Abstract
Sporadic colorectal cancer is one of the most common and lethal cancers worldwide. The locations and functions of immune cells in the colorectal tumor microenvironment are complex and heterogeneous. T-helper (Th)1 cell–mediated responses against established colorectal tumors are associated with better outcomes of patients (time of relapse-free or overall survival), whereas Th17 cell–mediated responses and production of interleukin 17A (IL17A) have been associated with worse outcomes of patients. Tumors that develop in mouse models of colorectal cancer are rarely invasive and differ in many ways from human colorectal tumors. However, these mice have been used to study the mechanisms by which Th17 cells and IL17A promote colorectal tumor initiation and growth, which appear to involve their direct effects on colon epithelial cells. Specific members of the colonic microbiota may promote IL17A production and IL17A-producing cell functions in the colonic mucosa to promote carcinogenesis. Increasing our understanding of the interactions between the colonic microbiota and the mucosal immune response, the roles of Th17 cells and IL17 in these interactions, and how these processes are altered during colon carcinogenesis, could lead to new strategies for preventing or treating colorectal cancer.
Keywords: Inflammation, Microbiota, Mouse Models Colon Carcinogenesis
Graphical Abstract
Sporadic colorectal cancer (CRC) is a common, preventable cancer that accounts for nearly 10% of new cancers each year worldwide.1 Risk increases with age, so each generation eventually requires screening for prevention. CRC is a leading cancer in regions with a higher socioeconomic status, such as the United States and Western Europe. However, incidence varies among regions—nearly 50% of cases of CRC worldwide are in Asia.1 Concerning trends include increasing detection in young individuals and African Americans and a global increase in CRC incidence and mortality.2,3 The increase in mortality is predicted to continue until at least 2035, with a widening gap between lower- and higher-resource regions. Increasing our understanding of CRC pathogenesis could lead to new strategies for prevention and detection, as well as reduce the global burden of CRC.
We review the adaptive immune response to colorectal tumors, focusing on the contributions of the interleukin 17 (IL17) family members to CRC pathogenesis. We seek to integrate findings from mouse models with data from clinical investigations. Mouse models have been most commonly used to study the activities of immune cells during early stages of colon tumorigenesis—the most commonly used mouse models of CRC develop adenomas only. In contrast, CRC is late disease, after the transition from adenoma. In CRC, our understanding of immune cell infiltration of tumors, such as by IL17-producing cells, comes mostly from analyses of patient outcomes. We review data from mouse and human studies of the adaptive immune response to colon tumors and discuss how these findings might lead to new treatment approaches for patients.
Mouse Models
Mouse models are important for studying the mechanisms by which the mucosal immune system (including innate and adaptive responses) contributes to colorectal tumorigenesis. Despite the species’ gap, the mouse and human genomes are closely related and immune and carcinogenesis pathways are often conserved. Mouse models are therefore used to study how specific genes that regulate the immune response and other mechanisms contribute to CRC pathogenesis. Many mice develop colorectal tumors via genetic alterations associated with human colon carcinogenesis. One model involves disruption of the adenomatous polyposis coli gene (Apc). MinApc+/− mice develop multiple intestinal neoplasias. Loss of heterozygosity at APC is typically an early event in colon tumor formation and is observed in almost every sporadic CRC.4 Mutations in APC cause an inherited form of CRC called familial adenomatous polyposis. Although MinApc+/− mice are commonly used to study colon tumorigenesis5,6 (most frequently MinApc+/Δ850 or MinApc+/Δ716 mice), they develop spontaneous tumors in the small intestine but only rare tumors in the colon, likely limiting their relevance for studies of human colon tumorigenesis.
To address this limitation in MinApc+/− mice, Hinoi et al7 developed CDX2P-NLS Cre;Apc+/loxP mice. These mice develop tumors in the colon—most (approximately 66%) have histologic features of colon adenocarcinomas, although they do not usually progress (remain at stage 0–1).7 In CDX2P-CreERT2Apcflox/flox mice, tamoxifen administration induces biallelic deletion of Apc selectively in colon epithelial cells (CECs).8 Loss of Apc in these mice is paralleled by epithelial barrier dysfunction and bacterial translocation with promotion of pattern recognition receptor signaling (such as Toll-like receptors), leading to production of inflammatory cytokines and tumor growth.9
C57Bl/6 mice that are given azoxymethane to induce mutations in CECs, combined with dextran sodium sulfate (DSS) to induce colitis, are used as a model of colitis-induced carcinogenesis.10 Azoxymethane causes formation of O-methyl guanine and promotes base mispairing and accumulation of mutations, including in genes of the Wnt signaling pathway.11 Mice must be given several doses of azoxymethane to induce tumorigenesis, but only 1 dose after administration of DSS.12 Nonobese diabetic severe combined immunodeficient (NOD SCID) mice and Rag−/− mice develop colitis after administration of DSS, so DSS colitis at least does not require an adaptive immune response.13
Studies of mice with colitis-associated tumors (after administration of azoxymethane and DSS) and MinApc+/− mice have revealed roles of nuclear factor κB (NF-κB), signal transducer and activator of transcription 3 (STAT3), and inflammasome signaling pathways in colon carcinogenesis.9,11,14,15 Mouse models have also been used to study the mechanisms by which innate (IL17-producing γδT cells, innate lymphoid cells, and natural killer [NK] T cells) and adaptive (T helper [Th]17 cell–mediated) immune cell sources of IL17 promote early stages of colon tumor development (Table 1). For example, researchers have created mice that do not express IL23 or its receptor, IL17 or its receptor (IL17R), and mice that do not develop Th17 cells (CD4DStat3 mice) or T and B cells (Rag−/− mice).9,15–19 Findings from these mice indicate roles for IL17A in colon tumorigenesis and growth (Table 1). However, MinApc+/− mice or mice with azoxymethane and DSS-induced tumors do not develop invasive colon adenocarcinomas, so they cannot be used to study colorectal tumor progression. In contrast, CDX2P-NLS Cre;Apc+/loxP mice develop distal colon adenocarcinomas, and 16% of these invade the submucosa, more closely resembling human in situ carcinomas.7 Other models of colitis-induced cancer or CRC, such as Il10−/− or Muc2−/− mice, do not offer additional advantages.20,21
Table 1.
