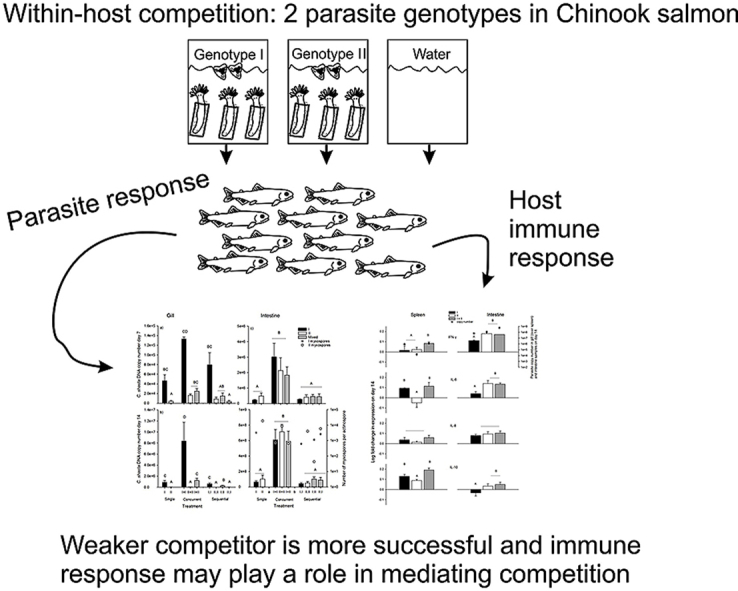
Abstract
Within-host competition can affect outcomes of infections when parasites occupy the same niche. We investigated within-host competition and infection outcomes in Chinook salmon exposed to two genotypes of Ceratonova shasta (myxozoan parasite). We assessed i) virulence (host mortality, median days to death), ii) within-host competition (abundance in host), and iii) success (spore production, proportion of myxospore-producing hosts) following concurrent and sequential exposures to single or mixed-genotype treatments. In single treatments, genotype-I replicated faster, and caused higher and earlier host mortality (higher virulence) but genotype-II produced more myxospores (higher success). In mixed treatments, costs of competition were observed for both genotypes evidenced by reduced replication or myxospore production following concurrent exposures, but only the less-virulent genotype suffered costs of competition when hosts were exposed to genotypes sequentially. To understand potential host effects on competition outcomes, we characterized systemic (spleen) and local (intestine) cytokine and immunoglobulin expression in single and mixed infections. We observed delayed systemic and immunosuppressive responses to the virulent genotype (I), rapid, localized and non-suppressive responses to the less-virulent genotype (II), and a combination of responses to mixed-genotypes. Thus, competition outcomes favoring the virulent genotype may be partially explained by the localized response to genotype-II that facilitates myxospore production (success) offsetting the systemic response to genotype-I that results in early inflammation and immunosuppression (that increases onset of mortality). This evidence for different but simultaneous responses to each genotype suggests selection should favor the exclusion of the weaker competitor and the evolution of increased virulence in the stronger competitor because the outcome was generally more costly for the less-virulent genotype. With caveats, our results are relevant for understanding infection outcomes in commercially and ecologically important salmonids in C. shasta endemic regions where mixed infections are commonplace.
Keywords: Asymmetric competition, Ceratonova shasta, Chinook salmon, Coinfection, Competitive suppression, Superinfection
Graphical abstract
Highlights
-
•
Competition between two genotypes of Ceratonova shasta was asymmetric in Chinook salmon hosts.
-
•
Genotype I was more virulent but genotype-II was more successful (produced more myxospores).
-
•
Costs of competition differed between genotypes, may be mediated by host immune response.
-
•
Host immune response to genotype-I was delayed systemic and immunosuppressive.
-
•
Host immune response to genotype-II was rapid, localized and non-suppressive.
1. Introduction
Infections involving more than one parasite or parasite strains in a single host are common (Read and Taylor, 2001; Pedersen and Fenton, 2007), and interactions between parasites can alter infection and disease dynamics (Cox, 2001; Gower and Webster, 2005; Bell et al., 2006; Mideo, 2009; Telfer et al., 2010). For closely related parasites with overlapping niches, interactions can be costly. For example, competition between strains of Plasmodium chabaudi Landau, Trypanosoma brucei Plimmer and Bradford and Schistosoma mansoni Sambon, altered infection outcomes by reducing within-host replication and mature parasite production (de Roode et al., 2005a, b; Gower and Webster, 2005; Balmer et al., 2009).
Mechanisms (e.g., competition, co-existence) driving the outcomes of parasite interactions can be independent of effects on hosts (e.g. virulence) and understanding the nature of competitive interactions can help predict infection outcomes. When competitors exhibit differences in within-host replication or transmission rates, competition is asymmetric and the outcome should favor the better (faster) competitor irrespective of virulence (May and Nowak, 1995; van Baalen and Sabelis, 1995; Frank, 1996; Mosquera and Adler, 1998). For example, in hosts infected with mixed strains of P. chabaudi and T. brucei, the more virulent strains replicated faster than less virulent strains and were thus the better competitors (de Roode et al., 2005a, b; Balmer et al., 2009). In contrast, in snail (Biomphalaria glabrata Say) hosts infected with multiple S. mansoni strains the less-virulent strain replicated faster than its more-virulent competitor (Gower and Webster, 2005).
The order of infection can also affect outcomes of mixed infections if competitively inferior parasites gain a competitive advantage when they are the first-infecting competitor (Lello, 2012). For example, replication of P. chaubaudi strains was suppressed when they were the second infecting competitor compared with the first, independent of strain virulence (de Roode et al., 2005b). A competitive advantage for the first-infecting strain was also observed in Daphnia magna Straus infected with Pasteuria ramosa Metchnikoff (Ben-Ami et al., 2008) and serotypes of Dengue virus in a mosquito (Pepin et al., 2008). In both of these systems, the first-infecting bacterial strain or viral serotype produced more spores and virus, respectively, and outcompeted the later infecting strain/serotype.
Salmon become infected by a variety of parasites in their natural habitats. We examined within-host competition between two genotypes of the myxozoan parasite Ceratonova shasta Noble, 1950, and outcomes of infections in Chinook salmon (Oncorhynchus tshawytscha Walbaum). Chinook salmon are ecologically and economically important and declines in returning adults have been attributed to C. shasta in at least one major river in the U.S. Pacific Northwest (Fujiwara et al., 2011). Within-host competition likely influences parasite dynamics because C. shasta genotypes coexist, virulence varies among genotypes, and mixed infections are common (Hurst et al., 2014). Chinook salmon may encounter C. shasta strains concurrently or sequentially in situ, thus exposure sequence could be important in predicting the outcome of mixed infections. To examine these interactions and outcomes, we experimentally infected Chinook salmon hosts with two genotypes of C. shasta in single and mixed, concurrent and sequential genotype exposures. We assessed i) host mortality and median days to death as measures of parasite virulence, ii) parasite abundance in host tissues as measures of parasite replication, and iii) production of mature parasite stages (myxospores) in hosts as measures of parasite success.
We expected to observe competition between C. shasta genotypes following concurrent and sequential exposures (Table 1). For concurrent mixed treatments, we hypothesized we would observe i) increased virulence, measured as higher and earlier host mortality, ii) competition due to parasite replication, measured as parasite abundance in host tissue, and iii) reduced parasite success, measured as lower myxospore production overall and lower proportions of myxospore-producing hosts, when compared with single-genotype treatments. We expected the virulent genotype (I) would suppress the less-virulent genotype (II), resulting in relatively higher measures of parasite success for the more virulent genotype compared with the less-virulent genotype (we expected measures of parasite success to be lower in mixed than single-genotype treatments, as stated in ii, above). For sequential-mixed treatments, we hypothesized we would observe i) virulence and ii) abundance similar to those of the first-infecting genotype, and iii) success (myxospore production) equal to single-genotype treatments for the first-infecting genotype but reduced for the second-infecting genotype. We expected the first-infecting genotype would suppress the second-infecting genotype resulting in higher relative measures of parasite success (myxospore production) for the first-infecting genotype.
Table 1.
Hypothesized and observed (in parentheses) responses of competition metrics in concurrent and sequential treatments. Predictions included: H = high, M = moderate, L = low. We hypothesized mixed concurrent treatments would exhibit lower virulence and parasite success, and higher replication (evidence of within host competition) relative to single treatments. We hypothesized sequential treatments would exhibit evidence of prior resident advantage and be similar to corresponding single treatments.
| Genotype/Response (metric) | Concurrent |
Sequential |
|||||
|---|---|---|---|---|---|---|---|
| I | II | Mixed | I | II | Mixed (prior resident = I) I.II | Mixed (prior resident = II) II.I | |
| Host mortality (Virulence) | H (H) | L (M) | M (H) | H (H) | L (M) | H (H) | M (H) |
| Copy number (Replication) | H (H) | L (M) | M (H) | H (H) | L (M) | H (H) | L (H) |
| Myxospore production (Parasite success) | H (L) | M (H) | L (L) | H (L) | M (H) | H (M) | L (H) |
Because the host immune system can play a significant role in infection outcomes by mediating competition between parasites, we characterized the host immune response in a subset of single and mixed-genotype treatments. Chinook salmon respond to genotype-I infection by recruiting Ig + cells to the site of replication and upregulating pro-inflammatory cytokines IFNγ and IL-6, and the regulatory cytokine IL-10 (Bjork et al., 2014) but the immune response to genotype II and mixed infections had not been characterized. We hypothesized that genotype-I would elicit a specific immune response, which would confer an advantage to genotype II in mixed infections (i.e., the non-target strain should evade the host immune response when the response is highly specific, Råberg et al., 2006).
2. Materials and methods
2.1. Host-Parasite system
During the C. shasta life cycle (Bartholomew et al. 1997), the salmon host becomes infected after the waterborne actinospore attaches to gill epithelia and invades the gill blood vessels. The parasite proliferates in the blood and migrates to the intestine, where replication continues, culminating in maturation of the myxospore (Bjork and Bartholomew, 2010). Myxospores are primarily released after host decomposition but low numbers may also be shed in fecal casts by live hosts. The life cycle is completed after myxospores infect polychaete hosts, Manayunkia sp. Leidy, which subsequently release actinospores (Bartholomew et al. 1997).
Genetically variable C. shasta lineages (“genotypes”) co-exist in river basins in the US Pacific Northwest (Atkinson and Bartholomew, 2010a, b; Hallett et al., 2012; Stinson, 2012; Stinson et al, 2018 In Press). Salmonids are infected by all genotypes, but host responses are genotype specific. Parasite-associated mortality in sympatric Chinook salmon is associated with genotype-I infection (Hurst and Bartholomew, 2012) whereas genotype-II infections are non-lethal (Atkinson and Bartholomew, 2010b; Hurst and Bartholomew, 2012). Polychaete cultures infected with genotypes I or II were established in our laboratory making controlled investigation of competition between C. shasta genotypes possible. Despite this advance, experiments on sympatric salmonid strains remain constrained by high infectious dose thresholds and limitations of actinospore production under laboratory conditions. We therefore conducted the current study using an allopatric (not exposed to C. shasta in wild) strain of Chinook salmon (Salmon River Hatchery, Otis, OR, USA), in which dose threshold is lower and parasite associated mortality is higher than in a sympatric strain.
