Abstract
Advances in technology enable increasing amounts of data collection from individuals for biomedical research. Such technologies, for example, in genetics and medical imaging, have also led to important scientific discoveries about health and disease. The combination of multiple types of high-throughput data for complex analyses, however, has been limited by analytical and logistic resources to handle high-dimensional data sets. In our previous EU Joint Programme–Neurodegenerative Disease Research (JPND) Working Group, called HD-READY, we developed methods that allowed successful combination of omics data with neuroimaging. Still, several issues remained to fully leverage high-dimensional multimodality data. For instance, high-dimensional features, such as voxels and vertices, which are common in neuroimaging, remain difficult to harmonize. In this Full-HD Working Group, we focused on such harmonization of high-dimensional neuroimaging phenotypes in combination with other omics data and how to make the resulting ultra-high-dimensional data easily accessible in neurodegeneration research.
Keywords: Neuroimaging, High-dimensional, Voxels, Omics, Genetics, Voxel-based morphometry
1. Introduction
Biological data can be acquired on a large scale because of the ongoing innovations in technical fields, giving researchers the power to perform big data analyses to gain meaningful insights in human pathophysiology [1]. Sometimes termed “omics,” this field of biomedical data analysis incorporates various lines of research such as genomics, metabolomics, proteomics, and also large-scale data sets from medical imaging (“radiomics”). Although each of these approaches has resulted in important discoveries individually, the integration of data from all these modalities has not yet been fully exploited [2], in part, because the high-dimensional nature of such analyses makes them challenging or not even feasible to perform.
In the HD-READY consortium (our previous EU Joint Programme–Neurodegenerative Disease Research [JPND] working group), we specifically focused on the computational and statistical requirements for analyzing high-dimensional data, tacking several problems that require infrastructural capabilities far beyond that available at single sites. The work performed in HD-READY was very successful, resulting in two key publications of novel methods and a software package (“HASE”) that overcame these hurdles [3], [4]. For example, associating 1.5 million neuroimaging phenotypes with 9 million genetic variants using the HASE software is now possible in several hours instead of years, with great reductions in the size of data to transfer (gigabytes instead of terabytes).
Tools delivered in HD-READY are tailor-made to tackle challenges posed by high-dimensional data. However, especially for large-scale imaging data sets, an important outstanding issue is the urgent need to establish a framework for harmonization. Variations in data collection and processing pipelines complicate comparisons in neuroimaging studies in general, but this is especially important in the case of high-dimensional data. For example, although gross hippocampal volumes obtained using different methods can still be compared to some extent, it becomes impractical to compare a certain hippocampal voxel with one from another data set that was acquired or processed differently.
Laudable prior efforts from the JPND program—such as STRIVE [5], METACOHORTS [6], and HARNESS—aimed to harmonize, either qualitatively or quantitatively, vascular imaging markers. These initiatives followed decades of research using heterogeneous methods. Such efforts focused primarily on aggregated neuroimaging measures, whereas voxelwise or vertexwise harmonization remained largely elusive. The field of high-dimensional research is relatively young, but growing rapidly, and can greatly benefit from such a harmonization effort early on. The Full-HD working group was therefore set up to address two research needs:
-
1.
To harmonize high-dimensional neuroimaging data, so that it can be combined with other omics data. As a wealth of neuroimaging data have already been acquired using different scanners, field strengths, and acquisition protocols, we set out to not only define a general framework to harmonize currently available high-dimensional phenotypes but also determine requirements for future/novel neuroimaging phenotypes. Voxelwise and vertexwise phenotypes had a central focus.
-
2.
To harmonize ultra-high-dimensional neuroimaging-by-omics data for neurodegeneration research. We foresee these neuroimaging-by-omics data sets becoming useful tools for neurodegeneration research. For example, if a particular brain atrophy pattern is detected in certain patients, it could be interesting to examine whether there are genetic variants giving rise to a similar pattern. Thus, it is essential that any framework for harmonization of such high-dimensional data should take the ease of use for other researchers into account.
2. Methods
2.1. Full-HD group composition
The Full-HD working group was supported by the international Joint Programme for Neurodegenerative Diseases initiative (www.neurodegenerationresearch.eu/). The aim of the 2016 call was to address harmonization of neuroimaging biomarkers that are relevant for neurodegenerative diseases.
