Abstract
Although the pathogenesis of Parkinson’s disease (PD) remains unclear, mutations in leucine-rich repeat kinase 2 (Lrrk2) are among the major causes of familial PD. Most of these mutations disrupt Lrrk2 kinase and/or GTPase domain function, resulting in neuronal degeneration. However, the signal pathways underlying Lrrk2-induced neuronal degeneration are not fully understood. There is an expanding body of evidence that suggests a link between Lrrk2 function and MAP kinase (MAPK) cascades. To further investigate this link in vivo, genetic RNAi screens of the MAPK pathways were performed in a Drosophila model to identify genetic modifier(s) that can suppress G2019S-Lrrk2-induced PD-like phenotypes. The results revealed that the knockdown of hemipterous (hep, or JNKK) increased fly survival time, improved locomotor function and reduced loss of dopaminergic neurons in G2019S-Lrrk2 transgenic flies. Expression of the dominant-negative allele of JNK (JNK-DN), a kinase that is downstream of hep in G2019S-Lrrk2 transgenic flies, elicited a similar effect. Moreover, treatment with the JNK inhibitor SP600125 partially reversed the G2019S-Lrrk2-induced loss of dopaminergic neurons. These results indicate that the hep pathway plays an important role in Lrrk2-linked Parkinsonism in flies. These studies provide new insights into the molecular mechanisms underlying Lrrk2-linked PD pathogenesis and aid in identifying potential therapeutic targets.
Keywords: Lrrk2, Parkinson’s disease, JNK, neuronal degeneration, hep, dopamine neuron, Drosophila model
INTRODUCTION
Parkinson’s disease (PD) is one of the most common neurodegenerative disorders and is characterized by the selective loss of dopaminergic neurons and the presence of Lewy bodies. Clinical features of PD include tremors, bradykinesia/akinesia, rigidity, and postural instability resulting from the loss of dopamine neurons in the substantia nigra(Dawson and Dawson 2003). The pathogenesis of PD remains incompletely understood. Lrrk2 mutations contribute to 5–13% of familial PD cases and 1–5% of sporadic PD cases and have become the most common known cause of PD(Kumari and Tan. 2009; Li et al. 2011).
The Lrrk2 protein (2527 aa, ~280 kDa) contains several domains including a leucine-rich repeat (Lrr) domain, a Ras of complex proteins (ROC) domain followed by its associated C terminal of ROC (COR) domain, a kinase domain, and a C-terminal WD40 domain. Lrrk2 is expressed in neurons, microglia, astrocytes and some immune cells (Galter et al. 2006) and can be detected in Lewy bodies(Zhu et al. 2006). The biological functions of LRRK2 is incompletely understood but it have been involved in multiple cellular pathways including neuronal outgrowth and guidance(MacLeod et al. 2006; Cookson 2010), cytoskeletal organization, mitochondrial dynamics, endocytosis, trafficking, autophagy, and protein interactions (Arranz et al. 2015; Esteves and Cardoso 2016; Roosen and Cookson 2016). Lrrk2 possesses GTPase and serine/threonine protein kinase activities (West et al. 2005; Smith et al. 2006; West et al. 2007; Li et al. 2014). Lrrk2 can undergo autophosphorylation and phosphorylate the generic substrate, myelin basic protein (MBP) (Smith et al. 2005; West et al. 2005). Increasing studies suggest the various candidates for Lrrk2 substrates including 4E-BP, Rab family proteins, s15, synaptojanin, EndoA, ezrin, radixin, and moesin, etc.(Parisiadou et al. 2009; Kumar et al. 2010; Beilina et al. 2014; Martin et al. 2014; Arranz et al. 2015; Soukup et al. 2016). However, these candidates still await further verification whether or not they are substrates of Lrrk2.
The G2019S mutation is the most common mutation in Lrrk2 and has been reported to increase its kinase activity (Smith et al. 2005; West et al. 2005; MacLeod et al. 2006). In cellular and animal models, overexpression of disease-causing mutations in Lrrk2 increases its kinase activity and induces neuronal degeneration, while inhibition of Lrrk2 kinase activity by genetic alteration or pharmacological inhibitors, reduce neuronal degeneration(Smith et al. 2005; Zhu et al. 2006; Lee et al. 2010; Liu et al. 2011; Li et al. 2014; Li et al. 2015; Atashrazm and Dzamko 2016; Thomas et al. 2016; Kang and Marto 2017), which suggests that Lrrk2 kinase activity plays a critical role in PD pathogenesis. However, the Lrrk2 kinase signaling cascades underlying neuronal degeneration are unclear.
Lrrk2 has been proposed to interact with various proteins, including mitogen-activated protein kinases (MAPKs)(Gloeckner et al. 2009; Hsu et al. 2010; Li et al. 2011). Recent studies also suggest that Lrrk2 may be involved in regulation of MAPK signaling cascades(West et al. 2007; Gloeckner et al. 2009; Chen et al. 2012; Bravo-San Pedro et al. 2013; Boon et al. 2014; Verma et al. 2014).
MAPKs are important signaling proteins that link extracellular stimuli to cellular responses(Bonny et al., 2005, Liu and Lin, 2005). There are three members in MAPK family: extracellular signal-regulated kinase (ERK), p38, and c-Jun N-terminal kinase (JNK). The ERK pathway plays a major role in regulation of cell growth and differentiation. JNK and p38 are activated in response to various stress stimuli (e.g., oxidative stress)(Bonny et al. 2005; Liu and Lin 2005), are associated with the induction of apoptosis and have implications in neurodegeneration during PD pathogenesis(Borsello and Forloni 2007). To further understand the roles of Lrrk2 and MAPK pathways in PD-induced neuronal degeneration, RNAi screens of MAPK pathway proteins in Lrrk2 transgenic flies were conducted using a PD Drosophila model that we have recently generated(Liu et al. 2008). In this model, overexpression of the human G2019S-Lrrk2 variant results in an increase in kinase activity and robust dopaminergic neurodegeneration that resembles the key features of Parkinsonism. The findings from our current study provide novel insights into the molecular mechanisms underlying Lrrk2-induced neuronal degeneration.
METHODS
Materials and Lrrk2 transgenic fly stocks
Fly stocks, including UAS-Lrrk2–1, UAS-G2019S-2, and DOPA decarboxylase (ddc)-Gal4 and elav-Gal4 flies were maintained in our laboratory on standard cornmeal medium at 25 °C as described previously(Liu et al. 2008). All UAS-RNAi fly lines targeting the major players in the MAPK pathways were from VDRC (Vienna Drosophila RNAi Center, Table. 1). UAS-JNK-DN fly stock was from Bloomington Stock Center (Bloomington, IN, USA). Anti-Flag and anti-actin antibodies were obtained from Sigma (St. Louis, MO, USA). Anti-tyrosine hydroxylase (TH), anti-JNK, anti-phospho-JNK, anti-MKK6, anti-MKK7 and anti-c-Jun antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). p38 and anti-MEK-1 were purchased from Santa Cruz Biotech. Anti-MEKK1 was purchased from Abcam. SP600125, a JNK inhibitor, was purchased from BioMol.
