Abstract
BACKGROUND
Recently, gut microbiota has been associated with various diseases other than intestinal disease. Thus, there has been rapid growth in the study of gut microbiota. Considering the numerous factors influencing gut microbiota such as age, diet, etc., area-based research is required. Indonesia has numerous different tribes and each of these tribes have different lifestyles. Hence, it is expected that each tribe has a specific gut microbiota. A deeper insight into the composition of gut microbiota can be used to determine the condition of gut microbiota in Indonesians and to consider which treatment may be suitable and effective to improve health status.
AIM
To investigate the gut microbiota of Indonesian subjects represented by Javanese and Balinese tribes by analyzing fecal samples.
METHODS
Fecal samples were collected from a total of 80 individuals with 20 in each of the young groups ranging from 25-45 years and the elderly group aged 70 years or more from two different regions, Yogyakarta and Bali. Fecal sample collection was performed at the end of the assessment period (day 14 ± 1 d) during which time the subjects were not allowed to consume probiotic or antibiotic products. The quantification of various Clostridium subgroups, Lactobacillus subgroups, Enterococcus, Streptococcus, Staphylococcus, Bacteroides fragilis group and Prevotella, Bifidobacterium and Atopobium cluster, Enterobacteriaceae and Pseudomonas was performed using the Yakult intestinal flora-scan (YIF-SCAN).
RESULTS
The bacterial population in younger subjects’ feces was higher than that in the elderly population, with a total of approximately 10.0 – 10.6 log10 bacterial cells/g feces. The most abundant bacteria in all groups were Clostridium, followed by Prevotella, Atopobium, Bifidobacterium and Bacteroides. In the elderly, an increase in Enterobacteriaceae, Coliform and Escherichia coli was found. In terms of bacterial counts in Yogyakarta, total bacteria, Clostridium coccoides (C. coccoides) group, Bifidobacterium, Prevotella, Lactobacillus plantarum subgroup, and Streptococcus were significantly higher (P < 0.05) in younger than elderly subjects, while the Lactobacillus gasseri subgroup, Lactobacillus casei subgroup, and Lactobacillus reuteri subgroup counts were significantly lower (P < 0.05) in younger subjects. In Balinese subjects, total bacteria, C. coccoides group, Clostridium leptum subgroup, Bacteroides fragilis group, and Prevotella were significantly higher (P < 0.05) in younger compared to elderly individuals, while the Lactobacillus ruminis subgroup, and Enterobacteriaceae were significantly lower (P < 0.05) in younger subjects. The results also revealed that, besides the C. coccoides group and Clostridium leptum group being the most abundant gut microbiota in both Yogyakarta and Balinese people, the latter was indicated by a higher Clostridium perfringens count, which was almost 10 times that of Yogyakarta subjects. This may be a response to different lifestyles in the different tribes; however, this phenomenon requires further extensive study.
CONCLUSION
Bacterial populations were higher in younger than in elderly subjects. Most abundant bacterial groups were Clostridium, Prevotella, Atopobium, Bifidobacterium, and Bacteroides. The level of Clostridium perfringens in Yogyakarta subjects was lower than that in Balinese subjects.
Keywords: Gut microbiota, Indonesian, Elderly, Young people, Enterotype
Core tip: Research on gut microbiota has been growing rapidly due to their relationship with various diseases. Two factors influencing gut microbiota are age and location. Indonesia has numerous different tribes. Hence, it is expected that each tribe will have a specific gut microbiota. This research aimed to investigate the gut microbiota of Indonesians represented by Javanese and Balinese tribes. The results showed that bacterial populations were higher in younger than elderly subjects. The most common bacterial groups were Clostridium, Prevotella, Atopobium, Bifidobacterium, and Bacteroides. The level of Clostridium perfringens between the tribes was different, which might be associated with diet and lifestyle.
INTRODUCTION
Indonesia has a population of 262 million people, with children (aged 0-14 years) comprising 25%, young-adults (aged 15-64 years) comprising 68%, and the elderly (aged 65 and over years) comprising 7% of the population, as suggested by Index Mundi (https://www.indexmundi.com/indonesia/age_structure. html). Moreover, Indonesia has hundreds of tribes each of which has its own lifestyle which has resulted in an expected variety of gut microbiota. The development of gut microbiota composition starts when an infant is born; however, it changes markedly when an infant learns to eat, followed by a stable state starting from teenager and adult age and finally begins to undergo other changes in the elderly. In the child age group, fecal microbiota is not as complicated as that in advanced age. This was indicated by a decrease in Bifidobacteria species diversity and an increase in Bacteroidetes species diversity[1]. These microbial composition changes may affect gut microbiota, especially the individual’s metabolic capacity, and have an important role in health. In addition, degenerative transformation effects on the physiology and function of the gut are possibly due to aging and associated with changes in colonic ecosystem composition and metabolic activities[2]. Previous studies have also found that Bifidobacteria is a protective intestinal microorganism, and decreases in the elderly group age. With regard to putatively detrimental microorganism populations, notably Clostridia and Enterobacteriaceae, these increase in the elderly age group[3].
Three principal human enterotypes were proposed based on a genus or group of specific bacteria[4]. These three enterotypes, which are Bacteroides, Prevotella, and Ruminococcus, were based on 39 individuals (22 European, 13 Japanese, 4 American) that were not nation or continent specific. The study showed that all three enterotypes were largely driven by a certain species composition, although most species are not obligated to provide a plethora of molecular functions. This indicates that functional analysis is needed to understand more about microbial composition. In addition[5], a total of 98 individuals were observed to detect how gut microbial composition is affected by diet, and supported the previously mentioned enterotypes[4]. However, the study showed that the dominant enterotypes, which are Bacteroides and Prevotella, better described gut composition. Both enterotypes have their own role on gut microbiota composition. The former enterotype was related to a diet containing high protein and animal fat, while the latter enterotype was associated with a high carbohydrate diet.
According to a study of 303 Asian children in which the subjects’ gut microbiota community profiles were investigated, Prevotella (P-type) or Bifidobacterium/ Bacteroides (BB-type) are able to encourage two classifications of enterotype-like clusters[6]. The P-type is mostly found in subjects from Indonesia and Thailand, while the BB-type is mainly found in Japan, Taiwan and China. Subjects with high P-type have diets rich in resistant starch. This explained the low bile acid biosynthesis and high carbohydrate digestion suggested by predictive metagenomics.