2.1.1. Establishment of infection
All animal challenges, husbandry and sampling were conducted at the John L. Fryer Aquatic Animal Health Laboratory at Oregon State University, Corvallis, Oregon. Fish were exposed to actinospores in single (I.W, W.I, II.W or W.II), concurrent (I + I, I + II, or II + II), and sequential (I.I, I.II, II.I, or II.II) genotype treatments (Fig. 1, Table 2). Treatment groups consisted of 90 allopatric juvenile Chinook salmon (∼10 g), with 30 individuals per replicate (n = 3), with the following exceptions: negative control (W) replicates (n = 2) and concurrent treatment (I.I and II.II) replicates (n = 3) consisted of 25 individuals per replicate because of fish availability. Each treatment received two 24 h static “exposures” with the first on day 0 and the second on day 6. Specific-pathogen-free (SPF) well water (“W”) was used as a negative control and a mock exposure on days 0 or 6 in treatments that received no actinospores on those exposure dates (Fig. 1). The target doses were 5 actinospores fish−1 on a single exposure date for single (positive control) treatments, 10 actinospores fish−1 on a single exposure date for concurrent treatments, and 5 actinospores fish−1 on the first exposure, plus 5 actinospores fish−1 on the second exposure date, for a total target dose of 10 actinospores fish−1 in sequential treatments.
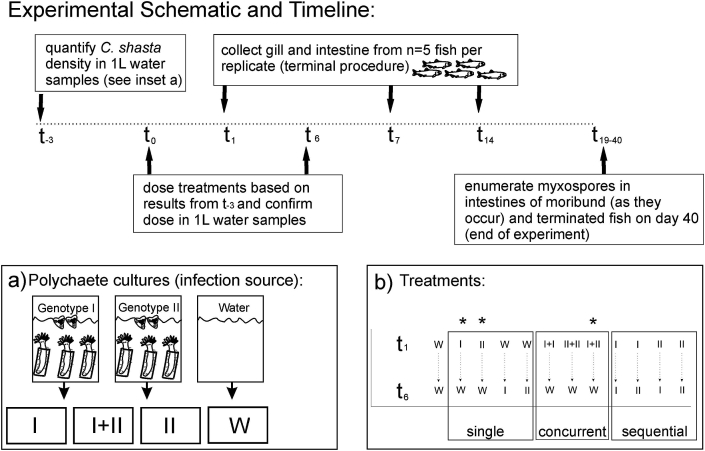
Fig. 1.
Experimental schematic and timeline. Timeline begins at t-3 when density of parasites in polychaete cultures (inset a) was estimated in replicate water samples to calculate dose administered on t0 and t6. Specific-pathogen-free (SPF) well water (“W”) was used as a negative control and a mock exposure t0 and t6 in treatments that received no parasites on those exposure dates “W”- denotes water, “I: denotes genotype-I and “II” denotes genotype-II (inset b). * denote treatments used for cytokine and immunoglobulin assays (b).
Table 2.
Experimental doses and measured response variables for virulence (percent morality and median day to death) and success (myxospores per actinospore and total myxospores per fish). Juvenile Chinook salmon were exposed to genotypes I (“I”) and/or II (“II”), or specific-pathogen free well water (“W”). Exposures were conducted either concurrently on day 0 or sequentially on days 0 and 6. “.” indicates separation between dose dates (e.g., I.W = exposed to genotype-I on day 0, exposed to water on day 6). Values are means ± 1 S.E. Target doses were as follows: 5 actinospores fish−1 administered in a single exposure (positive controls), 10 actinospores fish−1 administered in a single exposure (concurrent treatments), and 10 actinospores fish−1 administered over two exposure periods (5 per exposure, sequential treatments). Actual doses varied from target doses.
| Treatment type | Treatment | Dose per fish | Percent Mortality | Median Day to Death | Myoxspores per actinospore | Total myxospores per fish |
|---|---|---|---|---|---|---|
| Negative controls |
– |
0 |
0 |
N/A |
. |
0 |
| Positive controls (target dose 5 actinospores fish−1) | I.W | 2.9 | 100 ± 0 | 23 ± 0 | 3249 ± 1690 | 5800 ± 402 |
| W.I | 2.2 | 100 ± 0 | 23 ± 0 | 1939 ± 181 | 4264 ± 795 | |
| II.W | 3.0 | 58 ± 6 | 40 ± 1 | 18795 ± 3792 | 59166 ± 16189 | |
| W.II |
5.3 |
56 ± 11 |
43 ± 2 |
23042 ± 20572 |
68705 ± 52379 |
|
| Concurrent (target dose 10 actinospores fish−1; 5 each per genotype, in a single exposure) | I + I.W | 5.6 | 93 ± 7 | 21 ± 0 | 715 ± 105 | 3872 ± 576 |
| I + II.W | 3.6(I), 2.4(II) | 98 ± 2 | 22 ± 0 | 747 ± 398(I), 646 ± 284(II) | 2538 ± 1196(I), 1445 + 597(II) |
|
| II + II.W |
2.9 |
40 ± 15 |
36 ± 5 |
8548 ± 4122 |
20896 ± 7260 |
|
| Sequential (target dose 10 actinospores fish−1; 5 each per genotype, administered over 2 exposures) | I.I | 10.1 (7.3 + 2.8) | 100 ± 0 | 22 ± 1 | 617 ± 191 | 6237 ± 2071 |
| I.II | 4.8(I), 3.7(II) | 98 ± 2 | 23 ± 0 | 1137 ± 275(I), 43 ± 17(II) |
5161 ± 1045(I), 183 ± 84(II) |
|
| II.I | 1.7(I), 4.0(II) | 100 ± 0 | 30 ± 0 | 2620 ± 1816(I), 5859 ± 2544(II) |
2843 ± 1079(I), 17821 ± 10356(II) |
|
| II.II | 7.0 (2.3 + 4.7) | 40 ± 4 | 40 ± 1 | 4024 ± 2952 | 31579 ± 23953 |
Positive controls included single-genotype treatments that were exposed to actinospores on day 0, and SPF water on day 6 (I.W and II.W) and that were exposed to SPF water on day 0 and actinospores on day 6, (W.I and W.II) to control for potential differences in actinospore dose and viability between exposure days. Concurrent single-genotype exposures (I + I, II + II) were conducted on day 0 to control for total actinospore dose (a target of 10 parasites fish−1) in the mixed concurrent treatment (a target of 5 actinospores fish−1 of each genotype, 10 total). Exposures to the same genotype sequentially (I.I and II.II) were conducted to serve as controls for mixed-sequential treatments.
2.1.2. Parasite dose
Experimental doses were obtained from infected laboratory polychaete cultures producing genotype I or II actinospores. Density of actinospores in polychaete cultures was estimated prior to the experiment by filtering 3 replicate 1 L water samples through a 5 μm nitrocellulose membrane and measuring the quantity of actinospores [number of actinospores in each sample by a C. shasta-specific qPCR assay (Hallett and Bartholomew, 2006)]. Based on the density of actinospores in each culture, we calculated the volume of polychaete culture water required to deliver the target dose of 5 actinospores per fish for 1x treatments (e.g. single and sequential) and 10 actinospores per fish for 2x treatments (concurrent) (Fig. 1, Table 2). Target doses were mixed into buckets and were standardized to 23 L (polychaete culture water + well water) prior to administering to replicates. Impellers were used to mix the water in the fish tanks during exposures, and all fish within a replicate were assumed to experience equal exposure (same age and stock). Because actinospore densities in polychate cultures vary daily but can't be measured in real-time, actual experimental doses were measured in 1 L water samples collected from each treatment replicate at the time of dosing and quantified, as above. Following exposure, replicates were maintained on specific pathogen-free well water at 18 °C and fed a commercial fish diet daily (Bio-Oregon, Longview, WA, USA). Salmon were monitored daily for clinical disease signs and euthanized if moribund.
2.2. Virulence metrics
We calculated percent mortality and median days to death (MDD) for each replicate as measures of parasite virulence. Fish that exhibited terminal disease signs were euthanized and that date was recorded and used for MDD calculations. The MDD was adjusted for delayed single-genotype exposures (W.I and W.II) because parasite exposure first occurred on day 6 (i.e. if MDD for W.I was 56, adjusted MDD would be 50). Percent mortality was calculated for each replicate at the end of the experiment as total number of euthanized or dead fish [“morts”/(total number of fish – fish sampled on days 1, 7 and 14) x 100].
2.3. Competition metrics
Five salmon from each replicate (except W.I and W.II treatments) were sampled on days 1, 7 and 14, for a total of 15 fish per replicate for parasite abundance assays. Fish were euthanized using an overdose of tricaine methanesulfonate (MS-222, Argent Chemical Laboratories, Redmond, WA, USA), and the left gill (days 1, 7 and 14) and 0.25 mg of the posterior intestine (days 7 and 14) were collected and stored in individual tubes at −20 °C until processing. Intestine samples were not collected on day 1 because parasites are not detected in intestinal tissue early in the infection (Bjork and Bartholomew, 2010). To estimate parasite abundance, we measured the quantity of C. shasta DNA in gill and intestine by quantitative polymerase chain reaction (qPCR). DNA was extracted, purified (Hurst et al., 2014) and assayed (18 S, Hallett and Bartholomew, 2006). Samples were run in triplicate and tested for inhibition (IPC); samples were diluted and re-assayed if inhibited (Hallett and Bartholomew, 2009). A standard curve was generated for estimating parasite DNA copy number in 0.1 g of host gill and intestinal tissue by assaying ten-fold serial dilutions of a synthetic parasite template (Hallett and Bartholomew, 2006) spiked with intestinal or gill tissue. A standard curve for either gill (y = −3.35x + 38.40; r2 = 0.997; Hurst et al., 2014) or intestine (y = −3.54x + 40.08; r2 = 0.999) was then used to calculate parasite copy number in the sample. To determine genotype proportion for each of the above samples, DNA (ITS1) was amplified by PCR and 3 μL from each fish was sequenced and analyzed as in Hurst et al. (2014).
2.4. Parasite success metrics
We calculated estimates of myxospore production as measures of parasite success including i) total number of myxospores produced per actinospore (“myxospore production”), and ii) proportion of myxospore-producing hosts. To count myxospores, intestines were excised and 1 mL of tap water was used to flush mature parasites (myxospores) from the tissue into a microcentrifuge tube. Myxospores in 3 1 μL aliquots were counted on a hemocytometer and counts were extrapolated to determine total myxospores produced per fish (mean count/hemocytometer square x 10ˆ4) and 20 μL aliquots were retained for genotyping. For single-genotype treatments, myxospores from 10 randomly selected fish were assayed to confirm genotype. For mixed-genotype treatments, all fish were assayed and genotype proportion was calculated, as above. All fish were examined for myxospores but sampled fish (days 1, 7, 14) were excluded from myxospore production calculations because they were terminated before myxospore production occurs. Myxospore production (number of myxospores produced for each actinospore) was calculated as [mean total myxospores per fish/genotype specific actinospore dose per fish]. Proportion of myoxpore-producing hosts was calculated as [total number of myxospore-producing fish that died or were terminated at the end of the experiment/(total number of fish – fish sampled on days 1, 7 and 14)].
2.5. Host responses to single and mixed infections
Samples for measuring cytokine and immunoglobulin expression were collected from a subset of treatments including genotype-I (I.W), genotype-II (II.W), genotypes I and II (I + II.W) and an uninfected control (W.W) for a total of 330 samples. A 25 mg portion of the intestine (the parasite's target tissue, used to measure the local response) and spleen (used to measure the systemic response) were collected, preserved in RNALater (Qiagen) and frozen at −80 °C. RNA was extracted from tissues using the Roche High Resolution RNA Tissue Kit and was then converted to cDNA using the Roche Transcriptor First Strand cDNA Synthesis Kit with oligo DT primers and amplified using SYBRgreen. Samples were analyzed on an Applied Biosystems Step One Plus Real-time PCR system and cycle threshold determined using instrument provided software.
