Abstract
BACKGROUND
Head and neck squamous cell carcinoma (HNSCC) is considered to be a progressive disease resulting from alterations in multiple genes regulating cell proliferation and differentiation like receptor tyrosine kinases (RTKs) and members of the fibroblast growth factor receptors (FGFR)-family. Single-nucleotide polymorphism (SNP) Arg388 of the FGFR4 is associated with a reduced overall survival in patients with cancers of various types. We speculate that FGFR4 expression and SNP is associated with worse survival in patients with HSNCC.
AIM
To investigate the potential clinical significance of FGFR4 Arg388 in the context of tumors arising in HNSCC, a comprehensive analysis of FGFR4 receptor expression and genotype in tumor tissues and correlated results with patients’ clinical data in a large cohort of patients with HNSCC was conducted.
METHODS
Surgical specimens from 284 patients with HNSCC were retrieved from the Institute of Pathology at the Ludwig-Maximilian-University in Germany. Specimens were analyzed using immunohistochemistry and polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). The expression of FGFR4 was analyzed in 284 surgical specimens of HNSCC using immunohistochemstry. FGFR4 polymorphism was detected by PCR-RFLP. Patients’ clinical data with a minimum follow-up of 5 years were statistically evaluated with a special emphasis on survival analysis employing Kaplan-Meier estimator and Cox regression analysis.
RESULTS
Concerning the invasive tumor areas the intensity of the FGFR4 expression was evaluated in a four-grade system: no expression, low expression, intermediate and high expression. FGFR4 expression was scored as “high” (+++) in 74 (26%), “intermediate” (++) in 103 (36.3%), and “low” (+) in 107 (36.7%) cases. Analyzing the FGFR4 mutation it was found in 96 tumors (33.8%), 84 of them (29.6%) having a heterozygous and 12 (4.2%) homozygous mutated Arg388 allele. The overall frequency concerning the mutant alleles demonstrated 65% vs 34% mutated alleles in general. FGFR4 Arg388 was significantly associated with advanced tumor stage (P < 0.004), local metastasis (P < 0.0001) and reduced disease-free survival (P < 0.01). Furthermore, increased expression of FGFR4 correlated significantly with worse overall survival (P < 0.003).
CONCLUSION
In conclusion, the FGFR4 Arg388 genotype and protein expression of FGFR4 impacts tumor progression in patients with HNSCC and may present a useful target within a multimodal therapeutic intervention.
Keywords: : Fibroblast growth factor receptor 4, Single-nucleotide polymorphism, Head and neck squamous cell carcinoma, Reduced survival, Cancer progression, Polymerase chain reaction, Immunohistochemistry, Outcome
Core tip: Single-nucleotide polymorphism Arg388 of the fibroblast growth factor receptor-4 (FGFR4) is associated with a reduced overall survival in patients with various cancers. Here, the potential clinical significance of FGFR4 Arg388 was investigated in the context of head and neck carcinoma in 284 patients using immunohistochemistry and polymerase chain reaction. Advanced tumor stage and local metastasis was significantly associated with reduced disease-free survival in mutant FGFR4 Arg388 carriers. Increased expression of FGFR4 correlated significantly with worse overall survival impacting tumor progression in patients with head and neck squamous cell carcinoma and may present a useful target within a multimodal therapeutic intervention.
INTRODUCTION
The genes encoding fibroblast growth factor receptors (FGFR) 1-4 constitute the FGFR and are structurally related to receptor tyrosine kinases (RTK). FGFR are involved in a variety of cellular processes including angiogenesis, wound healing, tissue repair, and tumorigenesis[1]. Several aberrations and abnormalities in genes of FGFR family members, including point mutations, gene fusions, splice variations, and single nucleotide polymorphisms (SNP), have been the focus of studies in the past[2]. Oncogenic effects of FGFR and their ligands, more than 20 in numbers, include initiation of DNA synthesis, enhancement of cell growth, invasion, and metastatic potential. Molecular abnormalities and overexpression of FGFR have been described in carcinomas of ovary[3], bladder[4], pancreas[5], and breast[6]. Several studies have also reported that high levels of FGF could inhibit cell transformation[7]. In some cancers, FGF signaling has no effect whatsoever in tumor progression and is not indicative of disease-free survival time[8].
Head and neck squamous cell carcinoma (HNSCC) account for 3% of new cancer cases[9], representing the fifth most common cancer worldwide[10] with more than 500000 patients affected globally every year[11]. Irrespective of the progress made in surgery, chemotherapy, immunotherapy, and radiation, the overall survival of HNSCC patients remains poor with approximately 50% of patients succumbing to disease. Schulze-Osthoff and colleagues[12] studied the cellular distribution of basic FGF (bFGF) in situ in HNSCC. The expression of bFGF was heterogenous in malig-nancies with respect to the cell type, i.e., tumour cells, endothelia, and infiltrating immune cells, as well as overall protein levels. Strikingly, in vascular tumours that have obvious neoangiogenic capacity, bFGF was not detected[12].
Here, we assessed a potential correlation between the expression of FGFR4 polymorphic alleles and clinical outcome, histological grading, rate of metastasis, prognosis, and tumor size in 284 patients with HNSCC. Both increased expression of FGFR4 and mutant FGFR4 correlated with a tumor progression and local metastasis status and disease progression.
MATERIALS AND METHODS
Patients and tissue specimens
Tumor tissue was collected between 1984 and 1998 at the Institute of Pathology derived from the Department of Otorhinolaryngology, LMU University of Munich, Medical Center, from 350 patients with pathologically proved HNSCC. Forty-nine patients were excluded because of an incomplete follow-up survival period or inadequate histological tissue quality. Further, 17 patients were excluded owing to lack of adequate data. The study was approved by the local ethics committee at the Ludwig-Maximilian University in Germany.
The analyzed cohort including 284 patients scoring as well differentiated (n = 116), moderately differentiated (n = 141), poorly differentiated (n = 11), and non-differentiated carcinomas (n = 16). For clinic-pathological correlation, TNM disease stages were evaluated according to the “International Union against Cancer” classification (UICC)[13]. Median follow-up time was 84 mo (N1/M, lymph node metastasis) and up to 105 mo (N0, without lymph node metastasis). Clinic-pathological data is summarized in Table 1. None of the patients had received chemotherapy or radiotherapy before tissue collection. After surgical extirpation, specimens were formalin-fixed, paraffin embedded, and cut into sections of 3 μm to 4 μm.
Table 1.
