 3. Neutrophil deficient LysMCre;Mcl1fl/fl mice have previously been shown to have higher attachment and decreased eradication of pathogenic bacteria in the colon 19. To assess for tumor-associated bacteria, we performed fluorescence in situ hybridization (FISH) using a universal bacterial probe (EUB338). Relative to AOM/DSS tumors from LysMCre;Mcl1wt/wt mice, PMN-deficient tumors have an increase in tumor-associated bacteria (Fig. 5B & C). Bacteria can induce intra-tumoral genetic instability by promoting DNA double-stranded breaks 24. PMN-deficient tumors had significantly more phosphorylated histone p-H2AX indicative of increased intra-tumoral genetic instability (Fig. 5D & E). High activation of inflammatory pathways such as NF-κB and STAT3 were also observed (Fig. S6A). IL17 was identified as a highly-expressed cytokine in RNA-seq analysis in LysMCre;Mcl1fl/fl colon tumors (Fig. 5A & F). Colon tumor-associated microbes drive tumor progression through activation of IL17 expression 25. Moreover, epithelial attachment of pathogenic microbes can directly induce an IL17 response 26.
Figure 5. Neutrophils limit tumor-associated bacteria.
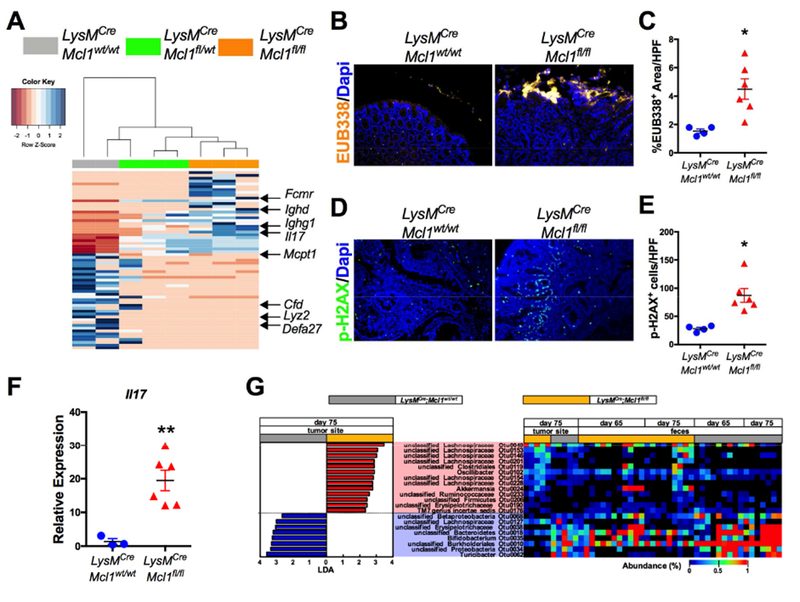
(A) Heat map of RNA-seq analysis of colon tumors from LysMCre;Mcl1wt/wt, LysMCre;Mcl1fl/wt, and LysMCre;Mcl1fl/fl mice. (B) Representative images fluorescence in situ hybridization (FISH) using universal bacteria EUB338 probe labeled with Cy3 in colon tumors from LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice. (C) Quantification of EUB338 positive area per high powered field (HPF). *p<0.05. (D) Representative images and (E) quantification of immunofluorescence staining of p-H2AX in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl colon tumors. **p<0.05. (F) qPCR analysis of IL17 expression in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl colon tumors. **p<0.01. (G) Heat map of significantly altered tumor-associated (Day 75) and fecal bacterial OTU abundances (Day 65 & Day 75) between LysMCre;Mcl1fl/fl and LysMCre;Mcl1wt/wt mice. Microbiota composition was analyzed by 16S rRNA Illumina sequencing and the OTUs that show significant difference (p<0.05) in tumor-associated microbiota compositions between LysMCre;Mcl1fl/fl and LysMCre;Mcl1wt/wt mice were determined by LEfSe analysis.
We next assessed alterations in microbiota by bacterial sequencing of fecal and primary tumor samples from LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice after AOM/DSS induced colon tumorigenesis. No significant changes in tumor-associated and fecal bacterial diversity were detected (Fig. S6B-D). However, several bacteria species were either over or under-represented in tumors and feces of LysMCre;Mcl1fl/fl mice (Fig. 5G). Interestingly, Akkermansia, a mucinolytic bacteria previously shown to be associated with human colon tumors was significantly increased in tumors from LysMCre;Mcl1fl/fl mice 27. Furthermore, Proteobacteria was significantly decreased in tumors from LysMCre;Mcl1fl/fl mice, which has previously been suggested to be decreased in human colon tumors 28. Collectively, these data suggest that PMNs play an important role in restricting bacterial dysbiosis in colon tumors.
Colon microbiota are essential for increased tumor growth in PMN-deficient mice
The colon microbiota is strongly associated with initiation and progression of colon tumors 29. Several bacterial species have been proposed to have a direct causal role in colon cancer30. To evaluate the contribution of bacteria to tumor growth, LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl were induced by AOM/DSS and beginning on day 14 after AOM treatment LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice were randomized into no antibiotics (Ctrl) and antibiotics treatment (Abx) groups (Fig. 6A). LysMCre;Mcl1fl/fl bone marrow chimeras had significantly reduced PMNs one-day prior to AOM administration (Fig. 6B). Whereas, LysMCre;Mcl1fl/fl mice developed dramatically larger colon tumors than LysMCre;Mcl1wt/wt mice, antibiotic treatment decreased tumor size and decreased tumor burden with no change in total tumor number in LysMCre;Mcl1fl/fl mice (Fig. 6C-F). Antibiotics treatment reduced PMN-deficient tumor-associated genetic instability (Fig. S7A). Furthermore, antibiotics reduced tumor invasion in LysMCre;Mcl1fl/fl mice (Fig. 6G-H). These data define an important contribution of colon microbiota in enhancing tumor growth and progression observed in PMN-deficient animals.
