Abstract
Supramolecular systems have applications in areas as diverse as materials science, biochemistry, analytical chemistry, and nanomedicine. However, analyzing such systems can be challenging due to the wide range of time scales, binding strengths, distances, and concentrations at which non-covalent phenomena take place. Due to their versatility and sensitivity, Förster resonance energy transfer (FRET)-based techniques are excellently suited to meet such challenges. Here, we detail the ways in which FRET has been used to study non-covalent interactions in both synthetic and biological supramolecular systems. Among other topics, we examine methods to measure molecular forces, determine protein conformations, monitor assembly kinetics, and visualize in vivo drug release from nanoparticles. Furthermore, we highlight multiplex FRET techniques, discuss the field’s limitations, and provide a perspective on new developments.
Graphical Abstract
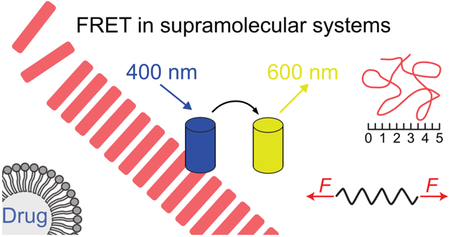
This review details the ways Förster resonance energy transfer (FRET) can be used to study natural and synthetic supramolecular systems.
1. Introduction
Supramolecular chemistry describes the self-organization of molecules through non-covalent interactions. In biology, such interactions play a vital role in cellular structures and signaling processes. For example, nucleotide complexation into DNA and its subsequent packing into chromatin depend strongly on hydrogen bonding,1 but also vesicular transport2 and enzymatic catalysis3 rely heavily on non-covalent interactions. In addition to its relevance in biology, supramolecular chemistry plays a dominant role in the design of synthetic systems. For example, stimuli-responsive catalysts are frequently based on metal coordination,4,5 while molecular switches often rely on mechanical bonds.6 On a larger scale, synthetic supramolecular systems are applied in smart materials,7,8 organic electronics,9 and biomaterials.10 Lastly, synthetic supramolecular structures are increasingly being used for in vivo applications, for instance as biomaterials or drug carriers.11 In the latter case, non-covalent interactions within the carrier, as well as those between the carrier and both blood constituents and cellular components, are vital for optimizing drug delivery.12,13 Closely related is the exploitation of supramolecular systems for bioimaging approaches ranging from intravital microscopy of intracellular processes to the study of in vivo cell migration by whole body optical imaging.14
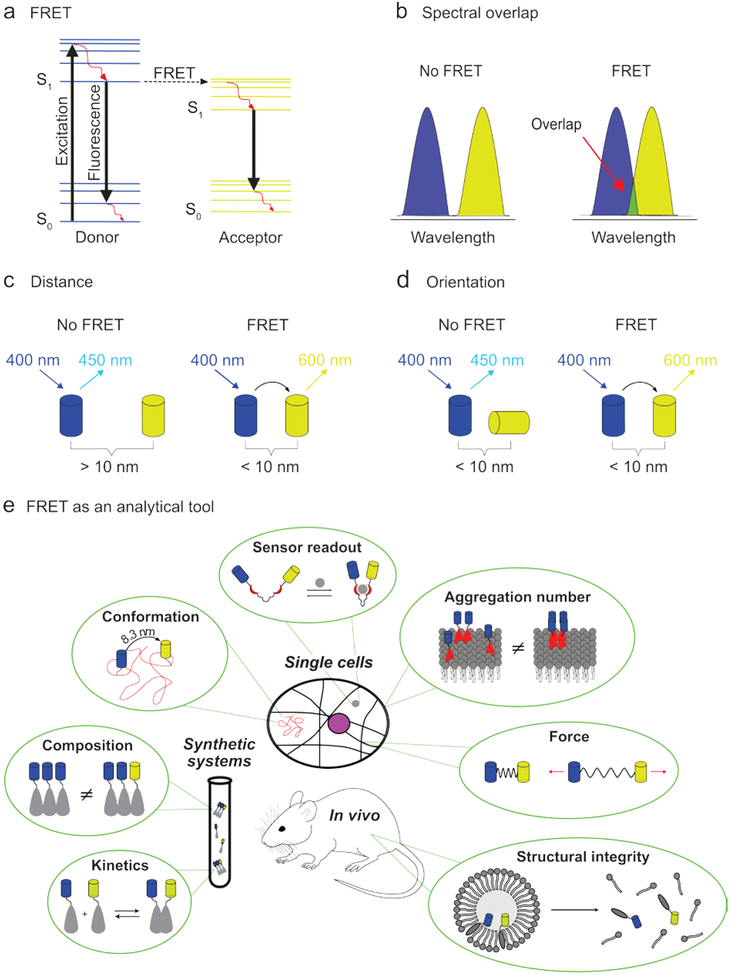
The high levels of dynamicity and adaptability required for such applications are often difficult to achieve using purely covalent chemistry. However, the wide range of energies, time-scales, and concentrations associated with non-covalent interactions present unique challenges for their characterization. Not surprisingly, this has led to a large number of dedicated analytical techniques. Among these, Förster resonance energy transfer (FRET)-based approaches have been particularly successful, largely due to their versatility, high sensitivity, and non-invasiveness. Building upon earlier work by Jean and Francis Perrin, Theodor Förster provided the first quantitative description of the non-radiative energy transfer between fluorophores in 1948.15,16 In this process, an excited donor fluorophore transfers its energy non-radiatively to a nearby acceptor molecule, which typically relaxes through the emission of light (Fig. 1a). As this process originates from dipole-dipole interactions, certain criteria need to be met in order for it to take place. First and foremost, the process requires sufficient spectral overlap between the donor emission and acceptor absorption fluorophore. Secondly, the distance between the fluorophores needs to be less than approximately 10 nm.17 Finally, for efficient transfer, the fluorophores’ dipoles should be orientated favorably toward each other, as defined by the orientation factor κ2 (Fig. 1b–d, see ref18 for more information on κ2).19,20 Once these criteria are met, the FRET efficiency - defined as the percentage of photons absorbed by donor fluorophores that contribute to FRET21 - can provide highly sensitive, temporally specific information on molecular distance and orientation. This high sensitivity is a direct result of the strong R−6 distance dependence of the transfer probability associated with most FRET processes (R being the donor-acceptor distance). Quantitative information can be obtained when spectral cross-talk and the acceptor:donor ratio are also taken into account.17,22 These aspects make FRET excellently suited to study a wide-range of supramolecular phenomena (Fig. 1e).
Fig. 1.
The principle of FRET and its application in the study of supramolecular systems. (a) Jablonski diagram illustrating the energy levels and transitions associated with FRET: solid black arrows indicate radiative transitions; red arrows indicate non-radiative transitions. For FRET to occur, certain criteria need to be met. Requirements include: (b) sufficient spectral overlap between the emission spectrum of the donor fluorophore (blue) and the excitation spectrum of the acceptor fluorophore (yellow), (c) less than approximately 10 nm distance between the fluorophores, and (d) favorable inter-fluorophore orientation. The depicted excitation and emission wavelengths serve as an example. (e) FRET can be used to study many aspects of supramolecular systems, such as: kinetics, aggregate composition, molecular conformation, sensor incitement, aggregation number (using homo-FRET), applied force, and structural integrity.
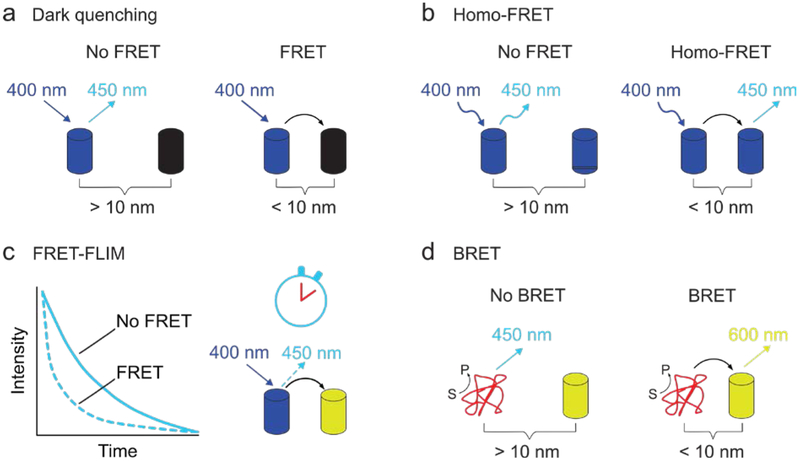
In addition to the traditional form of FRET – one type of fluorophore gets excited and transmits its energy to a second type of fluorophore whose emission intensity is measured – several variations have been developed. In one variation, the acceptor is non-fluorescent and therefore releases the transferred energy non-radiatively (a dark quencher, Fig. 2a).23 Energy transfer is in this case observed by monitoring the decrease in fluorescence intensity of the donor fluorophore. This approach has the advantage that the part of the spectrum that would be occupied by the emission of the acceptor fluorophore becomes available for other FRET pairs. In another variation, termed homo-FRET, energy is transferred between two identical fluorophores. Here, the acceptor fluorophore is excited by polarized light and FRET can be observed as a reduction in anisotropy in the emitted light. This technique can be used to quantify the number of monomers in an aggregate, such as the number of proteins in a protein cluster vide infra (Fig. 2b).24 Alternatively, not the emission intensity, but the donor’s fluorescence lifetime can monitored (Fig. 2c). As FRET provides additional de-excitation routes it will enhance the decay of the donor’s excited state and thereby reduce its fluorescence life-time. This technique is most commonly used in microscopy, where it is termed FRET fluorescence-lifetime imaging microscopy (FRET-FLIM) and which has the significant advantage of being insensitive to variations in fluorophore concentration.25 Lastly, it can sometimes be difficult to irradiate a donor fluorophore in vivo. To circumvent this problem the donor fluorophore can be replaced by a bioluminescent protein (Fig. 2d).26,27 To discriminate from traditional FRET, this process is termed bioluminescence resonance energy transfer (BRET).
Fig. 2.
Different variations on traditional FRET where: (a) the acceptor releases the obtained energy non-radiatively (dark quenching), (b) the FRET pair consists of identical fluorophores (homo-FRET), (c) the fluorescence lifetime of the donor fluorophore is used to monitor energy transfer (FRET-FLIM), or (d) the donor fluorophore is replaced by a bioluminescent protein (red) to thereby eliminate the need for external illumination (BRET). The depicted excitation and emission wavelengths serve as an example.