Table of chi-square test of association between fibroblast growth factor receptor 4 genotype (Gly/Gly, Gly/Arg or Arg/Arg) and other pathological variables
|
Fibroblast growth factor receptor 4 genotype |
P-value | ||||
| Gly388 | Arg388 | n | |||
| Sex | Female | 36 | 12 | 48 | 0.157 |
| Male | 152 | 84 | 236 | ||
| Age, mean (SD); sample size | 54.20 (10.47); (188) | 53.42 (10.46); (96) | 284 | 0.55291 | |
| Survey outcome | Alive | 80 | 44 | 124 | 0.598 |
| Dead | 108 | 52 | 160 | ||
| Histological grading | High | 71 | 45 | 116 | 0.177 |
| Moderate | 95 | 46 | 141 | ||
| Low | 10 | 1 | 11 | ||
| No difference | 12 | 4 | 16 | ||
| Progression status | Positive | 92 | 38 | 130 | 0.135 |
| Negative | 96 | 58 | 154 | ||
| Recurrence status | Negative | 134 | 60 | 194 | 0.133 |
| Positive | 54 | 36 | 90 | ||
| Tumor size | T1 | 21 | 20 | 41 | 0.004 |
| T2 | 62 | 29 | 91 | ||
| T3 | 48 | 24 | 72 | ||
| T4 | 57 | 23 | 80 | ||
| Lymph Nodes | N0 | 66 | 26 | 92 | 0.507 |
| N1 | 44 | 28 | 72 | ||
| N2 | 53 | 30 | 83 | ||
| N3 | 25 | 12 | 37 | ||
| Metastases | M0 | 182 | 92 | 274 | 0.673 |
| M1 | 6 | 4 | 10 | ||
P-value from t-test for equality of means. NS indicates that the test result is not statistically significant.
A control group of healthy Caucasian individuals without a personal history of cancer was chosen to determine the distribution of the FGFR4 polymorphism in the general population.
Immunohistochemistry
Immunohistochemical staining was performed on paraffin sections, which were dewaxed in xylene and hydrated through graded alcohols to water, then subsequently placed in PBS. To optimize immunostaining with the antibody, endogenous peroxidase was quenched by incubation in 1.0% hydrogen peroxide for 10 min. After rinsing in PBS buffer, sections were incubated in citrate buffer (pH 6) twice for 6 min in a microwave at 700 W. Sections were rinsed in PBS again and incubated with 1:100 pre-diluted FGFR4 primary antibodies (clone 16; Santa Cruz, Santa Cruz, CA, United States). Before incubation with the Vectastain Elite ABC solution for 30 min at room temperature, sections were incubated with biotinylated goat anti-rabbit antibody for 30 min (Santa Cruz, CA). Antigen was visualized with 3-3’-diaminobenzidine (Sigma, St. Louis, MO) as dark brown precipitates. Sections were counterstained with Mayer’s hematoxylin and mounted. Staining intensity was differentiated into three groups: (1) + (weak); (2) ++ (moderate); and (3) +++ (high).
In order to obtain a reliable immunohistochemical scoring of FGFR4 expression, in the present study we have scored HNSCC tumor samples into three im-munohistochemical classes on the basis of the distribution and intensity of the staining reaction. FGFR4 immunostaining was blindly assessed by direct microscopic analysis with no knowledge of either the clinical outcome or other clinicopathological data. The staining intensity was scored into three classes by a senior pathologist (SI). To test the interobserver reproducibility of the scoring system, two other investiga-tors (EW and CB) evaluated the same slides without knowledge of the former classification. The classification of the senior pathologist was used in the statistical analysis.
Polymerase Chain Reaction Analysis
Genomic DNA was extracted from paraffin-embedded tissue sections as described previously[14]. To determine the distribution of FGFR4 Arg388 and FGFR4 Gly388 alleles in head and neck cancer patients, following primers were used: 5’-GAC CGC AGC AGC GCC CGA GGC CAG GTA TAC G– 3’ (sense) and 5’–AGA GGG AAG CGG GAG AGC TTC TGC ACA GTG G–3’ (antisense). G to A transition in codon 388 creates a new BstNI restriction site (New England Biolabs, Beverly, MA), which was used for discrimination of both alleles. PCR-beads (Ready To GoTM, Amersham Pharmacia Biotech, Freiburg, Germany) were applied in a 25 μL total PCR reaction volume. Annealing temperature was 70°C. PCR amplicons were digested with BstNI and separated in a 12% non-denaturing polyacrylamide gel (260 V, 4 h) and DNA was visualized by 10 min ethidium bromide staining. Two fragments of 80 and 29 bp characterized the FGFR4 Arg388 allele, while a single visible band of 109 bp characterized the FGFR4 Gly388 allele.
Statistical analysis
Statistical evaluations were performed using SPSS 12.0 (Chicago, IL). Frequencies of genotypes amongst different subgroups were calculated by the chi-quadrate test and P-values to assess the association between allele and malignancy in the patients. Because of their small number (Arg/Arg: n = 12), FGFR4 Arg388 homozygous alleles were grouped together with heterozygous FGFR4 Arg388 alleles (Gly/Arg: n = 84). Actuarial Kaplan-Meier survival curves of disease-free survival were plotted and compared using log-rank statistics due to the levels of the respective covariates. Comprehensive statistical analysis included univariate and multivariate Cox regression tests. All tests were performed at a significance level of α = 0.05.
RESULTS
FGFR4 genotypes in head and neck cancer patients and controls
The biologic role of the recently reported SNP in the FGFR4 allele leading to the FGFR4 Arg388 substitution still remains unknown. This is despite the fact that high FGFR4 gene expression levels are present in about 30% of all non-neoplastic epithelial cells[15]. We investigated the potential correlation of FGFR4 and its SNP with HNSCC outcome. Transition in the DNA codon for glycine to arginine at amino acid position 388 creates a new restriction site for the enzyme BstNI. Genotyping of tissue from cancer patients was conducted by PCR-RFLP analysis, where FGFR4 Gly388 was seen as a single 109 bp band and FGFR4 Arg388 as two bands of 80 bp and 29 bp (Figure 1). When adding heterozygous (n = 84) and homozygous allele cases (n = 12), the FGFR4Arg388 SNP represented one third of patients (n = 96/284, 33.8%) with 29.6% heterozygous and 4.2% homozygous cases, while it made up to 55% FGFR4Arg388 allele frequency in healthy controls (Table 2). Hence, the FGFR4Arg388 allele frequency was reduced in malignancy. Further comparison of allele distribution between controls and carcinomas revealed a concurrent and significantly increased occurrence of FGFR4Gly388 in cancer patients (45% and 66.2%, respectively).
Figure 1.
