Abstract
Background
DNA damage repair (DDR) alterations are important events in cancer initiation, progression, and therapeutic resistance. However, the involvement of DDR alterations in glioma malignancy needs further investigation. This study aims to characterize the clinical and molecular features of gliomas with DDR alterations and elucidate the biological process of DDR alterations that regulate the cross talk between gliomas and the tumor microenvironment.
Methods
Integrated transcriptomic and genomic analyses were undertaken to conduct a comprehensive investigation of the role of DDR alterations in glioma. The prognostic DDR-related cytokines were identified from multiple datasets. In vivo and in vitro experiments validated the role of p53, the key molecule of DDR, regulating M2 polarization of microglia in glioma.
Findings
DDR alterations are associated with clinical and molecular characteristics of glioma. Gliomas with DDR alterations exhibit distinct immune phenotypes, and immune cell types and cytokine processes. DDR-related cytokines have an unfavorable prognostic implication for GBM patients and are synergistic with DDR alterations. Overexpression of MDK mediated by p53, the key transcriptional factor in DDR pathways, remodels the GBM immunosuppressive microenvironment by promoting M2 polarization of microglia, suggesting a potential role of DDR in regulating the glioma microenvironment.
Interpretation
Our work suggests that DDR alterations significantly contribute to remodeling the glioma microenvironment via regulating the immune response and cytokine pathways.
Fund
This study was supported by: 1. The National Key Research and Development Plan (No. 2016YFC0902500); 2. National Natural Science Foundation of China (No. 81702972, No. 81874204, No. 81572701, No. 81772666); 3. China Postdoctoral Science Foundation (2018M640305); 4. Special Fund Project of Translational Medicine in the Chinese-Russian Medical Research Center (No. CR201812); 5. The Research Project of the Chinese Society of Neuro-oncology, CACA (CSNO-2016-MSD12); 6. The Research Project of the Health and Family Planning Commission of Heilongjiang Province (2017–201); and 7. Harbin Medical University Innovation Fund (2017LCZX37, 2017RWZX03).
Keywords: DNA damage repair, Microglia, Glioma microenvironment, p53, Midkine
Abbreviations: BER, base excision repair; CCL2, C-C motif chemokine ligand 2; CGGA, The Chinese Glioma Genome Atlas dataset; ChIP, Chromatin immunoprecipitation; CNV, copy number variation; DAB, Diaminobenzidine; DAPI, 4′,6-diamidino-2-phenylindole; DDR, DNA damage response; DMEM, Dulbecco's Modified Eagle Medium; DR, direct repair; DSBR, DNA double-strand break repair; ELISA, Enzyme-Linked Immunosorbent Assay; EM/PM classification, EGFR module / PDGFRA (platelet derived growth factor receptor A) -based molecular classification; FA, Fanconi anemia; FBS, fetal bovine serum; FC, fold change; GBM, glioblastoma; GCM, glioma-conditioned medium; GME, glioma microenvironment; GO, gene ontology; GSEA, gene set enrichment analysis; HR, homologous recombination; IF, immunofluorescence; IGV, Integrative Genomics Viewer; IHC, immunohistochemistry; KEGG, Kyoto Encyclopedia of Genes and Genomes; LGG, lower grade glioma; MDK, midkine; MEM, Minimum Essential Media; MGMT, O6-methylguanine-DNA methyltransferase; MMR, mismatch repair; NER, nucleotide excision repair; NFκB, nuclear factor kappa B; NHEJ, nonhomologous end joining; PBS, phosphate buffered saline; Pen/Strep, penicillin/streptomycin; PI, protease inhibitor; qRT-PCR, Quantitative real-time reverse transcription-polymerase chain reaction; SNP, single nucleotide polymorphism; SSBR, DNA single-strand break repair; ssGSEA, single sample gene set enrichment analysis; TBST, Tris-buffered Saline with Tween 20; TCGA, The Cancer Genome Atlas dataset; TLS, trans-lesion synthesis; TMZ, temozolomide; TNFA, tumor necrosis factor alpha; γH2AX, phosphorylated on serine 139 on H2A histone family member X; AKT, protein kinase B; ATM, ataxia telangiectasia-mutated gene; TP53, tumor protein P53; ATR, Ataxia Telangiectasia And Rad3-Related Protein; C5, Complement C5; IL6, interleukin 6; TNFSF4, TNF superfamily member 4; SAA1, serum amyloid A1; VEGFA, vascular endothelial growth factor A; HRP, horseradish peroxidase; Arg1, arginase 1; Fizz1, found in inflammatory zone 1; Mrc1, mannose receptor C-type 1; IDH, isocitrate dehydrogenase (NADP(+)); ATRX, alpha thalassemia/mental retardation syndrome X-linked; EGFR, epidermal growth factor receptor; PTEN, phosphatase and tensin homolog; NADP, nicotinamide adenine dinucleotide phosphate;; SWI/SNF, SWItch/Sucrose Non-Fermentable.
