Abstract
Background
Acute T-cell mediated rejection (TCMR) is usually indicated by alteration in serum-creatinine measurements when considerable transplant damage has already occurred. There is, therefore, a need for non-invasive early detection of immune signals that would precede the onset of rejection, prior to transplant damage.
Methods
We examined the RT-qPCR expression of 22 literature-based genes in peripheral blood samples from 248 patients in the Kidney Allograft Immune Biomarkers of Rejection Episodes (KALIBRE) study. To account for post-transplantation changes unrelated to rejection, we generated time-adjusted gene-expression residuals from linear mixed-effects models in stable patients. To select genes, we used penalised logistic regression based on 27 stable patients and 27 rejectors with biopsy-proven T-cell-mediated rejection, fulfilling strict inclusion/exclusion criteria. We validated this signature in i) an independent group of stable patients and patients with concomitant T-cell and antibody-mediated-rejection, ii) patients from an independent study, iii) cross-sectional pre-biopsy samples from non-rejectors and iv) longitudinal follow-up samples covering the first post-transplant year from rejectors, non-rejectors and stable patients.
Findings
A parsimonious TCMR-signature (IFNG, IP-10, ITGA4, MARCH8, RORc, SEMA7A, WDR40A) showed cross-validated area-under-ROC curve 0.84 (0.77–0.88) (median, 2.5th–97.5th centile of fifty cross-validation cycles), sensitivity 0.67 (0.59–0.74) and specificity 0.85 (0.75–0.89). The estimated probability of TCMR increased seven weeks prior to the diagnostic biopsy and decreased after treatment. Gene expression in all patients showed pronounced variability, with up to 24% of the longitudinal samples in stable patients being TCMR-signature positive. In patients with borderline changes, up to 40% of pre-biopsy samples were TCMR-signature positive.
Interpretation
Molecular marker alterations in blood emerge well ahead of the time of clinically overt TCMR. Monitoring a TCMR-signature in peripheral blood could unravel T-cell-related pro-inflammatory activity and hidden immunological processes. This additional information could support clinical management decisions in cases of patients with stable but poor kidney function or with inconclusive biopsy results.
Abbreviations
| ABMR | antibody mediated rejection |
| AR | acute rejection |
| AUC | area under the ROC curve |
| ATG | anti-thymocyte globuline |
| ATS | American Transplantation Society |
| BKVN | BK-virus nephropathy |
| BTS | British Transplantation Society |
| CV.AUC | cross-validated AUC |
| eGFR | estimated glomerular filtration rate |
| ESOT | European Society of Organ Transplantation |
| GLMM | generalised linear mixed-effects models |
| HLA | Human Leucocyte Antigen |
| IS | immunosuppression |
| KTR | kidney transplant recipients |
| Parsimonious | The answer that makes the fewest assumptions; in this manuscript the smallest set of genes showing a satisfactory predictive performance |
| PB | peripheral blood |
| ROC | receiver operator characteristics curve |
| RT-qPCR | Real time – quantitative Polymerase Chain Reaction |
| SCr | Serum Creatinine |
| TCMR | T cell mediated Rejection |
| TTS | The Transplantation Society |
Research in context.
Evidence before the study
Patients with kidney transplants are at significant risk of transplant failure, risking return to renal replacement therapy or having another kidney transplant. Apart from HLA variants mismatches, specific genetic features that are responsible for kidney transplant failure have not been identified thus far.
It remains unknown which of the large number of patients with kidney transplants will get worsening of their kidney function with time. Molecular analysis of peripheral samples from transplant recipients potentially would allow surveillance of immune activation enabling earlier detection and treatment of rejection.
Our literature search has been primarily focused on PubMed and Scopus searches and through information received in and around transplantation meetings (BTS, ATC, ESOT and TTS) where preliminary work of ours and other groups has been presented and discussed.
Previous studies in kidney transplant recipients have identified a number of genes in blood and urine samples which correlate with acute rejection; many of which are involved in cytotoxic T lymphocyte function and cell trafficking. These include Granzyme B, Perforin, Fas-ligand, FoxP3 and CXCL10 and interleukins. However, single genes have lacked the sensitivity and specificity to translate early acute rejection detection into clinical practice. In urine, a three-gene signature has been found which was also able to predict the clinical episode by some weeks. In blood microarray studies have identified gene-sets capable of distinguishing acute rejection. These, however, have not been analysed in a serial fashion to allow for determination of their predictive value and they do not examine the effects of anti-rejection therapy. In cardiac transplantation a commercially available 11 gene set has been shown to reduce the need to perform biopsies and led to greater patient satisfaction.
Most recently, the multi-centre AART study from the US has identified a 17 gene set in blood with an AUC of 0.94 and show a predictive value up to 3 months before detection by biopsy, but further clinical validation is still awaiting.
Added value of this study
This is the first European study to comprehensively analyse serial blood samples from renal transplant recipients. We collected samples from 450 consecutive adult recipients at regular intervals over their first year post-transplant. This has allowed us to perform both cross sectional and longitudinal analysis. Patients selected for the discovery phase all received a similar anti-rejection protocol. Importantly this included induction therapy with an IL-2R blocking antibody (Basiliximab) rather than a lymphocyte depleting antibody, the latter being more common practice in the US. Given that some of the genes are lymphocyte expressed, the induction agent might have a significant effect on lymphocyte gene expression, which we have observed. In longitudinal analysis we have demonstrated for the first time the significant intra patient variability over time and a relationship to changes in anti-rejection therapy. Here we describe a parsimonious (the one that makes the fewest assumptions) T cell mediated rejection (TCMR) signature using the expression of seven genes in peripheral blood.
We have also been able to demonstrate the predictive value of our signature, with detection of acute rejection demonstrable up to two months before the clinical event. We have subsequently carried out validation in a separate cohort of patients. All in all the number of samples analysed throughout our study nearly doubles the numbers of samples used in the AART study, including therefore a more comprehensive longitudinal picture of the gene measurements.
In order to assist the differential diagnosis with BK-virus nephropathy (BKVN), which has the same clinical presentation as T cell mediated rejection (TCMR), but requires the opposite therapy, namely immunosuppression reduction, we have additionally developed a six-gene signature of BKVN. Further, we have examined patients with alternative induction regimens. Patients treated with Rituximab showed similar gene-expression patterns to patients treated with Basiliximab, whilst patients receiving Alemtuzumab treatment showed both, high TCMR and high BKVN positivity.
Implications of all the available evidence
Information from gene expression in peripheral blood samples from transplant recipients could provide valuable information to clinicians for more personalised management and finally provide some information on the recipient's immune status.
Potential benefits include earlier detection and treatment of acute rejection as well as separation from other causes of graft dysfunction, something which the presently used non-invasive monitoring tool, namely serum creatinine is unable to do. It may also allow reduction of anti-rejection therapy in other patients, minimising side effects, that may further allow personalised precision medicine.
A trial of these biomarkers for evaluation in clinical practice is now needed.
We believe the potential of the analysis strategy we applied could be used in other biomarker signatures where longitudinal evaluation is critical and this warrants the scrutiny by the wider readership.
Alt-text: Unlabelled Box
1. Introduction
Kidney transplantation remains the optimal treatment for patients with end-stage kidney disease but requires life-long anti-rejection therapy, which is a major contributor to morbidity and mortality in kidney transplant recipients (KTRs). Balancing the level of immune suppression in each recipient remains a major challenge, and occurs in a reactive fashion in response to clinical events.
Monitoring of allograft function presently relies on serum creatinine (SCr) values. SCr is not a sensitive marker, as it often changes only after a considerable graft damage, and is not a specific marker either, as it can be affected by several factors other than rejection and patients further require a percutaneous biopsy to diagnose the cause of transplant dysfunction. A biopsy, however, is an invasive procedure carrying risks and, being prone to sampling error, could potentially fail to adequately uncover the cause of transplant dysfunction, with many cases reported as “borderline suspicious for acute cellular rejection” [1]. Further, a biopsy is usually carried out only when there is clear evidence of transplant dysfunction, at which point irreversible tissue damage may already have occurred. Studies from centres carrying out routine biopsies at defined time-intervals have also demonstrated a significant amount (10–30%) of rejection in the presence of unchanged renal function.
As molecular events precede the development of the immune response, they provide an ideal opportunity to detect host responses before significant damage to the transplant has occurred. While such changes can be detected in tissue from biopsies, the ability to detect a signal in non-invasive samples such as peripheral blood and urine has the added practical advantage of allowing collection of serial samples. Monitoring of gene-expression signatures in peripheral blood and urine samples offers the opportunity for surveillance of the recipient immune system and earlier detection of acute rejection (AR), of diverse aetiology.
In fact, previous studies have identified in both, blood and urine, a number of mRNAs associated with AR [7]. These have included molecules associated with cytotoxic lymphocyte function, such as Perforin, Granzyme B, Fas-ligand and FoxP3. Single genes, however, lack the sensitivity and specificity to translate into clinical practice, and could hardly capture the complexity of the rejection process. Technological advances now allow reliable and cost-effective analysis of multiple genes in a single sample. In urine, a three-gene signature of AR has been described with an area under the curve (AUC) of 0.85 (sensitivity 79%, specificity 78%) and an increase in gene expression detected up to 20 days before a clinically-evident AR [2]. In cardiac transplantation, the use of an 11-gene panel has been studied and compared against the standard approach of routine biopsies. Use of the panel resulted in fewer performed biopsies and greater patient satisfaction [3].
A critical differential diagnosis of AR is polyoma BK-virus nephropathy (BKVN) [4]. This is manifested, similarly to AR, with graft dysfunction and mononuclear infiltrates in biopsy samples but, unlike AR, is the result of immunosuppression (IS) that maybe excessive for the requirements of the individual. Importantly, the treatment of BKVN (reduction of IS medication) is opposite to that of AR and the definitive diagnosis relies on a specialised immunohistochemistry staining of a biopsy sample [5]. While a reasonable inter-laboratory agreement in detection of BKVN was found in a Banff quality assurance initiative [6], focal lesions may become responsible for a false-negative biopsy. Taking all evidence into account, there is still a need for an alternative non-invasive biomarker of clinically-relevant BKVN.