This working group brought together 17 experts from 5 countries, of which 4 are JPND member states (the Netherlands, Germany, Austria, Australia; the United States is not). Unlike the HD-READY consortium, where over 40 investigators were involved, we deliberately focused on a key set of collaborators. These include principle investigators of some of the largest neuroimaging cohorts and consortia worldwide (Study of Health In Pomerania [SHIP], Rotterdam Study, Austrian Stroke Prevention Study [ASPS], Brain Imaging Genetics [BIG], ENIGMA Consortium, Framingham Heart Study), giving us access to over 25,000 magnetic resonance imaging images, which ensured that recommendations and methods developed in the working group could be readily tested, fine-tuned, and applied to real data sets. Furthermore, we included not only experts on high-dimensional neuroimaging but also experts on omics data and neurodegeneration research.
2.2. Mode of operation
Over the course of 6 months, the working group held several teleconferences, one face-to-face meeting, and several outreach activities to disseminate our findings. Outreach included (1) presenting our work at scientific meetings in poster and oral sessions, that is, the Alzheimer Association International Conference, the VasCog conference, and the Organization for Human Brain Mapping [OHBM] meeting; (2) contacting organizers of various teaching courses and summer schools on statistical and imaging methods with the request to include our methodology in their course work, that is, Erasmus Summer Programme, Neuroepiomics, and the Cognomics Radboud Summer School Programme; and (3) publishing the results and recommendations from this working group in high-impact journals with a focus on open access publishing.
3. Results
3.1. Full-HD methodological framework
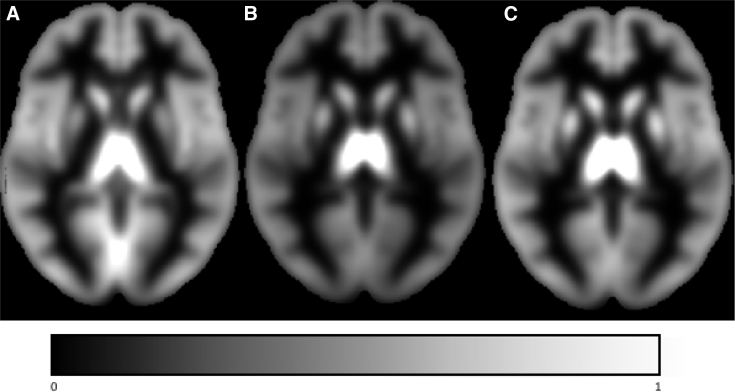
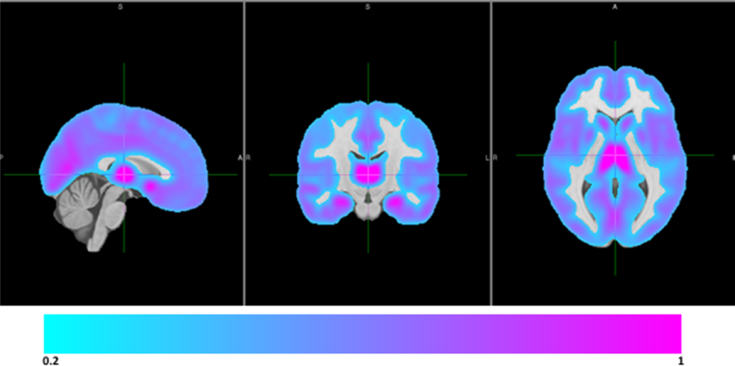
Given the high-dimensional origin of the omics and neuroimaging phenotypes, we developed and integrated quality control and harmonization methods for such data into the HASE software [4]. This framework relies heavily on the partial derivatives approach [3], that is, the proposed meta-analysis algorithm (developed during HD-READY), which allows for more insight into the data compared with classical meta-analysis and thus also more quality control. To illustrate this, we show that for voxelwise analyses, it is possible to generate mean gray matter density maps per cohort without access to individual-level data (Fig. 1). This makes it possible to verify that imaging processing pipelines used were consistent between cohorts and that all brain regions were included in the analysis. During the pilot phase of Full-HD, we were able to detect, among other errors, incorrect modulation of images, incorrect masking of images, incorrect normalization of phenotypes, and even in one case incorrect phenotypes themselves. Most errors would not have been detected using the quality control used in conventional meta-analysis. In addition, this framework allows for reduction of noise and false-positive errors. Specifically, based on such mean maps, researchers can screen and if required exclude phenotypes (e.g., voxels) that have little variation (which may be unexpected, or possibly erroneous) and create a mask for the phenotype analysis space (Fig. 2); this approach has some similarities to the approach commonly used for genetics data when filtering variants based on their minor allele frequency [7]. Importantly, this approach would also be applicable for quality control and harmonization of epigenetic data, gene expression data, metabolomics, and the microbiome.
Fig. 1.
