Abstract
Background
Cancer cells use specific V-ATPase subunits to activate oncogenic pathways. Therefore, we investigated V-ATPase deregulation in aggressive gliomas and associated signaling.
Methods
V-ATPase genes expression and associated pathways were analyzed in different series of glioma available from public databases, as well as in patients' cohort. Activation of pathways was analyzed at gene and protein expression levels. A genetic model of glioma in Drosophila melanogaster and mice with GBM patients-derived orthotopic xenografts were used as in vivo models of disease.
Findings
GBM and recurrent gliomas display a specific V-ATPase signature. Such signature resolves the heterogeneous class of IDH-wild type lower-grade gliomas, identifying the patients with worse prognosis independently from clinical and molecular features (p = 0·03, by Cox proportional-hazards model). In vivo, V-ATPase subunits deregulation significantly impacts tumor growth and proliferation. At the molecular level, GBM-like V-ATPase expression correlates with upregulation of Homeobox genes.
Interpretation
Our data identify a V-ATPase signature that accompanies glioma aggressiveness and suggest new entry points for glioma stratification and follow-up.
Fund
This work was supported by Fondazione Cariplo (2014–1148 to VV), Fondazione IRCCS Ca' Granda, and Fondazione INGM Grant in Molecular Medicine 2014 (to VV).
Keywords: V-ATPase, IDHwt/lower-grade glioma, Homeobox genes, Glioma stem cells
Research in context.
Evidence before this study
Molecular characterization of gliomas has significantly improved patients' stratification. However, IDH-wild type lower-grade gliomas remain a heterogeneous class with variable outcome. Recent evidence highlighted preferential use of specific vacuolar H + -ATPase pump subunits by cancer cells to activate oncogenic pathways and overexpression of specific subunits in cancer stem cells.
Added value of this study
We show that the expression of specific V-ATPase subunits by gliomas determines cancer aggressiveness in vivo and predicts patient prognosis, also in IDH-wild type lower-grade gliomas. The oncogenic V-ATPase profile associates with homeobox-containing genes overexpression.
Implications of all the available evidence
Evaluation of the level of expression of selected V-ATPase subunits in IDH-wild type lower-grade gliomas could complement routine molecular characterization to identify patients with most aggressive forms of the disease. Moreover, our data suggest that V-ATPase could be a novel interesting therapeutic target in a fraction of gliomas.
Alt-text: Unlabelled Box
1. Introduction
The vacuolar ATPase (V-ATPase) is a multisubunit proton pump that plays a role in multiple processes in eukaryotic cells. It comprises a membrane-embedded V0 sector, which regulates proton permeability, and an enzymatic V1 ATPase sector. Regulated assembly of the V1 sector on the V0 sector, along with modulation of ATPase activity, are the main determinants of pump efficiency. The core function of V-ATPase is acidification of endosomes and lysosomes, two organelles crucial for proteostasis and metabolism of cellular nutrients. In addition, in bone, kidney and gut cells, a plasma membrane localized V-ATPase acidifies the extracellular milieu and enables specific functions. The pump localization is regulated by the use of specific subunits alternative to the ones present in intracellular compartments [1].
Altered V-ATPase activity is associated with several human diseases [2]. In cancer, the role of V-ATPase is complex and likely context-dependent. Tumor cells are exquisitely sensitive to V-ATPase inhibition, indicating that V-ATPase activity is more limiting in cancer than in non-cancer cells. Upon V-ATPase inhibition, changes in cytosolic pH stabilize proapoptotic proteins, alter trafficking of extracellular nutrients, or reverse V-ATPase-induced drug resistance [3]. Overexpression of V-ATPase occurs in a number of cancer cell lines and tumor samples. V-ATPase may also be involved in modulating the activity of endocytic factors such as Rac1 and EGFR, which are crucial for cell motility [4]. Invasive cancer cells gain expression of V-ATPase at the plasma membrane, possibly to facilitate low pH-induced activation of proteases that modify the extracellular matrix. Delivery of V-ATPase specifically to the plasma membrane of breast cancer cells relies on overexpression of the V0A3 subunit, which is normally specific to osteoclasts; this suggests that changes in pump subunit composition support cancer-specific functions [5]. Despite the fact that the role of V-ATPase in cancer is diverse and insufficiently understood, the emerging evidence strongly suggests that V-ATPase could be a promising target for anticancer therapy.
Glial tumors are among the most difficult to profile and treat. The 2016 WHO classification of gliomas changed disease diagnosis and patient stratification markedly, shifting from a morphological view to a molecular-based classification [6]. In this new context, mutated isocitrate dehydrogenase 1 or 2 enzymes (IDHmut) are a major classifier of disease, as well as being key genetic events during gliomagenesis. IDH wild-type (IDHwt) tumors have a dismal outcome and are generally regarded as glioblastoma (GBM), even when they are histologically classified as lower-grade grade II and III gliomas. Nevertheless, a recent study shows that adult IDHwt lower grade gliomas (LGG/IDHwt) are prognostically and molecularly heterogeneous, meaning that not all are characterized by a poor, GBM-like outcome [7].
We showed previously that the G1 subunit of V-ATPase V1 sector (V1G1) is upregulated in primary stem cell-enriched cultures of GBM neurospheres (NS), and that higher expression of this subunit identifies glioma patients with shorter disease-free and overall survival independent of clinical or molecular variables [8]. These data suggest that changes in V-ATPase composition, and possibly activity, promote GBM aggressiveness and maintain the cancer stem cell niche. However, the significance of changes in V-ATPase subunits in GBM is unknown.
Therefore, to identify the mechanisms underlying glioma aggressiveness, along with novel and clinically relevant markers, we examined all V-ATPase subunits and associated signaling pathways, focusing on the less characterized LGG/IDHwt class.
2. Materials and methods
2.1. Patients' series
TCGA dataset: Data from the TCGA glioma cohort was downloaded from TCGA portal (Jay 2015 release; https://tcga-data.nci.nih.gov/docs/publications/lgggbm_2015/). Briefly, the lower-grade gliomas (LGG) and glioblastoma (GBM) dataset consisted in 1032 diffuse gliomas and 12,717 genes. Of these, we included in our study only tumors for which RNAseqV2 analysis was performed and with clinical and molecular annotation [9]. Cases with mutated IDH1 or 2 were considered IDHmutant (IDHmut). Therefore we included 481 patients, of whom 330 were LGG/IDHmut, 73 were de novo GBM (i.e. IDHwt cases only) and 76 were LGG/IDHwt cases. LGG/IDHmut and GBM cases constitute our training set, whereas LGG/IDHwt cases represent the test set. Detailed patients' information are reported in Tables S1,S2.
Primary and recurrent LGG cohort: This series consisted in 23 primary and treatment-naïve LGG patients and in 21 unmatched relapsed gliomas, which evolved from LGG disease to higher grade glioma. Patients' characteristics are detailed in Table S3. All patients received a gross total resection of the tumor at Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico between 2013 and 2016. None of the patients included in the study received a biopsy or neoadjuvant therapy. The study was approved by an Institutional Review Board (IRB#275/2013) and signed informed consent was obtained from all patients.
Matched primary and recurrent gliomas (n = 28; TCGA_LGG data source) were also analyzed through the GlioVis portal, (http://gliovis.bioinfo.cnio.es/) for V-ATPase expression.
Gravendeel dataset: Gene expression data (19,944 genes, Affymetrix GeneChip Human Genome U133 Plus 2.0 Array) and clinicopathological correlates from LGG/IDHmut (n = 72), GBM (n = 159) and LGG/IDHwt (n = 41) cases [10,11] were retrieved from the GlioVis portal. Only IDH1 mutation status was available for this dataset. Grade I, pilocytic astrocytomas (n = 4), were used only for heatmap generation and were excluded from ROC analysis. As for the TCGA cohort, LGG/IDHmut and GBM constituted the training set (Table S4), whereas the LGG/IDHwt cases constituted the validation set (Table S5).
