Abstract
Hepatocellular carcinoma (HCC) is a lethal human malignancy and a leading cause of cancer‐related death worldwide. Patients with HCC are often diagnosed at an advanced stage, and the prognosis is usually poor. The multikinase inhibitor sorafenib is the first‐line treatment for patients with advanced HCC. However, cases of primary or acquired resistance to sorafenib have gradually increased, leading to a predicament in HCC therapy. Thus, it is critical to investigate the mechanism underlying sorafenib resistance. Transactivation response element RNA‐binding protein 2 (TARBP2) is a multifaceted miRNA biogenesis factor that regulates cancer stem cell (CSC) properties. The tumorigenicity and drug resistance of cancer cells are often enhanced due to the acquisition of CSC features. However, the role of TARBP2 in sorafenib resistance in HCC remains unknown. Our results demonstrate that TARBP2 is significantly downregulated in sorafenib‐resistant HCC cells. The TARBP2 protein was destabilized through autophagic–lysosomal proteolysis, thereby stabilizing the expression of the CSC marker protein Nanog, which facilitates sorafenib resistance in HCC cells. In summary, here we reveal a novel miRNA‐independent role of TARBP2 in regulating sorafenib resistance in HCC cells.
Keywords: cancer stem cells, hepatocellular carcinoma, miRNA, Nanog, sorafenib resistance, TARBP2
Abbreviations
- BFA
bafilomycin A1
- CHX
cycloheximide
- CQ
chloroquine
- CSC
cancer stem cell
- HCC
hepatocellular carcinoma
- miRNA
microRNA
- MTT
3‐(4,5 dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide
- NH4Cl
ammonium chloride
- PKR
dsRNA‐dependent protein kinase
- RISC
RNA‐induced slicing complex
- SEM
standard error of the mean
- SR
sorafenib‐resistant
- TARBP2
transactivation response element RNA‐binding protein 2
1. Introduction
Liver cancer is the second leading cause of cancer death, accounting for 9% of all types of cancer, and the number of new cancer cases has increased to more than 780 000 annually; thus, liver cancer remains a major health problem worldwide (El‐Serag, 2011; Torre et al., 2015). Hepatocellular carcinoma (HCC) is the most common primary malignancy among all primary liver cancers, contributing to ~ 80–90% of cases (El‐Serag, 2011). HCC occurs predominantly in patients with cirrhosis and chronic liver disease, which are chiefly caused by hepatitis B and C viral infections and other risk cofactors, including alcohol consumption, aflatoxin intake, smoking, and metabolic disorders (Balogh et al., 2016). Approximately 30–40% of all HCC patients are diagnosed at early stages and receive potentially curative treatments (Llovet et al., 2008). Patients in the early stage of HCC are treated with curative treatment options, including liver resection, radiation therapy, and chemotherapy (El‐Serag et al., 2008). Chemotherapy drugs, such as floxuridine, cisplatin, and doxorubicin, are the most common treatments, with 5‐year survival rates reaching 75% in HCC patients (Lin et al., 2012a; Raza and Sood, 2014). However, HCC is often diagnosed at advanced stages due to the asymptomatic features of early HCC, and the life expectancy of patients with advanced HCC is poor, with a mean survival of 7 months (Giannini et al., 2015; Pinter et al., 2012). A leading cause of the high mortality rate in HCC patients is the lack of effective therapeutic options, specifically for patients in advanced stages (Colagrande et al., 2015). The treatment options for patients with unresectable advanced HCC are extremely limited. Sorafenib is the current recommended standard treatment for advanced HCC (Colagrande et al., 2015).
Sorafenib (Nexavar®, BAY 43‐9006, Bayer, Whippany, NJ, USA), an FDA‐approved systemic drug, is the first‐line treatment for improving the survival rate of patients with advanced HCC (Cervello et al., 2012; Gauthier and Ho, 2013). As a multikinase inhibitor, sorafenib blocks a broad spectrum of malignant phenotypes, including cell proliferation, tumor angiogenesis, and metastasis of HCC, by repressing the activity of tyrosine kinase receptors to inhibit PI3K/AKT and the Ras/Raf/MAPK pathway (Carlomagno et al., 2006; Wellbrock et al., 2004; Wilhelm et al., 2004). Although sorafenib provides a survival advantage of 2–3 months, the therapeutic effects of sorafenib are temporary, and the response rate in HCC is quite low (Keating and Santoro, 2009). Sorafenib treatment is characterized by high‐level heterogeneity in individual responses, and only ~ 30% of patients benefit from sorafenib treatment. Most patients acquire sorafenib resistance within 6 months, increasing the risk of distant metastasis and cancer recurrence (Chow et al., 2013; Keating and Santoro, 2009; Liang et al., 2013; Llovet et al., 2008; Paez‐Ribes et al., 2009). Therefore, it is critical to identify a novel molecular mechanism underlying sorafenib resistance.
Transactivation response element RNA‐binding protein 2 (TARBP2), a well‐known double‐stranded RNA‐binding protein, stabilizes the microRNA (miRNA) biogenesis factor Dicer to induce miRNA maturation, thereby governing the translation of mRNA (Gatignol et al., 1991). Additionally, TARBP2 acts as a dsRNA‐dependent protein kinase (PKR) inhibitor to suppress the phosphorylation of eIF2α, thus enhancing cell mitosis and destabilizing transcripts to promote cancer metastasis, exhibiting miRNA‐independent properties in regulating cancer (Garcia et al., 2006; Goodarzi et al., 2014; Kim et al., 2014). Alternatively, the expression of genes related to cancer stem cells (CSCs) in HCC can induce malignant features to promote tumorigenic ability, drug resistance, and metastasis. The CSC markers SOX2, Nanog, and OCT4 are highly expressed in solid tumors and breast, liver, colon, and lung cancers (Dai et al., 2013; Karoubi et al., 2009; Leis et al., 2012; Sun et al., 2013), and these markers are associated with drug resistance to tamoxifen, gefitinib, and paclitaxel (Di and Zhao, 2015; Singh and Settleman, 2010; Vinogradov and Wei, 2012). Downregulation of TARBP2 increases Nanog and OCT4 expression and contributes to clonogenicity and tumor growth in Ewing sarcoma family tumors, suggesting that TARBP2 might exhibit potential as a central mediator in regulating the properties of CSC (De Vito et al., 2012). Nevertheless, the role of TARBP2 in sorafenib resistance in HCC remains unidentified. Thus, an improved understanding of the underlying molecular mechanism is required to resolve the predicament of current sorafenib therapy. Here, we demonstrate an miRNA‐independent mechanism of TARBP2, in which downregulation of the TARBP2 protein promotes sorafenib resistance in HCC cells through stabilization of Nanog expression.
2. Materials and methods
2.1. Cell culture
Huh7, Huh7/sorafenib‐resistant (SR), and PLC5 cells were kindly provided by D‐L Ou, Graduate Institute of Oncology, College of Medicine, National Taiwan University, Taipei, Taiwan. PLC5/SR cells were established by long‐term exposure to sorafenib (LC Laboratories, Woburn, MA, USA) at a low dose (5 μm), which was increased to a higher dose (20 μm) over 3 months. HEK‐293T cells were obtained from the American Type Culture Collection. The HEK‐293T, Huh7, PLC5, Huh7/SR, and PLC5/SR cells were grown in Dulbecco's modified Eagle's medium (DMEM)/F12 (HiMedia, MUM, IND). Sorafenib (5 μm) was added to maintain sorafenib resistance in the Huh7/SR and PLC5/SR cells. These cells were supplemented with 10% fetal bovine serum (Gibco, Waltham, MA, USA) and 1% penicillin/streptomycin (GeneDireX, Las Vegas, NV, USA) at 37 °C under 5% CO2.
2.2. Western blot analysis
The TARBP2‐expressing plasmid from D‐Y Jin was obtained from Addgene (Kok et al., 2007). Plasmids expressing Myc‐TARBP2, TARBP2 ΔC4, and Dicer were kindly provided by A. Gatignol (Daniels et al., 2009). TARBP2 or TARBP2 ΔC4 was transfected into HCC cells for 48 h using jetPRIME (Polyplus‐transfection, New York, NY, USA and HyFect™ DNA transfection reagent (Leadgene Biomedical, Tainan, Taiwan)). To harvest total proteins, the cells were washed with PBS buffer and lysed in RIPA buffer [Tris‐base (50 mm), NaCl (150 mm), NP‐40 (10%), Na3VO4 (200 mm), EDTA (100 mm), sodium deoxycholate (0.1%), SDS (1%)]. The cell lysates were sonicated using an ultrasonic processor, and the supernatant was collected after centrifugation at 18 000 g for 30 min at 4 °C. An equal quantity of protein was resuspended in gel sample buffer and was separated via SDS/PAGE. The proteins separated in the SDS/PAGE were transferred to a polyvinylidene difluoride membrane at 400 mA for 2 h. The membrane was blocked with TBST buffer (0.02 m Tris‐base, 0.15 m NaCl, 5 mL Tween 20, pH 7.5) containing 5% nonfat milk for 1 h at room temperature. After blocking, the membrane was incubated with a specific primary antibody overnight at 4 °C. After washing with TBST buffer, the membrane was hybridized with a horseradish peroxidase‐conjugated secondary antibody for 1 h at room temperature. The membrane was then washed with TBST buffer. Protein expression was visualized using enhanced chemiluminescence (PerkinElmer, Waltham, MA, USA). The blots were exposed to autoradiography film to obtain the results.