Table.1.
List of UAS-RNAi fly lines of MAPK pathways in the screen
| Human gene | Fly gene | RNAi stock(VDRC) |
|---|---|---|
| MEK1/2 | D-Sor (CG15793) | 40026 |
| Elk1 | pointed(CG17077) | 7171 |
| MEKK1 | mekk1(CG7717) | 25528, 25529 |
| MKK3/6 | licorne(CG12244) | 20166 |
| p38MAPK | mpk2 (CG5475) | 52277 |
| MKK7 | hep (CG4353) | 47507, 47509 |
| SAPK/JNK1,2,3 | basket (CG5680) | 34138, 34139 |
| JUN | Jra (CG2275) | 10835 |
Western blot analysis
For western blot analyses, fly head homogenates were subjected to 4–12% NuPAGE Bis-Tris gels and transferred onto polyvinylidene difluoride membranes (Invitrogen). The membranes were blocked in 5% nonfat milk blocking buffer and then probed with various primary antibodies followed by the secondary detection antibodies. Proteins in the blots were detected using enhanced chemiluminescence (ECL) reagents. NIH ImageJ software was used for quantification.
Fly survival assay
Cohorts of 60 flies from each group were subjected to fly survival assays weekly as previously described(Liu et al. 2008). For SP600125 treatment experiments, SP600125 was added to the food from day 1 of eclosion and continued for the lifetimes of the flies. Fresh food with SP600125 was provided every 3–4 days. Survival experiments were repeated three times. Mortality was analyzed using Kaplan–Meier survival analysis. A p value <0.05 was considered significant. For the median lifespan we counted the time for half fly survival in each tube and the data were analyzed using one-way ANOVA followed by Turkey post hoc test with GraphPad Prism software.
Climbing assay
To assess locomotor activity, fly climbing assays (negative geotaxis assay) were employed weekly as described previously(Liu et al. 2008). Tested flies were placed in a plastic cylinder (length, 25 cm; diameter, 1.5 cm). Cohorts of 60 flies from each group were used. Flies were tapped to the bottom of the cylinder. Flies that were able to climb to or above the median line of the cylinder within 10 seconds were counted. Fly climbing abilities were calculated based on three repeats of the assay each week.
Whole-mount immunostaining of dissected fly heads and dopamine (DA) neuron counting
Whole-mount dissected adult brains at 4–6 weeks of age were subjected to fluorescent immunostaining using anti-TH (DA neuron marker) antibodies as described previously(Liu et al. 2008). Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 568 goat anti-mouse IgG (Invitrogen) were used as the secondary detection antibodies. There were eight flies in each experimental immunostaining group. DA neurons in 12 out of 13 clusters were counted under fluorescent (Zeiss LSM 250) or confocal microscopy (Zeiss LSM 510). There are too many DA neurons in the PAM, which results in a high density of fluorescence in this area. Thus, neurons in the PAM cluster were not included in our assays. Our previous study demonstrated that Flag-Lrrk2-linked immunofluorescence colocalized with anti-TH immunostaining in neurons of all DA clusters(Liu et al. 2008).
Data analysis
Quantitative data were expressed as arithmetic means ± SEM based on three separate experiments. Statistically significant differences between groups were analyzed by one-way ANOVA using the GraphPad Prism statistical software. A p value <0.05 was considered significant.
RESULTS
RNAi screen of MAPK pathway proteins in G2019S-Lrrk2 transgenic flies
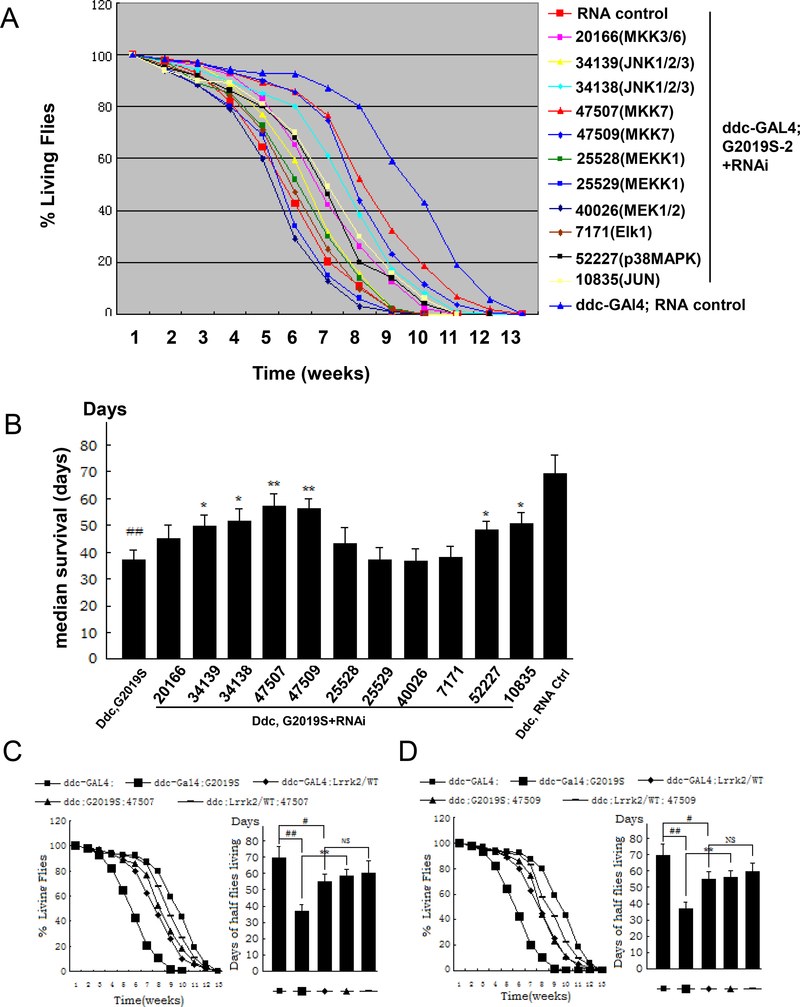
To investigate the roles of MAPK pathway in Lrrk2-induced neuronal degeneration in vivo, RNAi screens of MAPK pathway proteins (Table 1) were conducted using UAS-G2019S-Lrrk2 transgenic flies. Specific USA-RNAi targeting MAPK pathway players were co-expressed with the mutant UAS-G2019S-Lrrk2 variant in dopaminergic neurons in ddc-Gal4 promoter driver flies. Knockdown of the hemipterous (hep, RNAi lines 47507 and 47509) kinase protected against G2019S-Lrrk2-induced early death and elicited the strongest effect (a protection rate of greater than 30% at ages of 5 to 7 weeks) among the genes that we screened (Fig. 1A and 1B). Knockdown of the hep downstream players basket (JNK, RNAi lines 34138 and 34139) and Jra (Jun, RNAi line 10835) also exhibited modest protective effects (Fig. 1A and 1B). Expression of RNAi 52227 targeting p38 slightly increased the fly survival time, but the effect was more modest (less than 25% relative to the G2019S-Lrrk2 transgenic flies). Expression of RNAi 40026 and 25529 targeting MEK1/2 and MEKK1 of the ERK pathway slightly enhanced G2019S-Lrrk2-induced PD-like phenotypes, but the differences were not significant (Fig. 1A and 1B). Western blot analysis showed that all RNAi lines could significantly knockdown the corresponding protein levels (Data is not shown here).