Highlights
-
•
Gliomas with DNA damage repair alterations had distinct genomic variation spectrum.
-
•
DDR alterations exhibit distinct immune phenotypes, cytokine processes and immune cell types in glioma.
-
•
DDR-related cytokines in GME have an unfavorable prognostic implication for GBM patients.
-
•
P53-mediated midkine expression derived from glioma cells promotes M2 polarization of microglia.
Research in context.
Evidence before this study
DNA damage repair (DDR) pathways regulate cell stress responses, epithelial cell fate, and tissue integrity to remodel the microenvironment. The maintenance of genome stability is a major challenge faced by cells continually exposed to endogenous and exogenous factors that generate DNA damage. Glioma cells have evolved mechanisms to recognize and repair DNA damage, and defects in this process can lead to disease. Treatment of human cells with anticancer drugs that promote double strand DNA breaks could lead to the induction of immune response cytokines by impact on immune cells function. The DDR alterations induced by intrinsic or extrinsic factors can affect a variety of immune response, such as innate immunity, lymphocyte development and humoral immunity. However, the involvement of DDR alterations in glioma malignancy needs further investigation.
Added value of this study
We conducted a comprehensive investigation of DDR alterations in glioma based on integrated transcriptomic and genomic analyses and explored the clinical and molecular characteristics of gliomas with DNA damage repair alterations. In glioma, DDR alterations were associated with clinical and molecular characteristics, exhibiting distinct immune phenotypes, cytokine processes and immune cell types. DDR-related cytokines have an unfavorable prognostic implication for GBM patients. The DDR key molecule p53 transcriptionally up-regulated MDK expression to promote M2 polarization of microglia, suggesting a potential role of DDR in regulating the glioma microenvironment.
Implications of all the available evidence
These results suggest that DDR alterations significantly contribute to remodeling the glioma microenvironment via regulating the immune response and cytokine pathways.
Alt-text: Unlabelled Box
1. Introduction
Gliomas remain the most common malignant primary brain tumors in the central nervous system and are characterized by multiple somatic mutations and aberrant activation of inflammatory responses [1]. Despite aggressive treatment with surgical resection, radiotherapy, and chemotherapy, the tumors tend to ultimately recur [2]. Most of the current treatments cause different kinds of DNA damage in glioma cells. As cells are continually exposed to endogenous and exogenous factors that generate DNA damage, the maintenance of genome stability is a major challenge faced by the cells. Glioma cells have evolved mechanisms to recognize and repair DNA damage, known as the DNA damage response (DDR), and defects in this process can lead to disease [3]. DDR is composed of eight canonical pathways: mismatch repair (MMR), nucleotide excision repair (NER), base excision repair (BER), nonhomologous end joining (NHEJ), homologous recombination (HR), Fanconi Anemia (FA), trans-lesion synthesis (TLS) and direct repair (DR) [4]. The DDR can affect a variety of immune response, such as innate immunity, lymphocyte development and humoral immunity [5,6].