During the last decades, novel fluorophores based on organic molecules, nanocrystals, polymers, nanometals, and quantum dots have been developed and made commercially available.28–32 Additional advances include highly advanced techniques such as atomic force microscopy supplemented with FRET (AFM-FRET)33 and time-resolved measurements with nanosecond resolution.34 These innovations have made FRET an easily accessible – and widely applicable – technique for studying molecular interactions. Here, we review the various ways in which FRET can be deployed to study supramolecular systems. We will focus on the specific aspects of supramolecular systems that can be investigated using FRET and the approaches used in such studies. In addition, we examine current developments in the field and discuss how these will expand our understanding of cellular processes and help advance synthetic supramolecular systems and their in vivo applications.
2. Monitoring the kinetics of supramolecular systems
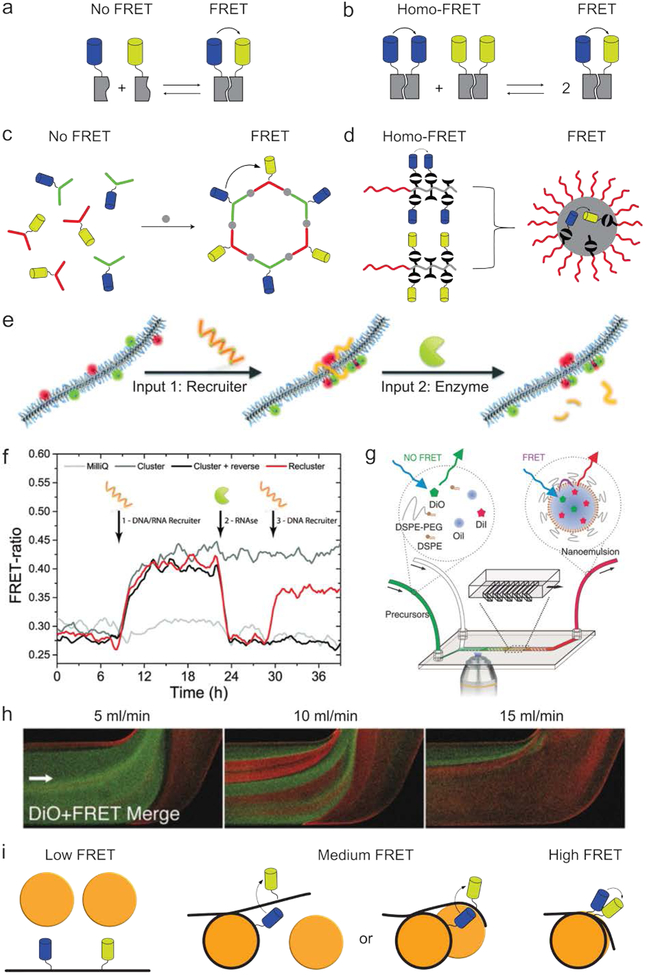
Quantitative insight into assembly kinetics is vital for a thorough mechanistic understanding of supramolecular systems.35–37 Due to their high temporal resolution and low invasiveness in systems analysis, FRET-based techniques are among the most commonly used methods. In a typical experiment, the different building blocks are either inherently fluorescent or labeled with fluorophores, and the FRET efficiency is monitored throughout the assembly process (Fig. 3a). Conversely, self-assembled systems’ stability can be used to monitor disassembly kinetics and provide quantitative information.38 Additionally, exchange rates can be obtained by judiciously labeling different aggregate populations (Fig. 3b).39 A logical prerequisite for such FRET-based kinetic measurements is that the fluorophores themselves must not affect the assembly process.40,41 In addition, the technique should be able to capture the full time range over which the supramolecular processes takes place, which can be anywhere from picoseconds to multiple weeks.42–44 While FRET has been successfully used to study assembly processes taking place on picosecond timescales,43 fluorophore bleaching compromises the technique’s ability to continuously monitor processes that take longer than several minutes.45,46
Fig. 3.
Monitoring kinetics using FRET. (a) By labeling monomers with a FRET pair (blue and yellow), the observed FRET efficiency can be used to monitor the assembly of building blocks (grey) in a time-resolved manner. (b) FRET can be used to monitor monomer exchange between different aggregate populations, each functionalized with only one type of fluorophore. (c) Labeling both monomer types with either a FRET donor or acceptor allows the kinetics of a coordination-driven self-assembly process to be monitored. Image based on ref47. (d) Labeling identical polymers with either a FRET donor or acceptor allowed their assembly into micelles to be studied at concentrations as low as 10−7 M. Image based on ref48. (e) A supramolecular polymer consisting mainly of neutral monomers and a small amount of fluorophore-labeled positively charged monomers was synthesized. Upon the addition of a negatively charged ssDNA-RNA hybrid, the fluorophore-labeled monomers were brought in close proximity, resulting in increased FRET efficiency. The nucleotide strand could be fragmented through the addition of the enzyme RNAse, thereby largely restoring the system to its initial state. (f) FRET data associated with the experiments shown in e. Image e and f are both reprinted from ref49 with permission from the Royal Society of Chemistry, Copyright 2015. (g) The microfluidic-mediated formation of nanoemulsions could be monitored in real-time using optical microscopy.50 (h) Representative images obtained using the set-up in g, showing the influence of flow rate on particle formation kinetics. Green depicts emission from the FRET donor, red indicates FRET and thereby formation of the nanoparticles. Images g and h adapted with permission from ref50. Copyright 2017, Wiley-VCH. (i) The kinetics of nucleosome core particles (orange) were studied by labeling DNA (black) with a FRET pair (blue and yellow). Three different populations could be observed based on the observed FRET efficiency. Image based on ref1.
Many synthetic supramolecular constructs consist of a small number of molecules positioned in a well-defined spatial arrangement. A typical example are structures consisting of several molecules held together by metal-ligand coordination (Fig. 3c). The assembly kinetics of such systems are often difficult to determine unambiguously, due to the formation of various intermediates and structural analogues. By labelling the monomers with fluorophores, FRET can be used to monitor the assembly process and thus elucidate their assembly kinetics.47 Although such insight can, in theory, also be obtained with other techniques, such as mass spectrometry or NMR, FRET’s distance dependency makes it possible to noninvasively observe exchange processes between structurally similar aggregates at low concentrations and with superior time resolution. A second process that is notoriously difficult to study is block-copolymers’ assembly into micelles, as their critical micelle concentration is typically in the low micromolar range and therefore below the detection limit of most analytical techniques.51 By labeling block-copolymers with fluorophores, micelle formation and exchange dynamics have been successfully monitored using FRET at polymer concentrations as low as 10−7 M (Fig. 3d).48
Besides studying molecule assembly and exchange, FRET can also be used to monitor changes within existing aggregates. In a system reported by Albertazzi et. al., a supramolecular polymer consisting of neutral benzene-1,3,5-tricarboxamide monomers was doped with a small amount of positively charged, fluorophore-labeled monomers (Fig. 3e).49 As the fluorophore-functionalized monomers were initially randomly distributed over the length of the polymer, their relative distance was relatively large, so that FRET was inefficient. However, with the addition of a negatively charged ssDNA-RNA hybrid, the fluorophore-functionalized monomers were recruited into clusters, leading to increased FRET efficiency. This process could be reversed through the addition of RNAse, which cleaved the nucleotide strand and largely returned the system to its initial state. As a result of FRET’s unique properties, these changes in intra-aggregate monomer distribution could be quantified in a time-resolved manner (Fig. 3f).
Because its emitted light is typically in the visible spectrum, FRET can also monitor assembly kinetics using optical microscopy. This is particularly interesting in a microfluidic set-up, in which temporal aggregate formation can be studied in a spatial dimension. Using this approach, the formation of several types of nanoparticle platforms was studied in real time by adding an apolar FRET donor and acceptor to the nanoparticles’ building blocks (Fig. 3g).50 As the FRET pair will only be in close proximity upon formation of the nanoparticles, the FRET signal is used to observe this process. Amongst other information, this methodology provided insight into the influence of microfluidic flow rate on particle formation kinetics (Fig. 3h, green indicates free dye, red indicates FRET and thereby assembled nanoparticles).
Lastly, FRET is also very well suited to monitor the kinetics of biological structures. For example, the processes through which DNA is read from chromatin are poorly understood due to the significant challenges of monitoring this in cells. Through the site-specific labeling of DNA with FRET pairs, the assembly of nucleosome core particles,1,52 as well as their compaction into chromatin,53 has been studied in a time-resolved manner (Fig. 3i). Interestingly, these studies revealed that large sections of DNA buried in nucleosomes temporally unwind (on a timescale of seconds) and thereby become available for interaction with other biomolecules. Collectively, FRET-based techniques can reveal the assembly kinetics of a broad scope of supramolecular systems, information that would be difficult to obtain using alternative techniques.
3. Determining molecular conformation
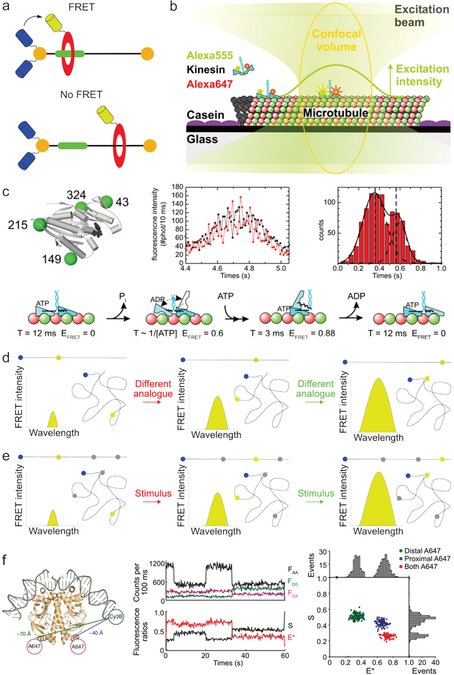
Upon molecular assembly, a product’s exact conformation dictates many of its characteristics, including stability, chirality, density, color, and polarity. While techniques such as crystal structure analysis, AFM, and TEM can be used to elucidate an aggregate’s conformation, they require drying or freezing the sample and therefore do not provide insight into the dynamics of the system. In addition, sample processing steps can in some cases lead to structural changes in the product. Conformations can be accurately determined in solution using NOESY-NMR; however, this approach is limited to length-scales below ≈ 0.5 nm.54 Not surprisingly, FRET’s ability to transfer energy over larger distances has made it a valuable tool for elucidating molecular conformations in solution.55–59 Knowledge of conformational changes is especially relevant for molecules containing mechanical bonds, as their conformation often responds to external factors, including pH, light, and redox potential.6,60,61 For example, rotaxanes consist of a one-dimensional stator threaded through a mobile circular rotor, which can change position upon certain stimuli (Fig. 4a).60 By labeling the stator and rotor with fluorophores, FRET has been used not only to confirm the compound’s interlocked structure, but also to monitor the rotor’s movement along the stator.62 A second and more complex example of conformational analysis is the study of motor proteins. While synthetic molecular motors are a relatively new research field,63 such systems are an extensively studied mode of intracellular transport and whole cell movement.64 Due to the challenges associated with synchronizing multiple motors, they are best studied using single molecule techniques, at high temporal resolution and with low invasivity, as has been achieved using FRET microscopy.64 Kinesin movement along a microtubule is one of the best studied phenomena.65–67 One approach investigated kinesin movement by labeling its two domains with a FRET pair (Fig. 4b).65 As the movement of kinesin induced changes in its conformation, and thereby in the distance of the attached fluorophores, the rate and mechanism of kinesin movement could be estimated from temporal fluctuations in the FRET efficiency (Fig. 4c). This was achieved by studying kinesin analogs in which fluorophores were site-specifically attached on different positions. By monitoring the acceptor and donor emission of these analogs during the movement of kinesin, different FRET populations and their occurrence over time could be monitored. By extensive analysis of these data, detailed insight into the gait of kinesin could be obtained.