We assayed inflammatory (TNFα, IL-1β, IFNγ), and regulatory cytokines (TGFβ, IL-10), IL-6 and IL-8 (both inflammatory and regulatory) as well as β-actin as a reference and immunoglobulins IgT and IgM. Cytokines were selected based on Bjork et al. (2014), IgM based on Schouten et al. (2013) and IgT based on Dolan et al. (2016) for characterizing the early immune response of Chinook salmon to C. shasta genotypes and identifying potential differences between mixed and single-genotype infections.
2.6. Analyses
We designed a fully factorial experiment (as 1x, 2x doses, Fig. 1) for analysis by ANOVA but dosing challenges resulted in non-standardized dose applications so we instead used general linear models and included genotype-specific dose in the model. We tested for fixed effects of genotype (3 levels: I, II, mixed), interactions between genotype and treatment (4 levels: Single, Concurrent, and 2 types of sequential; QI - genotype-I exposure first, QII- genotype-II exposure first), and dose on (i) virulence (mortality, median days to death), (ii) competition (parasite abundance on days 1, 7 and 14 in gill tissue, days 7, 14 in intestinal tissue), and (iii) success (myxospore production and proportion of myxospore-producing hosts) responses. We used type III sums of squares obtained using the GLM procedure in SAS (version 9.4; SAS Institute, Cary, North Carolina) and significant results were analyzed by Tukey's Honestly-Significant Difference tests (Tukey's HSD) for pairwise comparisons. We excluded genotype-specific dose as a fixed factor in myxospore production models because this metric was used to calculate the response. In order to balance the design we excluded delayed single-genotype treatments (W.I and W.II) from the analyses after detecting no significant differences in dose or response metrics when compared with I.W or II.W.
Cytokine and immunoglobulin expression were measured by qPCR, as above. The log fold change in expression was normalized to controls (the mean of 2 technical replicates from uninfected fish samples was divided by the mean of 2 technical replicates from each experimental fish sample) and differences among treatments and days were examined by two-way ANOVAs (day, treatment, and treatment*day). Values were considered significantly different from controls at p < 0.008 (to account for multiple comparisons- 2 tissues, 3 cytokine types-regulators, dual or inflammatory).
3. Results
3.1. Parasite dose
We aimed for target doses of approximately 5 (1x) and 10 (2x) spores per fish, but actual parasite doses were highly variable and ranged from 1 to 8 (I) and 1–7 (II) parasites per fish on day 0 and from 1 to 4 (I) and 2–7 (II) on day 6 resulting in doses of 2–3 (single treatments), 3–6 (concurrent) and 6–10 (sequential) parasites per fish (Table 2). Sequencing results confirmed the appropriate genotypes had been administered to each replicate. No parasites were detected in samples from “W” treatments from either exposure.
3.2. Virulence
Genotype-I was more virulent than genotype-II and outcomes of mixed infections for fish were most similar to genotype-I infections, independent of whether they were single or mixed, concurrent or sequential. Mortality was higher in mixed and genotype-I only treatments than in genotype-II only treatments (genotype effect: F2 = 44.2, p < 0.0001), ranging from 98 to 100% in genotype-I only and most mixed-genotype treatments and from 37 to 58% in genotype-II only treatments (Table 2, interactions between genotype and treatment, genotype specific dose not significant,p values > 0.05).
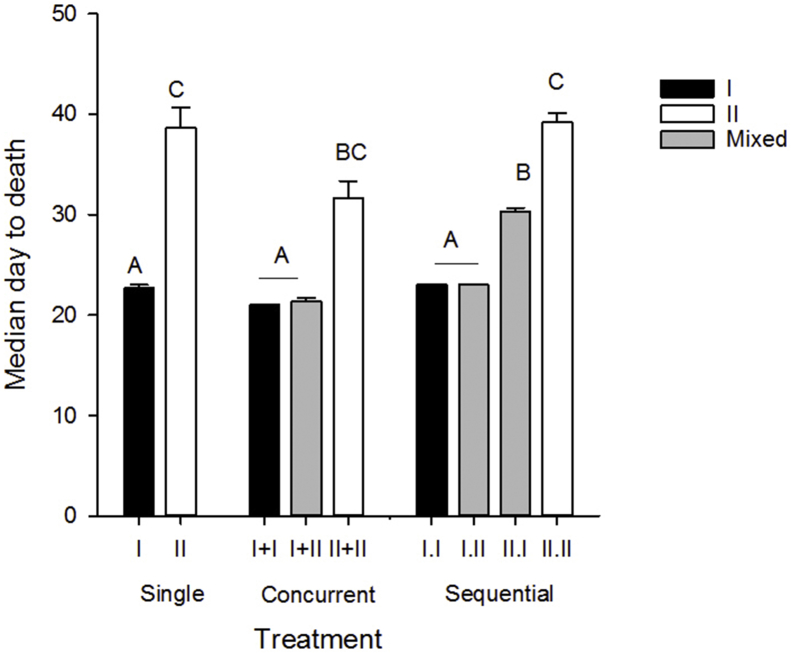
Median day to death (MDD) ranged from 21 to 40 days post exposure, and like mortality, variation in MDD was explained by parasite genotype (F2 = 31.8, p < 0.0001). However, interactions between genotype and treatment were significant (F6 = 11.1, p < 0.0001) and explained by the effects of delayed exposure to genotype-I in sequential treatments II.I. The MDD was shortest in genotype-I only (I.W, I + I, I.I), mixed concurrent I + II and sequential I.II treatments, and was intermediate in sequential II.I treatments, reflecting the delayed exposure to Genotype-I. MDD was also intermediate in II + II treatments and latest in genotype-II only (II.W, II.II) treatments (Fig. 2, Table 2). Surprisingly, effects of genotype specific doses were not significant (p values > 0.05).
Fig. 2.
Median day to death, as a measure of parasite virulence, in treatment groups. Black bars denote genotype-I only, white denote genotype-II only, and grey denote mixed-genotype treatments. Letters indicate treatments that differed (Tukey's HSD tests, α = 0.05).
3.3. Competition
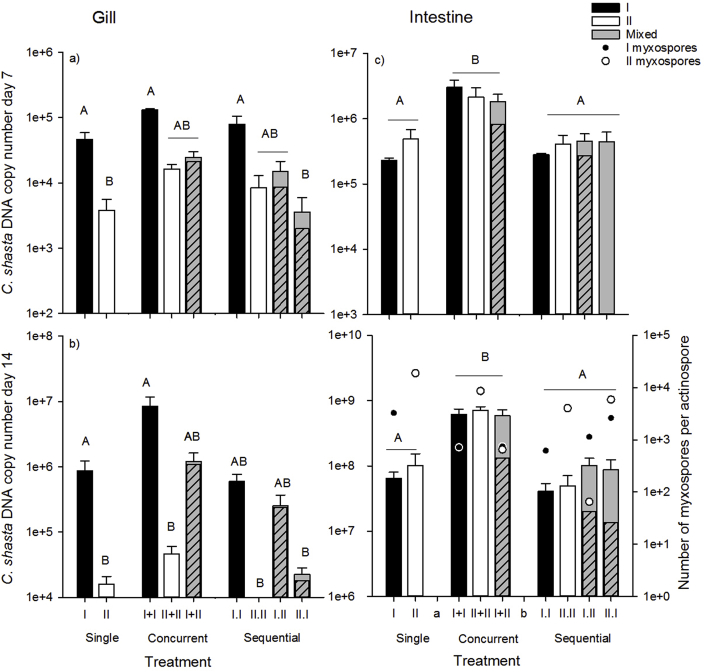
Parasite copy numbers increased over time in gill and intestine samples indicating parasite replication was occurring (Table 3, Supporting information, Fig. S1a), but we observed different patterns in the tissues. In general, parasite copy numbers in gill samples were lower and primarily comprised of genotype-I DNA, whereas copy numbers were higher in intestine samples, and the proportion of genotypes was highly variable (Fig. 3). In gill tissue, genotype-I only treatment copy numbers were high compared to genotype-II only and mixed treatment copy numbers (genotype effect) on days 7 (F2 = 4.11, p = 0.036), and 14 (F2 = 6.8, p = 0.007), and in mixed treatments, genotype I comprised the majority of the parasite DNA (Fig 3ab, not significant on day 1). Samples collected on day 1 (t1) from gill tissue may have been collected too early in the infection because only II.W differed (was lower) from other treatments (treatment x genotype F6 = 4.25, p = 0.009, effect of dose not significant). Samples collected from gill tissue on days 7 and 14 are suggestive that competition was occurring in mixed treatments, with both genotypes incurring costs. Total copy numbers were highest in genotype-I only treatments and similar in mixed and genotype-II only treatments in gill tissue samples collected on day 7 (t7) (Fig. 3a, Table 3, treatment x genotype F6 = 2.99, p = 0.0374, genotype specific dose effects not significant). Although parasite copy numbers in mixed-genotype treatments were only marginally lower than their single counterparts, genotype-I DNA comprised the majority of the mixed treatment samples, suggesting genotype-II replication was much reduced in mixed-treatments. Trends in parasite copy number on day 14 (t14) in gill tissue samples were similar to day 7 (Fig. 3b, Table 3 treatment x genotype F6 = 12.95, p < 0.0001, genotype specific dose effects not significant), again demonstrating genotype-I DNA comprised the majority of the sample, and genotype-II replication was significantly reduced. Interestingly, among the mixed-genotype treatments parasite copy numbers were high in the (concurrent) I + II, intermediate in the I.II (sequential), and lower in the II.I (sequential) treatments, suggesting that the outcome differed depending upon when a competitor is introduced.
Table 3.
Abundance estimates (parasite copy number), in gill and intestinal tissues including the proportion of genotype I in mixed treatments shown in parentheses (proportion of genotype II = 1-proportion of genotype I), a measure of competition (mixed treatments only).
|
Treatment type |
Gills |
Intestine |
||||
|---|---|---|---|---|---|---|
| Treatment | Day 1 | Day7 | Day 14 | Day7 | Day 14 | |
| Negative controls |
W.W |
0 |
0 |
0 |
0 |
0 |
| Positive controls | I.W | 1.6 E+03 | 4.7 E+04 | 8.7 E+05 | 2.3 E+05 | 6.4E+07 |
| W.I | . | . | . | . | . | |
| II.W | 5.1 E+02 | 3.8 E+03 | 1.6 E+04 | 4.9 E+05 | 1E+08 | |
| W.II |
. |
. |
. |
. |
. |
|
| Concurrent | I + I.W | 6.6 E+03 | 1.3 E+05 | 8.4 E+06 | 3.0 E+06 | 6.1E+08 |
| I + II.W | 2.9 E+03 (0.93) | 2.5 E+04 (0.86) | 1.2 E+06 (0.91) | 1.8 E+06 (0.45) | 5.9E+08 (0.66) | |
| II + II.W |
2.6 E+03 |
1.6 E+04 |
4.6 E+04 |
2.1 E+06 |
7.1E+08 |
|
| Sequential | I.I | 3.0 E+03 | 7.9 E+04 | 6.0 E+05 | 2.8 E+05 | 4.1E+07 |
| I.II | 2.3 E+03 (1.00) | 1.5 E+04 (0.58) | 2.5 E+05 (0.95) | 4.6 E+05 (0.59) | 1E+08 (0.94) | |
| II.I | 1.3 E+03 (0.00) | 3.6 E+03 (0.56) | 2.3 E+04 (0.80) | 4.4 E+05 (0.00) | 8.8E+07 (0.81) | |
| II.II | 1.7 E+03 | 8.5 E+03 | 7.9 E+03 | 4.1 E+05 | 5E+07 | |
Fig. 3.