The critical links between the microenvironment and the DDR pathways regulate cell stress responses, epithelial cell fate, and tissue integrity [7]. DDR-pathways inhibition together with inactivation of homologous recombination mediated by Rad50 might be a reasonable strategy for improving the effectiveness of temozolomide (TMZ) treatment in malignant glioma [8]. Our previous study showed that after treatment with the DDR pathways inhibitor KU-55933, TMZ induced higher levels of γH2AX, indicating more TMZ-related DNA damage [9]. DNA damage repair protein ATM controls insulin-mediated signaling, which in turn regulates AKT signaling and promotes glioblastoma (GBM) cell growth [10]. p53 gain-of-function and the downstream phosphorylation of ATM in the DNA damage process upregulate C—C motif chemokine ligand 2 (CCL2) and tumor necrosis factor alpha (TNFA) expression via nuclear factor kappa B (NFκB) signaling. Consequently, microglia and monocyte-derived immune cell infiltration increases, and this positively correlates with tumor-associated immunity in patients with glioblastoma [11].
In the present study, we conducted a comprehensive investigation of DDR alterations in glioma based on integrated transcriptomic and genomic analyses. We observed that the clinical and molecular characteristics of gliomas with DNA damage repair alterations exhibit distinct immune phenotypes, immune cell types and cytokine processes in gliomas. DDR-related cytokines have an unfavorable prognostic implication for GBM patients and are synergistic with DDR alterations. Overexpression of MDK mediated by p53, the key transcriptional factor in DDR pathways, remodels the GBM immunosuppressive microenvironment by promoting M2 polarization of microglia, suggesting a potential role of DDR in regulating the glioma microenvironment.
2. Materials and methods
2.1. DDR gene set compilation
The gene set of the canonical DDR pathways, including DR, MMR, NER, BER, HR, NHEJ and FA factors along with checkpoint genes, was built using the Molecular Signature Database [12,13], the KEGG pathway-specific genes and the updated table of DDR-pathways genes listed at http://sciencepark.mdanderson.org/labs/wood/dna_repair_genes.html. Additionally, the trans-lesion DNA synthesis (TLS) pathway genes were left out from the current analysis because of their ambiguous role in SSBR and DSBR [14].
2.2. Data collection and tissue specimens
Three transcriptome datasets from patients diagnosed with glioma (WHO II-IV) were used. The datasets used were The Chinese Glioma Genome Atlas (CGGA, 545 samples) dataset (http://www.cgga.org.cn), The Cancer Genome Atlas (TCGA, 702 samples) dataset (http://cancergenome.nih.gov/) and the Rembrandt dataset (https://caintegrator.nci.nih.gov/rembrandt/, 444 samples). The copy number variation (CNV) profile and somatic mutation data were obtained online (http://cancergenome.nih.gov/). The ChIP-seq data were obtained from the GSM2944126, GSM2944127, GSM2296271 and GSM2296277 datasets. The TP53 microarray expression dataset was obtained from the GSE60813 dataset. The clinical samples were confirmed by two pathologists. Informed consent was obtained from patients involved in this study, and the study protocol was approved by the Clinical Research Ethics Committee of the Second Affiliated Hospital of Harbin Medical University. The clinical and molecular characteristics of samples in the TCGA, Rembrandt and CGGA datasets are recorded in Table S1.
2.3. Cells and reagents
The human microglial clone 3 cell line, HMC3 (Dr. J. Pocock, University College London), was established in the laboratory of Prof. Tardieu in 1995 [15]. HMC3 expresses microglial and macrophage surface markers and shows a distinct response of cytokines and chemokines in contact to pathogens [[16], [17], [18]]. The cells were cultured in Minimum Essential Media (MEM) (Thermo Fisher Scientific, Darmstadt, Germany) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, Taufkirchen, Germany) and 100 units/ml (U/ml) penicillin/streptomycin (Pen/Strep, Invitrogen, Darmstadt, Germany) in T-75 flasks (PRIMARIA™ Tissue Culture Flask, Becton Dickinson, Heidelberg, Germany). The cells were passaged at a confluency of 80%. For experiments, cells were plated in 24-well plates (10,000 cells/well) (Sarstedt, Nümbrecht, Germany) 24 h before coculture experiments or treatment with pharmacological substances. The LN229 human GBM cells were cultured in DMEM/F12 medium with 10% FBS. The BV-2 mouse microglial cell line was cultured in Dulbecco's Modified Eagle Medium (DMEM) with 10% FBS. The GL261 tumor cells were maintained in DMEM supplemented with 10% FBS, 2 mM l-glutamine, and 1% penicillin–streptomycin (Solarbio, China).