Fig. 1. RNAi screens of MAPK pathway proteins in ddc-Gal4;G2019S-2 transgenic flies.
A. Using fly survival as a readout, 11 UAS-RNAi lines combined with ddc-Gal4;G2019S-2 flies were screened as listed in Table 1. ddc-Gal4 or UAS-G2019S-2 flies served as equal non- target protein expression control and showed only one result here. A scrambled RNAi sequence served as an RNA control. Fly survival was recorded throughout the flies’ lifetimes (n=60). Survival data were analyzed via Kaplan–Meier log rank survival analysis. B. In median lifespan analysis we counted the time until half of the remaining flies survived in each tube. The 47507 and 47509 lines exhibited a better protective effect than 34138, 34139, 10835 and 52227 RNAi lines. The other RNAi lines didn’t alter fly survival compared to ddc-Gal4;G2019S-2 flies(n=60). C and D. Two hep RNAi lines, 47507 (C) and 47509 (D), were crossed to ddc-Gal4;Lrrk2/WT and ddc-Gal4;G2019S flies. The survival curves and the median lifespan were analyzed(n=60). * p<0.05, ** p<0.01 indicates statistically significant differences between RNAi knockdown and ddc-Gal4;G2019S flies. ## p<0.01 indicates statistically significant differences between the ddc-Gal4;G2019S and ddc-Gal4 control lines. NS denotes no significance.
To further validate the hep RNAi result, the survival experiment was repeated. Knockdown of hep by both 47507 and 47509 significantly increased survival in transgenic G2019S-Lrrk2 flies (Fig. 1C and 1D). Expression of WT-Lrrk2 slightly decreased survival compared with non-transgenic control flies, however knockdown of hep in WT-Lrrk2 expressing flies did not change survival compared with WT-Lrrk2 expressing flies alone (Fig. 1C, 1D).
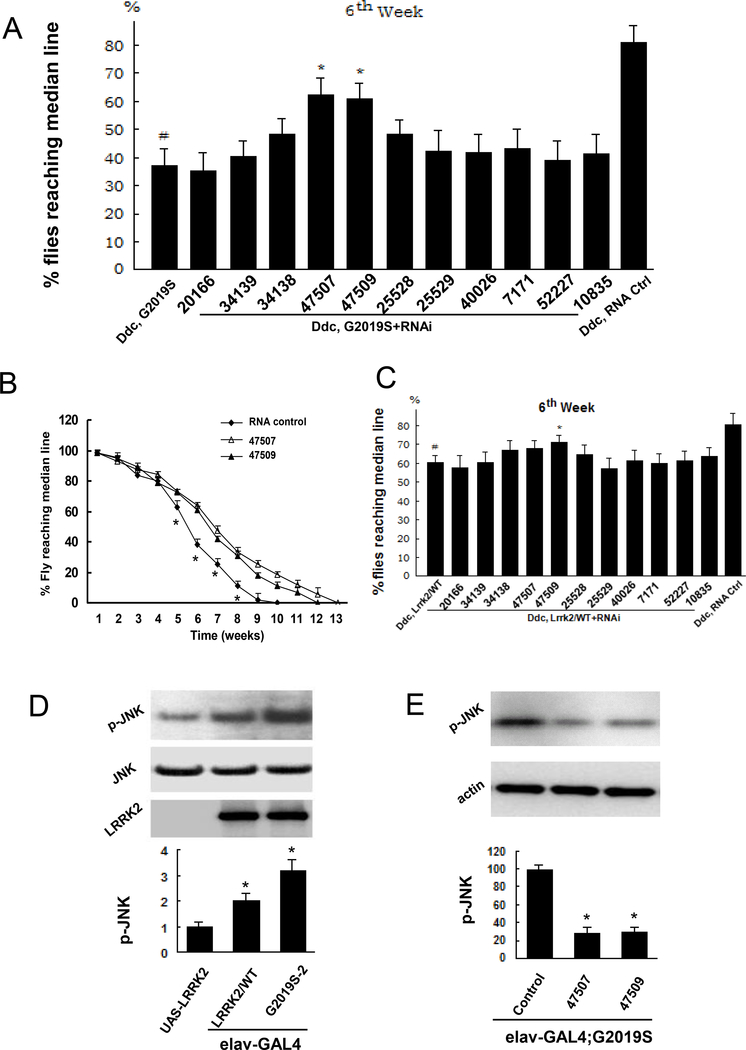
Fly climbing assays exhibited Lrrk2/G2019S induced locomotor dysfunction in ddc-Gal4;G2019S-2 flies (Fig. 2A). Knockdown of hep by both 47507 and 47509 RNAi inddc-Gal4;Lrrk2/G2019S improved locomotor activity (Fig. 2A and 2B). Expression of WT-Lrrk2 slightly decreased locomotor activity compared with non-transgenic control flies. Knockdown of hep in ddc-Gal4; Lrrk2/WT flies completely prevented this (Fig. 2C).
Fig. 2. Lrrk2 increased basket (d-JNK) phosphorylation in Lrrk2 transgenic flies.
A and B. Fly climbing assay was performed weekly. A. Data from the flies at 6 weeks old. Flies expressing 47507 and 47509 RNAi andG2019S-2 exhibited improved motor function compare with ddc-Gal4;G2019S-2 flies. A scrambled RNAi sequence served as an RNA control. B. Data from the whole life of the flies. Knockdown of hep in ddc-Gal4;G2019S flies improved locomotor impairment beginning at 5 weeks of age. C. Data from the flies at 6 weeks old. Fly climbing assay was performed weekly and the data were from the flies at 6 weeks of age. Coexpressing 47509 RNAi and Lrrk2/WT slightly improved the locomotor function compared with flies expressing Lrrk2/WT expression alone. The other RNAi lines didn’t alter motor function in ddc-Gal4; Lrrk2/WT flies. In figure A, B and C, (n=60) * p<0.05 indicates statistically significant differences between hep knockdown and ddc-Gal4;G2019S flies. In figure A and C, # p<0.05 indicates statistically significant differences between the ddc-Gal4;G2019S and ddc-Gal4 non-transgenic control lines. D. Fly head homogenates were subjected to western blot analysis. Expression of Lrrk2/WT and G2019S increased drosophila basket (p-JNK) phosphorylation in fly brains(n=6). *p<0.05 by ANOVA, indicating statistically significant differences between Lrrk2 transgenic flies and control, and between Lrrk2/WT and G2019S-2 transgenic flies. E. Knockdown of hep by RNAi lines 47507 and 47509 significantly decrease p-JNK in elav-Gal4;G2019S(n=6). *p<0.05 by ANOVA, indicating statistically significant differences between hep knockdown and control RNA flies.