2.3. Isolation of RNA and quantitative real‐time PCR
Total RNA was isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Total mRNA (200 ng) was reverse‐transcribed into cDNA using reverse transcriptase, random primers, dNTPs, and an RNase inhibitor. The parameters for reverse transcription were as follows: 25 °C for 10 min, 42 °C for 45 min, and 70 °C for 15 min. The cDNA was amplified using SYBR™ Green Master Mix (Invitrogen) and gene‐specific primers. The amplified replication signal was detected using the (Applied Biosystems, Waltham, MA, USA) Step One real‐time PCR system according to the manufacturer's protocols. The PCR cycling parameters were as follows: 95 °C for 3 min and 40 cycles of 95 °C for 15 s, 60 °C for 1 min and 75 °C for 15 s. The primers used to detect the specific sequences were as follows: TARBP2 (F: 5′‐GGG CTC TTG GGT TCT GTA GT‐3′; R: 5′‐GTT TGT AAT ACC GTC CCG CC‐3′), Nanog (F: 5′‐ATA GCA ATG GTG TGA CGC AG‐3′;R: 5′‐ACC AGG TCT GAG TGT TCC AG‐3′), GAPDH (F: 5′‐ACC CAC TCC TCC ACC TTT GAC‐3′; R: 5′‐TCC ACC ACC CTG TTG CTG TAC‐3′). GAPDH was used as an endogenous control to normalize TARBP2 and Nanog expression.
2.4. Cell viability analysis
Cell viability was determined using the 3‐(4,5 dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) assay. The cells were seeded in triplicate at a density of 3500 cells per well in 96‐well plates. After 24 h, the cells were treated with the indicated concentrations of sorafenib for 48 h. The cells were then treated with MTT solution (5 mg·mL−1) for 2 h. Next, the medium was removed, and 100 μL of DMSO was added to each well to dissolve the insoluble purple formazan product. The absorbance of the colored solution was measured at 570 nm using a spectrophotometer. All experiments were performed in triplicate.
2.5. shRNA‐packaged lentivirus knockdown
pCMVΔR8.91, pMD.G, TARBP2, Nanog, and GFP short hairpin‐constructed plasmids were purchased from the National RNAi Core Facility Platform located at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica. For lentivirus production, HEK‐293T cells were cotransfected with a constructed short hairpin‐carrying plasmid (1 μg), pCMVΔR8.91 (5 μg), and pMD.G (5 μg). After transfection for 24 h, the supernatant was collected and filtered through a 0.45‐μm filter (Millipore, Billerica, MA, USA). HCC cells were seeded in 10‐cm dishes containing DMEM/F12. The lentivirus and polybrene (1 μg·mL−1) were added to the cells, followed by incubation for 48 h at 37 °C under 5% CO2. The medium was replaced with fresh medium supplemented with 1 μg·mL−1 puromycin to select stable clones. After 48 h of selection, the culture medium was removed and replaced with fresh medium containing 0.5 μg·mL−1 puromycin to maintain the gene knockdown of stable clones.
2.6. Sphere formation
Cells were trypsinized and suspended to generate single cells, for seeding at a density of 1000 cells per well in nonadherent plates in serum‐free DMEM/F12 medium, with epidermal growth factor (50 ng·mL−1), basic fibroblast growth factor (50 ng·mL−1; R&D Systems, Minneapolis, MN, USA), and 1× B27 supplement (Invitrogen) for 14 days. Quantification of sphere formation was performed by directly counting the number of spheres per well in plates.
2.7. HCC xenograft model of acquired resistance to sorafenib
The protocol for the xenograft experiments was approved by the Institutional Animal Care and Use Committee of the College of Medicine, National Taiwan University. All animal experiments were performed according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institutes of Health. Male BALB/c athymic (nu+/nu+) mice were inoculated subcutaneously with Huh7 cells. When tumors reached a 100 mm3 volume, mice were treated with sorafenib or placebo. Sorafenib (10 mg·kg−1·day−1) was administered daily via gavage. Tumor volume and body weight were recorded every 7 days. At the end of sorafenib treatment, the tumor samples were grouped into two groups: SR (tumor volume > 1000 mm3) and sorafenib‐sensitive (SS; tumor volume < 1000 mm3) tumors. The tumor samples were collected for western blotting (Hsu et al., 2016).
2.8. Bioinformatics analysis
Oncomine was used for the analysis and visualization of the TGCA liver cancer datasets (http://www.oncomine.org) (Rhodes et al., 2004). SurvExpress is a biomarker validation tool and database for the integration of cancer gene expression and clinical outcome data (http://bioinformatica.mty.itesm.mx/SurvExpress) (Aguirre‐Gamboa et al., 2013). PRECOG is a bioinformatics tool for analyzing the associations between genomic profiles and cancer outcomes (http://precog.stanford.edu/) (Fernandez‐Ricaud et al., 2016).
2.9. Statistical analysis
All data are expressed as the mean ± standard error of the mean (SEM) from at least three individual experiments. One‐way ANOVA was used to analyze statistically significant differences among multiple groups. Two‐way ANOVA was used to analyze multiple groups with two categorical variables. All analyses were performed using prism 6.0 software (GraphPad Software Inc., San Diego, CA, USA). P values of < 0.05 were considered statistically significant. *P < 0.05, **P < 0.01, or ***P < 0.005.
3. Results
3.1. TARBP2 downregulation correlates with a poor outcome in patients with HCC and enhances sorafenib resistance in HCC cells
To determine the significance of TARBP2 in the clinical outcome of patients with HCC, TARBP2 was validated via bioinformatics database analysis. Data on TARBP2 mRNA expression in normal and tumor tissues were collected from 441 unique analyses in the Oncomine database (Rhodes et al., 2004). TARBP2 expression was observed to be significantly elevated in most cancer types. In particular, TARBP2 expression was relatively decreased in liver cancer and pancreatic cancer, suggesting that TARBP2 levels were suppressed in liver and pancreatic tumors (Fig. 1A). The prognostic index of TARBP2 in liver cancer patients was analyzed using the SurvExpress database (Aguirre‐Gamboa et al., 2013), and the patients were categorized into low‐ and high‐risk groups based on their survival time and status. TARBP2 was downregulated in the high‐risk group, which was associated with a poor prognosis in patients with HCC (Fig. 1B; P = 1.25e‐25). Additionally, to correlate TARBP2 expression with the survival of HCC patients, we mined the PRECOG database to collect survival rates from groups of 50 (Fig. 1C; HR = 0.26; 95% CI: 0.07–0.95, P = 2.37e‐02) and 91 patients (Fig. 1D; HR = 0.52; 95% CI: 0.29–0.96, P = 3.27e‐02) with liver cancer for Kaplan–Meier survival analysis (Fernandez‐Ricaud et al., 2016). These clinical results showed that downregulation of TARBP2 was correlated with a poor prognosis and survival rate of patients with HCC. Based on clinical evidence revealing the significance of TARBP2 downregulation in HCC patients, we further investigated the molecular mechanism underlying SR in HCC cells. We first established two SR HCC cell lines, from Huh7 and PLC5 parental cells, via repeated long‐term exposure of the cancer cells to sorafenib at increasing dose concentrations (5–20 μm) for 3 months. To examine the level of sorafenib resistance in the HCC cell lines, the cells were treated with sorafenib in a dose‐dependent manner for 48 h, and cell viability was measured using the MTT assay. The inhibitory concentrations (IC50) of sorafenib in Huh7 and Huh7/SR cells were 4.72 ± 0.67 and 9.66 ± 0.99 μm, respectively (Fig. 1E). The IC50 values of sorafenib in PLC5 and PLC5/SR cells were 9.63 ± 0.98 and 14.84 ± 1.17 μm, respectively (Fig. 1F). These results indicated that the HCC cells were stably resistant to sorafenib, and these paired cell lines were used for further investigations. To determine the role of TARBP2 in sorafenib resistance in HCC cells, TARBP2 protein expression was analyzed using western blotting. The TARBP2 protein was significantly downregulated in SR HCC cells (Fig. 1G,H). TARBP2 is suppressed in HCC/SR cells, suggesting that downregulation of TARBP2 enhances sorafenib resistance in HCC cells.
Figure 1.
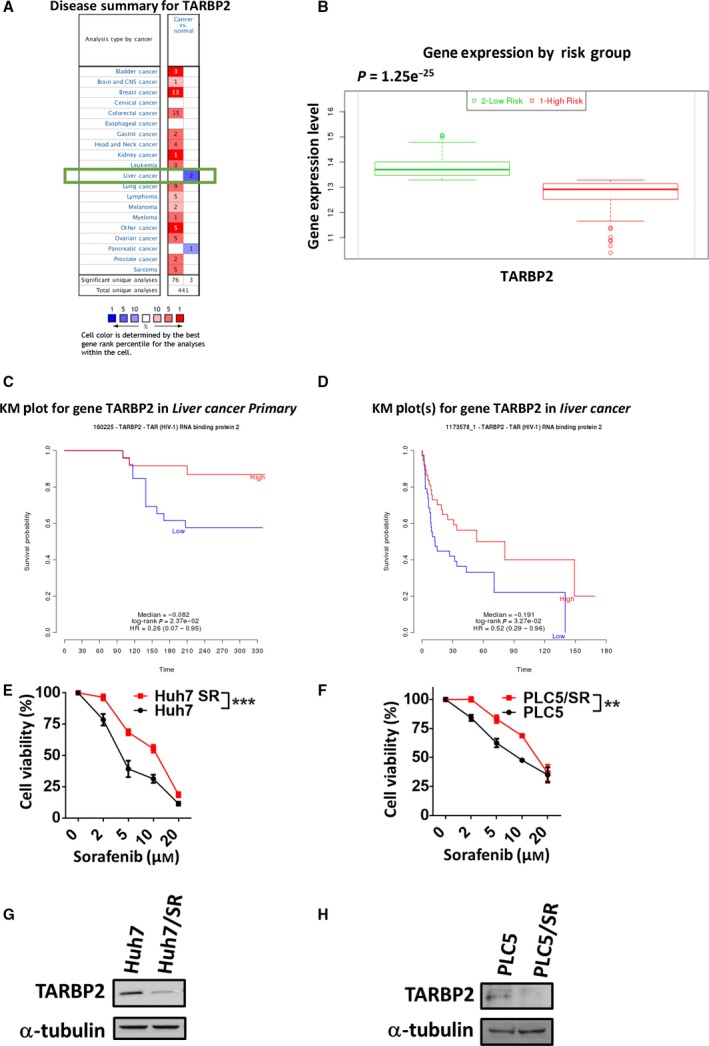
Downregulation of TARBP2 correlates with a poor outcome of patients with HCC and facilitates SR in HCC cells. (A) TARBP2 mRNA expression data were collected from the Oncomine database with thresholds of a P value ≤ 0.01 and gene rank ≤ 10%. The numbers in the colored cells represent the number of analyses. The red cells indicate increased TARBP2 mRNA expression in tumor tissues; the blue cells indicate reduced TARBP2 mRNA expression in tumor tissues. (B) The prognostic index of TARBP2 in 162 liver cancer patients was analyzed from the SurvExpress database and categorized into low‐ and high‐risk groups (x‐axis). The expression of TARBP2 is presented along the y‐axis. (C, D) Kaplan–Meier curves were generated from the PRECOG database. The data were collected from groups of 50 (GSE364; C) and 91 (GSE1898; D) liver cancer patients. (E, F) Expression of TARBP2 in paired HCC cell lines. Huh7 and Huh7/SR (E) or PLC5 and PLC5/SR (F) cells were treated with the indicated concentrations of sorafenib for 48 h. Cell viability was measured using the MTT assay. Data are presented as mean ± SEM, with at least n = 3 per group. Multigroup comparisons were analyzed by two‐way ANOVA with Tukey's post hoc test. P values < 0.05 were considered statistically significant. **P < 0.01; or ***P < 0.005. (G and H) The expression of TARBP2 in Huh7 (G) and PLC5 (H) cells was determined via western blot analysis.