Members of the IL17 Family and Effects
| Cytokine | Receptor42 | Cell source42 | Model | Possible roles in development of CRC | Reference no. |
|---|---|---|---|---|---|
| IL17A (IL17, CTLA8) | IL17RA–IL17RC | Th17, Tc17, γδT, NKT, and NK cells; innate lymphoid cells; monocytes; neutrophils; B cells; epithelial cells; and Paneth cells |
CDX2P-NLS Cre;Apc+/loxP and CDX2P-CreERT2; Apcflox/flox mice; Villin-CreERT2;Apcflox/flox organoids CRC cell lines |
Activate NF-κB, which regulates transcription of genes that protect transformed CECs from apoptosis Promote angiogenesis |
47, 48 |
| IL17B | IL17RB | Gene expression in pancreas, small intestine, stomach, chondrocytes, intestinal epithelial cells, and neurons | Mice with DSS-induced acute colitis or Citrobacter rodentium infection | Not described in CRC Competes with IL17E for binding of IL17RB Potential antagonist of endogenous IL17E |
63 |
| IL17C | IL17RA–IL17RE | Epithelial cells, endothelial cells, keratinocytes, and leukocytes | Mice with azoxymethane and DSS colitis-associated colorectal tumors and MinApc+/− mice. | Promotes expression of BCL2 and BCLXL, in an autocrine manner that reduces apoptosis of transformed intestinal epithelial cells | 64 |
| IL17D | Unknown | Unknown | Mice with sarcoma induced by 3-methylcholanthrene and air pouch–induced inflammation | Not described in CRC Levels increased in immunogenic tumors, whereas nonimmunogenic tumors have decreased levels Activation of Nrf2 in tumors induces expression of IL17D and NK cell-dependent tumor regression |
65, 66 |
| IL17E (IL25) | IL17RA–IL17RB | Dendritic cells, alveolar macrophages, T cells, eosinophils, IgE-activated mast cells, basophils, microglia, Paneth cells, and epithelial cells | Mice with acute colitis after administration of oxazolone, 2,4,6-trinitrobenzene sulfonic acid or DSS Cells from patients with Crohn’s disease |
Endogenous IL17E promotes tumor development, whereas exogenous IL17E reduces tumor development Commensal-dependent expression of IL25 by intestinal epithelial cells limits the expansion of Th17 cells in the intestine by inhibiting expression of macrophagederived IL23 |
67–69 |
| IL17F | IL17RA–IL17RC | Th17, Tc17, γδT, NKT, and NK cells; innate lymphoid cells, monocytes; neutrophils; B cells; epithelial cells; and Paneth cells | Human colon cancer cells and xenograft tumors, in mice with azoxymethane and DSS colitis-associated colorectal tumors and an in vitro angiogenesis study | Overexpression delayed tumor growth, whereas knockout mice have accelerated tumorigenesis Recombinant IL17F regulates angiogenesis |
46, 49 |
Mice with conditional disruption of Msh2 in intestinal epithelial cells or crypt stem cells are studied as models of Lynch syndrome (Villin-Cre:Msh2loxp/loxp and Lgr5-CreERT2;Msh2flox/−, respectively).22,23 Although these mice develop tumors with microsatellite instability and inactivation of Apc, these tumors again form preferentially in the small intestine, whereas patients with Lynch syndrome (hereditary nonpolyposis CRC) develop colon and extracolonic tumors.22 Studies of the tumor immune microenvironment in these mice have not been published.
Studies of germ-free or conventional mice colonized with a single bacterial species have provided strong evidence to support the role of select microbes in colon tumor initiation and progression. Wu et al17 showed that endogenous Th17 cells contribute to colon tumorigenesis in MinApc+/Δ716 mice colonized with enterotoxigenic Bacteroides fragilis (ETBF). Mouse models are therefore useful for studying the contribution of immune responses, including those mediated by IL17-producing cells, to early stages of carcinogenesis, based on quantification of microadenoma or macroadenoma numbers and tumor burden. However, these mice imperfectly recapitulate sporadic CRC in humans, who present with invasive adenocarcinoma in >85% of cases.
