Abstract
CYP2D6 metabolizes ~25% of all clinically used drugs, with numerous genetic polymorphisms affecting enzyme activity and drug response. Clinical utility of current CYP2D6 genotyping is partially compromised the unresolved complex haplotype structure of the CYP2D6 locus. We have identified a distal enhancer SNP rs5758550 that robustly increases CYP2D6 expression whereas rs16947 (CYP2D6*2), previously considered inert, reduces correct mRNA splicing and expression, thereby affecting presumed activity of other alleles on the *2 haplotype.
Objectives:
This study aims to determine the structure and frequency of haplotypes containing either rs5758550 or rs16947, or both, together with other relevant CYP2D6 alleles, assigning predictive enzyme activity scores to each, and addressing ambiguities in estimating diplotypes in different populations.
Methods:
The structure and frequency of haplotypes containing rs5758550 and/or rs16946 in different populations were determined by using phased genotype data from ‘The 1000 Genomes Project’. The assigned haplotype-phenotype relationship was tested by associating assigned CYP2D6 activity score with CYP2D6 enzyme activity in a cohort of 122 human liver microsomes.
Results:
Addition of enhancer SNP rs5758550 and *2 to a CYP2D6 panel improves prediction of CYP2D6 activity. Moreover, the haplotype containing rs5758550 and rs16947 predict extensive CYP2D6 activity more accurately than CYP2D6*2A, a surrogate marker for extensive activity.
Conclusion:
With further studies, the results support possible incorporation of rs5758550 and rs16947 into CYP2D6 biomarker panels for more accurate prediction of CYP2D6 metabolizer status.
Keywords: CYP2D6, metabolizer status, rs5758550, rs16946, CYP2D6*2, biomarker
Introduction
Cytochrome P450 2D6 (CYP2D6) metabolizes ~25% of currently used medications [1]. Large inter-person variability in CYP2D6 enzyme activity, caused by genetic polymorphisms, influences drug efficacy, drug dosage, and adverse drug effects [2]. Numerous variants in the transcribed CYP2D6 region have been identified and some result in non-functional or reduced function of CYP2D6 enzyme (PharmVar: https://www.pharmvar.org/genes). In vivo phenotyping studies led to classification of subjects into poor (PM), intermediate (IM), normal (NM), and ultrarapid (UM) CYP2D6 metabolizers [3,4]. Listed in the US Food and Drug Administration’s Table of Pharmacogenomics Biomarkers in Drug Labels (https://www.fda.gov/Drugs/ScienceResearch/ucm572698.htm), CYP2D6 variants are currently used as biomarker panels for predicting CYP2D6 metabolizer status. The Clinical Pharmacogenetics Implementation Consortium (CPIC) has published 13 guidelines for CYP2D6/codeine, CYP2D6/antidepressants and CYP2D6/tamoxifen gene/drug pairs to facilitate clinical implementation of CYP2D6 biomarker testing [5–9](see URL link www.CPICpgx.org/guidelines)). The CPIC guideline lists 11 alleles as having normal (wild-type) activity, 12–14 with reduced activity, and 35–38 abolishing activity [6,8]. The most common *2 allele (rs16947, MAF ~40%) is considered to convey normal activity, while *41 and *29 are considered to have reduced activity, present on the same haplotype as *2. Among all variants, *2, *9, *10, *17, *29, *41, *3, *4, *5, *6 are commonly reported alleles, while others are rare with minor allele frequencies less than 1%, or only presence in certain populations.
Recently, we identified a enhancer SNP rs5758550, which resides within a critical enhancer region directing CYP2D6 expression, located 115kb downstream of the CYP2D6 promoter [10,11]. In addition, we demonstrated that the signature variant of the *2 allele, rs16947, previously thought to convey normal enzyme activity, in fact reduces CYP2D6 mRNA expression 2-fold by affecting exon 6 splicing [11]. The new downstream enhancer SNP rs5758550 is in high linkage disequilibrium (LD) with *2 rs16947, but with lower minor allele frequency (~20% versus ~40%) compared to rs16947. Therefore, the combination of *2 and rs5758550 affect overall CYP2D6 mRNA expression. In a pediatric cohort, rs16947 alone (on a haplotype lacking rs5758550, ~20% MAF) is associated with reduced CYP2D6 enzyme activity, whereas rs5758550 alone (on a haplotype lacking rs16947, ~2% MAF) resulted in increased CYP2D6 activity [11]. Haplotypes containing both rs16947 and rs5758550 minor alleles were similar to the wild-type [11].
Interactions between the gain-of-function enhancer rs5758550 and the reduced function *2 allele resolve conflicting results associated with CYP2D6*2 [12–14]. For example, the SNP defining *41, rs28371725, an intronic SNP currently used as a biomarker predicting reduced CYP2D6 enzyme activity, does not substantially affect CYP2D6 expression by itself [11]. Rather *41 marks a *2 haplotype containing rs16947 lacking the enhancer SNP rs5758550 in Caucasian; however, this haplotype structure is weakened in other ethnic groups[11], so that*41 fails to accurately predict reduced CYP2D6 activity in African Americans or Mexicans[15,16].
Current biomarker panels use the surrogate markers CYP2D6*41 and CYP2D6*2A. Moreover, co-existence of other non-functional or reduced function variants [17] or structural variants [18] also affect genotype interpretation, as do CYP2D6 gene duplications carrying rs16947 [19] but lacking a separate downstream enhancer region – a gene duplication involving the *2 allele without duplicating the downstream enhancer region could have limited effect. Here we assess CYP2D6 haplotype structure and utility of biomarker panels in predicting CYP2D6 metabolizer status, taking advantage of phased haplotype structures of CYP2D6 from the 1,000 genome sequencing project (phase 3). The goal is to enhance the predictive value of CYP2D6 biomarker panels.