Figure 6. Microbiota and IL17 promotes tumor progression in PMN-deficient mice.
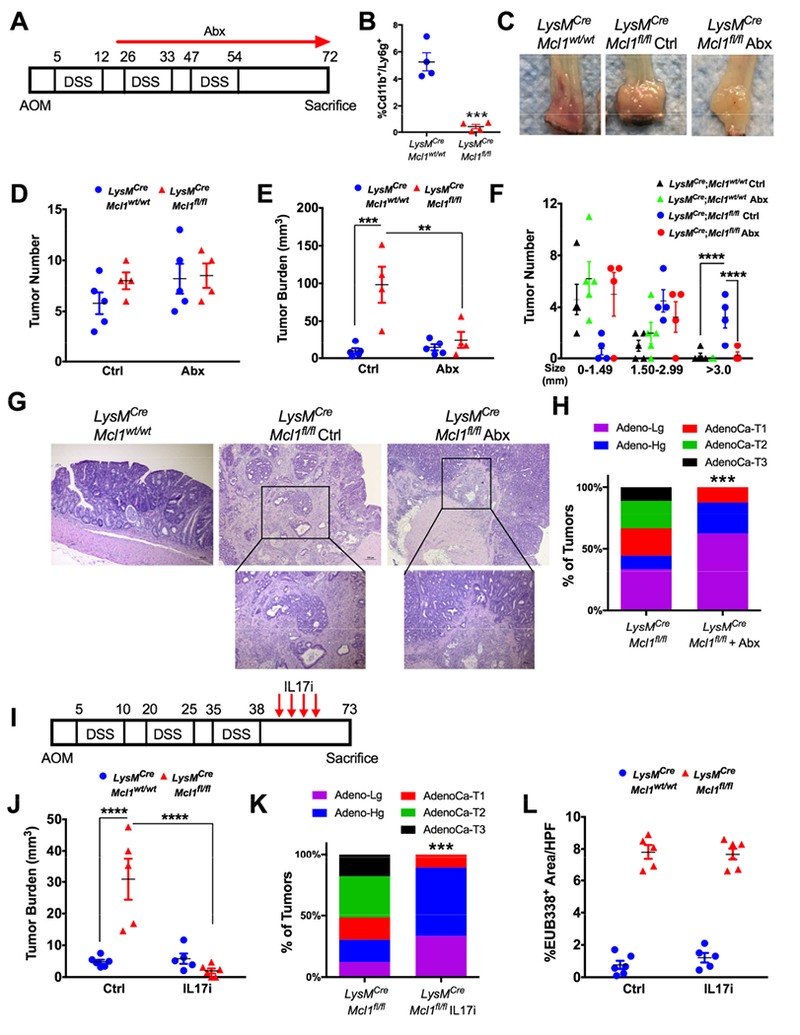
(A) Schematic of AOM/DSS-induced colon tumorigenesis (n=4-5). (B) Quantification of circulating Cd11b+/Ly6g+ cells in indicated mice. ***p<0.001 (C) Representative images of whole mount specimens, (D) tumor number, (E) tumor burden and (F) tumor size of colon tumors in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl treated with antibiotics (Abx) or no antibiotics (Ctrl). ****p<0.0001, ***p<0.001, **p<0.01. (G) Representative H&E analysis and (H) quantification of colon tumor progression from LysMCre;Mcl1fl/fl, and LysMCre;Mcl1fl/fl mice treated with antibiotics. Quantification of colon tumor stage in indicated mice was assessed in at least 20 individual tumors. ***p<0.001 relative to LysMCre;Mcl1wt/wt. Statistical analysis was performed with Mann-Whitney U test. (I) Schematic of AOM/DSS-induced colon tumorigenesis and IL17 inhibition (n=5-6). (J) Tumor burden and (K) quantification of colon tumor progression in LysMCre;Mcl1fl/fl, and LysMCre;Mcl1fl/fl mice treated with IL17 inhibition. (L) Quantification of tumor-associated bacteria with Cy3 labeled EUB338 FISH probe in indicated mice. Statistical analysis was performed with student’s t-test and two-way ANOVA followed by Tukey’s multiple comparisons test.
We identified the pro-tumorigenic and bacteria-dependent cytokine IL17 as being highly upregulated in PMN-deficient colon tumors. To determine if microbiota enhance colon tumorigenesis through IL17-dependent mechanism a neutralizing antibody that efficiently decreased systemic and local IL17 was used (S7B). Colon tumorigenesis was induced in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl by AOM/DSS. A shortened DSS cycle was used to maintain high survival in the LysMCre;Mcl1fl/fl mice. Immediately following the third cycle of DSS, mice were randomized to treatment with control IgG or an IL17 depleting antibody (Fig. 6I) 31. Similar to antibiotics, IL17 inhibition reversed tumor growth and progression in neutrophil deficient animals (Fig. 6J-K & S7C). IL17 inhibition had no impact on tumor-associated bacteria (Fig. 6L). These data suggest a pro-inflammatory loop through which colon-tumor associated PMNs restrict microbial outgrowth and IL17 expression to decrease tumor progression.