2.2. Primary, patients-derived or commercial cell cultures
GBM neurospheres (NS, n = 33) or differentiated cultures were obtained from fresh tumor tissue of GBM patients (Table S6) as previously described [8]. All patients were enrolled at the Division of Neurosurgery and glioma was diagnosed at the Division of Pathology of the Fondazione IRCCS Ca' Granda-Ospedale Maggiore Policlinico (Milan, Italy). All cases were reviewed and graded by senior pathologist (SB) according to the new classification of the World Health Organization [6]. Briefly, tissues underwent enzymatic and mechanical dissociation digestion in serum-free media using the Tumor Dissociation kit (Miltenyi). Then, cells suspensions were washed twice with HBSS (Gibco-Invitrogen, Thermo Fisher Scientific, Waltham, MA USA) and either seeded in NeuroCult (NC) media supplemented with growth factors (Proliferation Supplement, bFGF and EGF, all from Voden) to promote growth of stem cells (undifferentiated condition) or in RPMI supplemented with 10% Exo-FBS (System Biosciences) to promote cell differentiation.
2.3. DNA purification and targeted sequencing
We analyzed the mutation status of 28 cancer-related genes in our primary NS by targeted sequencing using a custom designed panel (n. of amplicons: 657; coverage 99%; Table S7). Briefly, genomic DNA was purified from 8 NS with high (namely NS#27,91,92,97) or low (namely NS#51,53,88,130) V-ATPase G1 expression using the DNeasy Blood & Tissue Kits (Qiagen, Hilden, Germany). Libraries for Illumina sequencing were constructed from 100 ng of total DNA with the TruSeq Custom Amplicon Low Input Library Prep Kit v2 (Illumina, San Diego, CA, USA). Paired-end sequencing (2 × 150) was then performed on an Illumina miSeq instrument. Reads were processed and aligned against the GRCh37Hg19 human reference genome by the MiSeqReporter software (Illumina). Variant calling file (VCF) for each sample were then generated and imported in Variant Studio (Illumina) for variant annotation and filtering using the ANNOVAR package (http://annovar.openbioinformatics.org/en/latest/). Thresholds for somatic mutation calling were: an allele depth value ≥30 reads; a variant frequency > 0·03; the variant should not be detected in >5% of healthy population (available from ExAC portal; http://exac.broadinstitute.org/) [12].
2.4. Generation of GBM patients' derived orthotopic mouse model
Animal experiments were carried out in compliance with the institutional guidelines for the care and use of experimental animals (European Directive 2010/63/UE and the Italian law 26/2014), authorized by the Italian Ministry of Health and approved by the Animal Use and Care Committee of the University of Milan. Female NOD/SCID mice (7–8 weeks of age, Envigo, Huntingdon, UK) were kept in the appropriate cages in an environment of 23 ± 1 °C and 50 ± 5% humidity, with a 12 h light/dark cycle and fed ad libitum. The orthotopic murine model was obtained by stereotaxic injection (coordinates: 1.5 mm lateral to the bregma, 0 mm behind and 3·0 mm ventral to the dura) of 1 × 10^5 patients derived NS stably transduced with a luciferase construct in 2 μl of phosphate-buffered saline (PBS) as described [13]. Following surgery, mice were monitored for recovery until complete awakening. For bioluminescence imaging, mice were anaesthetized with mixture of tiletamine/zolazepam (40 mg/kg, Zoletil) and xylazine (8 mg/kg, Xilor), and then 150 mg/kg of luciferin (Beetle Luciferin Potassium Salt, Promega) was injected intraperitoneally. After biodistribution, bioluminescence signal (BL) was acquired with the IVIS SPECTRUM/CT instrument (Perkin Elmer), always using the same parameters. Finally, all images were scaled and quantified by applying Regions of Interest (ROI) on BL signals using the Living Image Software. Data are expressed as BL counts. All animals were sacrificed at day 109 and their brain was recovered for histological examination.
2.5. Drosophila melanogaster experiments
Fly strains were kept and raised in vials containing standard food medium. All crosses were performed at 25 °C. To assess larval brain overgrowth phenotypes, larvae were grown for 120–140 h, and wandering 3rd instar larvae were selected for dissection. The genotypes of the Drosophila lines used in the study are: a); Repo-Gal4, UAS-CD8GFP, TM2; b) UAS-Pi3K92E-CAAX; Repo-Gal4, UAS-CD8GFP, UAS-ΔhEGFR,TM6C TubGal-80 (kindly provided by Renee Read); c) UAS-VhaPPA1–1 RNAi (v47188, from the Vienna Stock Center). The immunofluorescence analysis was performed as follows: larval brains were fixed using 4% PFA for 30 min at room temperature. Permeabilization was performed with 1% Triton X-100 diluted in PBS, for 30 min. Samples were blocked in 4% BSA, 0·1% Triton X-100 diluted in PBS for 30 min at room temperature. The primary antibody anti-GFP (1:1000; 13,970, Abcam) was incubated on at 4 °C. The Alexa Fluor 488-conjugated (1:500; A-11039, Thermo Fisher Scientific) secondary antibody was incubated at RT for 2 h. Tissues were mounted in glycerol and images were taken with a TCS SP5 microscope (Leica, Heidelberg, Germany) using 20×/NA 0·5, oil-immersion lenses and measurements were carried out with ImageJ software (https://imagej.nih.gov/ij/). To extract RNA from Drosophila L3 larval brains we used Maxwell RSC simplyRNA Tissue Kit and the Maxwell RSC Instrument. The concentration of extracted RNA was measured using the NanoDrop 1000 Spectrophotometer. Complementary DNA (cDNA) synthesis was performed SuperScript VILO cDNA Synthesis Kit and qPCR was carried out on the ABI/Prism 7900 HT Sequence Detector System (Thermo Fisher Scientific). The following primers (5′-3′) were selected from the Universal Probe Library (Roche): VhaPPA1–1_fwd: 5′ atcttcggttcggccatc 3′ VhaPPA1–1_rev: 5′ ataatggagtggcgaaggac 3′; RPL32_fwd: 5′ cggatcgatatgctaagctgt 3′ RPL32 rev: 5′ cgacgcactctgttgtcg 3′.
2.6. RNA purification and genes expression analyses
Total RNA was purified from cell cultures using Master Pure RNA purification kit (Epicentre Biotechnologies, Illumina; Madison, WI, USA) followed by DNA digestion with DNase I, Amplification Grade (Thermo Fisher Scientific). Then, 300 ng of DNA-free total RNA from cell cultures were reverse transcribed using the SuperScript II Reverse Transcriptase, random hexamers and TaqMan assays (listed in Table S8) (all from Thermo Fisher Scientific) in 20 μl or reaction mix. Targets gene expression was quantified by qPCR using TaqMan assays together with the reference genes β2microglobulin and/or 18S RNA using the 2^(−Delta Ct) formula. Then, targets relative quantities (RQ) were median normalized and log2-transformed.
2.7. Antibodies and reagents
Cathepsin L (941) was from ImmunoChemistry Technologies (Bloomington, MN, USA). The following primary antibodies were used for immunohistochemistry, immunofluorescence, and immunoblotting (IB): V-ATPase G1 (16143–1-AP, Proteintech), V-ATPase V0A2 (SAB2100187, Sigma Aldrich), Vinculin (V9131, Sigma Aldrich), Ki67 (ab15580, Abcam), Podoplanin (D2–40; Ventana Medical Systems, Roche), MMP2 (10373–2-AP, Proteintech), POU3F2 (12,137, Cell Signaling), HOXA10 (TA590263, Origen), HOXA7 (ab211521, Abcam), V-ATPase G2 (25316–1-AP, Proteintech).