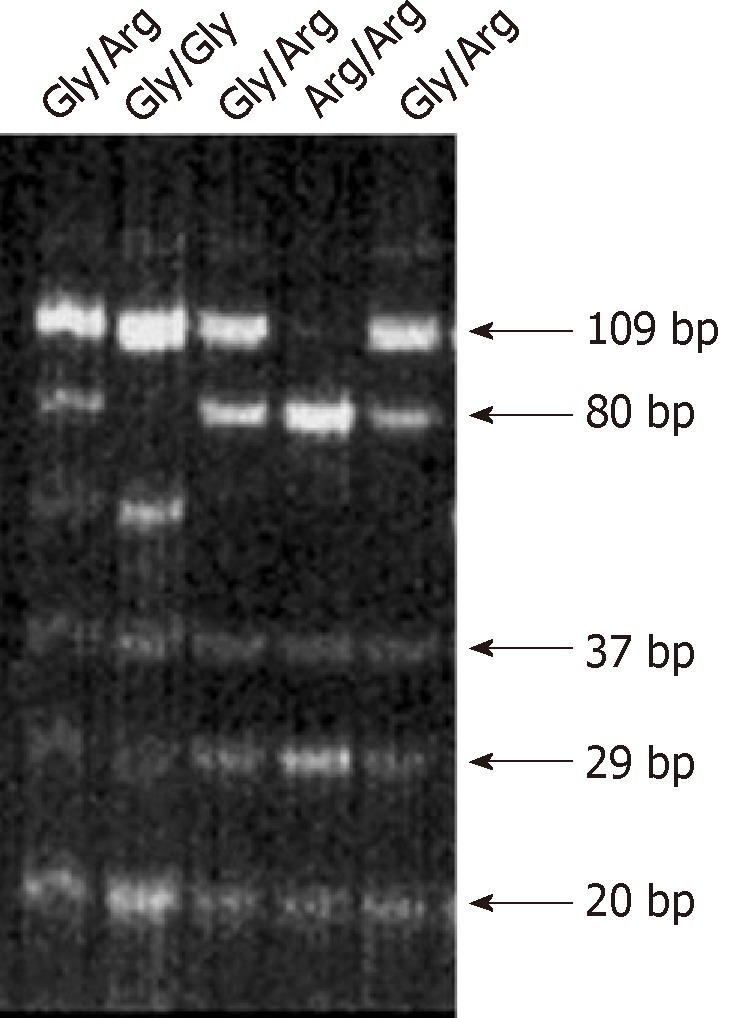
Polymerase chain reaction-restriction fragment length polymorphism analysis of fibroblast growth factor receptor 4 G388 gene. Five individual tissue specimen of Head and Neck Squmous Cell Carcinoma are presented. Line 2 shows a homozygote carrier for the Glycine allele, the dominant form of the fibroblast growth factor receptor 4 (FGFR4). Lane 1, 3 and 5 show a heterozygote carrier of the FGFR4 with both alleles glycine388 and arginine388. Lane 4 shows a homozygous carrier for the arginine allele.
Table 2.
Genotypes in patients and controls of fibroblast growth factor receptor 4 and calculated probability of gene distribution in second rows n (%)
| Fibroblast growth factor receptor 4 genotypes | Cancer cases (n = 284) | Controls (n = 123) |
| Gly/Gly | 188 (66.2) | 55 (45) |
| 0.65 | 0.47 | |
| Gly/Arg | 84 (29.6) | 60 (49) |
| 0.3 | 0.42 | |
| Arg/Arg | 12 (4.2) | 8 (6) |
| 0.03 | 0.09 |
FGFR4 protein expression in tumor tissue of patients with HNSCC and clinical characteristics
FGFR4 expression is widely shown in tumor specimen, and numerous reports indicate that high FGFR4 expression is associated with poorer outcome in patients with cancer of different entities[16-21]. We therefore wanted to determine, whether the level of FGFR4 protein was associated with oropharyngeal cancer progression and recurrences. Of the 284 cases that underwent immunohistochemical analysis, 74 (26%) harbored a strong expression, 103 specimens (36.3%) showed moderate expression levels, and 107 cases (36.7%) were detected with low expression of the FGFR4. Immunoreactivity of FGFR4 was found both in normal epithelium and in cancer epithelial cells (Figure 2). Analysis of FGFR4 protein expression in tumor tissues and distribution of FGFR4 genotypes in respective samples from patients with oropharyngeal cancer revealed no significant association. Results from multivariate Cox model including all the pathological variables is presented with three variables FGFR4 expression, lymph nodes and chemotherapy being significant (Table 3).
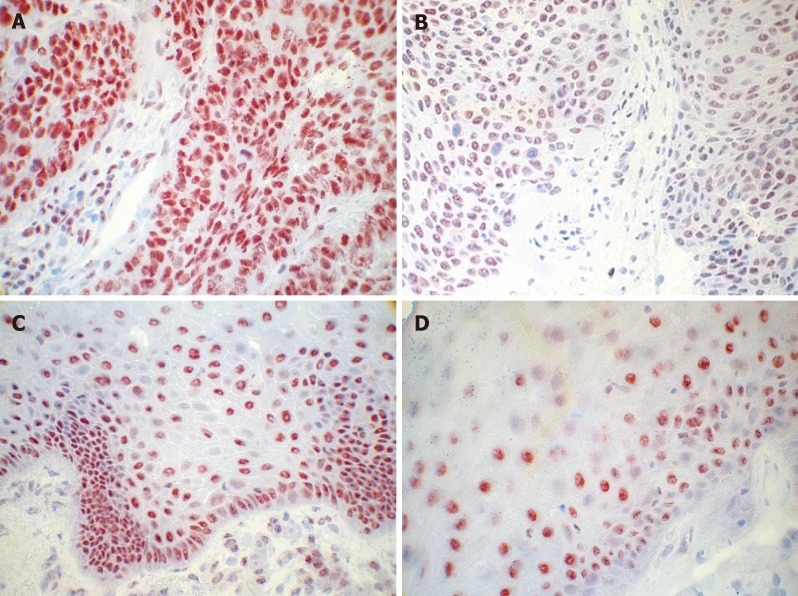
Figure 2.
Immunohistochemical analysis of fibroblast growth factor receptor 4 in primary head and neck squamous cell carcinoma tissue. A: High expression of fibroblast growth factor receptor 4 (FGFR4) in tissue of oropharyngeal carcinoma; B: Medium expression of FGFR4 in tissue of oropharyngeal carcinoma; C: Low expression of FGFR4 in tissue of oropharyngeal carcinoma; D: Expression of FGFR4 in healthy tissue of oropharyngeal mucosa (Mag. ×40).
Table 3.
Table of chi-square test of association correlating fibroblast growth factor receptor 4 expression, lymph nodes and therapy
|
Fibroblast growth factor receptor 4 expression |
|||||
| low | high | n | P-value | ||
| Lymph nodes | N0, N1 and N2 | 479 | 459 | 938 | 0.0001 |
| N3 | 117 | 58 | 175 | ||
| Chemotherapy | No | 403 | 269 | 672 | 0.0001 |
| Yes | 193 | 248 | 441 | ||
Association between FGFR4 polymorphism and clinical characteristics
As Bange et al[22] have shown, a significant correlation exists for the presence of the Arg388 allele and decreased disease-free survival in breast and colon cancer patients, we examined a large cohort of HNSCC patients’ tissues to identify the effect of this variant in these cases. We compared pathological and clinical characteristics with genotype distribution and receptor expression levels as shown in Table 4. Combining the homozygous and the heterozygous mutants, we found an association between the FGFR4 Arg388 allele and advanced tumor stages (P < 0.004). All other clinical variables remained without significant difference between the FGFR4 genotypes.