The Adaptive Immune Response to Colorectal Cancer
Human CRC is late disease, whereas most mouse models develop adenomas only, representing early disease. In patients, infiltration of colorectal tumors by immune cells has been associated with tumor progression and clinical outcomes (relapse-free survival or overall survival). Analyses of patient outcomes are complex because they are affected by disease stage, treatment, and environmental and genetic factors.
Landmark studies performed by Galon et al24 showed that the intra-tumor adaptive immune response affects clinical outcomes, including reduced tumor recurrence.25 CD8+ cytotoxic T cells that recognize colon tumor antigens have been identified, expanded ex vivo, and studied in adoptive transfer experiments. These studies have shown that adaptive immune responses can promote regression of colorectal tumors.26
Patients with metastatic colorectal tumors with mismatch repair deficiency and a high density of cytotoxic T cells in the primary tumor have a high rate of response to checkpoint blockade-based immunotherapy.27,28 So, cytotoxic lymphocytes present in the tumor that are inhibited by immunosuppressive ligands in the microenvironment can be activated by checkpoint blockade therapy to kill tumor cells.27,28 However, some patients with metastatic CRC do not respond to immunotherapy, even though the primary colon tumors are infiltrated by cytotoxic T cells. Other features of the immune infiltrate, including the quality of the adaptive anti-tumor immune response, could affect tumor progression and response to immunotherapy.
The adaptive immune response to CRC has been well studied—researchers have investigated the roles of tumor-infiltrating T-regulatory cells (Treg), CD8+ T cells, Th1 cells, Th17 cells, and T-follicular helper (Tfh) cells in CRC progression (Table 2). The balance between gene expression patterns of Th1 vs Th17 cells within tumors has been associated with patient outcomes, but findings have not always been consistent.25,29–33 Almost two-thirds of primary sporadic CRCs were estimated to have increased expression of IL17A (RNA and/or proteins), whereas others have increased expression of interferon gamma (IFNG) or a combination of IL17 and IFNG expression.29,31
Table 2.
Cells of the Adaptive Immune System and Association With Colorectal Cancer
| Cell type | Role in CRC pathogenesis | Reference no. |
|---|---|---|
| CD3+ memory T cells (CD4+, CD8+, CD45RO) | Prognostic value of type, density, and location of cells | 24 |
| Gene expression pattern associated with Th1 cells in tumors and higher numbers of CD8+ T cells are associated with better patient outcomes (such as fewer metastases) | 70 | |
| Immunoscore: Numbers of Th1 cells in tumors, effector memory T cells, and cytotoxic T cells correlate with better outcomes of patients with CRC and longer survival times of patients with metastatic CRC | 71 | |
| CD8+Foxp3+ T cells | Number of intratumor CD8+CD25+Foxp3+ T cells (Treg) correlate with the stage and the micro-invasive status of CRC | 72 |
| CD3+Foxp3+ T cells | Density of Foxp3+T cells in lymphoid follicles in normal mucosa might reduce survival times | 33 |
| Number of Foxp3+ T cells in CRC associates with increased survival times of patients with mismatch repair–proficient, but not mismatch repair– deficient, CRC | 32 | |
| Low ratio of intraepithelial CD3+ T cells to Foxp3+ T cells associates with poor outcomes | 73 | |
| Higher numbers of Treg in tumors, lower numbers of IL17-producing cells, associated with reduced metastasis | 74 | |
| CD3+CD8+ T cells | CD3+ and CD8+ tumor-infiltrating lymphocytes might increase survival times, but effects are confounded by CRC stage | 75 |
| Cytotoxic T lymphocytes, Th cells (Th1, Th2, Th17, Treg) | Th17 cell gene expression pattern in CRCs associated with poor outcomes Th1 cell and cytotoxicity expression pattern in CRCs associate with better outcomes |
29 |
| T follicular helper (Tfh) cells | Intratumor density of Tfh increased with tumor progression | 25 |
| Gene polymorphisms of Th17 cells (IL17A, IL17F, IL23 receptor) | Polymorphisms in genes expressed by Th17 cells associate with susceptibility to CRC and poor outcomes | 36–38 |
| Th17 and γδT17 cells | Detection of IL17+ T cells in human CRC specimens Most colorectal tumors contain Th17 and γδT17 cells | 16, 31, 39 |
NOTE. Data were selected from publications on the adaptive intra-tumor immune response.