2.8. Immunofluorescence and immunohistochemistry
For immunofluorescence experiments, NS were cytospinned on charged slides for 3 min at 900 rpm (Thermo Scientific, Waltham, MA, USA). Cells were first fixed in PFA 4% for 15 (monolayer cultures) or 30 min (NS), quenched for green auto-fluorescence using glycine 20 mM for 20 min and then permeabilized with Triton 0·5% for 30 min at room temperature. Then, samples were saturated with BSA 10% for 1 h at RT. Primary antibodies (1:100 in PBS-BSA 10%) were incubated overnight at 4 °C whereas secondary antibodies (1:1000) were incubated for 1 h at room temperature. Finally, nuclei were stained with Hoechst 3342 (Cell Signaling) for 5 min at RT and slides were mounted using the Prolong reagent (ThemoFisher Scientific). For negative controls, one slide per target protein was incubated with all reagents but the primary antibody. Confocal images were generated with a Leica TCS SP5 Confocal microscope (Leica Microsystems) with a magnification of 40×, and z-stack images were captured every 0·46 μm. Fluorescence quantification was calculated on full cell stacks after setting the threshold on control samples, and the mean intensity of the fluorochrome was calculated using ImageJ software. Immunohistochemistry (IHC) on formalin-fixed paraffin-embedded samples was performed using a Ventana Benchmark instrument and the Ultraview DAB or Red Detection kits as described [8].
2.9. Immunoblotting
Protein extracts were prepared in RIPA buffer supplemented with phosphatase and protease inhibitors (Roche). After lysis, samples were sonicated five times on ice (Bioruptor Plus sonication device, Diagenode), pelleted at 15·000rcf to remove debris and quantified using microBCA kit (Thermo Fisher Scientific). Proteins (40 μg) were boiled for 5 min at 95°, resolved by SDS-PAGE, transferred to nitrocellulose membrane, blocked in 5% not-fat powdered milk in PBS-T (1% Tween-20) and probed with antibodies. Proteins bands were then visualized using the Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare) and the LAS-3000 imager (Fujifilm).
2.10. Statistical analyses
Gene expression data filtering: For RNASeq data, we used previously normalized counts available from the TCGA portal. We included transcripts whose log2 expression level (normalized counts) was >5 in the top tenth percentile of the patients dataset (n = 7321 out of 12,717, 58%). For the Gravendeel dataset (Affymetrix platform) we used previously normalized and log2 transformed data available in GlioVis portal.
Gene expression data visualization: Transcripts' levels of V-ATPase subunits and of differentially expressed genes were visualized in gliomas using the ComplexHeatmap package available in Bioconductor (https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html). Principal component analysis (PCA) plots were generated in R (https://cran.r-project.org/) to visualize gliomas distribution according to specific genes signatures (V-ATPase or differentially expressed genes as indicated in figures' legends). Notched box-and-whiskers plot (MedCalc software) was used to show expression of homeobox genes in Gravendeel LGG/IDHwt cases belonging to the two V-ATPase classes. Briefly, all data points are shown as dots along with the central box (values from the 25 to 75 percentile), the middle line (median) and the extremes (min-to-max values) unless otherwise specified.
Identification of prognostic V-ATPase or “other” genes signatures: All V-ATPase transcripts expressed above the threshold (n = 15) were analyzed for differential expression in LGG/IDHmut and GBM cases of the TCGA training series using the Kolmogorov–Smirnov test and computing the fold-change (Log2Ratio, L2R) between the two classes. The accuracy of the data was assessed by a permutation-based estimate of the FDR. For each pair of chosen Kolmogorov–Smirnov p values and FC thresholds, the FDR was computed reshuffling 1000 times. Genes with an adj p < 10–5 and a L2R ≥ |0·3| were considered significant (n = 8). Then, to identify which V-ATPase gene or set of genes best discriminated LGG/IDHmut from GBM cases (training set) we performed a Receiver Operating Characteristic (ROC) analysis survey with significantly different V-ATPases or the combined top 3 or top 4 down- or u-regulated subunits. For cut-off identification the Youden's J statistic was used as described [8].
The adj p < 10–5 and a L2R ≥ |0·5| was used when the whole transcriptome (n = 3417) was queried for genes differentially expressed between the two gliomas' classes and for the Gravendeel series of gliomas. ROC analyses were performed using the ROCR package available within the R environment (http://rocr.bioinf.mpi-sb.mpg.de/). The identified cut-offs (J index) from the best ROCs were then applied to categorize the LGG/IDHwt patients as LGG/IDHmut-like or GBM-like.
Survival analysis: The impact of clinicopathological or molecular variables or of the V-ATPase classification on overall survival status of LGG/IDHwt patients was analyzed by univariate analysis using the Cox proportional-hazards regression model. Only variables with a p < 0·05 were included in the multivariate model. Kaplan-Meier survival curves were then generated with LGG/IDHwt patients sorted into the two V-ATPase classes. All analyses were performed using MedCalc software.
Identification and functional annotation of genes associated to V-ATPase signatures: Genes associated with each of the V-ATPase class were searched in LGG/IDHwt series and then verified in LGG/IDHmut and GBM cases. Genes were compared using the Kolmogorov–Smirnov test followed by a permutation-based estimate of the FDR. A gene was considered differentially expressed if with an FDR adjusted p < 10–5 and a L2R ≥ |2|. Lists of genes enriched in the V-ATPase LGG/IDHmut-like or GBM-like class were created and analyzed using DAVID functional annotation tool (https://david.ncifcrf.gov/). Networks of top pathways were generated using ConsensusPathDB (http://cpdb.molgen.mpg.de/) selecting gene set analysis/enrichment analysis terms. The search was performed in all databases.
Analysis of functional experiments: Data from functional experiments were imported in GraphPad Prism software (La Jolla, CA, USA) and analyzed using the non-parametric Mann-Whitney U and Kruskal-Wallis tests for two-sample or more than two-sample comparisons, respectively. Data are presented as mean ± SEM and experiments were performed at least three times and in technical duplicate unless otherwise specified. In particular, experiments involving patients' derived NS were performed with five V1G1High- and five V1G1Low- NS in triplicate. Categorical data were analyzed using chi square test. P values <0·05 were considered statistically significant.
3. Results
3.1. GBM are characterized by expression of specific V-ATPase pump subunits
Previously, we showed that V1G1 is enriched in primary cultures of GBM neurospheres (NS), and that higher expression of this subunit identifies glioma patients with shorter disease-free and overall survival, independent of other clinical or molecular variables. To confirm our previous data on differential V-ATPase subunit expression [8], we first assessed the expression of V1G1 NS and found that it correlates with expression of genes related to stemness (Fig. S1a), ability of primary GBM cultures to form NS with high proliferative activity (Fig. S1b), increased expression of the proteolytic enzyme MMP2 (Fig. S1c) and of cathepsin L activity (Fig. S1d).