Parasite copy number, as a measure of parasite competition in mixed-genotype treatments, in a) gill tissue sampled at 7d (t7), b) gill tissue sampled at 14d (t14) c) intestinal tissue sampled at 7d, and d) intestinal tissue sampled at 14d. Black bars denote genotype-I only, white denote genotype-II only, and grey denote mixed-genotype treatments. Inset striped grey bars represent total genotype I copy numbers, based on the proportion of genotype I in sequenced DNA samples (genotype II comprises the remainder-the solid grey bar). Letters indicate treatments that differed (Tukey's HSD tests, α = 0.05). Total number of genotype-I (black circles) and genotype-II (white circles) myxospores produced per actinospore, as a measure of parasite success in fish overlaid on parasite copy number in intestinal tissue sampled at 14d.
As in gill tissue, C. shasta parasite copy number increased over time in intestinal tissue (Table 3, Fig. S1b), and we again observed evidence of competition between the genotypes, with genotype-I comprising the majority of parasite DNA in mixed infections. In contrast to patterns observed in gills, on day 7 parasite abundance in intestinal tissue was high in concurrent treatments regardless of genotype treatment (F6 = 8.99, p = 0.0002, effects of genotype and genotype specific dose were not significant), and low in single and sequential treatments (Fig. 3c). Interestingly, the proportion of genotype-II was higher in this tissue, though typically still lower than the proportion of genotype-I. However, all II.I treatment samples were comprised entirely of genotype-II DNA, which was interesting because genotype-II DNA comprised a low proportion of the DNA in the sequential treatment in which it was the second-infecting genotype (I.II). We observed similar trends in intestinal tissue on day 14, (treatment x genotype F6 = 8.3, p = 0.003, Fig. 3d), but with genotype-II comprising lower proportions of the DNA than genotype-I in all mixed-genotype treatments.
3.4. Parasite success
Measures of parasite success including myxospore production and proportion of myxospore-producing hosts differed between genotypes and provided evidence of competition. In general, fewer myxospores per actinospore were produced in fish from genotype-I only and mixed-genotype treatments than genotype-II only treatments except with genotype-II was the first infecting genotype in sequential treatments (Fig. 3, Fig. 4a,b). The number of genotype-I myxospores produced per genotype-I actinospore did not differ between mixed (747–2620 myxospores per actinospore) and single (617–3249) genotype treatments (treatment x genotype and dose effects >0.05, Fig. 4a). In contrast, number of genotype-II myxospores per actinospore varied by almost 3 orders of magnitude among treatments (Table 2). The number of myxospores produced per actinospore were approximately 2–4x higher in II.W (18,795–23,042) than in the II + II, II.II, and II.I treatments, and 30x (I + II) to almost 500x (I.II) higher than in the remaining mixed treatments (genotype F1 = 31.4, p = 0.0001, treatment x genotype F4 = 9.7, p = 0.001 Fig. 4b).
Fig. 4.
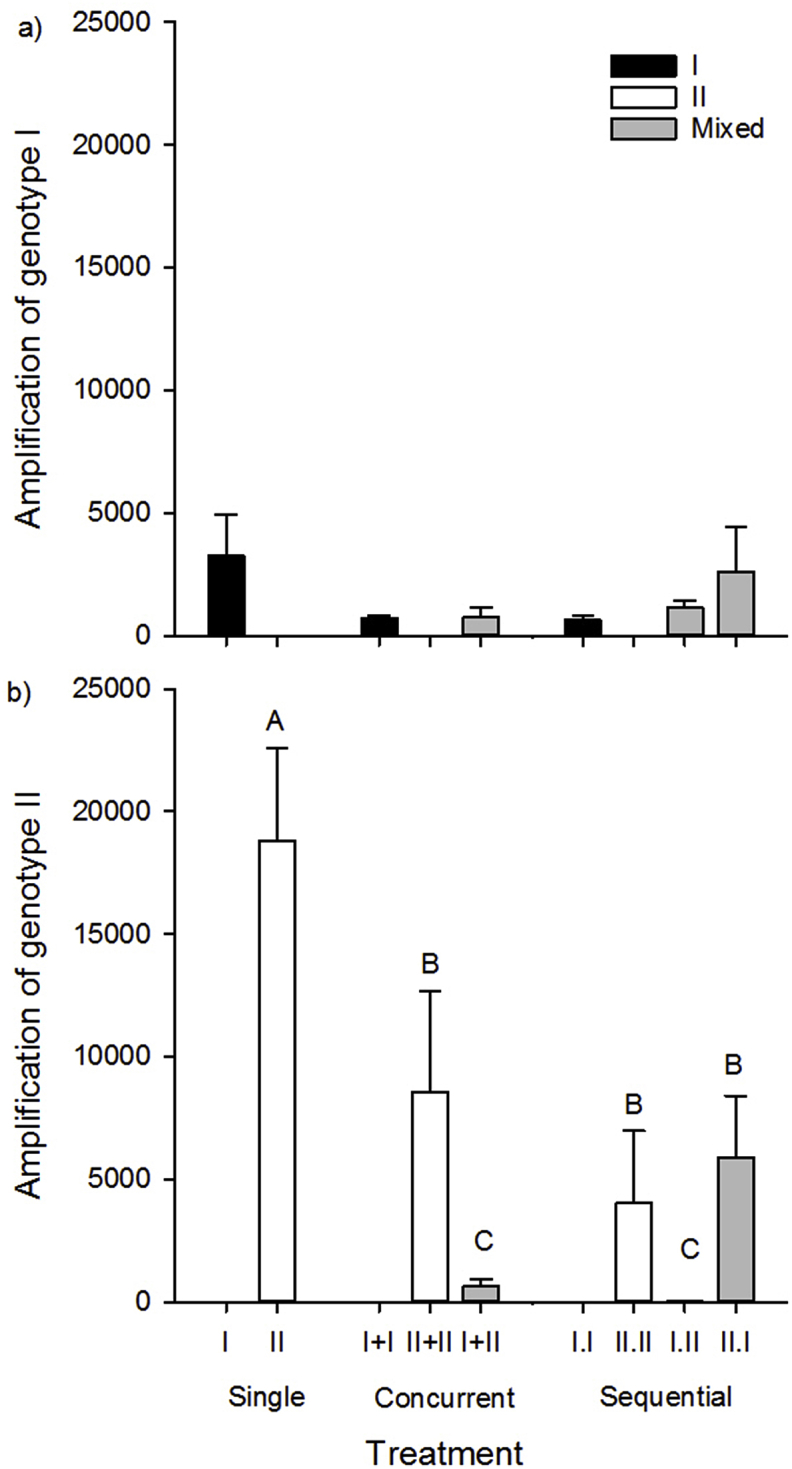
Total number of a) genotype-I and b) genotype-II myxospores produced per actinospore, as a measure of parasite success, in fish from single and mixed-genotype treatments. Black bars denote genotype-I only, white denote genotype-II only, and grey denote mixed-genotype treatments. Letters indicate treatments that differed (Tukey's HSD tests, α = 0.05).
The proportion of myxospore-producing hosts was lower in genotype-II only (0.30–0.44) treatments than in mixed (0.76–0.87) and genotype-I only (0.80–0.87) treatments (genotype effect: F2,16 = 16.0, p = 0.002, Fig. S4). Interactions between genotype and treatment, or effects of genotype dose were not significant (p values > 0.05). Despite the lower proportion of myxospore-producing hosts, genotype-II only treatments still produced more myxospores overall [myxospores/fish*(mortalities-(myxospore-producinghosts)), mean 197,580 myxospores of genotype-II versus 60,295 genotype-I].
3.5. Host responses to single and mixed infections
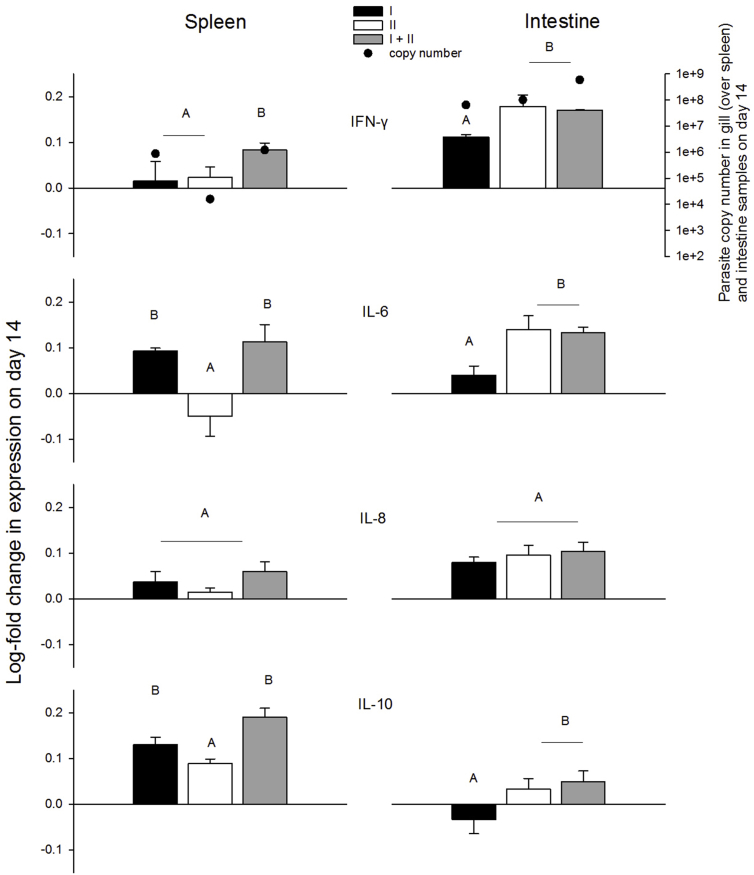
The expression of pro-inflammatory cytokines relative to controls differed among tissues and treatments (Fig. 5, Table 4). Several cytokines, such as TNF-α, TGF-β, and IL-1β showed no remarkable differences among treatments, nor changes over the course of the infection. IFN-γ was generally upregulated in spleen samples, and was higher in mixed compared with single-genotype treatments on all sampling dates. However, in intestine samples, IFN-γ was initially downregulated in all treatments, then increased over time with upregulation being significantly higher on day 14 in genotype-II and mixed treatments than in genotype-I treatments. The expression of IL-6 and IL-8 cytokines differed between tissues and genotypes. In the spleen, IL-6 was upregulated in genotype-I and mixed-genotype treatments (day 14), but not in genotype-II treatments. In contrast, in intestine samples, IL-6 was upregulated (day 14) in genotype-II and mixed treatments. In the spleen, IL-8 did not differ among treatments or days (F8,17 = 1.7, p = 0.17). In intestine samples, IL-8 also did not differ among treatments, however it was upregulated in samples collected on day 14 (day: F2 = 110.7, p < 0.001).
Fig. 5.