The HG7 cells were obtained from a female adult patient with GBM. The tumor tissue was washed in phosphate buffered saline (PBS) and minced to 1 mm3 [9]. Then, the tumor tissue was enzymatically dissociated with 0.05% trypsin. Finally, the tumor cells were suspended in culture medium. All cells were cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 (Corning, Armonk, NY, USA) supplemented with 10% fetal bovine serum (FBS, BD Biosciences, San Jose, CA, USA) and 1% antibiotics (Sigma, St. Louis, MO, USA) at 37 °C in a humidified atmosphere with 5% CO2 and 95% air.
2.4. Cell transfection
Cells for transfection were seeded in 6-well plates at 70–80% confluence. For human MDK overexpression, plasmid containing the human MDK sequence (NM_001270550, Genechem, Shanghai, China) was transfected into LN229 and HG7 cells with using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. For mouse Mdk overexpression, the plasmid contained the mouse Mdk sequence (NM_001012335, Genechem, Shanghai, China) was transfected into GL261 cells. After 6 h the supernatant was removed and changed to fresh culture medium and cells were selected by neomycin for 7 days. The overexpression of human MDK and mouse Mdk was validated by Western blot assays. For TP53 knockdown, siRNAs targeting human TP53 (stQ0002017–1, RIBOBIO, China) were transfected into LN229R and HG7R cells with using Lipofectamin 2000 as previously described.
2.5. Establishment of TMZ-resistant cells
The LN229, HG7 and GL261 cells were seeded into 96-well plates at 6000 cells per well, and the half maximal inhibitory concentration (IC50) of TMZ was determined. TMZ was then added to the cell culture medium at an IC50 1/50 concentration (LN229/0.83 ± 0.26 mM, HG7/1.48 ± 0.14 mM) to cultures of LN229, HG7 and GL261 cells in 6-well plates. After the cells grew stably, the drug dose was increased in multiples. Each dose was maintained for 15 days until the end of the fifth month. The induced TMZ-resistant cells were named LN229R, HG7R and GL261R.
2.6. Microarray analysis
RNA expression profiling was performed using Agilent custom human lncRNA and mRNA microarrays (Biotechnology Corporation, Shanghai, China). The raw data were normalized using the quantile algorithm from the limma package in R. Heatmaps representing differentially regulated genes were generated using Cluster 3.0 and R.
2.7. Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed using the Chromatin Immunoprecipitation Assay Kit (Millipore, 17–295) and the anti-p53 antibody (Abcam, ab1101) following the manufacturer's protocol. Briefly, 4 × 107 cells were fixed by 1% formaldehyde for 15 min, then crosslinking was stopped by adding 0.125 M glycine for 5 min. The cells in each tube were resuspended in 1 ml of cell lysis buffer (with PI), vortexed every 3 min for a total of 15 min and centrifuged at 300 ×g for 5 min, and then the supernatant was aspirated. Nuclei were resuspended in 2000 μl of Nuclei Lysis buffer (with PI) and sonicated on ice until chromatin fragments were approximately 250 to 500 bp in size, as detected with agarose gel electrophoresis. A 50 μl aliquot of sonicated samples was mixed with ChIP dilution buffer and PI in a total volume of 500 μl. Five μl of each sample was transferred into a new tube to be used as 1% input and stored at −80 °C. Three μg of anti-p53 primary antibody and 25 μl of magnetic protein A/G beads were added to each sample before overnight rotation at 4 °C. The DNA was purified and rehydrated using the kit, and then the sample was analyzed by qPCR.