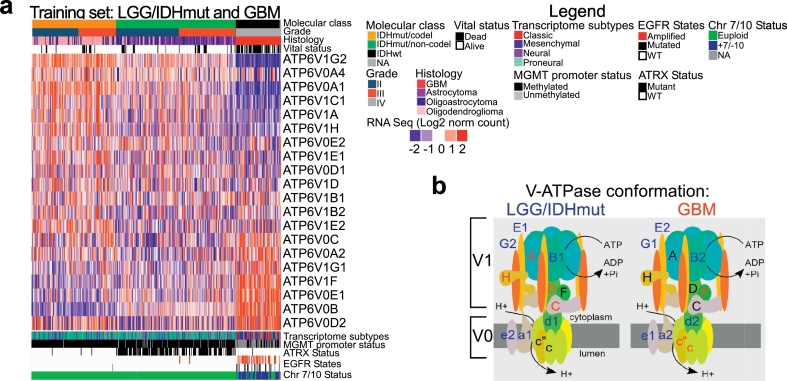
Interestingly, no specific mutation in key glioma genes was associated with V1G1 overexpression in NS (Table S9). We then sought to understand which V-ATPase subunits are deregulated in aggressive GBM, when compared with indolent IDH-mutant lower grade gliomas (LGG/IDHmut). Using data available from the TCGA database, we analyzed expression of V-ATPase genes (training set; Fig. 1a). As hypothesized, V-ATPase subunits varied greatly between the two glioma types. Interestingly, we could observe an alternative expression of pump subunits by aggressive GBM relative to LGG. GBM expressed ATP6V1G1 (encoding V1G1) rather than ATP6V1G2 (V1G2); ATP6V0A2 (V0A2) rather than ATP6V0A1 (V0A1) or ATP6V0A4 (V0A4); and ATP6V0E1 (V0E1) rather than ATP6V0E2(V0E2). In addition, some core and regulatory subunit genes, such as ATP6V1A (V1A), ATP6V1H (V1H), and ATP6V1C (V1C), were downregulated in GBM, whereas ATP6V0B (V0B) was upregulated (Fig. 1a). These changes in gene expression denoted glioma type-specific pump configurations (Fig. 1b).
Fig. 1.
V-ATPase pump conformation distinguishes aggressive gliomas from indolent LGG a) Expression (RNAseq data) of genes encoding all V-ATPase pump subunits (n = 20) in primary low-grade/IDH-mutated gliomas (LGG/IDHmut; n = 330) and glioblastoma (GBM; n = 75) from the TCGA dataset (training set). b) Cartoon showing V-ATPase pump conformation in LGG/IDHmut or GBM based on gene expression data determined in a.V0B encodes for subunit c”.
3.2. Deregulation of V-ATPase affects gliomagenesis in vivo
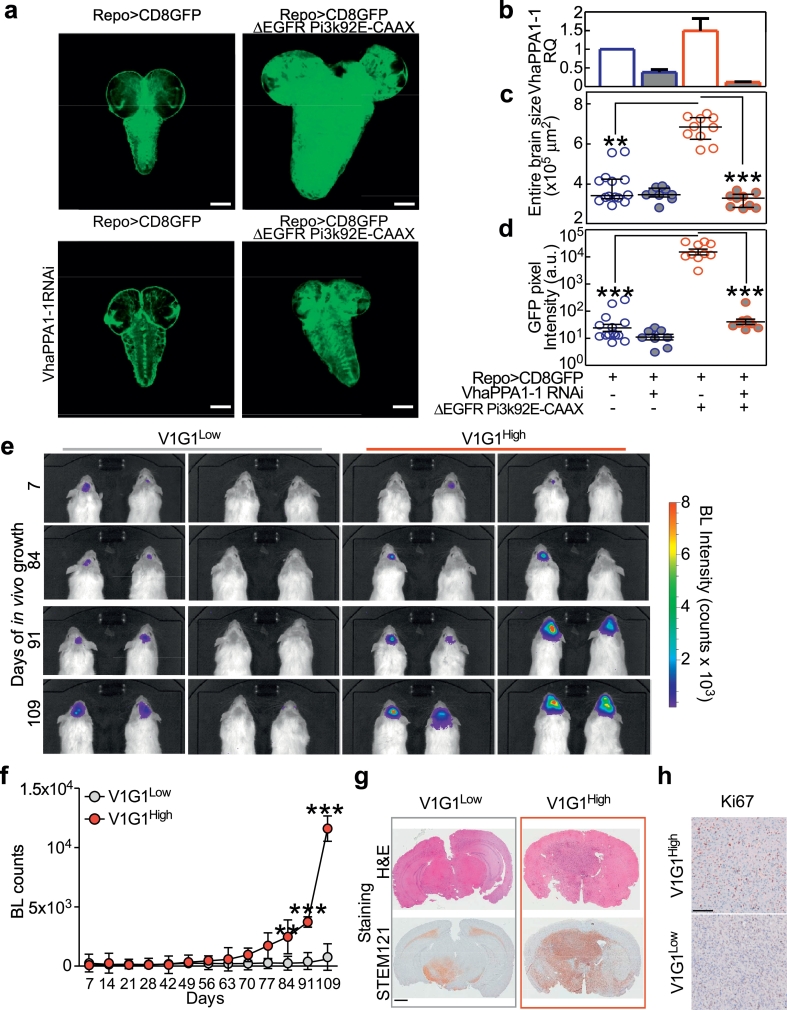
To test whether V-ATPase activity is required to support gliomagenesis in vivo, we first used a Drosophila glioma model based on overexpression of constitutively active forms of Drosophila PI3K (Pi3K92E-CAAX [14];) and of the human Epidermal Growth Factor Receptor (ΔEGFR) in larval glial cells (Fig. 2a). In this model, we found that down-regulation of V-ATPaseV0 c”, encoded by VhaPPA1–1 (the fly homolog of ATP6V0B;Fig. 2a,b) reduced tumor size up to control levels without affecting growth on an otherwise wild-type genetic background (Fig. 2c, d).
Fig. 2.
V-ATPase levels determine glioma growth in vivo. a–d) Down-regulation of VhaPPA1–1 prevents tumor growth in brains of Drosophila larvae. a) Single confocal medial sections of whole brains from 3rd instar larvae. Dorsal view, anterior up. Glial tissue is labeled by a membrane-targeted form of GFP (CD8GFP; green) expressed under the glial specific promoter of reverse polarity (repo) gene. Note the increase in CD8GFP expression and brain size in larvae expressing ΔEGFR and Pi3K92E-CAAX under repo (repo> CD8GFP ΔEGFR Pi3K92E-CAAX), which is prevented by downregulating VhaPPA1–1 (repo> CD8GFP ΔEGFR Pi3K92E-CAAX VhaPPA1–1). Scale bar, 100 μm. b) VhaPPA1–1 mRNA was measured in brains of 3rd instar larvae by qPCR. Bars, means ± s.d. of three independent experiments. c,d) Surface area (c) or GFP pixel intensity (d) quantification of brain sections (>6 brains per genotype). Bars, mean ± SEM; **, p < 0·01; ***, p < 0·001 (Kruskal-Wallis test with Dunn's post-test). e, f) GBM-derived neurospheres (NS) with low or high levels of V-ATPase V1G1 were stably transfected with a luciferase construct and injected intracranially into nude mice (n = 8 per group). Tumor growth was monitored over time (up to Day 109) by measuring luciferase emission (BL counts). **, p < 0·01; ***, p < 0·001 (Kruskal-Wallis test with Dunn's post-test). Dots, mean ± SEM (n = 13 per group). g) Representative hematoxylin and eosin (H&E) and STEM121 stains of brains from mice injected with V-ATPase V1G1 low- (left panels) or high- (right panels) NS at the time of harvesting (Day 109). h) Glioma cell proliferation (Ki67 staining) in the brains of indicated mice. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Next, we confirmed these data in an orthotopic mouse model of glioma. Importantly, we found that NS showing endogenous high expression of V-ATPaseG1 (V1G1High) were more tumorigenic in vivo than NS V1G1Low (Fig. 2e, f). Gliomas generated from V1G1High NS completely invaded the normal brain parenchyma of mice (as shown by STEM121 staining; Fig. 2g) and had a higher proliferation index (Ki67; Fig. 2h) than tumors derived from NS V1G1Low. Overall, these data indicate that the levels of V-ATPase expression affect glioma growth in vivo.