In this study we have performed a comprehensive analysis in serial peripheral blood samples from KTRs of a set of 22 candidate genes with reported association with T-cell-mediated rejection (TCMR) in the literature (Supplementary Table S1). We have identified a robust gene-expression signature for TCMR and have examined longitudinally gene expression and the effect of different anti-rejection therapies. We subsequently tested the performance of our signature in a validation set of patients and an independent cohort.
This information could finally provide clinicians with some insight into the status of a recipient's immune system and be used as part of the complex clinical management process, when deciding whether or not to perform a biopsy and in evaluating the level of anti-rejection therapy required by a particular individual.
2. Methods
2.1. Patients
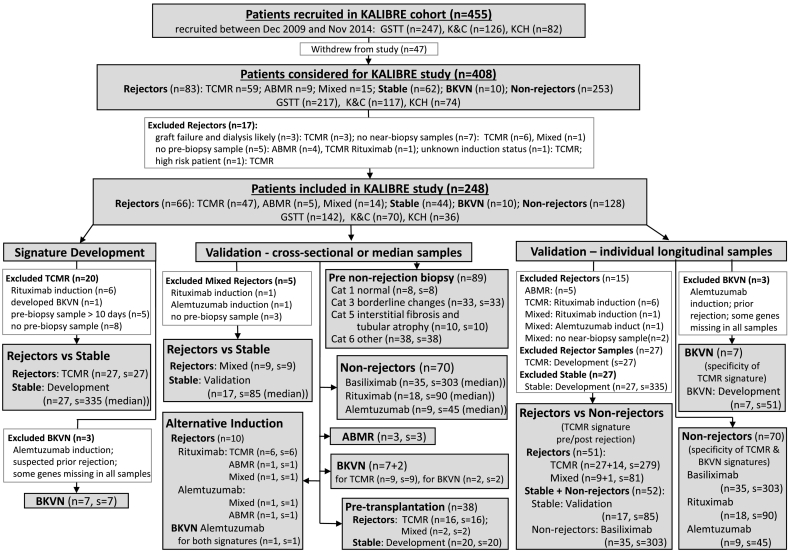
Blood samples were collected serially from 455 consecutive KTRs, transplanted at a single regional transplant centre (Guy's Hospital) in the Kidney Allograft Immunological Biomarkers of Rejection (KALIBRE) study. Patients were followed up at three independent Renal units (Guy's, King's College, and Kent & Canterbury Hospitals). Samples were collected at 26 time-points during clinic visits over the first post-transplant year. A total of 1464 samples from 248 patients were used in the study, including 66 patients with an episode of rejection (Supplementary Fig. S1). Patient flow-chart is shown in Fig. 1.
Fig. 1.
Flow diagram of patients from the KALIBRE study.
Biopsy categories were defined according to Banff ‘09 classification: GSTT – Guy's and St Thomas' NHS Foundation Trust (London, UK); KCH – King's College Hospital NHS Foundation Trust (London, UK); K&C – East Kent Hospitals University NHS Foundation Trust (Canterbury, UK); Cat – Banff category; ABMR – antibody-mediated rejection (category 2); TCMR – T-cell-mediated rejection (category 4); Mixed – histological features of both, ABMR and TCMR; BKVN – BK virus nephropathy (confirmed with a specialised histological staining); unless specifically indicated, patients received Basiliximab induction; Inclusion and exclusion criteria for patients in categories Rejector, Stable and BKVN for the discovery dataset are listed in Table 1; n – number of patients; s – number of samples; median – the median predicted probability from all samples of the same stable patient was used as representative for each stable patient in the signature development and cross-sectional validation.
All patients contributing to the signature-development training dataset (inclusion/exclusion criteria listed in Table 1) had received treatment according to an anti-rejection protocol including Basiliximab induction followed by maintenance therapy with Tacrolimus or Cyclosporine, Mycophenolate Mofetil and Prednisolone. Histological criteria followed the Banff ‘09 classification [8], as this was the most updated version at the beginning of recruitment and it was maintained for consistency throughout the study. Patients were categorised as Stable (when their SCr levels were within 20% of baseline), antibody-mediated rejection (category 2, ABMR); T-cell-mediated rejection (category 4, TCMR); mixed rejection (histological features of both, ABMR and TCMR) (mixed); and BK virus nephropathy (BKNV). Patient demographics are summarised in Table 2A and their immunological risk stratification in Table 2B. External validation KTRs (nine rejectors, 15 non-rejectors, one BKVN) were provided by patients from Guy's Hospital (UK) participating in the EMPIRIKAL trial [9] (EUdraCT: 2011-000958-30). We also included healthy controls (n = 14), previously recruited as part of the GAMBIT study [10].
Table 1.
Inclusion and exclusion criteria for patients in the signature-development dataset.
| Type | Inclusion criteria | Exclusion criteria |
|---|---|---|
| Rejector |
|
|
| Stable |
|
|
| BKVN |
|
|
The specialised BKVN staining has become a routine procedure in more recent years, but for the largest part of the KALIBRE study it would mainly have been be performed if a histological differential diagnosis from TCMR was questionable.
Table 2A.
Patient demographics. All cohorts.
| Group | Number patients | Age at transplantation | Gender Female (%) | Donor type living (%) | Time to biopsy (days) |
|---|---|---|---|---|---|
| Total patients used from KALIBRE study | |||||
| KTRs (all) | 248 | 47 (17–73) | 92 (37) | 138 (56) | – |
| Rejectors (all) | 66 | 48 (18–71) | 25 (38) | 38 (58) | 68 (4–384) |
| Signature development patients (KALIBRE study) | |||||
| Rejectors (TCMR) | 27 | 39 (19–68) | 9 (33) | 16 (59) | 103 (5–364) |
| Stable (Discovery)* | 27 | 41 (19–70) | 9 (33) | 17 (63) | – |
| BKVN* | 7 | 54 (36–69) | 1 (14) | 4 (57) | 195 (51–369) |
| Cross-sectional validation patients (KALIBRE study) | |||||
| Category 1 | 8 | 42 (24–64) | 2 (25) | 4 (50) | 87 (25–189) |
| Category 3 | 33 | 49 (25–73) | 14 (42) | 10 (30) | 49 (7–196) |
| Category 5 | 10 | 53 (33–70) | 4 (40) | 5 (50) | 58 (7–185) |
| Category 6 | 38 | 54 (18–71) | 14 (37) | 15 (39) | 56 (6–336) |
| Rejectors (Mixed-type) | 9 | 52 (24–64) | 4 (44) | 2 (22) | 87 (5–349) |
| Stable (Validation)⁎ | 17 | 43 (25–68) | 5 (29) | 10 (59) | – |
| Rejectors (pre-transplantation) | 18 | 38 (18–64) | 5 (28) | 16 (89) | 99 (4–384) |
| Stable (pre-transplantation) | 20 | 41 (19–70) | 8 (40) | 16 (80) | – |
| Rejectors (Rituximab/Alemtuzumab) | 10 | 42 (30–69) | 3 (30) | 10 (100) | 21 (4–367) |
| Longitudinal validation patients (KALIBRE study) | |||||
| Rejectors (TCMR + Mixed) | 51 | 48 (18–71) | 20 (39) | 26 (51) | 78 (4–384) |
| Stable (Discovery + Validation) | 44 | 42 (19–70) | 14 (32) | 27 (61) | – |
| Non-rejectors (Basiliximab) | 35 | 46 (18–73) | 14 (40) | 24 (69) | – |
| Non-rejectors (Alemtuzumab) | 9 | 43 (36–65) | 4 (44) | 8 (89) | – |
| Non-rejectors (Rituximab) | 18 | 47 (17–72) | 8 (44) | 18 (100) | – |
| External validation patients (EMPIRIKAL trial) | |||||
| Rejectors (TCMR + Mixed) | 9 | 57 (23–79) | 4 (44) | 0 | 8 (7–69) |
| Non-rejectors (External) | 15 | 50 (20–76) | 4 (27) | 0 | – |
| Healthy controls (GAMBIT study) | |||||
| Healthy Controls | 14 | 45 (23–72) | 3 (21) | – | – |
Patients contributed all individual samples for longitudinal validation, while for cross-sectional comparisons and signature development contributed a summary sample with median values per patient; Age at transplantation and Time to biopsy – summarised with median (minimum – maximum) per group; Longitudinal samples – covered the period between days four and 400 post transplantation; Rejector – a patient with a biopsy-proven rejection; TCMR – T-cell-mediated rejection; Mixed – histological features of TCMR and antibody-mediated rejection; Stable - patients fulfilling the inclusion/exclusion criteria in Table 1; Non-rejector – patients not fulfilling the selection criteria in Table 1 but without clinical or histological evidence of TCMR, ABMR or BKVN up to day 400 post-transplantation; BKVN – patients with biopsy-proven BK-virus nephropathy fulfilling the criteria in Table 1; Biopsy categories – non-rejection features, according to Banff’09 classification (category 1 – normal, category 3 – borderline changes, category 5 - interstitial fibrosis and tubular atrophy, category 6 – other non-rejection histology).
Table 2B.