2.8. Luciferase assay
Genomic DNA fragments of the human midkine gene, spanning from +1 to −3000 relative to the transcription initiation site, were generated by PCR and inserted into pGL3-Basic vectors (denoted as pGL3–midkine). LN229, LN229R, HG7 and HG7R cells were transfected with midkine reporters according to the manufacturer protocol. Transfection was done with pRL construct containing Renilla reniformis luciferase gene, which was used as normalizing control. Luciferase assays were performed using Dual Luciferase Assay System (Promega). Relative luciferase activity was defined as the ratio of firefly luciferase activity to R. reniformis luciferase activity. Error bars represented standard deviation of the three experiments.
2.9. Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted using TRIzol reagent (TaKaRa, Otsu, Japan). The cDNAs were synthesized with a PrimeScript RT reagent kit (TaKaRa) according to manufacturer's instructions. The endogenous levels of MDK mRNA and DNA in the p53-precipitated protein/DNA complex were determined using the SYBR PrimeScript RT-PCR Kit (Roche, Roswell, GA, USA). The qRT-PCR data were analyzed using the 2-△△Ct method. PCR primers were designed and synthesized using a primer designing tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/), and the primer sequences are listed in Table S2.
2.10. Western blot assay
Cells were scraped and collected in Thermo Fisher Scientific RIPA buffer (Solarbio) with 1% protease inhibitors. After centrifugation at 140,000 rpm for 30 min at 4 °C, the total cellular protein concentration was measured with a NanoDrop 2000C spectrophotometer (NanoDrop Products) according to the manufacturer's instructions. All samples were subjected to 7.5%/10%/12.5% sodium dodecyl sulfate polyacrylamide gel (EpiZyme Scientific) electrophoresis, and the gels were transferred onto PVDF membranes (Millipore, USA). The membranes were blocked in a 5% milk-TBST solution and incubated separately with rabbit anti-ATR (1:1000, Abcam, ab2905), rabbit anti-ATM (1:1000, Abcam, ab78), mouse anti-p53 (1:1000, Abcam, ab26), mouse anti-RAD50 (1:1000, Abcam, ab89), rabbit anti-γH2AX (1:1000, Abcam, ab2893), rabbit anti-C5 (1:500, ABclonal, A8104), rabbit anti-IL6 (1:500, ABclonal, A0286), rabbit anti-TNFSF4 (1:500, Abcam, ab156285), rabbit anti-human SAA1 (1:1000, Abcam, ab207445), rabbit anti-mouse SAA1 (1:1000, Abcam, ab199030), rabbit anti-MDK (1:1000, Abcam, ab170820), mouse anti-VEGFA (1:500, Abcam, ab1316) and mouse anti-GAPDH (1:1000, Abcam, ab8245). Following incubation with HRP-labeled mouse IgG secondary antibodies (1:4000, Invitrogen, #31430) and HRP-labeled rabbit IgG secondary antibodies (1:4000, Invitrogen, #31460), the protein bands were detected with the SuperEnhanced chemiluminescence detection reagents (Applygen Technologies Inc) in a ChemiDocTM MP Imaging System (BioRad).
2.11. Enzyme-linked immunosorbent assay (ELISA)
The levels of human C5 (Abcam, ab125963), SAA1 (Abcam, ab100635), IL6 (Abcam, ab46027), TNFSF4 (Abcam, ab213842), MDK (Abcam, ab193761) and VEGFA (Abcam, ab119566) were measured in the supernatant of human glioma cells using ELISA kits. The levels of mouse C5 (Abcam, ab193718), Saa1 (Abcam, ab157723), Il6 (Abcam, ab100712), Tnfsf4 (Abcam, ab193729), Mdk (LifeSpan BioSciences, LS-F5765) and Vegfa (Abcam, ab119565) were measured in the supernatant of mouse glioma cells using ELISA kits according to the manufacturer's instructions.