Previous results[6] revealed that gut microbiota in Indonesian schoolchildren was represented by two unique populations, which were quite distinct from those in other countries. Yogyakarta and Bali regions were selected to represent the Javanese tribe, the most abundant and widespread throughout the Indonesian archipelago and the Balinese tribe which is only concentrated in Bali Island. Both tribes have different lifestyles and diets as are strongly associated with different religions and beliefs. Based on these facts, Yogyakarta and Bali were selected as the sites for the current study as in the previous study[6]. Currently, various methods have been used to study gut microbiota populations. In this research, culture methods on various selective media reflecting living microorganisms are rarely used and replaced by culture independent methods, including quantitative real-time polymerase chain reaction (PCR). The objectives of the present study were to establish a baseline microbiota composition in both healthy young and elderly Indonesian subjects living in Yogyakarta and Bali. This baseline was then used to further study the correlation between gut microbiota and several diseases, such as obesity, type II diabetes, and liver diseases. Therefore, Yogyakarta and Bali were selected due to their different lifestyles and diet habits.
MATERIALS AND METHODS
Subject characteristics and ethic statement
The study subjects were selected from two different sites, Yogyakarta and Bali, Indonesia. Elderly subjects (aged 70 years and above) were recruited from Elderly Houses, while younger subjects (aged from 25-45 years) were volunteer university students and employees. All subjects signed an informed consent before the study. During pre-screening of the study subjects, they were instructed not to consume fermented milk products, probiotics, or prebiotics for the entire study (1-14 d). In addition, information on medical history, physical examination, demographic parameters and vital signs were obtained from each subject (Table 1). A pregnancy test was performed in women, where applicable. Ethical clearance was approved by the Medical and Health Research Ethics Committee, Faculty of Medicine, Universitas Gadjah Mada-Dr. Sardjito General Hospital (Ref: KE/FK/988/EC/2016), on 2 September 2016.
Table 1.
General characteristics of the subjects (n = 80)
| n (%) | |
| Gender | |
| Male | 30 (37.50) |
| Female | 50 (62.50) |
| Ethnicity | |
| Javanese | 34 (42.50) |
| Balinese | 40 (50.00) |
| Chinese | 3 (3.75) |
| Ambonese | 1 (1.25) |
| Bugis | 1 (1.25) |
| Melayu | 1 (1.25) |
| Weight (kg) (mean ± SD) | 55.65 (14.35) |
| Height (cm) (mean ± SD) | 156.07 (10.54) |
| Body mass index (kg/m2) (mean ± SD) | 22.57 (4.54) |
Sample collection
During the assessment period (1-14 d), information on the intestinal microbiota and intestinal environment were collected. Before fecal collection, each subject was given a stool kit and the procedure was explained. On day 14 (± 1 d), the subjects were asked to defecate upon a trail paper (Eiken Chemical Co., Ltd) which was immediately transferred into a fecal tube containing RNAlater (Thermo Fischer Scientific). If the fecal samples were collected during the day, the subjects delivered the sample to the clinical center as soon as possible. However, if fecal samples were collected during the evening, the subjects stored the samples in an icebox containing ice bags and brought them to the clinical center the following morning. In the elderly group, the study team was involved in fecal sample collection and questionnaire completion.
Intestinal microbiota
The Yakult intestinal flora-scan (YIF-SCAN), an intestinal flora analysis system, was used to measure the intestinal microbiota. The basic principle of YIF-SCAN is the quantitative reverse transcription PCR method and it was conducted in Yakult Central Institute, Japan. The samples were brought to Japan by the investigating team from Indonesia. The gut microbiota composition was analyzed using a previously published method[7,16] as follows: Total Bacteria: Clostridium coccoides group, Clostridium leptum subgroup, Bacteroides fragilis group, Bifidobacterium, Atopobium cluster, Prevotella, Clostridium perfringens, Clostridium difficile, Enterobacteriaceae, Lactobacillus (6 subgroups and 3 species); Lactobacillus casei subgroup, Lactobacillus gasseri subgroup, Lactobacillus plantarum subgroup, Lactobacillus reuteri subgroup, Lactobacillus ruminis subgroup, Lactobacillus sakei subgroup, Lactobacillus brevis, Lactobacillus fermentum, Lactobacillus fructivorans, Enterococcus, Staphylococcus, Streptococcus and Pseudomonas. The primer sets used for C. difficile and Streptococcus were previously described[8,9].
Culture method was also used to detect the composition of intestinal microbiotia in this study. By using selective media, this enabled bacterial growth and detection that may be missed. Therefore, the researchers used two methods and compared the results of total Lactobacillus to anticipate different results. Culture method was performed to detect the population of yeast and mold in general using the selective medium Malt Extract Agar (MEA), the population of Lactobacillus plantarum was calculated using Lactobacillus plantarum Selective Media (LPSM), while the population of Escherichia coli, and coliform non E. coli were calculated using Brilliance E. coli/ Coliform Selective Agar from Oxoid.
Questionnaire
The subjects were given a questionnaire at the screening period (15-23 August 2016). The questionnaire was designed to obtain stool frequency (number of stools per day) and stool consistency. For stool consistency parameters, the Bristol Stool Form Scale[10] was used as the measurement scale.
Data analysis
Microbiota composition data were tabulated into a spreadsheet (Microsoft Excel, 2016) as log10 bacterial cells/g feces mean ± SD (detection rate %), and for demographic parameters as [median (min-max)], unless stated otherwise. All under limit detection data were excluded from statistical analysis. All statistical analyses were conducted using (SPSS/PC + 4.0, Chicago, IL, United States). A comparison of continuous variables was conducted with the Student’s t-test for normally distributed data and the Mann Whitney test. Statistical analysis of the YIF-SCAN data was performed using the number of bacterial cells and the detection rate in the four groups (young subjects in Yogyakarta, elderly subjects in Yogyakarta, young subjects in Bali, elderly subjects in Bali), with the Mann-Whitney U-test and chi-square test, respectively. We used R software for the Mann-Whitney U-test and the chi-square test. The statistical methods and techniques mentioned were appropriate for the research.