Gray matter density maps for three cohorts generated from the partial derivatives. Mean gray matter density maps for three cohorts (the Rotterdam Study, SHIP, and ADNI), generated without access to individual-level data. This makes it possible to ensure that imaging processing pipelines were consistent between cohorts and all brain regions were included in the analysis. Maps of the local variation could also be derived. Abbreviations: ADNI, Alzheimer's Disease Neuroimaging Initiative; SHIP, Study of Health In Pomerania.
Fig. 2.
Selection of harmonious phenotypes for further meta-analysis. Masking of high-dimensional neuroimaging phenotypes for meta-analysis. Here, based on the mean maps, phenotypes with “low frequency” (i.e., not distributed evenly) can be excluded. This is similar to the approach common for genetic data where filtering of variants is performed based on the minor allele frequency. Sagittal (left), coronal (middle), and transversal (right) sections of the mean maps.
3.2. Full-HD logistical framework
In the HASE software, the computational burden is already shifted almost entirely to the meta-analysis stage, making it possible for a small cohort at a site with modest computational capacity to join in with multisite efforts. For the access to the resulting ultra-high-dimensional data sets, we recommend a similar solution with centralized storage of such data, to reduce the logistic burden for individual sites. Two approaches are put forward. First, it is possible to store the ultra-high-dimensional data on a storage server, which, depending on the type of analysis, would require several terabytes. In this case, a database format such as hdf5 [8], as used in HASE, will provide rapid access within such huge data sets. The second approach would be not to store the results of ultra-high-dimensional analyses but rather the partial derivatives. These partial derivatives are much smaller in size, but some additional computation would be needed to obtain the final results. In this approach, a storage server would not be sufficient but would need to be combined with processing power for the necessary computations. An online portal providing intuitive interaction with the data would likely be most suited for everyday researchers and clinicians aiming to query the data [9], [10]. Those who would want to do more in-depth research with raw data could be granted access.
The full results of these analyses are outside the scope of the current report and will be described in a separate article.
4. Discussion
In this JPND working group, we aimed to develop a framework for harmonizing ultra-high-dimensional imaging genetics analyses. Key features included dealing with voxelwise and vertexwise neuroimaging phenotypes. Importantly, other high-dimensional -omics technologies, such as genomics, proteomics, and metabolomics, among others, are also important for neurodegenerative disease and pose similar challenges as neuroimaging. Therefore, the recommendations and methods from this working group may be suitable to be incorporated by researchers working with other -omics technologies.
Exploiting these novel, ultra-high-dimensional technologies in research on neurodegenerative disease will require these data to be easily accessible to researchers who do not regularly work with high-dimensional data [11], [12]. However, the size and nature of these data make typical ways of sharing data (e.g., results tables or download links to the raw data) impractical. The working group provides concrete recommendations for such infrastructural challenges, that is, where to store the data and how to make it easily retrievable in a useful manner. Having each study acquire its own computational infrastructure is not feasible. Central computing, for example, cloud computing or cluster computing [13], is an emerging solution but should adhere to legal and ethical requirements [14], [15]. Again, we emphasize that recommendations from the working group pertaining to imaging allow for translation to other -omics technologies.
To actively follow up on the findings of the HD-READY and Full-HD working groups, the Uncovering Neurodegenerative Insights Through Ethnic Diversity (UNITED) consortium was initiated (see www.theunitedconsortium.com). Although the initial JPND working groups only included a limited number of members, the UNITED consortium aims to broaden the collaboration to a larger scale. This will be done by actively recruiting collaborators through visibility at international conferences and journals, while taking a particular interest in underrepresented populations to increase diversity.
4.1. Links
Framework for efficient high-dimensional association analyses (HASE): https://github.com/roshchupkin/HASE/.
Description of the framework and protocol for meta-analysis: www.imagene.nl/HASE.
The Uncovering Neurodegenerative Insights Through Ethnic Diversity (UNITED) consortium: www.theunitedconsortium.com.
Research in context.
-
1.
Systematic review: The authors searched the literature and inquired within their networks for methods potentially suitable for high-dimensional analyses of omics data. Although field-specific approaches exist to handle big data, cross-investigations between various types of data have been limited. These require either data reduction or infrastructure beyond current capabilities.
-
2.
Interpretation: Our review indicates there is a need for a method for jointly analyzing high-dimensional data within the omics fields.
-
3.
Future directions: In this report, we propose a framework for analyzing high-dimensional data. Still, this is only an initial step. Future research should focus on the interpretation of the results of such analyses and how the results themselves can be made easily accessible to other researchers, both inside and outside of the omics field.