Methods:
CYP2D6 haplotype structure:
Genotype data were obtained from the ‘1000 genomes project phase 3’, keeping the phasing information in 1092 individuals (2184 alleles) (Supplemental Table 1, Supplemental Digital Content 1, http://links.lww.com/FPC/B333). Eleven commonly reported CYP2D6 variants (referred to as common variants in this study) were selected (Supplemental Table 2, Supplemental Digital Content 1, http://links.lww.com/FPC/B333). Haplotypes containing enhancer SNP rs5758550 and/or *2 SNP rs16947, together with other functional SNPs, were manually assigned for each individual. Minor allele frequency and linkage disequilibrium (LD) plots were generated using Haploview. Activity scores were assigned on the basis of CPIC guidelines[5], and with proposed modifications for rs5758550 and rs16947 containing haplotypes [10,11] and estimates for gene duplications (not contained in the 1000 Genome Browser) that need further validation.
Human Liver Microsomes (HLMs):
Individual Human Liver Microsomes (HLMs) and tissue lysates were purchased from XenoTech LLC (https://www.xenotech.com). Genomic DNA was prepared from tissue lysates and genotyped for CYP2D6 common variants using GeneSight assay platform. Demographical information of the individual HLMs is shown in Supplemental Table 3, Supplemental Digital Content 1, http://links.lww.com/FPC/B333.
CYP2D6 enzyme activity:
HLMs (0.05 mg protein/mL protein) were incubated at 37°C for 10 minutes with 80 μM dextromethorphan (Sigma D9684) and 1 mM cofactor (NADPH). To terminate the reactions, 20 μL aliquots were dispensed into 80 μL acetonitrile containing stable-labeled dextrorphan-d3 as an internal standard. The samples were centrifuged Supernatants obtained after incubation were analyzed for levels of dextromethorphan and CYP2D6-dependent O-demethylated product dextrophan (pmol/mg protein/min) on an AB Sciex 4000/5000/5500 triple quadrupole mass spectrometer. Stably-labeled d3-dextrorphan served as internal standard.
Results and Discussion:
1. Minor allele frequency of common functional CYP2D6 SNPs and their linkage disequilibrium in different populations
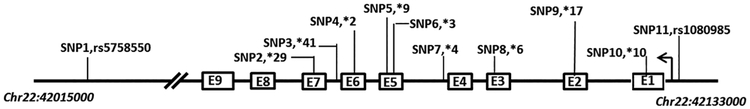
We used phased genotype data from the ‘1000 genomes project phase 3’, with DNA samples from 1092 individuals of different ancestries (Supplemental Table 1, Supplemental Digital Content 1, http://links.lww.com/FPC/B333), divided into four major populations, European (EUR), Asian (ASN), African (AFR) and American (AMR). Enhancer SNP rs5758550, *41 (rs28371725), *2 (rs16947), promoter SNP rs1080985 and other common CYP2D6 SNPs (*3, *4, *6, *9, *10, *17, *29) were selected in this study (Figure 1 and Supplemental Table 2, Supplemental Digital Content 1, http://links.lww.com/FPC/B333). CYP2D6*3, *4, and *6 do not produce functional CYP2D6 enzyme, due to nucleotide deletions (*3 and *6) [20,21] or splicing defects (*4) [20,22], while *9 [23], *17 [24,25] and *10 [26] reduce enzyme activity because of amino acid substitutions or deletion (*9). The function of *29 (rs59421388) [27,28] is uncertain, likely be a surrogate marker, because the *29 alleles always contains rs16947 (*2). The minor allele frequencies of these SNPs are shown in Table 1. While *2 (rs16947) is the most frequent SNP in EUR, AFR and AMR, *10 (rs1065852) is the most frequent SNP in ASN. CYP2D6*29 and *17 are only present in AFR and AMR, whereas *9 and *3 are absent in ASN, indicating large differences in minor allele frequencies in different populations. Also, large differences in linkage disequilibrium (LD) exist among these SNPs in the four populations (Supplemental Figure 1, Supplemental Digital Content 1, http://links.lww.com/FPC/B333). The LD structure of EUR is more similar to that of ASN than to AFR and AMR. Enhancer SNP rs5758550 and *2 SNP rs16947 are in high LD in EUR, ASN and AMR, but show lower LD in AFR. CYP2D6 *4 (rs3892097) alleles contain rs1065852 (*10) in all populations, but substantially differ in minor allele frequency in ASN (Table 1).
Figure 1.
Structure of the CYP2D6 gene locus on chromosome 22. The location of SNPs included in this study is shown, together with the numbering used here and either the * allele designation or the rs number (see Table 1). The transcription start site is indicated with an arrow on the right.
Table 1.
Minor allele frequencies of SNPs in different populations.
| SNP # | Allele name | rs number | EUR | ASN | AFR | AMR |
|---|---|---|---|---|---|---|
| SNP1 | enhancer | rs5758550 | 0.244 | 0.100 | 0.394 | 0.235 |
| SNP2 | *29 | rs59421388 | 0 | 0 | 0.124 | 0.006 |
| SNP3 | *41 | rs28371725 | 0.090 | 0.028 | 0.018 | 0.077 |
| SNP4 | *2 | rs16947 | 0.340 | 0.152 | 0.413 | 0.315 |
| SNP5 | *9 | rs5030656 | 0.021 | 0 | 0.006 | 0.017 |
| SNP6 | *3 | rs35742686 | 0.021 | 0 | 0.002 | 0.008 |
| SNP7 | *4 | rs3892097 | 0.191 | 0.002 | 0.061 | 0.152 |
| SNP8 | *6 | rs5030655 | 0.018 | 0.002 | 0.010 | 0.006 |
| SNP9 | *17 | rs28371706 | 0 | 0 | 0.213 | 0.006 |
| SNP10 | *10 | rs1065852 | 0.204 | 0.483 | 0.089 | 0.171 |
| SNP11 | promoter | rs1080985 | 0.249 | 0.124 | 0.067 | 0.215 |
2. Haplotypes containing rs5758550 and/or rs16947 in different populations
We assessed the frequency of haplotypes containing enhancer SNP rs5758550, rs16947 (*2), or both in four populations. The overall frequencies of alleles harboring rs5758550 and/or rs16947 range from 17% in ASN to 67% in AFR, indicating a high but variable prevalence in the four populations (Table 2). Also provided in Table 2 are predicted activity scores (on the basis of mRNA expression in liver [11]) with the wild-type CYP2D6 reference allele assigned a score of 1. The minor alleles of rs5758550 and rs16947 are assigned activity scores of 2 and 0.5, respectively, while scores for the other variants are taken from CPIC guidelines [5]. We grouped the different combinations of rs5758550 and rs16947 into three haplotypes: H1, rs5758550 without rs16947; H2, rs5758550 with rs16947; and H3, rs16947 without rs5758550. H1, H2 and H3 were further divided into sub-haplotypes based on the presence of other functional SNPs (*3, *4, *6, *9, *10 and *17, Table 2), forming a total of 12 haplotypes with different CYP2D6 activity (see Supplemental Figure 2, Supplemental Digital Content 1, http://links.lww.com/FPC/B333 for haplotype structure map). Because of different LD between SNPs in different populations, the frequencies of each haplotype differ greatly among the four populations (Table 2).