Lrrk2 increased d-JNK phosphorylation in transgenic flies
To assess whether the JNK (also termed basket or d-JNK, a substrate kinase of hep) pathway was activated in the brains of the Lrrk2 transgenic flies, the phosphorylation of d-JNK in Lrrk2 fly brains was assessed by western blot analysis using a specific anti-phosphorylation JNK antibody. Overexpression of either wild-type human Lrrk2/WT or mutant G2019S-2 Lrrk2 in neurons of fly brains with the elav-Gal4 driver increased d-JNK phosphorylation (Fig. 2D). Although Lrrk2/WT and G2019S-2 displayed equivalent protein expression, they increased the d-JNK phosphorylation by approximately 2- and 3.2-fold compared with the non-transgenic flies elav-Gal4 and UAS-Lrrk2, respectively (Fig. 2D).. Importantly, the expression of either 47507 RNAi or 47509 RNAi significantly reduced d-JNK in the fly brains compared with the elav-Gal4;G2019S transgenic flies (Fig. 2E).
Knockdown of hep reduced G2019S-Lrrk2-induced dopaminergic neuronal degeneration
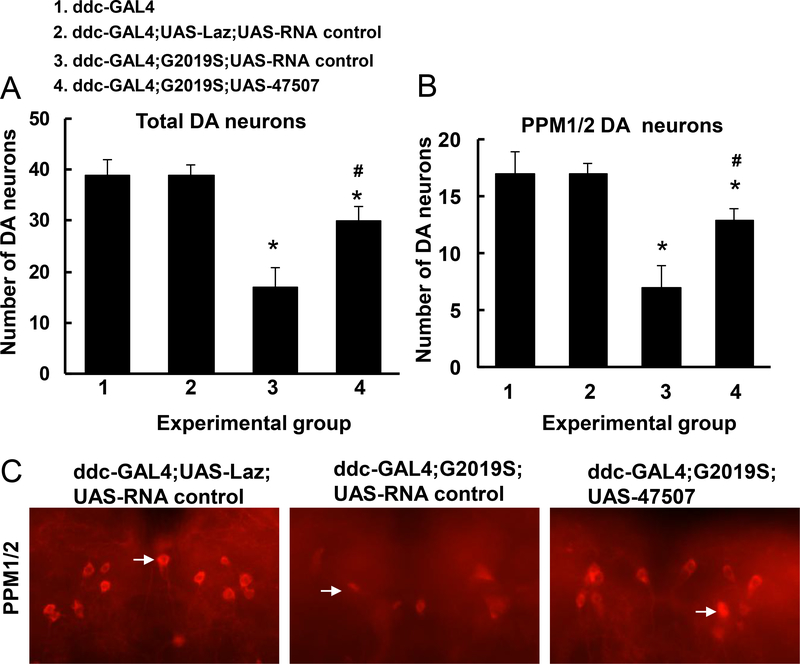
To further study the role of the hep pathway in Lrrk2-linked DA neuron degeneration, UAS-G2019S-Lrrk2 transgenic flies were crossed with UAS-hep-47507 RNAi flies under ddc-Gal4 promoter. Six-week-old flies were sacrificed, and the heads were subjected to whole-mount immunostaining using anti-TH antibodies. The number of DA neurons was significantly reduced in ddc-Gal4; G2019S flies compared with non-transgenic controls. In contrast, knockdown of hep by 47507 RNAi in G2019S-Lrrk2 transgenic flies rescued G2019S-LRRK2-induced TH-positive neuron loss (Fig. 3A, 3B, 3C).
Fig. 3. Knockdown of hep attenuated G2019S-Lrrk2-induced DA neuron loss and behavior change.
A. Quantitation of total DA neurons in all clusters except the PAM cluster. Six-week-old flies were sacrificed, and the brains were dissected. The entire brain was subjected to immunofluorescent staining using anti-TH antibodies. DA neurons were counted in all clusters except the PAM cluster(n=6). B. Quantitation of TH-positive neurons in the PPM1/2 clusters(n=6). * p<0.05 indicates statistically significant differences between non-transgenic and G2019S-Lrrk2 transgenic flies. # p<0.05 indicates statistically significant differences between ddc-GAL4;G2019S;UAS-47507 and G2019S expression alone flies. C. Representative images of anti-TH staining in PPM1 and PPM2 clusters at 6 weeks of age in various experimental groups. Arrows indicate examples of TH-positive neurons in each condition.