3.2. TARBP2 suppressed sorafenib resistance of HCC cells is miRNA‐independent
To determine the function of TARBP2 in sorafenib resistance in HCC cells, TARBP2‐overexpressing HCC cells were treated with sorafenib at the indicated concentrations (0, 2, 5, 10, and 20 μm) for 48 h (Fig. 2A,B). The MTT assay demonstrated that TARBP2 significantly sensitized the Huh7/SR cells to sorafenib treatment (Fig. 2B). Accordingly, overexpression of TARBP2 decreased the level of sorafenib resistance in PLC5/SR cells (Fig. 2C,D), suggesting that TARBP2 functions as a tumor suppressor by sensitizing = HCC cells to sorafenib treatment. To further confirm the function of TARBP2 in the parental HCC cells, TARBP2 was knocked down using two specific TARBP2‐CDS‐targeting short hairpin RNAs in Huh7 and PLC5 cells, which expressed higher levels of TARBP2. The TARBP2‐knockdown HCC cells were treated with the indicated concentrations (0, 2, 5, 10, and 20 μm) of sorafenib for 48 h (Fig. 2E,F). Knockdown of TARBP2 significantly enhanced sorafenib resistance in Huh7 cells (Fig. 2F). Inhibition of TARBP2 expression promoted sorafenib resistance in PLC5 cells (Fig. 2G,H), indicating that downregulation of TARBP2 facilitates sorafenib resistance in HCC cells. TARBP2 is an essential biogenesis factor in the RNA‐induced slicing complex (RISC) for miRNA biogenesis (Gatignol et al., 1991). C4‐domain‐truncated TARBP2 was used to disrupt the binding between TARBP2 and Dicer and block miRNA biogenesis (Daniels et al., 2009) to investigate whether the TARBP2‐enhanced sensitivity of HCC cells to sorafenib treatment is miRNA‐dependent (Fig. 2I,J). Cell viability was decreased in Huh7/SR cells expressing wild‐type TARBP2 and those expressing TARBP2 ΔC4 (Fig. 2J), suggesting that TARBP2‐mediated sensitization of HCC cells to sorafenib treatment is miRNA‐independent.
Figure 2.
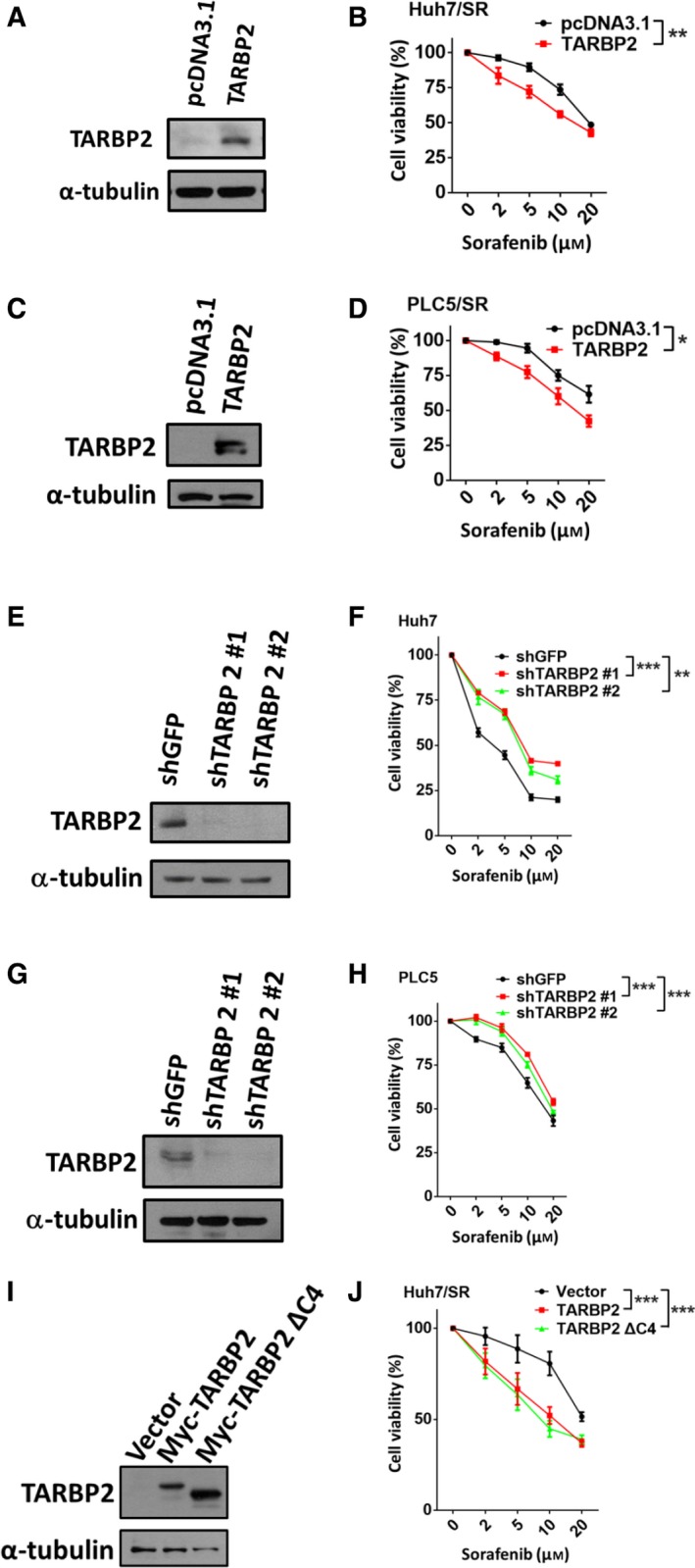
Downregulation of TARBP2 enhances SR in HCC cells. (A–H) Effect of TARBP2 expression on SR in HCC cells. TARBP2 was overexpressed in Huh7/SR cells (A) and PLC5/SR cells (C) for 48 h. TARBP2 was knocked down in Huh7 (E) and PLC5 (G) cells. TARBP2 protein expression was determined via western blot analysis. Cell viability was measured using the MTT assay (B, D, F, and H). (I, J) Effect of TARBP2 ΔC4 expression on SR in Huh7/SR cells. TARBP2 and truncated C4 TARBP2 protein expression was determined via western blot analysis (I). Cell viability was measured via the MTT assay (J). Data are presented as mean ± SEM, with at least n = 3 per group. Multigroup comparisons were analyzed by two‐way ANOVA with Tukey's post hoc test. P values < 0.05 were considered statistically significant. *P < 0.05; **P < 0.01; or ***P < 0.005.
3.3. TARBP2 protein is destabilized in sorafenib‐resistant HCC cells
After confirming that the TARBP2 protein is suppressed in HCC/SR cells, we next determined whether TARBP2 was downregulated via transcriptional regulation. Quantitative RT/PCR analysis demonstrated that TARBP2 mRNA levels remained unchanged in both Huh7/SR and PLC5/SR cells (Fig. 3A,B), indicating transcription‐independent regulation of the downregulation of TARBP2 in HCC/SR cells. To further evaluate whether downregulation of the TARBP2 protein occurred through translational or post‐translational regulation, the stability of the protein in the two pairs of HCC cell lines was determined via treatment with the protein synthesis inhibitor cycloheximide (CHX). Huh7 and Huh7/SR cells were treated with CHX for the indicated time periods (Fig. 3C), and the data showed that TARBP2 protein expression was stably maintained for 8 h but was dramatically reduced in Huh7/SR cells starting at 2 h after treatment with CHX (Fig. 3D). Similar effects were observed in the paired PLC5 cell lines (Fig. 3E,F), demonstrating that TARBP2 is downregulated through destabilization of the TARBP2 protein in HCC/SR cells.
Figure 3.