Immunological Risk stratification in test and validations cohorts.
| Group | Number patients | Low % patients |
Standard % patients |
High % patients |
|---|---|---|---|---|
| Total patients used from KALIBRE study | ||||
| KTRs (all) | 248 | 26.6 | 56.9 | 16.5 |
| Rejectors (all) | 66 | 22.7 | 63.6 | 13.6 |
| Signature development patients (KALIBRE study) | ||||
| Rejectors (TCMR) | 27 | 37.0 | 55.6 | 7.4 |
| Stable (Discovery)* | 27 | 33.3 | 66.7 | 0 |
| BKVN* | 7 | 14.3 | 85.7 | 0 |
| Cross-sectional validation patients (KALIBRE study) | ||||
| Category 1 | 8 | 50.0 | 37.5 | 12.5 |
| Category 3 | 33 | 27.3 | 57.6 | 15.2 |
| Category 5 | 10 | 30.0 | 60.0 | 10.0 |
| Category 6 | 38 | 42.1 | 50.0 | 7.9 |
| Rejectors (Mixed-type) | 9 | 0 | 88.9 | 11.1 |
| Stable (Validation)* | 17 | 29.4 | 58.8 | 11.8 |
| Rejectors (Rituximab/Alemtuzumab) | 10 | 0 | 60·0 | 40.0 |
| Longitudinal validation patients (KALIBRE study) | ||||
| Rejectors (TCMR + Mixed) | 51 | 27.5 | 64.7 | 7.8 |
| Stable (Validation) + Non-rejectors (Basiliximab) | 52 | 34.6 | 57.7 | 7.8 |
| Non-rejectors (Basiliximab) | 35 | 37.1 | 57.1 | 5.7 |
| Non-rejectors (Alemtuzumab) | 9 | 0 | 11.1 | 88.9 |
| Non-rejectors (Rituximab) | 18 | 0 | 50.0 | 50.0 |
| External validation patients (EMPIRIKAL trial) | ||||
| Rejectors (TCMR + Mixed) | 9 | N/A | ||
| Non-rejectors (External) | 15 | N/A | ||
Percent of patients assigned to each immunological risk level per group within the cohorts. As per local centre protocol patients deemed to be of Low immunological risk were: recipients without HLA antibodies or recipients receiving a first transplant kidney from a HLA identical sibling. Standard immunological risk: recipients with HLA antibodies; and the following groups (regardless of presence or absence of HLA antibodies): Husband to Wife, Child to Mother, Second or subsequent kidney transplant, Black recipient. High immunological risk: recipients who are cross match negative by flow-cytometry but who have a current or historic antibody which is directed against the new organ, and has arisen following exposure to this antigen from a previous solid organ transplant or pregnancy. Recipients who are cross-match positive by flow-cytometry are deemed HLA Antibody Incompatible (HLAi) and receive Alemtuzumab (Campath®) induction and may also undergo pre-operative antibody removal.
The induction agent for patients in the KALIBRE study was Basiliximab, unless otherwise specified. Patients in the EMPIRIKAL study all received induction with Basiliximab, and 2/3 of the donor grafts would have been treated with an experimental complement inhibitor right before transplantation (unblinding had not been available at the time of submission).
2.2. Ethics statement
Approval from research ethics committees was obtained for all included studies: KALIBRE - Research Ethics No: 09/H0711/58; GAMBIT - Research Ethics No: 09/H0713/12; EMPIRIKAL - Research Ethics No: 12/LO/1334. Written informed consent was obtained from all patients participating in each of those studies.
2.3. Gene-expression analysis
Peripheral blood was collected into Tempus™ Blood RNA Tubes (Life-Technologies) and stored at −20 °C. RNA isolation, cDNA synthesis and RT-qPCR conditions have been previously described in detail [10]. We analysed 22 genes (Supplementary Table S2a–b). Relative gene expression values were calculated with the –ΔCt method, detecting the difference with hypoxanthine-phosphoribosyltransferase (HPRT) as a house-keeping gene. An in-house quality control (QC) sample was included in every analytical batch, which showed very low between-run variability (coefficients of variation between 0·19% and 1·09%, median 0·48%). Missing data was minimal (below 0·5%).
2.4. Sample size
Sample size for signature development was determined by patient availability. We included all recipients with T-cell-mediated rejection (TCMR) (n = 27) and BKVN (n = 7) fulfilling the inclusion/exclusion criteria (Table 1) and the same number of stable patients (n = 27), matched to rejectors in age, sex, and donor type, with no biopsy performed and <20% SCr change after achieving baseline. Power calculation (using an exponential approximation to estimate AUC variance) [11], showed that with 27 patients in each group, we could estimate a 95% confidence interval with half-width 0·103 for an expected AUC of 0·85 and with better precision for higher AUC.
2.5. Statistical analysis
Statistical analysis was performed in R version 3.2.2 [12]. Non-parametric Wilcoxon-Mann-Whitney test was used for univariate class comparisons. Association between continuous variables was evaluated with Spearman correlation coefficient (r). Outliers were recoded to the next highest or lowest value for multivariable analysis. Missing gene-expression data were imputed with K-nearest neighbour for microarrays (impute package) [13]. Missing values were first imputed in a 22-gene matrix of longitudinal samples, including samples collected from day 4 to rejection in training rejectors (n = 201) and between days 4 and 400 post-transplantation from stable patients (n = 335, Supplementary Fig. S2). The complete training matrix was then used to impute missing gene-expression for test samples, one at a time and based only on the genes included in the examined model.
To account for the dependency of samples from the same patient, serial samples were analysed with generalised linear-mixed effects models (GLMM) with a linear, quadratic and cubic term for the fixed and random effects of time.
To account for dependency of gene-expression on time post-transplantation we generated time-adjusted gene-expression values, individually for each gene, as the residuals of cubic GLMM linear regression models with the –ΔCt values, based on serial samples from training stable patients (residuals for all other patients were generated using these training models).
To develop a TCMR signature, we compared samples from TCMR rejectors (a single pre-rejection sample per patient, zero to nine, median: three days pre-biopsy) and stable patients (serial samples of ten to 20, median: 12 per patient; total: 335, summarised with the median time-adjusted expression for each gene per patient). To develop a gene-expression signature of BKVN, we compared BKVN-positive patients (a single sample per patient, within seven (median zero) days of a diagnostic biopsy) with the combined group of TCMR rejectors and stable patients, to secure simultaneous discrimination from non-BKVN KTRs.
To select a parsimonious gene-expression signature, i.e. the smallest set of genes showing a satisfactory predictive performance, we used penalised logistic regression with an elastic net penalty [14] (glmnet package [12]). Elastic net enables gene selection by shrinking the regression coefficients of genes statistically non-informative for discrimination and, hence, retaining only genes, which are statistically-important based on the data used in the model. For the penalty parameters, we selected the alpha (tested in increments of 0·1), which enabled retaining a satisfactory model performance with the minimum number of strong predictors (i.e. those gene remaining without shrinkage at high values of alpha). The penalty parameter lambda was optimised as the median of 200 seven-fold cross-validation repeats of the cv.glmnet function. The final signature models were based on imputation, time-adjustment and elastic net regression performed in the complete signature-development dataset.
To evaluate model performance, we used the AUC (95% De Long confidence interval) and calculated sensitivity and specificity for a cut-off that optimised both for TCMR and specificity only for BKVN, but retaining sensitivity above 0·70 (pROC package) [15].
To compare the pre- and post-rejection trajectories of the probability of TCMR in rejectors and non-rejectors, we used GLMM linear regression with an interaction term for group and time. We used as outcome the predicted log-odds of rejection, which, unlike probability, has an unrestricted continuous scale. As a reference time-point in rejectors we used the day of the diagnostic biopsy. In non-rejectors, after demonstrating the time-independence of the predicted probability of TCMR, we assigned a time with respect to the reference point at random. This ensured that the distribution of samples from non-rejectors matched the pattern of rejectors with respect to time post-transplantation (Supplementary Fig. S3). Samples contributing to signature development, i.e. the 27 pre-biopsy samples for patients with TCMR and the 335 samples from the training stable patients, were excluded from the longitudinal analysis. Although the remaining pre-rejection samples from the 27 training rejectors were included in the imputation matrix, they did not contribute to elastic net regression (i.e. gene selection and regression coefficients) and, with a missingness below 0·5%, they would not have materially influenced signature development.
2.6. Validation strategy
It should primarily be noted, that obtaining the 22 initial genes from literature reports and not from a statistical analysis of microarrays performed in our own dataset meant that our study provided a validation dataset for already published findings.
Further, to evaluate the performance of the selected parsimonious gene-expression signature with unseen data, we used the following approaches:
First, we used cross-validation within the signature-development dataset. In the cross-validation cycles all steps of signature development (including the linear regression models generating time-adjusted residuals, the imputation of missing data and the elastic net regression models performing the inherent to them gene selection (starting from the complete list of 22 genes for each model) and the required optimisation of the lambda parameter), were performed with the training subset. The left-out test subset was used solely for model validation (see Note 1 in Supplementary Discussion for further details). A cross-validation AUC (CV.AUC) was determined for each of 50 repeats of seven-fold cross-validation cycles, along with sensitivity and specificity at the fixed cut-off determined as optimal for the final signature model. Model performance measures obtained in the 50 cross-validation cycles were summarised with median (2.5th – 97.5th centile).
Second, we performed cross-sectional validation in unseen test patients, using mixed-type rejectors (with histological features of both, TCMR and antibody-mediated rejection (ABMR)) and new (test) stable patients. We further examined samples collected prior to non-rejection biopsies with different histological categories, pre-rejection samples from patients with ABMR and from rejectors treated with alternative immunosuppression induction agents (Alemtuzumab and Rituximab), near-biopsy samples from patients with BKVN, and samples from healthy controls.
Third, we performed validation in longitudinal samples. To test signature specificity we used the individual longitudinal samples from the new test stable patients and also from other unseen test non-rejectors with more compromised renal function (with or without a for-cause biopsy during the first post-transplant year) and from non-rejectors with alternative immunosuppression induction (the median sample per patient participated in the cross-sectional validation). Specificity of the TCMR signature was further examined in longitudinal samples from BKVN patients. In addition, we compared serial samples from rejectors with the combined group of the non-rejectors and the new stable patients. Rejectors included independent test rejectors (with TCMR and mixed-type rejection) and only the pre and post rejection samples from the 27 training rejectors with TCMR, which were unseen in the elastic net regression defining the signature model.