Neutrophils restrict expansion of colon tumor-associated B cells
RNA-seq data also showed many genes associated with B-cells were higher in LysMCre;Mcl1fl/fl colon tumors. Immunofluorescence and flow cytometric analysis of tumors showed robust infiltration of B-cells into primary tumors from PMN-depleted mice compared to LysMCre;Mcl1wt/wt mice (Fig. S8A & B). No changes in circulating B-cells or fecal IgA were observed in untreated LysMCre;Mcl1fl/fl mice (Fig. S8C-E). In addition to heightened B-cell infiltration into the primary tumor, B-cells were also directly associated with highly invasive areas in colitis-associated and sporadic tumors from LysMCre;Mcl1fl/fl mice (Fig. 7A and Fig S8F). Interestingly IL17 depletion reversed intratumoral B-cells, suggesting a link between tumoral IL17 expression and B-cells (Fig. 7B). Likewise, neutrophil-deficient animals treated with antibiotics had significantly reduced tumor-associated B-cells (Fig. 7C). To define the functional role of B-cells downstream of bacteria and IL17 in LysMCre;Mcl1fl/fl mice, we decided to target B-cells using anti-B220 antibody-mediated B-cell inhibition 32. To evaluate the ability of anti-B220 therapy to decrease B-cells from colonic tissue during inflammation, mice were treated with 2.5% DSS and were randomized to Ctrl or anti-B220 antibody (400ug I.P.) treatment in acute DSS-colitis (Fig. S9A). Circulating B-cells and intracolonic B-cells were significantly decreased after three doses anti-B220 antibody (Fig. S9B & C). This treatment did not have a significant effect on the acute inflammatory response in colitis (Fig. S9D-F).
Figure 7. B-cells are important in PMN-deficient colitis-associated colon tumor model.
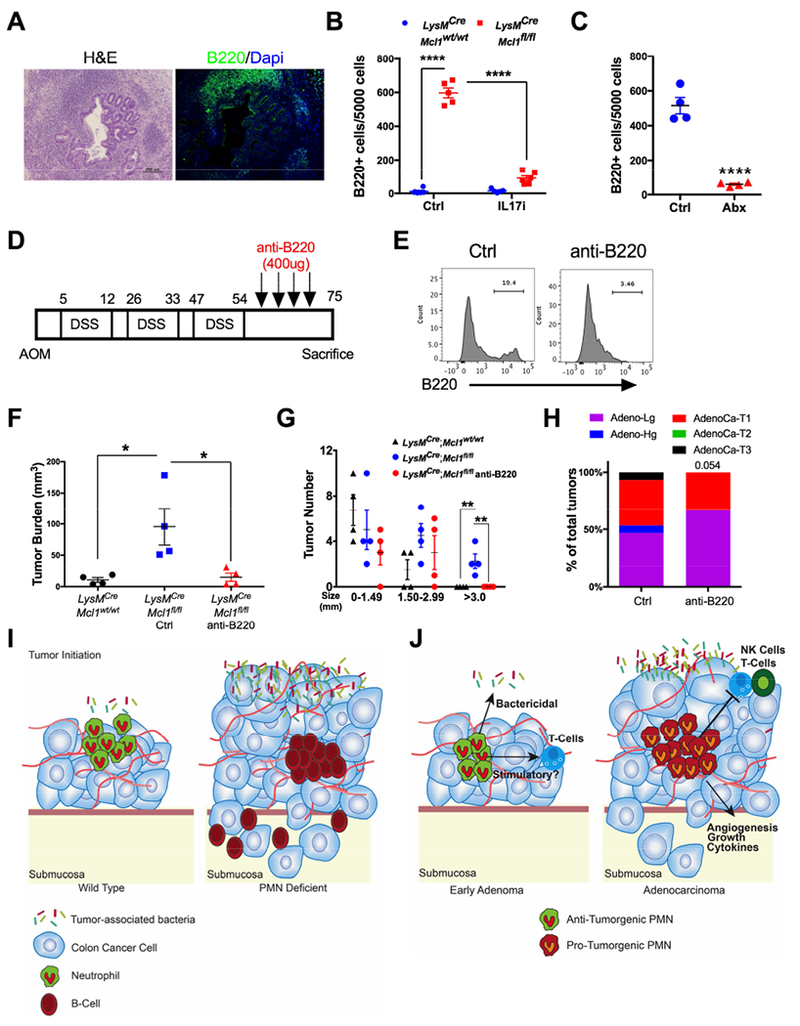
(A) Representative H&E and B220 staining from LysMCre;Mcl1fl/fl tumor tissue of invasive adenocarcinoma. (B) B220+ cell quantification from tumors from indicated mice treated with IL17 or control antibody shown as total positive cells per 5000 counted cells. ****p<0.0001. (C) B220+ cell quantification from tumor tissue from Ctrl and anti-B220 treated LysMCre;Mcl1fl/fl mice. ****p<0.0001. (D) Schematic of AOM/DSS-induced colon tumorigenesis. Anti-B220 treatment was initiated at day 54 with 400ug/mouse treatment every fourth day (n=4). (E) Representative flow cytometric analysis of B220 staining in colon tissue from Ctrl and anti-B220 treated LysMCre;Mcl1fl/fl mice. (F) Tumor burden, (G) tumor size, and (H) quantification of colon cancer stage in indicated mice. with anti-B220 antibody or control. *p<0.05 relative to LysMCre;Mcllwt/wt and LysMCre;Mcl1fl/fl mice treated with or without anti-B220. ** ##p<0.001 relative to LysMCre;Mcl1fl/fl + anti-B220, and **p<0.01 relative to all groups >3.0mm tumor size. Statistical analysis performed by either student’s t-test or one-way ANOVA followed by Tukey’s multiple comparison’s test or Mann-Whitney U test for tumor invasiveness. (I) Neutrophils restrict colon tumor progression and invasion by restricting outgrowth of tumor-associated microbiota and limiting infiltration of B-cells through IL17. (J) We propose a model in which PMNs in the earliest stage colon tumors inhibit tumorigenesis through repression of tumor-associated bacteria as well as other potential mechanisms such as stimulation of T-cells. In later stage colon tumors, PMNs acquire pro-tumorigenic function and inhibit anti-tumor immunity, directly increase tumor growth, secrete inflammatory cytokines, and promote angiogenesis.