Table 2.
Frequencies of haplotypes containing different combinations of rs5758550 and rs16947 in four populations. Reference genotype, activity score=1; *9, *10, *17, activity score =0.5; *3, *4, *6, activity score=0. N represents the total allele counts in each population.
| Name | SNPs in the haplotype | EUR N=758 |
ASN N=572 |
AFR N=492 |
AMR N=362 |
Predictedactivity score | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele count* | % | Allele count* | % | Allele count* | % | Allele count* | % | ||||
| rs5758550, no rs16947 (H1) | H1a | rs5758550 | 9 | 1.2 | 5 | 0.9 | 42 | 8.5 | 7 | 1.9 | 2 |
| H1b | rs5758550, rs3892097 (*4), rs1065852 (*10) | 5 | 0.7 | 0 | 0 | 1 | 0.2 | 2 | 0.6 | 0 | |
| H1c | rs5758550, rs35742686 (*3) | 1 | 0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1d | rs5758550. rs1065852 (*10) | 0 | 0 | 4 | 0.7 | 0 | 0 | 0 | 0 | 1 | |
| rs5758550 and rs16947 (H2) | H2a | rs5758550, rs16947 | 170 | 22.4 | 48 | 8.4 | 59 | 12 | 72 | 19.9 | 1 or 1.5 |
| H2b | rs5758550, rs16947, rs28371706 (*17) | 0 | 0 | 0 | 0 | 92 | 18.7 | 2 | 0.6 | 0.5 | |
| H2c | rs5758550, rs16947, 1065852 (*10) | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.6 | 0.5 | |
| rs16947, no rs5758550 (H3) | H3a | rs16947 | 88 | 11.6 | 36 | 6.3 | 118 | 24 | 38 | 10.5 | 0.5 |
| H3b | rs16947, rs1065852 (*10) | 0 | 0 | 3 | 0.5 | 0 | 0 | 0 | 0 | <0.5 | |
| H3c | rs16947, rs28371706 (*17) | 0 | 0 | 0 | 0 | 13 | 2.6 | 0 | 0 | <0.5 | |
| H3d | rs16947, rs5030656 (*9) | 0 | 0 | 0 | 0 | 3 | 0.6 | 0 | 0 | <0.5 | |
| H3e | rs16947, rs5030655 (*6) | 0 | 0 | 0 | 0 | 4 | 0.81 | 0 | 0 | 0 | |
| Total | 273 | 36.0 | 96 | 16.8 | 332 | 67.5 | 123 | 33.9 | |||
Minor allele count
H1 haplotype----containing only enhancer SNP rs5758550 without rs16947 (*2)
The H1a haplotype (lacking *3, *4 and *10; MAF 1.2% in EUR; Table 2) is projected to convey increased CYP2D6 activity [11]. However, the co-existence of other functional SNPs may change the activity of CYP2D6 (H1b, 1c and 1d). In EUR, the alleles most frequently present in this haplotype are non-functional *4 and *3 (H1b or H1c; MAF 0.66% and 0.13%), with 40% of H1 alleles also containing *4 or *3, resulting in no CYP2D6 activity (Table 2). In all EUR, *4 and *3 SNPs (rs3892097 and rs1065852), if present, are always on the same haplotype as H1, forming H1b or H1c haplotypes (Supplemental Table 2, Supplemental Digital Content 1, http://links.lww.com/FPC/B333). In contrast, this is uncertain in AFR and AMR with the odds of *4 SNP rs3892097 present on H1 being 25% and 60%, respectively (Supplemental Table 4, Supplemental Digital Content 1, http://links.lww.com/FPC/B333), making it ambiguous to assign the correct haplotype or diplotype. However, because of the relatively low allele frequency of *4 in AFR, the frequency of H1 also carrying *4 (H1b) is low in AFR (0.2%) (Table 2), indicating that a majority of H1 alleles in AFR are H1a alleles (frequency 8.5%) with CYP2D6 activity score of 2. In ASN, H1a alleles are rare, with 44% of H1 alleles also containing rs1065852 (*10), forming H1d (Table 2). *10 (rs1065852) occur on the same haplotype with rs5758550 in all ASN heterozygous carriers (Supplemental Table 4, Supplemental Digital Content 1, http://links.lww.com/FPC/B333), conveying an estimated CYP2D6 activity score of 1.
After excluding alleles also containing other functional SNPs, the minor allele frequencies of rs5758550-alone haplotype (H1a) are 1.2%, 0.9%, 8.5% and 1.9% in EUR, ASN, AFR and AMR, respectively (Table 2). Since H1a is associated with more than two-fold increases in CYP2D6 mRNA expression [11], similar to or even greater than CYP2D6*1 gene duplication (see discussion below), the prevalence of H1a in all populations, especially in AFR, is expected to contribute significantly to an ultra-rapid metabolizer status.