Fourth, we performed external validation with samples from independent rejectors with TCMR features and non-rejectors from the EMPIRIKAL trial (a pre-rejection sample for rejectors and longitudinal samples for non-rejectors).
2.7. Data sharing
Research data will be made available through application to the Biobank “Transplantation, Immunology and Nephrology Tissue and Information Nexus” (TIN-TIN) based at King's College London, London UK. Provisional Ethics Ref: 17/LO/0220.
2.8. Role of the funding sources
The study sponsors had no involvement in the study design, the collection, analysis, and interpretation of data, in the writing of the report, and in the decision to submit the paper for publication.
3. Results
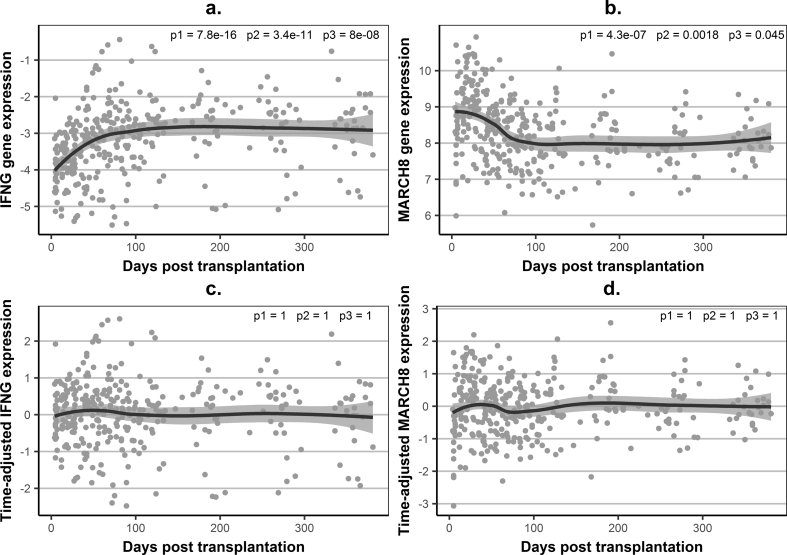
Examining gene expression in longitudinal samples of training stable patients demonstrated high within-patient variability and systematic trajectory changes over the first four months post-transplantation (Fig. 2a–b). The expression of 19 of the 22 studied genes was significantly associated with time (Supplementary Table S3). After accounting for prednisolone dose, which is systematically reduced during the first post-transplant months (Supplementary Fig. S4), the association of gene-expression and time was retained, independently of prednisolone, for 10 of the 22 genes (Supplementary Table S3), whilst eight genes showed an association with prednisolone, independent of time. Consequently, we generated time-adjusted gene-expression levels (Fig. 2c–d) and used these in signature development. This ensured that differences between stable patients and rejectors were accounted for by rejection and not by time-related post-transplantation changes.
Fig. 2.
Examples of association between gene-expression levels and time in stable patients.
(a) – observed (unadjusted) –ΔCt values for IFNG, as an example of a gene with increasing expression levels; (b) – observed (unadjusted) –ΔCt values for MARCH8, as an example of a gene with decreasing expression levels; (c) – time-adjusted gene-expression levels for IFNG, illustrating abolition of time-dependence for gene expression; (d) – time-adjusted gene-expression levels for MARCH8; Gene expression – observed ΔCt values, relative to hypoxanthine phosphoribosyltransferase (HPRT), in individual samples (grey dots) and a loess average curve with 95% confidence interval; Time-adjusted gene expression – fold difference on log2 scale (same scale as gene expression), derived as the difference between the observed –ΔCt values and the predicted values using generalised linear mixed-effects models (GLMM) linear regression, based on all serial samples (total n = 335) from the training stable kidney transplant recipients (n = 27) (a cubic relationship between gene expression and time post transplantation was modelled for the fixed and random effects); p1, p2, p3 - p-values for the linear, quadratic and cubic fixed-effects terms for time in GLMM (in (a) and (b) GLMM was based on the observed –ΔCt values and the p-values confirm a statistically significant association between gene-expression and time, in (c) and (d) GLMM was based on time-adjusted gene expression values and p-values demonstrate the time-independence of the time-adjusted gene-expression levels).
Using penalised logistic regression with elastic net penalty, we developed a parsimonious signature of TCMR, retaining seven genes with non-zero regression coefficients and, hence, referred to as a “seven-gene” signature (Supplementary Fig. S5/S6a/S7, Tables S4/S5), which showed improved predictive performance in cross-validation (CV.AUC 0.84 (0.77–0.88)) compared to the 22-gene model (Table 3, Fig. 3a). This suggests that many of the 22 original genes contribute more variability and noise to the 22-gene model than information facilitation the discrimination and, therefore, they could not be validated in our dataset (Supplementary Fig. S5). It should also be noted that a comparison with eGFR as a diagnostic marker is not appropriate, as SCr has been used as a selection criterion (see Note 2 in Supplementary Discussion).
Table 3.
Gene-expression signatures.
| Event | Genes | Data | AUC | Sens | Spec |
|---|---|---|---|---|---|
| TCMR | All genes | Training data | 0.96 (0.92–1.00) | 0·85 | 0·89 |
| Cross-validation | 0.80 (0.75–0.83) | 0.67 (0.59–0.70) | 0.81 (0.71–0.88) | ||
| TCMR | IFNG, IP-10, ITGA4, MARCH8, RORc, | Training data | 0.95 (0.91–1.00) | 0.85 | 0.93 |
| SEMA7A, WDR40A | Cross-validation | 0.84 (0.77–0.88) | 0.67 (0.59–0.74) | 0.85 (0.75–0.89) | |
| BKVN | All genes | Training data | 0.93 (0.85–1.00) | 0.71 | 0.93 |
| Cross-validation | 0.68 (0.64–0.73) | 0 (0–0.25) | 0.91 (0.85–0.94) | ||
| BKVN | IL-15, IL1R2, MARCH8, PDCD1, | Training data | 0.95 (0.90–1.00) | 0.71 | 0.91 |
| TGFB, WDR40A | Cross-validation | 0.73 (0.66–0.80) | 0.43 (0·29–0.57) | 0.89 (0.83–0.91) |
Signature-development (training) data comprised the following groups of patients defined according to the criteria in Table 1: Rejectors (n = 27) with T-cell mediated rejection providing a single sample per patient, collected between zero and nine days (median three days) prior to the date of the diagnostic biopsy; Stable (n = 27) patients providing a summary (median) of the predicted probabilities for all samples from a given patient (10 to 20 samples per patient, median 12, total 335 samples); BKVN (n = 7) patients with biopsy-proven BK-virus nephropathy providing a single sample per patient collected within seven days before or after a diagnostic biopsy (median zero days); Gene-expression signatures resulted from gene selection based on penalised logistic regression with elastic net penalty (Supplementary Fig. S5 and Fig. S11); AUC – area under the receiver operating characteristics (ROC) curve with 95% DeLong confidence interval; Sens – sensitivity; Spec – specificity; Cut-offs for Sens and Spec– probability of TCMR 0·5, probability of BKVN 0·2; Cross-validation – summary of model performance measures based on the predicted probabilities for unseen data obtained in each of 50 repeats of seven-fold cross-validation cycles – median (2.5th – 97.5th centile).
Highlighted in bold the most relevant AUC.
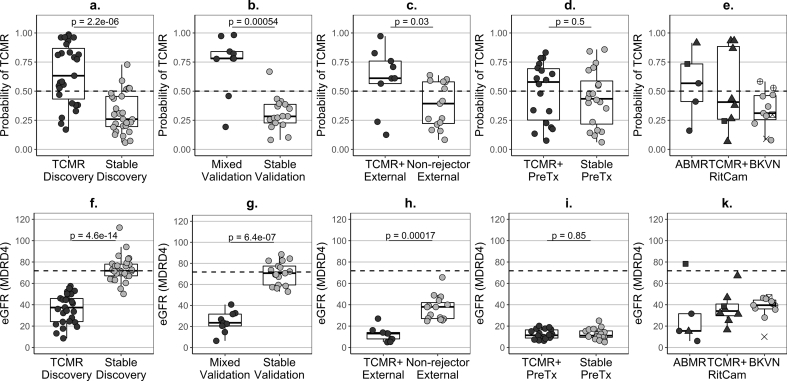
Fig. 3.
Predicted probability of TCMR and kidney function in rejectors and stable or non-rejector controls and in patients with BK virus nephropathy.
a – median of the set of probabilities of T-cell-mediated rejection (TCMR) estimated for each patient of the development dataset in the 50 repeats of the cross-validation cycles (Step 12 of Note 1 in Supplementary Discussion); (b–e) – predicted probability of TCMR, based on the newly-developed seven-gene signature (Supplementary Fig. S5); (f–k) – estimated glomerular filtration rate (eGFR), based on the MDRD4 equation; ABMR – antibody-mediated rejection (category 2 according to Banff’09 classification); TCMR – T-cell-mediated rejection (category 4); Mixed – histological features of both, ABMR and TCMR; TCMR+ – patients with TCMR alone and with mixed-type histology; BKVN – BK virus nephropathy (confirmed with a specialised histological staining); Stable – patients fulfilling the inclusion and exclusion criteria listed in Table 1; Non-rejector – patient not fulfilling the selection criteria in Table 1 but without evidence of TCMR, ABMR or BKVN up to day 400 post-transplantation; dot colour: black – rejectors, grey – stable or non-rejector patients; dot shape – induction agent in KALIBRE patients: circle – Basiliximab, triangle – Rituximab, square – Alemtuzumab; dotted lines – cut-off 0·5 for dichotomous classification of the probability of TCMR (a-e) and median eGFR (71.8 mL/min/1.73m2) of the summary of stable patients in the discovery signature-development dataset (F—K); samples: rejectors – single pre-biopsy sample, BKVN – single near-biopsy sample, stable/non-rejector patients – summary (median per patient) of longitudinal samples covering between days four and 400 post transplantation; Discovery (a,f) – training patients used in TCMR signature development (TCMR (n = 27) fulfilling the inclusion and exclusion criteria listed in Table 1 and stable patients (n = 27)); Validation (b,g) – test patients from the KALIBRE study used for TCMR signature validation (Mixed-type rejectors (n = 9) and new stable patients (n = 17)); External (c,h) – patients from the independent trial EMPIRIKAL used for TSMR signature validation (rejectors (n = 9) with TCMR or Mixed-type histology, non-rejectors (n = 15)); PreTx (d,i) – cross-sectional pre-transplantation samples – one per patient from rejectors with TCMR (n = 16) and Mixed-type (n = 2) histology and training stable patients from the signature development dataset (n = 20)); e,h – ABMR rejectors (n = 5), RitCam rejectors with alternative induction and TCMR (n = 6) or Mixed-type histology (n = 2); BKVN patients: grey dots BKVN (n = 7) included in the BKVN signature development dataset, crossed dots BKVN: +dot (n = 2) excluded from the training dataset due to prior suspected TCMR or some genes missing, x-dot (n = 1) Alemtuzumab induction, x (n = 1) patient from the EMPIRICAL trial; p-values derived from a Wilcoxon-Mann-Whitney tests.