Table 4.
Table of chi-square test of association between fibroblast growth factor receptor 4 genotypes (Gly/Gly and Gly/Arg or Arg/Arg), Lymph nodes, tumor stage, and recurrence status
|
Fibroblast growth factor receptor 4 genotype |
|||||
| Gly388 | Arg388 | n | P-value | ||
| Positive lymph nodes | N0, N1 and N2 | 644 | 295 | 939 | 0.010 |
| N3 | 75 | 17 | 92 | ||
| Tumor stage | T1 and T2 | 491 | 86 | 577 | 0.0001 |
| T3 and T4 | 228 | 226 | 454 | ||
| Recurrence status | 0 | 574 | 208 | 782 | 0.0001 |
| 1 | 145 | 104 | 249 | ||
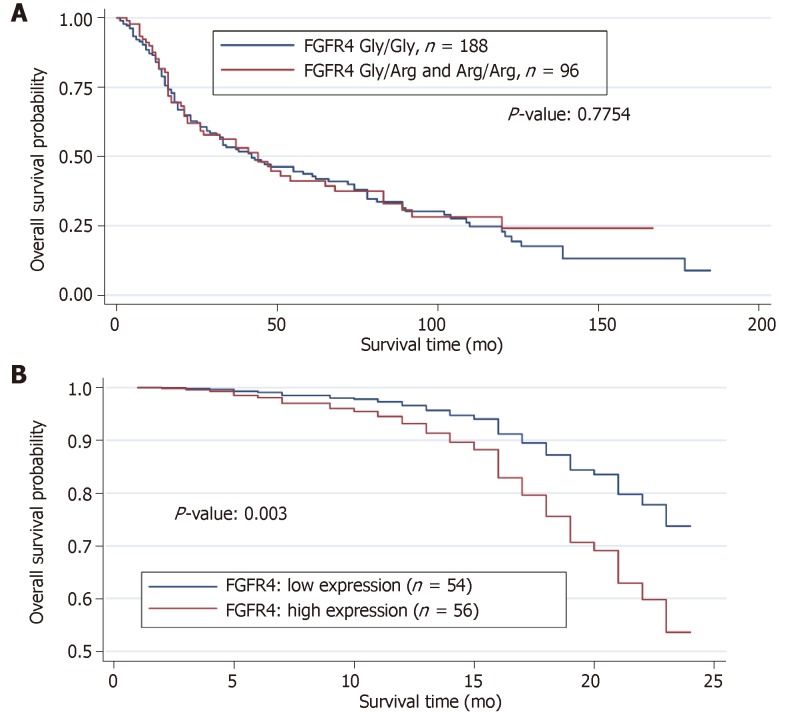
152 patients died within the follow-up period, and one third of them carried the mutated FGFR4 Arg388 allele. When compared with survivors, we did not detect any significant differences in the genotype distribution. The results of both the log rank test and univariate Cox model revealed no significant effect of FGFR4 genotype on overall survival. Several models using all the clinical variables including the FGFR4 genotype was used to discern possible effects of FGFR4 genotype in reducing the overall survival of the HNSCC patients. No significant effect of the FGFR4 genotype in overall survival was found. We performed a Kaplan-Meier survival analysis, which confirmed these observations (Figure 3A).
Figure 3.
Estimated survival function of head and neck squamous cell carcinoma patients for fibroblast growth factor receptor 4 genotype and expression. A: Wildtype fibroblast growth factor receptor 4 (FGFR4) Gly388 and mutant FGFR4 Arg388 are compared. The P-value reported is the result of the Log-rank test for comparison of survivals; B: Weak expression vs high expression are shown within the first 24 mo of survival study.
The estimated survival curve from the Cox model for the low and high FGFR4 expressions is presented in Table 3. The results from Cox model showed that high expression of FGFR4 yielded a significant reduction in the overall survival by about 130% (relative risk = 2.304, P < 0.003; Figure 3B). In contrast, application of chemotherapy reduces the risk of dying by about 70% (relative risk = 0.304, P < 0.0001) in patients with FGFR4 expression compared with those showing low or no expression.
Disease progression
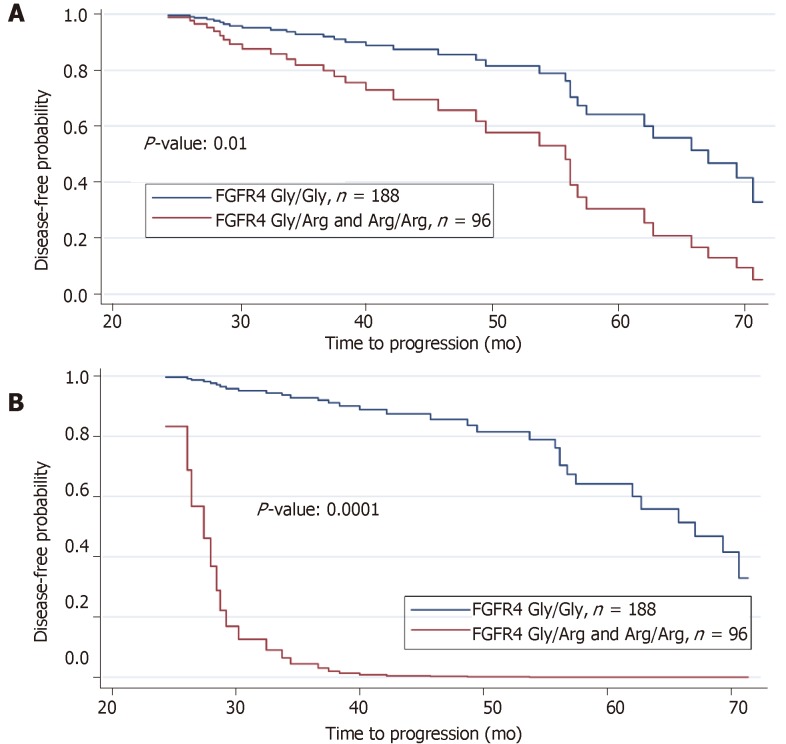
A reduction in disease-free survival was revealed in association with the FGFR4 Arg388 genotype as well as the interactive effects of lymph node (N3), tumor stage (T3 and T4) and recurrence status on prolonged (2-6 years) follow-up. The disease-free survival curve demonstrated a reduction in disease-free period due to mutant FGFR4 genotype Gly/Arg or Arg/Arg relative to wild type alleles Gly/Gly (Figure 4A, P < 0.01). The disease-free survival curve for FGFR4 genotype with the effects of all other three significant clinical variables (lymph node, tumor stage, and recurrence status) were then incorporated and the joint effects of these three variables on FGFR4 genotype is remarkably shown in Figure 4B (P < 0.0001). When the logistic regression model is fitted on FGFR4 genotype using lymph node, tumor stage and recurrence status as covariates, it reveals a remarkable relationship between these three clinical variables and FGFR4 genotype. The results of the respective logistic regression model we applied are presented in Table 5. In aggregate, positive lymph nodes, tumor stages and recurrence status are all related to FGFR4 genotype and result in a reduced disease-free survival in those harboring the variant allele.