JNK inhibitor, SP600125, suppressed G2019S-Lrrk2-induced PD-like phenotypes in flies
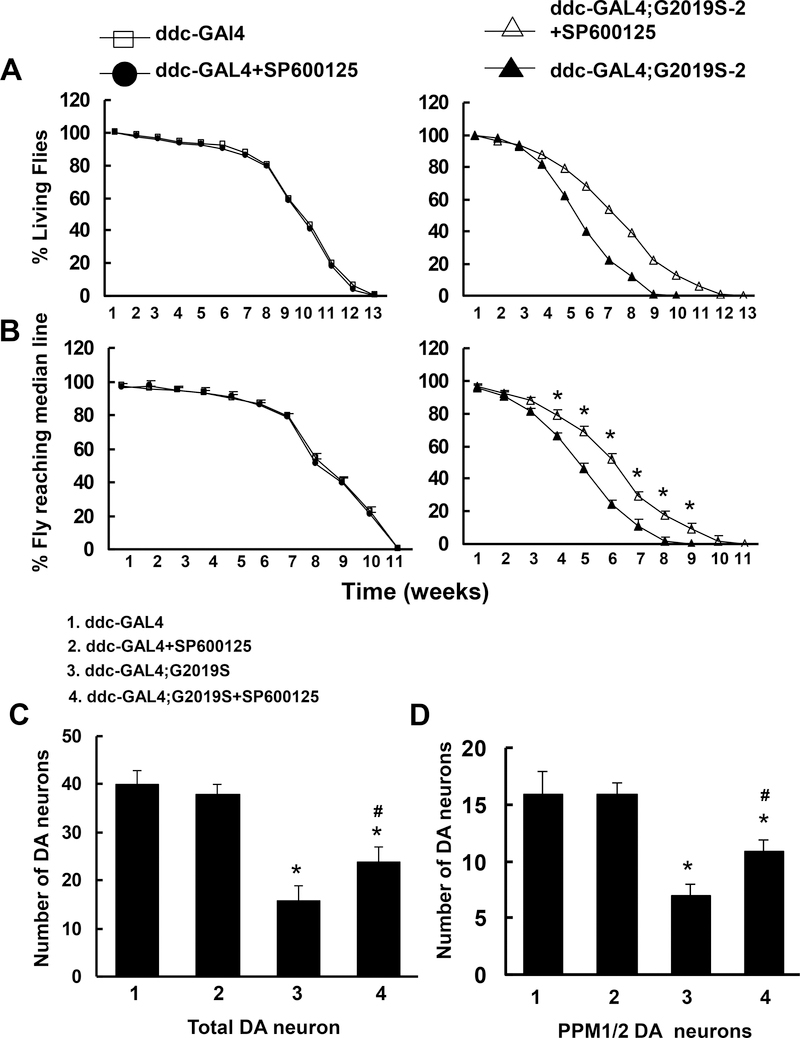
To further study the role of the hep downstream kinase JNK in DA neuron degeneration, the JNK inhibitor SP600125 (1 μM) was added to the food of the ddc-Gal4;G2019S flies beginning on the first day of eclosion and continuing for the flies’ lifetimes. SP600125 significantly improved locomotor function and increased survival time in ddc-Gal4;G2019S-2 flies but had no effect on these parameters in non-transgenic control flies (Fig. 4A and 4B). Moreover, whole-mount immunostaining examinations revealed that SP600125 reduced the loss in TH-positive neurons compared with the vehicle control (Fig. 4C and 4D).
Fig. 4. SP600125 attenuated the neurodegenerative changes in mutant G2019S-Lrrk2 transgenic flies.
SP600125 (1 μM) was added to the fly food for the lifetimes of non-transgenic flies (ddc-Gal4 or UAS-Lrrk2) and ddc-Gal4;G2019S-2 transgenic flies. A and B showed that SP600125 increased survival time and improved locomotor function in mutant G2019S-Lrrk2 flies. A. Survival curves for the flies (n=60). Survival data were analyzed with Kaplan–Meier log rank survival analysis. p<0.05 indicates statistically significant differences between non-transgenic and G2019S-Lrrk2 transgenic lines and between vehicle and SP600125-treated transgenic flies. B. Cohorts of 60 flies from each experiment were subjected to climbing assays weekly. *p<0.05 indicates statistically significant differences vs. the untreated ddc-Gal4;G2019S-2 transgenic flies. C and D. SP600125 attenuated G2019S-Lrrk2-induced DA neuron loss. C. Summary of total DA neuron counts in all clusters except the PAM cluste(n=6)r. D. Summary of TH-positive neuron counts in the PPM1/2 clusters(n=6). * p<0.05, compared with Gal4 control. # p<0.05, compared with vehicle.
Expression of dominant-negative JNK-DN reduced G2019S-Lrrk2-induced Parkinsonism in flies
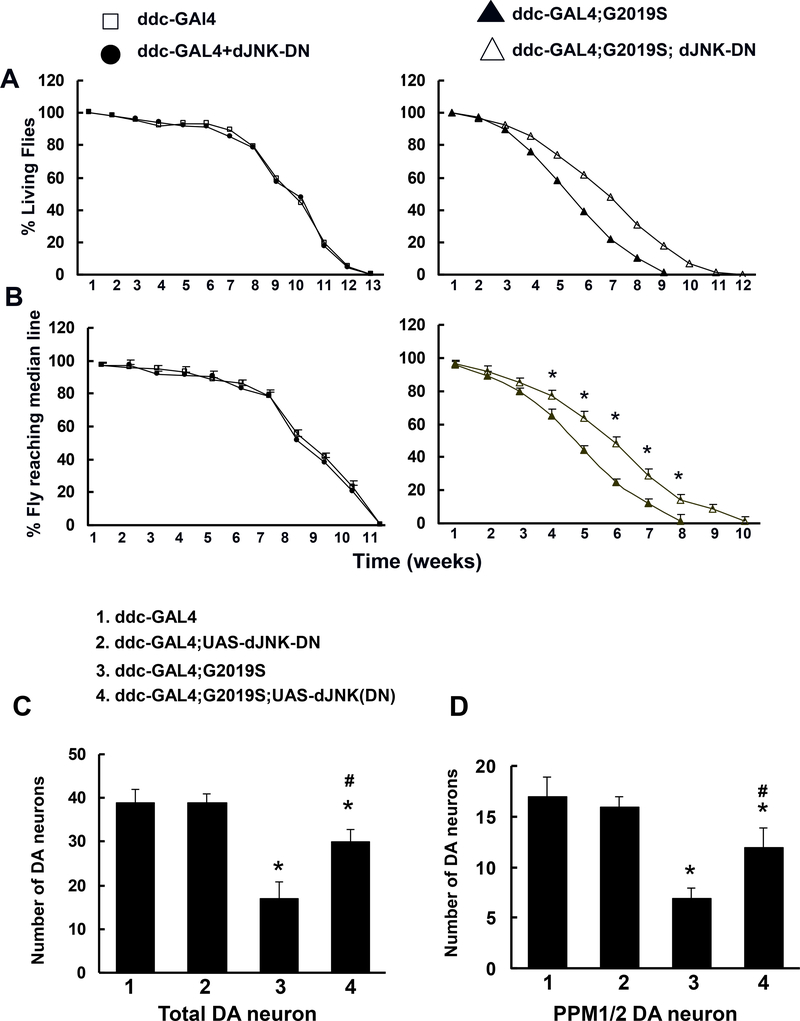
To further validate the roles of JNK in Lrrk2-induced PD-like phenotypes, ddc-Gal4;G2019S flies were crossed with UAS-JNK-DN (a dominant-negative form of JNK), which led to JNK-DN and G2019S-Lrrk2 co-expression in DA neurons. The expression of JNK-DN increased the survival times and improved the locomotor functions in G2019S-Lrrk2 transgenic flies (Fig. 5A and 5B). The JNK-DN also significantly prevented G2019S-Lrrk2-induced DA neuron degeneration (Fig. 5C and 5D).