Gene expression profiling studies of CRC specimens associated increased gene expression patterns of Th1 cells (IFNG and TBX21) and cytotoxic T cells (PRF1 and GZMB) with better outcomes of patients. In contrast, increased levels of IL17A and RORC messenger RNAs, expressed by Th17 cells, are associated with worse outcomes for patients.29 However, there is debate about the prognostic value of detecting IL17A-producing innate lymphoid, γδT cells, or NK cells vs Th17 cells in the tumor microenvironment. Overall, immunohistochemical analysis of a large microarray of primary CRCs (1151 samples) did not reveal an association between the density of IL17+ cells and overall survival or relapse-free survival times.31 In this study, however, the intra-epithelial (but not intra-stromal) density of IL17+ cells correlated with longer times of relapse-free survival. In contrast, a separate study associated reduced disease-free survival times of 125 patients with CRC with a Th17-cell gene expression pattern.29
Intra-epithelial Th17 cells produce CCL5 and CCL20, which promote localization of CCR5+CCR6+CD8+ cytotoxic T cells, presumably with anti-tumor activity, to tumor tissues.31 However, IL17A also induces production of factors by the stroma that sustain tumor cell proliferation, survival, and angiogenesis.31,34 Levels of IL17A are increased in the colorectal tumor stroma and intestinal epithelia of patients with colon adenomas, and all stages through carcinoma.35 Polymorphisms in genes encoding IL17A, IL17E, and IL23 receptor, which are expressed during Th17 cell differentiation, have been associated with increased risk of CRC36 and poor outcomes.37,38
These findings provide evidence that Th17 cells and IL17A contribute to CRC development and progression.39 However, studies are needed to distinguish the role of IL17A from the roles of cells that produce IL17A, and other cytokines that might contribute to tumor progression or, alternatively, to tumor cell death. It will also be important to consider tumor features (histopathology and stage) in evaluating associations between IL17, Th17 cells, and patient outcomes.40
In patients with melanoma, those who did not respond to anti-PD1 therapy were found to have increased numbers of RORγt+ Th17 cells in tumors.41 Furthermore, intestinal microbiota from the nonresponders were found to promote a Th17 cell–mediated immune response in spleens and transplanted tumors of germ-free recipient mice.41 These findings indicate that an inability to induce an effective anti-tumor immune response could, at least in part, involve activation of Th17 cells or IL17A polarization of cells in the tumor microenvironment by components of the microbiota. These findings might apply to CRC development. The Food and Drug Administration recently approved anti-PD1 therapy for patients with mismatch repair–deficient CRCs. Investigating the colorectal tumor immune microenvironment could facilitate studies to test the association between adaptive immune (Th1 cell, Th17 cell, and T-regulatory) cell responses, the microbiota, and responses of patients to immunotherapy.
The Interleukin 17 Family
Members of the IL17 family (IL17A, IL17B, IL17C, IL17D, IL17E, and IL17F) have immune regulatory functions and signal via a heterodimeric receptor.42 This receptor comprises a combination of IL17RA and another subunit, which determines cytokine binding and signaling specificity. IL17A and IL17F have the highest degree of structural homology (55%) and function via binding to the IL17RA–IL17RC heterodimer. IL17A and IL17F are secreted by Th17 cells, CD8+ T cells (Tc17), γδT17 cells, and innate lymphoid cells type 3 cells, which have been identified in human colorectal tumors and in mouse models (Table 1).16,17,39,43–45 Binding of IL17A or IL17F to the IL17RA–IL17RC heterodimer recruits the adaptor proteins ACT1 and TRAF6 to activate signaling pathways that include mitogen-activated protein kinase and NF-κB.42 IL17RA–IL17RC binds homodimers and heterodimers of IL17A and IL17F with distinct affinities—it binds IL17A–IL17A with the highest affinity.
Il17a−/− mice develop fewer colon tumors, whereas Il17f−/− mice develop more tumors, compared with wild-type mice, after administration of azoxymethane and DSS.46 By activating NF-κB, IL17RA signaling induces the production of IL6 by epithelial cells, which, in turn, activates STAT3 in epithelial cells to promote their survival and proliferation.47 In MinApc+/Δ716 mice colonized with ETBF to induce colon tumorigenesis, Th17 and γδT17 cell–derived IL17 induces production of CXCL1 by CECs, via activation of NF-κB. This chemokine recruits myeloid cells, which further promote proliferation and tumorigenesis.15
Infiltration of human colorectal tumors by γδT17 cells was associated with recruitment of myeloid-derived suppressor cells and increased invasiveness.39 Increased levels of IL17A in colorectal tumors has also been associated with increased expression of vascular endothelial growth factor and microvessel density, indicating that IL17A associates with angiogenesis.48 In contrast, human CRC samples have reduced levels of IL17F messenger RNA and IL17F protein, compared with parallel non-tumor tissues from the same patient. Overexpression of IL17F in HCT116 colon cancer cells slowed their growth as xenograft tumors in immunocompromised mice; IL17F-overexpressing xenograft tumors produced lower levels of vascular endothelial growth factor and contained fewer CD31+ cells than xenograft tumors grown from control HCT116 cells.46,49 Although these findings could indicate that IL17F slows development of colorectal tumors, little is known about the differential effects of IL17A and IL17F signaling via different receptors, dimerization of IL17A and IL17F, or variations in expression patterns of IL17RA and IL17RC on colorectal tumorigenesis.