H2 haplotypes----containing both enhancer SNP rs5758550 and rs16947 (*2)
The haplotype H2a containing both rs5758550 and rs16947 conveys normal or slightly increased enzyme activity[11]. In EUR and ASN, such haplotype appears to exclude other functional SNPs. However, in AFR and AMR, 60% and 5% of such alleles also contain *17 or *10, respectively (H2b and H2c, Table 2), causing reduced enzyme activity. After excluding haplotypes also containing other functional SNPs, the frequencies of haplotypes containing both rs16947 and rs5758550 (H2a) are 22%, 8.4%, 12% and 20% in EUR, ASN, AFR and AMR, respectively (Table 2), corresponding to nearly 50% of all rs16947 containing alleles in each population.
It is noted that a majority of alleles contained in H2a also harbor promoter region SNP rs1080985, owing to high LD between enhancer SNP rs5758550 (SNP1) and promoter SNP rs1080985 (SNP11) (Supplemental Figure 1, Supplemental Digital Content 1, http://links.lww.com/FPC/B333). This LD relationship accounts for the prediction of normal CYP2D6 activity using surrogate marker *2A (presence of both rs16947 and promoter SNP rs1080985 variant), or using presence of rs16947 plus promoter SNP −1584C to predict reduced CYP2D6 activity in Mexicans [16]. Although surrogate marker *2A is always concordant with haplotype containing both rs5758550 and rs16947, predicting normal CYP2D6 activity in ASN, EUR and AMR (Table 3a), the rate of concordance is only 44% in AFR, caused by lower allele frequency of the promoter SNP rs1080985 in AFR. Therefore, CYP2D6*2A cannot predict normal CYP2D6 activity in all populations.
Table 3a.
Prediction of normal activity haplotype (H2) by *2A (rs16947 and rs1080985) in four populations
| Haplotype | Frequency in population (%) | |||
|---|---|---|---|---|
| EUR | ASN | AFR | AMR | |
| rs5758550 +rs16947 (H2) | 22.4 | 8.2 | 12 | 19.9 |
| rs5758550 + rs16947 with rs1080985 (mark by *2A) | 21.7 | 8.2 | 5.5 | 19.1 |
| % rs5758550 + rs16947 marked by *2A | 96 | 100 | 44 | 95 |
H3 haplotypes----containing only rs16947 (*2) without enhancer SNP rs5758550
Haplotypes containing only rs16947 convey reduced enzyme activity (score 0.5) (Table 2) [11]. In EUR and AMR, such haplotypes exclude most other common functional SNPs. Yet in ASN and AFR, they also contain *10, *17, *9 and *6 (H3b, H3c, H3d and H3e, Table 2), resulting in strongly reduced CYP2D6 activity or no activity (H3e). In ASN, rs1065852 (*10) occurs on the same haplotype with rs16947 in less than 10% of individuals heterozygous for both H3 and 10*, accounting for only 7% of all H3 alleles. On the other hand, in AFR rs2837106 (*17), rs5030656 (*9) or rs5030655 (*6) occur together with rs16947 in all heterozygous carriers (Supplemental Table 4, Supplemental Digital Content 1, http://links.lww.com/FPC/B333), with 14% of H3 alleles carrying also these other reduced-function SNPs (H3c, H3d and H3e, Table 2). After excluding the co-existence of other functional SNPs, the frequencies of haplotype H3a with only rs19647 (reduced activity score 0.5) are 12%, 6.3%, 24 and 10.5% in EUR, ASN, AFR and AMR, corresponding to nearly one third of all rs16947 containing alleles in each population.
In EUR and AMR, a majority of H3 alleles also contains *41 SNP rs28371725, rendering *41 a reasonable surrogate marker for the H3a haplotype, conveying reduced CYP2D6 activity (Table 3b) [11]. Shown in Table 3b, over 70% of H3 alleles also contain *41 in EUR and AMR. Because of low allele frequency of *41 in ASN and AFR, only 44% and 4% of H3a alleles also contain *41, indicating that a significant portion of H3 alleles (up to 96% in AFR) with reduced CYP2D6 activity cannot be predicted with *41 (rs28371725) as a marker (Table 3b). In contrast, a few alleles containing both rs5758550 and rs16947 are marked by *41 (3 in EUR, 4 in AFR, and 1 in AMR), also leading to false prediction of reduced activity by *41. Similarly, CYP2D6*29 (rs59421388) marks the H3a haplotype in AFR but appears to be only a surrogate marker. CYP2D6*29 is exclusively co-existent with the H3 haplotype in AFR, but because of the lower allele frequency of *29 compared to H3 in AFR (12.4% vs 28%), only 38% of H3 alleles are marked by*29. Therefore, CYP2D6*29 also cannot serve as a surrogate marker for the H3 haplotype nor predict reduced CYP2D6 activity in AFR.
Table 3b.
Prediction of reduced activity haplotype (H3) by *41 in four populations
| Haplotype | Frequency in the population (%) | |||
|---|---|---|---|---|
| EUR | ASN | AFR | AMR | |
| rs16947 without rs5758550 (H3) | 11.6 | 6.3 | 23.9 | 10.5 |
| rs16947 without rs5758550 marked by *41 | 8.6 | 2.8 | 1 | 7.5 |
| % rs16947 without rs5758550(H3) marked by *41 | 74 | 44 | 4 | 71 |
3. Considerations for CYP2D6 genotyping panels and interpretation of genotyping results
Because surrogate markers cannot accurately predict CYP2D6 enzyme activity in all populations, and in view of evidence of functionality for rs16947 and downstream enhancer SNP rs5758550, we propose the possibility to remove surrogate markers (*41 and *29), change activity scores for *2 on the basis of the reduced activity of rs16947, and incorporate enhancer rs5758550 into CYP2D6 genotyping panels. This results in a streamlined genotyping panel (Supplemental Table 5, Supplemental Digital Content 1, http://links.lww.com/FPC/B333), which includes only 9 common single nucleotide polymorphisms and copy number variation (*5 and gene duplications), as well as other less frequent variants listed in CPIC guidelines [6].