3.3. A specific V-ATPase subunits expression pattern characterizes recurrent LGG and is prognostic in IDHwt low-grade gliomas
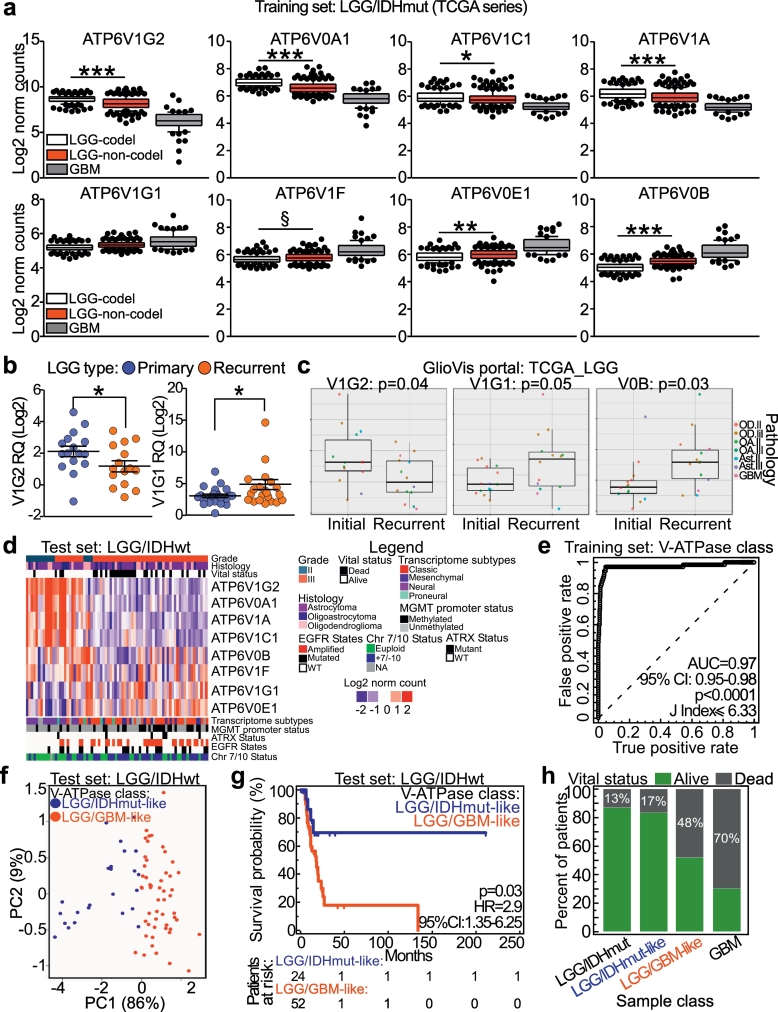
We then testes which V-ATPase subunits were differentially expressed between LGG/IDHmut and GBM cases. Statistical analysis revealed that eight V-ATPase genes were significantly deregulated in LGG/IDHmut, when compared with GBM (Table S10; Fig. S2). Considering only LGG/IDHmut cases, V1G2, V0A1, V1C1, and V1A were also downregulated in cases without 1p/19q co-deletion, whereas V1F and V0B were upregulated (Fig. 3a).
Fig. 3.
V-ATPase pump conformation resolves the heterogeneous class of LGG/IDHwt. a) Eight V-ATPase subunits were differentially expressed in LGG/IDHmut compared with GBM (p < 0·00001, and absolute Log2 ratio value >0·3; see Supplemental Table 10). Moreover, differential expression of these subunits was examined in LGG/IDHmut samples with respect to the presence/absence of the 1p19q co-deletion. ***, p < 0·0001; **, p = 0·001; *, p = 0·04; §, p = 0·006 (Kolmogorov–Smirnov test). Data are presented as box plots (lower, median and upper quartile) with whiskers indicating the 10 and 90 percentiles. c) Expression of V-ATPase subunit genes ATP6V1G2 and ATP6V1G1 in primary or recurrent LGG samples. Each dot represents one sample. *, p = 0·04 (Mann-Whitney U test). d) Processed RNAseq data from matched initial and recurrent disease of 28 LGG were retrieved from the GlioVis portal and V-ATPase subunit expression was examined. Significance of pairwise comparisons was obtained by Tukey's Honest Significant test. Data are presented as box plots (lower, median and upper quartile) with whiskers indicating the minimum and maximum values. e) Heatmap showing eight V-ATPase subunits in LGG/IDHwt patients (n = 76; TCGA dataset). f) ROC curve analysis coupled with the Youden's J statistic was used to identify the best classifiers for sorting LGG/IDHmut from GBM cases (training set). Simultaneous down-regulation of ATP6V1G2, ATP6V1C1, and ATP6V0A1 provided the best classification (sensitivity, 97%; specificity, 95%; see Table S11). g) LGG/IDHwt cases were annotated as LGG/IDHmut-like or GBM-like according to the ROC-generated cut-off determined in f. Principal component analysis (PCA) of V-ATPase genes in LGG/IDHwt cases from the two V-ATPase classes. h) Survival analysis of LGG/IDHwt patients according to V-ATPase class. i) Percentage of dead patients according to pathomolecular (LGG/IDHmut and GBM) and V-ATPase class (for LGG/IDHwt cases). p < 0·0001 (Chi-square test). Mean survival times were 26·7, 19·2, 13·9 and 11·1 months, respectively.
Next, we analyzed these eight pump subunits in our series of primary or relapsed LGG cases (Fig. 3b), and in LGG cases for which expression data regarding matched primary and recurrent disease were available in the GlioVis portal (Fig. 3c) [10]. In both analyses, relapsed tumors showed down-regulation of V1G2 and upregulation of V1G1 (Fig. 3b, c). Furthermore, imbalances between the two catalytic subunits in terms of sphere-forming efficiency were observed in GBM tissues used to generate primary cultures (Fig. S2a), and between primary GBM cultures maintained in a differentiated or undifferentiated (NS) state (Fig. S2b). These data suggest the existence of a striking tumor type-specific V-ATPase signature.
A gray area in glioma clinical management is the identification of patients with a GBM-like outcome among those with IDHwt non-co-deleted LGG (LGG/IDHwt). Although previous studies categorized most LGG/IDHwt tumors as other types based on complex molecular analysis [15], their pathobiology, clinical behavior, and, consequently, prognosis remain highly variable.
Based on the data obtained from GBM and relapsed gliomas, we analyzed the eight V-ATPase subunits in LGG/IDHwt patients available from the TCGA database (n = 76; Fig. 3d). Consistent with the possibility that the LGG/IDHwt patient group is heterogeneous, we noted a clear difference in expression of pump subunits in some cases. Therefore, we used ROC curve analysis to examine combinations of V-ATPase subunits to identify those that better classify the training set. The best classifier was provided by a combination of V1G2, V0A1, and V1C1, all of which were downregulated in GBM (Table S11; Fig. 3e). LGG/IDHwt patients were then categorized into two classes, LGG/IDHmut-like or LGG/GBM-like, based on the ROC-generated cut-off value (Youden J index≤6·33; Table S11). Principal component analysis (PCA; Fig. 3f) performed using the three subunit signature showed that the distribution of LGG/IDHwt samples was influenced significantly by V-ATPase classification, with 95% of sample variance explained by the first two components. Although V-ATPase classification of LGG/IDHwt patients was associated with a number of clinico-pathological and molecular features, such as tumor grade, derangement of chromosomes 7/10, methylation and transcriptome subtypes (Table S12), stratification by V-ATPase expression distinguished LGG/IDHwt patients with a poor prognosis (Fig. 3g), independent of clinical variables (Table 1 and Table S13). The ability of V-ATPase-based classification to identify patients within the LGG/IDHwt cohort with a poorer prognosis was also evident by looking at the percentage of deceased patients in the LGG/IDHmut-like and LGG/GBM-like groups, that was 17% and 48%, respectively (p < 0·0001 by Chi-square test; Fig. 3h).