Figure 4.
Prolonged followup of patients reveals importance of variant fibroblast growth factor receptor 4 allele. A: Estimated disease-free survival of Head and Neck Squamous Cell Carinoma patients by fibroblast growth factor receptor 4 (FGFR4) genotype showing a reduced disease-free survival due to mutant FGFR4 Arg388 relative to wildtype FGFR4 Gly388 after two years of progression study; B: Estimated disease-free survival (disease progression) of HNSCC patients by FGFR4 genotype with the interactive effects of lymph node, recurrence and tumor status incorporated in the reduction of disease-free survival due to mutant FGFR4 Arg388 relative to the predominant FGFR4 Gly388 allele after two years of study.
Table 5.
Results from Logistic regression of lymph node, tumor stage and recurrence status on fibroblast growth factor receptor 4 genotype (P < 0.0001)
| Fibroblast growth factor receptor 4 genotype | Coeff. | Std. Err. | z | P >|z| | Odds ratio | 95% CI |
| Recurrence status | 0.440 | 0.1693179 | 2.6 | 0.009 | 4.37 | 0.1080489-0.7717629 |
| positive lymph nodes | 1.972 | 0.1585958 | 12.44 | 0.0001 | 14.43 | 1.661438-2.283122 |
| Tumor stage | -1,721 | 0.2903174 | -5.93 | 0.0001 | 0.24 | -2.29047-1.152446 |
| constant | -1.826 | 0.1234243 | -14.79 | 0.0001 | -2.067534-1.58372 |
DISCUSSION
In recent years, many investigators have found polymorphisms in RTK and members of the FGF-family as well, that correlated with poor prognosis, increased tumor incidence, tumor growth, and lymph node metastasis[23]. In 1990 Schulze-Osthoff et al[12] detected bFGF in tissue of patients with squamous cell carcinoma of the head and neck region and localized them to cancer cells. Based on those previous studies, we hypothesized that FGFR4 is overexpressed in HNSCC and looked further for any correlations between the occurrence of the SNP of the FGFR4 Arg388 allele and clinical outcome and prognosis. Recently, we have demonstrated that a SNP in the transmembrane domain of the FGFR4 is associated with poor outcome in 104 patients with HNSCC[14]. These results were in accordance with the study of da Costa, which examined the role of this SNP in head and neck cancer in 75 cases[24] and in the study of Choi et al[25], who assessed it in 24 cases with oral carcinoma. Given the relatively small number of cases, which da Costa, Choi and we included in this analysis, and because of some contradictory results in other studies[8,26], we further examined these parameters on a homogenous and larger cohort of 301 patients with oropharyngeal cancer with respect to a correlation with cancer prognosis. Our results are in accordance with Dutra et al[27], who recently examined the role of FGFR4 SNP in patients with oral carcinoma in a smaller cohort of 122 patients. The presence of allele Arg388 was associated with lymphatic invasion and with disease related death. In addition, they showed that low expression of FGFR4 was related to lymph node positivity and relapse of disease, as well as disease related death[27]. Farnebo et al[28] reported in 2013 a positive correlation of the presence of FGFR4 Arg388 and improved survival in patients with HNSCC, but this analysis consisted of only 13 vs 27 patients (Arg388 vs wildtype). Importantly, Ansell et al[29] have demonstrated a role for Arg388 in increased chemosensitivity for cisplatin therapy in patients with HNSCC. This aspect has been underlined by Marmé et al[30], who found a relationship between Arg388 and pathological complete response in breast cancer with advanced disease.
Other investigators have demonstrated a role for FGFR4 SNP in entities like Non-Hodgkins lymphoma[17], in early stage gastric cancer[31], in prostate cancer with increased tumor risk and reduced survival with odds ratio 1.34 for Arg388 compared to Gly388[32], in hepatocellular carcinoma[33] and in lung cancer[34]. In 2011, Frullanti et al[35] conducted a comprehensive meta-analysis of 6817 cancer cases looking at the FGFR4 Gly388Arg SNP as a cancer prognostic factor. This study consisted of cases comprising 9 different entities of cancer showing a statistically significant association between the Arg388Arg genotype and nodal involvement (odds ratio = 1.33, 95% confidence interval 1.01-1.74) indicating a relevance for disease progression. Also, Ipenburg et al[36] carried out a thorough meta-analysis in 2016, where they evaluated the impact of FGFR4 Gly388 SNP in 8 studies comprising 1159 patients with high rates of FGFR4 Gly388Arg polymorphisms (32.5%-54.2%) and FGFR4 protein overexpression (16%-35%) correlating with worse overall and disease-free survival[37].
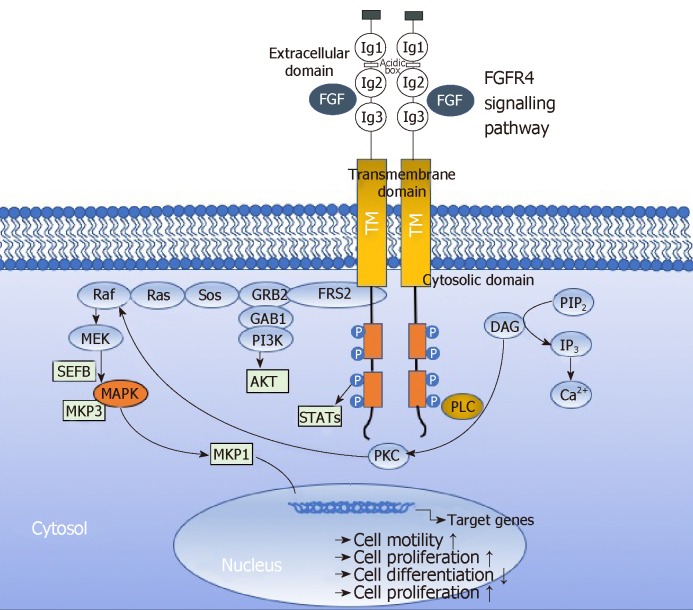
Activation of FGFRs has been shown to activate AKT, ERK1/2 and STAT3 signaling[38,39]. Activated FGFRs phosphorylates FRS2 on several sites, allowing the recruitment of the adaptor proteins son of sevenless (SoS) and growth factor receptor bound 2 (GRb2) to activate RAS and the downstream RAF and MAPK pathways. A separate complex involving GRb2-associated-binding protein 1 (GAb1) recruits a complex, which includes PI3K, and this activates an AKT-dependent anti-apoptotic pathway. Activation of FGFRs also phosphorylates and activates STAT3 directly, independent of FRS2. In aggregate, this supports the implication of FGFR signaling in several oncogenic behaviours, including proliferation, survival, migration, invasion and angiogenesis. Recently, Quintanal-Villalonga, Quintanal-Villalonga et al[37] reported on the pro-oncogenic role of the FGFR4-388Arg variant in lung cancer; this variant correlated with greater STAT3 and MAPK activation and higher expression of EMT markers in vitro. This pro-tumorigenic role was mediated by the induction of N-cadherin expression, which required STAT3 overactivation[37]. These results suggested that FGFR4-388Arg increases STAT3 activation, which consequently upregulates N-cadherin expression. Then, N-cadherin upregulation increases AKT and MAPK signaling, which may be ultimately responsible for the pro-oncogenic characteristics reported in tumor cells. Furthermuore, Gao et al[40] reported Fibroblast growth factor 19 amplification, which was reported to promote tumorigenic growth in several cancer types, corresponded with constitutive activation of FGFR4-dependent ERK/AKT-p70S6K-S6 signaling activation in HNSCC cells in a orthotopic knock-out mouse model of HNSCC underlining the implication of signaling via FGFR4 in tumor progression. Figure 5 depicts the FGFR4 signalling pathways in HNSCC sche-matically.