2.12. Confocal immunofluorescence (IF) microscopy
Cells were grown on coverslips overnight. Then cover cells to a depth of 2–3 mm with 4% formaldehyde diluted in warm PBS for 15 min in room temperature. Then aspirate fixative, rinse three times in 1 × PBS for 5 min each. After treating with 0.5% Triton-X100 (ThermoFisher, USA) diluted in warm PBS and blocking specimen in blocking buffer (5% Bovine Serum Albumin diluted in warm PBS, BioFroxx, Guangzhou, China) for 60 min in room temperature, primary antibodies were applied for 4 C overnight. AlexaFluor488- conjugated anti-mouse IgG (1:1000, Molecular Probes, A11029), AlexaFluor488-conjugated anti-rabbit IgG (1:1000, Molecular Probes, A11034) or AlexaFluor594-conjugated anti-rabbit IgG (1:1000, Molecular Probes, A11037) secondary antibodies were used in room-temperature for 1 h. Nuclei were counterstained with DAPI (1 μg/ml, Molecular Probes, D1306). Protein subcellular localization was analyzed under a Zeiss 510 META or Leica TCS-SP2 confocal laser scanning microscope. The concentration of primary antibody was rabbit anti-CD68 (1:200, Abcam, ab125212), rabbit anti-CD163 (1:200, Proteintech, 16,646–1-AP), rabbit anti-CD204 (1:200, Abcam, ab123946), rabbit anti-CD206 (1:200, Abcam, ab64693), mouse anti-Arg1 (1:200, Abcam, ab239731), rabbit anti-Mrc1 (1:200, Abcam, ab64693), rabbit anti-Fizz1 (1:200, Abcam, ab39626).
2.13. Tumor xenograft study
Six- to seven-week-old female wild-type C57BL/6 mice were purchased from the Chinese Academy of Medical Science (Beijing, China) Animal Center and housed in conventional pathogen-free conditions.
GL261, GL261R, GL261-Scr and GL261-OE cells were injected into the intracranial site of female nude mice as described [19,20]. All mice were sacrificed 6 weeks after tumor formation. These procedures were carried out following approval by the Harbin Medical University Institutional Animal Care and Use Committee.
2.14. Immunohistochemistry (IHC) assay
We obtained 10 paraffin-embedded GBM tissues from patients who provided informed consent under an Institutional Ethics Committee-approved study from the Second Affiliated Hospital of Harbin Medical University. The IHC assay was described in our previous work [21]. Briefly, the slices were incubated with primary antibody rabbit anti-C5 (1:100, ABclonal, A8104), rabbit anti-IL6 (1:200, ABclonal, A0286), mouse anti-TNFSF4 (1:200, Abcam, ab89896), rabbit anti-TNFSF4 (1:200, ThermoFisher, #MA5–17910), rabbit anti-SAA1 (1:200, Abcam, ab207445), rabbit anti-SAA1 (1:200, Abcam, ab199030), rabbit anti-MDK (1:200, Abcam, ab170820), mouse anti-VEGFA (1:200, Abcam, ab1316), rabbit anti-CD68 (1:200, Abcam, ab125212), rabbit anti-CD163 (1:200, Proteintech, 16,646–1-AP), rabbit anti-CD204 (1:200, Abcam, ab123946), rabbit anti-CD206 (1:200, Abcam, ab64693), mouse anti-Arg1 (1:200, Abcam, ab239731), rabbit anti-Fizz1 (1:200, Abcam, ab39626) and rabbit anti-Mrc1 (1:200, Abcam, ab64693) for 12 h at 4 °C. Then the slides were incubated with HRP labeled anti-mouse/anti-rabbit IgG secondary antibody (Boster, SV0004) at 37 °C for 30 min. After washing with PBS three times for 5 min each, the slices were stained with Diaminobenzidine (DAB, ZSGB-BIO, ZLI9018) for 2 min, rinsed in PBS and counterstained with hematoxylin.
Quantitative evaluation was performed by examining each section using at least 10 different high-power fields with the most abundant stained cells as our previous description [22,23]. Each stained slide was individually reviewed and scored by the two independent neuropathologists in a double-blinded fashion. The proportion of stained cells counts per field for each patient was used for statistical analysis. Staining was scored using a 4-point scale from “-” to “+++”, with “-” if there was no staining or very little staining, “+” if <10% of cells stained positively, “++” if 10%–30% of cells stained positively, and “+++” if >30% of cells stained positively [24]. The staining scores of cytokines were classified into four categories (“-”, “+”, “++” and “+++”) based on the staining intensity (none, weak, moderate, or strong) [25]. The representative imaged field were determined by the average method.
