Abstract
Background
Transcriptional dysregulation drives cancer formation but the underlying mechanisms are still poorly understood. Renal cell carcinoma (RCC) is the most common malignant kidney tumor which canonically activates the hypoxia-inducible transcription factor (HIF) pathway. Despite intensive study, novel therapeutic strategies to target RCC have been difficult to develop. Since the RCC epigenome is relatively understudied, we sought to elucidate key mechanisms underpinning the tumor phenotype and its clinical behavior.
Methods
We performed genome-wide chromatin accessibility (DNase-seq) and transcriptome profiling (RNA-seq) on paired tumor/normal samples from 3 patients undergoing nephrectomy for removal of RCC. We incorporated publicly available data on HIF binding (ChIP-seq) in a RCC cell line. We performed integrated analyses of these high-resolution, genome-scale datasets together with larger transcriptomic data available through The Cancer Genome Atlas (TCGA).
Findings
Though HIF transcription factors play a cardinal role in RCC oncogenesis, we found that numerous transcription factors with a RCC-selective expression pattern also demonstrated evidence of HIF binding near their gene body. Examination of chromatin accessibility profiles revealed that some of these transcription factors influenced the tumor's regulatory landscape, notably the stem cell transcription factor POU5F1 (OCT4). Elevated POU5F1 transcript levels were correlated with advanced tumor stage and poorer overall survival in RCC patients. Unexpectedly, we discovered a HIF-pathway-responsive promoter embedded within a endogenous retroviral long terminal repeat (LTR) element at the transcriptional start site of the PSOR1C3 long non-coding RNA gene upstream of POU5F1. RNA transcripts are induced from this promoter and read through PSOR1C3 into POU5F1 producing a novel POU5F1 transcript isoform. Rather than being unique to the POU5F1 locus, we found that HIF binds to several other transcriptionally active LTR elements genome-wide correlating with broad gene expression changes in RCC.
Interpretation
Integrated transcriptomic and epigenomic analysis of matched tumor and normal tissues from even a small number of primary patient samples revealed remarkably convergent shared regulatory landscapes. Several transcription factors appear to act downstream of HIF including the potent stem cell transcription factor POU5F1. Dysregulated expression of POU5F1 is part of a larger pattern of gene expression changes in RCC that may be induced by HIF-dependent reactivation of dormant promoters embedded within endogenous retroviral LTRs.
Keywords: Transcription factors, Kidney cancer, Renal cell carcinoma, Cancer epigenetics, Cancer stem cell, Regulatory genomics
Reseach in context.
Evidence before this study
The most common kidney malignancy, renal cell carcinoma (RCC), canonically stabilizes the hypoxia inducible factor (HIF) family of transcription factors early during its oncogenesis. The HIFs are potent transcription factors that initiate a gene expression program that promotes angiogenesis and metabolic derangements in RCC cells. Prior to this study, the genome-wide epigenetic changes that predicate these gene expression changes had not been characterized. It had also been known that additional transcription factors collaborate with HIF to direct RCC's epigenetic landscape, but their regulatory relationship to HIF had remained unclear.
Added value of this study
This study reports the generation and integrated analysis of nucleotide-resolution functional genomic datasets (chromatin accessibility and gene expression) on patient-matched tumor and normal primary cultures of RCC and its cell of origin – renal cortical tubule cells. Several transcription factors with increased expression in RCC show evidence of HIF binding near their gene body. Many of these same transcription factors show enrichment of their DNA binding motifs in open chromatin regions in the RCC samples. One of these, the stem cell transcription factor, POU5F1 is consistently upregulated in tumor cells both in this study and the larger The Cancer Genome Atlas (TCGA) cohort. Using 5′-RACE, the authors identified a novel HIF-responsive POU5F1 transcript initiating from an endogenous retroviral long terminal repeat (LTR) element. Rather than being unique, the authors found that several other endogenous retroviral LTRs in the RCC genome exhibit HIF binding and transcriptional activity thus providing an epigenomic mechanism for recurrent transcriptional signatures seen in RCC.
Implications of all the available evidence
This study and its associated datasets enrich our understanding of the complex gene regulatory programs that lie downstream of HIF activation in RCC. The use of patient-matched tumor-normal sample pairs greatly increases the robustness of genomic signals. HIF-dependent upregulation of POU5F1 and other genes induced in RCC may be influenced by exaptation of promoters embedded within usually dormant endogenous retroviral LTRs. Taken together, these data provide a novel epigenetic mechanism of gene dysregulation in RCC with immediate implications for patient prognosis.
Alt-text: Unlabelled Box
1. Introduction
Development of new therapeutic strategies for cancer treatment depends on identification of critical mechanisms and pathways utilized by tumor cells. Numerous insights have been gleaned from large tumor consortium programs such as The Cancer Genome Atlas (TCGA), which has extensively catalogued somatic mutations and selected phenotypic features from thousands of tumor and normal tissue samples across a variety of human cancers. To some extent, insights from such broad-based studies are intrinsically limited by tumor heterogeneity (including presence of non-tumor cell types) and general sample variability, which may collectively obscure sensitive and robust detection of subtle changes in cellular pathways such as transcription factor regulatory networks that define and govern the malignant state [1]. Epigenomic mapping of tumors in large consortium-driven projects has generally focused on DNA methylation analysis (TCGA, Roadmap Epigenomics Project) and targeted histone modification profiling using ChIP-seq (Roadmap). These systematic approaches leverage the fact that patterns of regulatory DNA (i.e. promoters, enhancers, insulators) activation and organization are extensively disrupted in cancer [1,2]. Generic identification of regulatory DNA is best achieved by open chromatin profiling methods such as DNase-seq [3] and ATAC-seq [4]. However, the complexity of these deep epigenomic mapping methods has focused their initial application to mouse tissues [5], cultured human cell lines [6], whole adult and fetal human tissues [7], hematopoietic neoplasms (where both malignant and normal cells of origin are readily obtained [8,9]), and a limited number of epithelial malignances [2]. When deploying sensitive epigenomic methods, matched normal tissues of origin provide the best control for patient genotype and environmental exposure but they are often discarded or unavailable at the time of tumor resection. Even very recent large-scale pan-cancer chromatin accessibility profiling projects have focused on detecting patterns across hundreds of tumor samples with heterogenous cellular composition and have omitted analysis of matched normal tissue controls [10]. Taken together, these hurdles have limited the characterization of primary human epithelial malignancies together with their patient-matched normal cells-of-origin.
In this regard, clear cell renal cell carcinoma (RCC), the most common and lethal kidney malignancy, is an ideal model cancer system for high-resolution functional genomic analyses for several reasons. First, RCC tissues are readily available since the standard of care is surgical removal of the often-large tumor mass, frequently with plentiful adjacent, non-neoplastic tissue. Second, the tumor cells and their cells-of-origin – proximal tubule epithelial cells [11] – are readily isolated at high purity, grow well in short-term primary cultures and maintain their genomic and phenotypic characteristics in vitro [12]; this removes the obstacle of contaminating non-relevant cell populations. Third, the majority of spontaneously arising tumors utilize a common oncogenic pathway: stereotypic loss of chromosome 3p, resulting in loss of heterozygosity for the VHL tumor suppressor gene combined with inactivation of the remaining allele of VHL [13]. While it is well understood that loss of functional VHL protein leads to constitutive stabilization of two DNA-binding transcription factors, hypoxia-inducible factors 1α and 2α (HIF1α, HIF2α) [14], the precise nature of genomic dysregulation downstream of HIF pathway activation that drives oncogenesis remains poorly understood. Given that RCC has an annual incidence of >60,000 and mortality of >14,000 in the United States alone (NCI SEER database), additional insights are urgently needed to develop new treatments.
Here, using a combination of DNase I-hypersensitivity mapping (DNase-seq) and transcriptome profiling (RNA-seq) of primary tumor and normal cell cultures derived from three patients, we uncover a high degree of concordance in the epigenomic landscape of RCC. Analyses of these high-resolution reference maps in conjunction with publicly available datasets [[15], [16], [17]] revealed unexpected insights into the genome dysregulation that influences the RCC phenotype. This approach provides a general framework for the analysis of other solid tumors for which matched malignant and normal cells can be isolated at high purity, and greatly amplifies the utility of cancer -omics catalogs.
2. Materials and methods
2.1. Patient tissue sample procurement and primary cell culture
Malignant and normal kidney tissues were obtained from patients undergoing radical nephrectomy for clear cell renal cell carcinoma with informed consent for DNA sequencing obtained prior to the surgery. The study (#1297) and consent forms were approved by the University of Washington's IRB. Patient 1's cultures were derived from an 80-year-old woman; Patient 2's cultures were derived from a 62-year-old man and Patient 3's cultures were obtained from a 63-year-old man. At the time of surgery, all patients presented with localized disease (stage 1). Approximately 1cm3 portions of tumor (from a central, non-necrotic location) and uninvolved kidney cortex (usually from the pole furthest from the tumor mass) were harvested and transported in RPMI medium on ice. These tissues were then minced with a sterilized razor blade and the resulting fragments were placed in 20mls of pre-warmed RPMI medium (without serum) supplemented with Accutase (Sigma, diluted 1:10), collagenase P (Roche, 100 μg/ml) and trypsin/EDTA (Gibco, 0.25% solution diluted 1:10). The tissue fragments were digested at 37 °C for 20 min with vigorous agitation. After digestion, the tissue fragments were spun down and macerated with a sterile plunger from a 5-ml syringe. These softened tissue fragments were then transferred into tissue culture flasks with pre-warmed culture medium (RPMI supplemented with 10% fetal bovine serum and ITS+ supplement, Corning). After 3–4 days (for tubule cultures) and 7–10 days (for RCC cultures), the tissue fragments were decanted and the adherent cells were fed with fresh medium. At this stage, primary tubule cells grew rapidly and had an epithelioid morphology, while primary RCC cells grew slowly, were larger and exhibited frequent cytoplasmic vacuoles typical of adenocarcinoma. Cells were sub-cultured 1:4 when they reached 80% confluence and used within two passages for all experiments.
2.2. 786-O and ACHN cell culture
The VHL-null 786-O (CRL-1932) and VHL-wildtype ACHN (CRL-1611) renal cell carcinoma cell lines were obtained from ATCC. Cells were cultured in RPMI medium supplemented with 10% fetal bovine serum, non-essential amino acids, glutamine and penicillin/streptomycin. Cells were sub-cultured 1:10 when they reached 80% confluence using Accutase to disaggregate adherent cells.
2.3. Processing of cell cultures for DNase-seq
Primary tubule and RCC cultures, 786-O and ACHN cells were subjected to DNase I treatment, small DNA fragment isolation and double-stranded library construction per published ENCODE protocols or a recently described low-input single-stranded library construction protocol [18,19]. Libraries were subjected to paired-end (2x36bp) sequencing. The majority of datasets used in this study were deemed of high quality (signal portion of tags, SPOT > 0.4) [6]. See Supplemental Table 1 for cell input, quality metrics and other sequencing metadata.
2.4. Processing of cell cultures for RNA-seq
Disaggregated cells from primary tubule or renal cell carcinoma cultures, 786-O and ACHN cells were washed once in PBS and stabilized in RNALater (Ambion). Total RNA was extracted using a mirVana RNA isolation kit (Ambion). Illumina sequencer compatible libraries were constructed using a TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold (Illumina) and subjected to paired-end (2x76bp) sequencing. See Supplemental Table 1 for cell input, quality metrics and other sequencing metadata.
2.5. Karyotyping of primary cell cultures
G-band karyotyping of the primary renal cell carcinoma cultures was performed by the University of Washington Cytogenetics and Genomics Laboratory in the Department of Laboratory Medicine.
2.6. Assessing VHL status of primary cell cultures
Genomic DNA from 200,000 cells from each of the primary cultures was extracted using an ArchivePure DNA purification kit from 5Prime. Oligonucleotide primers covering exons 1–3 of the VHL gene (VHL_exon1_F1, GCGCGAAGACTACGGAGGTC; VHL_exon1_R1, CGTGCTATCGTCCCTGCT; VHL_exon2_F1, TCCCAAAGTGCTGGGATTAC; VHL_exon2_R1, TGGGCTTAATTTTTCAAGTGG; VHL_exon3_F1, TGTTGGCAAAGCCTCTTGTT; VHL_exon3_R1, AAGGAAGGAACCAGTCCTGT) were used to amplify genomic sequence using KAPA HiFi Taq polymerase (Kapa Biosystems). The resulting PCR products were separated on an agarose gel, purified and subjected to Sanger sequencing (EuroFins Scientific).