Beyond IL17A and IL17F, it is not clear whether other IL17 family members contribute to CRC development (Table 1). IL17C binds to IL17RA–IL17RE and contributes to the integrity of the intestinal barrier.42 IL17E (also called IL25), binds to IL17RA–IL17RB and is produced by epithelial or myeloid cells; IL17E might contribute to development of Th2 cells and activation of type 2 innate lymphoid cells during anti-parasitic mucosal immune responses.42
The Microbiota and Interleukin 17
Mouse models of CRC provide evidence for the interaction among innate and adaptive mucosal immune responses and the colonic microbiota in colorectal tumorigenesis.50 It is not clear which members of the microbiota and which mucosal immune responses might promote sporadic or hereditary CRCs. Often cited candidate organisms include Streptococcus gallolyticus, ETBF, Fusobacterium nucleatum, polyketide synthase-positive (pks+) Escherichia coli, and Enterococcus faecalis. Adaptive and innate immune responses contribute to colon carcinogenesis in mice colonized with ETBF, whereas myeloid populations appear to contribute to colorectal tumor formation in mice colonized with F nucleatum and E faecalis.15,16,51
Drewes et al52 analyzed available 16S ribosomal RNA gene sequences through a single computational pipeline and associated 5 microbes with sporadic CRC: B fragilis, F nucleatum, Peptostreptococcus stomatis, Gemella morbilliform, and Parvimonas micra. F nucleatum, P stomatis, G morbilliform, and P micra are often found in the human oral microbiome. Members of the oral microbiome have increasingly been associated with CRC. However, the effects of these bacteria on mucosal immune responses and tumorigenesis have not been studied in mice. Dejea et al53,54 associated mucus-invasive bacterial biofilms with sporadic CRC and tumors in patients with familial adenomatous polyposis. Polymicrobial biofilms were observed on sporadic colorectal tumors and nontumor colon tissues from the same patients and associated with carcinogenic changes in CECs.53 In patients with familial adenomatous polyposis, biofilms were found on nonmalignant colon polyps and normal mucosa; >50% of the microbial population comprised ETBF and pks+ E coli. In C57Bl/6 mice given azoxymethane (without DSS) and co-colonized with ETBF and pks+ E coli, increased expression of IL17A and ETBF-mediated increases in adherence of pks+ E coli to the colonic mucosa were required for colon tumor development.54
The concept that microbe-induced IL17A mucosal adaptive immune responses can be carcinogenic is supported by observations that Helicobacter pylori–associated gastritis, which precedes most cases of gastric cancer, requires IL17A.55 The discovery that segmented filamentous bacteria, a mouse microbe, induces expression of IL17 in mouse small intestine led to the concept that specific microbes are required for induction of intestinal IL17-mediated immune responses.56 Segmented filamentous bacteria rarely, if ever, have pathologic effects in humans. However, studies are underway to identify bacteria that induce IL17 production in the human colon beyond the initial association of IL17A production with ETBF colonization. Other bacteria reported to induce intestinal production of IL17 in germ-free mice, similar to or greater than that induced by segmented filamentous bacteria, include Staphylococcus saprophyticus and Bifidobacterium adolescentis.57 Atarshi et al58 showed that a response of Th17 cells in the intestine to bacteria requires physical contact between the bacterium and intestinal epithelial cells. These results indicated that the mucosal Th17 cell–mediated response requires microbe-associated molecular patterns and physical proximity of the microbe to the epithelial cells. Further, the authors identified 20 bacteria in humans, including enterohemorrhagic E coli that increase numbers of Th17 cells in the mucosa.58
Cremonesi et al59 have associated specific bacteria and communities with production of chemokines in cells and mice, possibly including IL17. Colonization of germ-free mice with the Propionibacterium strain UF1 correlated with increases in intestinal Th17 cells and mitigation of necrotizing enterocolitis–like disease in neonatal mice.60 These findings support the function of IL17A in antibacterial host defense and epithelial repair and regeneration, including limiting epithelial permeability by inducing the expression of the tight junctional protein occludin.61 Intestinal mucosal IL17A, therefore, protects the intestine from pathogens but somehow also can transition to promote colon tumor development—studies are needed to determine the mechanisms of these protective and carcinogenic processes.
Future Directions
Development of CRC involves a series of genetic mutations in CECs, along with alterations to the colon tissue environment and microbiota. Mucosal innate and adaptive immune responses contribute to CRC.62 Studies of mice and patients have shown that excess mucosal IL17A, particularly from adaptive immune cell sources (and possibly from innate immune cells), along with increased production of cytokines and chemokines by myeloid cells, alter CECs to promote colon carcinogenesis and tumor progression.15,17 In contrast, mucosal production of IFNG and cytotoxic activities of Th1 cells appear to limit colon carcinogenesis (Figure 1).
Figure 1.