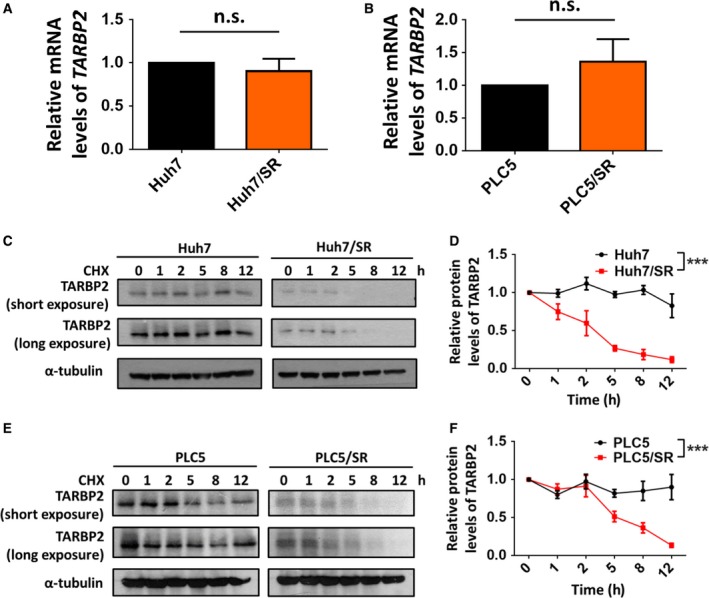
TARBP2 protein stability is decreased in HCC/SR cells. (A, B) The expression of TARBP2 mRNA was determined via real‐time PCR in paired Huh7 (A) or PLC5 (B) cell lines. Multigroup comparisons were analyzed by one‐way ANOVA with Tukey's post hoc test. (C–F) The TARBP2 protein was destabilized in HCC/SR cells. Huh7 and Huh7/SR (C) or PLC5 and PLC5/SR (E) cells were treated with CHX (100 μg·mL−1) for the indicated time periods. The relative quantity of the depicted proteins was analyzed through three independent experiments (D, F). Data are presented as mean ± SEM, with at least n = 3 per group. Multigroup comparisons were analyzed by two‐way ANOVA with Tukey's post hoc test. P values < 0.05 were considered statistically significant. ***P < 0.005.
3.4. The TARBP2 protein is suppressed though autophagic–lysosomal proteolysis in sorafenib‐resistant HCC cells
In eukaryotic cells, the ubiquitin‐proteasome and lysosome proteolytic pathways are two major pathways of protein degradation and are critical in regulating cancer progression (Glickman and Ciechanover, 2002; Mizushima, 2007). Therefore, we investigated whether the TARBP2 protein is degraded through the proteasome pathway in HCC/SR cells. Huh7/SR cells were treated with MG132 to inhibit proteasome activity. However, inhibition of proteasome‐mediated protein degradation did not prevent TARBP2 downregulation in Huh7/SR cells, suggesting that the degradation of the TARBP2 protein in HCC/SR cells is proteasome independent (Fig. 4A,B). Next, we determined whether the TARBP2 protein was degraded through the lysosomal pathway. Huh7/SR cells were treated with the lysosome inhibitors ammonium chloride (NH4Cl) and chloroquine (CQ) to inhibit the activity of lysosomal enzymes through neutralizing the lysosomal pH (Choi, 2012; Hart and Young, 1991). The results demonstrated that TARBP2 protein expression was restored through treatment with NH4Cl and CQ in Huh7/SR cells (Fig. 4C,D), indicating that TARBP2 is downregulated through lysosome‐mediated proteolysis. This observation prompted us to further investigate whether autophagy is involved in lysosome‐mediated TARBP2 protein degradation. Autophagy is an intracellular, bulk degradation process that delivers cytoplasmic components to the lysosomes for protein degradation via autophagosomes (Choi, 2012; Mizushima, 2007). To investigate whether the TARBP2 protein is degraded through the autophagic–lysosomal pathway, Huh7/SR cells were treated with bafilomycin A1 (BFA) to inhibit fusion between autophagosomes and lysosomes (Yamamoto et al., 1998). TARBP2 protein levels were reconstituted through BFA treatment in Huh7/SR cells (Fig. 4E,F). An increase in LC3B‐II/LC3B‐I conversion indicated the accumulation of autophagosomes. These data suggested that the degradation of TARBP2 occurs via the autophagic–lysosomal pathway in HCC/SR cells. ATG5 induces the formation of a torus‐shaped structure through expanding phagophores for autophagosome formation (Jung et al., 2016; Klionsky et al., 2003). To further clarify whether the autophagic–lysosomal pathway contributes to TARBP2 protein degradation, ATG5 was knocked down in Huh7/SR cells to inhibit autophagosome biogenesis. Consistent results indicated that the TARBP2 protein level was restored in ATG5‐knockdown Huh7/SR cells (Fig. 4G,H). These results demonstrated that TARBP2 protein degradation occurs through autophagic–lysosomal proteolysis in HCC/SR cells.
Figure 4.
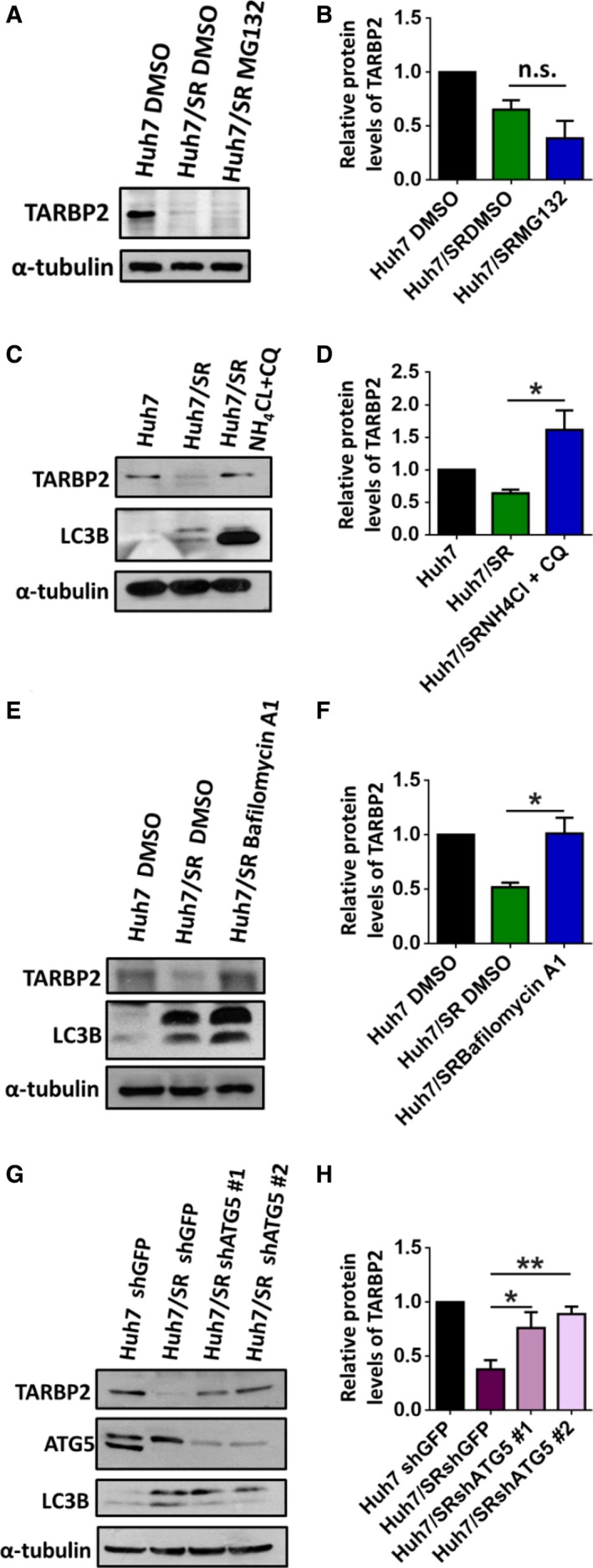
The TARBP2 protein is destabilized through the autophagic–lysosomal pathway. Effects of the proteolytic pathways on TARBP2 downregulation in HCC/SR cells. (A, B) Huh7/SR cells were treated with MG132 (5 μm) for 24 h. (C, D) Huh7/SR cells were treated with NH 4Cl (10 mm) and CQ (200 μm) for 48 h. (E, F) Huh7/SR cells were treated with BFA (100 nm) for 48 h. (G, H) ATG5 was knocked down in the Huh7/SR cells. The TARBP2, ATG5, and LC3B proteins were quantified via western blot analysis. Data are presented as the mean ± SEM from at least three independent experiments. Multigroup comparisons were analyzed by one‐way ANOVA with Tukey's post hoc test. P values < 0.05 were considered statistically significant. *P < 0.05; **P < 0.01.