2.7. 5′-RACE for novel POU5F1 transcripts
Total RNA was extracted from 7 × 106 786-O cells using the RNeasy Mini kit (QIAGEN cat #74104) according to manufacturer's protocol. We then used 9 μg total RNA input for RLM-RACE (ThermoFisher Scientific First-Choice RLM-RACE, cat# AM1700), following the manufacturer's “standard scale” 5′-RACE protocol, which ligates an adapter to the 5′ end of full-length, capped mRNA molecules. The primary PCR reaction was carried out using a common forward primer recognizing the 5′-RACE adapter and reverse primer located in each of the first five coding exons of POU5F1 (“R2” primers), using cycling conditions 94 °C 3 min, 35 cycles of 94 °C 3 min/60 °C 30s/72 °C 2 min, 72 °C 7 min. Of the 50 μl primary PCR, 2 μl was used for a secondary PCR with nested primers in the 5′-RACE adapter and within each of the five POU5F1 coding exons (“R1” primers), using the same cycling conditions as the primary PCR. Secondary PCRs were run on an agarose gel, the bands were excised and purified using a MinElute Gel Extraction kit (QIAGEN cat #28604) according to the manufacturer's protocol, and were sequenced from both ends using Sanger sequencing.
2.8. RT-PCR for canonical and novel POU5F1 transcripts
A clone of the VHL-null 786-O RCC cell line stably transduced with VHL (786-O + VHL) and an empty vector (786-O + EV) control line [20] were obtained from Dr. William Kaelin's laboratory (Dana-Farber Cancer Institute, Boston, MA). Approximately 200,000,786-O + EV and 786-O + VHL cells were exposed in triplicate to hypoxia (2% O2) or normoxia for 24 h. RNA was extracted using the RNeasy Plus Mini Kit (Qiagen, Valencia, CA), cDNA was synthesized using random hexamers and the Superscript IV First-Strand Synthesis Kit and was used to seed triplicate real-time PCR reactions using SYBR Green and standard cycling conditions for the Applied Biosystems 7900HT thermocycler. Primers were canonical OCT4 (5′-GAGCAAAACCCGGAGGAGT-3′ and 5′-TTCTCTTTCGGGCCTGCAC-3′); novel OCT4 (5′-GCTTGGCAAATTGCTCGAGTT-3′ and 5′-TGGAGTCCGGACATCTGAAAC-3′), and ACTB (5′-TCCCTGGAGAAGAGCTACG-3′ and 5′-GTAGTTTCGTGGATGCCACA-3′). A single peak was observed in the dissociation curve analysis for all genes and the sequence of the novel OCT4 PCR product was confirmed by Sanger sequencing using the same primers. Cycle threshold (Ct) values were determined using Applied Biosystems Sequence Detection software. Relative quantification was calculated as 2−delta Ct, where delta Ct values were determined by subtracting the ACTB mean Ct values from the target gene Ct values.
2.9. OCT4/POU5F1 immunohistochemistry
A tissue microarray (TMA) composed of cores of 102 cases of localized clear cell RCC, 25 cases of advanced/metastatic RCC, 62 cases of papillary RCC, 50 cases of chromophobe RCC/oncocytic neoplasms and 25 normal kidney controls was prepared with institutional IRB approval (study 9138). Twenty randomly selected RCC specimens (5 in each ISUP grade 1–4) were identified by a third-party honest broker, Northwest Biotrust at the University of Washington. One TMA section or a single section from each of the tumor mass and adjacent uninvolved kidney cortex were subjected to antigen retrieval with HIER ER1 buffer for 20 min (ER1 = Epitope Retrieval Buffer 1, Citrate based pH 6.0 solution). Immunohistochemistry for OCT4/POU5F1 was performed using a 1:250 dilution of the OCT-3/4 (C-10) mouse monoclonal antibody (catalog # sc5279 from Santa Cruz Biotechnology).
2.10. DNase-seq data
Sequence reads from our DNase-seq libraries were subjected to an in-house uniform data processing pipeline, which we have used previously for ENCODE DNase-seq datasets [6]. Briefly, read pairs passing quality filters are trimmed of adapter sequences and aligned to the reference human genome (GRCh37/hg19) using BWA [21]. Genomic regions with a significant enrichment of DNase I cleavages were identified using our hotspot algorithm [6] and were further refined to fixed-width, 150-base-pair regions (“peaks”) containing the highest cleavage density (referred to as DNase I hypersensitive sites, DHSs). Hotspot (FDR 1%) and peak calling were performed using both full-depth and uniformly sub-sampled (to 3.8 × 107 aligned read pairs) data. Also see Supplemental Table 1. As indicated for specific analyses, previously published DNase-seq data (e.g. H1 human embryonic stem cells) were accessed via the ENCODE data portal.
2.11. HIF ChIP-seq data
We downloaded sequence reads from ChIP-seq experiments for HIF-1α, HIF-2α and HIF-1β [16] from GEO (accession GSE67237), aligned them to the reference human genome (GRCh37/hg19) using BWA and identified peak summit locations using the macs2 algorithm [22].
2.12. RNA-seq data
RNA-seq libraries were aligned to the reference human genome (GRCh37/hg19) using TopHat 2.0.13 [23] and assigned to known transcript models (GENCODE v19 basic set) using Cufflinks 2.1.1 [24]. Also see Supplemental Table 1. Processed RNAseqV2 expression tables from TCGA Research Network (http://cancergenome.nih.gov/) were downloaded for frozen tissue samples from organ sites with matched normal and tumor tissues available for comparison. Patient annotations (e.g. tumor stage, metastasis status) for TCGA patient samples were obtained using the UCSC Xena browser tool [25]. As indicated for specific analyses, previously published RNA-seq data (e.g. H1 human embryonic stem cells) were accessed via the ENCODE data portal.
2.13. General data processing
Data analyses were carried out using custom R scripts that utilized Bioconductor (http://www.bioconductor.org) packages for analyzing high-throughput sequencing data, custom Python scripts, and the BEDOPS [26] suite of tools, as well the publicly available tools GoRILLA [27], GREAT [28], GENScan [29] and BDGP neural net promoter prediction [30] where indicated.
2.14. Generation of DHS master list
To facilitate comparisons at the same genomic locus across multiple samples, we created a “master list” of non-overlapping (i.e. non-redundant) 150 bp DHSs. FDR 1% peak calls from all primary tubule and RCC 38 million-tag-subsampled datasets were merged by keeping positions covered by peaks from at least three datasets. Regions where multiple overlapping peaks produced a large contiguous stretch of peak coverage were resolved to multiple, non-overlapping 150-bp segments using a sliding-window approach to find the 150-bp segments of highest coverage within the larger contiguous region.
2.15. Copy-number correction of DNase data
We utilized the “copynumber” package in R to identify genomic regions likely to be subject to copy-number alterations in our RCC samples, with the goal of correcting DNase cleavage counts accordingly so that differences between RCC and TUB samples were more likely to be driven by changes in TF occupancy than by altered copy number. Using the log2-normalized fold-change (RCC/TUB) of DNase tag densities within master list DHSs, we segmented the genomes of all three patient samples (discontinuity parameter gamma = 140). We classified regions whose absolute fold-change were at least twice the median as copy-number variable (Patient 1 = 22 regions, Patient 2 = 26, Patient 3 = 32), and used the mean value of the segment as a scaling factor for raw DNase read counts in those regions for the RCC samples. This analysis detected both 3p loss and 5q gain (confirmed by karyotyping of these patient samples) as well as several focal copy number changes.
2.16. Identification of differential DHSs
We utilized the DESeq2 software package [31] in R to identify DHSs with significant differences in accessibility between replicate tumor and normal samples, analyzing each patient separately. Copy-number-corrected tag counts meeting a minimum threshold in at least one sample (25) within the master-list DHSs were used as input for DESeq2, and sites that met an FDR threshold of 1% were considered differential DHSs.
2.17. Calling of HIF1/HIF2 binding sites and identification of HIF-occupied DHSs
We used macs2 peaks (FDR 1%) from HIF-1α, −1β, and −2α ChIP-seq performed in 786-O cells to classify HIF1 and HIF2 binding sites genome-wide. We classified HIF1 binding sites as HIF-1α peaks that overlapped (by at least 50 bp) a HIF-1β peak (1820 sites) and HIF2 binding sites as HIF-2α peaks that overlapped (by at least 50 bp) a HIF-1β peak (1243 sites). DHSs in our master list were classified as HIF-positive if they overlapped a HIF1 or HIF2 binding site by at least 37 bp (25% of DHS width).
2.18. Calculation of gene expression changes and GO term enrichment
Gene expression fold-changes were calculated as the log2 ratio of FPKM values for RCC/TUB (0.001 was added to each FPKM value to control for zero values). For each patient, genes with FPKM ≥1 in fold-change ≥1.5 in RCC were classified as ‘up-regulated’, the converse criteria were used to classify genes as ‘down-regulated’. All other genes were classified as ‘non-changing’, except those with FPKM <1 in both TUB and RCC, which were considered ‘non-expressed’. Shared (across all three patients) up- or down-regulated gene sets were used (along with the shared non-changing gene list as a background set) as input for the GoRILLA gene ontology enrichment tool.
2.19. Comparisons of regulatory landscapes and differential DHSs among patients
Principal components analysis was performed on log10-transformed DNase I tag densities within master list DHSs (or on FPKM values for RNA-seq data) using the “prcomp” function of R (with center = TRUE and scale = TRUE). Because the master list of DHSs was used to compute differential DHSs for each patient, the DESeq2 calls (FDR 1%) at each site were used to classify the directionality of change at the same genomic locations across all three patients.
2.20. Connection of HIF binding sites to neighboring differentially expressed genes
We were interested in which genes might be regulated by HIF binding events, and considered clusters of HIF+ DHSs as prime candidates for such connections. To this end, we systematically located clusters of HIF+ DHSs arbitrarily within 12.5 kb of one another, merging neighboring clusters, and examined a 1 Mb region centered on each cluster for genes with altered expression (≥1.5 fold-change) in either our patient samples or TCGA RNA-seq data.
2.21. Survival analyses
Survival analysis based on POU5F1 expression levels in the legacy TCGA RNA-seq expression data (split evenly into high- and low-expressing groups at the median expression level) was performed using the UCSC Xena web interface [25].
2.22. Uncovering candidate TF drivers of regulatory landscape alterations
Transcription factor motif models were curated from TRANSFAC (version 11) [32], JASPAR [33], and a SELEX-derived collection [34]. Instances of transcription factor recognition sequences in the human genome were identified by scanning the genome with these motif models using the FIMO tool [35] from the MEME Suite version 4.6 [36] with a 5th order Markov model generated from the 36 bp “mappable” genome used as the background model. Instances with a FIMO P < 10−4 were retained and used for subsequent analyses.
To obtain a “family-level” representation of TF recognition sequences, individual motif models used in the genome-wise FIMO scans were compared in a pairwise fashion using the TOMTOM [37] tool from the MEME Suite version 4.6 [36] with the parameters “-dist kullback -query-pseudo 0.1 -target-pseudo 0.1 -text -min-overlap 0 -thresh 1” and the same 5th order Markov model described above as background. Pairwise comparisons were then hierarchically clustered using Pearson correlation as a distance metric and complete linkage. The resulting trees were cut at a height of 0.1 to select clusters of highly similar motifs.
Motif enrichments were calculated by using a custom Python script to count the number of DHSs that contain a “family” motif (i.e. contained an instance of any motif model within a cluster of highly similar motif models). For a given analysis, these counts were compared between a “foreground” set of DHSs (e.g. shared DHSs with increased accessibility in RCC) and a “background” set (e.g. all other DHSs) and significance was determined using the hypergeometric distribution and subsequent Bonferroni correction of p-values.
Because motif enrichment was computed using family-level representations of TF recognition sequences, we aimed to uncover which member(s) of the POU family might be driving changes in the regulatory landscape of RCC by examining our and TCGA's RNA-seq data for all members of the POU family with a significant enrichment signal.
2.23. Enrichment of repetitive elements in HIF-occupied DHSs
The RepeatMasker annotation of the reference human genome (GRCh37/hg19) was downloaded from the UCSC Genome Browser and compared to our annotations of HIF+ DHSs. We classified a repeat element as coinciding with a DHS (HIF+ or otherwise) if they overlapped by at least 37 bp (25% of DHS width). To calculate enrichments of HIF sites at particular repetitive elements, we calculated frequency of overlap between each repeat family and HIF+ DHSs. A background distribution of expected overlaps was generated by permuting the identity of HIF+ DHSs within our master list of DHSs and repeating the frequency calculation five hundred times (this controlled for any bias of DHSs in general to coincide with particular repeat families). We calculated empirical p-values using a two-sided t-test and the Benjamini-Hochberg correction for multiple testing.
2.24. Assessment of promoter-like behavior at HIF-bound LTR elements
To determine whether HIF-bound LTR elements generally acted as novel promoters, we assessed the strand-specific transcription signal emanating from these elements in each patient. A 1 kb window downstream of each HIF+ LTR element (each LTR has a directionality) was used to count RNA-seq reads in both tubule (TUB) and tumor (RCC) samples mapping to both the positive and negative DNA strands. We then calculated the log2 fold-change (RCC/TUB) for each patient and clustered the data in a heatmap. We identified transcriptional activity at these HIF+ LTRs if they produced RNA transcripts in the same direction as the LTR element (i.e. transcripts induced only on the plus strand, not on the opposite strand, for a plus-strand-oriented LTR element). We also identified the nearest differentially expressed gene in our samples for each HIF+ LTR using the GENCODE v19 Basic annotation set.