RESULTS
Stool frequency and consistency
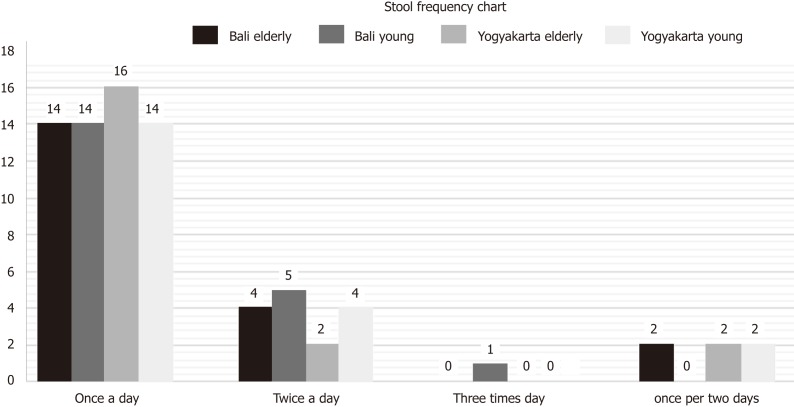
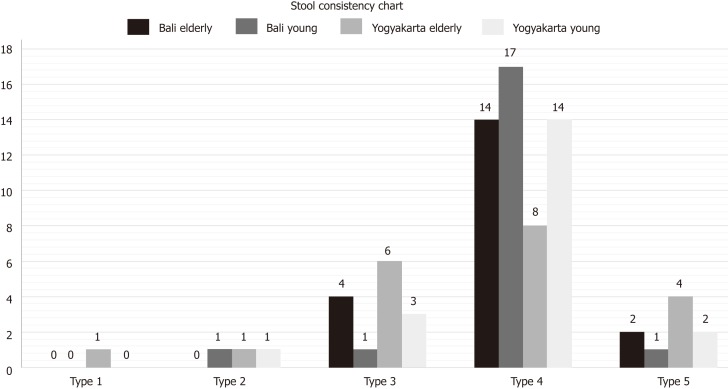
The data retrieved from the questionnaires regarding stool frequency and consistency are illustrated in Figures 1 and 2.
Figure 1.
Data on stool frequency in young and elderly subjects from Bali and Yogyakarta.
Figure 2.
Data on stool consistency in young and elderly subjects from Bali and Yogyakarta.
Gut microbiota composition of healthy Indonesians
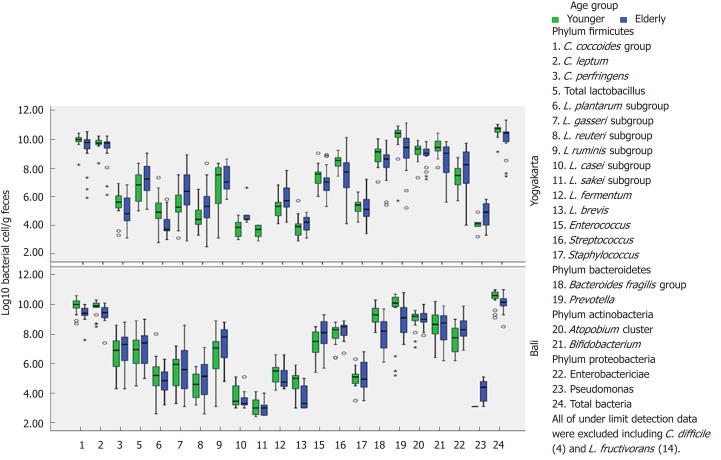
Before conducting gut microbiota composition analysis, subject characteristics consisting of gender, ethnicity, weight, height and body mass index (BMI) were obtained and are described in Table 1. Microbiota profiles (based on the YIF-SCAN) in young and elderly subjects living in Yogyakarta and Bali, Indonesia are presented in Table 2 and Figure 3. Populations of bacteria in the feces of young subjects were higher than in the elderly group. The total amount was approximately log10 10.0-10.6 bacterial cells/g feces, and the most abundant bacteria in all groups were Clostridium, followed by Prevotella, Atopobium, Bifidobacterium, and Bacteroides. These bacteria are obligate anaerobic bacteria. Interestingly, even though Prevotella showed the second highest population following Clostridium, its prevalence was not 100%. Furthermore, its prevalence in the elderly group from Yogyakarta was only 75%.
Table 2.
Microbiota profile comparison (based on Yakult intestinal flora-scan)
| No |
Log10 bacterial cells/g feces mean ± SD (detection rate %) |
||||
| Type of Bacteria |
Yogyakarta |
Bali |
|||
| Young | Elderly | Young | Elderly | ||
| Phylum Firmicutes | |||||
| 1 | Clostridium coccoides group | 9.9 ± 0.5 (100) | 9.3 ± 1.2a (100) | 9.9 ± 0.5 (100) | 9.4 ± 0.5c (100) |
| 2 | Clostridium leptum subgroup | 9.7 ± 0.4 (100) | 9.3 ± 1.1 (100) | 9.6 ± 0.5 (100) | 9.4 ± 0.6c (100) |
| 3 | Clostridium perfringens | 5.6 ± 0.9 (100) | 5.0 ± 1.1 (85) | 6.7 ± 1.2e (100) | 6.9 ± 1.4f (95) |
| 4 | Clostridium difficile | - (0) | - (0) | - (0) | - (0) |
| 5 | Total Lactobacillus | 6.7 ± 1.1 (95) | 7.2 ± 1.2 (95) | 6.8 ± 1.1 (100) | 7.1 ± 1.2 (100) |
| 6 | Lactobacillus plantarum subgroup | 5.0 ± 1.0 (100) | 4.1 ± 0.8a (85) | 5.1 ± 1.1 (100) | 4.8 ± 0.8f (100) |
| 7 | Lactobacillus gasseri subgroup | 5.4 ± 1.1 (100) | 6.4 ± 1.6a (90) | 5.5 ± 1.2 (100) | 5.6 ± 1.7 (90) |
| 8 | Lactobacillus reuteri subgroup | 4.6 ± 0.8 (95) | 5.3 ± 1.4a (90) | 4.5 ± 0.9 (90) | 4.9 ± 1.3 (100) |
| 9 | Lactobacillus ruminis subgroup | 6.8 ± 1.6 (45) | 7.6 ± 0.9 (35) | 6.4 ± 1.7 (80e) | 7.4 ± 1.3c (70f) |
| 10 | Lactobacillus casei subgroup | 3.7 ± 0.6 (40) | 4.9 ± 1.0a (25) | 3.8 ± 0.8 (40) | 3.6 ± 000.8f (35) |
| 11 | Lactobacillus sakei subgroup | 3.6 ± 0.5 (35) | 0b | 3.1 ± 0.6 (55) | 3.0 ± 0.6 (30f) |
| 12 | Lactobacillus fermentum | 5.3 ± 0.8 (75) | 5.9 ± 1.1 (80) | 5.3 ± 0.8 (55) | 5.1 ± 0.8f (60) |
| 13 | Lactobacillus brevis | 3.9 ± 0.8 (50) | 4.0 ± 0.9 (15a) | 4.7 ± 0.9 (35) | 3.7 ± 0.8 (40) |
| 14 | Lactobacillus fructivorans | - (0) | - (0) | - (0) | - (0) |
| 15 | Enterococcus | 7.4 ± 0.7 (90) | 7.1 ± 1.0 (90) | 7.4 ± 0.9 (95) | 7.9 ± 1.1f (95) |
| 16 | Streptococcus | 8.4 ± 0.5 (95) | 7.5 ± 1.6a (75) | 8.0 ± 0.7 (95) | 8.2 ± 0.7 (45d) |
| 17 | Staphylococcus | 5.3 ± 0.5 (100) | 5.1 ± 1.1 (90) | 5.0 ± 0.6 (100) | 5.2 ± 1.0 (100) |
| Phylum Bacteroidetes | |||||
| 18 | B. fragilis group | 8.9 ± 0.7 (100) | 8.3 ± 1.1 (100) | 9.3 ± 0.6 (100) | 7.9 ± 1.1c (100) |
| 19 | Prevotella | 10.0 ± 1.2 (85) | 9.1 ± 1.6a (75) | 9.4 ± 1.9 (80) | 9.0 ± 1.1c (85) |
| Phylum Actinobacteria | |||||
| 20 | Atopobium cluster | 9.1 ± 0.6 (100) | 8.9 ± 0.7 (100) | 8.9 ± 0.7 (100) | 9.0 ± 0.5 (100) |
| 21 | Bifidobacterium | 9.4 ± 0.6 (100) | 8.4 ± 1.3a (100) | 8.6 ± 0.9f (100) | 8.4 ± 1.2 (100) |
| Phylum Proteobacteria | |||||
| 22 | Enterobacteriaceae | 7.3 ± 0.8 (100) | 7.8 ± 1.6 (100) | 7.6 ± 0.9 (100) | 8.4 ± 0.8c (100) |
| 23 | Pseudomonas | 4.1 ± 0.6 (25) | 4.7 ± 1.0 (30) | 3.1 ± 0.0 (10) | 4.2 ± 0.8 (55d) |
| 24 | Total Bacteria | 10.6 ± 0.4 (100) | 10.0 ± 1.0a(100) | 10.5 ± 0.5 (100) | 10.1 ± 0.5c (100) |
P < 0.05 Yogyakarta Young vs Yogyakarta Elderly.