Fig. 5. Expression of dominant-negative JNK-DN attenuated the neurodegenerative changes in mutant G2019S-Lrrk2 transgenic flies.
ddc-Gal4;G2019S flies were crossed with UAS-JNKDN (a dominant negative form of JNK). A. Survival data were analyzed with Kaplan–Meier log rank survival analysis(n=60). p < 0.05 indicating statistically significant differences between non-transgenic and Lrrk2 transgenic lines and between ddc-Gal4;G2019S;JNK-DN and ddc-Gal4;G2019S flies. B. Locomotor activity as assessed by climbing assays(n=60). *p<0.05 indicating statistically significant differences between non-transgenic and Lrrk2 transgenic lines and between ddc-Gal4;G2019S;JNK-DN and ddc-Gal4;G2019S flies. C. Quantitation of total DA neuron counts in all clusters except the PAM cluster with anti-TH immunostaining(n=6). D. Quantitation of TH-positive neurons in the PPM1/2 clusters(n=6). * p<0.05 indicating statistically significant differences between non-transgenic and G2019S-Lrrk2 transgenic flies. # p<0.05 indicating statistically significant differences between ddc-Gal4;G2019S;JNK-DN and ddc-Gal4;G2019S flies.
DISCUSSION
The main finding of this study was that the hep pathway mediated Lrrk2-induced neuronal degeneration and was responsible for the PD-like phenotype in this Drosophila model. Our data revealed that the expression of wild-type and mutant G2019S-Lrrk2 in flies increased JNK phosphorylation (activation). The knockdown of hep (d-JNKK) and its downstream players, d-JNK and d-Jun, suppressed Lrrk2-induced DA neuron loss and improved locomotor function in transgenic flies. Moreover, both expression of dominant-negative JNK-DN and treatment with the JNK inhibitor SP600125 also attenuated DA neuron degeneration. These results indicate that activation of the hep/d-JNK/d-jun pathway plays a critical role in Lrrk2-linked DA neuron degeneration and thus may be a therapeutic target for PD.
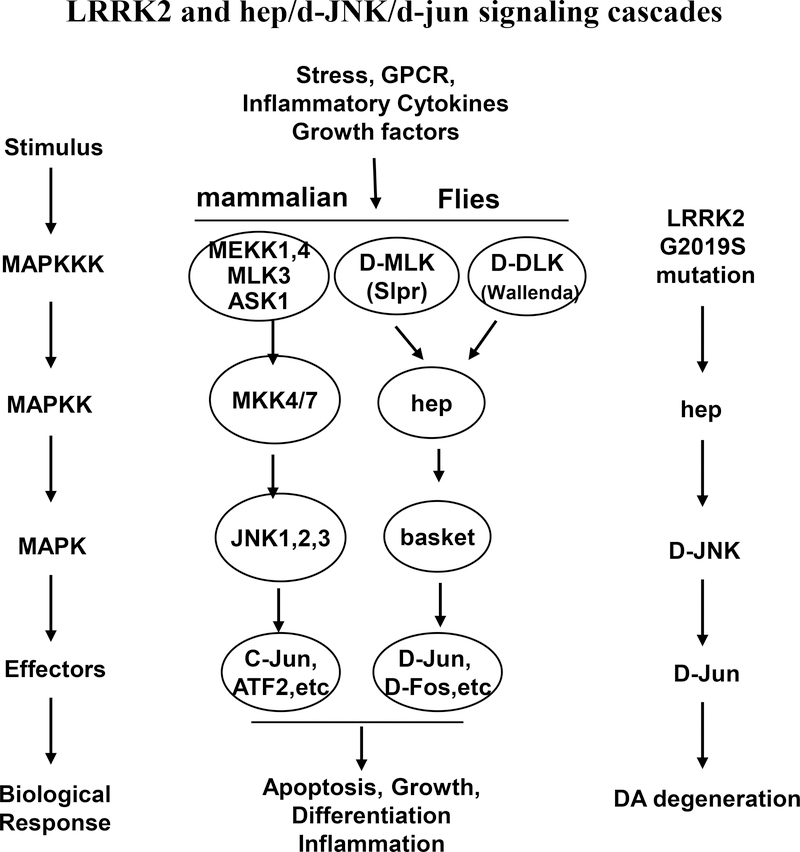
The JNK pathway is an established mediator of stress-induced apoptosis with implications for neurodegeneration in the pathogenesis of PD(Borsello and Forloni, 2007). The kinases and effectors of JNK pathways in mammals and Drosophila are summarized in Fig. 6 (Wang et al. 2012; Shaukat et al. 2016). Previous reports have demonstrated that phosphorylation and activation of the JNK pathway mediates neuronal death in mammalian models of neurodegenerative diseases(Burke 2007). For example, the JNK pathway has been found to be involved in the death of substantia nigra dopaminergic neurons caused by 6-hydroxydopamine injection(Ries et al. 2008). Our results demonstrated that Lrrk2 significantly increased JNK phosphorylation in the neurons of fly brains; the PD-linked mutant G2019S variant increased JNK activation by approximately 3-fold.
Fig. 6. The kinases and effectors of JNK pathways in mammals and drosophila.
MAPK pathway is important signaling pathway that is activated in response to various stress stimuli. The activation of JNK signaling pathway is associated with the induction of apoptosis and neurodegeneration in PD pathogenesis in mammals and in drosophila. The PD-linked mutant G2019S variant works as an MAPKKK and activates its substrate hep in drosophila. The activated hep starts activation of hep/d-JNK/d-jun signaling pathway and induces the neurodegeneration in drosophila with Lrrk2/ G2019S mutation.
The different methods can decrease the protein expression in flies include expression of UAS-RNAi, expression of dominant-negative genes and treatment with inhibitors. Our data indicated knockdown of hep (JNKK, MKK) expression, expression of dominant-negative JNK-DN, and treatment with JNK inhibitor SP600125 significantly suppressed Lrrk2-induced DA neuron loss in flies. These findings demonstrate that Lrrk2 activates a stress-response kinase cascade that includes the hep/d-JNK pathway and that the activation of this pathway mediates Lrrk2-induced neuron degeneration. Consistent with our results, a recent study with transgenic mice revealed that the phosphorylation of MKK4, JNK, and c-jun are upregulated in the substantia nigra of G2019S-Lrrk2 transgenic mice compared with non-transgenic or wild-type Lrrk2 transgenic mice(Chen et al. 2012). However, this study did not determine whether reducing the phosphorylation of these proteins could prevent the PD-linked phenotypes. Thus, our study is the first in vivo study to provide genetic and pharmacological evidence that inhibition of Lrrk2-induced JNK activation suppresses PD-like phenotypes in flies.