After assessing the haplotype structure containing rs16947 and rs5758550 in different populations, we further refine the interpretation of CYP2D6 genotype to reflect haplotypes with assigned CYP2D6 activity scores in different populations. Specifically, individuals heterozygous for H3 or H1 and heterozygous for *3, *4, *9, *10 or *17 can be assigned different diplotypes and CYP2D6 activity scores, resulting in metabolic phenotype predictions in different populations (Table 4a and b). For example, individuals heterozygous for H1 and *4 should be assigned H1b/*1 in EUR with an activity score of 1, while this is uncertain for other populations, with AFR more likely being H1a/*4 with activity score 2. Similarly, individuals heterozygous for H1 and *10 should be assigned diplotype of H1d/*1 in ASN and H1a/*10 in other populations, with activity score of 2 and 2.5, respectively. *6 is more likely to be on the H3 haplotype in AFR, resulting in diplotype of H3e/*1 with an activity score of 1 in heterozygous carriers, while H3a/*6 (activity score of 0.5) can be assigned in other populations. Moreover, *17 and *9 reside on the H3 haplotype in AFR, resulting in H3c or H3d. In contrast, H3 is unlikely to include other reduced activity alleles in EUR and AMR, resulting in H3a (score 0.5), while in ASN, there is a 10% chance that *10 is on the same haplotype with H3. Therefore, an individual’s racial information is useful when assigning diplotype from genotype data. Information on other less frequent alleles can be found in CPIC guidelines [5,6] and two recent review papers [19,29].
Table 4a.
Predicted activity scores of CYP2D6 diplotypes (allele combinations). Activity scores are the sum of the activity scores of each of the two alleles in a given subject.
| *1 | rs16947 (H3) | rs16947 and rs5758550(H2) | rs5758550 (H1) | *3, *4, *6 and other null allelese | *9, *10, *17 and other reduced activity allelesf | *1×2 | |
|---|---|---|---|---|---|---|---|
| *1 | 2 | ||||||
| rs16947 (H3) | 1.5 | 1 | |||||
| rs16947 and rs5758550 (H2) | 2 | 1.5 | 2 | ||||
| rs5758550 (H1) | 3 | 2.5 | 3 | 4 | |||
| *3, *4, *6, *5 and other null alleles | 1 | 0.5 or 1a | 1 | 1 or 2b | 0 | ||
| *9, *10, *17 and other reduced activity alleles | 1.5 | 1 or 1.25c | 1.5 | 2 or 2.5d | 0.5 | 1 | |
| *1 × 2 | 2.5 | 2 | 2.5 | 3.5 | 1.5 | 2 | 3 |
Table 4b.
Predicted metabolizer phenotypes based on CYP2D6 diplotypes (allele combinations). Metabolizer phenotypes are determined by activity scores in Table 4a. PM, activity scores=0; IM: activity scores=0.5–1; NM: activity scores= >1 to 2; UM: activity scores >2.
| *1 | rs16947 (H3) | rs16947 and rs5758550(H2) | rs5758550 (H1) | *3, *4, *6, *5 and other null allelese | *9, *10, *17 and other reduced activity allelesf | *1×2 | |
|---|---|---|---|---|---|---|---|
| *1 | NM | ||||||
| rs16947 (H3) | NM | IM | |||||
| rs16947 and rs5758550 (H2) | NM | NM | NM | ||||
| rs5758550 (H1) | UM | UM | UM | UM | |||
| *3, *4, *6, *5 and other null alleles | IM | IMa | IM | IM or NMb | PM | ||
| *9, *10, *17 and other reduced activity alleles | NM | IM or NMc | NM | NM or UMd | IM | IM | |
| *1 × 2 | UM | NM | UM | UM | NM | NM | UM |
Note:
Because H3 is on the same haplotype as *6 in AFR, the activity score for *6 and H3 combination is 1 (H3e/*1, IM) for AFR. The activity score in other populations or H3 with other null allele combinations in AFR are 0.5 (IM).
Because H1 is on the same haplotype as *4 and *3 in EUR, the activity score for combination of *4 or *3 and H1 is 1(IM, H1b/*1 or H1c/*1) in EUR. H1 has 25% and 60% chance of being on the same haplotype with *4 in AFR and AMR, respectively, the activity score for combination of *4 and H1 could be 1 or 2 in AFR or AMR. The activity score for H1 and *4 or *3 combination in ASN or for H1 and other null allele combinations are 2 (NM).
Because H3 is on the same haplotype as *17 in AFR, the activity score for combination of H3 and *17 is 1.25 (NM, H3a/*1) in AFR. The activity score in other populations or the combination of H3 and other reduced activity alleles are 1 (IM).
Because H1 is on the same haplotype as *10 in ASN, the activity score for combination of H1 and *10 is 2 (NM, H1d/*1) in ASN. The activity score in other populations or the combination of H1 with other reduced activity alleles are 2.5 (UM).
Other null alleles listed in CPIC guidelines, including *5, *7, *8, *11, *12, *13, *14, *15, *16, *18, *19, *20, *21, *31, *36, *38, *40, *42, *44, *47, *51, *56, *57 and *62.
Other reduced activity alleles listed in CPIC guideline, including *49, *50, *54, *55, *59, *69 and *72.