Fig. 4.
Determining molecular conformations using FRET. (a) Schematic depiction of a rotaxane consisting of a rotor (red) and stator (black and green). By labeling the rotor and stator with a FRET pair, the rotor’s position along the stator can be monitored. Image based on ref62. (b) Using FRET to monitor kinesin movement along a microtubule by labeling the two domains of kinesin (blue) with a fluorophore (green and red). Image adapted from ref65. (c) Schematic of a kinesin motor domain functionalized with 4 cystine residues facilitating site-specific labeling with fluorophores. By monitoring the fluorescence intensity of a FRET-pair labeled kinesin over time (black line donor fluorescence intensity, red line acceptor fluorescence intensity), two populations of FRET efficiency could be observed corresponding to different stages in kinesin’s step. By repeating such experiments using differently labeled kinesin analogues, detailed information of kinesin’s gait could be obtained. Images b and c are both adapted from ref65. Copyright 2009, National Academy of Sciences. (d) In a typical FRET conformational mapping experiment, various analogues with fluorophores at predefined positions are synthesized and analyzed. The obtained FRET intensities allow subsequent reconstruction of the molecular conformation. Blue and yellow dots represent the donor and acceptor fluorophore, respectively; black lines represent the molecule in its stretched and folded conformation. (e) In switchable FRET the molecule of interest is labeled with one donor fluorophore (blue) and multiple stimuli responsive acceptor fluorophores (yellow when on, gray when off). By sequentially measuring the FRET efficiency between the acceptor and each donor fluorophore, multiple distances can be determined using the same compound. (f) Example of switchable FRET results, used to determine molecular conformation. The following fluorescence intensities were monitored: FAA = acceptor emission upon acceptor irradiation, FDD = donor emission upon donor irradiation, FDA = acceptor emission upon donor irradiation. The acceptor fluorophores were switched in the following manner: 5 – 20 seconds, only proximal A647 on; 20 – 35 seconds, both acceptors on; 35 – 60 seconds, only distal A647 on). Image adapted with permission from ref68. Copyright 2010, Nature Publishing Group.
Besides monitoring basic conformational changes, such as sliding and bending, FRET can also be used to map the entire conformation of large molecules, such as proteins. In a typical experiment, a fluorophore (e.g., a FRET donor) is attached to a fixed and predefined site on the molecule, while a second fluorophore’s position (in this example the FRET acceptor) varies between different analogues (Fig. 4d). Because both fluorophores’ positions in the molecule are known, the observed FRET intensities are a measure for their distance and thus the molecule’s conformation. Comparisons with computational and X-ray data indicate that the results obtained using this approach are highly accurate.22,69 However, care should be taken when analyzing molecules that contain rigid sections or bulky fluorophores, as these influence fluorophore orientation and therefore FRET efficiency.70 Despite their obvious merit in determining molecular conformations, FRET mapping approaches are often experimentally demanding because of the need to synthesize a different analogue for each distance measurement. To overcome this inconvenience, Kapanidis et al. developed a switchable FRET methodology in which the energy of a single donor fluorophore can be selectively transferred among several photo-switchable acceptor fluorophores (Fig. 4e).68 By sequentially activating and deactivating the acceptor fluorophores, this approach can determine multiple distances using the same compound. In an example of such a procedure, the conformation of a single dsDNA-peptide complex was analyzed by labeling the dsDNA with a Cy3B donor fluorophore and the peptide with two A647 acceptor fluorophores (Fig. 4f, left). Subsequently, the donor and acceptor were alternatingly irradiated (μs timescales) and their emission monitored over time (Fig. 4f, middle). Using the obtained data, the apparent FRET efficiency (E*) - reporting the distance between the active FRET pair - and the ratio between the continuously active donor and the number of active acceptors (S) could be determined. By optically switching the acceptors on or off, three different populations could be observed (Fig. 4f, right, see caption for details). By comparing the FRET efficiency between both FRET pairs their relative distance can be determined. As FRET-mapping techniques work in solution, they also allow for time-resolved studies and can therefore provide insight into the rates and intermediates through which species interconvert.71 Such studies have been especially successful in unraveling protein dynamics,72–74 and nanosecond-level temporal resolutions are no exception.75,76
4. Determining aggregate configuration and composition
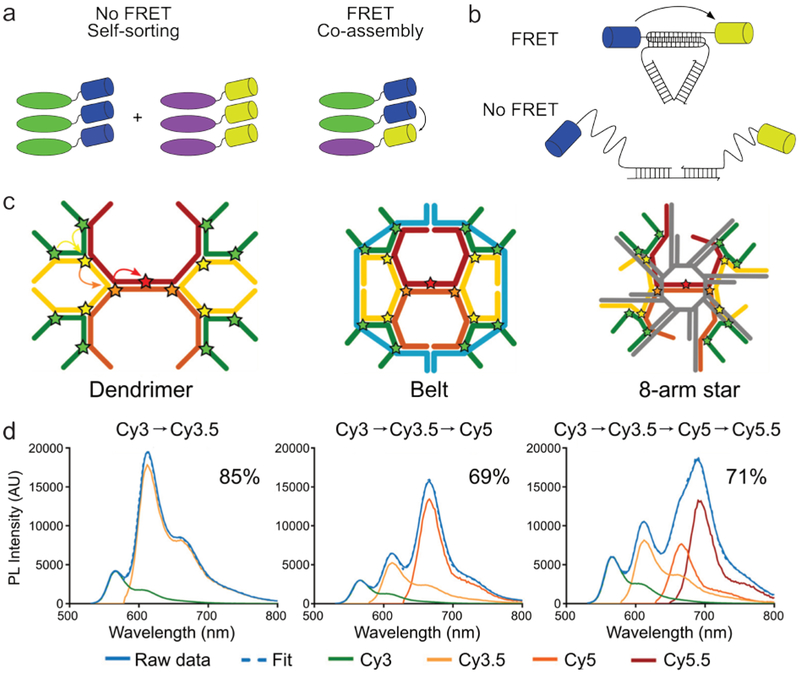
In addition to determining molecular conformation, FRET can also be used to investigate the structure of aggregates.8 Information about the number of monomers in a supramolecular structure and how these are arranged is vital to fully understand a supramolecular system. For example, the building blocks in many assembly processes are chiral and therefore allow for the formation of either co-assembled or enantiomerically pure assemblies. While circular dichroism spectroscopy can discriminate between these outcomes, this technique is less suited to samples in which many other compounds are present, as typically encountered in in vivo studies. It is therefore desirable to be able to monitor assembly behavior in a highly specific manner. To achieve such specificity, the two enantiomers of a supramolecular monomer can be labeled with either a FRET donor or acceptor (Fig. 5a). As a result, co-assembly can be observed by the presence of FRET, whereas self-sorting would not give rise to energy transfer.39,77 While the monomers in this case can only form one-dimensional stacks, many other complex compounds, such as DNA and proteins, can interact in a variety of ways. Accordingly, when designing supramolecular structures using such complex building blocks, it is vital to verify whether they interact in the conceived manner. Such an evaluation can be achieved by labeling the monomers with a FRET pair in a way that maximizes the difference in interfluorophore distance between the correct and misassembled configurations (Fig. 5b).78 A prime example of this approach was reported by C. W. Brown et al., who analyzed three types of DNA structures functionalized with FRET cascades i.e., a series of fluorophores able to sequentially transfer energy (Fig. 5c).79 By analyzing FRET in derivatives containing two, three, or all four fluorophores in the FRET cascade, the integrity of the different structures could be confirmed (Fig. 5d, depicting the fluorescence spectra of the dendrimer analogues as an example). Furthermore, the three different types of DNA structures could be discriminated by comparing FRET efficiencies for each step, e.g, while FRET in the dendrimer showed efficiencies of 85 %, 69 %, and 71 % for the three FRET events, the Belt structure displayed efficiencies of 89 %, 76 %, and 82 %.
Fig. 5.
Investigating aggregate composition and configuration using FRET. (a) Schematic depiction of a supramolecular system in which FRET allowed discrimination between the co-assembly and self-sorting of two stereoisomers (green and purple). Image based on ref77. (b) By judiciously functionalizing DNA structures with a FRET pair (blue and yellow), it becomes possible to distinguish between correctly and misassembled analogues. Image based on ref78. (c) Three different DNA based structures labeled with fluorophores (stars), allowing FRET to take place from the outside to the inside of the structure. (d) The photoluminescence intensity of the dendrimer construct with two, three and four fluorophores in the FRET cascade (black arrows depicted FRET). The FRET efficiency for each added step in the cascade is depicted as well and allows for discrimination between the three types of structures. Image c and d adapted with permission from ref79. Copyright 2017, American Chemical Society.