P < 0.01 Yogyakarta Young vs Yogyakarta Elderly.
P < 0.05 Bali Young vs Bali Elderly.
P < 0.01 Bali Young vs Bali Elderly.
P < 0.05 Yogyakarta Young vs Bali Young.
P < 0.05 Yogyakarta Elderly vs Bali Elderly.
Mann-Whitney U-test for bacterial cells/g feces, chi-square test for detection rate.
Figure 3.
Comparison of gut microbiota composition in young and elderly subjects from Bali and Yogyakarta.
Among the analyzed Clostridia, the most dominant was the Clostridium coccoides group, followed by Clostridium leptum subgroup and Clostridium perfringens, whereas the pathogenic Clostridium difficile was not found in any group. Enterobacteriaceae (phylum Proteobacteria), which is a facultative anaerobe had a population of log10 7.3 ± 0.8 to 8.4 ± 0.8 bacterial cells/g feces, less than the other obligate anaerobic phylum.
In terms of the bacterial counts in Yogyakarta subjects, total bacteria, Clostridium coccoides group, Bifidobacterium, Prevotella, Lactobacillus plantarum subgroup, and Streptococcus were significantly higher (P < 0.05) in younger than in elderly subjects, while the Lactobacillus gasseri subgroup, Lactobacillus casei subgroup, and Lactobacillus reuteri subgroup counts were significantly lower (P < 0.05) in younger subjects. On the other hand, in Balinese subjects, the younger groups had significantly higher (P < 0.05) total bacteria count, Prevotella subgroup, Clostridium leptum subgroup, Bacteroides fragilis group and Clostridium coccoides group. The counts for Lactobacillus ruminis subgroup and Enterobacteriaceae were significantly lower (P < 0.05) in younger subjects.
A comparison within young subjects showed that the Bifidobacterium count in Yogyakarta was significantly higher (P < 0.05) than that in Bali, while the Clostridium perfringens count was significantly lower (P < 0.05) in Yogyakarta. On the other hand, a comparison of elderly subjects from both sites showed significantly low (P < 0.05) counts for Clostridium perfringens, Lactobacillus plantarum subgroup and Enterococcus in Yogyakarta, while the counts for Lactobacillus casei subgroup and Lactobacillus fermentum were significantly higher (P < 0.05) in Yogyakarta. Counts for Lactobacillus plantarum subgroup were significantly higher (P < 0.05) in young subjects, while the counts for Lactobacillus reuteri subgroup and Enterobacteriaceae were significantly lower (P < 0.05) in young subjects.
In a comparison of young and elderly, younger subjects tend to have higher concentrations of microbiota based on the YIF-SCAN, compared to the elderly as shown in Table 3 and Figure 3, and a significant difference between younger and elderly subjects was found. Younger subjects had significantly higher (P < 0.05) total bacteria, Clostridium coccoides and Bacteroides fragilis groups, and Clostridium leptum, Bifidobacterium, Prevotella, and Lactobacillus plantarum subgroups, while Lactobacillus reuteri subgroup and Enterobacteriaceae were significantly lower (P < 0.05) in younger subjects. No significant difference was found in the distribution of total Lactobacillus. Additionally, Atopobium cluster showed a constant number.
Table 3.
Microbiota profile (culture method) in young and elderly subjects
| Microbiota (Log10 CFU/g feces) |
mean ± SD (detection rate %) |
P value | |
| Younger (n = 40) | Elderly (n = 40) | ||
| Coliform | 6.80 ± 0.83 (100) | 7.20 ± 0.73 (100) | 0.028 |
| Escherichia coli | 6.87 ± 0.74 (100) | 7.29 ± 0.78 (100) | 0.016 |
| Yeast | 4.17 ± 0.51 (53) | 4.28 ± 0.50 (43) | 0.411 |
| Mold | 0,0 ± 0,0 (0) | 3.68 ±0.27 (8) | - |
| Lactobacillus plantarum | 4.16 ± 0.85 (8) | 4.16 ± 0.03 (5) | 1.000 |
| Total LAB | 7.03 ± 0.83 (100) | 7.67 ± 1.23 (100) | 0.008 |
Comparison of continuous variable was carried out using the Independent T test for normally distributed data and the Mann Whitney test for non-normally distributed data.
In terms of the bacterial counts in Yogyakarta subjects, total bacteria, Clostridium coccoides group, Bifidobacterium, Prevotella, Lactobacillus plantarum subgroup, and Streptococcus were significantly higher (P < 0.05) in young subjects compared to the elderly, while the counts for Lactobacillus gasseri, Lactobacillus casei, and Lactobacillus reuteri subgroups were significantly lower (P < 0.05) in young subjects. Bacterial counts in Balinese younger subjects were significantly higher (P < 0.05) including total bacteria, Clostridium coccoides, Clostridium leptum, Bacteroides fragilis, and Prevotella compared to elderly subjects, while the counts for Lactobacillus ruminis subgroup and Enterobacteriaceae were significantly lower (P < 0.05) in younger subjects.