2.15. Statistical analysis
The differences in variables were assessed by Student's t-test for two group comparisons or one-way analysis of variance (one-way ANOVA) for comparisons of at least three groups (WHO grade and histological types as the basal levels, age at diagnosis and gender as other conditions). Comparisons of binary and categorical patient characteristics between subgroups were performed via the Chi-squared test. Single-sample GSEA (ssGSEA) [26] was used to calculate the enrichment score of every gene set for every sample. In this manner, ssGSEA projects a single sample's genes expression profile from the space of single genes onto the space of every gene set [27,28]. Each enrichment score represents the degree to which the genes in every gene set are coordinately up- or down-regulated within a sample. The Kaplan–Meier survival curve and log-rank tests were used to describe the survival distributions and assess statistical significance between the two groups. Cox regression analysis was performed with SPSS 24.0. GISTIC2.0 was used to assess CNVs associated with the DDR clusters. A GISTIC value of less than −1 or more than 1 was defined as a deletion or amplification, respectively. The gene ontology (GO) analysis was performed when the genes (r > 0.4) were submitted to the DAVID website (http://david.abcc.ncifcrf.gov/home.jsp). The cytokine genes were annotated on the Molecular Signature Database of the GSEA website (http://software.broadinstitute.org/gsea/index.jsp). Cytoscape [29] was used to visualize the significance of genes associated with prognostic survival. The Integrative Genomics Viewer (IGV) was used to visualize the ChIP-seq data [30]. R packages, such as consensus, ComplexHeatmap, pHeatmap, ggplot2 and corrplot, were used to produce figs. P values <.05 were considered statistically significant.
3. Results
3.1. Gliomas with DDR alterations exhibit distinct clinicopathological characteristics, immune phenotypes and cytokine processes
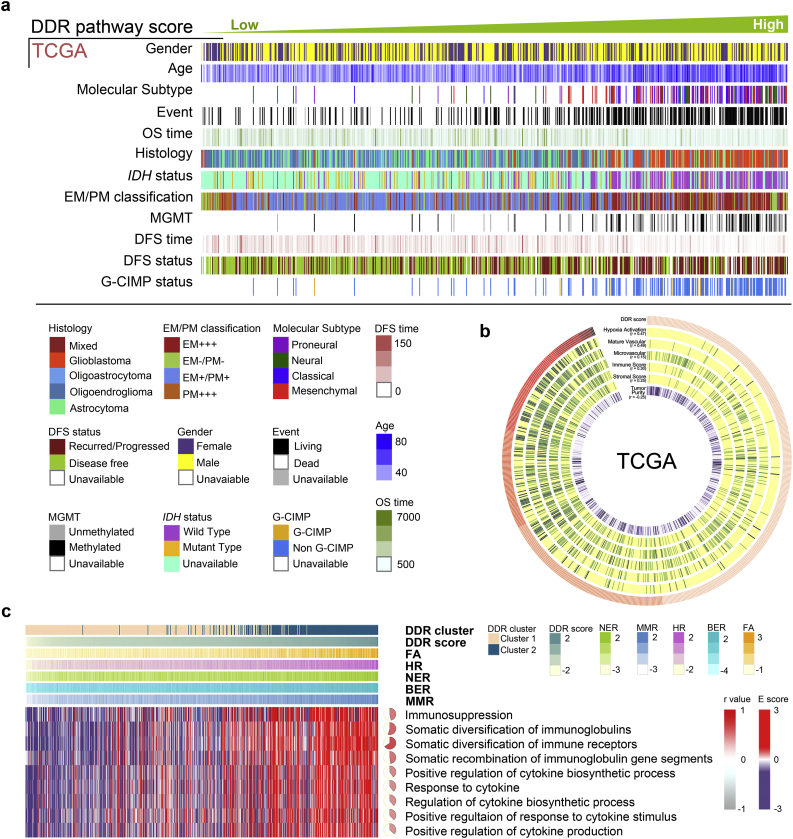
The clinical and molecular characteristics of samples in the TCGA (Fig. 1a), Rembrandt and CGGA datasets (Fig. S1) were arranged according to the ascending order of their DDR scores. Low DDR scores indicated better prognosis (P = 4.25E-25, HR = 0.25), lower WHO grades (P = 6.36E-262) and IDH mutant status (P = 7.00E-142). In contrast, high DDR scores were associated with an unfavorable prognosis, higher WHO grades and IDH wild-type status. For a better demonstration, samples were divided into two groups within each dataset: lower grade gliomas (LGG) and GBM (Fig. S2). In the TCGA LGG group (530 samples), low DDR scores were still correlated with better prognosis (P = 2.16E-7, HR = 0.41), lower WHO grades (P = 8.02E-100) and IDH mutant status (P = 7.98E-07). In the TCGA GBM group (172 samples), low DDR scores were correlated with MGMT methylated status (P = 1.99E-07) and EM/PM classification (P = 3.10E-12). The results were validated in the analyses of the Rembrandt and CGGA datasets.