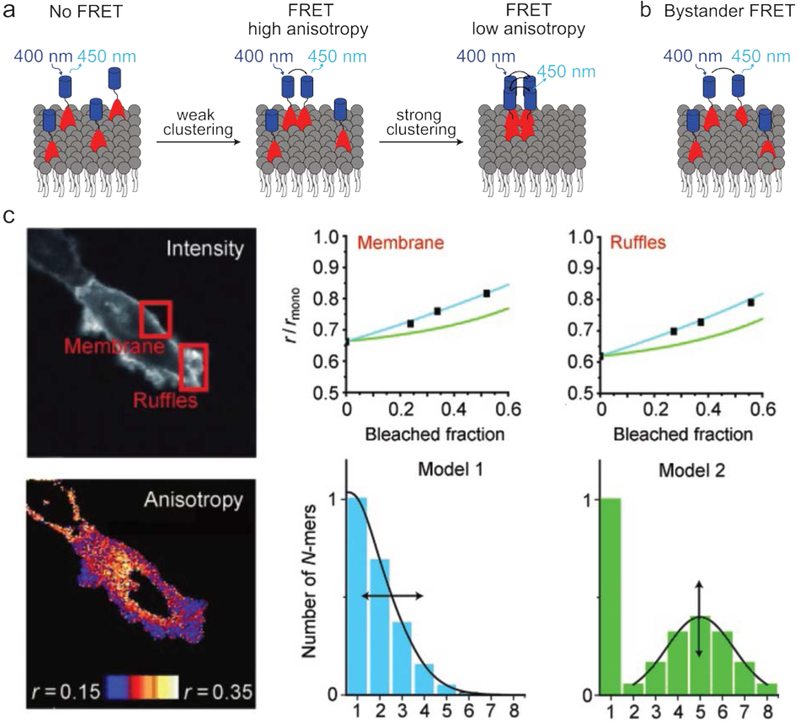
In addition to aggregates’ configuration and composition, the exact number of their building blocks also plays an important role in many systems. For example, cell signaling by membrane proteins depends strongly on the number of monomers in the formed structures.80,81 Such membranes consist of a fluid lipid surface through which various membrane proteins diffuse. Due to the high dynamicity of this environment, it can be challenging to monitor interactions between membrane proteins in a time-resolved and non-invasive manner. Interestingly, such observations have been achieved using homo-FRET, which can quantify the number of monomers in an aggregate (e.g., the number of proteins in a protein cluster) by monitoring anisotropy reduction in the emitted FRET signal (Fig. 6a).82 This technique can thus reveal the role of multivalency in signaling networks.24 In addition to forming homo-aggregates, membrane protein analysis is further complicated by system dimensionality. The two-dimensional confinement of the membrane proteins significantly increases the chance of FRET (and the corresponding loss of anisotropy) arising from fluorophores that are in close proximity as a result of random diffusion, rather than their associated proteins interacting. This phenomenon is termed by-stander FRET, and it must be taken into account when studying such systems (Fig. 6b).83,84 An example of the use of homo-FRET to determine the number of monomers in an aggregate is depicted in Fig. 6c. Here, cells expressing a fluorophore-labeled transmembrane protein were monitored at two different areas, i.e., a smooth and ruffled part of the cellular membrane. The observed reduction in anisotropy (denoted by r), indicates the degree of homo-FRET at both locations. While this reduction is by itself sufficient to determine the average cluster size, partial bleaching of the fluorophores was used to determine the cluster size distribution. This is possible because aggregates with different cluster sizes show a different decrease in anisotropy upon reduction of the number of active fluorophores.85
Fig. 6.
Investigating membrane protein clustering using homo-FRET. (a) Interactions between membrane proteins (red) can be obtained by labeling them with fluorophores (blue); due to the challenges associated with labeling such proteins they are often studied using homo-FRET. Additionally, with this method the aggregate number can be inferred from the anisotropy of the emitted FRET signal. (b) Schematic depiction of bystander FRET, in which FRET occurs as a result of two fluorophore-labeled molecules being in close proximity due to diffusion rather than interactions between the functionalized molecules. The depicted excitation and emission wavelengths serve as an example. (c) Example of the use of homo-FRET to determine aggregate cluster sizes. Cells expressing a fluorophore-labeled transmembrane protein were imaged to determine the spatial reduction in anisotropy (r) of the light emitted upon excitation with polarized light. By performing this experiment after various degrees of bleaching, the protein cluster size distribution could be determined. Image adapted with permission from ref24. Copyright 2011, Wiley-VCH.
5. Reading supramolecular sensors
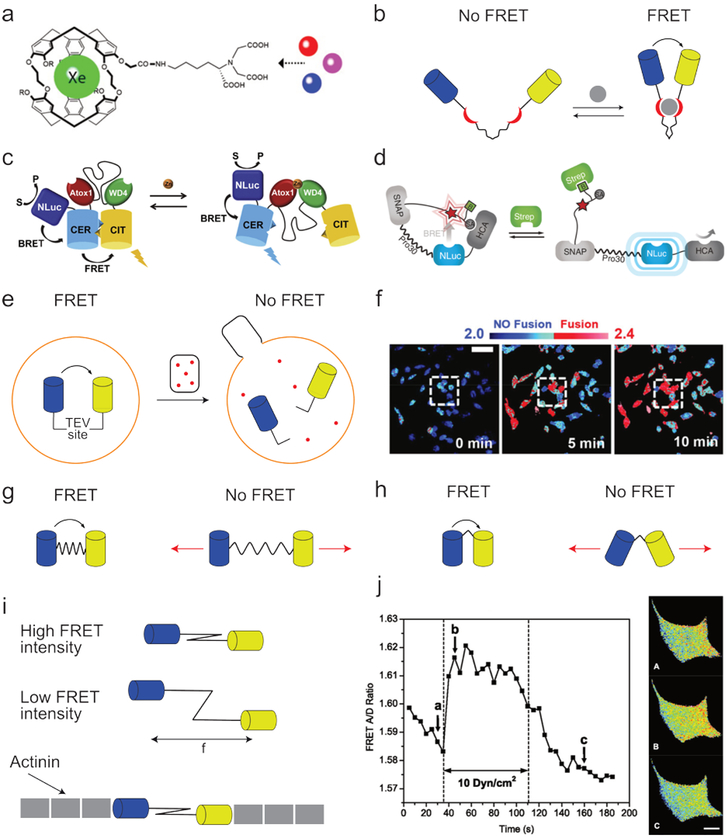
Sensors for chemical compounds or physical conditions are an increasingly popular application of synthetic supramolecular systems. Amongst others, sensors for metals,86 biomolecules,87 charge,88 pH,89 force,90 and temperature91 have been developed. While the range of sensor designs and substrates is beyond the scope of this review, the manner in which their activation can be monitored using FRET deserves some attention. In numerous molecular sensors, substrate binding is observed as a direct change in the sensor’s physiochemical properties, such as its chemical shift92 or fluorescence intensity93,94 (Fig. 7a). As these types of sensors do not rely on conformational changes, they are typically small and robust. However, the necessarily close sensor-substrate proximity makes them less appropriate for detecting larger molecules. Alternatively, FRET-based sensors are often more widely applicable as they rely on conformational changes induced by activating the sensor, and the FRET pair is thus not directly involved in the binding process (Fig. 7b).95
Fig. 7.
Examples of FRET sensors. (a) Example of a non-FRET-based metal sensor with which the presence of different metals can be monitored using 129Xe NMR. Image adapted with permission from ref107. Copyright 2010, Wiley-VCH. (b) Schematic depiction of a FRET-based molecular sensor. When the ligands (red) bind to the substrate (grey), the fluorophores (blue and yellow) are brought in close proximity, thereby increasing FRET efficiency. (c) A dual readout BRET/FRET sensor for measuring in vivo zinc concentrations. Images adapted with permission from ref108. Copyright 2017, Elsevier. (d) A combined sensor and switch in which adding streptavidin leads to reduced BRET and increased human carbonic anhydrase activity. Image adapted with permission from ref102. Copyright 2015, Nature Publishing Group. (e) The fusion of HIV-1 virions with living cells was studied using a covalently bound FRET pair connected by a linker containing a Tobacco Etch Virus protease-cleavable site. This FRET pair was incorporated into cells (orange), while HIV-1 virions (black) containing Tobacco Etch Virus protease (red) were added. As a result, virus capsule fusion with the cells resulted in linker cleavage and reduced FRET efficiency. (f) Example of the results obtained from the system described under e, showing the time-resolved fusion between HIV-1 virions and cells. Blue indicates shorter donor fluorophore lifetimes (2.0 ns) and thus an intact TEV site; red indicates longer donor fluorophore lifetimes (2.4 ns) and thus cleavage of the TEV site. Image adapted with permission from ref106. Copyright 2015, Nature Publishing Group. (g) Schematic depiction of a distance-based FRET force sensor consisting of a FRET pair (blue and yellow) connected through a spring-like linker. Because both the inter-fluorophore distance and the relation between fluorophore distance and FRET efficiency are known, the intensity of the applied force can be inferred from the intensity of the FRET signal. (h) Schematic depiction of an orientation-based FRET force sensor where the angle between the fluorophores changes as a result of the applied force, thereby resulting in altered FRET efficiency. Image based on ref109. (i) Example of the use of a FRET-based force sensor comprising a spring-like spectrin linker functionalized with two fluorophores. When a force pulls the fluorophores apart, the FRET efficiency decreases. The FRET sensor was functionalized with actinin and (j) incorporated in cells, allowing real-time measurements of cytoskeletal stresses. Red indicates lower tension; blue indicates higher tension. Cell pictures were taken at the times indicated with the corresponding letters in the graph. Image adapted with permission from ref109. Copyright 2011, American Physiological Society.
While initial resonance energy transfer sensors relied on FRET, advancements in bioluminescent proteins have sharply increased the number of bioluminescence resonant energy transfer (BRET)-based sensors, which eliminate the need for external illumination and are therefore better suited for in vivo applications. Interestingly, instead of developing entirely new BRET sensors, it is common for existing FRET sensors to be functionalized with a bioluminescent protein (Fig. 7c).96 This approach averts sensor re-optimization and provides both fluorescent and bioluminescent readouts. Although these sensors do not match the high tissue penetration possible with other techniques, such as magnetic resonance imaging (MRI; millimeters97 vs. meters,98 respectively), FRET and BRET do allow for much higher spatial and temporal resolutions (≈ 200 nanometers and picoseconds for FRET microscopy99,100 vs. millimeters and seconds for MRI101). Consequently, FRET or BRET sensors are typically used for detailed cell studies, while MRI is used for tissue or in vivo analysis.
Besides serving as sensors, such compounds are increasingly finding additional applications. In one example, a structure acted as both sensor and switch.102 Here, the bioluminescent protein NanoLuc (NLuc) was covalently attached to the enzyme human carbonic anhydrase (HCA), as well as an acceptor fluorophore functionalized with biotin (B) and an HCA inhibitor (SA, Fig. 7d). Due to a cleaver molecular design, adding streptavidin (Strep) led to SA displacement from HCA as well as an increase in BRET pair separation. Consequently, the observed BRET intensity measured not only the streptavidin concentration but also served as a reporter for the HCA activity.
In addition to physical conditions or small molecules, FRET can also monitor large-scale (e.g. cellular) processes. One important factor in the study of a given pathogen is the cell infection rate. While various microscopy techniques can track pathogen uptake, they do not provide real-time results, and it can be difficult to discriminate between pathogen uptake and adhesion.103–105 Although several optical methods do not have these limitations, they are often technically challenging and invasive.106 A FRET-based approach has been developed to overcome these problems (Fig. 7e).106 Here, a FRET pair was connected by a Tobacco Etch Virus protease (TEVp)-cleavable linker and introduced into the cells of interest. Additionally, TEVp was introduced into the pathogen. As a result, cell infection leads to TEVp uptake, FRET pair cleavage, and reduced FRET efficiency (Fig. 7f). In this example the FRET efficiency was not monitored by comparing emission intensities, but by measuring the donor’s fluorescence lifetime (FRET-FLIM). In intact structures FRET can take place and a relatively short lifetime of 2.0 ns was observed. After fusion and cleavage of the linker, the donor can no longer transfer its energy, resulting in a lengthening of the donor fluorescence lifetime to 2.4 ns.