Figure 5.
Fibroblast growth factor receptor 4 signalling pathway in cancer. Binding of fibroblast growth factors (FGF) leads to receptor activation, which leads to activation of several downstream signalling pathways, including RAS-RAF-MAPK, PI3K-AKT, signal transducer and activator of transcription (STAT) and phospholipase Cγ (PLCγ). Negative feedback signalling regulate at several levels by induction of negative regulators, including MAPK1 and 3 phosphatase (MKP1 and MKP3). Regulators modulate intracellular signalling through MAP Kinase pathways. These regulations eventually lead to altered cell biology including promotion of cell motility, cell proliferation or inhibition of cell differentiation. DAG: Diacylglycerol; FRS2α: FGFR substrate 2α; GRB2: Growth factor receptor-bound 2; IP3: Inositol triphosphate; P: Phosphorylation; PIP2: Phosphatidylinositol-4,5-biphosphate; PKC: Protein kinase C; Sos: Son of sevenless.
Despite the enormous efforts in evaluating the impact of FGFR4 SNP on progression and survival in various tumors, the field still lacks basic molecular analyses of the FGFR4 signaling in tumor cells. Those experiments could provide insights, which eventually lead to the identification of downstream signaling check points. These could be targeted with small molecules for cancer therapy in multimodal strategies.
In aggregate, the variant allele, FGFR Arg388, is associated with worst outcome in patients with head and neck cancer as well as several other cancer entities. To this end, it would be of significant importance to evaluate the biologic relevance of aberrant FGFR4 signaling in cancer cells and its significance for a potential biomarker role as well as its consequences in cancer therapy.
ARTICLE HIGHLIGHTS
Research background
Head and neck squamous cell carcinoma (HNSCC) is considered to be a progressive disease resulting from alterations in multiple genes regulating cell proliferation and differentiation like Receptor Tyrosine Kinases (RTKs) especially members of the Fibroblast Growth Receptor (FGFR)-family. Single-nucleotide polymorphism (SNP) Arg388 allele of the Fibroblast Growth Factor Receptor 4 (FGFR4) is associated with a reduced overall survival in patients with cancers of various types. We speculate that FGFR4 expression and Gly388Arg polymorphism are associated with worse survival in patients with HSNCC. To investigate the potential clinical significance of the FGFR4 Arg388 allele in the context of tumors arising in HNSCC, a comprehensive analysis of FGFR4 receptor expression and genotype in tumor tissues and correlated results with patients’ clinical data in a large cohort of patients with HNSCC was conducted.
Research motivation
We undertook the study to investigate the impact of FGFR4 expression in tumor tissue of HNSCC and the FGFR4 SNP Glycine to Arginine at position388 with regards to clinical pathological characteristics in a large patient cohort. Our previous study revealed significant results in a smaller group of patients with HNSCC, where this SNP was associated with reduced survival and tumor progression whilst strong evidence for implication of this SNP was demonstrated in other tumor entities. In the context of exploring future tumor therapeutic interventions we aimed at providing strong evidence a possibly relevant useful target.
Research objectives
The aim of this study was to evaluate the relevant impact of FGFR4 expression and FGFR4 SNP Gly/Arg388 in tumor progression of patients with HNSCC utilizing a large cohort analysis for the respective aspects and correlate it with the patients’ clinical data with special emphasis on patients’ overall survival and disease progression.
Research methods
We analyzed tumor specimens of 284 patients with HNSCC and used Immunhistochemistry to assess FGFR4 expression in tumor tissues and PCR-RLFP to identify FGFR4 genotype in the same tumor samples. Obtained data was correlated in a comprehensive statistical analysis, including multivariant analysis and logistic regression models.
Research results
We have shown that FGFR4 expression was almost evenly distributed among 3 groups with strong, mediate and low expression levels. FGFR4 polymorphism Arg388 was prevalent in 33.8% of patient. Strong FGFR4 expression in tumor cells was significantly associated with worse overall survival. Furthermore, FGFR4 Arg388 genotype was strongly associated with tumor progression, lymph node metastasis and reduced disease-free survival.
Research conclusions
Our results show the relevant impact of FGFR4 signaling for tumor progression and worse survival in patients with HNSCC. These results confirm previous small-scale studies with similar outcomes, now providing statistically robust results in order to underline the potential to develop new target therapies in HNSCC.
Research perspectives
Facing the complexity of tumor biology with implications of multiple alterated pathways in tumor development while therapy success in cancer therapy is still limited and therapies are mostly limited to surgery and radiochemotherapy, there is still a strong need for new targets in order to improve therapies in a multimodal setting. Obviously, further investigations are mandatory to better understand the mechanisms how altered signaling pathways in cancer affect tumor biology for future development of new target therapies.
ACKNOWLEDGEMENTS
We thank Prof. Axel Ullrich, Director of Max-Planck-Institute for Molecular Biology in Martinsried/Germany for providing research resources, space and his professional guidance throughout the study and to Michael T. Lotze at the University of Pittsburgh for careful review of the manuscript. Furthermore, we thank Dr. Waheed Babatunde Yahya, Head of Department of Statistics at the University of Ilorin, Nigeria, who relevantly helped performing the statistical analysis of the data during his time at the Technische Universität, Munich, Germany.
Footnotes
Institutional review board statement: The study was reviewed and approved by the Ludwig-Maximilian University Institutional Review Board.
Institutional animal care and use committee statement: Not applicable to this study.
Conflict-of-interest statement: The authors declare that they have no conflicts of interest.
Data sharing statement: Dataset available from the corresponding author at Christoph.bergmann@uk-essen.de.
ARRIVE statement: The authors have read the ARRIVE guidelines, and the manuscript was prepared and revised according to the ARRIVE guidelines.