2.25. Data availability
All primary and uniformly processed sequence data generated in this study are available at the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE117324. We recently performed a separate and non-overlapping analysis of the tubule data sets included in this study in comparison to human kidney glomerular outgrowth cultures and cultured podocytes [38]. Those data have also been deposited at GEO with accession number GSE115961.
3. Results
3.1. RCC regulatory landscapes are highly concordant across individual tumors
Using RCC as a model system, we first sought to reduce or eliminate the contribution of non-relevant cell types by generating primary cultures of RCC and proximal tubules (cell of origin for RCC) from three patients. In culture, tumor cells were large, grew slowly and frequently contained intracellular vacuoles, typical of adenocarcinoma. In contrast, proximal tubule cells were epithelioid in morphology and grew rapidly (Fig. 1A). Previous work has demonstrated that primary RCC cultures preserve the cytogenetic profile of their originating tumor [12]. In line with this, we found that the primary tumor cultures revealed characteristic karyotype abnormalities associated with RCC: all three patients' tumors carried a loss of the short arm of chromosome 3 (chr3p-) and a gain of the long arm of chromosome 5 (chr5q+) (Fig. 1B and Supplemental Fig. 1A). The VHL gene is located on chr3p, and Sanger sequencing of the remaining allele identified inactivating missense mutations in all three tumor samples (Supplemental Fig. 1B). Taken together with the loss of heterozygosity on chromosome 3p, this indicated that all three patients' tumors were VHL-null, typical of the majority of sporadic RCC [15].
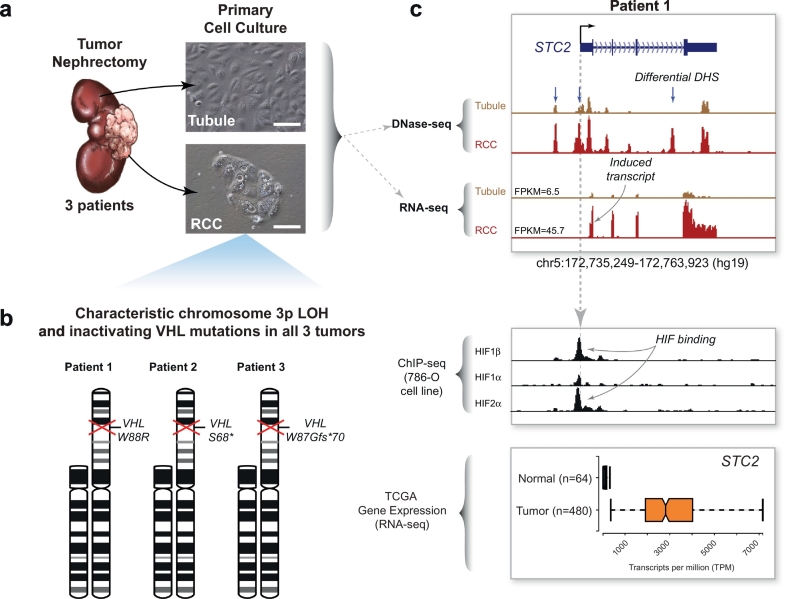
Fig. 1.
Overview of patient samples and data sets used for integrated analyses.
(a) Primary culture of tumor and matched-normal tubule cells from three patients. Renal cell carcinoma tumor nephrectomies from three patients were used to derive primary cultures of proximal tubules and renal cell carcinoma. Microscope images scale bar = 100 μm.
(b) Cytogenetic analysis of primary tumor cultures. Karyotype analysis of the carcinoma cultures revealed loss of the short of arm of chromosome 3 in all three patient samples. Sanger sequencing of the VHL gene in these same samples identified inactivating mutations in the remaining copy.
(c) Example of integrated analysis at the STC2 gene locus. DNase-seq and RNA-seq datasets were also generated from the primary tubule and carcinoma cultures and compared to HIF ChIP-seq datasets from the 786-O renal cell carcinoma cell line and RNA-seq expression data from TCGA. STC2, a canonical HIF target gene, exhibited several differential DHS (blue arrows), some of which coincided with HIF binding determined by ChIP-seq. Compared to normal tubules, the STC2 transcript was strongly induced in the primary tumor cultures and in the TCGA tumor samples (depicted with 10% outlier trim for clarity).
Next, we generated high-quality DNase-seq datasets in duplicate from each patient's primary RCC and tubule cultures. Windowed aggregation of DNase-seq tags again corroborated chromosome arm-level gains and losses delineated by conventional karyotyping (Supplemental Fig. 1C). Globally, accessible chromatin regions appear as DNase-hypersensitive sites (DHSs, called at FDR 1%) and most of these were located >5 kilobases (kb) from known transcription start sites, a feature typical of distal regulatory elements such as enhancers (Supplemental Fig. 1D). In parallel, we generated gene expression profiles (RNA-seq) from these cultures and compared them to TCGA RNA-seq data generated from 72 normal kidney tissues and 534 RCC specimens [15]. Lastly, we cross-referenced our DNase-seq and RNA-seq datasets with publicly available ChIP-seq data for HIF components (HIF1α, HIF2α, HIF1β) from the VHL-null 786-O RCC cell line [16]. As an example of such comparison, STC2, a well-known HIF-induced target gene [39], had several differentially accessible DHSs near its promoter in the RCC samples which correlated with increased STC2 gene expression in our own data and in the larger TCGA data set (Fig. 1C). Some of the induced DHSs near the STC2 promoter overlapped HIF ChIP-seq peaks, consistent with HIF binding at these regulatory elements. However, other induced DHSs do not appear to be bound by HIF, implicating a role for other transcription factors (TFs) in opening nuclear chromatin at these sites.
Genome-wide chromatin accessibility patterns define the regulatory landscape of each primary patient sample. Globally, the regulatory landscapes of the primary tubule cultures showed substantial overlap among the three patients (Fig. 2A). In contrast, while each tumor specimen retained a proportion of DHSs from its tubule of origin, the remainder of its landscape was composed of de novo DHSs. A proportion of these de novo DHSs was shared among the tumor samples, and together with the tubule-derived DHSs retained in the tumors, they defined the shared regulatory landscape of RCC. The similarity of the tubule regulatory landscapes was also evident in the tight clustering of these samples in principal component analysis whereas the RCC samples (and the 786-O RCC cell line) localized to distinct positions in the regulatory space (Fig. 2B).
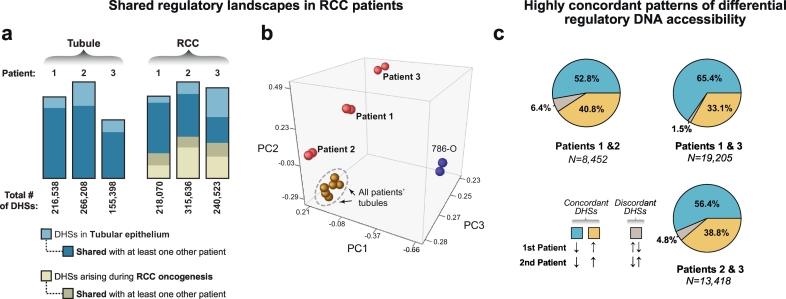
Fig. 2.
Shared regulatory landscapes in tubules and matched renal cell carcinomas from three patients.
(a) Comparison of the shared regulatory landscape among patient samples. The three tubule samples shared a significant proportion of DHSs. Each tumor's landscape of DHSs incorporated a different fraction of DHSs from its tubule of origin and activates de novo DHSs. In the tumors, most of the tubule-derived DHSs were shared with tubule-derived DHSs from other patients. In contrast, a smaller fraction of RCC-derived de novo DHSs was shared among patient tumors.
(b) Comparison of DNase-seq data by principal component analysis. While the tubule cultures from all three patients (brown spheres, in replicate) were tightly clustered, each tumor (red spheres, in replicate) and the 786-O cell line (blue spheres, in replicate) occupied a unique position in regulatory space.
(c) Differential DHSs show highly concordant patterns of accessibility across patient samples. In pairwise comparisons, the shared differential DHSs were classified as concordantly upregulated in the tumor samples (gold), downregulated in the tumor samples (blue) or discordant in the two patient samples being compared (grey). The majority (>95%) of shared differential DHS showed concordant up- or downregulation.
After obtaining a global picture of regulatory landscape similarities based on presence or absence of individual DHS peak calls, we identified accessibility changes between each patient's normal and tumor cells at a common set of DHSs, and then compared the behavior of those differentially accessible sites across the three patients. This analysis identified between 24,976–61,072 differential DHSs (dDHSs, FDR 1%; see Methods) in each patient (roughly equally split between sites with increased and decreased accessibility in tumor cells), representing ~8–20% of all sites examined (Supplemental Fig. 1E). At least 35% of these dDHSs were shared by at least 2 patients. Most strikingly, we found that 93.6–98.5% of dDHSs shared between any two patients displayed highly concordant directional accessibility changes in the tumor samples (Fig. 2C). In total, we identified 6080 dDHSs with concordant accessibility changes across all three patients.
The above results show that primary cultures of proximal tubules and RCC can be generated at high purity and provide an ideal platform for functional genomic methodologies. While the regulatory landscape of each patient's tumor cells was in part unique, the shared DHSs showed highly convergent accessibility changes across all three patients and therefore defined the core regulatory program of RCC.
3.2. Convergent gene expression landscapes
Examination of gene expression profiles for genes changing by >1.5× in all three patient samples revealed consistently increased expression of RCC-associated genes (including VEGFA, CA9, EGLN3, etc.) in tumor cultures with concomitant downregulation of normal tubule-associated transcripts (e.g. CDH1, ANPEP) (Supplemental Fig. 2A). Some tubule-derived genes did not change significantly in the RCC samples (e.g. MME). For subsequent analyses, we chose to anchor on genes that were expressed in our primary tumor cultures since the TCGA RNA-seq dataset is derived from whole kidney and tumor tissue and contains transcripts derived from non-tumor and non-tubule cell types (e.g. circulating immune cells, stromal cells, endothelial cells). Of genes that were expressed at a minimum threshold (FPKM≥1) in our samples, 1072 genes were upregulated and 1207 genes were downregulated across all three patient tumor samples compared to their respective tubule controls. Gene ontology analysis identified pathways characteristically dysregulated in RCC, such as genes related to the hypoxic response (e.g. VEGFA), organic ion transport (e.g. CA9) and lipid metabolism (e.g. FABP6), which were enriched in the upregulated gene set. Genes related to cell cycle regulation (e.g. AURKA, TOP2B) and chromatin organization (e.g. HMGA1) were consistently transcriptionally downregulated (Supplemental Fig. 2B). Thus, the gene expression landscapes of our primary cultures were largely concordant across patient samples and recapitulated the key transcriptional signatures of RCC.
3.3. Concordant tumor regulatory landscapes expose transcription factor drivers of RCC
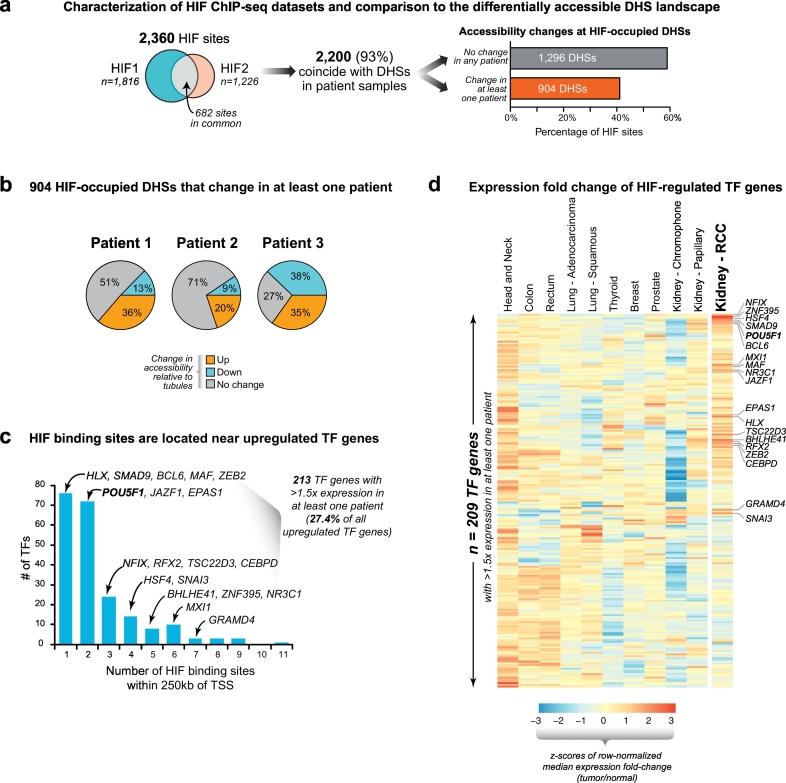
Chromatin accessibility profiling methodologies such as DNase-seq uniquely provide insight into the transcription factor drivers of oncogenesis [1]. Since HIF is canonically dysregulated in RCC, we next explored its role and that of other transcription factors (TFs) in driving the chromatin accessibility changes we observed in the regulatory landscapes of the patients' tumor samples. Even though most (>93%) HIF binding sites coincided with DHSs, ~70% of these DHSs showed no significant change in accessibility between tubule and RCC (Fig. 3A). Even the HIF-bound DHSs that showed significant accessibility changes in one tumor-normal pair often did not show differential DHS accessibility in the other patient samples (Fig. 3B). This suggested that HIF alone does not broadly reprogram the regulatory landscape of RCC, but did not exclude the possibility that it may regulate other TFs that contribute to the process of malignant transformation. 213/776 of the TFs that were upregulated (≥1.5×) in at least one patient RCC-tubule pair had a HIF-occupied DHS within 250 kb of their transcription start site (TSS) (Fig. 3C). A subset of these 213 TFs showed evidence of selective transcriptional induction in RCC compared to multiple somatic tumors for which matched normal tissues were available for comparison in the TCGA expression data (Fig. 3D). We rationalized that since the majority of RCC samples exhibit HIF activation, the TF gene subset that was consistently induced in the TCGA data is more likely to contain TFs truly subject to HIF regulation in RCC. The fact that only a subset of the putative HIF-regulated TFs in our primary culture system showed selective expression in the TCGA RCC RNA-seq data may reflect the contaminating effect of non-tumor cell types in TCGA samples that can obscure small changes in transcription factor genes that are typically expressed at low levels.