Lastly, many biochemical assemblies have been shown to respond to mechanical force.110,111 In addition, non-covalent interactions themselves often induce molecule or aggregate movement, such as cytoskeletal rearrangements,112 kinesin motor movement,113 or integrin adhesion.114 FRET-based force sensors can reveal the forces associated with such phenomena.90 In a typical configuration, two fluorophores are connected through a spring-like linker, so that the distance or angle between them depends on the force applied (Fig. 7g–h).90,115,116 After calibration using computational models or single-molecule measurements, such sensors can be used for quantitative measurements.117,118 To the best of our knowledge, FRET-based force sensors have not been used to study entirely synthetic supramolecular systems. However, they have provided unprecedented insight into mechanical forces’ role in biochemical processes, under both in vitro and in vivo conditions.90,119,120 An elegant study of non-covalent interactions using such sensors was reported by S. Hua et. al.109 Here, the authors created a force sensor composed of a subcloned spectrin linker functionalized with a FRET pair. This sensor was subsequently functionalized with actinin and incorporated into the actinin filaments of a living cell, where it provided real-time information about cytoskeletal stresses (Fig. 7i–j).
In any supramolecular system studied by FRET, unreliable results can arise from interfluorophore interactions, or interactions between the investigated structure and the fluorophores used to study it. Arguably, consideration of fluorophore interactions is especially important in sensors, as such interactions will influence both the orientation of the fluorophores (and thereby the FRET efficiency) as well as the way the sensor responds to stimuli. Furthermore, many in vivo sensors make use of fluorescent proteins, which can be prone to oligomerize.121 However, it has been shown that interfluorophore interactions can sometimes be beneficial. In an example by C. Schultz, interfluorophore interactions increased a sensor’s sensitivity by inducing a favorable conformation.40 While the strengths of such fluorophore interactions are likely difficult to predict, they may thus provide an additional way to tune sensors’ behavior.
6. Monitoring the in vivo distribution and stability of nanotherapeutics
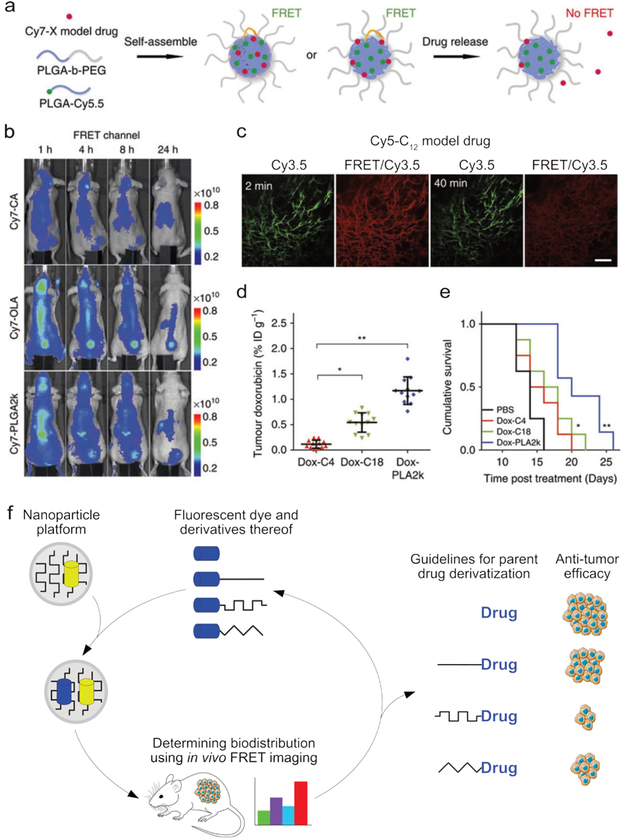
Synthetic supramolecular structures are increasingly exploited as drug delivery platforms and in vivo imaging agents.11 Understanding these structures’ biodistribution and stability under biological conditions is paramount to optimizing efficacy.122–124 As the complex and ‘hostile’ in vivo environment makes it difficult to reliably predict the relation between a nanotherapeutic’s structure and its behavior, insight must often be obtained experimentally. For example, by incorporating a FRET pair in a nanotherapeutic, its structural integrity can be monitored non-invasively in real time in vivo (Fig. 8a, where Cy7 and Cy5.5 form the FRET pair).125 Since it simultaneously elucidates the constructs’ spatial distribution, this approach can be used for imaging applications as well.126 The interpretation of FRET-facilitated in vivo stability measurements depends on a complex analysis that considers fluorophore effluence and assembly disintegration. The future success of nanoparticle therapeutics relies on methods to improve their design, preferably based on in vivo readouts. FRET techniques can be exploited to study nanotherapeutic in vivo stability and drug release. More specifically, FRET acceptor fluorophores that serve as model drugs can be incorporated in nanoparticle carriers that are labeled with a donor fluorophore. This system uses FRET imaging to determine the rate at which drugs leak from nanoparticle carriers.12,127,128 For example, two recent studies demonstrate how imaging can assist in determining nanotherapeutic tumor accumulation.
Fig. 8.
The in vivo stability and distribution of a supramolecular construct can be monitored using FRET. (a) The components of a polymeric nanoparticle were labeled with a FRET donor (Cy5.5), and a FRET acceptor (Cy7) was incorporated as a model drug. By measuring the decrease in FRET efficiency, the release of the model drug could be monitored. (b) Three formulations of the particles described in a were formed, each containing different derivatives of the Cy7 model drug, CA = carboxylic acid, OLA = oleylamine, PLGA = poly(lactic-co-glycolic acid). The biodistribution and structural integrity of the nanotherapeutic (i.e., the combined nanoparticle+model drug), could be visualized using FRET imaging. (c) Fluorescence images of a window chamber mouse injected with nanoparticles containing the Cy5-C12 model drug and Cy3.5 fluorophore, 2 and 40 minutes post injection. Scale bar = 100 μm injection (Cy3.5 was used instead of Cy7 to accommodate the microscopy protocol). (d) The tumor accumulation of doxorubicin observed in mice 24 hours after injection with nanoparticles containing one of three different doxorubicin derivatives, showing the strong influence of drug functionalization. (e) The cumulative survival curves corresponding to the experiments in d. Images a-e adapted with permission from ref12. Copyright 2016, Nature Publishing Group. (f) The biodistribution of a nanotherapeutic depends on the carrier’s size, composition, and compatibility with the incorporated drug. By labeling the carrier with a fluorophore and including a fluorescent model drug, the carrier’s stability and biodistribution can be monitored using FRET. In this way, the drug-carrier compatibility can be optimized, thereby enhancing nanotherapeutic efficacy.
Zhao et al. applied such a strategy to study drug derivatization’s effects on nanotherapeutic tumor delivery efficacy. Carrier nanoparticles were constructed from PEG-PLGA block copolymers, some of which were labeled with the near infrared (NIR) donor fluorophore Cy5.5. The NIR acceptor fluorophore Cy7 was chosen as a model drug (Fig. 8a). The latter dye was subsequently derivatized with different molecular units that increased Cy7’s compatibility with the Cy5.5-labeled PEG-PLGA nanoparticle carrier. In tumor-bearing mice (tumor in upper thigh), intravital microscopy and whole-body FRET imaging revealed that Cy7 derivatization dramatically improved the overall construct’s in vivo stability and enhanced tumor delivery (Fig. 8b–d).12 For example, little FRET was observed for the Cy7-CA injected mice, showing that this model drug quickly leaves the particle, while the high FRET efficicency observed for Cy7-OLA showed that this nanotherapeutic stayed intact much longer. These imaging-derived guidelines were subsequently applied to the clinically relevant cytotoxic agent doxorubicin. Similarly, albeit in a prodrug manner, this drug was derivatized to enhance its compatibility with the PEG-PLGA nanoparticle platform. Upon in vivo application in the same mouse model, dramatic differences in tumor accumulation and corresponding survival rates were observed (Fig. 8d). This iterative design process, in which FRET imaging provides guidelines that can be applied to improve the design of nanotherapeutics, is depicted in Fig. 8e. Currently, the nanomedicine field largely lacks the integration of such methods and the products that are translated to the clinic may therefore often be suboptimally designed. FRET imaging techniques’ ability to non-invasively monitor the stability of large constructs, as well as the effluence of small molecules, make them uniquely suited for such applications.
7. Future developments
Its low invasiveness and high sensitivity make FRET well suited to investigate processes on the molecular scale. However, the technique is not devoid of limitations and is therefore increasingly complemented by alternative techniques. One of the major drawbacks of FRET is the relatively short distance over which energy can be transferred (≈ 10 nm), which significantly constrains the study of larger structures. Fortunately, there are several ways to increase the FRET distance. One approach involves forming cascades, in which several FRET fluorophores with increasingly longer excitation wavelengths are organized into arrays to facilitate energy transfer over distances of 10 nm or more.129–133 Although originally mostly a scientific curiosity, such cascades are increasingly being used to analyze molecular conformations and dynamics, as shown in Fig. 5b.134 In addition to FRET cascades, much effort is focused on developing more efficient fluorophores. FRET’s heavy dependence on distance results from the R−6 decrease of transfer probability (R being the donor-acceptor distance). However, in case of fluorophores with sizes close to R or for favorable alignment of highly directional dipoles of organic fluorophores a weaker distance dependence is possible. For example, outside the regime of interaction between point dipoles (as assumed in Förster’s theory) a R−4 or R−2 distance dependence has been predicted,135 therefore (F)RET efficiency can often be increased to larger distances. For example, energy has been transferred over a distance of 20 nm from an organic fluorophore to a spherical nanometal where the metal nanoparticle, with strongly absorbing free electrons, acts as a surface rather than a point dipole.136 In addition, FRET is increasingly replaced by plasmon-enhanced or metal-induced resonance energy transfer, which have been shown to work over distances of 70 and 100 nm, respectively.137–139 An alternative class of fluorophores is formed by semiconductor nanoparticles or quantum dots. Besides having a higher efficiency, these generally do not blink and have a longer lifetime compared to organic fluorophores, making it possible to monitor molecules for extended periods of time.28,140 However, these nanocrystalline dyes are relatively large (typically >10 nm after the application of a coating), have unknown cytotoxic effects, and are less accessible to most organic functionalization chemistries. For these reasons, novel additives and conjugates are being developed to increase the stability of organic fluorophores.141,142
Another area of improvement for FRET is how the fluorophores are coupled to the molecules of interest. The chemical strategies used to introduce fluorophores are often incompatible with biological systems and thus limit the approach’s applicability.143 Further development of biocompatible labeling procedures, such as enzyme-mediated coupling,144 is expected to simplify in vivo studies and expand the types of systems that can be studied. Improved control over the exact location at which fluorophores are introduced is also highly desirable, for example in conformational protein mapping. Typically, such site-specific labeling proceeds by incorporating modified building blocks (e.g., unnatural amino acids) functionalized with a chemical moiety to which the fluorophores are coupled.143 However, these tagging procedures usually introduce only one or two fluorophores.145 Site-specific labeling with multiple fluorophores would enable simultaneous monitoring of multiple sections of a molecule and thereby increase the efficiency with which conformations and dynamics can be unraveled. Besides, the correlated movement of multiple molecular domains simply cannot be monitored using only two fluorophores.146 In addition to studying protein conformations, such multi-fluorophore assays will likely be highly relevant to the analysis of cellular signaling pathways,17,147 motor proteins,113 and microfluidic reactors.148 Yet using multiple fluorophores will also require additional insight into unequimolar donor and acceptor ratios,17 as well as further development of physical149 and mathematical150 spectral unmixing techniques.