The TCMR signature showed excellent discrimination between mixed-type rejectors and new stable patients in cross-sectional validation samples that had similar distribution of immunological risk pre-transplant stratification (Table 2B, Fig. 3b) (AUC 0.90 (0.70–1.00)). In the external validation dataset from the EMPIRIKAL trial, seven out of the nine rejectors (78%) were TCMR-positive near the diagnostic biopsy (Fig. 3c). EMPIRIKAL non-rejectors had distinctly worse kidney function compared to KALIBRE stable patients (Fig. 3f–g vs 3h), with eight out of 15 patients requiring dialysis in the first two weeks post-transplantation. Nevertheless, a discrimination could be achieved from TCMR (AUC 0.77 (0.53–1.00)). No discrimination could be achieved between TCMR and stable patients pre-transplantation (Fig. 3d) (AUC 0.57 (0.38–0.76)).
Whilst BKVN patients had low eGFR, similar to that of rejectors (Fig. 3k), they were TCMR-negative or only weakly positive (Fig. 3e). Five out of ten mixed-type rejectors treated with a different induction agent were TCMR-positive near the diagnostic biopsy (Fig. 3e). Three of the five patients with features only of ABMR in the first biopsy diagnostic of AR were TCMR-positive, but the one with the highest probability of TCMR showed features of mixed-type rejection in a subsequent biopsy, performed eight days after the collection of the sample shown in Fig. 3e.
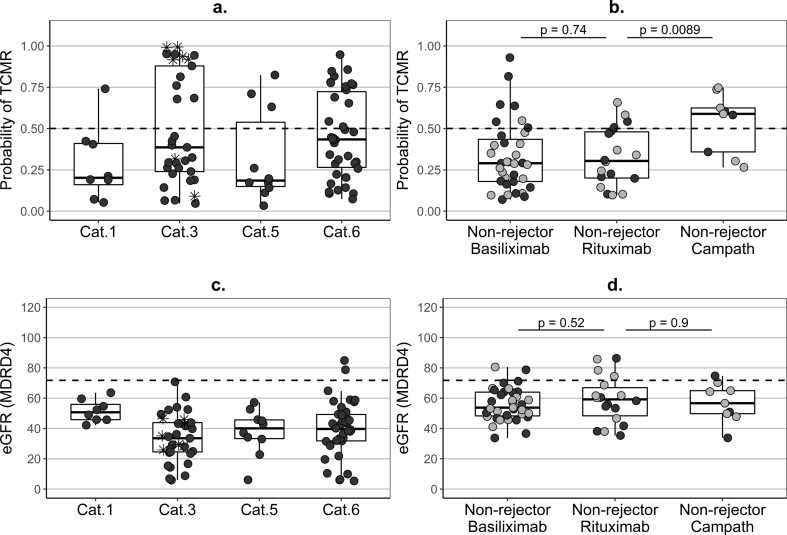
Preceding a for-cause biopsy without features of AR, seven out of eight KTRs with normal histology were TCMR-negative, but >30% of the patients with histological features of borderline changes, chronic rejection or other non-rejection alterations were TCMR-positive (Fig. 4a) and the predicted probability of TCMR was negatively correlated with eGFR (r = −0.40, p < 0.0001)(Fig. 4c). Half of the 14 healthy controls were TCMR-positive.
Fig. 4.
Predicted probability of TCMR and kidney function in non-rejectors.
(a–b) – predicted probability of T-cell-mediated rejection (TCMR), based on the newly-developed seven-gene signature (Supplementary Fig. S5); (c–d) – estimated glomerular filtration rate (eGFR), based on the MDRD4 equation; (a,c) – cross-sectional pre-biopsy samples (up to 15 days prior to the first post-transplantation biopsy (median two days) and a single one at 25 days) from patients which did not show features of rejection at least up to three months post biopsy; Cat – histological category according to Banff’09 classification (Cat.1 – normal (n = 8, one (13%) TCMR-positive), Cat.3 – borderline changes (n = 33, 13 (39%) TCMR-positive), Cat.5 – interstitial fibrosis and tubular atrophy (n = 10, three (30%) TCMR-positive), Cat.6 – other non-rejection histology (n = 38, 14 (37%) TCMR-positive)); star dots (*) – patients with histological features of borderline changes treated for rejection due to clinical considerations; (b,d)– summary of longitudinal validation follow-up samples covering one year post transplantation (median per patient) in non-rejectors with different induction agents (not fulfilling the strict inclusion/exclusion criteria set for stable patients in Table 1, but without clinical or histological evidence of rejection up to day 400 post tansplantation): Basiliximab induction (n = 35, 18 with biopsy, seven (20%) TCMR-positive median), Rituximab (n = 18, nine with biopsy, four (22%) TCMR-positive median), Alemtuzumab (n = 9, three with biopsy, six (67%) TCMR-positive median); black dots – non-rejection biopsy during the first post-transplant year; grey dots – no biopsy; dotted lines – cut-off 0·5 for dichotomous classification of the probability of TCMR (a,b) and median eGFR (71.8 mL/min/1.73m2) (c,d) from the summary samples of the stable patients in the discovery signature-development data; p-values derived from a Wilcoxon-Mann-Whitney tests.
In longitudinal samples from the stable patients used for signature development, the average predicted probability of TCMR remained constant with time post-transplantation, below the cut-off, and was not influenced by adjustment for prednisolone dose (Supplementary Fig. S8a). The probability of TCMR also remained below the cut-off for validation stable patients and non-rejectors (Supplementary Fig. S8b). Further, over the first post-transplant year, the TCMR signature demonstrated very good specificity (above 70%) in stable patients, non-rejectors, and BKVN patients (Table 4). Rituximab induction showed similarity to Basiliximab induction (Fig. 4b), but TCMR-signature positivity was higher following Alemtuzumab induction (71%), despite the comparable eGFR in alternative induction groups (Fig. 4d). Similarly, in non-rejectors of the EMPIRIKAL trial a larger proportion of the longitudinal samples were TCMR-positive (44%), with ten out of the 14 samples from the first post-transplant week being TCMR-positive.
Table 4.
Specificity of gene-expression signatures in longitudinal samples from non-rejection patients.
| Group | Patients | Samples total | Samples per patient* | TCMR negative(%) | BKVN negative (%) | Double negative (%) | Double positive (%) |
|---|---|---|---|---|---|---|---|
| Stable Discovery | 27 | 335 | 12 (10−20) | 262 (78) | 264 (79) | 192 (57) | 1 (0) |
| Stable Validation | 17 | 85 | 5 | 65 (76) | 58 (68) | 41 (48) | 3 (4) |
| Non-rejectors Basiliximab | 35 | 303 | 8 (5–20) | 216 (71) | 234 (77) | 149 (49) | 2 (1) |
| Non-rejectors Rituximab | 18 | 90 | 5 | 69 (77) | 68 (76) | 51 (57) | 4 (4) |
| Non-rejectors Alemtuzumab | 9 | 45 | 5 | 13 (29) | 15 (33) | 3 (7) | 20 (44) |
| Non-rejectors EMPIRIKAL | 15 | 77 | 5 (3–6) | 43 (56) | 63 (82) | 31 (40) | 2 (3) |
| BKVN | 7 | 51 | 7 (6–10) | 38 (75) | – | – | 1 (2) |
Cells – number (percentage) of longitudinal samples collected during the first post-transplant year; * – median (minimum – maximum); TCMR – gene-expression signature of T-cell-mediated rejection (Supplementary Fig. S5); BKVN – gene-expression signature of BK-virus nephropathy (six genes, Supplementary Fig. S11); negative – probability of event below the cut-off of 0·5 for TCMR and 0·2 for BKVN (equivalent to specificity); Positive – predicted probabilities of event above the cut-offs; Double positive/negative – predicted probabilities of both, TCMR and BKVN, for the same sample above/below the corresponding cut-offs; Stable – patients fulfilling the criteria in Table 1; Non-rejectors - patients with a more variable and compromised kidney function, which either did not require a biopsy during the first year post transplantation and, hence, were considered clinically stable, or had biopsies lacking features of T-cell, or antibody-mediated rejection, or BKVN.