3.5. TARBP2 reduces the sorafenib resistance of HCC through downregulation of the Nanog protein
Because TARBP2 protein downregulation occurred through the autophagic–lysosomal pathway and induced sorafenib resistance in HCC cells, we further investigated the components downstream of TARBP2 in sorafenib resistance in SR cells. CSCs, known as tumor‐initiating cells, exhibit a high ability to enhance tumorigenesis, metastasis, chemotherapy, and radiation resistance (Magee et al., 2012). The CSC markers SOX2, OCT4, and Nanog have been demonstrated to promote the resistance of cancer to drugs including sorafenib, tamoxifen, cisplatin, and paclitaxel (Di and Zhao, 2015; Shan et al., 2012; Singh and Settleman, 2010; Vinogradov and Wei, 2012). As previously described, TARBP2 inhibits CSC marker expression in Ewing sarcoma (De Vito et al., 2012). However, the role of TARBP2 in regulating CSC markers in HCC cells remains unclear. Thus, we examined the protein levels of Nanog, SOX2, and OCT4 in the paired HCC cells. Nanog protein expression was significantly increased in Huh7/SR cells (Fig. 5A). Next, we confirmed Nanog protein expression by manipulating TARBP2 expression in the paired Huh7 cell lines. Nanog protein expression was suppressed in TARBP2‐overexpressing Huh7/SR cells (Fig. 5B). To examine whether TARBP2‐mediated Nanog suppression is miRNA‐dependent, TARBP2 ΔC4 was overexpressed in Huh7/SR cells. Nanog remained suppressed in the TARBP2 ΔC4‐overexpressing Huh7/SR cells, suggesting that TARBP2‐mediated Nanog suppression is miRNA‐independent (Fig. 5C). Additionally, knockdown of TARBP2 increased the expression of the Nanog protein in the Huh7 cells (Fig. 5D). Supporting previous results, SOX2 and OCT4 expression exhibited no obvious difference in TARBP2‐overexpressing, TARBP2‐ΔC4‐overexpressing or TARBP2‐knockdown Huh7/SR and Huh7 cells (Fig. 5B–D). These results indicated that TARBP2‐mediated Nanog protein inhibition is miRNA independent. To further investigate the biological consequences of TARBP2‐induced Nanog suppression in HCC cells, TARBP2 and Nanog were co‐knocked down in Huh7 cells through sorafenib treatment to detect the level of sorafenib resistance (Fig. 5E,F). The MTT assay demonstrated that the knockdown of TARBP2 enhanced sorafenib resistance, whereas co‐knockdown of Nanog resensitized the Huh7 cells to sorafenib treatment (Fig. 5F). These results suggested that downregulation of TARBP2 enhances sorafenib resistance through stabilization of the Nanog protein in HCC/SR cells. To evaluate this mechanism in vivo, we generated an HCC xenograft model of acquired resistance to sorafenib. Male BALB/c athymic mice were subcutaneously injected with Huh7 cells. After tumor growth reached a volume of 100 mm3, the mice were treated with placebo or sorafenib (10 mg·kg−1·day−1) via gavage. At the end of sorafenib treatment, the tumor samples were classified into SR (tumor volume > 1000 mm3) and SS (tumor volume < 1000 mm3) groups. The individual tumor samples were collected for western blot analysis. The in vivo evidence showed that TARBP2 was significantly decreased in SR tumor tissues, whereas Nanog expression was upregulated (Fig. 5G,H). Nanog is a transcription factor that is activated in embryonic stem (ES) cells and has been demonstrated to play a role in the chemoresistance of liver CSCs (Chiba et al., 2006; Lee et al., 2011). Genetic changes regulate the cellular stemness of liver CSCs, where specific cell surface markers and functional markers are activated to maintain the features of these cells, including CD24, CD44, CD133, EpCAM, and ALDH1 (Ma et al., 2008; Yamashita and Wang, 2013; Yamashita et al., 2009). Having confirmed the role of TARBP2 in regulating Nanog expression, we next investigated whether TARBP2 regulated CSC marker expression. CD24, CD44, CD133, EpCAM, and ALDH1 were detected in the paired Huh7 cell lines, and we found that CD44, CD133, EpCAM, and ALDH1 were significantly increased in Huh7/SR cells, whereas these effects were reduced by restoration of TARBP2 (Fig. S1A). We consistently found that these markers were enhanced by the knockdown of TARBP2 in Huh7 cells (Fig. S1B). These results demonstrated that TARBP2 reduces CSC features. We further investigated the sphere formation of Huh7 cells as a representation of their CSC phenotypes (Visvader and Lindeman, 2008). The sphere number was increased in Huh7/SR cells, whereas sphere formation was abolished by the restoration of TARBP2 (Fig. S1C). An increased sphere‐forming capacity was observed in TARBP2‐knockdown Huh7 cells (Fig. S1D), supportively indicating that TARBP2 reduces CSC properties in HCC cells. These in vitro results suggested that downregulation of TARBP2 inhibits sorafenib resistance through stabilizing Nanog and maintaining CSC functions.
Figure 5.
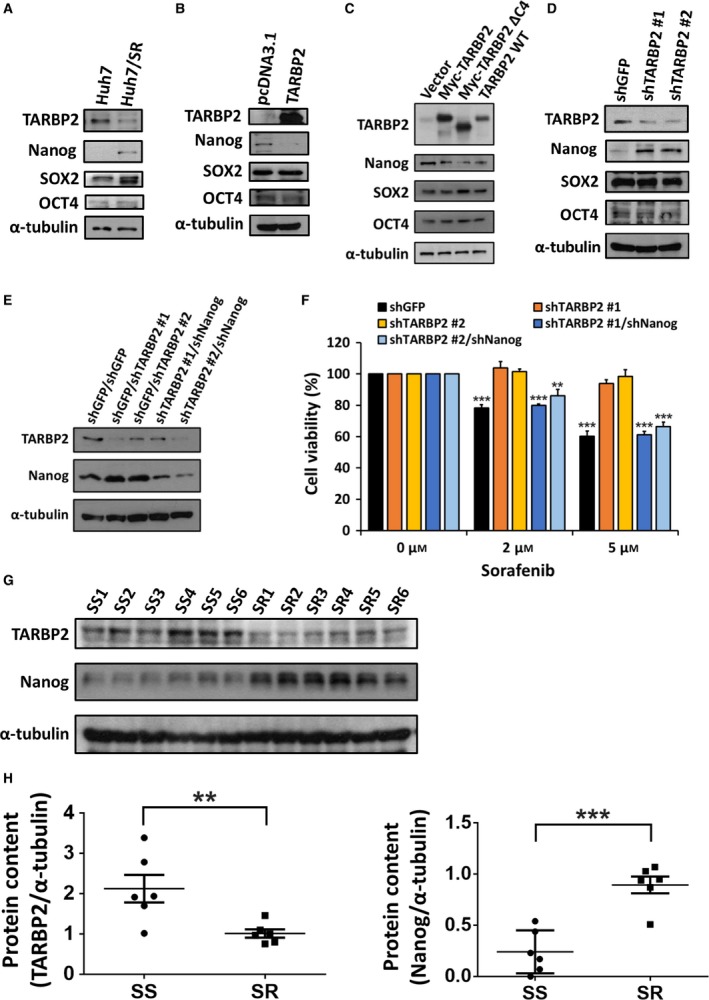
Downregulation of TARBP2 increases Nanog protein levels to promote SR. (A–D) The protein expression of Nanog, SOX2, and OCT4 in paired HCC cells was analyzed via western blot analysis (A). TARBP2 (B) or TARBP2 ΔC4 (C) was overexpressed in Huh7/SR cells for 48 h. TARBP2 was knocked down in Huh7 cells (D). TARBP2 and CSC markers were quantified via western blot analysis. (E, F) Effect of TARBP‐mediated Nanog downregulation on SR. TARBP2 and Nanog were co‐knocked down in Huh7 cells. TARBP2 and Nanog protein levels were determined via western blot analysis (E). The stable clones were treated with the indicated concentrations of sorafenib for 48 h. Cell viability was measured using the MTT assay (F). Data are presented as mean ± SEM, with at least n = 3 per group. (G, H) The protein lysates were homogenized from SS and SR tumor tissues (n = 6, G). TARBP2 and Nanog expression were analyzed by western blot analysis. TARBP2 and Nanog expression were normalized to α‐tubulin to quantify the protein content by using imagej software (National Institutes of Health, Bethesda, MD, USA) (H). Multigroup comparisons were analyzed by one‐way ANOVA with Tukey's post hoc test. P values < 0.05 were considered statistically significant. **P < 0.01; ***P < 0.005.
3.6. TARBP2 expression destabilizes Nanog protein
Because TARBP2 complements sorafenib treatment by suppressing Nanog expression in HCC cells, we next confirmed the mechanism underlying Nanog downregulation. The transcription level of NANOG was determined in the paired Huh7 cell lines. However, the mRNA expression of NANOG was decreased in the Huh7/SR cells (Fig. 6A). Owing to the contrast in Nanog protein and mRNA expression between Huh7 and Huh7/SR cells, we further verified whether NANOG mRNA levels were affected by this regulation and determined that NANOG mRNA expression was unaffected in TARBP2‐overexpressing Huh7/SR cells and TARBP2‐knockdown Huh7 cells (Fig. 6B,C). Despite the contrasting expression of Nanog protein and mRNA between Huh7 and Huh7/SR cells, further evidence demonstrated a transcription‐independent role of TARBP2 in mediating Nanog downregulation. Based on these results, we determined the protein stability of Nanog in the paired Huh7 cell lines with changes in TARBP2 expression (Fig. S2A,B). The degradation rate of the Nanog protein was enhanced in TARBP2‐overexpressing Huh/SR cells (Fig. 6D,E). Accordingly, knockdown of TARBP2 reduced the degradation rate of the Nanog protein (Fig. 6F,G), demonstrating that the stabilization of TARBP2 expression accelerates Nanog protein degradation. We further investigated whether TARBP2‐mediated Nanog protein degradation occurs through the proteasome pathway in Huh7 cells. NH4Cl and CQ were added to TARBP2‐overexpressing Huh7 cells to inhibit lysosome activity. Blocking the lysosome degradation pathway restored TARBP2‐mediated Nanog downregulation. Nevertheless, inhibition of proteasome‐mediated protein degradation could not prevent TARBP2‐mediated Nanog degradation, suggesting that the degradation of the TARBP2 protein in HCC/SR cells is lysosome‐dependent (Fig. 6H). Taken together, our results indicated that TARBP2 enhances sorafenib sensitization through acceleration of Nanog protein degradation in an miRNA‐independent manner. In SR HCC cells, TARBP2 is degraded through an autophagic–lysosomal pathway. Consequently, downregulation of TARBP2 enhances sorafenib resistance through stabilization of the Nanog protein in HCC cells and is correlated with poor clinical outcomes in HCC patients (Fig. 7).
Figure 6.