One way such physical unmixing techniques work is by altering the emission spectra of certain fluorophores to minimize spectral overlap.149 Secondly, increasingly more switchable fluorophores are being developed. Molecules and nanoparticles whose fluorescence can be switched on or off upon irradiation151–153 will surely benefit future conformational mapping studies, but are also finding applications in FRET-based switching and amplification of optical signals.154 In addition to physically changing the behavior of a fluorophore, spectral unmixing through advanced FRET signal observation technologies is a highly active field of research. Especially promising is time-gated FRET, in which the FRET signal is only recorded during a specific time window (e.g., 100 – 400 μs after excitation).155,156 This approach greatly reduces the influence of scattering and autofluorescence and helps to discriminate between fluorophores with different fluorescence decay times. Due to their long luminescence lifetimes, narrow emission spectra, high brightness, and easy color tunability, lanthanides and quantum dots are the most suitable fluorophores for such applications.157
Besides the development of FRET signal observation techniques, the current trend toward integrating FRET with other techniques will likely continue.158,159 For example, much effort is currently focused on developing super-resolution FRET to allow the phenomena monitored through FRET to be localized with unprecedented spatial resolution.160 In addition, FRET-AFM has shown great potential in simultaneously monitoring and manipulating molecules,33,161 and FRET-mass spectrometry techniques are revealing molecules’ gas-phase behavior.162 As with these microscopy and single-molecule techniques, in vivo FRET monitoring will likely also benefit from further integration with other techniques. Currently, such measurements are complicated by autofluorescence, poor fluorophore stability, and the limited penetration depth of the excitation and emitted light. Accordingly, in vivo FRET imaging is increasingly integrated with positron emission tomography (PET). Furthermore, integrating FRET with fluorescence molecular tomography has enormous potential to enhance the accuracy with which FRET can be localized in 3D.163 Lastly, the need to externally excite fluorophores will likely be reduced through the development of novel bioluminescent proteins164 and various synthetic alternatives, such as self-illuminating quantum dots.165
Combined, these developments are expected to broaden both the system types that can be studied using FRET and the insights that can be thus obtained. The ability to simultaneously monitor multiple fluorophores will greatly benefit the development of multicomponent synthetic systems and provide unprecedented knowledge of cellular reaction networks and dynamics. Furthermore, we expect that integrating FRET with various microscopy and imaging techniques will revolutionize the study of molecular interactions and improve the in vivo monitoring of nanotherapeutics and other supramolecular constructs.
8. Conclusions
The high sensitivity, broad temporal range, and non-invasiveness of FRET make it ideal for analyzing supramolecular systems. Rather than serving as a stand-alone technique, FRET has become a tool that can be implemented in a wide spectrum of analytical techniques and settings. The affordability and simplicity of basic FRET spectroscopy makes it accessible to any lab with a fluorescence spectrometer, while numerous specialized FRET techniques have been developed to address highly complex molecular questions. However, FRET also has several shortcomings that limit its applicability as an analytical tool: the relatively short distance range over which energy can be transferred, the instability of organic fluorophores, the limited number of FRET pairs that can be monitored simultaneously, and the challenge of obtaining quantitative results. Consequently, FRET is increasingly replaced or complemented by alternative techniques, such as plasmon-induced resonance energy transfer and super-resolution microscopy. Nonetheless, the relative ease with which FRET can provide insight into dynamic processes at the nanoscale guarantees its relevance to the analysis of supramolecular systems in the decades to come.
Supplementary Material
Acknowledgements
This work was supported by NIH grant R01HL125703.
Footnotes
Conflict of interest
There are no conflicts to declare.
Notes and references
- 1.Hazan NP, Tomov TE, Tsukanov R, Liber M, Berger Y, Masoud R, Toth K, Langowski J and Nir E, Biophys. J, 2015, 109, 1676–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonifacino JS and Glick BS, Cell, 2004, 116, 153–166. [DOI] [PubMed] [Google Scholar]
- 3.Warshel A, Sharma PK, Kato M, Xiang Y, Liu H and Olsson MHM, Chem. Rev, 2006, 106, 3210–3235. [DOI] [PubMed] [Google Scholar]
- 4.Mozhdehi D, Ayala S, Cromwell OR and Guan Z, J. Am. Chem. Soc, 2014, 136, 16128–16131. [DOI] [PubMed] [Google Scholar]
- 5.Blanco V, Leigh DA and Marcos V, Chem. Soc. Rev, 2015, 44, 5341–5370. [DOI] [PubMed] [Google Scholar]
- 6.van Dongen SFM, Cantekin S, Elemans JAAW, Rowan AE and Nolte RJM, Chem. Soc. Rev, 2014, 43, 99–122. [DOI] [PubMed] [Google Scholar]
- 7.Amabilino DB, Smith DK and Steed JW, Chem. Soc. Rev, 2017, 46, 2404–2420. [DOI] [PubMed] [Google Scholar]
- 8.Lou X-Y, Song N and Yang Y-W, Molecules, 2017, 22, 1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain A and George SJ, Mater. Today, 2015, 18, 206–214. [Google Scholar]
- 10.Webber MJ, Appel EA, Meijer EW and Langer R, Nat. Mater, 2015, 15, 3–26. [DOI] [PubMed] [Google Scholar]
- 11.Versluis F, van Esch JH and Eelkema R, Adv. Mater, 2016, 28, 4576–4592. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y, Fay F, Hak S, Manuel Perez-Aguilar J, Sanchez-Gaytan BL, Goode B, Duivenvoorden R, de Lange Davies C, Bjørkøy A, Weinstein H, Fayad ZA, Pérez-Medina C and Mulder WJM, Nat. Commun, 2016, 7, 11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahay G, Alakhova DY and V Kabanov A, J. Control. Release, 2010, 145, 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabral H, Nishiyama N and Kataoka K, Acc. Chem. Res, 2011, 44, 999–1008. [DOI] [PubMed] [Google Scholar]
- 15.Forster T, Ann. Phys, 1948, 2, 55–75. [Google Scholar]
- 16.Perrin J, Acad R. Sci, 1927, 184, 1097. [Google Scholar]
- 17.Bunt G and Wouters FS, Biophys. Rev, 2017, 9, 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Meer BW, Rev. Mol. Biotechnol, 2002, 82, 181–196. [DOI] [PubMed] [Google Scholar]
- 19.Haas E, in Intrinsically Disordered Protein Analysis: Volume 1, Methods and Experimental Tools, eds. Uversky VN and Dunker AK, Humana Press, Totowa, NJ, 2012, pp. 467–498. [Google Scholar]
- 20.Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T and Lilley DMJ, Proc. Natl. Acad. Sci , 2008, 105, 11176–11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou D, Piper JD, Abell C, Klenerman D, Kang D-J and Ying L, Chem. Commun, 2005, 4807–4809. [DOI] [PubMed] [Google Scholar]
- 22.Berney C and Danuser G, Biophys. J, 2003, 84, 3992–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Reste L, Hohlbein J, Gryte K and Kapanidis AN, Biophys. J, 2012, 102, 2658–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bader AN, Hoetzl S, Hofman EG, Voortman J, van Bergen en Henegouwen PMP, van Meer G and Gerritsen HC, ChemPhysChem, 2011, 12, 475–483. [DOI] [PubMed] [Google Scholar]
- 25.Wallrabe H and Periasamy A, Curr. Opin. Biotechnol, 2005, 16, 19–27. [DOI] [PubMed] [Google Scholar]
- 26.Dragulescu-Andrasi A, Chan CT, De A, Massoud TF and Gambhir SS, Proc. Natl. Acad. Sci, 2011, 108, 12060–12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfleger KDG and Eidne KA, Nat. Methods, 2006, 3, 165. [DOI] [PubMed] [Google Scholar]
- 28.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R and Nann T, Nat Meth, 2008, 5, 763–775. [DOI] [PubMed] [Google Scholar]
- 29.Wang L and Tan W, Nano Lett, 2006, 6, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tisler J, Reuter R, Lämmle A, Jelezko F, Balasubramanian G, Hemmer PR, Reinhard F and Wrachtrup J, ACS Nano, 2011, 5, 7893–7898. [DOI] [PubMed] [Google Scholar]
- 31.Ajayaghosh A, Vijayakumar C, Praveen VK, Babu SS and Varghese R, J. Am. Chem. Soc, 2006, 128, 7174–7175. [DOI] [PubMed] [Google Scholar]
- 32.Alemdaroglu FE, Alexander SC, Ji D, Prusty DK, Börsch M and Herrmann A, Macromolecules, 2009, 42, 6529–6536. [Google Scholar]
- 33.Vickery SA and Dunn RC, J. Microsc, 2001, 202, 408–412. [DOI] [PubMed] [Google Scholar]
- 34.Polley N, Singh S, Giri A, Mondal PK, Lemmens P and Pal SK, Sensors Actuators B Chem, 2015, 210, 381–388. [Google Scholar]