To assess B-cells in the progression of PMN-deficient tumors, colon tumorigenesis was induced in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice using AOM/DSS. Immediately following the third cycle of DSS, LysMCre;Mcl1fl/fl animals were randomized into Ctrl and anti-B220 treated groups and were treated beginning every fourth day from Day 50 through Day 75 (Fig. 7C). At 75-days post induction of colon tumorigenesis, anti-B220 treatment reduced colon B-cells (Fig. 7D). No change in tumor number was detected between any groups (Fig. S10A). However, tumor burden and tumor size were dramatically increased in LysMCre;Mcl1fl/fl mice relative to LysMCre;Mcl1wt/wt control animals (Fig. 7E & F). B-cell inhibition in LysMCre;Mcl1fl/fl mice improved the observed increase in tumor burden and tumor size (Fig. 7E & F). Furthermore, B-cell depletion led to a reduction in the percentage of invasive adenocarcinomas and increased percentage of non-invasive adenomas (Fig. 7G & S10B).
Discussion
PMNs are highly abundant immune cells in CRC. The recruitment of PMNs into early or chronic tumors are believed to occur through analogous mechanisms, which are largely mediated by CXCR2 ligands 2, 33. Many of these ligands are regulated by local hypoxia and HIF transcriptional activation in the tumor microenvironment 55. PMN and granulocytic-cell targeted therapeutics have been proposed for the treatment of several tumor types 4. Peptide-mimetics targeting the G-protein coupled receptor CXCR2 which mediates PMN and myeloid-derived suppressor cells (MDSCs) infiltration into tumors is efficacious in dampening colon tumorigenesis in both colitis-associated as well as sporadic colon tumor models 8. Peptide Fc-fusion proteins (peptibodies) targeting the S100 family of protein expressed on MDSCs and PMNs have also shown to dampen tumor growth in preclinical models 34. However, PMN infiltration into human colon tumors was associated with both positive and adverse clinical outcomes 35, 36. Experiments using mouse models further raise questions about the function of PMNs in the initiation and progression of colon tumors as conflicting data on the role of PMNs have been shown. Our work clearly demonstrates depletion of neutrophils prior to initiation of colon tumorigenesis profoundly enhances tumor growth, proliferation, and invasion.
Discrepancies between neutrophil function in our study relative to others could perhaps be explained by differences in models. Anti-Ly6g antibody depletion of PMNs is commonly used in tumor studies 37 Interestingly, flow cytometric analysis suggests Cd11b−/Ly6g+ exist, which could perhaps suggest non-PMN targets of this treatment may affect the robust reduction in colitis-associated tumors observed in this model 38. Although CXCR2 is not completely PMN selective, deletion of the chemokine receptor CXCR2 results in reduced Cd11b+/Ly6g+ infiltration into colon tumors and significant reduction in colon tumorigenesis 38. Strategies to inhibit CXCR2 may be effective in inhibiting a subset of neutrophils. Intratumoral PMN heterogeneity has been proposed to partly explain the differential effects of PMN depletion on tumorigenesis 39–42. Moreover, a landmark study showed that PMN plasticity can be driven towards pro-tumor “N2” PMNs or anti-tumor “N1” PMNs in a TGF-ß-dependent manner. Approaches to target all tumor-associated neutrophils as described in the present work potentiate disease progression. Other models of genetic neutrophil depletion have been used in the study of other cancers. The Csf3r knockout mouse fail to develop PMNs and enhance tumor growth in a murine uterine tumor model 43.
The colon microbiota and microbial dysbiosis drive colon tumorigenesis 29. This idea was highlighted with studies from germ-free mouse models which show decreased colon tumorigenesis 44. Furthermore, E. coli species were directly linked to tumor invasion 45. Deletion of PMNs profoundly increased tumor-associated bacteria and many known microbiota-dependent responses, including genetic instability and IL17 response, which are known to have critical roles in neoplastic progression. Importantly, antibiotics treatment reduced PMN-deficient tumor progression and invasion. However, antibiotics treatment did not impact tumorigenesis in WT animals, which is contrary to several published reports in germ-free animals 44. This may be explained by delayed treatment of antibiotics until two-weeks following colon tumor initiation. Our studies also identified IL17 as highly upregulated in colon tumors of neutrophil-deficient animals. Previous reports have shown IL17 knockout reduces tumor burden in AOM/DSS 46. IL17 inhibition using monoclonal antibodies reversed tumor progression in LysMCre;Mcllfl/fl mice suggesting IL17 is a key downstream mechanism linking microbiota expansion and increased tumor growth and invasion in genetic neutrophil depletion.
The precise function for PMNs in the progression of IBD is not well known. Chronic granulomatous disease (CGD), a primary deficiency in the NADPH oxidase complex resulting in inability of neutrophils to generate reactive oxygen species to kill pathogens, can predispose patients to development of IBD-like disease that resembles Crohn’s disease 47. Recently, it has been suggested that in addition to the pathogen killing function, decreased PMN-mediated O2-consumption through decreased ROS production in CGD reduces hypoxia-inducible transcription factor-1α (HIF-1α) activity, an essential regulator of colon epithelial barrier integrity in the TNBS colitis model 48. Several studies suggesting a prominent role for PMNs in promoting inflammatory responses have been reported 18. The data in our studies suggest that neutrophil depletion in LysMCre;Mcl1fl/fl mice increases the acute inflammatory response in DSS-colitis.