Manuscript source: Invited manuscript
Peer-review started: December 6, 2018
First decision: December 30, 2018
Article in press: January 30, 2019
Specialty type: Oncology
Country of origin: Germany
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Kok VC, Ozyigit G, Sukocheva OA S- Editor: Dou Y L- Editor: A E- Editor: Wu YXJ
Contributor Information
Eva Wimmer, Department of Otorhinolaryngology, Ludwig-Maximilians-University, Munich 81377, Germany.
Stephan Ihrler, Institute of Pathology, Ludwig-Maximilians-University, Munich 80337, Germany.
Olivier Gires, Department of Otorhinolaryngology, Ludwig-Maximilians-University, Munich 81377, Germany; Clinical Cooperation Group Personalized Radiotherapy of Head and Neck Tumors, Neuherberg 71083, Germany.
Sylvia Streit, Department of Molecular Biology, Max-Planck-Institute for Biochemistry, Planegg 82152, Germany.
Wolfgang Issing, Department of Otorhinolaryngology, Ludwig-Maximilians-University, Munich 81377, Germany; ENT Department, Freeman Hospital, Newcastle upon Tyne, NE7 7DN, United Kingdom.
Christoph Bergmann, Department of Otorhinolaryngology, Ludwig-Maximilians-University, Munich 81377, Germany; Department of Otorhinolaryngology, University of Duisburg-Essen, Essen 45147, Germany. christoph.bergmann@uk-essen.de.
References
- 1.Schlessinger J, Ullrich A. Growth factor signaling by receptor tyrosine kinases. Neuron. 1992;9:383–391. doi: 10.1016/0896-6273(92)90177-f. [DOI] [PubMed] [Google Scholar]
- 2.Jallal B, Schlessinger J, Ullrich A. Tyrosine phosphatase inhibition permits analysis of signal transduction complexes in p185HER2/neu-overexpressing human tumor cells. J Biol Chem. 1992;267:4357–4363. [PubMed] [Google Scholar]
- 3.Jaakkola S, Salmikangas P, Nylund S, Partanen J, Armstrong E, Pyrhönen S, Lehtovirta P, Nevanlinna H. Amplification of fgfr4 gene in human breast and gynecological cancers. Int J Cancer. 1993;54:378–382. doi: 10.1002/ijc.2910540305. [DOI] [PubMed] [Google Scholar]
- 4.Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, Chopin D, Thiery JP, Radvanyi F. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 5.Leung HY, Gullick WJ, Lemoine NR. Expression and functional activity of fibroblast growth factors and their receptors in human pancreatic cancer. Int J Cancer. 1994;59:667–675. doi: 10.1002/ijc.2910590515. [DOI] [PubMed] [Google Scholar]
- 6.McLeskey SW, Ding IY, Lippman ME, Kern FG. MDA-MB-134 breast carcinoma cells overexpress fibroblast growth factor (FGF) receptors and are growth-inhibited by FGF ligands. Cancer Res. 1994;54:523–530. [PubMed] [Google Scholar]
- 7.Korah RM, Sysounthone V, Golowa Y, Wieder R. Basic fibroblast growth factor confers a less malignant phenotype in MDA-MB-231 human breast cancer cells. Cancer Res. 2000;60:733–740. [PubMed] [Google Scholar]
- 8.Jézéquel P, Campion L, Joalland MP, Millour M, Dravet F, Classe JM, Delecroix V, Deporte R, Fumoleau P, Ricolleau G. G388R mutation of the FGFR4 gene is not relevant to breast cancer prognosis. Br J Cancer. 2004;90:189–193. doi: 10.1038/sj.bjc.6601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Audet N, Beasley NJ, MacMillan C, Jackson DG, Gullane PJ, Kamel-Reid S. Lymphatic vessel density, nodal metastases, and prognosis in patients with head and neck cancer. Arch Otolaryngol Head Neck Surg. 2005;131:1065–1070. doi: 10.1001/archotol.131.12.1065. [DOI] [PubMed] [Google Scholar]
- 10.Hussey DH, Latourette HB, Panje WR. Head and neck cancer: an analysis of the incidence, patterns of treatment, and survival at the University of Iowa. Ann Otol Rhinol Laryngol Suppl. 1991;152:2–16. doi: 10.1177/00034894911000s401. [DOI] [PubMed] [Google Scholar]
- 11.Gao YT, McLaughlin JK, Gridley G, Blot WJ, Ji BT, Dai Q, Fraumeni JF., Jr Risk factors for esophageal cancer in Shanghai, China. II. Role of diet and nutrients. Int J Cancer. 1994;58:197–202. doi: 10.1002/ijc.2910580209. [DOI] [PubMed] [Google Scholar]
- 12.Schulze-Osthoff K, Risau W, Vollmer E, Sorg C. In situ detection of basic fibroblast growth factor by highly specific antibodies. Am J Pathol. 1990;137:85–92. [PMC free article] [PubMed] [Google Scholar]
- 13.Pindbord JJ RP, Smith CJ, van der Waal I. Histological typing of cancer and precancer of the oral mucosa, 2nd ed. Berlin, New York: Springer; 1997. pp. 11–12. [Google Scholar]
- 14.Streit S, Bange J, Fichtner A, Ihrler S, Issing W, Ullrich A. Involvement of the FGFR4 Arg388 allele in head and neck squamous cell carcinoma. Int J Cancer. 2004;111:213–217. doi: 10.1002/ijc.20204. [DOI] [PubMed] [Google Scholar]
- 15.Hughes SE. Differential expression of the fibroblast growth factor receptor (FGFR) multigene family in normal human adult tissues. J Histochem Cytochem. 1997;45:1005–1019. doi: 10.1177/002215549704500710. [DOI] [PubMed] [Google Scholar]
- 16.Vokes EE, Weichselbaum RR, Lippman SM, Hong WK. Head and neck cancer. N Engl J Med. 1993;328:184–194. doi: 10.1056/NEJM199301213280306. [DOI] [PubMed] [Google Scholar]
- 17.Gao L, Feng Z, Li Q, Li L, Chen L, Xiao T. Fibroblast growth factor receptor 4 polymorphism is associated with increased risk and poor prognosis of non-Hodgkin's lymphoma. Tumour Biol. 2014;35:2997–3002. doi: 10.1007/s13277-013-1386-7. [DOI] [PubMed] [Google Scholar]
- 18.Przybylowska K, Smolarczyk K, Blasiak J, Kulig A, Romanowicz-Makowska H, Dziki A, Ulanska J, Pander B. A C/T polymorphism in the urokinase-type plasminogen activator gene in colorectal cancer. J Exp Clin Cancer Res. 2001;20:569–572. [PubMed] [Google Scholar]
- 19.Ameyaw MM, Tayeb M, Thornton N, Folayan G, Tariq M, Mobarek A, Evans DA, Ofori-Adjei D, McLead HL. Ethnic variation in the HER-2 codon 655 genetic polymorphism previously associated with breast cancer. J Hum Genet. 2002;47:172–175. doi: 10.1007/s100380200019. [DOI] [PubMed] [Google Scholar]