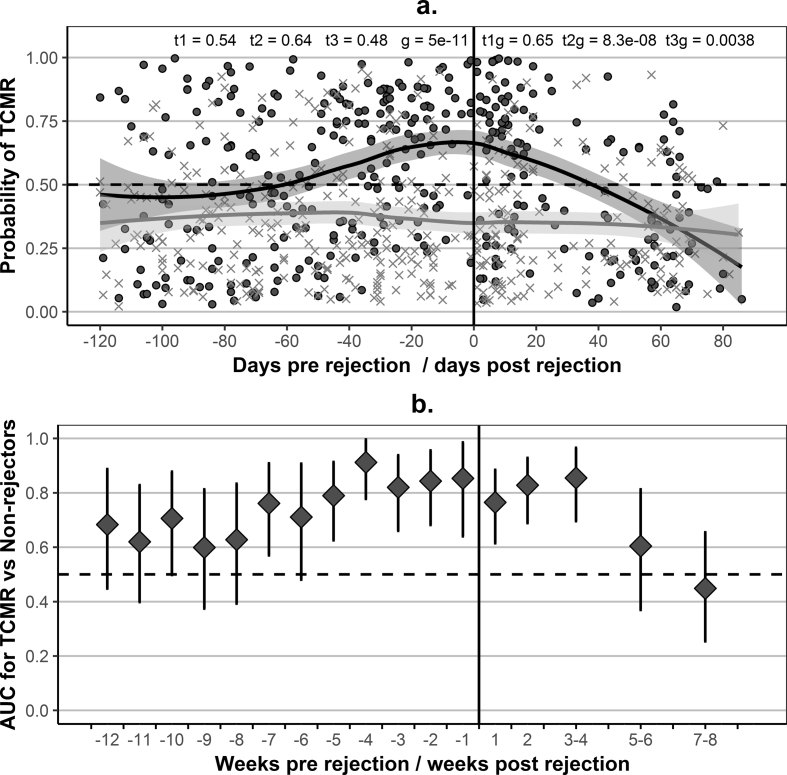
In longitudinal samples from rejectors, the probability of TCMR increased well ahead of rejection and decreased after treatment (Fig. 5a) following kidney function and not immunosuppression changes. There was a very clear difference between rejectors and non-rejectors at the time of rejection (p < 0.0001 for the group term in GLMM) and a clear difference between the average trajectories of the two groups (Supplementary Fig. S9). Discrimination between rejectors and non-rejectors was possible for at least five weeks before and four weeks after rejection. AUC remained near or above 0.80 for the five weeks preceding rejection and above 0.70 for weeks six and seven (Fig. 5b). It is not a common practice in the UK to use anti-thymocyte globuline (ATG) as an induction agent, but 11 of the rejectors had received it as a treatment for rejection. The probability of TCMR increased before the biopsy and remained above 0.70 within two weeks after administration of ATG and in some of the patients for considerably longer (Supplementary Fig. S10).
Fig. 5.
Longitudinal evaluation of the gene-expression signature of TCMR with respect to the diagnostic biopsy.
(a) – Evolution of the predicted probability of T-cell-mediated rejection (TCMR), based on the seven-gene signature of TCMR (Supplementary Fig. S5), with time pre and post rejection: individual samples and average loess trajectories for patient groups; Black dots – longitudinal samples from patients with TCMR: 279 samples from 41 patients (the 27 samples contributing to the signature-development dataset were excluded) and 81 samples from 10 patients with mixed-type rejection (antibody-mediated rejection (ABMR) and TCMR); Grey * – longitudinal samples from the 17 (test) stable patients included in the validation dataset (85 samples) (fulfilling the inclusion/exclusion criteria in Table 1) and the 35 non-rejectors with Basiliximab induction (303 samples) included in the validation dataset; Vertical line (zero days) – reference point; Reference point (time 0) – for rejectors, the day of the diagnostic biopsy (222 samples pre-biopsy (164 from TCMR and 58 from patients with mixed-type rejection) and 138 post rejection (115 TCMR, 23 mixed-type)); for stable patients and non-rejectors time was assigned at random, such that the samples were distributed along the timeline proportional to the number of samples from rejectors in the given week-wide time window, as well as proportional to the contribution of training and validation stable patients, and ensuring that a single sample per patient was included in each week-wide window (240 samples before (189 non-rejectors, 51 stable) and 148 after the reference point (114 non-rejectors, 34 stable)); samples from stable patients covered the period between days 4 and 400 post transplantation; t1, t2 and t3 – p-values for the linear, quadratic and cubic terms for time in a generalised linear mixed-effects model (GLMM) (linear regression) based on log-odds of TCMR as an outcome and including fixed and random effects for time, a group term (stable patients as baseline), and interaction terms between time and group; g – p-value for the group term in GLMM; t1g, t2g, and t3g – p-values for the interaction terms in GLMM; (b) – classification performance of the gene-expression signature of TCMR for weekly intervals (from 12 weeks pre to two weeks post rejection) or fortnightly intervals (from three to eight weeks post rejection), time limited to 120 days pre and 90 days post rejection; Diamonds – area under the receiver operating characteristics curve (AUC) for a comparison between unique samples per patient in each window (8 to 29 samples per window for rejectors (median 14, using 170 pre and 101 post-rejection samples) and 9 to 31 samples per window for stable patients and non-rejectors (median 13, using 184 samples assigned at random to pre and 109 samples assigned to post reference point, as described in A); Error bars – 95% DeLong confidence intervals for AUC.
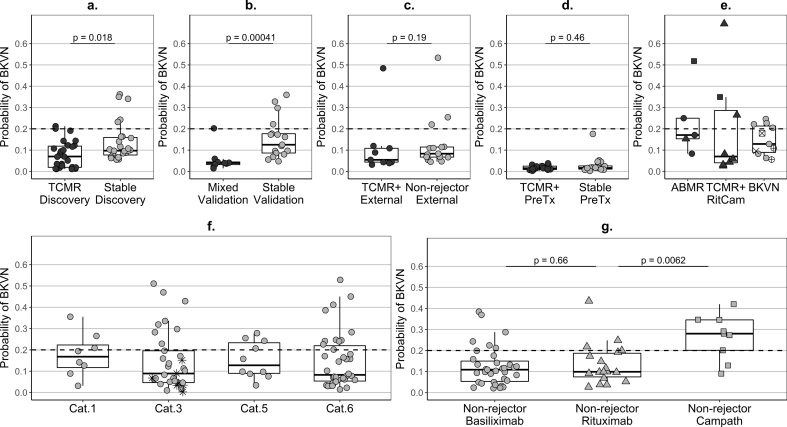
Given the histological similarities between BKVN and TCMR and the fact that they both represent some form of inflammation, it was important to explore whether the genes in the TCMR signature reflect BKVN activity. To assist differential diagnosis, we additionally developed a parsimonious six-gene signature of BKVN (Supplementary Fig. S11/S6b/S12, Tables S4/S5), showing (like the TCMR signature) an improved performance at cross-validation (CV.AUC 0.73 (0.66–0.80)) compared to the full 22-gene model (Table 3). Only MARCH8 and WDR40A genes were shared between the two signatures. These genes were strongly positively correlated (r = 0·96, p < 0.0001 in the joint signature-development group of rejectors, BKVN, and stable patients), but were lower in TCMR compared to BKVN (Supplementary Fig. S7) and were selected by the statistical algorithm as informative in both signatures because the signature for BKVN was trained to discriminate BKVN from TCMR, as well as from stable patients. Correspondingly, the signatures of BKVN and TCMR were negatively correlated (r = −0.45, p < 0.0001). Notably, the majority of TCMR and mixed-type rejectors were BKVN-negative pre-biopsy (Fig. 6a–c). The specificity of the BKVN signature in longitudinal samples from stable patients and non-rejectors was close or above 70% (Table 4), similarly in Basiliximab and Rituximab-induced patients (Fig. 6g), with virtually no double-positives for TCMR and BKVN (Table 4). On the contrary, 67% of the samples from Alemtuzumab-induced non-rejectors were BKVN-positive and, as high as 44%, were double-positive, with only a few samples being double-negative (Table 4). KTRs pre-transplantation (Fig. 6d), as well as healthy controls, were strongly BKVN-negative.
Fig. 6.
Predicted probability of BK-virus nephropathy in kidney transplant recipients.
Predicted probability – (a,e) calculated as the median of the set of probabilities estimated for each patient of the development dataset in the 50 repeats of the cross-validation cycles (Step 12 of Note 1 in Supplementary Discussion) or (b,c,d,f,g) based on the newly-developed six-gene signature (Supplementary Fig. S11); ABMR – antibody-mediated rejection (category 2 according to Banff’09 classification); BKVN – BK virus nephropathy (confirmed with a specialised histological staining); TCMR – T-cell-mediated rejection (category 4); Mixed – histological features of both, ABMR and TCMR; TCMR+ − patients with TCMR only or with mixed-type histology; Stable – patients fulfilling the inclusion and exclusion criteria listed in Table 1; Non-rejector – patients not fulfilling the selection criteria in Table 1 but without clinical or histological evidence of TCMR, ABMR or BKVN up to day 400 post-transplantation; dot colour: black – rejectors, grey – stable or non-rejector patients; dot shape - induction agent in KALIBRE patients: circle – Basiliximab, triangle – Rituximab, square – Alemtuzumab; dotted lines - cut-off 0.2 for dichotomous classification of the probability of BKVN; samples: rejectors - single pre-biopsy sample, BKVN –single near-biopsy sample, stable/non-rejector patients – summary (median per patients) of longitudinal samples covering between days four and 400 post transplantation; (a) Discovery – training patients used in TCMR signature development (TCMR (n = 27) fulfilling the inclusion and exclusion criteria listed in Table 1 and stable patients (n = 27)); (b) Validation – test patients from the KALIBRE study used for TCMR signature validation (Mixed-type rejectors (n = 9) and new stable patients (n = 17)); (c) External – patients from the independent trial EMPIRIKAL used for TSMR signature validation (rejectors (n = 9) with TCMR or Mixed-type histology and non-rejectors (n = 15)); (d) PreTx – cross-sectional pre-transplantation samples - one per patient from rejectors with TCMR (n = 16) and Mixed-type (n = 2) histology and training stable patients from the signature development dataset (n = 20); (e) ABMR rejectors (n = 5), RitCam rejectors with alternative induction and TCMR (n = 6) or Mixed-type (n = 2) histology; BKVN patients: grey dots BKVN (n = 7) included in the BKVN signature development dataset, crossed dots BKVN: +dot (n = 2) excluded from the training dataset due to prior suspected TCMR or some genes missing, x-dot (n = 1) Alemtuzumab induction, x (n = 1) a patient from the EMPIRICAL trial; (f) – cross-sectional pre-biopsy samples (up to 15 days prior to the first post-transplantation biopsy (median two days) and a single one at 25 days) from patients which did not show features of rejection at least up to three months post biopsy; Cat – histological category according to Banff’09 classification (Cat.1 – normal (n = 8, three (38%) BKVN-positive), Cat.3 – borderline changes (n = 33, eight (24%) BKVN-positive), Cat.5 - interstitial fibrosis and tubular atrophy (n = 10, four (40%) BKVN-positive), Cat.6 – other non-rejection histology (n = 38, 11 (29%) BKVN-positive)); star dots (*) – patients with histological features of borderline changes treated for rejection due to clinical considerations; (g) – summary (median per patient) of longitudinal validation follow-up samples from non-rejectors with different induction agents: Basiliximab induction (n = 35, six (17%) BKVN-positive median), Rituximab (n = 18, four (22%) BKVN-positive), Alemtuzumab (n = 9, seven (78%) BKVN-positive median); p-values derived from a Wilcoxon-Mann-Whitney tests.