Differences between Prevotella, Bifidobacterium, and Bacteroides and Enterobacteriaceae
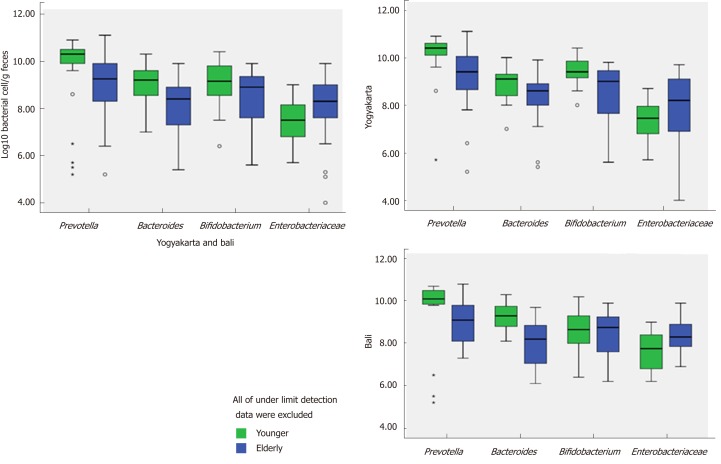
Populations of Prevotella, Bifidobacterium and Bacteroides fragilis group in young and elderly subjects in Yogyakarta and Bali are shown in Figure 4. From the figure, it can be seen that Prevotella was the most abundant bacteria in each group, and Prevotella enterotype was found in each group. It should also be noted that the populations of Prevotella, Bifidobacterium and Bacteroides fragilis group were fewer in elderly subjects. The bacteria which provide health benefits, namely Bifidobacterium were not widespread in elderly subjects. On the other hand, Enterobacteriaceae which has a disadvantageous effect on health was more common in the elderly.
Figure 4.
Populations of Prevotella, Bifidobacterium, Bacteroides and Enterobacteriaceae in young and elderly subjects from Yogyakarta and Bali.
The Lactobacillus group and others
The Lactobacillus subgroups which were dominant and had high prevalence (> 85%) were Lactobacillus plantarum, Lactobacillus gasseri, and Lactobacillus reuteri, while Lactobacillus ruminis although it showed a high population (6.4±1.7 – 7.6±0.9 log10 bacterial cells/g), its prevalence in Balinese subjects was only 70%-80% and as few as 35%-45% in Yogyakarta subjects. With regard to Lactobacillus casei subgroup, the number and prevalence were low.
From Table 2 and Table 3, it can be seen that in the elderly, there was an increase in Enterobacteriaceae (qPCR) together with Coliform and E. coli. Similar results were found for yeasts and molds, notably molds. Initially, molds were not found in younger subjects but were later found in the elderly. This may indicate that the elderly is more prone to mold infection. Also, it should be noted that Coliform and E. coli, which belong to Enterobacteriaceae were analyzed using culture methods, had a lower count in younger subjects compared to the elderly, and this was in accordance with the YIF-SCAN results. Using the culture method, the amount of yeast in feces was observed to be 4.17 log10 bacterial cells/g (younger subjects) and 4.28 log10 bacterial cells/g (elderly subjects), respectively, with a prevalence of 53% and 43%, respectively. Mold was not found in the feces of younger subjects, although a prevalence of 8% was found in the elderly with the highest number accounting for 3.68 log10 mold cells/g.
A comparison of Lactobacillus analysis between qPCR and culture method was conducted and the results are presented in Table 4. It was shown that no significant difference between the results of the two methods was detected.
Table 4.
Comparison between qPCR and culture method
| Lactobacillus (Log10 CFU/g feces) |
mean ± SD (detection rate %) |
|||
| Younger (n = 40) | Elderly (n = 40) | |||
| Bali | Yogyakarta | Bali | Yogyakarta | |
| Culture method | 7.04 ± 0.87 (100) | 7.01 ± 0.81 (100) | 8.17 ± 0.99b (100) | 7.17 ± 1.33a (100) |
| Yakult intestinal flora-scan | 6.7 ± 1.1 (95) | 7.2 ± 1.2 (95) | 6.8 ± 1.1 (100) | 7.1 ± 1.2 (100) |
P < 0.05 Yogyakarta Elderly vs Bali Elderly;
P < 0.01 Bali Young vs Bali Elderly.
DISCUSSION
Several factors are reported to have important roles that affect the normal gut microbiota[3,11]. These factors are: delivery type (vaginal or cesarean)[1]; infancy (breast milk or formula feeds) and adult (vegetable based or meat based) diet[2]; and antibiotics or antibiotic-like molecules derived from gut environment intake[3]. Of these, diet is the main factor affecting the change in gut microbiota following aging.
Changes in gut microbiota start when babies are weaned and consume solid foods, and these changes become stable in teenage and adult years[12-14]. Changes occur again in the elderly. Intestinal movements are weaker with age, extending transit time and changes in nutrient turn-over dynamics. The elderly possibly have reduced dentition and chewing strength affecting their diet that causes low support of microbial growth. Therefore, when compared, the elderly group was different from the younger group as the elderly have different physical aspects, such as reduced bowel function. This is in accordance with the results of the present study in subjects from Yogyakarta and Bali. The results showed that differences in gut microbiota were only seen between age groups, as some bacteria such as Bifidobacteria, Prevotella and Lactobacillus plantarum were reduced, while Enterobacteriaceae and Lactobacillus reuteri increased in elderly subjects. Both locations did not show a difference in gut microbiota composition, as rice is consumed as a staple carbohydrate source in both locations. Even though there was a slight difference in the main protein source, this did not result in a significant difference in the gut microbiota.
This difference in gut microbiota concentration may have been influenced by the difference in eating habits between younger and elderly subjects[15]. In terms of eating habit, most elderly subjects eat regularly (3 times a day) compared to younger subjects. The difference in daily eating frequency is significant. Younger people tend to consume probiotic and fruit or vegetable juices more than the elderly. However, milk, vegetables and fruits are more frequently consumed among younger subjects compared to the elderly. This might explain why there was a higher concentration of Bacteroides in the gut microbiota of younger subjects[15]. Our study had a limitation in study population heterogeneity; therefore, our subjects may not be representative of the Indonesian population. However, the subjects included in this study may provide a background description of the gut microbiota profile in young compared to elderly Indonesians.