One of the more surprising findings of our study is the enhanced tumor invasion in response to PMN depletion. Interestingly, 25% of tumors from LysMCre;Mcl1fl/wt animals were invasive and moderate albeit not statistically significant increases in tumor burden and size. These animals retain 10% of tumor-associated PMNs, which may confer modest protection in tumor growth and progression. PMNs are purported to increase tumor invasion by secreting enzymes such as matrix metalloproteinase 9 (MMP9) 49. In hepatocellular carcinoma, PMNs directly initiate an epithelial-mesenchymal transition in tumor cells 50. PMNs were recently shown to suppress Natural Killer (NK) cell-mediated breast cancer cell killing and promote tumor cell extravasation 51. Also in breast cancer, PMNs induced by IL17 producing T-cells were found to increase tumor metastasis through repression of CD8 T-cells 52. However, PMNs have also been shown to limit metastatic seeding of tumors in the lung through generation of H2O2 53. Our findings demonstrate in colon cancer that depleting neutrophils before initiation of tumors leads to cell invasion through all layers of the colon mucosa.
PMN-deficient tumors were highly infiltrated with B220+ B-cells which was IL17 and bacteria dependent. Analogous to PMNs, B-cell function in tumors is not well understood and could have both pro- and anti-tumor functions 54–57. Recently it was suggested that IL17 through NF-kB and STAT1 signaling is sufficient to promote B-cell infiltration into colon tumors 57. Bacteria are also known to induce colonic B-cell responses 26. Inhibition of B-cells decreased tumor size and decreased tumor invasion in PMN-deficient mice. Previous reports suggested that B-cells in colon lymphoid follicles were an early site of tumor cell invasion in rat and mouse models of colon cancer 58. It has also been suggested that human colon cancers are highly associated with B-cells and lymphoid follicles 59. In our model, B-cells were associated with invasive areas and promoted tumor growth and invasion.
Our data suggest a mechanism by which PMNs restrict tumor-associated bacteria and depletion of PMNs promotes a dramatic inflammatory response characterized by bacteria-dependent IL17 expression which drives B-cell infiltration, which are essential for colon tumor growth and progression (Fig. 7H). Clinical studies suggest PMNs are a positive prognostic indicator in early stage colon cancer 35. Studies in mice have proposed that PMNs in early stage tumors retain anti-tumor functions but a phenotypic switch to a more immunosuppressive state occurs throughout tumor progression 60. It is possible that similar mechanisms exist in colon cancer whereby the earliest tumor infiltrating PMNs serve to inhibit expansion of colon microbiota and B-cells to limit tumorigenesis and progression, whereas, in established tumors PMNs evolve a more pro-tumorigenic phenotype (Fig. 7I & J).
Supplementary Material
Figure S1. Macrophage infiltration in DSS colitis. (A) Representative flow cytometry dot plots and (B) quantification of F4/80+/Cd11b+ cells in DSS colitis time course. *p<0.05.
Figure S2. Characterization of LysMCre;Mcl1fl/fl mice. (A) Representative flow cytometry dot plots of F4/80+/Cd11b+ cells in peripheral blood. (B) Representative H&E staining and (C) Ki67 immunofluorescence from LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mouse colon tissue. (D) Cytokine array analysis from colon tissue lysates from LysMCre;Mcl1wt/wt (n=5) and LysMCre;Mcl1fl/fl (n=7).
Figure S3. Neutrophils are highly infiltrated in AOM/DSS colon tumors. (A) Representative flow cytometry dot plots and (B) quantification of Ly6g+/Cd11b+ cells in AOM/DSS colon tumors from WT mice. ****p<0.001. Statistical analysis performed by student’s t test.
Figure S4. Tumor infiltration of macrophages is unaltered in LysMCre;Mcl1fl/fl mice. (A) Representative flow cytometry dot plots and (B) quantification of F4/80+/Cd11b+ cells in colon tumors from LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice. Statistical analysis performed by student’s t test.
Figure S5. Monocytes are unchanged in Mrp8-Cre/Mcl1fl/fl mice. (A) Representative flow cytometry dot plots and (B) quantification of Ly6c+/Cd11b+ cells as a percentage of total myeloid cells from Mrp8Cre;Mcl1wt/wt and Mrp8Cre;Mcl1fl/fl mice. Statistical analysis performed by student’s t test.
Figure S6. PMN depletion increases microbiota-dependent inflammatory responses. (A) Western blot analysis of colon tumors from LysMCre;Mcl1wt/wt, LysMCre;Mcl1fl/wt, and LysMCre;Mcl1fl/fl mice. Statistical analysis was performed with student’s t-test. (B) NMDS plot based on θYC of LysMCre;Mcl1wt/wt (WT) and LysMCre;Mcl1fl/fl (KO) (C) β-diversity index, θYC of WT and KO mice. (D) Shannon’s α-diversity index of WT and KO mice.
Figure S7. Bacteria and IL-17 are important for neutrophil-deficient colon tumorigenesis. (A) Representative images of immunofluorescence staining of p-H2AX in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl colon tumors treated with Ctrl or antibiotics (Abx). (B) ELISA analysis of IL-17 protein in colon tissue explants and (C) serum of mice treated with Ctrl IgG or IL-17 IgG (IL17i). **** p<0.001. (D) Tumor size of colon tumors in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl treated with Ctrl IgG or IL-17 IgG (IL17i). ****p<0.0001.