Log-foldchange in cytokine expression relative to controls at day-14 in spleen and intestine samples. Parasite copy numbers measured in gill tissues is overlaid on IFN-gamma spleen plot and parasite copy numbers measured in intestine samples are overlaid on IFN-gamma intestine plot, but parasite copy number data are the same for, and apply to, all cytokine plots underneath. Letters denote treatments that differed (Tukey's HDS tests, α = 0.05).
Table 4.
Cytokine and immunoglobulin expression normalized to uninfected controls (+1. S.E.). Results are shown as log-fold-change by cytokine type and tissue (S = spleen, I = intestine).
| Type | Gene | Tissue | Day 1 |
Day 7 |
Day 14 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | |||||||||||
| I | II | Mixed | I | II | Mixed | I | II | Mixed | |||
| Pro-inflammatory | TNF-a | S | 0.02 (±0.01) | 0.01 (±0) | −0.02 (±0.01) | 0 (±0.03) | 0.05 (±0.02) | 0.03 (±0.01) | −0.01 (±0.04) | −0.02 (±0.02) | 0.02 (±0.01) |
| I | −0.02 (±0.01) | −0.01 (±0.02) | 0.05 (±0.02) | 0.02 (±0.02) | −0.03 (±0.01) | −0.03 (±0.01) | 0.01 (±0) | −0.01 (±0.01) | 0 (±0.02) | ||
| IL-1B | S | 0.06 (±0.03) | 0.07 (±0.02) | 0 (±0.03) | 0.03 (±0.02) | 0.07 (±0.02) | 0.08 (±0.02) | 0 (±0.02) | 0.02 (±0) | 0.03 (±0) | |
| I | −0.06 (±0.01) | −0.06 (±0.01) | −0.04 (±0) | −0.03 (±0) | −0.02 (±0) | 0 (±0.01) | −0.03 (±0) | −0.03 (±0) | −0.02 (±0) | ||
| IFN-g | S | 0.03 (±0.01) | 0.04 (±0.01) | 0.03 (±0.02) | 0.02 (±0.02) | 0.02 (±0.01) | 0.08 (±0.01) | 0.02 (±0.02) | 0.02 (±0.01) | 0.08 (±0.01) | |
| I |
−0.02 (±0.02) |
−0.05 (±0.03) |
−0.04 (±0.01) |
−0.02 (±0.01) |
0.02 (±0.01) |
0.03 (±0.01) |
0.11 (±0) |
0.18 (±0.01) |
0.17 (±0) |
||
| Inflammatory and regulatory | IL-6 | S | −0.01 (±0.02) | −0.01 (±0.02) | −0.04 (±0.02) | −0.02 (±0.02) | 0.01 (±0.01) | 0.06 (±0.02) | 0.09 (±0) | −0.05 (±0.03) | 0.1 (±0.03) |
| I | −0.01 (±0.01) | 0 (±0.02) | 0.06 (±0.02) | 0.03 (±0.01) | −0.01 (±0.01) | 0 (±0.01) | 0.04 (±0.01) | 0.14 (±0.02) | 0.13 (±0.01) | ||
| IL-8 | S | 0.03 (±0.01) | 0.01 (±0.01) | 0.01 (±0.01) | 0.03 (±0.03) | 0.06 (±0.02) | 0.04 (±0.01) | 0.01 (±0.01) | 0.01 (±0.01) | 0.05 (±0) | |
| I |
0.01 (±0) |
−0.03 (±0.01) |
−0.01 (±0.01) |
−0.01 (±0.01) |
0.01 (±0) |
0.01 (±0.01) |
0.08 (±0.01) |
0.1 (±0.01) |
0.09 (±0) |
||
| Regulatory | IL-10 | S | 0.04 (±0.02) | 0.05 (±0.03) | 0 (±0.02) | 0.01 (±0.03) | 0.02 (±0.02) | 0.03 (±0.02) | 0.13 (±0.01) | 0.09 (±0.01) | 0.18 (±0) |
| I | −0.02 (±0.01) | −0.03 (±0.01) | −0.02 (±0.01) | −0.07 (±0.02) | −0.07 (±0.01) | 0.01 (±0.01) | −0.03 (±0.02) | 0.03 (±0.01) | 0.04 (±0) | ||
| TgF B | S | 0.08 (±0.04) | 0.1 (±0.01) | 0.08 (±0) | 0.01 (±0.02) | 0.03 (±0.01) | 0.01 (±0.01) | 0.02 (±0.01) | 0 (±0.01) | 0.03 (±0.01) | |
| I |
−0.01 (+0.01) |
0 (+0.01) |
−0.01 (+0.01) |
0.03 (+0.01) |
0.05 (+0) |
0.05 (+0.01) |
0.04 (+0) |
0.06 (+0.01) |
0.06 (+0.03) |
||
| Reference | Actin | S | 0.08 (+0.02) | 0.11 (+0) | 0.04 (+0.03) | 0.01 (+0.03) | 0.02 (+0.01) | 0.03 (+0) | 0.02 (+0.02) | 0.03 (+0.01) | 0.05 (+0.01) |
| I | −0.01 (+0.01) | 0.02 (+0.01) | 0 (+0) | 0.03 (+0.01) | 0.03 (+0.02) | 0.03 (+0) | −0.01 (+0) | 0.02 (+0.01) | 0.05 (+0) | ||
We observed significant differences in gene expression for regulatory cytokines among treatments. In the spleen, IL-10 was upregulated over time and highest on day 14 in genotype-I and mixed-genotype treatments (no difference in genotype-II treatments over time) and differences among treatments were detected on day 14. IL-10 upregulation increased over time in intestine samples (day: F2 = 110.7, p < 0.001) and was highest on day 14 in genotype-II and mixed-genotype treatments (treatment*day F4 = 3.85, p = 0.02).
Immunoglobulin production of both IgM and IgT was relatively unchanged in the spleen during the course of the infection, but differences in immunoglobulin production were observed in the intestine depending on C. shasta genotype (Fig. 6, Table 4). Overall, fish infected with either genotype-II or a mixture of genotypes had increased IgM and IgT heavy chain transcripts compared to fish infected with genotype-I: IgM was higher in infected fish on day 14 samples from mixed and genotype-II treatments than genotype I treatments (genotype: F2 = 110.7, p 0.005). IgT was downregulated for all treatments at days 1 and 7, and for genotype-I treatments on day 14, but was upregulated in genotype-II and mixed-genotype treatments (not significant, day: p = 0.02 and genotype: p = 0.04).
Fig. 6.
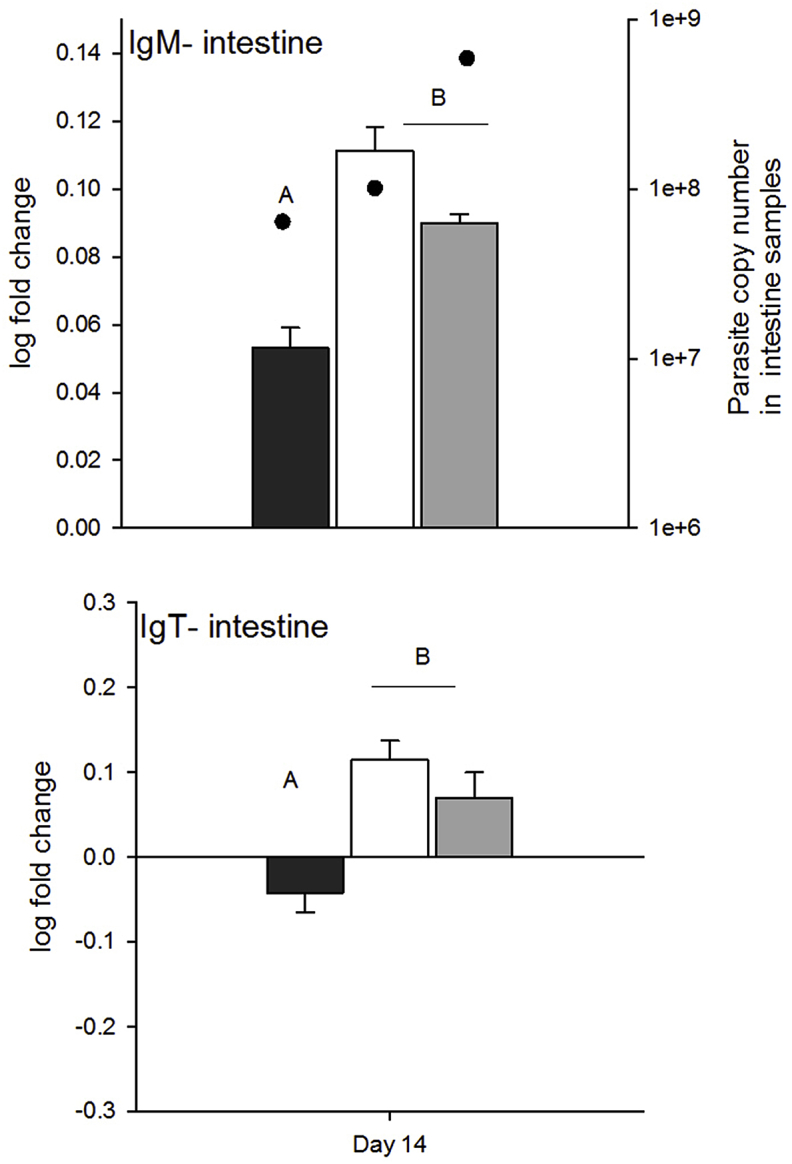
Log fold change in immunogloblulin expression relative to controls on day 14 in intestine samples. Parasite copy numbers measured in intestine samples are overlaid on IgM intestine plot (parasite copy number data are the same for the IgT plot). Letters denote treatments that differed (Tukey's HSD tests, α = 0.05).
4. Discussion
4.1. Does within-host competition occur?
We observed evidence of asymmetric competition between C. shasta genotypes in Chinook salmon. Although both genotypes incurred costs of competition, the less-virulent genotype (II) suffered disproportionately higher costs because replication and myxospore production (the ultimate measure of parasite success) were reduced in mixed-genotype treatments. Despite this asymmetry, the less-virulent genotype (II) was actually a superior competitor because it produced more myxospores overall in mixed treatments despite having lower virulence, slower replication rates, and a higher proportion of non-myxospore-producing hosts.
Knowledge of mechanisms driving competitive interactions is critical for understanding the progression and outcomes of infections. We examined a variety of metrics to test for evidence of competition through different mechanisms. We expected virulence measures would be highest in mixed-genotype treatments, because we assumed the host would incur additive costs from infection by multiple genotypes. However, percent mortality and median day-to-death were similar in mixed-genotype treatments to those of the virulent genotype (I; high percentage and early) treatments, suggesting that competition between the genotypes was not (additionally) harming, nor benefitting the host (Table 1).
We expected competition measures (replication) would demonstrate evidence of competitive suppression through reduced parasite DNA copy numbers (Table 1). We observed significant differences (in replication) between the genotypes, which may partially explain differences in virulence between the genotypes (see below), in addition to reduced DNA copy numbers in mixed-genotype treatments compared to the single-genotype treatments in both tissue types (Tables 1 and 3). In gill tissue, genotype specific parasite DNA copy numbers were lower in mixed than single-genotype treatments, which suggests both genotypes suffered from a reduction in proliferation when exposed along with a competitor. However, genotype-II appeared to suffer disproportionately high costs of competition in gill tissue samples; frequently making up less than 0.2 of the total DNA. This trend was also observed in intestinal tissue samples, but the proportion of genotype-II was higher than in the gill samples.