Table 1.
Multivariate analysis of LGG/IDHwta.
| Covariate | p | HRb | 95% CIc |
|---|---|---|---|
| Age diagnosis | 0·002 | 1·07 | 1·0–1·1 |
| V-ATPase Class (LGG/IDHmut-like) | 0·03 | 0·27 | 0·09–0·88 |
| LGmd (cluster 6) | 0·87 | 0·90 | 0·25–3·19 |
Overall model fit: p = .0008.
HR, Hazard Ratio.
CI, Confidence Interval.
Pan Glioma DNA Methylation Cluster (Ceccarelli M. Cell) [9].
To verify that identification of two classes of LGG/IDHwt patients with different prognoses was not a false-positive, and that other gene-based classifications were not as powerful as the V-ATPase-based classification, we repeated the analyses using as classifier the genes showing the greatest differential expression between LGG/IDHmut and GBM patients (n = 3417, L2R = p < 10−5; Fig. S3a), or transcripts significantly downregulated in GBM (n = 2035). Then, we calculated a ROC cut-off value that differentiated LGG/IDHwt patients into two groups (Table S11). We then performed PCA analysis to verify the accuracy of this classifier (called “Other genes”; Fig. S3b). In this case, sample groups in PCA were not as distinct as they were for the V-ATPase class (only 31% or 37% of sample variance was explained by the first two components; Fig. S3b). Most importantly, these two classifications were not prognostic (p = 0·08 and p = 0·25 by Log-Rank test; Fig. S3c, d).
Finally, we validated the three V-ATPase subunits' signature using a third series of gliomas for which molecular and histological classifications were available [11]. Again, we observed lower expression of V1G2, V0A1, and V1C1 in GBM than in the more indolent LGG/IDHmut cases (Fig. S4a). Expression of the three subunits in LGG/IDHwt and GBM cases was then used to build a ROC curve to identify a cut-off value for sorting LGG/IDHwt cases into the two V-ATPase classes. As before, the non-arbitrary Youden J Index was used (Fig. S4b). PCA analysis confirmed elevated accuracy of the classifier (89% of total sample variance explained by the first two components; Fig. S4c). Also, and most importantly, the V-ATPase classifier was prognostic for LGG/IDHwt patients (p = 0·0003, HR = 3·15, by Log-Rank test; Fig. S4d). Therefore, we conclude that down-regulation of ATP6V1G2, V0A1, and V1C1 subunits is critical to predict glioma aggressiveness.
3.4. The GBM-like V-ATPase profile is associated with de-differentiation and upregulation of homeobox genes
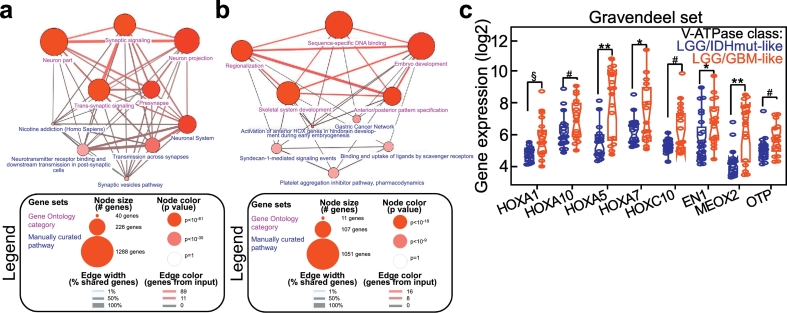
To gain insight into the molecular phenotype of LGG/IDHwt gliomas belonging to the two V-ATPase classes, we analyzed the gene signatures associated with the LGG/IDHmut-like or LGG/GBM-like classes (Fig. S5a, b). Three hundred and fifty-nine genes were differentially expressed between the two groups (Fig. S5a; Table S14), with the majority (n = 309, 86%) being downregulated. Interestingly, this transcriptional profile differentiated GBM from LGG/IDHmut cases (Fig. S5b). Functional annotation of this signature (Table S15) showed that genes related to neuronal processes and signaling were enriched in LGG/IDHmut-like cases (Fig. 4a). In sheer contrast, LGG/GBM-like cases were characterized by expression of genes involved in embryogenesis, cancer, and cell-cell signaling (Fig. 4b; Table S16). Interestingly, homeobox genes, mainly the HOX family, were significantly over-represented in LGG/GBM-like cases (38% and 24% of upregulated transcripts, respectively; Table S14).
Fig. 4.
The V-ATPase GBM-like signature associates with de-differentiation and upregulation of homeobox genes. a, b) DAVID analysis of pathways associated with LGG/IDHmut (a) or GBM-like (b) classes in LGG/IDHwt cases. The complete list of genes used to generate these plots is provided in Tables S15, S16. c) Heatmap showing epigenetic enzymes expression in LGG/IDHwt cases grouped according to the V-ATPase class. For each gene the p value (Kolmogorov-Smirnov test) is indicated. The distribution of homeobox genes expression in each sample is provided at the bottom. d) Selected homeobox genes were analyzed in an independent set of LGG/IDHwt patients grouped according to the V-ATPase class (Gravendeel series; n = 40). **, p = 0·001; *, p = 0·02; #, p = 0·03; §, p = 0·006; °, p = 0·04 (Mann-Whitney U test). Data are presented as notched box plots (lower, median and upper quartile) with whiskers indicating the minimum and maximum values. Each circle represents a sample.
Finally, homeobox genes deregulation was confirmed in a third series of gliomas (Gravendeel dataset; Fig. 4c).
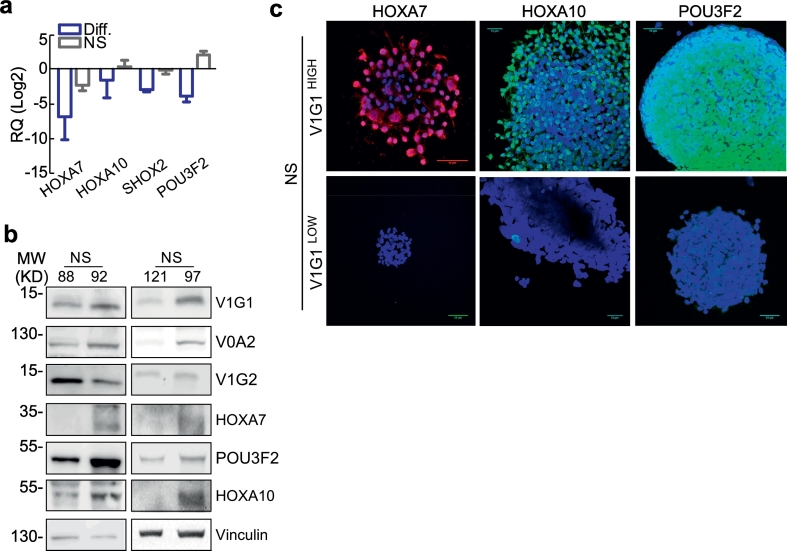
Homeobox genes are key players in glioma progression [16,17] and, most importantly, in transcriptional reprogramming of stem-like GBM cells [18] but the connection between homeobox genes and V-ATPase signaling was only partially explored in non-mammalian systems [19]. In mammalian systems, we observed that the neurodevelopmental homeobox transcription factor POU3F2 is regulated by the V-ATPase inhibitor Bafilomycin A1 [8]. Therefore, we examined expression of homeobox genes HOXA7, HOXA10, SHOX2, and POU3F2 in our patient-derived GBM primary cultures. All of these transcription factors were expressed at higher levels in GBM-derived NS than in differentiated cultures generated from NS (Fig. 5a). Consistent with our previous observation of specific V-ATPase subunit expression in aggressive glioma, the homeobox proteins were expressed at high levels in NS characterized by elevated V1G1 and low V1G2 levels (Fig. 5b, c and Fig. S6a–c).