Figure S8. Increased B-cells in PMN-deficient colon tumors. (A) Flow cytometric analysis and (B) representative images of B220 staining of colon tumor tissue from LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice. (C) Flow cytometric analysis of B220 staining and (D) quantification of peripheral blood B-cells. (E) IgA ELISA analysis from feces of LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice. (F) Representative immunofluorescence of B220 staining from WT tumor tissue and invasive adenocarcinoma tissue from LysMCre;Mcl1fl/fl mouse.
Figure S9. B-cell depletion does not exacerbate DSS colitis. (A) Schematic of 2.5% DSS treatment and anti-B220 injections (400ug/mouse I.P.) on days 1, 3, and 5. (B) Representative flow cytometric analysis of B220 staining in peripheral blood and colon tissue and (C) quantification B-cells of Ctrl and anti-B220 treated animals. (D) Representative H&E histologic analysis (E) colon length and (F) total inflammation score of treated mice. Statistical analysis performed by student’s t test.
Figure S10. B-cell depletion reduces PMN-deficient tumor progression. (A) Tumor number and (B) representative histologic analysis indicated tumor stage from Ctrl and anti-B220 treated LysMCre;Mcl1fl/fl mice. Statistical analysis was performed by one-way ANOVA followed by Tukey’s multiple comparisons test.
Table S1: Primers
Acknowledgments
Funding Information: This work was supported by NIH grants (CA148828 and DK095201 to Y.M.S., ES028802 to J.A.C.), the University of Michigan Gastrointestinal Peptide Center (Y.M.S.), a pilot grant from the University of Michigan GI Spore (CA130810 to Y.M.S.), D.T. was supported by NIH Grant (F30CA213664). S.K.R. was supported by NIH grant (K99DK110537). Crohn’s and Colitis Foundation of America Senior Award (N.I).
Abbreviations:
- Abx
antibiotics
- AdenoCa
adenocarcinoma
- Adeno-Hg
high grade adenoma
- Adeno-Lg
low grade adenoma
- AOM
azoxymethane
- CGD
chronic granulomatous disease
- DSS
dextran sulfate sodium
- FDR
false discovery rate
- FISH
fluorescent in situ hybridization
- HPF
high powered field
- IBD
inflammatory bowel disease
- OTU
operational taxonomic units
- PMN
polymorphonuclear neutrophils
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Conflict of Interests:
The authors declare no conflict of interest.
References
- 1.Terzić J, Grivennikov S, Karin E, et al. Inflammation and Colon Cancer. Gastroenterology 2010;138:2101–2114.e5. [DOI] [PubMed] [Google Scholar]
- 2.Triner D, Shah YM. Hypoxia-inducible factors: a central link between inflammation and cancer. The Journal of Clinical Investigation 2016;126:3689–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicolás-Ávila JÁ, Adrover JM, Hidalgo A. Neutrophils in Homeostasis, Immunity, and Cancer. Immunity;46:15–28. [DOI] [PubMed] [Google Scholar]
- 4.Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nature Reviews Cancer 2016;16:431. [DOI] [PubMed] [Google Scholar]
- 5.Houghton AM, Rzymkiewicz DM, Ji H, et al. Neutrophil elastase-mediated degradation of IRS-1 accelerates lung tumor growth. Nature Medicine 2010;16:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang T-t, Zhao Y-l, Peng L-s, et al. Tumour-activated neutrophils in gastric cancer foster immune suppression and disease progression through GM-CSF-PD-L1 pathway. Gut 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nature Medicine 2015;21:938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamieson T, Clarke M, Steele CW, et al. Inhibition of CXCR2 profoundly suppresses inflammation-driven and spontaneous tumorigenesis. The Journal of Clinical Investigation 2012;122:3127–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang K, Bai Y-P, Wang C, et al. Crucial Involvement of Tumor-Associated Neutrophils in the Regulation of Chronic Colitis-Associated Carcinogenesis in Mice. PLOS ONE 2012;7:e51848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ocana A, Nieto-Jiménez C, Pandiella A, et al. Neutrophils in cancer: prognostic role and therapeutic strategies. Molecular Cancer 2017;16:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dzhagalov I, St. John A, He Y-W The antiapoptotic protein Mcl-1 is essential for the survival of neutrophils but not macrophages. Blood 2007;109:1620–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y, Sentani K, Wiese A, et al. Sox9 Induction, Ectopic Paneth Cells, and Mitotic Spindle Axis Defects in Mouse Colon Adenomatous Epithelium Arising From Conditional Biallelic Apc Inactivation. The American Journal of Pathology 2013;183:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Németh T, Futosi K, Sitaru C, et al. Neutrophil-specific deletion of the CARD9 gene expression regulator suppresses autoantibody-induced inflammation in vivo. Nature Communications 2016;7:11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubé PE, Yan F, Punit S, et al. Epidermal growth factor receptor inhibits colitis-associated cancer in mice. The Journal of Clinical Investigation 2012;122:2780–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozich JJ, Westcott SL, Baxter NT, et al. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Applied and Environmental Microbiology 2013;79:5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Applied and Environmental Microbiology 2009;75:7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wéra O, Lancellotti P, Oury C. The Dual Role of Neutrophils in Inflammatory Bowel Diseases. Journal of Clinical Medicine 2016;5:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamada N, Sakamoto K, Seo S-U, et al. Humoral Immunity in the Gut Selectively Targets Phenotypically Virulent Attaching-and-Effacing Bacteria for Intraluminal Elimination. Cell host & microbe 2015;17:617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertis MD, Massi E, Poeta ML, et al. The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. Journal of Carcinogenesis 2011;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fearon ER. Molecular Genetics of Colorectal Cancer. Annual Review of Pathology: Mechanisms of Disease 2011;6:479–507. [DOI] [PubMed] [Google Scholar]