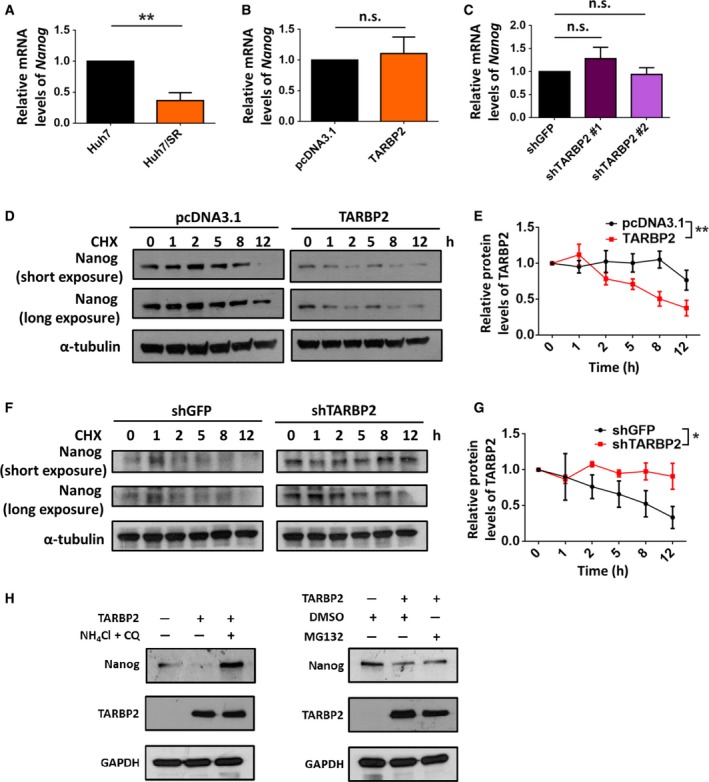
TARBP2 expression reduces Nanog protein stability. (A–C) Effect of TARBP2 on the mRNA expression of NANOG. CSC markers and NANOG mRNA expression in paired HCC cells were analyzed via real‐time PCR (A). TARBP2 was overexpressed in Huh7/SR cells for 48 h (B). TARBP2 was knocked down in Huh7 cells (C). NANOG mRNA expression was analyzed using real‐time PCR. Multigroup comparisons were analyzed by one‐way ANOVA with Tukey's post hoc test. (D–G) Effect of TARBP2 expression on Nanog protein stability. TARBP2 was overexpressed in Huh7/SR for 48 h. The cells were treated with CHX (100 μg·mL−1) for the indicated time periods (D). TARBP2 was knocked down in Huh7 cells. The cells were treated with CHX (100 μg·mL−1) for the indicated time periods (F). The relative quantity of the depicted proteins was analyzed through three independent experiments (E, G). (H) Effects of the proteolytic pathways on TARBP2‐mediated Nanog downregulation in Huh7 cells. TARBP2 was overexpressed in Huh7 cells for 24 h. The cells were treated with NH 4Cl (10 mm) and CQ (200 μm) for 48 h or with MG132 (5 μm) for 24 h. Data are presented as mean ± SEM. Data are presented as the mean ± SEM from at least three independent experiments. Multigroup comparisons were analyzed by two‐way ANOVA with Tukey's post hoc test. P values < 0.05 were considered statistically significant. *P < 0.05; **P < 0.01.
Figure 7.
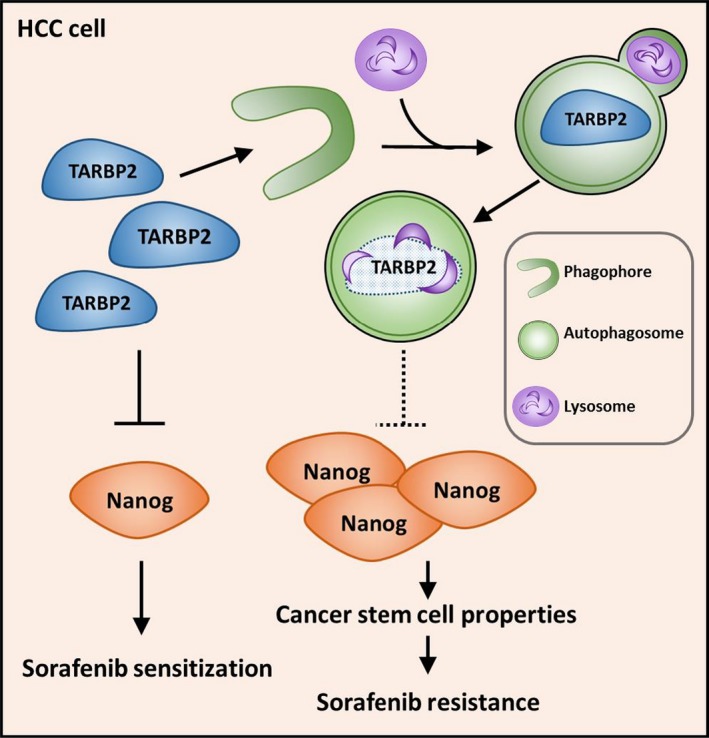
Schematic model of TARBP2‐mediated Nanog suppression in SR in HCC. TARBP2 was destabilized in HCC/SR cells through the autophagic–lysosomal proteolytic pathway. Downregulation of TARBP2 stabilized Nanog protein expression and increased CSC properties to enhance SR in HCC cells.
4. Discussion
Transactivation response element RNA‐binding protein 2 has been associated with CSC features. In the present study, we identified a novel role of TARBP2 in sorafenib resistance in HCC. The TARBP2 protein was destabilized in HCC/SR cells through autophagic–lysosomal proteolytic degradation. Downregulation of TARBP2 stabilized Nanog protein expression and enhanced sorafenib resistance in HCC cells. Among the components of miRNA biogenesis, downregulation of DGCR8, Dicer, p68, and p72 has been associated with a poor prognosis in HCC (Kitagawa et al., 2013). Our results demonstrated that suppression of TARBP2 expression promoted sorafenib resistance in HCC cells (Fig. 2E–H), and downregulation of TARBP2 was correlated with poor outcomes in patients with HCC (Fig. 1A–D), suggesting that miRNA biogenesis factors are globally repressed in HCC cancer progression. Additionally, TARBP2 expression is reduced in several cancers, including colorectal cancer, gastric cancer, and Ewing sarcoma (De Vito et al., 2012; Garre et al., 2010; Yu and Li, 2016). However, TARBP2 functions as an oncogene to contribute to malignant transformation and proliferation in cutaneous malignant melanoma and adrenocortical carcinoma, and its expression is associated with an unfavorable prognosis in patients with breast cancer (Caramuta et al., 2013; Lin et al., 2014; Sand et al., 2012). These reports suggest that the effects of alteration of TARBP2 expression on cancer development are tissue‐specific.
Nanog is a pivotal transcription factor involved in the self‐renewal of CSCs and maintenance of the undifferentiated state of pluripotent cells (Gawlik‐Rzemieniewska and Bednarek, 2016; Pan and Thomson, 2007). Upregulation of Nanog has been reported to contribute to oncogenesis in multiple types of cancer (Chiou et al., 2010; Lin et al., 2012b; Meng et al., 2010). Notably, although a high level of Nanog expression correlates with a poor prognosis and sorafenib resistance in HCC, how Nanog is induced through these regulatory mechanisms is unknown (Jeter et al., 2015; Shan et al., 2012). We observed that downregulation of TARBP2 enhances Nanog protein expression through stabilization of the Nanog protein to render HCC cells resistant to sorafenib (Fig. 5E,F). TARBP2‐mediated Nanog protein degradation occurs via the lysosome pathway (Fig. 6). High activation of PI3K/Akt has been reported to induce sorafenib resistance in HCC (Jeter et al., 2015; Shan et al., 2012), and this activation is sustained to promote OCT4 and Nanog expression for chemoresistance and EMT in cancer cells (Almozyan et al., 2017), suggesting that PI3K/Akt‐mediated phosphorylation stabilizes Nanog protein expression to facilitate sorafenib resistance. The prolyl isomerase Pin1 has been shown to interact with phosphorylated Nanog. This interaction prevents the proteolysis of Nanog through inhibiting its ubiquitination. Thus, stabilized Nanog promotes self‐renewal to maintain the pluripotency of ES cells (Moretto‐Zita et al., 2010). In HCC, the status of Erk phosphorylation in peripheral blood mononuclear cells has been indicated as a predictor of the efficacy of sorafenib plus octreotide LAR treatment outcomes in HCC patients. In HCC patients with resistance to sorafenib plus octreotide LAR, Erk activity was observed to gradually increase after 10 days from the beginning of treatment (Caraglia et al., 2011). This finding is suggested that hyperphosphorylated Nanog is stabilized to confer self‐renewal and CSC features in HCC for sorafenib resistance.
Cytosolic LC3 conversion (LC3‐I to LC3‐II) functions in autophagosome formation and cargo selection and is regarded as a marker of autophagosome biogenesis (Tanida and Waguri, 2010; Tanida et al., 2005). TARBP2 was shown to be downregulated through an autophagic–lysosomal pathway, and we observed that the HCC/SR cells exhibited greater autophagosome formation than the parental cells (Fig. 4C,E,G), suggesting that autophagy activity is increased in Huh7/SR cells to enhance TARBP2 protein degradation. Selective autophagy is the initial process in autophagy, in which specific cellular material or organelles tagged with ubiquitin are selectively recognized (Stolz et al., 2014). Upregulation of miR‐423‐5p secretion in sorafenib‐treated patients with HCC has been found to result in favorable progress in relief and stabilization of the disease. The increased miR‐423‐5p inhibits the cell cycle and promotes autophagy activity (Stiuso et al., 2015). The miRNA biogenesis factors Dicer and Ago2 are degraded by autophagy through recognition of the autophagy receptor to promote cancer progression (Gibbings et al., 2012; Lai et al., 2018). Both Dicer and Ago2 are ubiquitylated through an E3 ligase to specifically interact with the autophagy receptor. As a component of RISC with Dicer and Ago2, TARBP2 might be degraded in cancer cells via selective autophagy. It has been suggested that sorafenib treatment promotes autophagy to selectively degrade TARBP2 and that this effect may persist, leading to sorafenib resistance (Stiuso et al., 2015). However, the E3 ligase of TARBP2 and ubiquitination levels need to be determined for further investigation in cancer. Supporting this concept, the SUMOylation of TARBP2 stabilizes TARBP2 protein expression through reducing its ubiquitination to suppress tumor progression, indicating that the ubiquitination of TARBP2 is essential for cancer progression (Chen et al., 2015). Additionally, autophagy facilitates sorafenib resistance in HCC cells (Liu et al., 2013; Zhai et al., 2014), suggesting that inhibition of autophagy resensitizes HCC cells to sorafenib treatment through blocking the degradation of TARBP2. Thus, our study may offer a potential strategy for overcoming sorafenib resistance through inhibition of autophagy activity.