Fig. 3.
Concordant tumor regulatory landscapes expose key transcription factor drivers of RCC.
(a) HIF-binding only accounts for a small proportion of the differentially accessible RCC regulatory landscape. ChIP-seq datasets for HIF1A and HIF2A showed substantial overlap with each other and most of these peaks coincided with a DHS in the tubule and/or RCC DNase-seq datasets. Most HIF peaks in DHSs mapped to non-changing/constitutive DHS in the tubules and RCC.
(b) Differentially accessible HIF-bound DHSs show different patterns of accessibility across patient samples. Of the 904 HIF peaks that mapped to differentially accessible DHSs in at least one patient sample, most did not show significant change across the other patients' samples.
(c) Transcription factors with changing expression located near HIF binding sites. The expression levels of 213 transcription factors changed by >1.5× in at least one patient sample and exhibited at least one HIF-bound DHS within 250 kb of their transcription start site (TSS). Many of these contained numerous HIF binding sites in proximity to their TSS, including transcription factors linked to renal cell carcinoma (ZEB2, BHLHE41) and POU5F1.
(d) Selective expression of transcription factors in cancer. The transcription factors that were expressed (FPKM>1) and changing by at least 1.5-fold in any of the three patient samples (from panel C) were examined for differential expression in a wide range of tumors that have matched normal tissues available in the TCGA RNA-seq expression dataset (209 transcription factors are depicted; 4 factors are not mapped in the TCGA RNA-seq data). Transcription factors with RCC-selective increased expression are highlighted (e.g. HSF4, BHLHE41, ZEB2, POU5F1, etc.).
To uncover the identities of the TFs that are likely to be driving the regulatory program of RCC, we determined the relative enrichment of TF recognition sequences within the shared set of differential DHSs (discussed above) compared to a background of static DHSs. AP-1, ETS and E-box family recognition sequences were significantly enriched in DHSs with decreased accessibility in RCC (Fig. 4A). Motifs for basic helix-loop-helix (bHLH) family transcription factors (which include MYC, HIF and BHLHE41) were enriched in DHSs that do not change their accessibility in RCC, i.e. they remain constitutively accessible in both tubule and RCC samples. Recognition sequences for several TF families (including homeodomain, nuclear receptor and HNF1/POU) were enriched in DHSs with increased accessibility in RCC.
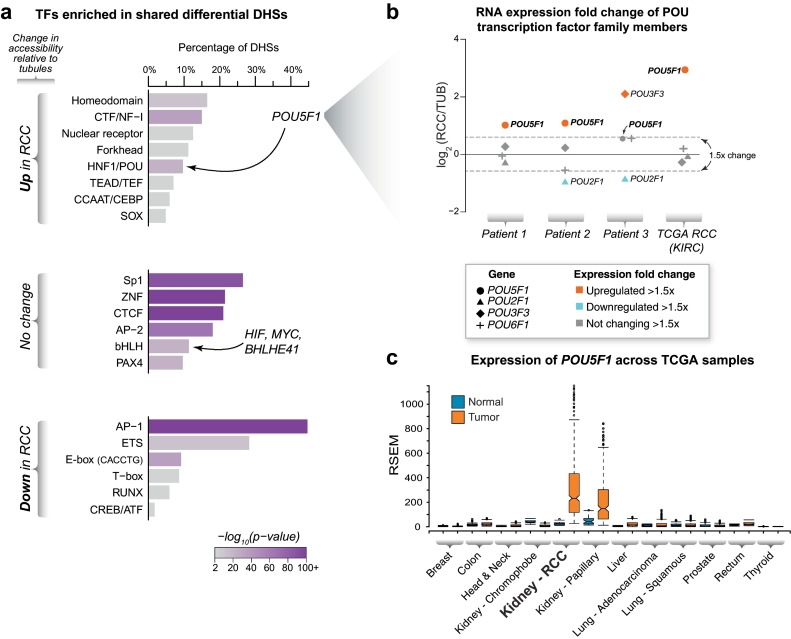
Fig. 4.
Correlation of DNA binding motif enrichments with gene expression identifies enrichment for POU5F1 in RCC.
(a) Transcription factor enrichment. Examination of differentially accessible or non-changing DHSs revealed different classes of transcription factors whose DNA binding recognition sequences were enriched in each category. The motif families containing transcription factors with genetic evidence linked to renal cell carcinoma susceptibility (i.e. MYC, BHLHE41, ZEB2 and HIF) and the stem cell related transcription factor POU5F1 (OCT4) are indicated.
(b) Examination of RNA expression identifies candidate POU-family transcription factors driving motif enrichments in the DHS landscape. Since multiple transcription factors within the POU family share redundant DNA binding motifs, examination of transcription factor expression patterns may identify specific family members that are driving motif enrichment signatures. Examination of the differential gene expression patterns of these family members in RCC vs. tubules in our primary cultures and in the TCGA RNA-seq dataset revealed upregulation of POU5F1 in RCC.
(c) Expression of POU5F1 in diverse somatic tumors. The mRNA expression levels of the stem cell related transcription factor POU5F1 (OCT4) in several non-germ cell tumors was compared to their matched normal tissue controls. The ends of the bar plots represent the 25th and 75th quartiles with whiskers representing 1.5× inter-quartile range (10% outlier trim applied for clarity).
Since several TF family members can recognize the same DNA binding recognition sequence, we next asked if the differential TF gene expression levels between tubules and RCC could help identify the specific family members that were contributing to the observed motif enrichment in the regulatory landscape. This analysis revealed that for the POU family transcription factors, only the stem cell related factor POU5F1 (also known as OCT4) was consistently expressed and upregulated in RCC compared to tubules (Fig. 4B). POU5F1 and some of the transcription factors which are associated with genetic risk for RCC and whose binding sequences were enriched in differentially accessible DHSs (e.g. BHLHE41) showed evidence of regulation by HIF (Fig. 3C). POU5F1 is normally expressed only in stem cells and germ cell-derived tumors but in the larger TCGA data set, it showed strikingly selective induction in RCC and papillary kidney cancer (both derived from proximal tubule cells) compared to normal kidney tissue (Fig. 4C). Other known cellular reprogramming transcription factor genes, namely SOX2, KLF4 and NANOG, were not induced in RCC (data not shown).
Taken together, these results suggest that instead of driving large-scale changes in chromatin accessibility by itself, HIF may have a broader impact on the regulatory landscape of RCC by activating the expression of other transcription factors. We sought to corroborate this notion by closer examination of the role of HIF in the regulation of POU5F1.
3.4. Expression of a novel POU5F1 transcript in RCC from an alternate adult human- and kidney-specific promoter
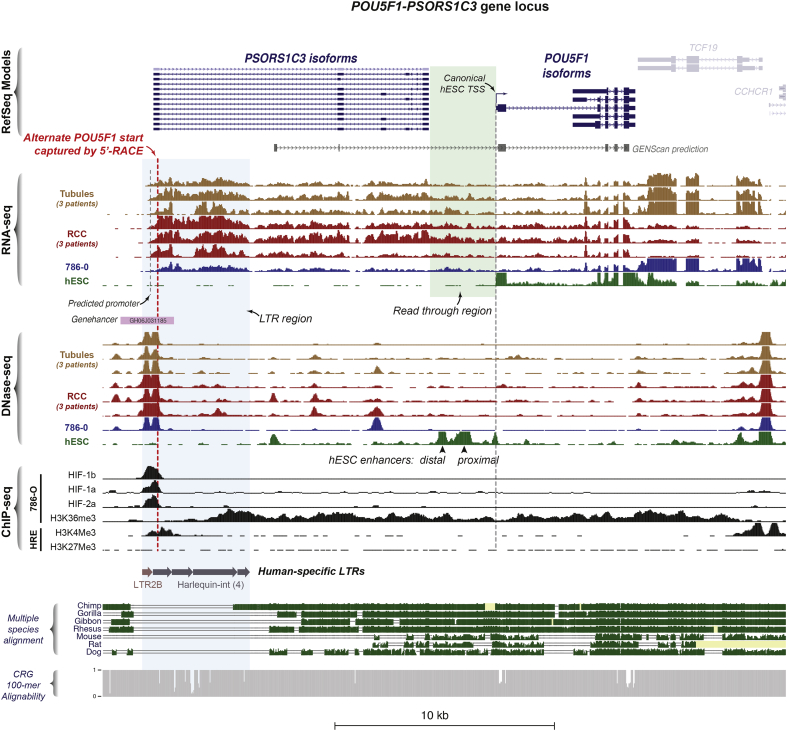
Close examination of the chromatin accessibility and RNA-seq data from our three patients revealed a stretch of RNA transcription starting from a bipartite DHS at 5′-end of the long non-coding RNA (lncRNA) gene PSORS1C3. These transcripts appeared to read through the PSORS1C3 gene and into the annotated POU5F1 transcript isoforms which lie on the same strand (Fig. 5). Like POU5F1, the PSORS1C3 gene is also selectively upregulated in the TCGA RCC data (Supplemental Fig. 3). These transcripts were also present in 786-O cells but were not detected in H1 human embryonic stem cells (hESCs). The initiation of these transcripts lay within a DHS ~16 kb upstream of the POU5F1 TSS, which was distinct from the well-characterized distal and proximal enhancers that regulate POU5F1 in hESCs [40]. Curiously, this DHS was only present in adult kidney tubule- and RCC-derived cells/cell lines and was not detected in hESCs, fetal kidney tissues or many other diverse cell types (Supplemental Fig. 4).
Fig. 5.
A novel human-specific promoter initiates long RNA transcripts through the PSORS1C3-POU5F1 locus in RCC.
Overview of the POU5F1-PSORS1C3 genomic locus (hg19 chr6:31,125,253-31,156,354). RNA-seq tracks for the primary patient samples and the RCC cell line 786-O revealed a novel transcript originating from a DHS ~16 kb upstream of the known human embryonic stem cell (hESC) TSS. This transcript reads through the PSORS1C3 lncRNA gene into the POU5F1 gene body (green shaded box). ChIP-seq in 786-O cells revealed binding of HIF components (HIF1α, HIF2α, HIF1β) to this DHS with evidence of histone modification typical of active transcription across the entire transcript (H3K36Me3). This DHS was also associated with histone modifications characteristic of an active promoter in human renal epithelial cells (HRE), i.e. positioned nucleosomes marked by H3K4Me3 and depletion of the repressive H3K27Me3 mark. Examination of sequence conservation showed that this alternate promoter lies within a complex tandem long terminal repeat (LTR) element that is unique to humans (blue shaded box). CRG, Center for Genomic Regulation.
FANTOM5 data suggested that this DHS acts as a promoter in the kidney: it coincided with a peak in the human renal epithelium (HRE) ChIP-seq signal for H3K4me3, which marks active promoters and lacked an H3K27me3 peak, a repressive chromatin mark. In 786-O cells, this DHS demarcated H3K36me3 signal, a mark associated with transcription elongation, the other end of which extended into annotated POU5F1 transcripts [41,42]. The GeneLoc algorithm, which integrates data from FANTOM, ENCODE, ENSEMBL and VISTA databases [43], also annotated this DHS as a potential promoter/enhancer (Genehancer ID: GH06J031185). A neural-network based eukaryotic promoter prediction algorithm [30] also identified a potential promoter within this DHS. Both of these lines of evidence are consistent with this DHS' location at the TSS of actively transcribed PSORS1C3 gene (Fig. 5). The PSORS1C3 gene is known to have numerous splice isoforms [47] and numerous expressed sequence tags (ESTs) are present at the POU5F1-PSORS1C3 locus (Supplemental Fig. 5). Still, given the presence of RNA-seq reads in the genomic interval between PSORS1C3 and POU5F1 (green shaded box, Fig. 5) and the fact that read through transcription is frequently seen in RCC [44], we sought to determine whether novel transcripts of POU5F1 were generated from the DHS 16 kb upstream of its canonical TSS in RCC. Knowing that the expression of POU5F1 may be confounded by that of its pseudogene, POU5F1B [45,46], we first examined chromatin accessibility and gene expression at the POU5F1B pseudogene locus in our samples, and did not detect significant amounts of either (Supplemental Fig. 6).