Fig. 5.
NS with elevated V1G1 levels are characterized by upregulation of Homeodomain-containing genes. a) The indicated transcripts were analyzed by qPCR in primary NS or differentiated cultures. Bars, mean ± SEM (n = 4). b,) Representative NS cultures with high (NS92 and 97) or low (NS88 and 121) V1G1 expression were analyzed for the indicated proteins expression by western blotting. Vinculin was a loading control. c) The homeodomain proteins HOXA10, HOXA7 or POU3F2 were analyzed by immunofluorescence in NS with Low or High V1G1 levels. Scale bars, 50 μm.
4. Discussion
Here, we show that the most aggressive adult glioma, GBM, exhibits a peculiar expression profile of V-ATPase genes that implies use of pump subunits different from that used by indolent tumors, such as IDHmut/1p19q co-deleted LGG. Interestingly, this subunit signature is also present in relapsed disease, in gliomas enriched in cancer stem cells and with higher tumorigenic potential in vivo. From a clinical standpoint, such an alternative use of V-ATPase subunits can identify the patients that have a worse prognosis within the LGG/IDHwt class, a provisional entity in the current WHO classification. Our approach to stratify the LGG/IDHwt group was confirmed using different public datasets of patients. At the transcriptional level, the GBM-like V-ATPase signature correlates with overexpression of developmental genes, such as the homeodomain-containing transcripts HOXA7, HOXA10, and POU3F2. Recently, classification of adult glioma has moved from a pure histological assessment to a molecular-based one. Currently, glioma classification relies on determination of the mutational status of IDH1 and 2, followed by co-deletion of 1p/19q, loss of ATRX, and TP53 mutation. The presence of IDH mutations and 1p/19q co-deletion defines the least aggressive subtype of glioma, the oligodendroglioma. By contrast, wild-type IDH and ATRX genes, and absence of the 1p/19 co-deletion, are typical of GBM [6]. While this classification has significantly improved the stratification of adult LGG, the “LGG/IDHwt” group is not fully resolved and requires further investigation. Former studies show that some of these cases should be reclassified as GBM, but not all patients have a uniformly poor prognosis and a GBM-like outcome [7]. Therefore, a more refined characterization of LGG/IDHwt is important for patient clinical management after surgery. In this context, the sub-typing of LGG/IDHwt cases according to the V-ATPase class might support personalization of the therapeutic approach, directing only GBM-like patients to intensive therapeutic protocols similar to that followed by GBM patients, such as combination of both radiation and chemotherapy.
Previously, we reported that V-ATPase G1 is enriched in glioma stem cells and identifies glioma patients with a poorer prognosis [8]. Here, we expanded our analysis to all V-ATPase subunits and we found that LGG/IDHmut samples express V-ATPase subunits associated with a more differentiated, excitatory phenotype (e.g., V0A1 and V1G2), whereas GBM and GBM-like LGG/IDHwt cases show upregulation of those associated with a more undifferentiated phenotype (e.g., V0A2 or V1G1). In mammalian cells, subunit V0A controls targeting of V-ATPase to different subcellular destinations [[20], [21], [22]]. In human, V0A1 is associated mainly with synaptic vesicles, V0A4 is associated mainly with the plasma membrane of non-neuronal cells, whereas V0A2 is overexpressed on the plasma membrane of ovarian cancer cells where it confers resistance to chemotherapy [3]. Thus, changes in expression of V0A subunits may correlate with either the differentiation status, or with tumor-specific subcellular activities. V0A subunits are not the only ones we found modulated, thus additional consequences on V-ATPase biology can be envisaged. To this regard, the interaction between V1G and V1C is known to aid dissociation of the V1 sector from the V0 sector [20] and V1H inhibits the ATPase activity of the catalytic V1 sector when not assembled on the membrane-embedded V0 pore [23,24]. While the effects of alternative subunits use on pump activity have not been tested, overall our data imply that aggressive gliomas might profit from changes in pump assembly, intracellular localization, or ATPase activity to grow, invade, and resist therapy [3,[25], [26], [27], [28]]. Considering this, the preferential use of a certain subunit isoform by aggressive gliomas could be exploited to develop anti-cancer therapeutics that selectively target “oncogenic” V-ATPase in cancer cells. This hypothesis is supported by evidence from preclinical models of tumorigenesis showing that inhibiting cancer-associated V-ATPases is not lethal [2,[29], [30], [31]].
The finding that the GBM-like pump is a marker of poor prognosis for LGG/IDHwt patients independent of age, grade, and molecular profile (including the chr7/10 alterations) suggests that the pattern of V-ATPase subunit expression in these tumors might play a functional role in their biology, perhaps contributing to dictate the number of cancer stem cells within the tumor mass. In gliomas harboring a GBM-like V-ATPase, such hypothesis is consistent with upregulation of homeobox-containing genes. Although preliminary, our data show overexpression of homeobox genes in clinical series and in GBM patient-derived neurospheres with high expression of V1G1. Very few data that connect V-ATPase and homeobox gene signaling are available. We showed previously that expression of the neurodevelopmental core transcription factor POU3F2 was repressed after impairment of V-ATPase activity using BafA1 [8]. Further, POU3F2 has been previously demonstrated to be a fundamental reprogramming transcription factor for GBM stem cells [18], and elevated expression of the homeobox signature genes in subgroups of GBM [32] or in GBM-derived NS [33] has been documented. Recently, the HOXA locus was identified as a driver for chromosome 7 gains in IDH wild-type GBM, thereby promoting tumor proliferation and radioresistance [16,34]. In agreement with this, we found that chromosome 7 gains were more common in LGG/IDHwt cases expressing the GBM-like pump than in LGG/IDHmut-like cases. Nevertheless, chr7/10 alterations were not prognostic per se (Tables S12 and S13); therefore, this event may not completely explain overexpression of HOXA genes and tumor aggressiveness in LGG/IDHwt cases. Concluding, our data provide novel insights into gliomagenesis and uncover a novel mechanism by which GBM and GBM-like LGG might use these factors to favors de-differentiation towards an embryonic-like pluripotent state. Further studies are needed to disclose the mechanisms underpinning homeodomain genes and V-ATPase interplay.
Author contributions statement
A.T. and I.B. conceived and designed the experiments and prepared the tables/figures; A.T. performed bioinformatics analyses; I.B. conducted in vitro experiments; C.M., M.F. and L.O. performed in vivo experiments and analyzed the data; G.G. and A.M.S. performed qPCR, in vitro experiments and analyzed data; A.D.C., M.C. provided patients' samples and clinical data; S.B. reviewed patients' series and performed IHC experiments and analyzed the data; T.V. and V.V. drafted and wrote the manuscript; All authors reviewed the manuscript.
Declaration of interest
There are no conflicts of interest to disclose.
Acknowledgments
This work was supported by Fondazione Cariplo (2014-1148 to VV), Fondazione IRCCS Ca' Granda, and Fondazione INGM Grant in Molecular Medicine 2014 (to VV). T.V. acknowledges the support of AIRC investigator grant 20661 and Worldwide Cancer Research grant 18-0399. AMS and MF were supported by a Fellowship from the Doctorate School in Molecular and Translational Medicine of Milan University. We are thankful to Dr. Annamaria Morotti from the Department of Pathophysiology and Transplantation (University of Milan) for help with immunoblotting and to the INGM Imaging Facility for scientific and technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.01.052.
Contributor Information
Thomas Vaccari, Email: thomas.vaccari@unimi.it.