The high DNA copy numbers of genotype-I in mixed and single-genotype treatments demonstrates genotype-I replication occurred quickly and may explain the early onset of mortality of all hosts infected with genotype-I in single or mixed treatments. The elevated cytokine expression responses measured in spleen samples suggest that high parasite copy number (e.g. I and I + II treatments) results in increased systemic inflammation, which may lead to damaging immune-induced pathology (e.g., Skugor et al., 2008). Thus, the early and rapid parasite replication by genotype-I could be stressful for the fish host, resulting in the high and rapid mortality associated with this genotype.
The production of genotype specific myxospores provides the most compelling evidence of competition between genotypes. As transmission stages, myxospores represent the best measure of parasite success that we can quantify in this type of experiment. Compared with their single counterparts, production of genotype-II myxospores was reduced in mixed-genotype treatments. In contrast, production of genotype-I myxospores did not differ between mixed or single-genotype treatements. We had expected to observe high copy numbers (replication signal) in intestine samples of fish from high myxospore producing treatments (genotype-II only, particularly II.W, see Table 2), but myxospore production and parasite abundance (copy number) in intestinal tissue samples were not correlated. We observed the highest parasite copy numbers in intestine samples from concurrent treatments, (intermediate; e.g., II + II, to low myxospore producers e.g., I + I and I + II). One explanation for this result is that competition occurring between the proliferative stages earlier in the infection (i.e., chemical war between genotypes, Massey et al., 2004) may negatively impact genotype-II myxospore production, particularly when the genotype-I dose was administered first (e.g. I.II) or was higher (e.g. I + II). Another possibility is that host mortality occurred prior to peak myxospore formation in treatments involving genotype-I. For example, if mortality in fish from I, I + I and I + II treatments had occurred later (as in genotype II only treatments), more myxospores could have been produced by treatments having high parasite copy numbers.
We have suggested above that i) genotype-II replicates more slowly than genotype-I, and follow this up with the conjecture that ii) in a highly susceptible host genotype-I causes mortality too rapidly for maximum parasite proliferation (myxospore production) to occur. Consequently, there's no loss to myxospore production for genotype-I in mixed infections, but genotype-II suffers from a loss in potential. We argue that maximum parasite proliferation (myxospore production) was achieved by genotype-II only in single-genotype treatments, and that the introduction of a competitor (genotype-I) resulted in decreased production unless genotype-II was the first-infecting parasite. This suggests that genotype-II suffers a disproportionately high cost of competition, rather than being a weaker competitor than genotype-I.
Costs of competition can occur through direct interference, resource limitation, or as a result of the host immune response (Read and Taylor, 2001, Råberg et al., 2006). We suggest the rapid host death caused by the virulent genotype (I) causes resource limitation for genotype-II because the hosts die from infection prior to achieving maximum myxospore production. Resource limitation (e.g. available red blood cells) also been proposed as a mechanism regulating within-host competition in a malarial system (Bell et al., 2006). Although genotype-I may trade off myxospore production with early mortality, it still comes out ahead because high proportions of its hosts produce myxospores (greater than 80%) compared with genotype-II (approximately 30–40%). This tradeoff may tip the balance of success to favor a genotype that consistently kills its hosts before it reaches full transmission potential and explain why genotype-I success (myxospore production) was similar across mixed and single-genotype treatments.
In natural system, such a tradeoff would have implications for persistence and the evolution of virulence. In our experimental conditions, genotype-I myxospores comprised only 2% of the entire “population” produced, which would drive selection quickly through direct impact on the next parasite generation, assuming variability in genotype distributions (majority of genotype-II myxospores were produced by single-genotype treatments). However, these results should be cautiously interpreted for wild salmonids and C. shasta because we used Chinook that are not exposed to C. shasta under natural conditions (“allopatric”), and outcomes of infections in strains of Chinook that have co-evolved with the parasite (“sympatric”) will differ. However, although the percent mortality response was high compared to what would be observed in sympatric Chinook exposed to similar doses of genotype-I, MDD was similar.
The host immune responses may also have implications for genotype persistence and virulence. The different responses could determine infection outcomes because they are elicited simultaneously. Cytokine and antibody expression suggest hosts mount i) a partially effective immune response to genotype-II resulting in a reduction in genotype-II systemically, effectively confining the damage to the intestine (allowing the host to live longer), and ii) an immunosuppressive response to genotype-I resulting in systemic proliferation, fast replication and high virulence. These responses may drive selection for the different traits expressed by each genotype (early systemic replication versus late and localized replication). Thus, these results may help explain differences in outcomes of Chinook infected with C. shasta genotypes-I and II, although we acknowledge that the immune responses to C. shasta genotypes may differ between allopatric and sympatric hosts.
4.2. Competition costs and benefits
Faster parasite replication rates frequently correlate with selection for higher virulence, as the parasite uses host resources more and more quickly (May and Nowak, 1995; van Baalen and Sabelis, 1995; Frank, 1996; Mosquera and Adler, 1998). However, the production of C. shasta transmission stages (myxospores) is negatively impacted when host death occurs too rapidly. Consequently, the more virulent genotype (I) initially appeared to be a better competitor because of the higher virulence and replication, yet it fell short of its less-virulent competitor when considered in terms of transmission potential (myxospore production), which is arguably, a better measure of success. In single-genotype infections, genotype-II produced five times more myxospores per actinospore and an order of magnitude more myxospores overall despite causing lower host mortality (survivors did not produce myxospores). In concurrent mixed-genotype treatment groups, we argue that the costs of competition were higher for genotype-II because genotype-I replicated faster and the host died from the infection prior to reach the peak period for genotype-II myxospore production. In contrast, genotype-I produced similar (if not higher) numbers of myxospores per actinospore in mixed and single-genotype treatments, suggesting competition had low to no effect on the outcome of infection.
4.3. Prior residency can reduce competitive suppression
The costs of competition for both genotypes were reduced when introduced first. When genotype-II was introduced first, infected fish produced similar numbers of genotype-II myxospores as fish exposed to genotype-II only, despite experiencing earlier and higher percent mortality. One mechanism to explain this result is that genotype-II responded in a facultative manner (e.g. Read and Taylor, 2001) to genotype-I. Gower and Webster (2005) also observed increased replication by a low virulence strain of S. mansoni in response to a high virulence strain. Genotype-I also appeared to benefit from this sequential encounter, producing the most myxospores of any mixed treatment. Perhaps the offset in the timing of this encounter (less-virulent followed by more virulent) of genotypes altered the trajectories of host immune response andresulted in less inflammation or immunosuppression, and facilitated myxospore production (cytokine and Ig profiles not measured in sequential treatments).
4.4. Host immune response influences infection outcome and implications for genotype dominance
In general, we observed systemic immune responses to genotype I infections that could be detrimental to the host, and localized (specific to the site of infection) responses to genotype-II infections that should be more detrimental to the parasite than to the host (Cox, 2001, Dickerson and Findly, 2014). Genotype-I infections were characterized by increased immune gene expression in the spleen, indicative of a systemic response to infection. Levels of the pro-inflammatory cytokine IL-6 and the anti-inflammatory cytokine IL-10 were elevated following infection, which may be indicative of a systemic inflammatory response, that could trigger a negative feedback mechanism to control runaway inflammation (Cox, 2001). Genotype-II infections were characterized increased immune gene expression in the gut (intestine), indicative of a localized immune response to infection. Levels of IL6, IFNγ, and IL10 were elevated compared to those measured in the genotype-I infected fish. Mixed-genotype cytokine profiles resembled that of genotype-I in the spleen, and that of genotype-II in the intestine. We also observed an increase in the production of antibodies (both IgM and IgT) at the site of infection (intestine) in genotype-II and mixed-genotype infected fish, but not in genotype-I infected fish.
Therefore, the host response to genotype-I is primarily systemic, which could be detrimental to the host and cause early mortality, while the host response to genotype-II is more specific to the site of infection, and may localize damage from myxospore production. During coinfection, we suspect these dual and conflicting responses could reduce the efficacy of the overall immune response, particularly against the virulent genotype (e.g., Dolan et al., 2016). Thus, the dual and conflicting immune responses to mixed infections could play arole in maintaining lower virulence (at least for genotype-I).
4.5. Competition influences infection outcome and genotype dominance
Outcomes of interactions between parasite genotypes can include i) coinfection, where both genotypes coexist within the host, ii) superinfection, where one genotype excludes the other and iii) single infection, where a host becomes immune to infection by other genotypes (Nowak and May 1994; May and Nowak, 1995; Mosquera and Adler, 1998). The production of both genotype-I and II myxospores following sequential and concurrent mixed infections demonstrates that coinfection occurred in our experimental system. Coinfection could drive selection for higher virulence in both genotypes over time (e.g. Mosquera and Adler, 1998). However, increased virulence in the form of more rapid mortality could have important fitness consequences for genotype-I if it already falls within the left tail of the myxospore production distribution, unless myxospore production timing also shifts earlier.
Although ours is not the first study to demonstrate within-host competition between parasite genotypes (de Roode et al., 2005a, b; Gower and Webster, 2005; Balmer et al., 2009), it is the first to do so in a vertebrate host from an outbred population. Consequently, our results may be relevant for heterogeneous populations. Host susceptibility is correlated with different outcomes in other experimental systems (e.g., coinfection in susceptible hosts, superinfection in resistant hosts, de Roode et al., 2004), thus the heterogeneity in susceptibility to C. shasta expressed among salmonid life stages and strains in natural systems could also result in variable outcomes following mixed infections. Juvenile sympatric Chinook salmon are highly resistant to C. shasta (Bartholomew, 1998), unlike the allopatric Chinook used in our study. Despite this resistance, in the Klamath River over 80% of field exposed juvenile Chinook salmon had mixed infections with parasite genotypes I and II (Hurst et al., 2014). However, those that subsequently became diseased produced only genotype-I myxospores (Atkinson and Bartholomew, 2010a, b; Hallett et al., 2012; Hurst et al., 2012, 2014). These data suggest genotype-I excludes genotype-II (superinfection) as the likely outcome of mixed infections in (sympatric) resistant juvenile Chinook. However, our results (coinfection) may be relevant for our understanding of disease dynamics in immunocompromised adult salmon (Robertson et al., 1961) or in naïve salmonids as the parasite's range expands.
Conflicts of interest
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome. We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us. We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property. We further confirm that any aspect of the work covered in this manuscript that has involved either experimental animals or human patients has been conducted with the ethical approval of all relevant bodies and that such approvals are acknowledged within the manuscript. We understand that the Corresponding Author is the sole contact for the Editorial process (including Editorial Manager and direct communications with the office). She is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs. We confirm that we have provided a current, correct email address which is accessible by the Corresponding Author (Alexander) and which has been configured to accept email from alexanju@science.oregonstate.edu.
Data accessibility
Data will be made available from the Dryad Digital Repository if published.
Acknowledgements
We thank the Salmon River Hatchery (Otis, OR) for providing Chinook salmon for the study. People that assisted with experimental set-up and sampling include: Rebecca Cull-Peterson, Ruth Milston-Clemens, Adam Ray, Peter Wong and Ryan Craig. Michelle Jordan. Nicholas Som and Adam Ray provided much appreciated statistical advice. We appreciate reviews of this manuscript by Laura Taggart-Murphy, Gema Alama Bermejo, Rhea Hanselmann and Anna Jolles prior to submission. We sincerely thank the journal's anonymous reviewers; their thoughtful input greatly improved the manuscript. Funding for this study was provided by The Flyfisher’s Club of Oregon, Oregon State University Department of Microbiology Tartar Award, William Q. Wick Marine Fisheries Award, and the National Oceanic and Atmospheric Administration's Graduate Sciences Program.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2019.03.008.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplemental figure S1.