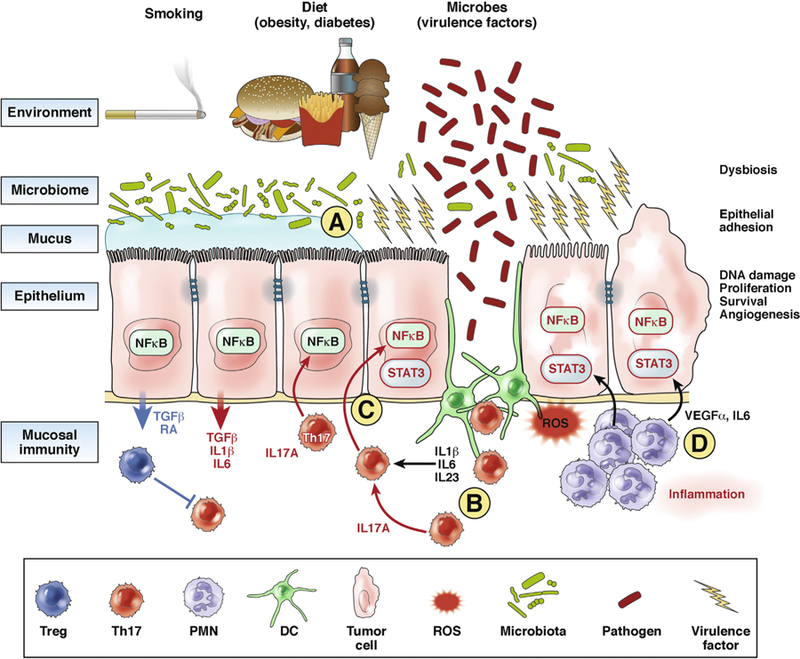
Roles of IL17 in colorectal tumorigenesis. Risk factors for CRC include obesity, diet, smoking, diabetes, and inflammatory bowel diseases, which all modify the colon microbiome. (A) Specific microbes or communities are likely to deliver specific virulence factors to CECs, which induce—perhaps through mucus modification or CEC signaling—changes in epithelial barrier function, leading to a low level of mucosal inflammation. (B) Increased permeability of the colon mucosal barrier leads to inflammation. In the case of IL17-mediated mucosal immune responses, access of the microbiota to the myeloid compartment, including dendritic cells (DCs), as well as epithelial cell production of IL1β, transforming growth factor β (TGFβ), and IL6, are required for activation of a Th17 cell–mediated response. IL17A likely contributes to colon tumor development (not clear whether other IL17 family members do as well). (C) Production of IL17A, along with activation of STAT3, NF-κB, and signaling pathways in the CEC, promote CEC transformation.15 (D) Myeloid cells produce a small amount of IL17 in the mucosa or in the tumor microenvironment, and larger amounts of tumor growth factors, such as vascular endothelial growth factor (VEGF) and IL6. These growth factors, along with mutations in CECs (caused by microbes, spontaneous errors in DNA replication, or inflammation byproducts such as reactive oxygen species [ROS] and nitric oxide species) lead to transformation of CECs. PMN, polymorphonuclear cells; RA, retinoic acid; Treg, T-regulatory cell.
It will be important to determine how IL17A, which mediates antibacterial and antifungal immune responses and promotes epithelial repair and regeneration, contributes to carcinogenesis in the colon and other tissues. It is possible that chronic mucosal production of IL17A, in response to specific microbes or community consortia, alters signaling pathways in CECs or induces changes in DNA structure or mutations that contribute to their transformation. Transformation of CECs often involves activation of STAT3 and NF-κB, but it is not clear how IL17A might facilitate these changes. Studies are needed to determine whether specific mutations in CECs or alterations in chromatin structure, combined with exposure to specific cytokines or immune system cells, induce transformation. It is important to determine whether specific microbes or microbial communities, and the immune responses they induce in the colonic mucosa, are associated with risk of CRC.
Acknowledgments
The authors thank Drew Pardoll for his editorial input and members of their laboratories for their scientific contributions.
Funding
This work was supported by National Institutes of Health grants R01CA196845 (CLS), R01CA179440 (CLS), R01GM111682 (FW), R01CA203891 (FH), Bloomberg-Kimmel Institute for Cancer Immunotherapy (CLS, FH), Commonwealth Pilot Project (CLS, FH), Hopkins-Bristol Myer Squibb Immunotherapy Collaboration (CLS, FH), Cancer Research Institute/Fight CRC (CLS, FH).
Abbreviations used in this paper:
- CEC
colonic epithelial cell
- CRC
colorectal cancer
- DSS
dextran sodium sulfate
- ETBF
enterotoxigenic Bacteroides fragilis
- IFNG
interferon gamma
- IL17
interleukin 17
- IL17R
interleukin 17 receptor
- NF-κB
nuclear factor-κB
- NK
natural killer
- pks
polyketide synthase
- STAT3
signal transducer and activator of transcription 3
- Tfh
T-follicular helper
- Th
T-helper
Footnotes
Conflicts of interest
The authors disclose the following: FH and CLS are supported, in part, by a research grant provided by Bristol Myer Squibb.