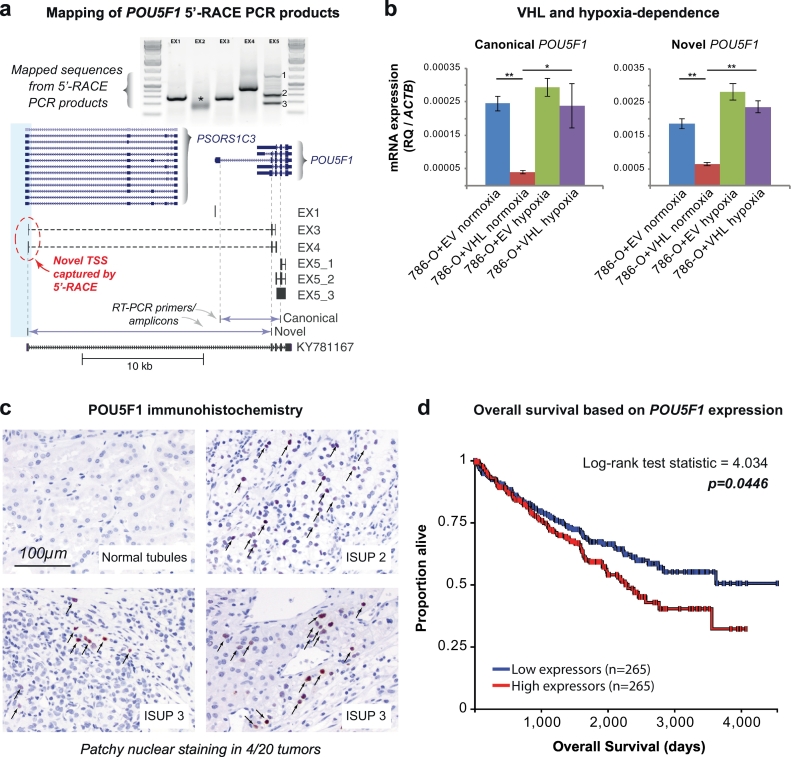
We then proceeded to unambiguously determine if the putative alternate promoter initiated transcription of a novel POU5F1 isoform. To do this, we performed 5’-RACE on cDNA isolated from the VHL-null 786-O RCC cell line and sequenced the resulting products (Fig. 6A). This captured a new transcription start site for POU5F1 originating within the specified DHS (Fig. 5). Several exon-exon combinations were observed in the 5-RACE reaction product suggesting a complex mixture of isoforms expressed in 786-O cells. Curiously, these putative isoforms were also distinct from the GENScan prediction [29] for exon-intron junctions for the long transcript (Fig. 5) and from the OCT4C/OCT4C1 variants (GenBank AB971680, AB971681) that have been recently described [47] (Supplemental Fig. 5). The closest match to this isoform's structure is the expressed sequence tag (EST) KY781167 (Fig. 6A), recently identified in breast cancer [48].
Fig. 6.
The novel transcript for POU5F1 exhibits HIF dependence and POU5F1 expression levels correlate with patient survival.
(a) Schematic mapping of POU5F1 5′-RACE PCR products. 5′-RACE performed on 786-O RNA captured a transcription start site originating in the -16 kb DHS (blue shaded box) therefore defining a novel isoform of POU5F1 that is produced following read through of the PSORS1C3 gene. Reverse primers in known POU5F1 exons (e.g. EX1 = reverse primer in exon 1) were used to amplify the 5′-ends of the cDNA molecule captured by 5′-RACE and sequence mapped to the genome. The exon 2 primer (*) failed to yield mappable sequence. The exon 5 primer yielded 3 different products (EX5-1, EX5-2, EX5-3). The location of RT-PCR primers to detect the canonical and novel POU5F1 transcript variants are also indicated. The inferred isoform structure is compared to EST KY781167.
(b) Canonical and novel POU5F1 transcripts exhibit HIF-dependence. RT-PCR primers were used to quantify the canonical and novel POU5F1 transcripts in 786-O cells stably transduced with VHL (786-O + VHL) or empty vector (786-O + EV) cultured in normoxia or hypoxia (2% O2) for 24 h. Expression levels (relative quantification, RQ) were calculated using the β-actin housekeeping gene (ACTB). Compared to the empty vector control, reintroduction of VHL protein into 786-O cells suppressed expression of both POU5F1 transcripts. Exposing 786-O + VHL cells to hypoxia induced both POU5F1 transcripts. N.B. expression scale differences between the canonical and novel transcripts. Error bars indicate standard deviations of three experimental replicates. *p < 0.05, **p < 0.005.
(c) Immunohistochemistry of POU5F1 protein in renal cell carcinoma samples. POU5F1 (OCT4) immunohistochemistry was performed on RCC samples from 20 patients (5 from each of ISUP grades 1–4) and showed patchy nuclear positivity (arrows) in a single random sample from 4 patients. No nuclear staining was seen in any of the matched normal renal parenchyma from the same patients.
(d) Overall survival as a function of POU5F1 expression in TCGA. Patients with POU5F1 expression data from TCGA (KIRC) were evenly divided into two groups split at the median expression level (233 RSEM normalized) and Kaplan-Meier curves for overall survival were plotted using the UCSC Xena browser tool.
Critically, the DHS located -16 kb upstream of the canonical POU5F1 TSS contained HIF binding motifs which coincided with strong HIF1α and HIF2α ChIP-seq signal in the 786-O cell line, suggesting that HIF is bound to this promoter element in RCC. We note that this HIF site is encoded by long-terminal repeat (LTR) elements of the Harlequin-int and LTR2B subfamilies of ERV1 endogenous retroviruses. This repeat configuration appeared to represent an evolutionarily recent insertion into the human genome as it was not conserved among higher primates or other mammals (Fig. 5). Good alignability [49] at this composite LTR reduced the possibility that degeneracy of viral repeat elements was confounding locus-specific mapping of short-read sequences.
Finally, we asked if the canonical and novel isoforms of POU5F1 exhibited dependence on VHL protein (stably reintroduced into the 786-O cell line) and/or hypoxia using isoform specific RT-PCR primers (Fig. 6A). Reintroduction of VHL protein into 786-O cells cultured in normoxia strongly suppressed expression of both canonical and novel POU5F1 transcripts (Fig. 6B). The presence of VHL protein also resulted in significant induction of canonical and novel POU5F1 transcripts when the 786-O + VHL cells were cultured in hypoxia (Fig. 6B). These transcripts did not change appreciably when 786-O cells (stably transduced with empty vector as a control, 786-O + EV) were shifted from normoxia to hypoxia, consistent with already maximal HIF-signaling in this VHL-null cell line. Taken together, these results established the presence of a kidney-specific promoter element that originated at this site by insertion of transcriptionally active endogenous retrovirus elements specific to the human lineage. This alternate promoter produces a novel transcript isoform of POU5F1 in RCC by read through transcription of the PSORS1C3 lncRNA gene.
3.5. POU5F1 transcript levels in RCC correlate with overall survival
Next, we sought to evaluate if increased POU5F1 transcription led to increased protein levels in human RCC specimens. The novel transcript identified by 5′-RACE did not contain a translation initiation codon and consistent with this, OCT4 protein was not readily detectable in 786-O cells by immunoblotting or mass spectrometry (not shown). However, this did not exclude the possibility that increased transcriptional activity from the alternate promoter could permit expression of canonical OCT4 protein in a subset of cells that in turn was responsible for the population-level POU-family motif enrichment seen in DHSs with increased accessibility in RCC. We decided to test this possibility on human RCC specimens using an antibody recognizing a C-terminal epitope of POU5F1 (OCT4) that is expected to be represented in all of the known isoforms of POU5F1. Initial experiments using a tissue microarray with 102 cases of localized RCC and 25 cases of advanced stage/metastatic RCC did not reveal significant POU5F1 (OCT4) expression in the tumor cells (data not shown). However, since the tissue cores for each individual tumor in the array are very small and may not be representative of the often large and heterogeneous RCC tumors [50,51], we decided to test POU5F1 (OCT4) expression in larger tissue sections from 20 different patient tumors alongside their matched normal kidney controls. In 4 out of 20 RCC tissue sections, patchy nuclear POU5F1 (OCT4) protein expression was readily detectable (Chi-squared p-value = 0.035, Fig. 6C). We did not observe POU5F1 (OCT4) expression in any of the normal kidney tissue sections examined. Therefore, even though POU5F1 transcript induction appears to be a consistent feature of RCC (Fig. 4C), POU5F1 (OCT4) protein is inconsistently detected, which may reflect focal or patchy expression in these large tumors. Lastly, we examined POU5F1 expression in the TCGA data set as a function of clinical staging parameters. The expression of POU5F1 did not correlate with metastasis status (Supplemental Fig. 7A), but was positively correlated with pathologic tumor stage, with higher stage tumors exhibiting greater expression of POU5F1 (Supplemental Fig. 7B). Strikingly, patients with high expression of POU5F1 exhibited lower overall survival compared to patients with lower expression levels (Fig. 6D). Interestingly, PSORS1C3 transcript levels were not correlated with overall survival (not shown). These results demonstrate that POU5F1 (OCT4) protein can be expressed in a patchy fashion in RCC tumors and that POU5F1 expression levels predict overall survival in patients with RCC.
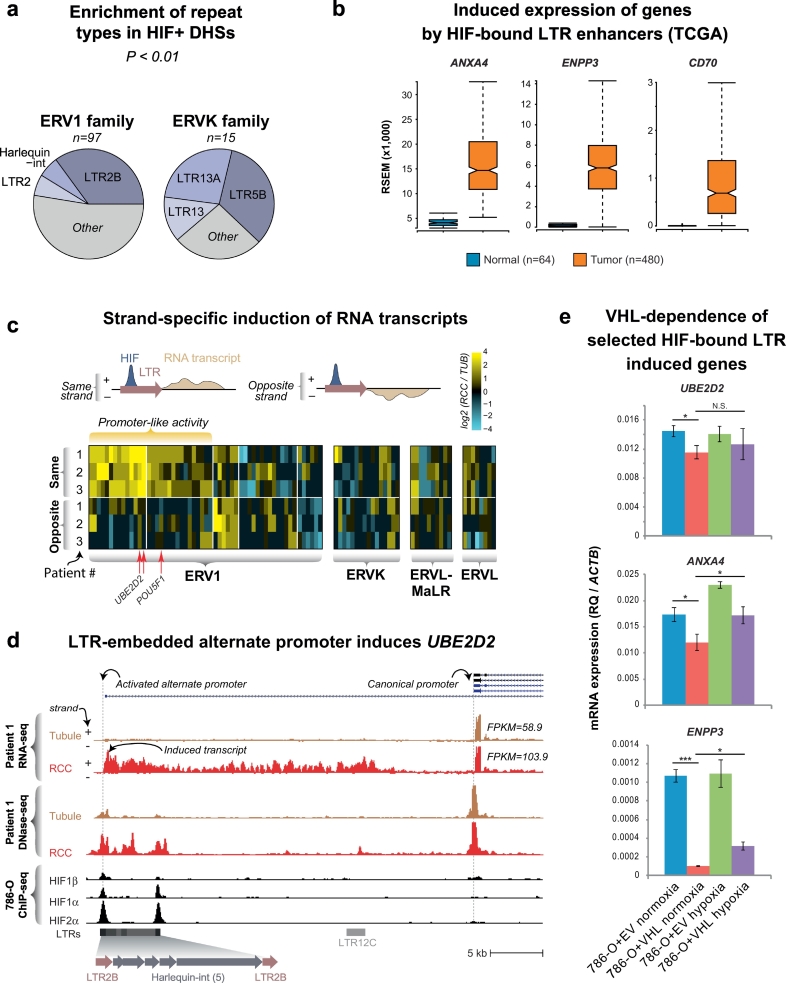
3.6. Generalized HIF-driven exaptation of LTRs in RCC
We next asked whether HIF binding of specific repetitive elements was a generalized phenomenon, and found that 178 out of the 2200 (8.1%) HIF-bound DHS overlapped an LTR element. Approximately 50% of these (90/178) were DHS that exhibited differential chromatin accessibility between the tubule and RCC samples consistent with active regulation at these sites. This specific localization to LTRs was significant for HIF-bound DHSs in ERV1 and ERVK LTR families (empirical P < 0.01), particularly with LTR2/2B and Harlequin-int type elements (Fig. 7A). We posited that HIF binding to LTRs might exapt their regulatory domains to influence the gene expression landscape of RCC. This could occur either with the HIF-bound LTRs acting as enhancers or as direct transcriptional activators/alternate promoters as we had observed for the PSORS1C3-POU5F1 locus. Investigating the first possibility, we found that of the 178 HIF-bound LTRs, 29 are within 250 kb of a gene induced ≥1.5× in the TCGA RNA-seq data (same criteria as used for Fig. 3) suggesting that HIF-bound LTRs may be influencing the gene expression program of RCC. This set of genes included ANXA4 [52], ENPP3 [53], and CD70 [54] that are invariably induced in RCC (Fig. 7B). To explore the second possibility, we tallied transcript production 1 kb downstream of HIF-bound LTRs as read counts on both the plus and minus DNA strands (Supplemental Table 2). We identified 72 transcriptionally active HIF-bound LTRs defined by transcript production (≥20 read counts) in at least one of our samples and strand-selective transcriptional induction (i.e. promoter-like activity) was most prominent for the ERV1 class (Fig. 7C). Some of these appeared to act as alternate promoters associated with upregulation of nearby genes such as for UBE2D2 (Fig. 7D). Similar to the alternate POU5F1 promoter, the UBE2D2 exapted LTR promoter had a tandem LTR2-Harlequin-int substructure. We next decided to test if some of these HIF-LTRs retained intact VHL-HIF axis responsiveness. We extracted RNA from 786-O empty vector or VHL-transduced cells exposed to normoxia or hypoxia (2% O2) as before. We tested the expression of two genes in which HIF-bound LTRs might act as enhancers (ANXA4, ENPP3) and one gene in which the HIF-bound LTRs might act as a direct transcriptional activator/alternate promoter (UBE2D2). Expression levels of these three genes were suppressed when VHL protein was reintroduced into VHL-deficient 786-O cells, consistent with a HIF-dependent mechanism of regulation (Fig. 7E). Taken together, these results suggest that in RCC, HIF stabilization and binding to regulatory elements embedded within LTR elements exapts latent regulatory elements that can act as promoters or enhancers of gene expression.