Based on 16S rRNA gene sequencing, Firmicutes is the most abundant bacteria living in the human gastrointestinal tract, followed by Bacteroidetes, Actinobacteria and Proteobacteria[16-19]. The results of the present study showed that the gut microbiota is dominated by Firmicutes, Bacteroidetes, and Actinobacteria (which are obligate anaerobic bacteria) and Proteobacteria (which is a facultative anaerobic bacterium). This was in line with the results of previous research. Based on the bacterial composition, it should be noted that each group had a Prevotella enterotype. From the recent gut microbiota profiling of children in Yogyakarta and Bali, it was stated that Prevotella was the most dominant bacteria[6]. In addition, Papua New Guineans, which is the neighboring country to Indonesia, showed a gut composition dominated by Prevotella over Bacteroides[20]. Therefore, it can be concluded that Indonesians, from children to the elderly had a Prevotella enterotype.
Bifidobacterium-Enterobacteriaceae. The low abundance of Proteobacteria in younger subjects was due to the strict anaerobic environment of the colon. The facultative species may represent 0.1% in this strict anaerobic environment[21]. Interestingly, during aging, the population of Enterobacteriaceae from phyla Proteobacteria increased, followed by a reduction in protective bacteria Bifidobacterium. This also agrees with the results of prior research, notably by[22] where even though there was a difference in population between Bacteroides, Prevotella and Bifidobacterium in each country and age group, in common was that higher proportions of Enterobacteriaceae were found in all elderly volunteers independent of the location.
Probiotic supplementation contributes to gut microbiota composition alterations from adults to the elderly[13]. As humans age, factors related to their diet are highly linked with changes in the gut microbiome. However, a connection between these factors has not been proven. An increase in medication intake is also prevalent in the elderly and can support reduced gut microbiota composition. On the other hand, the use of anti-diabetic drugs has been shown to have a positive impact on gut microbiota, which was shown by increasing populations of beneficial microbiota and their metabolites[23,24]. Several studies have reported that the gut microbiota has an important impact not only in colorectal cancer development, but also in the treatment of colorectal cancer. It was reported that the efficacy of anticancer agents is regulated by gut microbiota and improves the killing effect of 5-fluorouracil, an anticancer drug[25]. Nevertheless, chemotherapy treatments can disrupt the equilibrium of gut microbiota and cause damage to gut microbiota[26,27]. In specific diseases such as heartburn and other symptoms caused by gastroesophageal reflux disease and functional dyspepsia, proton pump inhibitors (PPIs) are used worldwide. However, it was shown that long-term use of PPIs lowered Faecalibacteria and increased Streptococci thought to be associated with enteric infection[28]. Alterations in gut microbiota following the use of pharmaceutical agents, which is associated with disease, possibly recovers after the administration of pre-and probiotics. The manipulation of gut microbiota to improve health status using foods composed of prebiotic oligosaccharide components is being investigated. A previous study found that ingestion of fermented milk containing probiotics was able to enhance stool quality and defecation frequency[29].
Commensal Clostridia-known to be the microbial with the highest population found in the gut, was represented by Clostridium cocoides and Clostridium leptum subgroup in the present study (Table 2). The number of Clostridia decreased in elderly subjects in both locations. The Gram-positive bacteria, Clostridia, start to colonize in the first month in breastfed infants, and populate a certain area of the intestinal mucosa within intestinal cells[30]. Therefore, Clostridia can play an important role in affecting the physiologic, metabolic and immune processes. On the other hand, Bifidobacterium is believed to be one of the first microbes to colonize the human gastrointestinal tract and provide beneficial health effects as they have the ability to produce butyric acid[31,32]. The Atopobium cluster belongs to phylum Actinobacteria alongside Bifidobacterium. The population of this bacterium in adults was the same as that of Bifidobacterium, approximately 109, and unlike Bifidobacterium which decreased in the elderly, Atopobium counts were constant[16].
Lactobacillus plantarum was found to have the highest prevalence among the higher-level species of Lactobacillus within the gut. This bacterium was related to high BMI and blood glucose. However, other groups and species of Lactobacillus have been found at various glucose levels in adults and the elderly[2]. Additionally, Lactobacillus plantarum is abundant in fermented Indonesian traditional food[33], and has been found to have probiotic potential[34,35].
A culture method was used in this research to determine some microorganisms. However, another culture-independent method based on cloned 16S rRNA sequence was introduced[21]. Moreover, with profiling culture enriched molecules, most human gut microbiota, especially bacteria, are able to be cultured[36]. In addition, using selective media will enable bacterial growth and detection that may be missed. The culture method which relied on various specific media was compared with molecular-based analysis in this research and a significant difference was not found between the two methods. With the culture method, elderly subjects tended to have higher concentrations of Coliform, E. coli and total lactic acid bacteria. Thus, our findings were not similar to those in previous studies in an Asian population[37,38]. However, these data were obtained from an East Asian population (Japanese) and the study was conducted in the early 1970s. Our study population, both young and elderly subjects showed a different pattern where these bacteria were found in relatively higher concentrations (Table 3). The different distribution of gut microbiota in our study showed that the gut microbiota may change over time, and the gut microbiota of Indonesians was unique.
Probiotics and prebiotics have the potential to maintain gut microbiota balance. Analysis of the gut microbiota was carried out to observe its potential to modulate and improve health, particularly in the elderly when protective bacteria (Bifidobacteria) declined and potential pathogenic Enterobacteriaceae increased. Numerous conditions could trigger the shift in elderly gut microbiota, such as weakened chewing ability, a decline in intestinal physiological condition to digest, lowered appetite, and limited ability to prepare food. Consumption of probiotics and prebiotics could be used as a way to retain the balance of microbiota in the elderly, in order to maintain health.
ARTICLE HIGHLIGHTS
Research background
Studies on gut microbiota are growing rapidly, as the gut microbiota has been associated with various diseases. There are numerous factors influencing gut microbiota composition; hence, further research is needed. Indonesia has a high diversity of tribes and lifestyles. Therefore, it is expected that each tribe has its specific gut microbiota. Thus, a deeper insight into the composition of gut microbiota can be further used to determine the condition of gut microbiota in Indonesians and assess which treatment may be suitable and effective to improve health status.
Research motivation
This research aimed to investigate gut microbiota composition in two different groups in relation to age and tribe. The results obtained may be further used to determine the condition of gut microbiota in Indonesians and to assess which treatment may be suitable and effective to improve health status.
Research objectives
The objective of this study was to compare gut microbiota composition in Indonesian subjects represented by two different age groups (elderly and young) from the Javanese and Balinese tribes, by the analysis of fecal samples. The results obtained may be further used to determine the condition of gut microbiota in Indonesians and to assess which treatment may be suitable and effective to improve health status.