References
- 1.Douaiher J, Ravipati A, Grams B, et al. Colorectal cancer-global burden, trends, and geographical variations. J Surg Oncol 2017;115:619–630. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 3.Williams R, White P, Nieto J, et al. Colorectal cancer in African Americans: an update. Clin Transl Gastroenterol 2016;7:e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–767. [DOI] [PubMed] [Google Scholar]
- 5.Su LK, Kinzler KW, Vogelstein B, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 1992; 256:668–670. [DOI] [PubMed] [Google Scholar]
- 6.Fodde R, Edelmann W, Yang K, et al. A targeted chain-termination mutation in the mouse Apc gene results in multiple intestinal tumors. Proc Natl Acad Sci U S A 1994;91:8969–8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinoi T, Akyol A, Theisen BK, et al. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res 2007;67:9721–9730. [DOI] [PubMed] [Google Scholar]
- 8.Feng Y, Sentani K, Wiese A, et al. Sox9 induction, ectopic Paneth cells, and mitotic spindle axis defects in mouse colon adenomatous epithelium arising from conditional biallelic Apc inactivation. Am J Pathol 2013; 183:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grivennikov SI, Wang K, Mucida D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012; 491:254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okayasu I, Ohkusa T, Kajiura K, et al. Promotion of colorectal neoplasia in experimental murine ulcerative colitis. Gut 1996;39:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greten FR, Eckmann L, Greten TF, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 2004;118:285–296. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka T, Kohno H, Suzuki R, et al. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci 2003;94:965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dieleman LA, Ridwan BU, Tennyson GS, et al. Dextran sulfate sodium-induced colitis occurs in severe combined immunodeficient mice. Gastroenterology 1994; 107:1643–1652. [DOI] [PubMed] [Google Scholar]
- 14.Hu B, Elinav E, Huber S, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci U S A 2013;110:9862–9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung L, Thiele Orberg E, Geis AL, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 2018;23:203–214 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Housseau F, Wu S, Wick EC, et al. Redundant innate and adaptive sources of IL17 production drive colon tumorigenesis. Cancer Res 2016;76: 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 2009; 15:1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chae WJ, Gibson TF, Zelterman D, et al. Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proc Natl Acad Sci U S A 2010; 107:5540–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyun YS, Han DS, Lee AR, et al. Role of IL-17A in the development of colitis-associated cancer. Carcinogenesis 2012;33:931–936. [DOI] [PubMed] [Google Scholar]
- 20.Arthur JC, Perez-Chanona E, Muhlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012;338:120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velcich A, Yang W, Heyer J, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 2002;295:1726–1729. [DOI] [PubMed] [Google Scholar]
- 22.Kucherlapati MH, Lee K, Nguyen AA, et al. An Msh2 conditional knockout mouse for studying intestinal cancer and testing anticancer agents. Gastroenterology 2010;138:993–1002 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wojciechowicz K, Cantelli E, Van Gerwen B, et al. Temozolomide increases the number of mismatch repair-deficient intestinal crypts and accelerates tumorigenesis in a mouse model of Lynch syndrome. Gastroenterology 2014;147:1064–1072 e5. [DOI] [PubMed] [Google Scholar]
- 24.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006; 313:1960–1964. [DOI] [PubMed] [Google Scholar]
- 25.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013; 39:782–795. [DOI] [PubMed] [Google Scholar]
- 26.Tran E, Ahmadzadeh M, Lu YC, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015;350:1387–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015; 372:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tosolini M, Kirilovsky A, Mlecnik B, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res 2011;71:1263–1271. [DOI] [PubMed] [Google Scholar]
- 30.Wagsater D, Lofgren S, Hugander A, et al. Expression of interleukin-17 in human colorectal cancer. Anticancer Res 2006;26:4213–4216. [PubMed] [Google Scholar]
- 31.Amicarella F, Muraro MG, Hirt C, et al. Dual role of tumour-infiltrating T helper 17 cells in human colorectal cancer. Gut 2017;66:692–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frey DM, Droeser RA, Viehl CT, et al. High frequency of tumor-infiltrating FOXP3(+) regulatory T cells predicts improved survival in mismatch repair-proficient colorectal cancer patients. Int J Cancer 2010;126:2635–2643. [DOI] [PubMed] [Google Scholar]
- 33.Salama P, Stewart C, Forrest C, et al. FOXP3+ cell density in lymphoid follicles from histologically normal mucosa is a strong prognostic factor in early stage colon cancer. Cancer Immunol Immunother 2012; 61:1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung AS, Wu X, Zhuang G, et al. An interleukin-17-mediated paracrine network promotes tumor resistance to anti-angiogenic therapy. Nat Med 2013;19: 1114–1123. [DOI] [PubMed] [Google Scholar]
- 35.Cui G, Yuan A, Goll R, et al. IL-17A in the tumor micro-environment of the human colorectal adenoma-carcinoma sequence. Scand J Gastroenterol 2012; 47:1304–1312. [DOI] [PubMed] [Google Scholar]
- 36.Bedoui SA, Barbirou M, Stayoussef M, et al. Association of interleukin-17A polymorphisms with the risk of colorectal cancer: a case-control study. Cytokine 2018; 110:18–23. [DOI] [PubMed] [Google Scholar]
- 37.Omrane I, Medimegh I, Baroudi O, et al. Involvement of IL17A, IL17F and IL23R polymorphisms in colorectal cancer therapy. PLoS One 2015;10:e0128911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omrane I, Benammar-Elgaaied A. The immune microenvironment of the colorectal tumor: involvement of immunity genes and microRNAs belonging to the TH17 pathway. Biochim Biophys Acta 2015;1856:28–38. [DOI] [PubMed] [Google Scholar]