- 22.Xue X, Taylor M, Anderson E, et al. Hypoxia-Inducible Factor-2α Activation Promotes Colorectal Cancer Progression by Dysregulating Iron Homeostasis. Cancer Research 2012;72:2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abram CL, Roberge GL, Hu Y, et al. Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. Journal of immunological methods 2014;408:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nougayréde J-P, Homburg S, Taieb F, et al. <em>Escherichia coli</em> Induces DNA Double-Strand Breaks in Eukaryotic Cells. Science 2006;313:848–851. [DOI] [PubMed] [Google Scholar]
- 25.Grivennikov SI, Wang K, Mucida D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL17-mediated tumour growth. Nature 2012;491:254–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atarashi K, Tanoue T, Ando M, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell;163:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J, Kato I. Gut microbiota, inflammation and colorectal cancer. Genes & Diseases 2016;3:130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Z, Guo B, Gao R, et al. Microbiota disbiosis is associated with colorectal cancer. Frontiers in Microbiology 2015;6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennan CA, Garrett WS. Gut Microbiota, Inflammation, and Colorectal Cancer. Annual Review of Microbiology 2016;70:395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor immune microenvironment. Cell host & microbe 2013;14:207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coffelt SB, Kersten K, Doornebal CW, et al. IL17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature 2015;522:345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olkhanud PB, Damdinsuren B, Bodogai M, et al. Tumor-evoked regulatory B cells promote breast cancer metastasis by converting resting CD4(+) T cells to T regulatory cells. Cancer research 2011;71:3505–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Triner D, Xue X, Schwartz AJ, et al. Epithelial Hypoxia-Inducible Factor 2α Facilitates the Progression of Colon Tumors through Recruiting Neutrophils. Molecular and Cellular Biology 2017;37:e00481–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin H, Lerman B, Sakamaki I, et al. Generation of a novel therapeutic peptide that depletes MDSC in tumor-bearing mice. Nature medicine 2014;20:676–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wikberg ML, Ling A, Li X, et al. Neutrophil infiltration is a favorable prognostic factor in early stages of colon cancer. Human Pathology 2017;68:193–202. [DOI] [PubMed] [Google Scholar]
- 36.Haram A, Boland MR, Kelly ME, et al. The prognostic value of neutrophil-to-lymphocyte ratio in colorectal cancer: A systematic review. Journal of Surgical Oncology 2017;115:470–479. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Wang K, Han GC, et al. Neutrophil infiltration favors colitis-associated tumorigenesis by activating the interleukin-1 (IL-1)/IL-6 axis. Mucosal Immunology 2014;7:1106. [DOI] [PubMed] [Google Scholar]
- 38.Katoh H, Wang D, Daikoku T, et al. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer cell 2013;24:631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fridlender ZG, Sun J, Kim S, et al. Polarization of Tumor-Associated Neutrophil Phenotype by TGF-β: “N1” versus “N2” TAN. Cancer Cell;16:183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sagiv Jitka Y, Michaeli J, Assi S, et al. Phenotypic Diversity and Plasticity in Circulating Neutrophil Subpopulations in Cancer. Cell Reports;10:562–573. [DOI] [PubMed] [Google Scholar]
- 41.Ostanin DV, Kurmaeva E, Furr K, et al. Acquisition of Antigen-Presenting Functions by Neutrophils Isolated from Mice with Chronic Colitis. The Journal of Immunology 2012;188:1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mishalian I, Bayuh R, Levy L, et al. Tumor-associated neutrophils (TAN) develop pro-tumorigenic properties during tumor progression. Cancer Immunology, Immunotherapy 2013;62:1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blaisdell A, Crequer A, Columbus D, et al. Neutrophils Oppose Uterine Epithelial Carcinogenesis via Debridement of Hypoxic Tumor Cells. Cancer cell 2015;28:785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Kundu P, Seow SW, et al. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APC Min/+ mice. Carcinogenesis 2012;33:1231–1238. [DOI] [PubMed] [Google Scholar]
- 45.Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science 2012;338:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hyun YS, Han DS, Lee AR, et al. Role of IL17A in the development of colitis-associated cancer. Carcinogenesis 2012;33:931–6. [DOI] [PubMed] [Google Scholar]
- 47.Schappi M, Smith V, Goldblatt D, et al. Colitis in chronic granulomatous disease. Archives of Disease in Childhood 2001;84:147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell EL, Bruyninckx WJ, Kelly CJ, et al. Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity 2014;40:66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kessenbrock K, Plaks V, Werb Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell 2010;141:52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou S-L, Zhou Z-J, Hu Z-Q, et al. CXCR2/CXCL5 axis contributes to epithelial-mesenchymal transition of HCC cells through activating PI3K/Akt/GSK-3β/Snail signaling. Cancer Letters 2015;358:124–135. [DOI] [PubMed] [Google Scholar]