4. Discussion
We present out the most comprehensive analysis of potential non-invasive biomarkers of AR following kidney transplantation to date. Notably, we have conducted longitudinal, as well as cross sectional analysis, considering changes in gene expression over time post-transplantation. We have also examined the effect of immunosuppressive agents (type of induction agent and prednisolone reduction) and have shown separation from BKVN, a different form of allograft inflammation. We accept, however, that a limitation of our study is the relatively small number of independent validation patients with TCMR, the lack of diagnostically difficult patients and the very limited number of BKVN patients. It should also be noted that AR is not a simplified present/absent condition and has various degrees of severity, so the clinical value of AUCs and other performance measures for binary outcomes should be evaluated with caution. Further, the number of patients with features only of ABMR in the first biopsy diagnostic of AR was limited, so we could not reliably evaluate whether our six-gene signature could discriminate TCMR from ABMR, or whether it would have the same predictive value for ABMR. However, we believe that our signature is relevant to TCMR because it includes genes that have been associated with TCMR in completely different datasets and the statistical algorithm was trained to discriminate TCMR from non-rejection.
Two previous similarly-sized studies have identified gene panels in non-invasive samples to detect AR. The assessment of AR in renal transplantation (AART study) involved 436 adult renal transplant recipients from eight transplant centres in the United States (US), Spain and Mexico and used the 17 gene panel kidney solid organ response test (kSORT) to detect patients at high risk of AR [16]. However, this study collected only cross-sectional samples with lower number of samples analysed, from a heterogeneous population of both, adult and paediatric recipients, from different countries and without a standard immunosuppression regimen. Given that the majority of the centres were in the US, it is likely they received depleting antibody induction therapy. This might explain why the statistical selection of genes for their signature did not favour any of the literature-based genes selected by the statistical algorithm in our signature, when it is highly likely that the microarrays informing gene selection in the AART study would have contained these genes. As some of the genes involved are derived from lymphocytes, which are killed by depleting induction therapy, it is not surprising that we have found an effect of this therapy on our biomarker performance. Potentially, differences in the statistical approaches for selection may have also contributed to the lack of overlap. The CTOT-04 study collected serial urine samples from 485 recipients from multiple centres across the US, and identified a three-gene signature predictive of AR [2]. However, the study highlighted the difficulty of QC of urine samples. Analysis of urine is not possible in many patients with delayed graft function and anuria. This signature was also positive in BKVN.
Our study is the only one to date to demonstrate and display the pronounced within-patient gene-expression variability, systematic changes post-transplantation and association with prednisolone dose. It also uniquely examines individual patient trajectories. Our QC samples demonstrated very low between-batch variability, indicating that the high within-person variability is driven by biological, rather than analytical factors. TCMR-positivity in non-rejectors with poorer kidney function was similar to that in stable KTRs with good kidney function (Table 4), illustrating that our TCMR signature provides information on the underlying immunological response, independent of kidney function. Further, half of the samples from healthy controls, expected to show vigilant immunological response to everyday environmental triggers, were TCMR-positive, indicating an association of our TCMR signature with active host-defence mechanisms.
Evidence that the TCMR signature genes reflect pro-inflammatory immunological pathways stems from the fact that IFNG and IP10, both coding cytokines generated after Th1-cell activation [17], were up-regulated in TCMR (Supplementary Fig. S5). Further, SEMA7A gene, included in the Allomap signature of cardiac AR [18] and strongly negatively associated with heart function [19], showed the highest positive regression coefficient, equivalent to the largest fold-increase in TCMR (Supplementary Fig. S5). Its product Sema7A, a membrane bound semaphorine, is a potent pro-inflammatory monocyte [20] and macrophage stimulator [21].
Our signature emerges four weeks earlier than that shown by a three-gene urine-based signature of TCMR [2], which shares with our signature IP10 up-regulation. Similarly Perforin, GranzymeB, CXCR3 and TGFB were statistically excluded as not relevant to TCMR discrimination (Supplementary Fig. S5). A criticism of the three-gene signature has been the lack of discrimination from BKVN [22]. We, however, additionally provide a six-gene BKVN signature, negatively associated with the predicted probability of TCMR, to complement the differential diagnosis (Supplementary Fig. S11). Only in the case of Alemtuzumab induction there was high positivity for both, TCMR and BKVN (Table 4), the latter likely stemming from the vigorous immunosuppression. All healthy controls and KTRs pre-transplantation (Fig. 6d) were strongly BKVN-negative, further supporting that our BKVN signature reflects BKV activation kept tightly under control in individuals without immunosuppression.
In support of a mechanistic involvement of BKVN signature-genes (Supplementary Fig. S11) in the immune response to viral pathogens, TGFB gene expression in peripheral blood mononuclear cells and transplants has been found positively associated with BKV viremia and BKVN in KTRs [[23], [24], [25]]. Further, IL15 gene, the product of which is instrumental to NK-cell activation in response to viral infections [26] and is implicated in the expansion of BKV-specific T-cells [27], has been reported as downregulated in human endothelial cells infected with BKV [28]. In addition, MARCH8, has been identified as an antiviral factor involved in reduction of viral infectivity, with high expression in monocyte-derived macrophages [29]. Nevertheless, our BKVN signature would need further validation in a larger BKVN dataset.
While not able to replace the present biopsies as a gold standard to confirm AR, our panel may have a role in serial monitoring, providing the clinician with valuable extra information on immune system status to help manage KTRs. Serial monitoring with biopsies remains a high-risk, costly and impractical strategy. Clinical decision-making post-transplantation is complex and utilises a number of factors to determine a particular course of action, and this should remain the case. Potential clinical applications of our test could refine better the patients that may need a biopsy, it could include earlier detection and treatment of AR through earlier biopsy, help in interpretation of cases where the biopsy is reported as “borderline”, detection of sub-clinical rejection in a biopsy where there is no evidence of graft dysfunction based on SCr and separation of other causes of graft dysfunction such as BKVN. The panel could also be used to detect patients at low risk of rejection, thereby allowing reduction of immunosuppression, thus minimising side effects. Further prospective analysis is now required to determine whether or not the use of such a test can improve clinical outcomes.
Acknowledgments
Acknowledgements
SC, IRM, DS, and MHF have received funding from FP7-HEALTH-2012-INNOVATION-1 (project number 305147: BIO-DrIM), MHF also received funding from (grant agreement no HEALTH-F5–2010–260687). MHF, GL, TK and SS acknowledge funding from the Medical Research Council (grants G0600698 and MR/J006742/1; G0802068; MR/K002996/1 and G0801537/ID: 88245). MHF, GL and PC from Guy's and St Thomas' Charity (grants R080530 and R090782). SS and MHF received funding from Quest Diagnostic Collaborations.
All authors received support from the National Institute for Health Research Biomedical Research Centre at Guy's and St Thomas' and King's College London. Anonymised clinical data was also provided NIHR Health Informatics Collaborative. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. The NIHR Health Informatics Collaborative includes: Bradley JR, University of Cambridge; Channon KM, University of Oxford; Lord GM, King's College London; Weber J, Imperial College London; Williams B, University College London; Crichton C, University of Oxford; Davies J, University of Oxford; Welch J, University of Oxford; Woods K, Oxford University Hospitals NHS Foundation Trust. All NHS centres received service support through UK Clinical Research Network portfolio.
The study sponsors had no involvement in the study design, the collection, analysis, and interpretation of data, nor in the writing of the report and neither in the decision to submit the paper for publication.
We gratefully acknowledge the contribution from other clinicians and research nurses who helped in the sample and clinical data provision from KALIBRE study centres, as detailed:
Guy's & St Thomas´ NHS Foundation Trust: William Gage, Adrian Green, Rosalynn Austin, Jude Green, Darlene J Catalan, Ndi Ekwere, May Flor Rabuya, Susie Wen, Karen Williams and Golda-Grace Azanu.
King's College Hospital NHS Foundation Trust: Mohammead Yousaf and Fiona Sharples,
Kent and Canterbury University Hospitals NHS Trust: Sarah Knight, Gillian Eaglestone, Louis Watts, Joanne Carter and Hazel Broad.
The completeness of this project would not have been possible without very significant contributions from project managers Tom Lewis and Akiko Tsutsui and our data architect Syed Hassan; a number of students participated in clinical data input coordinated for a long while by Alexandra Dudek. In the definition and tracking of acute rejection Katie Brown helped with prior data and histopathologist Catherine Horsfield has been central for biopsy report records.