Valentina Vaira, Email: valentina.vaira@unimi.it.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Cotter K., Stransky L., McGuire C., Forgac M. Recent insights into the structure, regulation, and function of the V-ATPases. Trends Biochem Sci. 2015;40(10):611–622. doi: 10.1016/j.tibs.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stransky L., Cotter K., Forgac M. The function of V-ATPases in cancer. Physiol Rev. 2016;96(3):1071–1091. doi: 10.1152/physrev.00035.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulshrestha A., Katara G.K., Ginter J. Selective inhibition of tumor cell associated Vacuolar-ATPase 'a2' isoform overcomes cisplatin resistance in ovarian cancer cells. Mol Oncol. 2016;10(6):789–805. doi: 10.1016/j.molonc.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiedmann R.M., von Schwarzenberg K., Palamidessi A. The V-ATPase-inhibitor archazolid abrogates tumor metastasis via inhibition of endocytic activation of the Rho-GTPase Rac1. Cancer Res. 2012;72(22):5976–5987. doi: 10.1158/0008-5472.CAN-12-1772. [DOI] [PubMed] [Google Scholar]
- 5.Cotter K., Liberman R., Sun-Wada G. The a3 isoform of subunit a of the vacuolar ATPase localizes to the plasma membrane of invasive breast tumor cells and is overexpressed in human breast cancer. Oncotarget. 2016;7(29):46142–46157. doi: 10.18632/oncotarget.10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louis D.N., Perry A., Reifenberger G. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 7.Aibaidula A., Chan A.K., Shi Z. Adult IDH wild-type lower-grade gliomas should be further stratified. Neuro Oncol. 2017;19(10):1327–1337. doi: 10.1093/neuonc/nox078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Cristofori A., Ferrero S., Bertolini I. The vacuolar H+ ATPase is a novel therapeutic target for glioblastoma. Oncotarget. 2015;6(19):17514–17531. doi: 10.18632/oncotarget.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceccarelli M., Barthel F.P., Malta T.M. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman R.L., Wang Q., Carro A., Verhaak R.G., Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017;19(1):139–141. doi: 10.1093/neuonc/now247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gravendeel L.A., Kouwenhoven M.C., Gevaert O. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 12.Wang X., Teer J.K., Tousignant R.N. Breast cancer risk and germline genomic profiling of women with neurofibromatosis type 1 who developed breast cancer. Genes Chromosomes Cancer. 2018;57(1):19–27. doi: 10.1002/gcc.22503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo Dico A., Martelli C., Valtorta S. Identification of imaging biomarkers for the assessment of tumour response to different treatments in a preclinical glioma model. Eur J Nucl Med Mol Imaging. 2015;42(7):1093–1105. doi: 10.1007/s00259-015-3040-7. [DOI] [PubMed] [Google Scholar]
- 14.Read R.D., Fenton T.R., Gomez G.G. A kinome-wide RNAi screen in Drosophila Glia reveals that the RIO kinases mediate cell proliferation and survival through TORC2-Akt signaling in glioblastoma. PLoS Genet. 2013;9(2) doi: 10.1371/journal.pgen.1003253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuss D.E., Kratz A., Sahm F. Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol. 2015;130(3):407–417. doi: 10.1007/s00401-015-1454-8. [DOI] [PubMed] [Google Scholar]
- 16.Kurscheid S., Bady P., Sciuscio D. Chromosome 7 gain and DNA hypermethylation at the HOXA10 locus are associated with expression of a stem cell related HOX-signature in glioblastoma. Genome Biol. 2015;16:16. doi: 10.1186/s13059-015-0583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan R., Han L., Wang Q. HOXA13 is a potential GBM diagnostic marker and promotes glioma invasion by activating the Wnt and TGF-beta pathways. Oncotarget. 2015;6(29):27778–27793. doi: 10.18632/oncotarget.4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suva M.L., Rheinbay E., Gillespie S.M. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157(3):580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banreti A., Hudry B., Sass M., Saurin A.J., Graba Y. Hox proteins mediate developmental and environmental control of autophagy. Dev Cell. 2014;28(1):56–69. doi: 10.1016/j.devcel.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 20.Toei M., Saum R., Forgac M. Regulation and isoform function of the V-ATPases. Biochemistry. 2010;49(23):4715–4723. doi: 10.1021/bi100397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finnigan G.C., Cronan G.E., Park H.J., Srinivasan S., Quiocho F.A., Stevens T.H. Sorting of the yeast vacuolar-type, proton-translocating ATPase enzyme complex (V-ATPase): identification of a necessary and sufficient Golgi/endosomal retention signal in Stv1p. J Biol Chem. 2012;287(23):19487–19500. doi: 10.1074/jbc.M112.343814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breton S., Brown D. Regulation of luminal acidification by the V-ATPase. Physiology (Bethesda) 2013;28(5):318–329. doi: 10.1152/physiol.00007.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diab H., Ohira M., Liu M., Cobb E., Kane P.M. Subunit interactions and requirements for inhibition of the yeast V1-ATPase. J Biol Chem. 2009;284(20):13316–13325. doi: 10.1074/jbc.M900475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jefferies K.C., Forgac M. Subunit H of the vacuolar (H+) ATPase inhibits ATP hydrolysis by the free V1 domain by interaction with the rotary subunit F. J Biol Chem. 2008;283(8):4512–4519. doi: 10.1074/jbc.M707144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McConnell M., Feng S., Chen W. Osteoclast proton pump regulator Atp6v1c1 enhances breast cancer growth by activating the mTORC1 pathway and bone metastasis by increasing V-ATPase activity. Oncotarget. 2017;8(29):47675–47690. doi: 10.18632/oncotarget.17544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P., Chen H., Han L., Zou X., Shen W. Expression and role of V1A subunit of V-ATPases in gastric cancer cells. Int J Clin Oncol. 2015;20(4):725–735. doi: 10.1007/s10147-015-0782-y. [DOI] [PubMed] [Google Scholar]
- 27.Capecci J., Forgac M. The function of vacuolar ATPase (V-ATPase) a subunit isoforms in invasiveness of MCF10a and MCF10CA1a human breast cancer cells. J Biol Chem. 2013;288(45):32731–32741. doi: 10.1074/jbc.M113.503771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGuire C., Cotter K., Stransky L., Forgac M. Regulation of V-ATPase assembly and function of V-ATPases in tumor cell invasiveness. Biochim Biophys Acta. 2016;1857(8):1213–1218. doi: 10.1016/j.bbabio.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan N., Song L., Zhang S. Bafilomycin A1 targets both autophagy and apoptosis pathways in pediatric B-cell acute lymphoblastic leukemia. Haematologica. 2015;100(3):345–356. doi: 10.3324/haematol.2014.113324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Schwarzenberg K., Lajtos T., Simon L., Muller R., Vereb G., Vollmar A.M. V-ATPase inhibition overcomes trastuzumab resistance in breast cancer. Mol Oncol. 2014;8(1):9–19. doi: 10.1016/j.molonc.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kartner N., Manolson M.F. V-ATPase subunit interactions: the long road to therapeutic targeting. Curr Protein Pept Sci. 2012;13(2):164–179. doi: 10.2174/138920312800493179. [DOI] [PubMed] [Google Scholar]
- 32.Gallo M., Ho J., Coutinho F.J. A tumorigenic MLL-homeobox network in human glioblastoma stem cells. Cancer Res. 2013;73(1):417–427. doi: 10.1158/0008-5472.CAN-12-1881. [DOI] [PubMed] [Google Scholar]
- 33.Murat A., Migliavacca E., Gorlia T. Stem cell-related "self-renewal" signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 34.Cimino P.J., Kim Y., Wu H.J. Increased HOXA5 expression provides a selective advantage for gain of whole chromosome 7 in IDH wild-type glioblastoma. Genes Dev. 2018;32(7–8):512–523. doi: 10.1101/gad.312157.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2