Overexpression of TARBP2 induces tumor formation via inhibition of PKR phosphorylation and PKR‐mediated eIF2α phosphorylation (Benkirane et al., 1997; Kim et al., 2014). As an RNA‐binding protein, TARBP2 enhances invasion and metastatic colonization by directly binding APP and ZNF395 transcripts, thereby post‐transcriptionally enhancing their decay rate in breast cancer (Goodarzi et al., 2014). This evidence proves that TARBP2 exhibits an miRNA‐independent role in regulating cancer progression. By disrupting the interaction between Dicer and TARBP2, TARBP2 retains the ability to downregulate Nanog expression (Fig. 5C). Thus, the present study reveals another miRNA‐independent role of TARBP2 that destabilizes the Nanog protein and consequently resensitizes HCC cells to sorafenib treatment.
5. Conclusions
Transactivation response element RNA‐binding protein 2 is significantly downregulated in SR HCC cells. Restoration of TARBP2 expression resensitizes HCC/SR cells to sorafenib treatment. The TARBP2 protein is destabilized through autophagic–lysosomal proteolysis and thereby stabilizes the protein expression of the CSC marker Nanog to facilitate sorafenib resistance of HCC cells. TARBP2 expression inversely correlates with Nanog levels in SR HCC tumors. In present study, we reveal a novel miRNA‐independent role of TARBP2 in regulating sorafenib resistance in HCC cells.
Author contributions
HHL, CWL, DLO, and PSC conceived and designed all experiments. HHL conducted the experiments on TARBP2 ΔC4. DLO performed the animal experiment. CWL performed all other experiments in this study. CCH, DLO, HYS, CFC, and PSC discussed the data. HHL, CWL, DLO, CFC, HYS, and PSC wrote and revise the paper.
Conflicts of interest
The authors declare no conflict of interest.
Supporting information
Fig. S1. TARBP2 inhibits CSCs marker expression.
Fig. S2. Manipulation of TARBP2 expression for analysis of Nanog protein stability.
Acknowledgements
We thank Dr. Anne Gatignol for providing plasmids expressing Myc‐TARBP2 and TARBP2 ΔC4. We thank Dr. Dong‐Yan Jin for sharing the plasmids with Addgene. This study was supported by the Ministry of Science and Technology, Taiwan (MOST 105‐2320‐B‐006‐050, MOST 106‐2320‐B‐006‐038, MOST 106‐2320‐B‐006‐021, MOST 106‐2314‐B‐002‐229‐MY3, and MOST 107‐2320‐B‐006‐068).
Hui‐Huang Lai, Chih‐Wei Li equally contributed to this study.
Da‐Liang Ou has been a National Taiwan University YongLin Scholar since 2018.
Contributor Information
Da‐Liang Ou, Email: dlou@ntu.edu.tw.
Pai‐Sheng Chen, Email: bio.benson@gmail.com.
References
- Aguirre‐Gamboa R, Gomez‐Rueda H, Martinez‐Ledesma E, Martinez‐Torteya A, Chacolla‐Huaringa R, Rodriguez‐Barrientos A, Tamez‐Pena JG and Trevino V (2013) SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE 8, e74250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almozyan S, Colak D, Mansour F, Alaiya A, Al‐Harazi O, Qattan A, Al‐Mohanna F, Al‐Alwan M and Ghebeh H (2017) PD‐L1 promotes OCT4 and Nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int J Cancer 141, 1402–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balogh J, Victor D 3rd, Asham EH, Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM and Monsour HP Jr (2016) Hepatocellular carcinoma: a review. J Hepatocell Carcinoma 3, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkirane M, Neuveut C, Chun RF, Smith SM, Samuel CE, Gatignol A and Jeang KT (1997) Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA‐dependent protein kinase PKR. EMBO J 16, 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraglia M, Giuberti G, Marra M, Addeo R, Montella L, Murolo M, Sperlongano P, Vincenzi B, Naviglio S, Prete SD et al (2011) Oxidative stress and ERK1/2 phosphorylation as predictors of outcome in hepatocellular carcinoma patients treated with sorafenib plus octreotide LAR. Cell Death Dis 2, e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramuta S, Lee L, Ozata DM, Akcakaya P, Xie H, Hoog A, Zedenius J, Backdahl M, Larsson C and Lui WO (2013) Clinical and functional impact of TARBP2 over‐expression in adrenocortical carcinoma. Endocr Relat Cancer 20, 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlomagno F, Anaganti S, Guida T, Salvatore G, Troncone G, Wilhelm SM and Santoro M (2006) BAY 43‐9006 inhibition of oncogenic RET mutants. J Natl Cancer Inst 98, 326–334. [DOI] [PubMed] [Google Scholar]
- Cervello M, Bachvarov D, Lampiasi N, Cusimano A, Azzolina A, McCubrey JA and Montalto G (2012) Molecular mechanisms of sorafenib action in liver cancer cells. Cell Cycle 11, 2843–2855. [DOI] [PubMed] [Google Scholar]
- Chen C, Zhu C, Huang J, Zhao X, Deng R, Zhang H, Dou J, Chen Q, Xu M, Yuan H et al (2015) SUMOylation of TARBP2 regulates miRNA/siRNA efficiency. Nat Commun 6, 8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, Nakauchi H and Taniguchi H (2006) Side population purified from hepatocellular carcinoma cells harbors cancer stem cell‐like properties. Hepatology 44, 240–251. [DOI] [PubMed] [Google Scholar]
- Chiou SH, Wang ML, Chou YT, Chen CJ, Hong CF, Hsieh WJ, Chang HT, Chen YS, Lin TW, Hsu HS et al (2010) Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell‐like properties and epithelial‐mesenchymal transdifferentiation. Cancer Res 70, 10433–10444. [DOI] [PubMed] [Google Scholar]
- Choi KS (2012) Autophagy and cancer. Exp Mol Med 44, 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow AK, Ng L, Lam CS, Wong SK, Wan TM, Cheng NS, Yau TC, Poon RT and Pang RW (2013) The enhanced metastatic potential of hepatocellular carcinoma (HCC) cells with sorafenib resistance. PLoS ONE 8, e78675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colagrande S, Regini F, Taliani GG, Nardi C and Inghilesi AL (2015) Advanced hepatocellular carcinoma and sorafenib: diagnosis, indications, clinical and radiological follow‐up. World J Hepatol 7, 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Ge J, Wang X, Qian X, Zhang C and Li X (2013) OCT4 regulates epithelial‐mesenchymal transition and its knockdown inhibits colorectal cancer cell migration and invasion. Oncol Rep 29, 155–160. [DOI] [PubMed] [Google Scholar]
- Daniels SM, Melendez‐Pena CE, Scarborough RJ, Daher A, Christensen HS, El Far M, Purcell DF, Laine S and Gatignol A (2009) Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol Biol 10, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vito C, Riggi N, Cornaz S, Suva ML, Baumer K, Provero P and Stamenkovic I (2012) A TARBP2‐dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell 21, 807–821. [DOI] [PubMed] [Google Scholar]
- Di C and Zhao Y (2015) Multiple drug resistance due to resistance to stem cells and stem cell treatment progress in cancer (Review). Exp Ther Med 9, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Serag HB (2011) Hepatocellular carcinoma. N Engl J Med 365, 1118–1127. [DOI] [PubMed] [Google Scholar]
- El‐Serag HB, Marrero JA, Rudolph L and Reddy KR (2008) Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology 134, 1752–1763. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Ricaud L, Kourtchenko O, Zackrisson M, Warringer J and Blomberg A (2016) PRECOG: a tool for automated extraction and visualization of fitness components in microbial growth phenomics. BMC Bioinformatics 17, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C and Esteban M (2006) Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev 70, 1032–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garre P, Perez‐Segura P, Diaz‐Rubio E, Caldes T and de la Hoya M (2010) Reassessing the TARBP2 mutation rate in hereditary nonpolyposis colorectal cancer. Nat Genet 42, 817–818; author reply 818. [DOI] [PubMed] [Google Scholar]
- Gatignol A, Buckler‐White A, Berkhout B and Jeang KT (1991) Characterization of a human TAR RNA‐binding protein that activates the HIV‐1 LTR. Science 251, 1597–1600. [DOI] [PubMed] [Google Scholar]
- Gauthier A and Ho M (2013) Role of sorafenib in the treatment of advanced hepatocellular carcinoma: an update. Hepatol Res 43, 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawlik‐Rzemieniewska N and Bednarek I (2016) The role of NANOG transcriptional factor in the development of malignant phenotype of cancer cells. Cancer Biol Ther 17, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini EG, Farinati F, Ciccarese F, Pecorelli A, Rapaccini GL, Di Marco M, Benvegnu L, Caturelli E, Zoli M, Borzio F et al ; Italian Liver Cancer (ITA.LI.CA) group (2015) Prognosis of untreated hepatocellular carcinoma. Hepatology 61, 184–190. [DOI] [PubMed] [Google Scholar]
- Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P and Voinnet O (2012) Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol 14, 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH and Ciechanover A (2002) The ubiquitin‐proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82, 373–428. [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Zhang S, Buss CG, Fish L, Tavazoie S and Tavazoie SF (2014) Metastasis‐suppressor transcript destabilization through TARBP2 binding of mRNA hairpins. Nature 513, 256–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PD and Young MR (1991) Ammonium chloride, an inhibitor of phagosome‐lysosome fusion in macrophages, concurrently induces phagosome‐endosome fusion, and opens a novel pathway: studies of a pathogenic mycobacterium and a nonpathogenic yeast. J Exp Med 174, 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C, Lin LI, Cheng YC, Feng ZR, Shao YY, Cheng AL and Ou DL (2016) Cyclin E1 inhibition can overcome sorafenib resistance in hepatocellular carcinoma cells through Mcl‐1 suppression. Clin Cancer Res 22, 2555–2564. [DOI] [PubMed] [Google Scholar]