Acknowledgments
Funding was obtained through the Joint Programming in Neurodegenerative Diseases Initiative and the Netherlands Organisation for Health Research and Development (grant number 733051002).
Footnotes
B.F. has obtained educational speaking fees from Medice and Shire. H.J.G. has received travel grants and speaker's honoraria from Fresenius Medical Care and Janssen Cilag. He has received research funding from the German Research Foundation (DFG), the German Ministry of Education and Research (BMBF), the DAMP Foundation, Fresenius Medical Care, the EU Joint Programme–Neurodegenerative Disorders (JPND), and the European Social Fund (ESF). W.N. is a founder, shareholder, and scientific lead of Quantib BV. The other authors report no conflicts of interest or financial interests related to this work.
References
- 1.Alyass A., Turcotte M., Meyre D. From big data analysis to personalized medicine for all: challenges and opportunities. BMC Med genomics. 2015;8:33. doi: 10.1186/s12920-015-0108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medland S.E., Jahanshad N., Neale B.M., Thompson P.M. Whole-genome analyses of whole-brain data: working within an expanded search space. Nat Neurosci. 2014;17:791. doi: 10.1038/nn.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams H.H.H., Adams H., Launer L.J., Seshadri S., Schmidt R., Bis J.C. bioRxiv; 2016. Partial Derivatives Meta-analysis: Pooled Analyses When Individual Participant Data Cannot Be Shared; p. 038893. [Google Scholar]
- 4.Roshchupkin G.V., Adams H.H.H., Vernooij M.W., Hofman A., VanDuijn C.M., Ikram M.A. HASE: Framework for efficient high-dimensional association analyses. Scientific Rep. 2016;6:36076. doi: 10.1038/srep36076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wardlaw J.M., Smith E.E., Biessels G.J., Cordonnier C., Fazekas F., Frayne R. STandards for ReportIng Vascular changes on nEuroimaging (STRIVE v1). Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 2013;12:822–838. doi: 10.1016/S1474-4422(13)70124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.METACOHORTS Consortium. Dichgans M., Wardlaw J., Smith E., Zietemann V., Seshadri S., Sachdev P. METACOHORTS for the study of vascular disease and its contribution to cognitive decline and neurodegeneration: An initiative of the Joint Programme for Neurodegenerative Disease Research. Alzheimer's Demen. 2016;12:1235–1249. doi: 10.1016/j.jalz.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winkler T.W., Day F.R., Croteau-Chonka D.C., Wood A.R., Locke A.E., Mägi R. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9:1192. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Folk M., Heber G., Koziol Q., Pourmal E., Robinson D., in Proceedings of the EDBT/ICDT . (ACM); 2011. Workshop on Array Databases; pp. 36–47. [Google Scholar]
- 9.Roshchupkin G.V., Adams H.H., van der Lee S.J., Vernooij M.W., van Duijn C.M., Uitterlinden A.G. Fine-mapping the effects of Alzheimer's disease risk loci on brain morphology. Neurobiol Aging. 2016;48:204–211. doi: 10.1016/j.neurobiolaging.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 10.van der Lee S.J., Roshchupkin G.V., Adams H.H., Schmidt H., Hofer E., Saba Y. Gray matter heritability in family based and population based studies using voxel based morphometry. Hum Brain Mapp. 2017;38:2408–2423. doi: 10.1002/hbm.23528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahmud S., Iqbal R., Doctor F. Cloud enabled data analytics and visualization framework for health-shocks prediction. Future Generation Computer Syst. 2016;65:169–181. [Google Scholar]
- 12.Moskowitz A., McSparron J., Stone D.J., Celi L.A. Preparing a new generation of clinicians for the era of big data. Harv Med student Rev. 2015;2:24. [PMC free article] [PubMed] [Google Scholar]
- 13.Brody J.A., Morrison A.C., Bis J.C., O'Connell J.R., Brown M.R., Huffman J.E. Analysis commons, a team approach to discovery in a big-data environment for genetic epidemiology. Nat Genet. 2017;49:1560. doi: 10.1038/ng.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roski J., Bo-Linn G.W., Andrews T.A. Creating value in health care through big data: opportunities and policy implications. Health Aff. 2014;33:1115–1122. doi: 10.1377/hlthaff.2014.0147. [DOI] [PubMed] [Google Scholar]
- 15.Auffray C., Balling R., Barroso I., Bencze L., Benson M., Bergeron J. Making sense of big data in health research: towards an EU action plan. Genome Med. 2016;8:71. doi: 10.1186/s13073-016-0323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]