Fig. 7.
Activation of cryptic LTR-derived promoters in RCC.
(a) Enrichment of HIF-bound DHS in LTR families. The ERV1 and ERVK families of LTR show significant enrichment for HIF-bound DHSs. Of these, the LTR2B subfamily shows the greatest number of HIF-bound DHSs (n = 34).
(b) TCGA expression of selected genes putatively induced by HIF-bound cryptic promoters in LTRs. The ends of the bar plots represent the 25th and 75th quartiles with whiskers representing 1.5× inter-quartile range (10% outlier trim applied for clarity). All tumor-normal comparisons are significant (p < 1 × 10−10) by one-tailed t-test.
(c) HIF-bound DHSs in LTRs show strand-specific induction of RNA transcripts. Since LTRs are intrinsically directional, enumeration of RNA-seq reads up to 1 kb on either the same or opposite strand of the LTR identifies elements with HIF-dependent promoter-like activity (increased transcripts in RCC samples compared to tubules with the same directional orientation as the LTR). The heatmap represents the ratio of the RCC/tubule read counts for each patient on the indicated strand.
(d) A HIF-bound LTR is transcriptionally active and is associated with increased expression of the UBE2D2 gene. Similar to the alternate POU5F1 promoter, some of the HIF-bound LTRs that show promoter-like activity drive the expression of novel transcripts and increase the expression of nearby genes. Shown is the expression of UBE2D2 transcripts, which increases 1.76× in Patient 1's RCC compared to its matched tubule control.
(e) HIF-bound LTR-induced genes exhibit HIF-dependence. RT-PCR primers were used to quantify the indicated transcripts in 786-O cells stably transduced with VHL (786-O + VHL) or empty vector (786-O + EV) cultured in normoxia or hypoxia (2% O2) for 24 h. Expression levels (relative quantification, RQ) were calculated using the β-actin housekeeping gene (ACTB). N.B. expression scale differences between the canonical and novel transcripts. Error bars indicate standard deviations of three experimental replicates. *p < 0.05, ***p < 0.005, N.S. not significant (two-tailed t-test).
4. Discussion
Even for a well-studied tumor such as RCC, there is a notable deficit in the understanding of genome dysregulation that drives oncogenesis. Here we demonstrate that while each patient's tumor can exhibit its own unique epigenomic signature, subtraction of the genotype-matched cell-of-origin baseline and comparison across individuals can robustly identify the core regulatory landscape of cancer. Using high-resolution epigenomic mapping on primary tumors and matched normal cells from three patients, we identified multiple transcription factors with differential expression patterns and significant DNA binding motif enrichments that likely contribute to the tumor phenotype. Transcription factors that drive genome dysregulation in RCC have hitherto only been explored in piecemeal fashion. Besides the HIFs, other sequence-specific factors have been implicated individually in various aspects of RCC biology including PAX2 [[55], [56], [57], [58]], PAX8 [[59], [60], [61]], CEBPβ [62], NRF2 [63,64], FOXO [[65], [66], [67]], STAT3 [[68], [69], [70], [71], [72], [73], [74]], FOXM1 [75,76], POU5F1 (OCT4) [77,78], P53 [[79], [80], [81], [82]], TCF21 [83,84], HCF1 [85], HNF1/2 [[86], [87], [88]] and most recently BHLHE41 [89,90] and ZNF395 [91,92]. Here, we show that many of these transcription factors may in fact be regulated by HIF and influence the regulatory landscape in RCC.
One transcription factor that is frequently upregulated in RCC is the stem cell factor POU5F1, and we found that its DNA recognition sequence is enriched in the open chromatin regions of RCC. Our examination of the POU5F1 genomic locus identified an adult kidney-selective and hypoxia/HIF-responsive promoter that produces a novel transcript isoform for POU5F1 in RCC. This promoter is embedded in an endogenous retroviral LTR element appears to induce POU5F1 by read through transcription of the long non-coding RNA gene PSORS1C3, a phenomenon that is pervasive in RCC [44]. Hypoxia is a known stimulant of POU5F1 expression in embryonic stem and cancer cells [[93], [94], [95], [96]] and can even reprogram committed cells into a pluripotent state [97,98]. Given the unique kidney-specific activity of the LTR-embedded alternate promoter and the fact that VHL inactivation and constitutive HIF stabilization appear to be early events in sporadic RCC [99,100], future studies should focus on determining how VHL inactivation and/or hypoxia contribute to the regulation of POU5F1 expression in kidney tubule cells and RCC from both the canonical and LTR-embedded alternate promoters.
The novel POU5F1 transcript that we identified does not appear to contain a translation initiation codon. Perhaps due to this, we found that only a subset of cells in patients' tumors appear to produce POU5F1 (OCT4) protein. However, the expression of this potent transcription factor in even a subset of cancer cells may still be clinically relevant as this population may represent self-renewing RCC cancer stem cells [76]. Consistent with this idea, we found that higher POU5F1 transcript levels in RCC are associated with poor patient survival in the TCGA data set. Activation of stem cell-like epigenetic and transcriptional programs are associated with malignant transformation, though clear cell RCC appears to behave differently than other tumor types [101]. Our work suggests that further investigation of the role of POU5F1 in RCC tumor cells at single cell resolution [102], and especially in patients with advanced stage tumors, will shed light on the role of this transcription factor on the regulatory landscape and biology of this tumor.
Our analysis of the PSORS1C3-POU5F1 locus led us to uncover a broader epigenetic mechanism influencing the gene expression program in RCC. Rather than being unique to PSORS1C3-POU5F1, we found that in fact, several retroviral LTR elements are bound by HIF and exhibit an accessible chromatin profile in our samples. Some of these HIF-bound LTRs may function as distal enhancers inducing the expression of genes that are important therapeutic targets in RCC such as ENPP3 and CD70 [53,103,104]. Many of these genes also show transcriptional upregulation in the TCGA dataset and at the protein level in mass spectrometry-based profiling of RCC [105]. Other HIF-bound LTRs exhibit strand-specific promoter-like activity that may induce the expression of neighboring genes (e.g. UBE2D2, an E2-ubiquitin ligase, whose downstream substrates include P53 [106,107]) in a manner analogous to POU5F1. Repeat elements such as LTRs are enriched in primate-specific regulatory elements [108] and are known to influence transcription factor regulatory networks [109]. Exaptation of promoters embedded within LTRs is emerging as an important mechanism of genomic dysregulation during oncogenesis [110]. This phenomenon was first shown for expression of CSF1R [111] and IRF5 [112] in Hodgkin lymphoma. Activation of LTR-embedded promoters has also been linked to production of novel gene isoforms such as for ALK in melanoma [113] and FABP7 in diffuse large B cell lymphoma [114]. To our knowledge, this report represents the first description of retroviral LTR exaptation in RCC and the mechanism appears to be distinct from previous examples of this phenomenon. Since HIF activation is one of the earliest steps in RCC oncogenesis [99], it is likely that unmasking of HIF-responsive LTRs and exaptation of their potent regulatory elements influences the expression landscape of the tumor, most notably by upregulation of POU5F1.
The data generated and described here are freely available to provide a reference map upon which future functional genomic studies on RCC can be constructed and interpreted. Overall, our approach demonstrates the power of epigenomic analysis focused on small numbers of pure primary tumor and matched normal cell-of-origin cultures which can provide a clarifying lens through which to interpret inherently noisier large tumor-sequencing datasets. This general framework can reveal unanticipated insights into tumor biology and is readily applicable to other cancers in which tumor cells and matched normal cells-of-origin are available.
Conflict of interest disclosure
All authors have no competing financial conflicts of interests to disclose.
Author contributions
SA conceived of the project, procured and processed specimens, designed and performed experiments and analyzed data. KTS and CPM performed experiments and analyzed data. MT performed and interpreted the POU5F1 tissue microarray immunohistochemistry study. SA, KTS, JDV, AR, ER, and EH performed analyses, data interpretation and visualization. RS, AJ and JN processed and curated sequencing data and imported external datasets. DB, MD and DD processed samples for DNase-seq and RNA-seq. RS, MF, MB and RK collated sample metadata and submitted datasets to public repositories. JM and HR-B provided 786-O reagents, interpreted data and edited the manuscript and figures. YZ contributed to experiment design, interpreted data and edited the manuscript. JH supported the study in part, interpreted data and edited the manuscript. SA and KTS primarily wrote the manuscript and all authors edited the manuscript and figures for content and clarity.
Funding
SA was supported in part by a Damon Runyon Cancer Research Foundation Fellowship (DRG 114-13). We would like to thank John A. Stamatoyannopoulos whose ENCODE grant from NHGRI (U54HG007010) supported sequencing and data processing for this project. This project was also supported by NCATS grants to JH (5UH3TR000504 and 1UG3TR002158), a NCI Cancer Center Support Grant (P30CA015704) to the Fred Hutchinson Cancer Research Center/University of Washington Cancer Consortium and by an unrestricted gift from the Northwest Kidney Centers to the Kidney Research Institute.
Acknowledgments
We would like to thank Dr. Kimberly Muczynski and the Kidney Research Institute at the University of Washington for assistance with patient consenting and tissue procurement. We would like to thank Magdalena Skipper and John A. Stamatoyannopoulos for advice on data visualization and figure layout.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.01.063.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Stergachis A.B., Neph S., Reynolds A., Humbert R., Miller B., Paige S.L. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013 Aug 15;154(4):888–903. doi: 10.1016/j.cell.2013.07.020. (Elsevier Inc) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polak P., Karlić R., Koren A., Thurman R., Sandstrom R., Lawrence M.S. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015 Feb 10;518(7539):360–364. doi: 10.1038/nature14221. (Nature Publishing Group) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008 Jan 25;132(2):311–322. doi: 10.1016/j.cell.2007.12.014. 2008 ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013 Oct 6;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014 Nov 20;515(7527):355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E. The accessible chromatin landscape of the human genome. Nature. 2012 Sep 6;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kundaje A., Ernst J., Yen A., Zhang Z., Wang J., Ward L.D. Integrative analysis of 111 reference human epigenomes. Nature. 2015 Feb 18;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corces M.R., Buenrostro J.D., Wu B., Greenside P.G., Chan S.M., Koenig J.L. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016 Oct;48(10):1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu K., Zaba L.C., Satpathy A.T., Giresi P.G., Li R., Jin Y. Chromatin accessibility landscape of cutaneous T cell lymphoma and dynamic response to HDAC inhibitors. Cancer Cell. 2017 Jul;32(1) doi: 10.1016/j.ccell.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corces M.R., Granja J.M., Shams S., Louie B.H., Seoane J.A., Zhou W. The chromatin accessibility landscape of primary human cancers. Science. 2018 Oct 26;362(6413) doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen F., Zhang Y., Şenbabaoğlu Y., Ciriello G., Yang L., Reznik E. Multilevel genomics-based taxonomy of renal cell carcinoma. Cell Rep. 2016 Mar 15;14(10):2476–2489. doi: 10.1016/j.celrep.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cifola I., Bianchi C., Mangano E., Bombelli S., Frascati F., Fasoli E. Renal cell carcinoma primary cultures maintain genomic and phenotypic profile of parental tumor tissues. BMC Cancer. 2011 Jun 13;11(1):244. doi: 10.1186/1471-2407-11-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seizinger B.R., Rouleau G.A., Ozelius L.J., Lane A.H., Farmer G.E., Lamiell J.M. Von Hippel-Lindau disease maps to the region of chromosome 3 associated with renal cell carcinoma. Nature. 1988 Mar 17;332(6161):268–269. doi: 10.1038/332268a0. 1988 ed. [DOI] [PubMed] [Google Scholar]
- 14.Maxwell P.H., Wiesener M.S., Chang G.W., Clifford S.C., Vaux E.C., Cockman M.E. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999 May 20;399(6733):271–275. doi: 10.1038/20459. (1999 ed. Nature Publishing Group) [DOI] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research N Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013 Jul 4;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salama R., Masson N., Simpson P., Sciesielski L.K., Sun M., Tian Y.M. Heterogeneous effects of direct hypoxia pathway activation in kidney cancer. PloS One. 2015;10(8) doi: 10.1371/journal.pone.0134645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricketts C.J., De Cubas A.A., Fan H., Smith C.C., Lang M., Reznik E. The cancer genome atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep. 2018 Mar 31:1–43. doi: 10.1016/j.celrep.2023.113063. (Elsevier Company) [DOI] [PubMed] [Google Scholar]
- 18.Gansauge M.-T., Meyer M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat Protoc. 2013 Mar 14;8(4):737–748. doi: 10.1038/nprot.2013.038. [DOI] [PubMed] [Google Scholar]
- 19.Snyder M.W., Kircher M., Hill A.J., Daza R.M., Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016 Jan 14;164(1–2):57–68. doi: 10.1016/j.cell.2015.11.050. (Elsevier) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Q., Bartz S., Mao M., Li L., Kaelin W.G. The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol Cell Biol. 2007 Mar;27(6):2092–2102. doi: 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009 Jul 15;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9) doi: 10.1186/gb-2008-9-9-r137. (BioMed Central) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009 May 1;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013 Jan;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldman M, Craft B, Kamath A, Brooks AN, Zhu J, Haussler D. The UCSC Xena Platform for cancer genomics data visualization and interpretation. 2018.