- 51.Spiegel A, Brooks MW, Houshyar S, et al. Neutrophils suppress intraluminal NK-mediated tumor cell clearance and enhance extravasation of disseminated carcinoma cells. Cancer discovery 2016;6:630–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coffelt SB, Kersten K, Doornebal CW, et al. IL17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature 2015;522:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Granot Z, Henke E, Comen E, et al. Tumor entrained neutrophils inhibit seeding in the premetastatic lung. Cancer cell 2011;20:300–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuen GJ, Demissie E, Pillai S. B lymphocytes and cancer: a love-hate relationship. Trends in cancer 2016;2:747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shalapour S, Lin X-J, Bastian IN, et al. Inflammation-induced IgA+ cells dismantle antiliver cancer immunity. Nature 2017;551:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee K, Spata M, Bayne LJ, et al. Hif1α deletion reveals pro-neoplastic function of B cells in pancreatic neoplasia. Cancer discovery 2016;6:256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu R, Lu Z, Gu J, et al. MicroRNAs 15A and 16–1 Activate Signaling Pathways That Mediate Chemotaxis of Immune Regulatory B cells to Colorectal Tumors. Gastroenterology 2017. [DOI] [PubMed] [Google Scholar]
- 58.Nauss KM, Locniskar M, Pavlina T, et al. Morphology and Distribution of 1,2-Dimethylhydrazine Dihydrochloride-Induced Colon Tumors and Their Relationship to Gut-Associated Lymphoid Tissue in the Rat2. JNCI: Journal of the National Cancer Institute 1984;73:915–924. [PubMed] [Google Scholar]
- 59.Bronen RA, Glick SN, Teplick SK. Diffuse lymphoid follicles of the colon associated with colonic carcinoma. American Journal of Roentgenology 1984;142:105–109. [DOI] [PubMed] [Google Scholar]
- 60.Eruslanov EB, Bhojnagarwala PS, Quatromoni JG, et al. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. The Journal of Clinical Investigation 2014;124:5466–5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Macrophage infiltration in DSS colitis. (A) Representative flow cytometry dot plots and (B) quantification of F4/80+/Cd11b+ cells in DSS colitis time course. *p<0.05.
Figure S2. Characterization of LysMCre;Mcl1fl/fl mice. (A) Representative flow cytometry dot plots of F4/80+/Cd11b+ cells in peripheral blood. (B) Representative H&E staining and (C) Ki67 immunofluorescence from LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mouse colon tissue. (D) Cytokine array analysis from colon tissue lysates from LysMCre;Mcl1wt/wt (n=5) and LysMCre;Mcl1fl/fl (n=7).
Figure S3. Neutrophils are highly infiltrated in AOM/DSS colon tumors. (A) Representative flow cytometry dot plots and (B) quantification of Ly6g+/Cd11b+ cells in AOM/DSS colon tumors from WT mice. ****p<0.001. Statistical analysis performed by student’s t test.
Figure S4. Tumor infiltration of macrophages is unaltered in LysMCre;Mcl1fl/fl mice. (A) Representative flow cytometry dot plots and (B) quantification of F4/80+/Cd11b+ cells in colon tumors from LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice. Statistical analysis performed by student’s t test.
Figure S5. Monocytes are unchanged in Mrp8-Cre/Mcl1fl/fl mice. (A) Representative flow cytometry dot plots and (B) quantification of Ly6c+/Cd11b+ cells as a percentage of total myeloid cells from Mrp8Cre;Mcl1wt/wt and Mrp8Cre;Mcl1fl/fl mice. Statistical analysis performed by student’s t test.
Figure S6. PMN depletion increases microbiota-dependent inflammatory responses. (A) Western blot analysis of colon tumors from LysMCre;Mcl1wt/wt, LysMCre;Mcl1fl/wt, and LysMCre;Mcl1fl/fl mice. Statistical analysis was performed with student’s t-test. (B) NMDS plot based on θYC of LysMCre;Mcl1wt/wt (WT) and LysMCre;Mcl1fl/fl (KO) (C) β-diversity index, θYC of WT and KO mice. (D) Shannon’s α-diversity index of WT and KO mice.
Figure S7. Bacteria and IL-17 are important for neutrophil-deficient colon tumorigenesis. (A) Representative images of immunofluorescence staining of p-H2AX in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl colon tumors treated with Ctrl or antibiotics (Abx). (B) ELISA analysis of IL-17 protein in colon tissue explants and (C) serum of mice treated with Ctrl IgG or IL-17 IgG (IL17i). **** p<0.001. (D) Tumor size of colon tumors in LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl treated with Ctrl IgG or IL-17 IgG (IL17i). ****p<0.0001.
Figure S8. Increased B-cells in PMN-deficient colon tumors. (A) Flow cytometric analysis and (B) representative images of B220 staining of colon tumor tissue from LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice. (C) Flow cytometric analysis of B220 staining and (D) quantification of peripheral blood B-cells. (E) IgA ELISA analysis from feces of LysMCre;Mcl1wt/wt and LysMCre;Mcl1fl/fl mice. (F) Representative immunofluorescence of B220 staining from WT tumor tissue and invasive adenocarcinoma tissue from LysMCre;Mcl1fl/fl mouse.
Figure S9. B-cell depletion does not exacerbate DSS colitis. (A) Schematic of 2.5% DSS treatment and anti-B220 injections (400ug/mouse I.P.) on days 1, 3, and 5. (B) Representative flow cytometric analysis of B220 staining in peripheral blood and colon tissue and (C) quantification B-cells of Ctrl and anti-B220 treated animals. (D) Representative H&E histologic analysis (E) colon length and (F) total inflammation score of treated mice. Statistical analysis performed by student’s t test.
Figure S10. B-cell depletion reduces PMN-deficient tumor progression. (A) Tumor number and (B) representative histologic analysis indicated tumor stage from Ctrl and anti-B220 treated LysMCre;Mcl1fl/fl mice. Statistical analysis was performed by one-way ANOVA followed by Tukey’s multiple comparisons test.
Table S1: Primers