Declaration of conflict of interests
Grants have been declared above.
MR, PM, TLT, CD, CD, CD-V, YK, FD, RM, AS, SPK, BT and CF declare no conflict of interest.
MHF and IRB are currently employees of UCB Celltech, a pharmaceutical company based in the UK. Their involvement in the conduct of this research was solely in their capacity as academics whilst at King's College London.
In memoriam: David Grimwade was a central figure defining our RT-PCR strategy and gene selection, he was co-applicant in the Charity Grant. Terry Strom would have loved to see this manuscript finished, we were continuing his legacy, he was tremendously enthusiastic and supportive every single time he came to visit when we were updating him with the progress of the project.
Contributions
Sofia Christakoudi contributed to literature search, study design, data analysis and interpretation & manuscript preparation and revision. Manohursing Runglall contributed to literature search, study design, sample collection, data analysis and interpretation & manuscript preparation and revision. Paula Mobillo contributed to sample collection, data analysis and interpretation & manuscript preparation and revision. Tjir-Li Tsui contributed to sample collection, data analysis and interpretation & manuscript preparation and revision. Claire Duff contributed to sample collection, data analysis and interpretation & manuscript preparation and revision. Clara Domingo-Vila contributed to sample collection, data analysis and interpretation & manuscript preparation and revision. Yogesh Kamra contributed to sample collection, data analysis and interpretation & manuscript preparation and revision. Florence Delaney contributed to sample collection, data analysis and interpretation & manuscript preparation and revision. Rosa Montero contributed to literature search, sample collection, data analysis and interpretation & manuscript preparation and revision. Anastasia Spiridou contributed to sample collection, data analysis and interpretation & manuscript preparation and revision. Theodoros Kassimatis contributed to sample collection, data analysis and interpretation & manuscript preparation and revision. Sui Phin-Kon contributed to sample collection, data analysis and interpretation & manuscript preparation and revision. Beatriz Tucker contributed to sample collection, data analysis and interpretation & manuscript preparation and revision. Christopher Farmer contributed to sample collection, data analysis and interpretation & manuscript preparation and revision. Terry B Strom contributed to literature search, study design and grant provision/supervision. Graham M Lord contributed to manuscript preparation and revision and grant provision/supervision. Irene Rebollo-Mesa contributed to study design, data analysis and interpretation, manuscript preparation and revision and grant provision/supervision. Daniel Stahl contributed to study design, data analysis and interpretation, manuscript preparation and revision and grant provision/supervision. Steven Sacks contributed to literature search, study design, manuscript preparation and revision and grant provision/supervision. Maria P Hernandez-Fuentes contributed to literature search, study design, data analysis and interpretation, manuscript preparation and revision and grant provision/supervision. Paramit Chowdhury contributed to literature search, study design, data analysis and interpretation, manuscript preparation and revision and grant provision/supervision.
All authors agree to be accountable for the accuracy of the manuscript (except Terry Strom R.I.P.).
Funding
EU: FP7-HEALTH-2012-INNOVATION-1 (project-305147: BIO-DrIM) and FP7 grant agreement no HEALTH-F5–2010–260687.
Medical Research Council: G0600698, MR/J006742/1; G0802068; MR/K002996/1 and G0801537/ID: 88245.
NIHR Biomedical Research Centre at Guy's and St Thomas' and King's College London.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.01.060.
Contributor Information
Maria P. Hernandez-Fuentes, Email: maria.hernandez@kcl.ac.uk.
Paramit Chowdhury, Email: Paramit.Chowdhury@gstt.nhs.uk.
Appendix A. Supplementary data
Supplementary material
References
- 1.Hoffmann S.C., Hale D.A., Kleiner D.E. Functionally significant renal allograft rejection is defined by transcriptional criteria. Am J Transplant. 2005;5(3):573–581. doi: 10.1111/j.1600-6143.2005.00719.x. [DOI] [PubMed] [Google Scholar]
- 2.Suthanthiran M., Schwartz J.E., Ding R. Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. N Engl J Med. 2013;369(1):20–31. doi: 10.1056/NEJMoa1215555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng M.C. The AlloMap genomic biomarker story: 10 years after. Clin Transplant. 2017;31(3) doi: 10.1111/ctr.12900. [DOI] [PubMed] [Google Scholar]
- 4.Pan L., Lyu Z., Adam B. Polyomavirus BK nephropathy-associated transcriptomic signatures: a critical reevaluation. Transplant Direct. 2018;4(2):e339. doi: 10.1097/TXD.0000000000000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohl D.L., Brennan D.C. BK Virus Nephropathy and Kidney Transplantation. Clin J Am Soc Nephrol. 2007;2(Supplement 1):S36–S46. doi: 10.2215/CJN.00920207. [DOI] [PubMed] [Google Scholar]
- 6.Adam B., Randhawa P., Chan S. Banff Initiative for Quality Assurance in Transplantation (BIFQUIT): reproducibility of polyomavirus immunohistochemistry in kidney allografts. Am J Transplant. 2014;14(9):2137–2147. doi: 10.1111/ajt.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartono C., Muthukumar T., Suthanthiran M. Noninvasive diagnosis of acute rejection of renal allografts. Curr Opin Organ Transplant. 2010;15(1):35–41. doi: 10.1097/MOT.0b013e3283342728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sis B., Mengel M., Haas M. Banff '09 meeting report: antibody mediated graft deterioration and implementation of Banff working groups. Am J Transplant. 2010;10(3):464–471. doi: 10.1111/j.1600-6143.2009.02987.x. [DOI] [PubMed] [Google Scholar]
- 9.Kassimatis T., Qasem A., Douiri A. A double-blind randomised controlled investigation into the efficacy of Mirococept (APT070) for preventing ischaemia reperfusion injury in the kidney allograft (EMPIRIKAL): study protocol for a randomised controlled trial. Trials. 2017;18(1):255. doi: 10.1186/s13063-017-1972-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rebollo-Mesa I., Nova-Lamperti E., Mobillo P. Biomarkers of tolerance in kidney transplantation: are we predicting tolerance or response to immunosuppressive treatment? Am J Transplant. 2016;16(12):3443–3457. doi: 10.1111/ajt.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajian-Tilaki K. Sample size estimation in diagnostic test studies of biomedical informatics. J Biomed Inform. 2014;48:193–204. doi: 10.1016/j.jbi.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 12.RCoreTeam . R Foundation for Statistical Computing; Vienna, Austria: 2015. R: A language and environment for statistical computing.https://wwwR-projectorg/ [Google Scholar]
- 13.Troyanskaya O., Cantor M., Sherlock G. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17(6):520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 14.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 15.Robin X., Turck N., Hainard A. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roedder S., Sigdel T., Salomonis N. The kSORT assay to detect renal transplant patients at high risk for acute rejection: results of the multicenter AART study. PLoS Med. 2014;11(11) doi: 10.1371/journal.pmed.1001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luster A.D., Ravetch J.V. Biochemical characterization of a gamma interferon-inducible cytokine (IP-10) J Exp Med. 1987;166(4):1084–1097. doi: 10.1084/jem.166.4.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng M.C., Eisen H.J., Mehra M.R. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6(1):150–160. doi: 10.1111/j.1600-6143.2005.01175.x. [DOI] [PubMed] [Google Scholar]
- 19.Eleid M.F., Caracciolo G., Cho E.J. Natural history of left ventricular mechanics in transplanted hearts: relationships with clinical variables and genetic expression profiles of allograft rejection. JACC Cardiovasc Imaging. 2010;3(10):989–1000. doi: 10.1016/j.jcmg.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Holmes S., Downs A.M., Fosberry A. Sema7A is a potent monocyte stimulator. Scand J Immunol. 2002;56(3):270–275. doi: 10.1046/j.1365-3083.2002.01129.x. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki K., Okuno T., Yamamoto M. Semaphorin 7A initiates T-cell-mediated inflammatory responses through alpha1beta1 integrin. Nature. 2007;446(7136):680–684. doi: 10.1038/nature05652. [DOI] [PubMed] [Google Scholar]
- 22.Suthanthiran M., Muthukumar T. Urinary-cell mRNA and acute kidney-transplant rejection. N Engl J Med. 2013;369(19):1860–1861. doi: 10.1056/NEJMc1310006. [DOI] [PubMed] [Google Scholar]
- 23.Abend J.R., Imperiale M.J. Transforming growth factor-beta-mediated regulation of BK virus gene expression. Virology. 2008;378(1):6–12. doi: 10.1016/j.virol.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mannon R.B., Hoffmann S.C., Kampen R.L. Molecular evaluation of BK polyomavirus nephropathy. Am J Transplant. 2005;5(12):2883–2893. doi: 10.1111/j.1600-6143.2005.01096.x. [DOI] [PubMed] [Google Scholar]
- 25.Yin W.Y., Lee M.C., Huang H.B., Lu M.C. Increased gene expression of TGF-beta in peripheral blood mononuclear cells from renal transplant patients with polyomavirus BK viremia. Clin Transplant. 2016;30(4):393–398. doi: 10.1111/ctr.12698. [DOI] [PubMed] [Google Scholar]
- 26.Paolini R., Bernardini G., Molfetta R., Santoni A. NK cells and interferons. Cytokine Growth Factor Rev. 2015;26(2):113–120. doi: 10.1016/j.cytogfr.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Geyeregger R., Freimuller C., Stevanovic S. Short-term in-vitro expansion improves monitoring and allows affordable generation of virus-specific T-cells against several viruses for a broad clinical application. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0059592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grinde B., Gayorfar M., Rinaldo C.H. Impact of a polyomavirus (BKV) infection on mRNA expression in human endothelial cells. Virus Res. 2007;123(1):86–94. doi: 10.1016/j.virusres.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Tada T., Zhang Y., Koyama T. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins. Nat Med. 2015;21(12):1502–1507. doi: 10.1038/nm.3956. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material