Lrrk2 has been suggested to be a member of the MLK subfamily of MAPKKKs(West et al. 2005; West et al. 2007). Previous in vitro studies have demonstrated that Lrrk2 binds with and directly phosphorylates MKK3, −4, −6 and −7(West et al. 2007; Gloeckner et al. 2009; Hsu et al. 2010), however, the kinase dead-Lrrk2 construct does not(Gloeckner et al. 2009; Hsu et al. 2010). Furthermore, Lrrk2 does not bind with other MLKs or JNK as demonstrated in other studies and our unpublished data, indicating that JNK is not substrate of Lrrk2, but a downstream effector of Lrrk2. Knockdown of target gene expression with RNAi is a powerful tool for studying the functions of genes. A genome-wide library of Drosophila RNAi transgenes (VDRC, Austria and NIG-Fly, Japan) covers 88% of the predicted protein-coding genes in the Drosophila genome(Dietzl et al. 2007). In this library, RNAi transgenes consist of short gene fragments that are cloned as inverted repeats and are expressed using the binary GAL4/UAS system, which enables the conditional inactivation of gene functions in specific tissues of intact organisms(Dietzl et al. 2007). Recent cell culture studies suggested that Lrrk2 may activate MAPK(West et al. 2007; Gloeckner et al. 2009; Chen et al. 2012). To further investigate whether MAPK pathways play important roles in Lrrk2-induced neuronal degeneration in vivo, RNAi screens of MAPK pathway proteins (Table 1) were conducted using UAS-G2019S-Lrrk2 transgenic flies that display key features of human PD including the selective loss of dopaminergic neurons and locomotor impairment(Liu et al., 2008). Our RNAi screen data also proved that knockdown of the ERK pathway slightly shortened the survival time of G2019S-Lrrk2 transgenic flies, whereas knockdown of the p38 pathway did not significantly affect fly survival. Taken together, these findings suggest that hep (JNKK or MKK) is a predominant mediator of neuronal degeneration and is likely a substrate of Lrrk2 kinase. Mutant Lrrk2 activated the hep/d-JNK/d-jun pathway(Cha et al. 2005), which resulted in neuron degeneration (Fig. 6). Additionally, the expression of parkin suppresses the constitutively active form of hemipterous (hepCA) as well as JNK signaling by reducing bsk/JNK transcription(Hwang et al. 2010). The co-expression of human parkin in Lrrk2 G2019S flies provides protection against dopaminergic neurodegeneration(West et al. 2007; Gloeckner et al. 2009; Ng et al. 2009; Hsu et al. 2010). Taken together, these findings suggest that parkin and Lrrk2 may interact in the JNK pathway.
Although down-regulation of the hep/JNK pathway strikingly attenuated Lrrk2-induced Parkinsonism in flies, it did not completely rescue neuronal degeneration or locomotor impairment, which suggests that other signaling pathways may also be involved in Lrrk2-linked neuron loss. For example, disruption of the autophagy pathway has been suggested to be relevant in previous studies. Additionally, other Lrrk2 interaction partners (e.g., Fas-Associated Death Domain) or unidentified Lrrk2 substrate(s) may also play a role in Lrrk2 cellular pathways and associated toxicity(Ko et al. 2009). These aspects await further investigation.
In conclusion, our results demonstrate that the hep/d-JNK/d-Jun pathway partially mediates Lrrk2-induced neuronal degeneration and might be a potential therapeutic target for PD intervention.
Acknowledgments
We thank Drs. Christopher A. Ross and Craig Montell for their helpful discussion and suggestions. We thank the American Parkinson Disease Association, the NIH/NINDS grants R01NS093383 and R21NS096620, the National Natural Science Foundation of China (grant No. 81371392) and Natural Science Foundation of Jiangsu (grant No. BK20131161) for their support. All authors state that there are no conflicts of interest.
References
- Arranz AM, Delbroek L, Van Kolen K, Guimaraes MR, Mandemakers W, Daneels G, Matta S, Calafate S, Shaban H, Baatsen P, De Bock PJ, Gevaert K, Vanden Berghe P, Verstreken P, De Strooper B, Moechars D 2015. LRRK2 functions in synaptic vesicle endocytosis through a kinase-dependent mechanism. Journal of cell science 128:541–552. [DOI] [PubMed] [Google Scholar]
- Atashrazm F, Dzamko N 2016. LRRK2 inhibitors and their potential in the treatment of Parkinson’s disease: current perspectives. Clinical pharmacology : advances and applications 8:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilina A, Rudenko IN, Kaganovich A, Civiero L, Chau H, Kalia SK, Kalia LV, Lobbestael E, Chia R, Ndukwe K, Ding J, Nalls MA, International Parkinson’s Disease Genomics C, North American Brain Expression C, Olszewski M, Hauser DN, Kumaran R, Lozano AM, Baekelandt V, Greene LE, Taymans JM, Greggio E, Cookson MR 2014. Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proceedings of the National Academy of Sciences of the United States of America 111:2626–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonny C, Borsello T, Zine A 2005. Targeting the JNK pathway as a therapeutic protective strategy for nervous system diseases. Rev Neurosci 16:57–67. [DOI] [PubMed] [Google Scholar]
- Boon JY, Dusonchet J, Trengrove C, Wolozin B 2014. Interaction of LRRK2 with kinase and GTPase signaling cascades. Frontiers in molecular neuroscience 7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsello T, Forloni G 2007. JNK signalling: a possible target to prevent neurodegeneration. Curr Pharm Des 13:1875–1886. [DOI] [PubMed] [Google Scholar]
- Bravo-San Pedro JM, Niso-Santano M, Gomez-Sanchez R, Pizarro-Estrella E, Aiastui-Pujana A, Gorostidi A, Climent V, Lopez de Maturana R, Sanchez-Pernaute R, Lopez de Munain A, Fuentes JM, Gonzalez-Polo RA 2013. The LRRK2 G2019S mutant exacerbates basal autophagy through activation of the MEK/ERK pathway. Cellular and molecular life sciences : CMLS 70:121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke RE 2007. Inhibition of mitogen-activated protein kinase and stimulation of Akt kinase signaling pathways: Two approaches with therapeutic potential in the treatment of neurodegenerative disease. Pharmacol Ther 114:261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha GH, Kim S, Park J, Lee E, Kim M, Lee SB, Kim JM, Chung J, Cho KS 2005. Parkin negatively regulates JNK pathway in the dopaminergic neurons of Drosophila. Proceedings of the National Academy of Sciences of the United States of America 102:10345–10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Weng YH, Chien KY, Lin KJ, Yeh TH, Cheng YP, Lu CS, Wang HL 2012. (G2019S) LRRK2 activates MKK4-JNK pathway and causes degeneration of SN dopaminergic neurons in a transgenic mouse model of PD. Cell Death Differ 19:1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson MR 2010. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nat Rev Neurosci 11:791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL 2003. Molecular pathways of neurodegeneration in Parkinson’s disease. Science 302:819–822. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151–156. [DOI] [PubMed] [Google Scholar]