- 39.Wu P, Wu D, Ni C, et al. γδT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity 2014; 40:785–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Llosa NJ, Geis AL, Thiele-Orberg E, et al. Interleukin-17 and type 17 helper T cells in cancer management and research. Immunotargets Ther 2014;3:39–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018; 359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monin L, Gaffen SL. Interleukin 17 family cytokines: signaling mechanisms, biological activities, and therapeutic implications. Cold Spring Harb Perspect Biol 2018;10. [DOI] [PMC free article] [PubMed]
- 43.Kirchberger S, Royston DJ, Boulard O, et al. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med 2013; 210:917–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhuang Y, Peng LS, Zhao YL, et al. CD8(+) T cells that produce interleukin-17 regulate myeloid-derived suppressor cells and are associated with survival time of patients with gastric cancer. Gastroenterology 2012; 143:951–962 e8. [DOI] [PubMed] [Google Scholar]
- 45.Ivanov II, McKenzie BS, Zhou L, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 2006;126:1121–1133. [DOI] [PubMed] [Google Scholar]
- 46.Tong Z, Yang XO, Yan H, et al. A protective role by interleukin-17F in colon tumorigenesis. PLoS One 2012; 7:e34959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K, Kim MK, Di Caro G, et al. Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity 2014;41:1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Duan Y, Cheng X, et al. IL-17 is associated with poor prognosis and promotes angiogenesis via stimulating VEGF production of cancer cells in colorectal carcinoma. Biochem Biophys Res Commun 2011; 407:348–354. [DOI] [PubMed] [Google Scholar]
- 49.Starnes T, Robertson MJ, Sledge G, et al. Cutting edge: IL-17F, a novel cytokine selectively expressed in activated T cells and monocytes, regulates angiogenesis and endothelial cell cytokine production. J Immunol 2001;167:4137–4140. [DOI] [PubMed] [Google Scholar]
- 50.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014;15:317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiele Orberg E, Fan H, Tam AJ, et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol 2017;10:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drewes JL, White JR, Dejea CM, et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 2017;3:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dejea CM, Wick EC, Hechenbleikner EM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A 2014; 111:18321–18326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dejea CM, Fathi P, Craig JM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018; 359:592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bagheri N, Azadegan-Dehkordi F, Shirzad H, et al. The biological functions of IL-17 in different clinical expressions of Helicobacter pylori-infection. Microb Pathog 2015;81:33–38. [DOI] [PubMed] [Google Scholar]
- 56.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009;139:485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan TG, Sefik E, Geva-Zatorsky N, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci U S A 2016;113:E8141–E8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atarashi K, Tanoue T, Ando M, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 2015;163:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cremonesi E, Governa V, Garzon JFG, et al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut 2018;67:1984–1994. [DOI] [PubMed] [Google Scholar]
- 60.Colliou N, Ge Y, Sahay B, et al. Commensal Propioni-bacterium strain UF1 mitigates intestinal inflammation via Th17 cell regulation. J Clin Invest 2017;127:3970–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee JS, Tato CM, Joyce-Shaikh B, et al. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity 2015;43:727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Terzic J, Grivennikov S, Karin E, et al. Inflammation and colon cancer. Gastroenterology 2010;138:2101–2114.e5. [DOI] [PubMed] [Google Scholar]
- 63.Reynolds JM, Lee YH, Shi Y, et al. Interleukin-17B antagonizes interleukin-25-mediated mucosal inflammation. Immunity 2015;42:692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song X, Gao H, Lin Y, et al. Alterations in the microbiota drive interleukin-17C production from intestinal epithelial cells to promote tumorigenesis. Immunity 2014;40:140–152. [DOI] [PubMed] [Google Scholar]
- 65.Saddawi-Konefka R, Seelige R, Gross ET, et al. Nrf2 induces IL-17D to mediate tumor and virus surveillance. Cell Rep 2016;16:2348–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Sullivan T, Saddawi-Konefka R, Gross E, et al. Interleukin-17D mediates tumor rejection through recruitment of natural killer cells. Cell Rep 2014;7: 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Camelo A, Barlow JL, Drynan LF, et al. Blocking IL-25 signalling protects against gut inflammation in a type-2 model of colitis by suppressing nuocyte and NKT derived IL-13. J Gastroenterol 2012; 47:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McHenga SS, Wang D, Janneh FM, et al. Differential dose effects of recombinant IL-25 on the development of dextran sulfate sodium-induced colitis. Inflamm Res 2010;59:879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caruso R, Sarra M, Stolfi C, et al. Interleukin-25 inhibits interleukin-12 production and Th1 cell-driven inflammation in the gut. Gastroenterology 2009; 136:2270–2279. [DOI] [PubMed] [Google Scholar]
- 70.Mlecnik B, Bindea G, Angell HK, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 2016;44:698–711. [DOI] [PubMed] [Google Scholar]
- 71.Mlecnik B, Van den Eynde M, Bindea G, et al. Comprehensive intrametastatic immune quantification and major impact of immunoscore on survival. J Natl Cancer Inst 2018;110. [DOI] [PubMed]
- 72.Chaput N, Louafi S, Bardier A, et al. Identification of CD8+CD25+Foxp3+ suppressive T cells in colorectal cancer tissue. Gut 2009;58:520–529. [DOI] [PubMed] [Google Scholar]
- 73.Sinicrope FA, Rego RL, Ansell SM, et al. Intraepithelial effector (CD3+)/regulatory (FoxP3+) T-cell ratio predicts a clinical outcome of human colon carcinoma. Gastroenterology 2009;137:1270–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Q, Feng M, Yu T, et al. Intratumoral regulatory T cells are associated with suppression of colorectal carcinoma metastasis after resection through overcoming IL-17 producing T cells. Cell Immunol 2014; 287:100–105. [DOI] [PubMed] [Google Scholar]
- 75.Deschoolmeester V, Baay M, Van Marck E, et al. Tumor infiltrating lymphocytes: an intriguing player in the survival of colorectal cancer patients. BMC Immunol 2010;11:19. [DOI] [PMC free article] [PubMed] [Google Scholar]