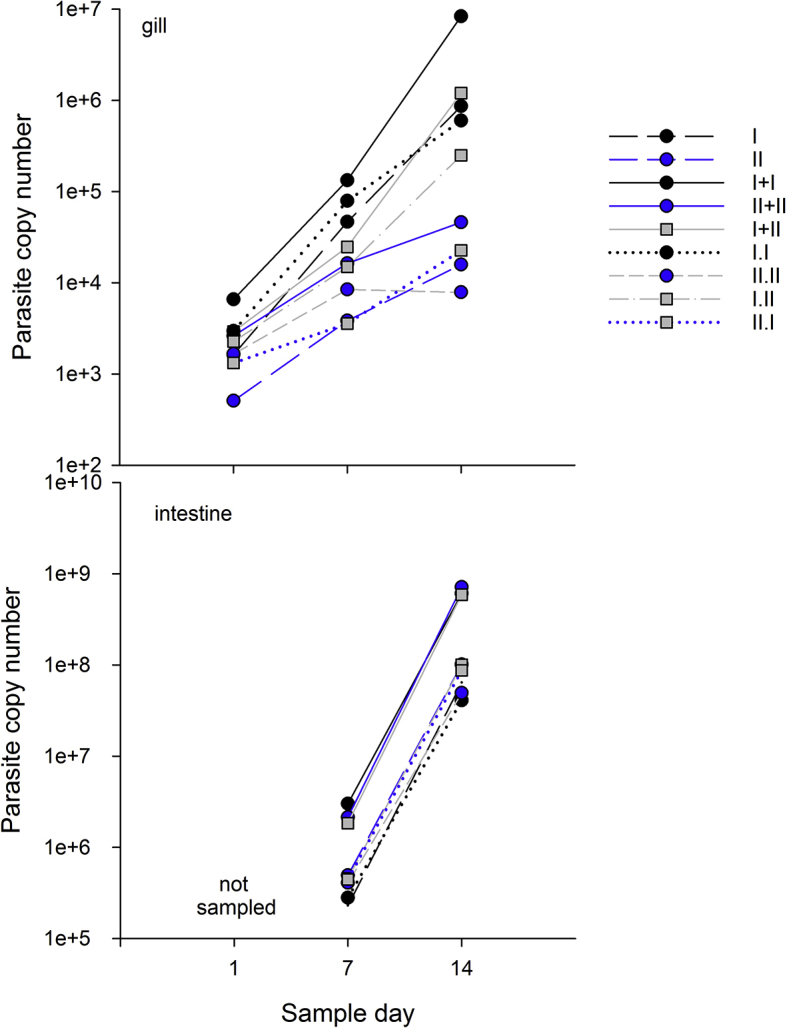
Total Ceratonova shasta DNA copy number in treatments in a) gill tissue and b) intestine tissue.
Supplemental figure S2.
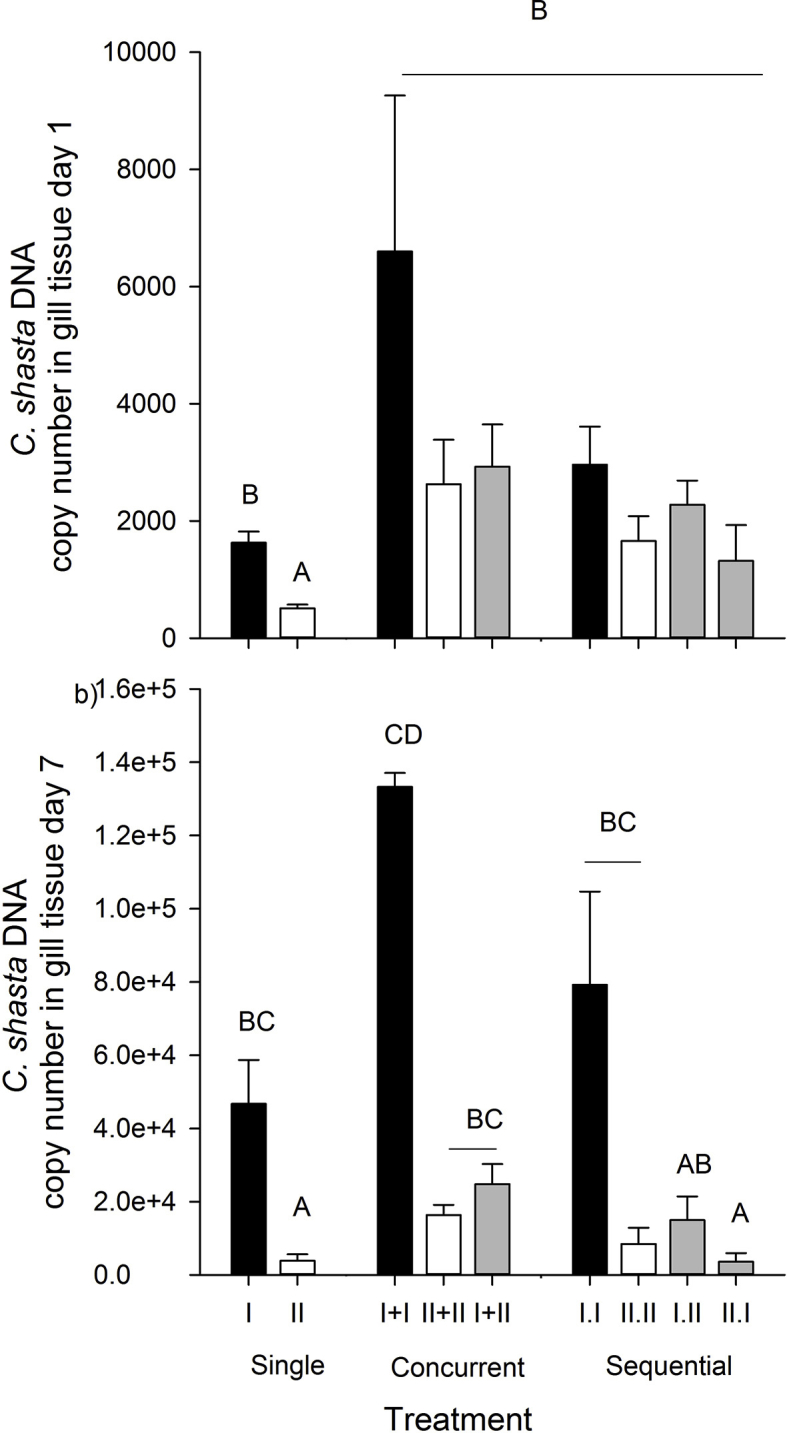
Total Ceratonova shasta DNA copy number in gill tissue sampled at 1d (t7) post exposure. Black bars denote genotype-I only, white denote genotype-II only, and grey denote mixed-genotype treatments. Letters indicate treatments that differed (Tukey's HSD tests, α = 0.05).
Supplemental figure S3.
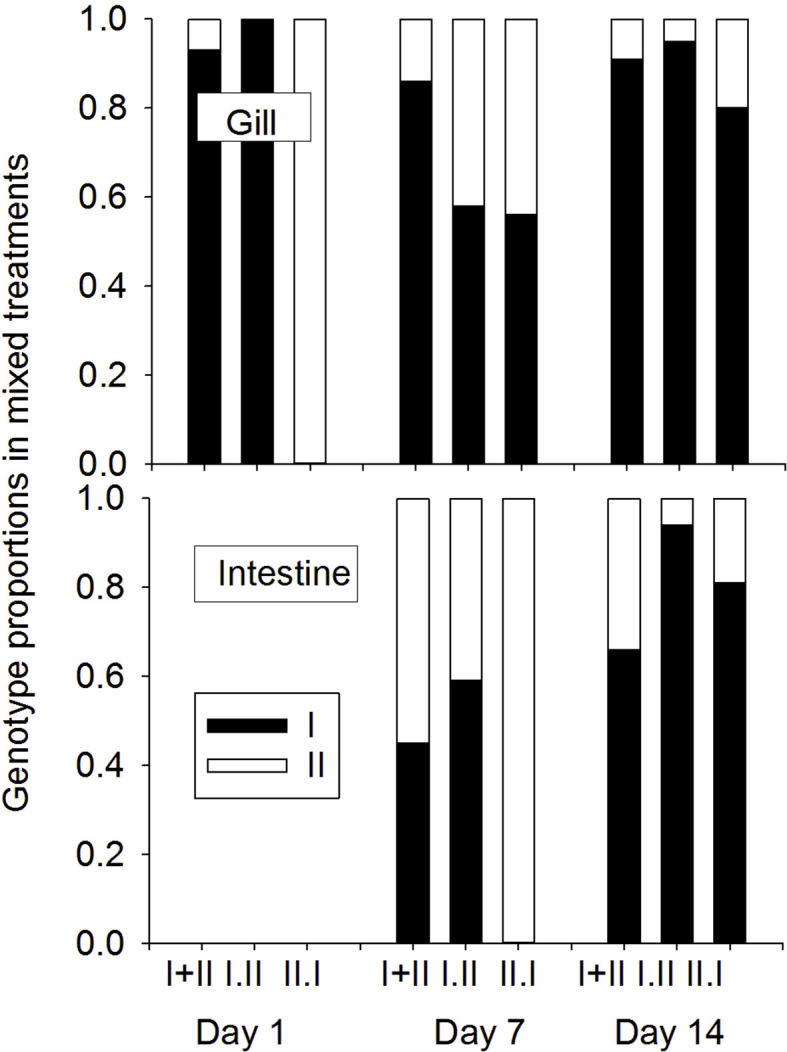
Proportions of genotype I and II DNA in tissue samples used to determine parasite copy number, as a measure of competition in mixed-genotype treatments, in gill tissue (top) sampled at 1, 7 and 14 days, and in intestine tissue (bottom) sampled at 7, and 14 days (not on day 1).
Black bars denote genotype-I, white denote genotype-I, and data are shown only for mixed-genotype treatments.
Supplemental figure S4.
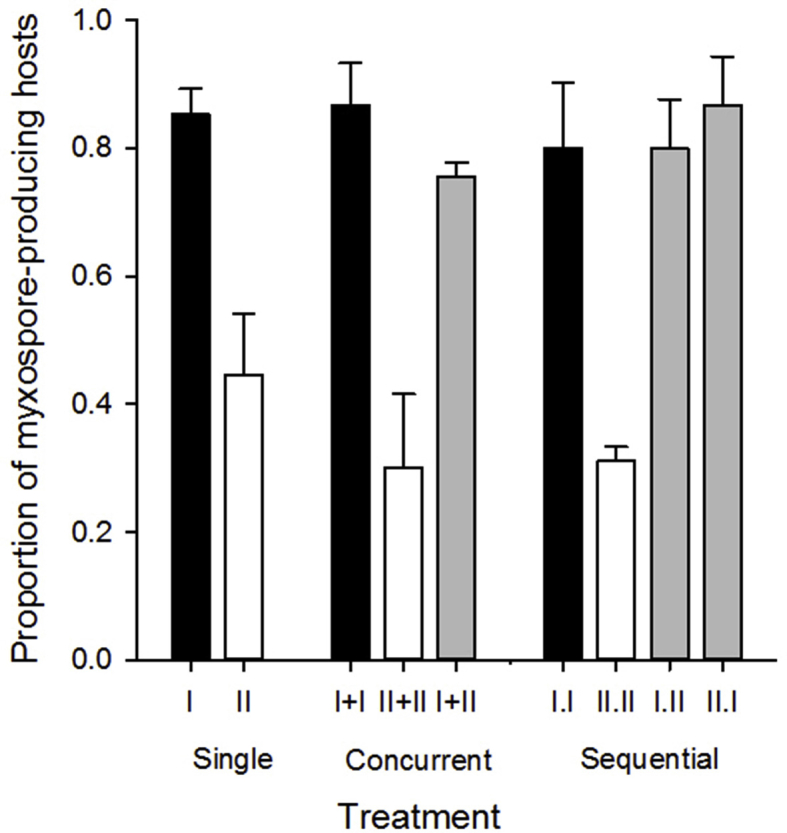
Proportion of myxospore producing hosts, fish in which myxospores were produced/observed. Letters indicate treatments that differed (Tukey's HSD tests, α = 0.05).