- 35.Smulders MMJ, Nieuwenhuizen MML, de Greef TFA, van der Schoot P, Schenning APHJ and Meijer EW, Chem. – A Eur. J, 2010, 16, 362–367. [DOI] [PubMed] [Google Scholar]
- 36.Korevaar PA, George SJ, Markvoort AJ, Smulders MMJ, Hilbers PAJ, Schenning APHJ, De Greef TFA and Meijer EW, Nature, 2012, 481, 492–496. [DOI] [PubMed] [Google Scholar]
- 37.Adelizzi B, Filot IAW, Palmans ARA and Meijer EW, Chem. – A Eur. J, 2017, 23, 6103–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azcarate JC, Diaz SA, Fauerbach JA, Gillanders F, Rubert AA, Jares-Erijman EA, Jovin TM and Fonticelli MH, Nanoscale, 2017, 9, 8647–8656. [DOI] [PubMed] [Google Scholar]
- 39.Sendai T, Biswas S and Aida T, J. Am. Chem. Soc, 2013, 135, 11509–11512. [DOI] [PubMed] [Google Scholar]
- 40.Jost CA, Reither G, Hoffmann C and Schultz C, ChemBioChem, 2008, 9, 1379–1384. [DOI] [PubMed] [Google Scholar]
- 41.Vira S, Mekhedov E, Humphrey G and Blank PS, Anal. Biochem, 2010, 402, 146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cordier P, Tournilhac F, Soulié-Ziakovic C and Leibler L, Nature, 2008, 451, 977. [DOI] [PubMed] [Google Scholar]
- 43.Mandal S, Kuchlyan J, Banik D, Ghosh S, Banerjee C, Khorwal V and Sarkar N, ChemPhysChem, 2014, 15, 3544–3553. [DOI] [PubMed] [Google Scholar]
- 44.Smith MM and Smith DK, Soft Matter, 2011, 7, 4856–4860. [Google Scholar]
- 45.Roy R, Hohng S and Ha T, Nat Meth, 2008, 5, 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diaspro A, Chirico G, Usai C, Ramoino P and Dobrucki J, Handbook of biological confocal microscopy, 3rd edn., 2006. [Google Scholar]
- 47.Huang C-B, Xu L, Zhu J-L, Wang Y-X, Sun B, Li X and Yang H-B, J. Am. Chem. Soc, 2017, 139, 9459–9462. [DOI] [PubMed] [Google Scholar]
- 48.Rajdev P, Basak D and Ghosh S, Macromolecules, 2015, 48, 3360–3367. [Google Scholar]
- 49.Albertazzi L, van der Veeken N, Baker MB, Palmans ARA and Meijer EW, Chem. Commun, 2015, 51, 16166–16168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanchez-Gaytan BL, Fay F, Hak S, Alaarg A, Fayad ZA, Pérez-Medina C, Mulder WJM and Zhao Y, Angew. Chemie [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mok MM, Thiagarajan R, Flores M, Morse DC and Lodge TP, Macromolecules, 2012, 45, 4818–4829. [Google Scholar]
- 52.Koopmans WJA, Brehm A, Logie C, Schmidt T and van Noort J, J. Fluoresc, 2007, 17, 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Llères D, James J, Swift S, Norman DG and Lamond AI, J. Cell Biol, 2009, 187, 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berliner NRKLJ, Biological Magnetic Resonance, Modern Techniques in Protein NMR, Kluwer Academic, 16th edn., 2002. [Google Scholar]
- 55.Sahoo H, J. Photochem. Photobiol. C Photochem. Rev, 2011, 12, 20–30. [Google Scholar]
- 56.Brunger AT, Strop P, Vrljic M, Chu S and Weninger KR, J. Struct. Biol, 2011, 173, 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woźniak AK, Schröder GF, Grubmüller H, Seidel CAM and Oesterhelt F, Proc. Natl. Acad. Sci , 2008, 105, 18337–18342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hillisch A, Lorenz M and Diekmann S, Curr. Opin. Struct. Biol, 2001, 11, 201–207. [DOI] [PubMed] [Google Scholar]
- 59.You X, Nguyen AW, Jabaiah A, Sheff MA, Thorn KS and Daugherty PS, Proc. Natl. Acad. Sci , 2006, 103, 18458–18463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tian H and Wang Q-C, Chem. Soc. Rev, 2006, 35, 361–374. [DOI] [PubMed] [Google Scholar]
- 61.Gil-Ramírez G, Leigh DA and Stephens AJ, Angew. Chemie Int. Ed, 2015, 54, 6110–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Onagi H and Rebek J Jr., Chem. Commun, 2005, 4604–4606. [DOI] [PubMed] [Google Scholar]
- 63.Kassem S, van Leeuwen T, Lubbe AS, Wilson MR, Feringa BL and Leigh DA, Chem. Soc. Rev, 2017, 46, 2592–2621. [DOI] [PubMed] [Google Scholar]
- 64.Prevo B, Acar S, Kruijssen DLH and Peterman EJG, Single-Molecule Spectroscopy of Motor Proteins, John Wiley & Sons, Ltd, 2006. [Google Scholar]
- 65.Verbrugge S, Lansky Z and Peterman EJG, Proc. Natl. Acad. Sci , 2009, 106, 17741–17746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Isojima H, Iino R, Niitani Y, Noji H and Tomishige M, Nat. Chem. Biol, 2016, 12, 290. [DOI] [PubMed] [Google Scholar]
- 67.Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA and Vale RD, Nature, 1999, 402, 778. [DOI] [PubMed] [Google Scholar]
- 68.Uphoff S, Holden SJ, Le Reste L, Periz J, van de Linde S, Heilemann M and Kapanidis AN, Nat. Methods, 2010, 7, 831. [DOI] [PubMed] [Google Scholar]
- 69.dos Remedios CG and Moens PDJ, J. Struct. Biol, 1995, 115, 175–185. [DOI] [PubMed] [Google Scholar]
- 70.Schuler B, Lipman EA, Steinbach PJ, Kumke M and Eaton WA, Proc. Natl. Acad. Sci, 2005, 102, 2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deniz AA, Dahan M, Grunwell JR, Ha T, Faulhaber AE, Chemla DS, Weiss S and Schultz PG, Proc. Natl. Acad. Sci , 1999, 96, 3670–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Michalet X, Weiss S and Jäger M, Chem. Rev, 2006, 106, 1785–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taraska JW, Puljung MC, Olivier NB, Flynn GE and Zagotta WN, Nat. Methods, 2009, 6, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nath A and Rhoades E, FEBS Lett, 2013, 587, 1096–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lidke DS and Wilson BS, Trends Cell Biol, 2009, 19, 566–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grupi A and Haas E, J. Mol. Biol, 2011, 411, 234–247. [DOI] [PubMed] [Google Scholar]
- 77.Sarkar A, Dhiman S, Chalishazar A and George SJ, Angew. Chemie Int. Ed, 2017, 56, 13767–13771. [DOI] [PubMed] [Google Scholar]
- 78.Surana S, Bhatia D and Krishnan Y, Methods, 2013, 64, 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brown CW, Buckhout-White S, Díaz SA, Melinger JS, Ancona MG, Goldman ER and Medintz IL, ACS Sensors, 2017, 2, 401–410. [DOI] [PubMed] [Google Scholar]
- 80.Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, Llaguno M, V Hollingsworth J, King DS, Banani SF, Russo PS, Jiang Q-X, Nixon BT and Rosen MK, Nature, 2012, 483, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lingwood D and Simons K, Science, 2010, 327, 46–50. [DOI] [PubMed] [Google Scholar]
- 82.Loura LMS and Prieto M, Front. Physiol, 2011, 2, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.King CR, Raicu V and Hristova K, Biophys. J, 2018, 110, 428. [Google Scholar]
- 84.Clayton AH and Chattopadhyay A, Biophys. J, 2014, 106, 1227–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sharma P, Varma R, Sarasij RC, Gousset Ira, K., Krishnamoorthy G, Rao M and Mayor S, Cell, 2004, 116, 577–589. [DOI] [PubMed] [Google Scholar]
- 86.Carter KP, Young AM and Palmer AE, Chem. Rev, 2014, 114, 4564–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Medintz IL, Trends Biotechnol, 2006, 24, 539–542. [DOI] [PubMed] [Google Scholar]
- 88.Abraham BG, Santala V, V Tkachenko N and Karp M, Anal. Bioanal. Chem, 2014, 406, 7195–7204. [DOI] [PubMed] [Google Scholar]
- 89.Modi S, M. G. S, Goswami D, Gupta GD, Mayor S and Krishnan Y, Nat. Nanotechnol, 2009, 4, 325. [DOI] [PubMed] [Google Scholar]
- 90.Cost A-L, Ringer P, Chrostek-Grashoff A and Grashoff C, Cell. Mol. Bioeng, 2015, 8, 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xie N, Huang J, Yang X, He X, Liu J, Huang J, Fang H and Wang K, Anal. Chem, 2017, 89, 12115–12122. [DOI] [PubMed] [Google Scholar]
- 92.Allouche-Arnon H, Tirukoti ND and Bar-Shir A, Isr. J. Chem, 2017, 57, 843–853. [Google Scholar]
- 93.Zhang L and Wang E, Nano Today, 2014, 9, 132–157. [Google Scholar]
- 94.Iverson NM, Barone PW, Shandell M, Trudel LJ, Sen S, Sen F, Ivanov V, Atolia E, Farias E, McNicholas TP, Reuel N, Parry NMA, Wogan GN and Strano MS, Nat. Nanotechnol, 2013, 8, 873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rowland CE, Lii CWB, Medintz IL and Delehanty JB, Methods Appl. Fluoresc [DOI] [PubMed] [Google Scholar]
- 96.Aper SJA, Dierickx P and Merkx M, ACS Chem. Biol, 2016, 11, 2854–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Crosignani V, Dvornikov A, Aguilar JS, Stringari C, Edwards R, Mantulin WW and Gratton E, J. Biomed. Opt, 2012, 17, 116023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Key J and Leary JF, Int. J. Nanomedicine, 2014, 9, 711–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tramier M, Gautier I, Piolot T, Ravalet S, Kemnitz K, Coppey J, Durieux C, Mignotte V and Coppey-Moisan M, Biophys. J, 2002, 83, 3570–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Llères D, Bailly AP, Perrin A, Norman DG, Xirodimas DP and Feil R, Cell Rep, 2017, 18, 1791–1803. [DOI] [PubMed] [Google Scholar]