It is challenging to translate CYP2D6 genotype into phenotype, in particular distinguishing between UM, NM and IM [19,29]. There are multiple ways to assign metabolic phenotype from activity scores. According to CPIC guidelines, the activity score for NM is 1–2, while activity score for IM is less than 1 [6]. The wide range of scores for IM and NM is at least in part a result of poor correlations between genotype and phenotype because of incomplete genotyping of all relevant variants (for example enhancer SNP rs5758550) and misinterpretation of the *2 variant. When considering *2 as carrying the reduced activity allele rs16947 and incorporating rs5758550 into genotyping panels, we can narrow the range of activity scores for NM. We modified the Dutch Pharmacogenetics Working Group (DPWG) phenotype classification system, with activity scores for NM >1 to 2, and for IM 1 or <1 [29]. Table 4b summarizes the metabolic phenotype assignment based on CYP2D6 diplotype. The different sub-haplotypes of H1 and H3 show different metabolic phenotypes, reflecting complex CYP2D6 genotype-phenotype relationships. In clinical use, these complex relationships can be resolved with algorithms that take ethnic background (self-reported) into consideration, or by applying recently developed software Stargazer to next-generation sequencing data [30], yielding refined estimates of UM, NM, and IM. Yet these technologies will require proper clinical trials to define validity and utility in all tested populations. Moreover, genotype predicted activity scores can be substrate specific as for codeine and tamoxifen [6,8].
4. Genotype-phenotype relationship in HLM
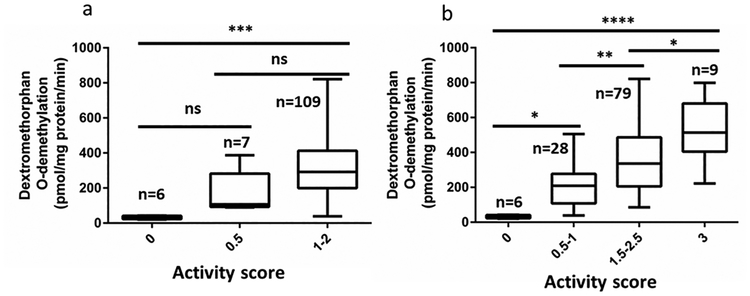
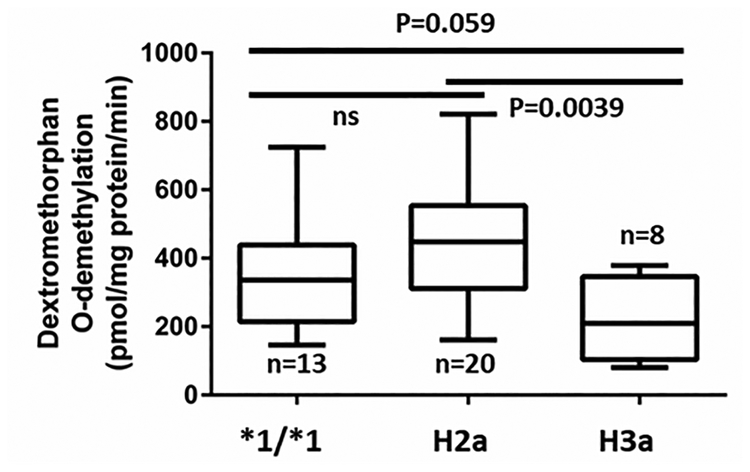
We then tested the performance of revised CYP2D6 genotyping panel and activity score system in a cohort of 122 human liver microsomes. CYP2D6 variants were detected using GeneSight assay platform (Supplemental Table 6, Supplemental Digital Content 1, http://links.lww.com/FPC/B333). Minor allele frequencies of SNPs tested are in Supplemental Table 7, Supplemental Digital Content 1, http://links.lww.com/FPC/B333. Shown in Figure 2, compared to traditional activity score system (panel a), the new activity score system (panel b) incorporating enhancer SNP rs5758550 and *2 yields better linear relationship between activity score and enzyme activity with statistically significant differences between each activity score group, consistent with our previous results[11]. Also consistent with previous results [11], *2 alone (H3a haplotype) has reduced enzyme activity compared to wild-type or *2 plus enhancer rs5758550 (H2a) (Figure 3). Moreover, our result shows that enhancer rs5758550 is better than CYP2D6*2A (promoter rs1080985 plus *2) in predicting normal or increased CYP2D6 activity associated with H2a haplotype (*2 plus enhancer SNP rs5758550), with F statistics and p value being [F=9.8, p=0.0001] and [F=11.3, p<0.0001] for *2A and enhancer rs5758550, respectively. Taken together, we have replicated our previously results[11] in an independent cohort, showing the incorporation of rs16947 and rs5758550 appears to predict CYP2D6 activity more accurately than a standard CYP2D6 panel. Additional clinical studies are needed to test these predictions in larger cohorts, taking particular consideration for testing a larger AFR cohort.
Figure 2.
Relationship between CYP2D6 enzyme activity in human liver microsomes and activity score derived from genotype data using CPIC standard score system (a) or new score system (b). Data are present as box plot. The box and horizontal lines show the 25th and 75th percentiles and mean, and whiskers show the minimum and maximum values. The differences between groups were analyzed by ANOVA.
Figure 3.
Comparing CYP2D6 enzyme activity in liver microsomes with wild-type (*1/*1), H2a (rs16947 plus rs5758550) or H3a (rs16947 alone) haplotypes. Data are present as box plot. The box and horizontal lines show the 25th and 75th percentiles and mean, and whiskers show the minimum and maximum values. The differences between groups were analyzed by ANOVA.