Research methods
Fecal samples were collected from a total of 80 subjects with 20 in each young group ranging from 25-45 years and elderly group ranging from 70 years or more from two different regions, Yogyakarta and Bali. The collection of fecal samples was performed at the end of the assessment period (day 14 ± 1 d) and during this period, the subjects were not allowed to consume probiotic and antibiotic products. The quantification of various Clostridium subgroups, Lactobacillus subgroups, Enterococcus, Streptococcus, Staphylococcus; Bacteroides fragilis group and Prevotella, Bifidobacterium and Atopobium cluster; Enterobacteriaceae and Pseudomonas was performed using the Yakult intestinal flora-scan (YIF-SCAN).
Research results
Bacteria populations in younger subjects’ feces were higher than in elderly subjects. The most abundant bacteria in both age groups were Clostridium, followed by Prevotella, Atopobium, Bifidobacterium and Bacteroides. In the elderly, an increase in Enterobacteriaceae, Coliform and E. coli was found. In terms of bacterial counts in Yogyakarta, total bacteria, Clostridium coccoides group, Bifidobacterium, Prevotella, Lactobacillus plantarum subgroup, and Streptococcus were significantly higher (P < 0.05) in younger than in elderly subjects, while the counts for Lactobacillus gasseri subgroup, Lactobacillus casei subgroup, and Lactobacillus reuteri subgroup were significantly lower (P < 0.05) in the younger group. In Balinese subjects, total bacteria, Clostridium coccoides group, Clostridium leptum subgroup, Bacteroides fragilis group, and Prevotella were significantly higher (P < 0.05) in younger compared to elderly subjects, while the counts for Lactobacillus ruminis subgroup and Enterobacteriaceae were significantly lower (P < 0.05) in younger subjects. The results also revealed that the Clostridium perfringens count in Balinese subjects was 10 times that in Yogyakarta subjects. This may have been a response to the lifestyles of the different tribes although this phenomenon requires further extensive study.
Research conclusions
Bacterial populations were higher in younger than in elderly subjects. The most dominant bacterial groups in these subjects were Clostridium followed by Prevotella, Atopobium, Bifidobacterium, and Bacteroides. The gut microbiota of the two tribes in this study were similar, but it was observed that Clostridium perfringens in Yogyakarta subjects was almost ten times lower than that in Balinese due to lifestyle and diet. However, the effect of lifestyle and diet on gut microbiota composition requires further investigation.
Research perspectives
These results proved that gut microbiota composition within different age groups and tribes is different. Different tribes may have different lifestyles and diets; therefore, future studies should investigate how different diets contribute to gut microbiota composition.
ACKNOWLEDGEMENTS
We would like to expression our gratitude to Shuta Yamamoto, Yukiko Kado, Akira Takahashi, Takashi Asahara and Takuya Takahashi for their participation in stool DNA extraction and intestinal microbiota analysis. We would also like to thank Akira Shigehisa for assisting with the protocol development. Indonesian Prodia Lab Services, specifically the CRO Division, is acknowledged for managing and field-organizing the research.
Footnotes
Institutional review board statement: All subjects agreed to participate in this study after informed consent and ethical permission was obtained.
Informed consent statement: All subjects agreed to participate in this study after informed consent and ethical permission was obtained.
Institutional animal care and use committee statement: No animals were included in this study.
ARRIVE guidelines statement: The authors have read the ARRIVE guidelines, and the manuscript was prepared and revised according to the ARRIVE guidelines.
Peer-review started: November 22, 2018
First decision: December 12, 2018
Article in press: January 26, 2019
Specialty type: Gastroenterology and hepatology
Country of origin: Indonesia
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Takagi T, Talebi Bezmin Abadi A, Shi JL S-Editor: Yan JP L-Editor: Webster JR E-Editor: Ma YJ
Contributor Information
Endang Sutriswati Rahayu, Department of Food and Agricultural Technology, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia. endangsrahayu@ugm.ac.id.
Tyas Utami, Department of Food and Agricultural Technology, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia.
Mariyatun Mariyatun, Department of Food and Agricultural Technology, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia.
Pratama Nur Hasan, Department of Food and Agricultural Technology, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia.
Rafli Zulfa Kamil, Department of Food and Agricultural Technology, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia.
Ryan Haryo Setyawan, Department of Food and Agricultural Technology, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia.
Fathyah Hanum Pamungkaningtyas, Department of Food and Agricultural Technology, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia.
Iskandar Azmy Harahap, Department of Food and Agricultural Technology, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia.
Devin Varian Wiryohanjoyo, Department of Food and Agricultural Technology, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia.
Putrika Citta Pramesi, Department of Food and Agricultural Technology, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia.
Muhammad Nur Cahyanto, Department of Food and Agricultural Technology, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia.
I Nengah Sujaya, Department of Public Health, Udayana University, Denpasar 80234, Indonesia.
Mohammad Juffrie, Department of Public Health, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia.