- 20.Balasubramanian SP, Brown NJ, Reed MW. Role of genetic polymorphisms in tumour angiogenesis. Br J Cancer. 2002;87:1057–1065. doi: 10.1038/sj.bjc.6600625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rieger-Christ KM, Mourtzinos A, Lee PJ, Zagha RM, Cain J, Silverman M, Libertino JA, Summerhayes IC. Identification of fibroblast growth factor receptor 3 mutations in urine sediment DNA samples complements cytology in bladder tumor detection. Cancer. 2003;98:737–744. doi: 10.1002/cncr.11536. [DOI] [PubMed] [Google Scholar]
- 22.Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, Schmitt M, Knyazeva T, Müller S, Gärtner S, Sures I, Wang H, Imyanitov E, Häring HU, Knayzev P, Iacobelli S, Höfler H, Ullrich A. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 2002;62:840–847. [PubMed] [Google Scholar]
- 23.Ho CL, Sheu LF, Li CY. Immunohistochemical expression of angiogenic cytokines and their receptors in reactive benign lymph nodes and non-Hodgkin lymphoma. Ann Diagn Pathol. 2003;7:1–8. doi: 10.1053/adpa.2003.50000. [DOI] [PubMed] [Google Scholar]
- 24.da Costa Andrade VC, Parise O, Jr, Hors CP, de Melo Martins PC, Silva AP, Garicochea B. The fibroblast growth factor receptor 4 (FGFR4) Arg388 allele correlates with survival in head and neck squamous cell carcinoma. Exp Mol Pathol. 2007;82:53–57. doi: 10.1016/j.yexmp.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Choi KY, Rho YS, Kwon KH, Chung EJ, Kim JH, Park IS, Lee DJ. ECRG1 and FGFR4 single nucleotide polymorphism as predictive factors for nodal metastasis in oral squamous cell carcinoma. Cancer Biomark. 2012;12:115–124. doi: 10.3233/CBM-130299. [DOI] [PubMed] [Google Scholar]
- 26.Spinola M, Leoni V, Pignatiello C, Conti B, Ravagnani F, Pastorino U, Dragani TA. Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J Clin Oncol. 2005;23:7307–7311. doi: 10.1200/JCO.2005.17.350. [DOI] [PubMed] [Google Scholar]
- 27.Dutra RL, de Carvalho MB, Dos Santos M, Mercante AM, Gazito D, de Cicco R, Group G, Tajara EH, Louro ID, da Silva AM. FGFR4 profile as a prognostic marker in squamous cell carcinoma of the mouth and oropharynx. PLoS One. 2012;7:e50747. doi: 10.1371/journal.pone.0050747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farnebo L, Tiefenböck K, Ansell A, Thunell LK, Garvin S, Roberg K. Strong expression of survivin is associated with positive response to radiotherapy and improved overall survival in head and neck squamous cell carcinoma patients. Int J Cancer. 2013;133:1994–2003. doi: 10.1002/ijc.28200. [DOI] [PubMed] [Google Scholar]
- 29.Ansell A, Farnebo L, Grénman R, Roberg K, Thunell LK. Polymorphism of FGFR4 in cancer development and sensitivity to cisplatin and radiation in head and neck cancer. Oral Oncol. 2009;45:23–29. doi: 10.1016/j.oraloncology.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Marmé F, Werft W, Benner A, Burwinkel B, Sinn P, Sohn C, Lichter P, Hahn M, Schneeweiss A. FGFR4 Arg388 genotype is associated with pathological complete response to neoadjuvant chemotherapy for primary breast cancer. Ann Oncol. 2010;21:1636–1642. doi: 10.1093/annonc/mdq017. [DOI] [PubMed] [Google Scholar]
- 31.Shen YY, Lu YC, Shen DP, Liu YJ, Su XY, Zhu GS, Yin XL, Ni XZ. Fibroblast growth factor receptor 4 Gly388Arg polymorphism in Chinese gastric cancer patients. World J Gastroenterol. 2013;19:4568–4575. doi: 10.3748/wjg.v19.i28.4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu B, Tong N, Chen SQ, Hua LX, Wang ZJ, Zhang ZD, Chen M. FGFR4 Gly388Arg polymorphism contributes to prostate cancer development and progression: a meta-analysis of 2618 cases and 2305 controls. BMC Cancer. 2011;11:84. doi: 10.1186/1471-2407-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Zhou Y, Lu M, An Y, Li R, Chen Y, Lu DR, Jin L, Zhou WP, Qian J, Wang HY. Association between fibroblast growth factor receptor 4 polymorphisms and risk of hepatocellular carcinoma. Mol Carcinog. 2012;51:515–521. doi: 10.1002/mc.20805. [DOI] [PubMed] [Google Scholar]
- 34.Matakidou A, El Galta R, Rudd MF, Webb EL, Bridle H, Eisen T, Houlston RS. Further observations on the relationship between the FGFR4 Gly388Arg polymorphism and lung cancer prognosis. Br J Cancer. 2007;96:1904–1907. doi: 10.1038/sj.bjc.6603816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frullanti E, Berking C, Harbeck N, Jézéquel P, Haugen A, Mawrin C, Parise O, Jr, Sasaki H, Tsuchiya N, Dragani TA. Meta and pooled analyses of FGFR4 Gly388Arg polymorphism as a cancer prognostic factor. Eur J Cancer Prev. 2011;20:340–347. doi: 10.1097/CEJ.0b013e3283457274. [DOI] [PubMed] [Google Scholar]
- 36.Ipenburg NA, Koole K, Liem KS, van Kempen PM, Koole R, van Diest PJ, van Es RJ, Willems SM. Fibroblast Growth Factor Receptor Family Members as Prognostic Biomarkers in Head and Neck Squamous Cell Carcinoma: A Systematic Review. Target Oncol. 2016;11:17–27. doi: 10.1007/s11523-015-0374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintanal-Villalonga Á, Ojeda-Márquez L, Marrugal Á, Yagüe P, Ponce-Aix S, Salinas A, Carnero A, Ferrer I, Molina-Pinelo S, Paz-Ares L. The FGFR4-388arg Variant Promotes Lung Cancer Progression by N-Cadherin Induction. Sci Rep. 2018;8:2394. doi: 10.1038/s41598-018-20570-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hynes NE, Dey JH. Potential for targeting the fibroblast growth factor receptors in breast cancer. Cancer Res. 2010;70:5199–5202. doi: 10.1158/0008-5472.CAN-10-0918. [DOI] [PubMed] [Google Scholar]
- 39.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 40.Gao L, Lang L, Zhao X, Shay C, Shull AY, Teng Y. FGF19 amplification reveals an oncogenic dependency upon autocrine FGF19/FGFR4 signaling in head and neck squamous cell carcinoma. Oncogene. 2018 doi: 10.1038/s41388-018-0591-7. [DOI] [PubMed] [Google Scholar]