5. Activity score for CYP2D6 gene duplication
CYP2D6 gene duplication is considered to double CYP2D6 expression and activity if the duplicated genes are *1 or *2 [6,19,29]. However, our previous results indicate that the downstream enhancer containing the rs5758550 wild-type allele is critical for normal CYP2D6 expression, since deletion of this enhancer region reduces CYP2D6 expression at least twofold in HepG2 cell expressing CYP2D6*1 [31]. CYP2D6 gene duplications exclude the enhancer region, located over 115kb downstream, so that only one CYP2D6 gene copy can be regulated by the enhancer. Moreover, CYP2D6 duplications often include the *2 allele, which is frequent in AFR and largely conveys reduced metabolic activity with our new score system. Therefore, gene copies not controlled by the enhancer may express at levels less than 50%. Whereas genotype PM is consistent with phenotype PM, the relationship between genotype UM, based on gene duplications, and phenotype UM remains uncertain [32]. Only one out of 6 individuals with *1/*1×2 or *1/*2×2 genotype assigned activity score of >2 showed UM phenotype [33]. Therefore, we propose to assign CYP2D6*1× 2 an activity score of 1.5 instead of 2 (Table 4a and 4b), and for CYP2D6*2 duplication, the activity score is expect to be close to 1 (score = 1.25). These relationships suggest that gene duplication may not contribute significantly to the UM phenotype, while enhancer SNP rs5758550 (H1a) could be a main source for the UM phenotype, consistent with reports that only 30–40% individuals with UM phenotype carried CYP2D6 gene duplication [33,34]. However, this requires further investigation in large cohorts, and multiple CYP2D6 gene duplications need to be evaluated further.
There are limitations in this study to consider. We only focused on 11 common functional variants (except for promoter SNP rs1080985 with uncertain activity). There are other rare variants with reduced enzyme activity that share haplotypes with the enhancer SNP, and thereby, could change the activity score assignment. For example, reduced activity allele *59 is part of the CYP2D6*2A haplotype (likely also containing the enhancer SNP), displaying reduced activity rather than normal activity [17]. Moreover, presence of structural variants can also confound assignment of activity scores [18]. In addition, because of the limited number of livers studied, the activity of haplotypes with different combinations of enhancer and *2 alleles cannot be fully evaluated, in particular those with the enhancer SNP allele lacking *2, expected to be more prevalent in people of African descent.
In conclusion, alleles containing rs16947 and rs5758550 can form different haplotypes with other common normal, non-functional, or reduced-function SNPs in different populations, resulting in different CYP2D6 activity. The downstream enhancer SNP rs5758550 without rs16947 (H1a haplotype) appears to convey higher CYP2D6 activity than CYP2D6 *1 gene duplication. We propose that panels incorporating enhancer SNP rs5758550 and rs16947 may predict CYP2D6 activity more accurately than current CYP2D6 genotyping panel as written in the CPIC guideline [6]. This proposal must be validated through further assessment of CYP2D6 haplotypes with larger, more diverse cohorts as well as testing modified panels for clinical utility linked to patient outcomes. Our results further point out a weakness of the *allele nomenclature when it does not accurately reflect the causative haplotype harboring more than one functional variant, or contain surrogate markers.
Supplementary Material
Acknowledgments
Source of Funding: This study was supported by National Institutes of Health Pharmacogenetics Research Network grant U01 GM092655 (WS) and the National Institute of Health grant R01GM120396 (DW)
Footnotes
Conflict of interest: The Ohio State University has applied for a patent covering the use of newly discovered CYP2D6 variants (inventors DW and WS). WS is member of the Scientific Advisory Board of AssureRx.
References:
- 1.Eichelbaum M, Gross AS. The genetic polymorphism of debrisoquine/sparteine metabolism--clinical aspects. Pharmacol Ther. 1990; 46 (3):377–394. [DOI] [PubMed] [Google Scholar]
- 2.Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA. 2001; 286 (18):2270–2279. [DOI] [PubMed] [Google Scholar]
- 3.Griese EU, Zanger UM, Brudermanns U, Gaedigk A, Mikus G, Morike K, et al. Assessment of the predictive power of genotypes for the in-vivo catalytic function of CYP2D6 in a German population. Pharmacogenetics. 1998; 8 (1):15–26. [DOI] [PubMed] [Google Scholar]
- 4.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008; 83 (2):234–242. [DOI] [PubMed] [Google Scholar]
- 5.Crews KR, Gaedigk A, Dunnenberger HM, Klein TE, Shen DD, Callaghan JT, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin Pharmacol Ther. 2012; 91 (2):321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crews KR, Gaedigk A, Dunnenberger HM, Leeder JS, Klein TE, Caudle KE, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther. 2014; 95 (4):376–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hicks JK, Swen JJ, Thorn CF, Sangkuhl K, Kharasch ED, Ellingrod VL, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther. 2013; acepted doi: 10.1038/clpt.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goetz MP, Sangkuhl K, Guchelaar HJ, Schwab M, Province M, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin Pharmacol Ther. 2018; 103 (5):770–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell GC, Caudle KE, Whirl-Carrillo M, Gordon RJ, Hikino K, Prows CA, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin Pharmacol Ther. 2017; 102 (2):213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D, Papp AC, Sun X. Functional characterization of CYP2D6 enhancer polymorphisms. Hum Mol Genet. 2015; 24 (6):1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, Poi MJ, Sun X, Gaedigk A, Leeder JS, Sadee W. Common CYP2D6 polymorphisms affecting alternative splicing and transcription: long-range haplotypes with two regulatory variants modulate CYP2D6 activity. Hum Mol Genet. 2014; 23 (1):268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu A, Kneller BM, Rettie AE, Haining RL. Expression, purification, biochemical characterization, and comparative function of human cytochrome P450 2D6.1, 2D6.2, 2D6.10, and 2D6.17 allelic isoforms. J Pharmacol Exp Ther. 2002; 303 (3):1291–1300. [DOI] [PubMed] [Google Scholar]