References
- 1.Hopkins MJ, Sharp R, Macfarlane GT. Variation in human intestinal microbiota with age. Dig Liver Dis. 2002;34 Suppl 2:S12–S18. doi: 10.1016/s1590-8658(02)80157-8. [DOI] [PubMed] [Google Scholar]
- 2.Štšepetova J, Sepp E, Kolk H, Lõivukene K, Songisepp E, Mikelsaar M. Diversity and metabolic impact of intestinal Lactobacillus species in healthy adults and the elderly. Br J Nutr. 2011;105:1235–1244. doi: 10.1017/S0007114510004770. [DOI] [PubMed] [Google Scholar]
- 3.Toole PWO, Claesson MJ. Gut microbiota : Changes throughout the lifespan from infancy to elderly. Int Dairy J. 2010;20:281–291. [Google Scholar]
- 4.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J MetaHIT Consortium, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakayama J, Watanabe K, Jiang J, Matsuda K, Chao SH, Haryono P, La-Ongkham O, Sarwoko MA, Sujaya IN, Zhao L, Chen KT, Chen YP, Chiu HH, Hidaka T, Huang NX, Kiyohara C, Kurakawa T, Sakamoto N, Sonomoto K, Tashiro K, Tsuji H, Chen MJ, Leelavatcharamas V, Liao CC, Nitisinprasert S, Rahayu ES, Ren FZ, Tsai YC, Lee YK. Diversity in gut bacterial community of school-age children in Asia. Sci Rep. 2015;5:8397. doi: 10.1038/srep08397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuda K, Tsuji H, Asahara T, Matsumoto K, Takada T, Nomoto K. Establishment of an analytical system for the human fecal microbiota, based on reverse transcription-quantitative PCR targeting of multicopy rRNA molecules. Appl Environ Microbiol. 2009;75:1961–1969. doi: 10.1128/AEM.01843-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuda K, Tsuji H, Asahara T, Takahashi T, Kubota H, Nagata S, Yamashiro Y, Nomoto K. Sensitive quantification of Clostridium difficile cells by reverse transcription-quantitative PCR targeting rRNA molecules. Appl Environ Microbiol. 2012;78:5111–5118. doi: 10.1128/AEM.07990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubota H, Tsuji H, Matsuda K, Kurakawa T, Asahara T, Nomoto K. Detection of human intestinal catalase-negative, Gram-positive cocci by rRNA-targeted reverse transcription-PCR. Appl Environ Microbiol. 2010;76:5440–5451. doi: 10.1128/AEM.03132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heaton K, Lewis S. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol. 2007;32:920–924. doi: 10.3109/00365529709011203. [DOI] [PubMed] [Google Scholar]
- 11.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, Abe F, Osawa R. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voreades N, Kozil A, Weir TL. Diet and the development of the human intestinal microbiome. Front Microbiol. 2014;5:494. doi: 10.3389/fmicb.2014.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conlon MA, Bird AR. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2014;7:17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar M, Babaei P, Ji B, Nielsen J. Human gut microbiota and healthy aging: Recent developments and future prospective. Nutr Healthy Aging. 2016;4:3–16. doi: 10.3233/NHA-150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorasin T, Hoyles L, McCartney AL. Dynamics and diversity of the 'Atopobium cluster' in the human faecal microbiota, and phenotypic characterization of 'Atopobium cluster' isolates. Microbiology. 2015;161:565–579. doi: 10.1099/mic.0.000016. [DOI] [PubMed] [Google Scholar]
- 17.Rajilić-Stojanović M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9:2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- 18.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 19.Vrieze A, Holleman F, Zoetendal EG, de Vos WM, Hoekstra JB, Nieuwdorp M. The environment within: how gut microbiota may influence metabolism and body composition. Diabetologia. 2010;53:606–613. doi: 10.1007/s00125-010-1662-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenhill AR, Tsuji H, Ogata K, Natsuhara K, Morita A, Soli K, Larkins JA, Tadokoro K, Odani S, Baba J, Naito Y, Tomitsuka E, Nomoto K, Siba PM, Horwood PF, Umezaki M. Characterization of the gut microbiota of Papua New Guineans using reverse transcription quantitative PCR. PLoS One. 2015;10:e0117427. doi: 10.1371/journal.pone.0117427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, Cresci A, Silvi S, Orpianesi C, Verdenelli MC, Clavel T, Koebnick C, Zunft HJ, Doré J, Blaut M. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72:1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montandon SA, Jornayvaz FR. Effects of Antidiabetic Drugs on Gut Microbiota Composition. Genes (Basel) 2017;8 doi: 10.3390/genes8100250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu Y, Wang X, Li J, Zhang Y, Zhong H, Liu R, Zhang D, Feng Q, Xie X, Hong J, Ren H, Liu W, Ma J, Su Q, Zhang H, Yang J, Wang X, Zhao X, Gu W, Bi Y, Peng Y, Xu X, Xia H, Li F, Xu X, Yang H, Xu G, Madsen L, Kristiansen K, Ning G, Wang W. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat Commun. 2017;8:1785. doi: 10.1038/s41467-017-01682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin C, Cai X, Zhang J, Wang W, Sheng Q, Hua H, Zhou X. Role of Gut Microbiota in the Development and Treatment of Colorectal Cancer. Digestion. 2018:1–7. doi: 10.1159/000494052. [DOI] [PubMed] [Google Scholar]
- 26.Mateeva E, Smilov I. Acid-base balance and blood gas composition in the mother, fetus and newborn infant following the use of central electroanalgesia during natural labor. Akush Ginekol (Sofiia) 1988;27:38–43. [PubMed] [Google Scholar]
- 27.van Vliet MJ, Tissing WJ, Dun CA, Meessen NE, Kamps WA, de Bont ES, Harmsen HJ. Chemotherapy treatment in pediatric patients with acute myeloid leukemia receiving antimicrobial prophylaxis leads to a relative increase of colonization with potentially pathogenic bacteria in the gut. Clin Infect Dis. 2009;49:262–270. doi: 10.1086/599346. [DOI] [PubMed] [Google Scholar]
- 28.Takagi T, Naito Y, Inoue R, Kashiwagi S, Uchiyama K, Mizushima K, Tsuchiya S, Okayama T, Dohi O, Yoshida N, Kamada K, Ishikawa T, Handa O, Konishi H, Okuda K, Tsujimoto Y, Ohnogi H, Itoh Y. The influence of long-term use of proton pump inhibitors on the gut microbiota: an age-sex-matched case-control study. J Clin Biochem Nutr. 2018;62:100–105. doi: 10.3164/jcbn.17-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utami T, Cahyanto MN, Juffrie M, Rahayu ES. Recovery of Lactobacillus Casei Strain Shirota (Lcs) From the Intestine of Healthy Indonesian Volunteers After Intake of Fermented Milk and Its Impact on the Enterobacteriaceae Faecal Microbiota. Int J Probiotics Prebiotics. 2015;10:77–84. [Google Scholar]
- 30.Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. doi: 10.1186/1757-4749-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Callaghan A, van Sinderen D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahayu ES. Lactic Acid Bacteria in Fermented Food of Indonesian Origin. Agritech. 2003;23:75–84. [Google Scholar]
- 34.Rahayu ES, Cahyanto MN, Windiarti L, Sutriyanto J, Kandarina T, Utami T. Effects of consumption of fermented milk containing indigenous probiotic lactobacillus plantarum dad-13 on the fecal microbiota of healthy Indonesian volunteers. Int J Probiotics Prebiotics. 2016;11:91–98. [Google Scholar]
- 35.Rahayu ES, Yogeswara A, Mariyatun, Windiarti L, Utami T, Watanabe K. Molecular Characteristic of Indigenous Probiotic Strain From Indonesia. Int J Probiotics Prebiotics. 2015;10:1–7. [Google Scholar]
- 36.Lau JT, Whelan FJ, Herath I, Lee CH, Collins SM, Bercik P, Surette MG. Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med. 2016;8:72. doi: 10.1186/s13073-016-0327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finegold SM, Attebery HR, Sutter VL. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am J Clin Nutr. 1974;27:1456–1469. doi: 10.1093/ajcn/27.12.1456. [DOI] [PubMed] [Google Scholar]
- 38.Mitsuoka T, Hayakawa K. The fecal flora in man. I. Composition of the fecal flora of various age groups. Zentralbl Bakteriol Orig A. 1973;223:333–342. [PubMed] [Google Scholar]