References
- Atkinson S.D., Bartholomew J.L. Disparate infection patterns of Ceratomyxa shasta (Myxozoa) in rainbow trout Oncorhynchus mykiss and Chinook salmon Oncorhynchus tshawytscha correlate with internal transcribed spacer-1 sequence variation in the parasite. Int. J. Parasitol. 2010;40:599–604. doi: 10.1016/j.ijpara.2009.10.010. [DOI] [PubMed] [Google Scholar]
- Atkinson S.D., Bartholomew J.L. Spatial, temporal and host factors structure the Ceratomyxa shasta (Myxozoa) population in the Klamath River basin. Infect. Genet. Evol. 2010;10:1019–1026. doi: 10.1016/j.meegid.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Balmer O., Stearns S.C., Schötzau A., Brun R. Intraspecific competition between co-infecting parasite strains enhances host survival in African trypanosomes. Ecology. 2009;90:3367–3378. doi: 10.1890/08-2291.1. [DOI] [PubMed] [Google Scholar]
- Bartholomew J.L., Whipple M.J., Stevens D.G., Fryer J.L. The life cycle of Ceratomyxa shasta, a myxosporean parasite of salmonids, requires a freshwater polychaete as an alternate host. J. Parasitol. 1997;83:859–868. [PubMed] [Google Scholar]
- Bartholomew J.L. Host resistance to infection by the myxosporean parasite Ceratomyxa shasta: a review. J. Aquat. Anim. Health. 1998;10:112–120. [Google Scholar]
- Bell A.S., de Roode J.C., Sim D., Read A.F. Within‐host competition in genetically diverse malaria infections: parasite virulence and competitive success. Evolution. 2006;60:1358–1371. [PubMed] [Google Scholar]
- Ben‐Ami F., Mouton L., Ebert D. The effects of multiple infections on the expression and evolution of virulence in a Daphnia‐endoparasite system. Evolution. 2008;62:1700–1711. doi: 10.1111/j.1558-5646.2008.00391.x. [DOI] [PubMed] [Google Scholar]
- Bjork S.J., Bartholomew J.L. Invasion of Ceratomyxa shasta (Myxozoa) and comparison of migration to the intestine between susceptible and resistant fish hosts. Int. J. Parasitol. 2010;40:1087–1095. doi: 10.1016/j.ijpara.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Bjork S.J., Zhang Y.A., Hurst C.N., Alonso-Naveiro M.E., Alexander J.D., Sunyer J.O., Bartholomew J.L. Defenses of susceptible and resistant Chinook salmon (Oncorhynchus tshawytscha) against the myxozoan parasite Ceratomyxa shasta. Fish Shellfish Immunol. 2014;37:87–95. doi: 10.1016/j.fsi.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox F.E.G. Concomitant infections, parasites and immune responses. Parasitology-Cambridge. 2001;122:S23–S38. doi: 10.1017/s003118200001698x. [DOI] [PubMed] [Google Scholar]
- de Roode J.C., Culleton R., Cheesman S.J., Carter R., Read A.F. Host heterogeneity is a determinant of competitive exclusion or coexistence in genetically diverse malaria infections. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2004;271:1073–1080. doi: 10.1098/rspb.2004.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roode J.C., Pansini R., Cheesman S.J., Helinski M.E., Huijben S., Wargo A.R., Bell A.S., Chan B.H.K., Walliker D., Read A.F. Virulence and competitive ability in genetically diverse malaria infections. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7624–7628. doi: 10.1073/pnas.0500078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roode J.C., Helinski M.E., Anwar M.A., Read A.F. Dynamics of multiple infection and within‐host competition in genetically diverse malaria infections. Am. Nat. 2005;166:531–542. doi: 10.1086/491659. [DOI] [PubMed] [Google Scholar]
- Dolan B.P., Fisher K.M., Colvin M.E., Benda S.E., Peterson J.T., Kent M.L., Schreck C.B. Innate and adaptive immune responses in migrating spring-run adult chinook salmon, Oncorhynchus tshawytscha. Fish Shellfish Immunol. 2016;48:136–144. doi: 10.1016/j.fsi.2015.11.015. [DOI] [PubMed] [Google Scholar]
- Dickerson H.W., Findly R.C. Immunity to Ichthyophthirius infections in fish: A synopsis. Dev. Comp. Immunol. 2014;43:290–299. doi: 10.1016/j.dci.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Frank S.A. Models of parasite virulence. QRB (Q. Rev. Biol.) 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- Fujiwara M., Mohr M.S., Greenberg A., Foott J.S., Bartholomew J.L. Effects of ceratomyxosis on population dynamics of Klamath fall-run Chinook salmon. Trans. Am. Fish. Soc. 2011;140:1380–1391. [Google Scholar]
- Gower C.M., Webster J.P. Intraspecific competition and the evolution of virulence in a parasitic trematode. Evolution. 2005;59:544–553. [PubMed] [Google Scholar]
- Hallett S.L., Bartholomew J.L. Application of a real-time PCR assay to detect and quantify the myxozoan parasite Ceratomyxa shasta in river water samples. Dis. Aquat. Org. 2006;71:109–118. doi: 10.3354/dao071109. [DOI] [PubMed] [Google Scholar]
- Hallett S.L., Bartholomew J.L. Development and application of a duplex QPCR for river water samples to monitor the myxozoan parasite Parvicapsula minibicornis. Dis. Aquat. Org. 2009;86:39–50. doi: 10.3354/dao02104. [DOI] [PubMed] [Google Scholar]
- Hallett S.L., Ray R.A., Hurst C.N., Holt R.A., Buckles G.R., Atkinson S.D., Bartholomew J.L. Density of the waterborne parasite Ceratomyxa shasta and its biological effects on salmon. Appl. Environ. Microbiol. 2012;78:3724–3731. doi: 10.1128/AEM.07801-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst C.N., Bartholomew J.L. Ceratomyxa shasta genotypes cause differential mortality in their salmonid hosts. J. Fish Dis. 2012;35:725–732. doi: 10.1111/j.1365-2761.2012.01407.x. [DOI] [PubMed] [Google Scholar]
- Hurst C.N., Holt R.A., Bartholomew J.L. Dam removal and implications for fish health: Ceratomyxa shasta in the Williamson River, Oregon, USA. N. Am. J. Fish. Manag. 2012;32:14–23. [Google Scholar]
- Hurst C.N., Wong P., Hallett S.L., Ray R.A., Bartholomew J.L. Transmission and persistence of Ceratomyxa shasta genotypes in Chinook salmon. J. Parasitol. 2014;100:773–777. doi: 10.1645/13-482.1. [DOI] [PubMed] [Google Scholar]
- Lello J. Co-infection: immunological consequences. In: Lamb T.J., editor. Immunity to Parasitic Infection. John Wiley Sons; West Sussex, UK: 2012. pp. 325–333. [Google Scholar]
- Massey R.C., Buckling A., ffrench-Constant R. Interference competition and parasite virulence. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2004;271:785–788. doi: 10.1098/rspb.2004.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May R.M., Nowak M.A. Coinfection and the evolution of parasite virulence. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1995;261:209–215. doi: 10.1098/rspb.1995.0138. [DOI] [PubMed] [Google Scholar]
- Mideo N. Parasite adaptations to within-host competition. Trends Parasitol. 2009;25:261–268. doi: 10.1016/j.pt.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Mosquera J., Adler F.R. Evolution of virulence: a unified framework for coinfection and superinfection. J. Theor. Biol. 1998;195:293–313. doi: 10.1006/jtbi.1998.0793. [DOI] [PubMed] [Google Scholar]
- Nowak M.A., May R.M. Superinfection and the evolution of parasite virulence. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1994;255:81–89. doi: 10.1098/rspb.1994.0012. [DOI] [PubMed] [Google Scholar]
- Pedersen A.B., Fenton A. Emphasizing the ecology in parasite community ecology. Trends Ecol. Evol. 2007;22:133–139. doi: 10.1016/j.tree.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Pepin K.M., Lambeth K., Hanley K.A. Asymmetric competitive suppression between strains of dengue virus. BMC Microbiol. 2008;8 doi: 10.1186/1471-2180-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Råberg L., de Roode J.C., Bell A.S., Stamou P., Gray D., Read A.F. The role of immune‐mediated apparent competition in genetically diverse malaria infections. Am. Nat. 2006;168:41–53. doi: 10.1086/505160. [DOI] [PubMed] [Google Scholar]
- Read A.F., Taylor L.H. The ecology of genetically diverse infections. Science. 2001;292:1099–1102. doi: 10.1126/science.1059410. [DOI] [PubMed] [Google Scholar]
- Robertson O.H., Krupp M.A., Favour C.B., Hane S., Thomas S.F. Physiological changes occurring in the blood of the Pacific salmon (Oncorhynchus tshawytscha) accompanying sexual maturation and spawning. Endocrinology. 1961;68:733–746. doi: 10.1210/endo-68-5-733. [DOI] [PubMed] [Google Scholar]
- Schouten J., Clister T., Bruce A., Epp L., Zwollo P. Sockeye salmon retain immunoglobulin-secreting plasma cells throughout their spawning journey and post-spawning. Dev. Comp. Immunol. 2013;40:202–209. doi: 10.1016/j.dci.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skugor S., Glover K.A., Nilsen F., Krasnov A. Local and systemic gene expression responses of Atlantic salmon (Salmo salar) to infection with the salmon louse (Lepeophtheirus salmonis) BMC Genomics. 2008;9:498. doi: 10.1186/1471-2164-9-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson M.E.T. Oregon State University; Corvallis, Oregon: 2012. Re-examining Ceratomyxa shasta in the Pacific Northwest; p. 100.http://hdl.handle.net/1957/28348 M. S. Thesis. [Google Scholar]
- Stinson M.E.T., Atkinson S.D., Bartholomew J.L. Widespread distribution of Ceratonova shasta (Cnidaria: myxosporea) genotypes indicates evolutionary adaptation to its salmonid fish hosts. J. Parasitol. 2018;104:645–650. doi: 10.1645/18-79. [DOI] [PubMed] [Google Scholar]
- Telfer S., Lambin X., Birtles R., Beldomenico P., Burthe S., Paterson S., Begon M. Species interactions in a parasite community drive infection risk in a wildlife population. Science. 2010;330:243–246. doi: 10.1126/science.1190333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baalen M., Sabelis M.W. The dynamics of multiple infection and the evolution of virulence. Am. Nat. 1995;146:881–910. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available from the Dryad Digital Repository if published.