- 13.Marcucci KA, Pearce RE, Crespi C, Steimel DT, Leeder JS, Gaedigk A. Characterization of cytochrome P450 2D6.1 (CYP2D6.1), CYP2D6.2, and CYP2D6.17 activities toward model CYP2D6 substrates dextromethorphan, bufuralol, and debrisoquine. Drug Metab Dispos. 2002; 30 (5):595–601. [DOI] [PubMed] [Google Scholar]
- 14.Sakuyama K, Sasaki T, Ujiie S, Obata K, Mizugaki M, Ishikawa M, et al. Functional characterization of 17 CYP2D6 allelic variants (CYP2D6.2, 10, 14A-B, 18, 27, 36, 39, 47–51, 53–55, and 57). Drug Metab Dispos. 2008; 36 (12):2460–2467. [DOI] [PubMed] [Google Scholar]
- 15.Gaedigk A, Ndjountche L, Leeder JS, Bradford LD. Limited association of the 2988g > a single nucleotide polymorphism with CYP2D641 in black subjects. Clin Pharmacol Ther. 2005; 77 (3):228–230; author reply 230–221. [DOI] [PubMed] [Google Scholar]
- 16.Luo HR, Gaedigk A, Aloumanis V, Wan YJ. Identification of CYP2D6 impaired functional alleles in Mexican Americans. Eur J Clin Pharmacol. 2005; 61 (11):797–802. [DOI] [PubMed] [Google Scholar]
- 17.Gaedigk A, Riffel AK, Leeder JS. CYP2D6 Haplotype Determination Using Long Range Allele-Specific Amplification: Resolution of a Complex Genotype and a Discordant Genotype Involving the CYP2D6*59 Allele. J Mol Diagn. 2015; 17 (6):740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Tredici AL, Malhotra A, Dedek M, Espin F, Roach D, Zhu GD, et al. Frequency of CYP2D6 Alleles Including Structural Variants in the United States. Front Pharmacol. 2018; 9:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaedigk A Complexities of CYP2D6 gene analysis and interpretation. Int Rev Psychiatry. 2013; 25 (5):534–553. [DOI] [PubMed] [Google Scholar]
- 20.Kagimoto M, Heim M, Kagimoto K, Zeugin T, Meyer UA. Multiple mutations of the human cytochrome P450IID6 gene (CYP2D6) in poor metabolizers of debrisoquine. Study of the functional significance of individual mutations by expression of chimeric genes. J Biol Chem. 1990; 265 (28):17209–17214. [PubMed] [Google Scholar]
- 21.Saxena R, Shaw GL, Relling MV, Frame JN, Moir DT, Evans WE, et al. Identification of a new variant CYP2D6 allele with a single base deletion in exon 3 and its association with the poor metabolizer phenotype. Hum Mol Genet. 1994; 3 (6):923–926. [DOI] [PubMed] [Google Scholar]
- 22.Gough AC, Miles JS, Spurr NK, Moss JE, Gaedigk A, Eichelbaum M, et al. Identification of the primary gene defect at the cytochrome P450 CYP2D locus. Nature. 1990; 347 (6295):773–776. [DOI] [PubMed] [Google Scholar]
- 23.Tyndale R, Aoyama T, Broly F, Matsunaga T, Inaba T, Kalow W, et al. Identification of a new variant CYP2D6 allele lacking the codon encoding Lys-281: possible association with the poor metabolizer phenotype. Pharmacogenetics. 1991; 1 (1):26–32. [DOI] [PubMed] [Google Scholar]
- 24.Masimirembwa C, Persson I, Bertilsson L, Hasler J, Ingelman-Sundberg M. A novel mutant variant of the CYP2D6 gene (CYP2D6*17) common in a black African population: association with diminished debrisoquine hydroxylase activity. Br J Clin Pharmacol. 1996; 42 (6):713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oscarson M, Hidestrand M, Johansson I, Ingelman-Sundberg M. A combination of mutations in the CYP2D6*17 (CYP2D6Z) allele causes alterations in enzyme function. Mol Pharmacol. 1997; 52 (6):1034–1040. [DOI] [PubMed] [Google Scholar]
- 26.Yokota H, Tamura S, Furuya H, Kimura S, Watanabe M, Kanazawa I, et al. Evidence for a new variant CYP2D6 allele CYP2D6J in a Japanese population associated with lower in vivo rates of sparteine metabolism. Pharmacogenetics. 1993; 3 (5):256–263. [DOI] [PubMed] [Google Scholar]
- 27.Wennerholm A, Johansson I, Hidestrand M, Bertilsson L, Gustafsson LL, Ingelman-Sundberg M. Characterization of the CYP2D6*29 allele commonly present in a black Tanzanian population causing reduced catalytic activity. Pharmacogenetics. 2001; 11 (5):417–427. [DOI] [PubMed] [Google Scholar]
- 28.Wennerholm A, Dandara C, Sayi J, Svensson JO, Abdi YA, Ingelman-Sundberg M, et al. The African-specific CYP2D617 allele encodes an enzyme with changed substrate specificity. Clin Pharmacol Ther. 2002; 71 (1):77–88. [DOI] [PubMed] [Google Scholar]
- 29.Hicks JK, Swen JJ, Gaedigk A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr Drug Metab. 2014; 15 (2):218–232. [DOI] [PubMed] [Google Scholar]
- 30.Lee SB, Wheeler MM, Patterson K, McGee S, Dalton R, Woodahl EL, et al. Stargazer: a software tool for calling star alleles from next-generation sequencing data using CYP2D6 as a model. Genet Med. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Papp AC, Sun X. Functional characterization of CYP2D6 enhancer polymorphisms. Hum Mol Genet. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.A LL, Naranjo ME, Rodrigues-Soares F, Penas LEM, Farinas H, Tarazona-Santos E. Interethnic variability of CYP2D6 alleles and of predicted and measured metabolic phenotypes across world populations. Expert Opin Drug Metab Toxicol. 2014; 10 (11):1569–1583. [DOI] [PubMed] [Google Scholar]
- 33.Montane Jaime LK, Lalla A, Steimer W, Gaedigk A. Characterization of the CYP2D6 gene locus and metabolic activity in Indo- and Afro-Trinidadians: discovery of novel allelic variants. Pharmacogenomics. 2013; 14 (3):261–276. [DOI] [PubMed] [Google Scholar]
- 34.Llerena A, Dorado P, Ramirez R, Gonzalez I, Alvarez M, Penas-Lledo EM, et al. CYP2D6 genotype and debrisoquine hydroxylation phenotype in Cubans and Nicaraguans. Pharmacogenomics J. 2012; 12 (2):176–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.