- 26.Neph S., Kuehn M.S., Reynolds A.P., Haugen E., Thurman R.E., Johnson A.K. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012 Jul 15;28(14):1919–1920. doi: 10.1093/bioinformatics/bts277. (2012 Ed.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinforma. 2009;10(1):48. doi: 10.1186/1471-2105-10-48. (BioMed Central) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010 May;28(5):495–501. doi: 10.1038/nbt.1630. (Nature Research) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997 Apr 25;268(1):78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 30.Reese M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem. 2001 Dec;26(1):51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 31.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. (BioMed Central) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bryne J.C., Valen E., Tang M.-H.E., Marstrand T., Winther O., da Piedade I. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008 Jan;36(Database issue):D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P. DNA-binding specificities of human transcription factors. Cell. 2013 Jan 17;152(1–2):327–339. doi: 10.1016/j.cell.2012.12.009. (Elsevier Inc) [DOI] [PubMed] [Google Scholar]
- 35.Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011 Apr 1;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009 Jul;37(Web Server issue) doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S. Quantifying similarity between motifs. Genome Biol. 2007;8(2):R24. doi: 10.1186/gb-2007-8-2-r24. (BioMed Central) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sieber K.B., Batorsky A., Siebenthall K., Hudkins K.L., Vierstra J.D., Sullivan S. Integrated Functional Genomic Analysis Enables Annotation of Kidney Genome-Wide Association Study Loci. J Am Soc Nephol. 2019;30(3):421–441. doi: 10.1681/ASN.2018030309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law A.Y.S., Wong C.K.C. Stanniocalcin-2 is a HIF-1 target gene that promotes cell proliferation in hypoxia. Exp Cell Res. 2010 Feb 1;316(3):466–476. doi: 10.1016/j.yexcr.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 40.Nordhoff V., Hübner K., Bauer A., Orlova I., Malapetsa A., Schöler H.R. Comparative analysis of human, bovine, and murine Oct-4 upstream promoter sequences. Mamm Genome. 2001 Feb 27;12(4):309–317. doi: 10.1007/s003350010279. [DOI] [PubMed] [Google Scholar]
- 41.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M. An atlas of active enhancers across human cell types and tissues. Nature. 2014 Mar 26;507(7493):455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.FANTOM Consortium and the RIKEN PMI and CLST (DGT), Forrest A.R.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J.L. A promoter-level mammalian expression atlas. Nature. 2014 Mar 27;507(7493):462–470. doi: 10.1038/nature13182. (Nature Research) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fishilevich S., Nudel R., Rappaport N., Hadar R., Plaschkes I., Iny Stein T. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017 Jan 1;2017:1217. doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grosso A.R., Leite A.P., Carvalho S., Matos M.R., Martins F.B., Vítor A.C. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. Elife. 2015 Nov 17;4:43. doi: 10.7554/eLife.09214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeda J., Seino S., Bell G.I. Human Oct3 gene family: cDNA sequences, alternative splicing, gene organization, chromosomal location, and expression at low levels in adult tissues. Nucleic Acids Res. 1992 Sep 11;20(17):4613–4620. doi: 10.1093/nar/20.17.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liedtke S., Enczmann J., Waclawczyk S., Wernet P., Kögler G. Oct4 and its pseudogenes confuse stem cell research. Cell Stem Cell. 2007 Oct 11;1(4):364–366. doi: 10.1016/j.stem.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Malakootian M., Azad F.M., Naeli P., Pakzad M., Fouani Y., Bajgan E.T. Novel spliced variants of OCT4, OCT4C and OCT4C1, with distinct expression patterns and functions in pluripotent and tumor cell lines. Eur J Cell Biol. 2017 Jun 1;96(4):347–355. doi: 10.1016/j.ejcb.2017.03.009. (Elsevier GmbH) [DOI] [PubMed] [Google Scholar]
- 48.Zhao F.-Q., Misra Y., Li D.-B., Wadsworth M.P., Krag D., Weaver D. Differential expression of Oct3/4 in human breast cancer and normal tissues. Int J Oncol. 2018 Jun;52(6):2069–2078. doi: 10.3892/ijo.2018.4341. [DOI] [PubMed] [Google Scholar]
- 49.Derrien T., Estellé J., Marco Sola S., Knowles D.G., Raineri E., Guigo R. Fast computation and applications of genome mappability. PloS One. 2012;7(1) doi: 10.1371/journal.pone.0030377. Ouzounis CA, editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerlinger M., Rowan A.J., Horswell S., Math M., Larkin J., Endesfelder D. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012 Mar 8;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerlinger M., Horswell S., Larkin J., Rowan A.J., Salm M.P., Varela I. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014 Feb 2;46(3):225–233. doi: 10.1038/ng.2891. (Nature Publishing Group) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wykoff C.C., Sotiriou C., Cockman M.E., Ratcliffe P.J., Maxwell P., Liu E. Gene array of VHL mutation and hypoxia shows novel hypoxia-induced genes and that cyclin D1 is a VHL target gene. Br J Cancer. 2004 Mar 22;90(6):1235–1243. doi: 10.1038/sj.bjc.6601657. (Nature Publishing Group) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doñate F., Raitano A., Morrison K., An Z., Capo L., Aviña H. AGS16F is a novel antibody drug conjugate directed against ENPP3 for the treatment of renal cell carcinoma. Clin Cancer Res. 2016 Apr 15;22(8):1989–1999. doi: 10.1158/1078-0432.CCR-15-1542. [DOI] [PubMed] [Google Scholar]
- 54.Junker K., Hindermann W., Voneggeling F., Diegmann J., Haessler K., Schubert J. CD70: A New Tumor Specific Biomarker For Renal Cell Carcinoma. J Urol. 2005 Jun;173(6):2150–2153. doi: 10.1097/01.ju.0000158121.49085.ba. [DOI] [PubMed] [Google Scholar]
- 55.Daniel L., Lechevallier E., Giorgi R., Sichez H., Zattara-Cannoni H., Figarella-Branger D. Pax-2 expression in adult renal tumors. Hum Pathol. 2001 Mar;32(3):282–287. doi: 10.1053/hupa.2001.22753. (2001st ed) [DOI] [PubMed] [Google Scholar]
- 56.Doberstein K., Pfeilschifter J., Gutwein P. The transcription factor PAX2 regulates ADAM10 expression in renal cell carcinoma. Carcinogenesis. 2011 Nov;32(11):1713–1723. doi: 10.1093/carcin/bgr195. (2011 Ed.) [DOI] [PubMed] [Google Scholar]
- 57.Gnarra J.R., Dressler G.R. Expression of Pax-2 in human renal cell carcinoma and growth inhibition by antisense oligonucleotides. Cancer Res. 1995 Sep 15;55(18):4092–4098. (1995 Ed.) [PubMed] [Google Scholar]
- 58.Luu V.D., Boysen G., Struckmann K., Casagrande S., Teichman Von A., Wild P.J. Loss of VHL and hypoxia provokes PAX2 up-regulation in clear cell renal cell carcinoma. Clin Cancer Res. 2009 May 15;15(10):3297–3304. doi: 10.1158/1078-0432.CCR-08-2779. (2009 Ed.) [DOI] [PubMed] [Google Scholar]
- 59.Hu Y., Hartmann A., Stoehr C., Zhang S., Wang M., Tacha D. PAX8 is expressed in the majority of renal epithelial neoplasms: an immunohistochemical study of 223 cases using a mouse monoclonal antibody. J Clin Pathol. 2012 Mar;65(3):254–256. doi: 10.1136/jclinpath-2011-200508. (2011 Ed.) [DOI] [PubMed] [Google Scholar]
- 60.Laury A.R., Perets R., Piao H., Krane J.F., Barletta J.A., French C. A comprehensive analysis of PAX8 expression in human epithelial tumors. Am J Surg Pathol. 2011 Jun;35(6):816–826. doi: 10.1097/PAS.0b013e318216c112. (2011 Ed.) [DOI] [PubMed] [Google Scholar]
- 61.Tong G.X., Memeo L., Colarossi C., Hamele-Bena D., Magi-Galluzzi C., Zhou M. PAX8 and PAX2 immunostaining facilitates the diagnosis of primary epithelial neoplasms of the male genital tract. Am J Surg Pathol. 2011 Oct;35(10):1473–1483. doi: 10.1097/PAS.0b013e318227e2ee. (2011 Ed.) [DOI] [PubMed] [Google Scholar]
- 62.Oya M., Horiguchi A., Mizuno R., Marumo K., Murai M. Increased activation of CCAAT/enhancer binding protein-beta correlates with the invasiveness of renal cell carcinoma. Clin Cancer Res. 2003 Mar;9(3):1021–1027. (2003rd Ed.) [PubMed] [Google Scholar]
- 63.Kinch L., Grishin N.V., Brugarolas J. Succination of Keap1 and activation of Nrf2-dependent antioxidant pathways in FH-deficient papillary renal cell carcinoma type 2. Cancer Cell. 2011 Oct 18;20(4):418–420. doi: 10.1016/j.ccr.2011.10.005. (2011 Ed.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ooi A., Dykema K., Ansari A., Petillo D., Snider J., Kahnoski R., Anema J., Craig D., Carpten J., Tech B.T., Furge K.A. CUL3 and NRF2 mutations confer an NRF2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res. 2013 Apr 1;73(7):2044–2051. doi: 10.1158/0008-5472.CAN-12-3227. Epub 2013 Jan 30. [DOI] [PubMed] [Google Scholar]
- 65.Cho D.C., Mier J.W. Dual inhibition of PI3-kinase and mTOR in renal cell carcinoma. Curr Cancer Drug Targ. 2013 Feb;13(2):126–142. doi: 10.2174/1568009611313020003. [DOI] [PubMed] [Google Scholar]
- 66.Gan B., Lim C., Chu G., Hua S., Ding Z., Collins M. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell. 2010 Nov 16;18(5):472–484. doi: 10.1016/j.ccr.2010.10.019. (2010 Ed.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu C., Jin B., Chen L., Zhuo D., Zhang Z., Gong K., Mao Z. MiR-30d induces apoptosis and is regulated by the Akt/FOXO pathway in renal cell carcinoma. Cell Signal. 2013 May;25(5):1212–1221. doi: 10.1016/j.cellsig.2013.01.028. Epub 2013 Feb 15. [DOI] [PubMed] [Google Scholar]
- 68.Bill M.A., Nicholas C., Mace T.A., Etter J.P., Li C., Schwartz E.B. Structurally modified curcumin analogs inhibit STAT3 phosphorylation and promote apoptosis of human renal cell carcinoma and melanoma cell lines. PloS One. 2012;7(8) doi: 10.1371/journal.pone.0040724. (2012 Ed.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horiguchi A., Oya M., Shimada T., Uchida A., Marumo K., Murai M. Activation of signal transducer and activator of transcription 3 in renal cell carcinoma: a study of incidence and its association with pathological features and clinical outcome. J Urol. 2002 Aug;168(2):762–765. (2002nd Ed.) [PubMed] [Google Scholar]
- 70.Horiguchi A., Asano T., Kuroda K., Sato A., Asakuma J., Ito K. STAT3 inhibitor WP1066 as a novel therapeutic agent for renal cell carcinoma. Br J Cancer. 2010 May 25;102(11):1592–1599. doi: 10.1038/sj.bjc.6605691. (2010 Ed.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jung J.E., Lee H.G., Cho I.H., Chung D.H., Yoon S.H., Yang Y.M. STAT3 is a potential modulator of HIF-1-mediated VEGF expression in human renal carcinoma cells. FASEB J. 2005 Aug;19(10):1296–1298. doi: 10.1096/fj.04-3099fje. 2005 ed. [DOI] [PubMed] [Google Scholar]
- 72.Li L., Gao Y., Zhang L.L., He D.L. Concomitant activation of the JAK/STAT3 and ERK1/2 signaling is involved in leptin-mediated proliferation of renal cell carcinoma Caki-2 cells. Cancer Biol Ther. 2008 Nov;7(11):1787–1792. doi: 10.4161/cbt.7.11.6837. (2008 Ed.) [DOI] [PubMed] [Google Scholar]