- 101.Uğurbil K, Xu J, Auerbach EJ, Moeller S, Vu AT, Duarte-Carvajalino JM, Lenglet C, Wu X, Schmitter S, Van de Moortele PF, Strupp J, Sapiro G, De Martino F, Wang D, Harel N, Garwood M, Chen L, Feinberg DA, Smith SM, Miller KL, Sotiropoulos SN, Jbabdi S, Andersson JLR, Behrens TEJ, Glasser MF, Van Essen DC and Yacoub E, Neuroimage, 2013, 80, 80–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schena A, Griss R and Johnsson K, Nat. Commun, 2015, 6, 7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stein BS, Gowda SD, Lifson JD, Penhallow RC, Bensch KG and Engleman EG, Cell, 1987, 49, 659–668. [DOI] [PubMed] [Google Scholar]
- 104.Jolly C, Kashefi K, Hollinshead M and Sattentau QJ, J. Exp. Med, 2004, 199, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Isberg RR, Science, 1991, 252, 934–938. [DOI] [PubMed] [Google Scholar]
- 106.Jones DM and Padilla-Parra S, Sci. Rep, 2015, 5, 13449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tassali N, Kotera N, Boutin C, Léonce E, Boulard Y, Rousseau B, Dubost E, Taran F, Brotin T, Dutasta J-P and Berthault P, Anal. Chem, 2014, 86, 1783–1788. [DOI] [PubMed] [Google Scholar]
- 108.Arts R, Aper SJA and Merkx M, in Enzymes as Sensors, eds. Thompson RB and T.-M. CAB in Fierke E, Academic Press, 2017, vol. 589, pp. 87–114. [Google Scholar]
- 109.Rahimzadeh J, Meng F, Sachs F, Wang J, Verma D and Hua SZ, Am. J. Physiol. - Cell Physiol, 2011, 301, 646–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vogel V and Sheetz MP, Curr. Opin. Cell Biol, 2009, 21, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang N, Tytell JD and Ingber DE, Nat. Rev. Mol. Cell Biol, 2009, 10, 75. [DOI] [PubMed] [Google Scholar]
- 112.Nédélec F, Surrey T and Karsenti E, Curr. Opin. Cell Biol, 2003, 15, 118–124. [DOI] [PubMed] [Google Scholar]
- 113.Prevo B and Peterman EJG, Chem. Soc. Rev, 2014, 43, 1144–1155. [DOI] [PubMed] [Google Scholar]
- 114.Morimatsu M, Mekhdjian AH, Adhikari AS and Dunn AR, Nano Lett, 2013, 13, 3985–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meng F and Sachs F, J. Cell Sci, 2012, 125, 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Meng F and Sachs F, J. Cell Sci, 2011, 124, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shroff H, Reinhard BM, Siu M, Agarwal H, Spakowitz A and Liphardt J, Nano Lett, 2005, 5, 1509–1514. [DOI] [PubMed] [Google Scholar]
- 118.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T and Schwartz MA, Nature, 2010, 466, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Freikamp A, Mehlich A, Klingner C and Grashoff C, J. Struct. Biol, 2017, 197, 37–42. [DOI] [PubMed] [Google Scholar]
- 120.Freikamp A, Cost A-L and Grashoff C, Trends Cell Biol, 2016, 26, 838–847. [DOI] [PubMed] [Google Scholar]
- 121.Zacharias DA, Sci. STKE, 2002, 2002, 23–23. [DOI] [PubMed] [Google Scholar]
- 122.Feiner-Gracia N, Buzhor M, Fuentes E, Pujals S, Amir RJ and Albertazzi L, J. Am. Chem. Soc, 2017, 139, 16677–16687. [DOI] [PubMed] [Google Scholar]
- 123.Chen H, Kim S, He W, Wang H, Low PS, Park K and Cheng J-XX, Langmuir, 2008, 24, 5213–7. [DOI] [PubMed] [Google Scholar]
- 124.Lee S-Y, Tyler JY, Kim S, Park K and Cheng J-X, Mol. Pharm, 2013, 10, 3497–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Priem B, Tian C, Tang J, Zhao Y and Mulder WJM, Expert Opin. Drug Deliv, 2015, 1–14. [DOI] [PubMed] [Google Scholar]
- 126.Yang P-P, Yang Y, Gao Y-J, Wang Y, Zhang J-C, Lin Y-X, Dai L, Li J, Wang L and Wang H, Adv. Opt. Mater, 2015, 3, 646–651. [Google Scholar]
- 127.Chen H, Kim S, Li L, Wang S, Park K and Cheng J-X, Proc. Natl. Acad. Sci , 2008, 105, 6596–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jiwpanich S, Ryu J-H, Bickerton S and Thayumanavan S, J. Am. Chem. Soc, 2010, 132, 10683–10685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Haustein E, Jahnz M and Schwille P, ChemPhysChem, 2003, 4, 745–748. [DOI] [PubMed] [Google Scholar]
- 130.Suresh M, Mandal AK, Suresh E and Das A, Chem. Sci, 2013, 4, 2380–2386. [Google Scholar]
- 131.Rowland CE, Delehanty JB, Dwyer CL and Medintz IL, Mater. Today, 2017, 20, 131–141. [Google Scholar]
- 132.Spillmann CM, Buckhout-White S, Oh E, Goldman ER, Ancona MG and Medintz IL, Chem. Commun, 2014, 50, 7246–7249. [DOI] [PubMed] [Google Scholar]
- 133.Olejko L, Cywinski PJ and Bald I, Nanoscale, 2016, 8, 10339–10347. [DOI] [PubMed] [Google Scholar]
- 134.Krainer G, Hartmann A and Schlierf M, Nano Lett, 2015, 15, 5826–5829. [DOI] [PubMed] [Google Scholar]
- 135.Saini S, Singh H and Bagchi B, J. Chem. Sci, 2006, 118, 23–35. [Google Scholar]
- 136.Yun CS, Javier A, Jennings T, Fisher M, Hira S, Peterson S, Hopkins B, Reich NO and Strouse GF, J. Am. Chem. Soc, 2005, 127, 3115–3119. [DOI] [PubMed] [Google Scholar]
- 137.Ray PC, Fan Z, Crouch RA, Sinha SS and Pramanik A, Chem. Soc. Rev, 2014, 43, 6370–6404. [DOI] [PubMed] [Google Scholar]
- 138.Sönnichsen C, Reinhard BM, Liphardt J and Alivisatos AP, Nat. Biotechnol, 2005, 23, 741. [DOI] [PubMed] [Google Scholar]
- 139.Chizhik AI, Rother J, Gregor I, Janshoff A and Enderlein J, Nat. Photonics, 2014, 8, 124. [Google Scholar]
- 140.Muir J, Arancibia-Carcamo IL, MacAskill AF, Smith KR, Griffin LD and Kittler JT, Proc. Natl. Acad. Sci, 2010, 107, 16679–16684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zheng Q, Juette MF, Jockusch S, Wasserman MR, Zhou Z, Altman RB and Blanchard SC, Chem. Soc. Rev, 2014, 43, 1044–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Campos LA, Liu J, Wang X, Ramanathan R, English DS and Muñoz V, Nat. Methods, 2011, 8, 143. [DOI] [PubMed] [Google Scholar]
- 143.Zhang G, Zheng S, Liu H and Chen PR, Chem. Soc. Rev, 2015, 44, 3405–3417. [DOI] [PubMed] [Google Scholar]
- 144.Liu J, Hanne J, Britton BM, Shoffner M, Albers AE, Bennett J, Zatezalo R, Barfield R, Rabuka D, Lee J-B and Fishel R, Sci. Rep, 2015, 5, 16883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sachdeva A, Wang K, Elliott T and Chin JW, J. Am. Chem. Soc, 2014, 136, 7785–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hohng S, Lee S, Lee J and Jo MH, Chem. Soc. Rev, 2014, 43, 1007–1013. [DOI] [PubMed] [Google Scholar]
- 147.Götz M, Wortmann P, Schmid S and Hugel T, in Single-Molecule Enzymology: Fluorescence-Based and High-Throughput Methods, eds. Spies M and Chemla YR, Academic Press, 2016, vol. 581, pp. 487–516. [DOI] [PubMed] [Google Scholar]
- 148.Varghese SS, Zhu Y, Davis TJ and Trowell SC, Lab Chip, 2010, 10, 1355–1364. [DOI] [PubMed] [Google Scholar]
- 149.Chen G, Song F, Wang J, Yang Z, Sun S, Fan J, Qiang X, Wang X, Dou B and Peng X, Chem. Commun, 2012, 48, 2949–2951. [DOI] [PubMed] [Google Scholar]
- 150.Zimmermann T, Marrison J, Hogg K and O’Toole P, in Confocal Microscopy: Methods and Protocols, ed. Paddock SW, Springer New York, New York, NY, 2014, pp. 129–148. [Google Scholar]
- 151.Schweighöfer F, Dworak L, Hammer CA, Gustmann H, Zastrow M, Rück-Braun K and Wachtveitl J, Sci. Rep, 2016, 6, 28638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Díaz SA, Gillanders F, Jares-Erijman EA and Jovin TM, Nat. Commun, 2015, 6, 6036. [DOI] [PubMed] [Google Scholar]
- 153.V Subach F, Zhang L, Gadella TWJ, Gurskaya NG, Lukyanov KA and V Verkhusha V, Chem. Biol, 2010, 17, 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jia S, Tuyoshi F, Jean-Pierre P, Tsunenobu O, Ryuju S, Hidetoshi O, Arnaud B, François B, Robert P, Keitaro N and Rémi M, Angew. Chemie Int. Ed, 55, 3662–3666. [Google Scholar]
- 155.Algar WR, Wegner D, Huston AL, Blanco-Canosa JB, Stewart MH, Armstrong A, Dawson PE, Hildebrandt N and Medintz IL, J. Am. Chem. Soc, 2012, 134, 1876–1891. [DOI] [PubMed] [Google Scholar]
- 156.Zwier JM and Hildebrandt N, in Reviews in Fluorescence 2016, ed. Geddes CD, Springer International Publishing, Cham, 2017, pp. 17–43. [Google Scholar]
- 157.Dos Santos MC and Hildebrandt N, TrAC Trends Anal. Chem, 2016, 84, 60–71. [Google Scholar]
- 158.Gomez-Godinez V, Preece D, Shi L, Khatibzadeh N, Rosales D, Pan Y, Lei L, Wang Y and Berns MW, Microsc. Res. Tech, 2015, 78, 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Swoboda M, Grieb MS, Hahn S and Schlierf M, in Fluorescent Methods for Molecular Motors, eds. Toseland CP and Fili N, Springer Basel, Basel, 2014, pp. 253–276. [Google Scholar]
- 160.Grecco HE and Verveer PJ, ChemPhysChem, 2011, 12, 484. [DOI] [PubMed] [Google Scholar]
- 161.He Y, Lu M, Cao J and Lu HP, ACS Nano, 2012, 6, 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Daly S, Poussigue F, Simon A-L, MacAleese L, Bertorelle F, Chirot F, Antoine R and Dugourd P, Anal. Chem, 2014, 86, 8798–8804. [DOI] [PubMed] [Google Scholar]
- 163.Charron DM and Zheng G, Nano Today, 2018, 18, 124–136. [Google Scholar]
- 164.Nat. Rev. Mol. Cell Bio, 2002, 3, 906. [DOI] [PubMed] [Google Scholar]
- 165.So M-K, Xu C, Loening AM, Gambhir SS and Rao J, Nat. Biotechnol, 2006, 24, 339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.