- Esteves AR, Cardoso SM 2016. LRRK2 at the Crossroad Between Autophagy and Microtubule Trafficking: Insights into Parkinson’s Disease. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. [DOI] [PubMed] [Google Scholar]
- Galter D, Westerlund M, Carmine A, Lindqvist E, Sydow O, Olson L 2006. LRRK2 expression linked to dopamine-innervated areas. Ann Neurol 59:714–719. [DOI] [PubMed] [Google Scholar]
- Gloeckner CJ, Schumacher A, Boldt K, Ueffing M 2009. The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. J Neurochem 109:959–968. [DOI] [PubMed] [Google Scholar]
- Hsu CH, Chan D, Wolozin B 2010. LRRK2 and the stress response: interaction with MKKs and JNK-interacting proteins. Neurodegener Dis 7:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Kim D, Choi G, An SW, Hong YK, Suh YS, Lee MJ, Cho KS 2010. Parkin suppresses c-Jun N-terminal kinase-induced cell death via transcriptional regulation in Drosophila. Mol Cells 29:575–580. [DOI] [PubMed] [Google Scholar]
- Kang UB, Marto JA 2017. Leucine-rich repeat kinase 2 and Parkinson’s disease. Proteomics 17. [DOI] [PubMed] [Google Scholar]
- Ko HS, Bailey R, Smith WW, Liu Z, Shin JH, Lee YI, Zhang YJ, Jiang H, Ross CA, Moore DJ, Patterson C, Petrucelli L, Dawson TM, Dawson VL 2009. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proceedings of the National Academy of Sciences of the United States of America 106:2897–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Greggio E, Beilina A, Kaganovich A, Chan D, Taymans JM, Wolozin B, Cookson MR 2010. The Parkinson’s disease associated LRRK2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation. PloS one 5:e8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari U, Tan EK 2009. LRRK2 in Parkinson’s disease: genetic and clinical studies from patients. FEBS J 276:6455–6463. [DOI] [PubMed] [Google Scholar]
- Lee BD, Shin JH, Van Kampen J, Petrucelli L, West AB, Ko HS, Lee YI, Maguire-Zeiss KA, Bowers WJ, Federoff HJ, Dawson VL, Dawson TM 2010. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson’s disease. Nat Med 16:998–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, He X, Thomas JM, Yang D, Zhong S, Xue F, Smith WW 2015. A novel GTP-binding inhibitor, FX2149, attenuates LRRK2 toxicity in Parkinson’s disease models. PloS one 10:e0122461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang D, Sushchky S, Liu Z, Smith WW 2011. Models for LRRK2-Linked Parkinsonism. Parkinsons Dis 2011:942412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang D, Zhong S, Thomas JM, Xue F, Liu J, Kong L, Voulalas P, Hassan HE, Park JS, MacKerell AD Jr., Smith WW 2014. Novel LRRK2 GTP-binding inhibitors reduced degeneration in Parkinson’s disease cell and mouse models. Human molecular genetics 23:6212–6222. [DOI] [PubMed] [Google Scholar]
- Liu J, Lin A 2005. Role of JNK activation in apoptosis: a double-edged sword. Cell Res 15:36–42. [DOI] [PubMed] [Google Scholar]
- Liu Z, Hamamichi S, Lee BD, Yang D, Ray A, Caldwell GA, Caldwell KA, Dawson TM, Smith WW, Dawson VL 2011. Inhibitors of LRRK2 kinase attenuate neurodegeneration and Parkinson-like phenotypes in Caenorhabditis elegans and Drosophila Parkinson’s disease models. Human molecular genetics 20:3933–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang X, Yu Y, Li X, Wang T, Jiang H, Ren Q, Jiao Y, Sawa A, Moran T, Ross CA, Montell C, Smith WW 2008. A Drosophila model for LRRK2-linked parkinsonism. Proceedings of the National Academy of Sciences of the United States of America 105:2693–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A 2006. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron 52:587–593. [DOI] [PubMed] [Google Scholar]
- Martin I, Kim JW, Lee BD, Kang HC, Xu JC, Jia H, Stankowski J, Kim MS, Zhong J, Kumar M, Andrabi SA, Xiong Y, Dickson DW, Wszolek ZK, Pandey A, Dawson TM, Dawson VL 2014. Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson’s disease. Cell 157:472–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CH, Mok SZ, Koh C, Ouyang X, Fivaz ML, Tan EK, Dawson VL, Dawson TM, Yu F, Lim KL 2009. Parkin protects against LRRK2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila. The Journal of neuroscience : the official journal of the Society for Neuroscience 29:11257–11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisiadou L, Xie C, Cho HJ, Lin X, Gu XL, Long CX, Lobbestael E, Baekelandt V, Taymans JM, Sun L, Cai H 2009. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience 29:13971–13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries V, Silva RM, Oo TF, Cheng HC, Rzhetskaya M, Kholodilov N, Flavell RA, Kuan CY, Rakic P, Burke RE 2008. JNK2 and JNK3 combined are essential for apoptosis in dopamine neurons of the substantia nigra, but are not required for axon degeneration. J Neurochem 107:1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosen DA, Cookson MR 2016. LRRK2 at the interface of autophagosomes, endosomes and lysosomes. Molecular neurodegeneration 11:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaukat Z, Liu D, Hussain R, Khan M, Gregory SL 2016. The Role of JNK Signalling in Responses to Oxidative DNA Damage. Current drug targets 17:154–163. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA 2006. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci 9:1231–1233. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB, Dawson VL, Dawson TM, Ross CA 2005. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proceedings of the National Academy of Sciences of the United States of America 102:18676–18681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup SF, Kuenen S, Vanhauwaert R, Manetsberger J, Hernandez-Diaz S, Swerts J, Schoovaerts N, Vilain S, Gounko NV, Vints K, Geens A, De Strooper B, Verstreken P 2016. A LRRK2-Dependent EndophilinA Phosphoswitch Is Critical for Macroautophagy at Presynaptic Terminals. Neuron 92:829–844. [DOI] [PubMed] [Google Scholar]
- Thomas JM, Li T, Yang W, Xue F, Fishman PS, Smith WW 2016. 68 and FX2149 Attenuate Mutant LRRK2-R1441C-Induced Neural Transport Impairment. Frontiers in aging neuroscience 8:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M, Steer EK, Chu CT 2014. ERKed by LRRK2: a cell biological perspective on hereditary and sporadic Parkinson’s disease. Biochimica et biophysica acta 1842:1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Pan J, Chen SD 2012. Kinases and kinase signaling pathways: potential therapeutic targets in Parkinson’s disease. Progress in neurobiology 98:207–221. [DOI] [PubMed] [Google Scholar]
- West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM 2005. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proceedings of the National Academy of Sciences of the United States of America 102:16842–16847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM 2007. Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Human molecular genetics 16:223–232. [DOI] [PubMed] [Google Scholar]
- Zhu X, Siedlak SL, Smith MA, Perry G, Chen SG 2006. LRRK2 protein is a component of Lewy bodies. Ann Neurol 60:617–618. [DOI] [PubMed] [Google Scholar]