- 73.Xin H., Zhang C., Herrmann A., Du Y., Figlin R., Yu H. Sunitinib inhibition of Stat3 induces renal cell carcinoma tumor cell apoptosis and reduces immunosuppressive cells. Cancer Res. 2009 Mar 15;69(6):2506–2513. doi: 10.1158/0008-5472.CAN-08-4323. (2009 Ed.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xin H., Herrmann A., Reckamp K., Zhang W., Pal S., Hedvat M. Antiangiogenic and antimetastatic activity of JAK inhibitor AZD1480. Cancer Res. 2011 Nov 1;71(21):6601–6610. doi: 10.1158/0008-5472.CAN-11-1217. (2011 Ed.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu X.R., Chen Y.H., Liu D.M., Sha J.J., Xuan H.Q., Bo J.J. Increased expression of forkhead box M1 protein is associated with poor prognosis in clear cell renal cell carcinoma. Med Oncol. 2013 Mar;30(1):346. doi: 10.1007/s12032-012-0346-1. (2012 Ed.) [DOI] [PubMed] [Google Scholar]
- 76.Xue Y.J., Xiao R.H., Long D.Z., Zou X.F., Wang X.N., Zhang G.X. Overexpression of FoxM1 is associated with tumor progression in patients with clear cell renal cell carcinoma. Journal of translational medicine. 2012;10:200. doi: 10.1186/1479-5876-10-200. (2012 Ed.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bussolati B., Bruno S., Grange C., Ferrando U., Camussi G. Identification of a tumor-initiating stem cell population in human renal carcinomas. FASEB J. 2008 Oct;22(10):3696–3705. doi: 10.1096/fj.08-102590. (2008 Ed.) [DOI] [PubMed] [Google Scholar]
- 78.Smith B.H., Gazda L.S., Conn B.L., Jain K., Asina S., Levine D.M. Three-dimensional culture of mouse renal carcinoma cells in agarose macrobeads selects for a subpopulation of cells with cancer stem cell or cancer progenitor properties. Cancer Res. 2011 Feb 1;71(3):716–724. doi: 10.1158/0008-5472.CAN-10-2254. (2011 Ed.) [DOI] [PubMed] [Google Scholar]
- 79.Oda H., Nakatsuru Y., Ishikawa T. Mutations of the p53 gene and p53 protein overexpression are associated with sarcomatoid transformation in renal cell carcinomas. Cancer Res. 1995 Feb 1;55(3):658–662. (1995 Ed.) [PubMed] [Google Scholar]
- 80.Reiter R.E., Anglard P., Liu S., Gnarra J.R., Linehan W.M. Chromosome 17p deletions and p53 mutations in renal cell carcinoma. Cancer Res. 1993 Jul 1;53(13):3092–3097. 1993rd ed. [PubMed] [Google Scholar]
- 81.Torigoe S., Shuin T., Kubota Y., Horikoshi T., Danenberg K., Danenberg P.V. p53 gene mutation in primary human renal cell carcinoma. Oncol Res. 1992;4(11−12):467–472. (1992nd ed) [PubMed] [Google Scholar]
- 82.Uhlman D.L., Nguyen P.L., Manivel J.C., Aeppli D., Resnick J.M., Fraley E.E. Association of immunohistochemical staining for p53 with metastatic progression and poor survival in patients with renal cell carcinoma. J Natl Cancer Inst. 1994 Oct 5;86(19):1470–1475. doi: 10.1093/jnci/86.19.1470. (1994 Ed.) [DOI] [PubMed] [Google Scholar]
- 83.Ye Y.W., Jiang Z.M., Li W.H., Li Z.S., Han Y.H., Sun L. Down-regulation of TCF21 is associated with poor survival in clear cell renal cell carcinoma. Neoplasma. 2012;59(6):599–605. doi: 10.4149/neo_2012_076. 2012 ed. [DOI] [PubMed] [Google Scholar]
- 84.Zhang H., Guo Y., Shang C., Song Y., Wu B. Urology. 2012 Dec;80(6) doi: 10.1016/j.urology.2012.08.013. (2012 Ed.) [DOI] [PubMed] [Google Scholar]
- 85.Peña-Llopis S., Vega-Rubín-de-Celis S., Liao A., Leng N., Pavía-Jiménez A., Wang S. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012 Jun 10;44(7):751–759. doi: 10.1038/ng.2323. (Nature Publishing Group) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Anastasiadis A.G., Lemm I., Radzewitz A., Lingott A., Ebert T., Ackermann R. Loss of function of the tissue specific transcription factor HNF1 alpha in renal cell carcinoma and clinical prognosis. Anticancer Res. 1999 May-Jun;19(3A):2105–2110. (1999 Ed.) [PubMed] [Google Scholar]
- 87.Rebouissou S., Vasiliu V., Thomas C., Bellanne-Chantelot C., Bui H., Chretien Y. Germline hepatocyte nuclear factor 1alpha and 1beta mutations in renal cell carcinomas. Hum Mol Genet. 2005 Mar 1;14(5):603–614. doi: 10.1093/hmg/ddi057. (2005 Ed.) [DOI] [PubMed] [Google Scholar]
- 88.Sel S., Ebert T., Ryffel G.U., Drewes T. Human renal cell carcinogenesis is accompanied by a coordinate loss of the tissue specific transcription factors HNF4 alpha and HNF1 alpha. Cancer Lett. 1996 Mar 29;101(2):205–210. doi: 10.1016/0304-3835(96)04136-5. (1996 Ed.) [DOI] [PubMed] [Google Scholar]
- 89.Bigot P., Colli L.M., Machiela M.J., Jessop L., Myers T.A., Carrouget J. Functional characterization of the 12p12.1 renal cancer-susceptibility locus implicates BHLHE41. Nat Commun Nat Res. 2016;7:12098. doi: 10.1038/ncomms12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grampp S., Schmid V., Salama R., Lauer V., Kranz F., Platt J.L. Multiple renal cancer susceptibility polymorphisms modulate the HIF pathway. PLoS Genet. 2017 Jul;13(7) doi: 10.1371/journal.pgen.1006872. Linehan M, editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rhie S.K., Guo Y., Tak Y.G., Yao L., Shen H., Coetzee G.A. Identification of activated enhancers and linked transcription factors in breast, prostate, and kidney tumors by tracing enhancer networks using epigenetic traits. Epigenetics Chromatin. 2016;9(1):50. doi: 10.1186/s13072-016-0102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao C., Wood C.G., Karam J.A., Maity T., Wang L. The role of ZNF395 in renal cell carcinoma proliferation, migration, and invasion. J Clin Oncol. 2016 Jan 10;34(2_suppl):592. [Google Scholar]
- 93.Ezashi T., Das P., Roberts R.M. Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci U S A. 2005 Mar 29;102(13):4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Westfall S.D., Sachdev S., Das P., Hearne L.B., Hannink M., Roberts R.M. Stem Cells and Development. 17(5) Mary Ann Liebert, Inc. publishers; 2008 Oct. Identification of oxygen-sensitive transcriptional programs in human embryonic stem cells; pp. 869–881. (140 Huguenot Street, 3rd FloorNew Rochelle, NY 10801-5215USA) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Forristal C.E., Wright K.L., Hanley N.A., Oreffo R.O.C., Houghton F.D. Hypoxia inducible factors regulate pluripotency and proliferation in human embryonic stem cells cultured at reduced oxygen tensions. Reproduction. 2009 Dec 15;139(1):85–97. doi: 10.1530/REP-09-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mathieu J., Zhang Z., Zhou W., Wang A.J., Heddleston J.M., Pinna C.M.A. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011 Jun 30;71(13):4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mathieu J., Zhang Z., Nelson A., Lamba D.A., Reh T.A., Ware C. Hypoxia induces re-entry of committed cells into pluripotency. Stem Cells. 2013 Sep;31(9):1737–1748. doi: 10.1002/stem.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mathieu J., Zhou W., Xing Y., Sperber H., Ferreccio A., Agoston Z. Hypoxia-inducible factors have distinct and stage-specific roles during reprogramming of human cells to pluripotency. Cell Stem Cell. 2014 May 1;14(5):592–605. doi: 10.1016/j.stem.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mitchell T.J., Turajlic S., Rowan A., Nicol D., Farmery J.H.R., O'Brien T. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell. 2018 Apr 19;173(3):611–617. doi: 10.1016/j.cell.2018.02.020. (Elsevier Inc) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Turajlic S., Xu H., Litchfield K., Rowan A., Horswell S., Chambers T. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell. 2018 Apr 19;173(3) doi: 10.1016/j.cell.2018.03.043. (Elsevier Inc) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Malta T.M., Sokolov A., Gentles A.J., Burzykowski T., Poisson L., Kamińska B. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018 Apr 5;173(2) doi: 10.1016/j.cell.2018.03.034. (Elsevier Inc) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Young M.D., Mitchell T.J., Vieira Braga F.A., Tran M.G.B., Stewart B.J., Ferdinand J.R. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018 Aug 10;361(6402):594–599. doi: 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shaffer D.R., Savoldo B., Yi Z., Chow K.K.H., Kakarla S., Spencer D.M. T cells redirected against CD70 for the immunotherapy of CD70-positive malignancies. Blood. 2011 Feb 8;117(16):4304–4314. doi: 10.1182/blood-2010-04-278218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Owonikoko T.K., Hussain A., Stadler W.M., Smith D.C., Kluger H., Molina A.M. First-in-human multicenter phase I study of BMS-936561 (MDX-1203), an antibody-drug conjugate targeting CD70. Cancer Chemother Pharmacol. 2015 Nov 14;77(1):155–162. doi: 10.1007/s00280-015-2909-2. Springer Berlin Heidelberg. [DOI] [PubMed] [Google Scholar]
- 105.Song Y., Zhong L., Zhou J., Lu M., Xing T., Ma L. Data-independent acquisition-based quantitative proteomic analysis reveals potential biomarkers of kidney cancer. Proteomics Clin Appl. 2017 Dec;11(11–12):1700066. doi: 10.1002/prca.201700066. [DOI] [PubMed] [Google Scholar]
- 106.Saville M.K., Sparks A., Xirodimas D.P., Wardrop J., Stevenson L.F., Bourdon J.-C. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J Biol Chem. 2004 Oct 1;279(40):42169–42181. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- 107.Lee J.-Y., Tokumoto M., Fujiwara Y., Hasegawa T., Seko Y., Shimada A. Accumulation of p53 via down-regulation of UBE2D family genes is a critical pathway for cadmium-induced renal toxicity. Sci Rep. 2016;6:21968. doi: 10.1038/srep21968. (Nature Publishing Group) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jacques P.-É., Jeyakani J., Bourque G. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 2013 May;9(5) doi: 10.1371/journal.pgen.1003504. Feschotte C, editor. (Public Library of Science) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bourque G., Leong B., Vega V.B., Chen X., Lee Y.L., Srinivasan K.G. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008 Nov;18(11):1752–1762. doi: 10.1101/gr.080663.108. (Cold Spring Harbor Lab) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Babaian A., Mager D.L. Endogenous retroviral promoter exaptation in human cancer. Mob DNA. 2016;7(1):24. doi: 10.1186/s13100-016-0080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lamprecht B., Walter K., Kreher S., Kumar R., Hummel M., Lenze D. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med. 2010 May;16(5) doi: 10.1038/nm.2129. (571–9–1pfollowing579) [DOI] [PubMed] [Google Scholar]
- 112.Babaian A., Romanish M.T., Gagnier L., Kuo L.Y., Karimi M.M., Steidl C. Onco-exaptation of an endogenous retroviral LTR drives IRF5 expression in Hodgkin lymphoma. Oncogene. 2016 May 12;35(19):2542–2546. doi: 10.1038/onc.2015.308. [DOI] [PubMed] [Google Scholar]
- 113.Wiesner T., Lee W., Obenauf A.C., Ran L., Murali R., Zhang Q.F. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature. 2015 Oct 15;526(7573):453–457. doi: 10.1038/nature15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lock F.E., Rebollo R., Miceli-Royer K., Gagnier L., Kuah S., Babaian A. Distinct isoform of FABP7 revealed by screening for retroelement-activated genes in diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2014 Aug 26;111(34):E3534–E3543. doi: 10.1073/pnas.1405507111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Data Availability Statement
All primary and uniformly processed sequence data generated in this study are available at the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE117324. We recently performed a separate and non-overlapping analysis of the tubule data sets included in this study in comparison to human kidney glomerular outgrowth cultures and cultured podocytes [38]. Those data have also been deposited at GEO with accession number GSE115961.