- Jeter CR, Yang T, Wang J, Chao HP and Tang DG (2015) Concise review: NANOG in cancer stem cells and tumor development: an update and outstanding questions. Stem Cells 33, 2381–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HJ, Seo I, Casciello F, Jacquelin S, Lane SW, Suh SI, Suh MH, Lee JS and Baek WK (2016) The anticancer effect of chaetocin is enhanced by inhibition of autophagy. Cell Death Dis 7, e2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoubi G, Gugger M, Schmid R and Dutly A (2009) OCT4 expression in human non‐small cell lung cancer: implications for therapeutic intervention. Interact Cardiovasc Thorac Surg 8, 393–397. [DOI] [PubMed] [Google Scholar]
- Keating GM and Santoro A (2009) Sorafenib: a review of its use in advanced hepatocellular carcinoma. Drugs 69, 223–240. [DOI] [PubMed] [Google Scholar]
- Kim Y, Yeo J, Lee JH, Cho J, Seo D, Kim JS and Kim VN (2014) Deletion of human tarbp2 reveals cellular microRNA targets and cell‐cycle function of TRBP. Cell Rep 9, 1061–1074. [DOI] [PubMed] [Google Scholar]
- Kitagawa N, Ojima H, Shirakihara T, Shimizu H, Kokubu A, Urushidate T, Totoki Y, Kosuge T, Miyagawa S and Shibata T (2013) Downregulation of the microRNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci 104, 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M et al (2003) A unified nomenclature for yeast autophagy‐related genes. Dev Cell 5, 539–545. [DOI] [PubMed] [Google Scholar]
- Kok KH, Ng MH, Ching YP and Jin DY (2007) Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J Biol Chem 282, 17649–17657. [DOI] [PubMed] [Google Scholar]
- Lai HH, Li JN, Wang MY, Huang HY, Croce CM, Sun HL, Lyu YJ, Kang JW, Chiu CF, Hung MC et al (2018) HIF‐1alpha promotes autophagic proteolysis of Dicer and enhances tumor metastasis. J Clin Invest 128, 625–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TK, Castilho A, Cheung VC, Tang KH, Ma S and Ng IO (2011) CD24(+) liver tumor‐initiating cells drive self‐renewal and tumor initiation through STAT3‐mediated NANOG regulation. Cell Stem Cell 9, 50–63. [DOI] [PubMed] [Google Scholar]
- Leis O, Eguiara A, Lopez‐Arribillaga E, Alberdi MJ, Hernandez‐Garcia S, Elorriaga K, Pandiella A, Rezola R and Martin AG (2012) Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 31, 1354–1365. [DOI] [PubMed] [Google Scholar]
- Liang Y, Zheng T, Song R, Wang J, Yin D, Wang L, Liu H, Tian L, Fang X, Meng X et al (2013) Hypoxia‐mediated sorafenib resistance can be overcome by EF24 through Von Hippel‐Lindau tumor suppressor‐dependent HIF‐1alpha inhibition in hepatocellular carcinoma. Hepatology 57, 1847–1857. [DOI] [PubMed] [Google Scholar]
- Lin T, Ding YQ and Li JM (2012b) Overexpression of Nanog protein is associated with poor prognosis in gastric adenocarcinoma. Med Oncol 29, 878–885. [DOI] [PubMed] [Google Scholar]
- Lin S, Hoffmann K and Schemmer P (2012a) Treatment of hepatocellular carcinoma: a systematic review. Liver Cancer 1, 144–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Wu M, Liu P, Wei F, Li L, Tang H, Xie X, Liu X, Yang L and Xie X (2014) Up‐regulation and worse prognostic marker of cytoplasmic TARBP2 expression in obstinate breast cancer. Med Oncol 31, 868. [DOI] [PubMed] [Google Scholar]
- Liu B, Cao Y, Jiang H and Mao A (2013) Autophagy facilitates the sorafenib resistance of hepatocellular carcinoma cells. West Indian Med J 62, 698–700. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A et al (2008) Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 359, 378–390. [DOI] [PubMed] [Google Scholar]
- Ma S, Chan KW, Lee TK, Tang KH, Wo JY, Zheng BJ and Guan XY (2008) Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res 6, 1146–1153. [DOI] [PubMed] [Google Scholar]
- Magee JA, Piskounova E and Morrison SJ (2012) Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell 21, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng HM, Zheng P, Wang XY, Liu C, Sui HM, Wu SJ, Zhou J, Ding YQ and Li J (2010) Over‐expression of Nanog predicts tumor progression and poor prognosis in colorectal cancer. Cancer Biol Ther 9, 295–302. [DOI] [PubMed] [Google Scholar]
- Mizushima N (2007) Autophagy: process and function. Genes Dev 21, 2861–2873. [DOI] [PubMed] [Google Scholar]
- Moretto‐Zita M, Jin H, Shen Z, Zhao T, Briggs SP and Xu Y (2010) Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci USA 107, 13312–13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez‐Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, Inoue M, Bergers G, Hanahan D and Casanovas O (2009) Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell 15, 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G and Thomson JA (2007) Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res 17, 42–49. [DOI] [PubMed] [Google Scholar]
- Pinter M, Hucke F, Graziadei I, Vogel W, Maieron A, Konigsberg R, Stauber R, Grunberger B, Muller C, Kolblinger C et al (2012) Advanced‐stage hepatocellular carcinoma: transarterial chemoembolization versus sorafenib. Radiology 263, 590–599. [DOI] [PubMed] [Google Scholar]
- Raza A and Sood GK (2014) Hepatocellular carcinoma review: current treatment, and evidence‐based medicine. World J Gastroenterol 20, 4115–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM (2004) ONCOMINE: a cancer microarray database and integrated data‐mining platform. Neoplasia 6, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand M, Skrygan M, Georgas D, Sand D, Gambichler T, Altmeyer P and Bechara FG (2012) The miRNA machinery in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases and benign melanocytic nevi. Cell Tissue Res 350, 119–126. [DOI] [PubMed] [Google Scholar]
- Shan J, Shen J, Liu L, Xia F, Xu C, Duan G, Xu Y, Ma Q, Yang Z, Zhang Q et al (2012) Nanog regulates self‐renewal of cancer stem cells through the insulin‐like growth factor pathway in human hepatocellular carcinoma. Hepatology 56, 1004–1014. [DOI] [PubMed] [Google Scholar]
- Singh A and Settleman J (2010) EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29, 4741–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiuso P, Potenza N, Lombardi A, Ferrandino I, Monaco A, Zappavigna S, Vanacore D, Mosca N, Castiello F, Porto S et al (2015) MicroRNA‐423‐5p promotes autophagy in cancer cells and is increased in serum from hepatocarcinoma patients treated with sorafenib. Mol Ther Nucleic Acids 4, e233. [DOI] [PubMed] [Google Scholar]
- Stolz A, Ernst A and Dikic I (2014) Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16, 495–501. [DOI] [PubMed] [Google Scholar]
- Sun C, Sun L, Jiang K, Gao DM, Kang XN, Wang C, Zhang S, Huang S, Qin X, Li Y et al (2013) NANOG promotes liver cancer cell invasion by inducing epithelial‐mesenchymal transition through NODAL/SMAD3 signaling pathway. Int J Biochem Cell Biol 45, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Tanida I, Minematsu‐Ikeguchi N, Ueno T and Kominami E (2005) Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1, 84–91. [DOI] [PubMed] [Google Scholar]
- Tanida I and Waguri S (2010) Measurement of autophagy in cells and tissues. Methods Mol Biol 648, 193–214. [DOI] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J and Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65, 87–108. [DOI] [PubMed] [Google Scholar]
- Vinogradov S and Wei X (2012) Cancer stem cells and drug resistance: the potential of nanomedicine. Nanomedicine (Lond) 7, 597–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE and Lindeman GJ (2008) Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer 8, 755–768. [DOI] [PubMed] [Google Scholar]
- Wellbrock C, Karasarides M and Marais R (2004) The RAF proteins take centre stage. Nat Rev Mol Cell Biol 5, 875–885. [DOI] [PubMed] [Google Scholar]
- Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M et al (2004) BAY 43‐9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res 64, 7099–7109. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R and Tashiro Y (1998) Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H‐4‐II‐E cells. Cell Struct Funct 23, 33–42. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E et al (2009) EpCAM‐positive hepatocellular carcinoma cells are tumor‐initiating cells with stem/progenitor cell features. Gastroenterology 136, 1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T and Wang XW (2013) Cancer stem cells in the development of liver cancer. J Clin Invest 123, 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X and Li Z (2016) The role of TARBP2 in the development and progression of cancers. Tumour Biol 37, 57–60. [DOI] [PubMed] [Google Scholar]
- Zhai B, Hu F, Jiang X, Xu J, Zhao D, Liu B, Pan S, Dong X, Tan G, Wei Z et al (2014) Inhibition of Akt reverses the acquired resistance to sorafenib by switching protective autophagy to autophagic cell death in hepatocellular carcinoma. Mol Cancer Ther 13, 1589–1598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. TARBP2 inhibits CSCs marker expression.
Fig. S2. Manipulation of TARBP2 expression for analysis of Nanog protein stability.