Fig. 1.
DDR alterations were associated with clinicopathological characteristics, immune phenotypes and cytokine processes in glioma.
(a) Samples from the TCGA dataset were arranged in ascending order according to their DDR scores. The relationship between the DDR scores and patients' characteristics were depicted. (b) The heatmap showed that TME signatures including glioma purity, stromal score, immune score, hypoxia activation, microvascular and mature vascular pathways were correlated with the DDR scores in the TCGA dataset. (c) Samples in the TCGA dataset were arranged in ascending order according to their DDR scores. The heatmap showed that the DDR scores were related to immunosuppression and cytokine-associated pathways. The Pearson correlation was calculated between the DDR scores and immunosuppression or cytokine-associated pathways.
DDR gene expression was used as a quantitative measure for determining two clusters within each dataset. With the consensus-based recommendations, samples were separated into two clusters (Fig. S3). The occurrence of the 1p/19q codeletion, a genomic hallmark of oligodendroglioma [31], decreased as DDR scores increased. However, Chr7 amplification accompanied by Chr10 loss, a representative event of GBM [32], was positively correlated with higher DDR scores (Fig. S4a). The raw copy number heatmap showed that the 1p/19q codeletion was more frequent in the DDR cluster1 samples compared to the DDR cluster2 samples, however the Chr7 amplification accompanied by Chr10 loss occurred more frequently in the DDR cluster2 samples than in the DDR cluster1 samples (Fig. S4b, c). These analytical results were also converted to q values, and DDR cluster1 and cluster2 were visualized separately with line plots according to chromosome order (Fig. S4b, Table S3, Table S4). We further examined the functional consequences of glioma mutations (Fig. S4d), including IDH1 (P = 5.19E−96), TP53 (P = 2.35E−03), ATRX (P = 6.53E−13), EGFR (P = 6.91E−38) and PTEN (P = 3.26E−21) [[33], [34], [35], [36], [37]], which might disrupt the DNA damage response, apoptosis, or senescence pathways such as altering histone methylation, leading to genetic instability, impairing nonhomologous end joining, and alternate lengthening of telomeres [9,[38], [39], [40]].
To further delineate the common principles of the microenvironment variation associated with alterations in DDR, we identified that the microenvironment gene signatures [41] included hypoxia activation signature (P < .0001 in TCGA, P < .0001 in Rembrandt, P = .0228 in CGGA-Agilent, P < .0001 in CGGA-RNAseq), mature vascular signature (P < .0001 in TCGA, P = .0037 in Rembrandt, P < .0001 in CGGA-Agilent, P = .285 in CGGA-RNAseq) and microvascular signature (P < .0001 in TCGA, P < .0001 in Rembrandt, P < .0001 in CGGA-Agilent, P < .0001 in CGGA-RNAseq) were positive correlated with the DDR scores (Fig. 1b, Fig. S5a). The DDR scores were positively correlated with immunosuppression (Fig. 1c, r = 0.47, P < .0001), somatic diversification of immune receptors (r = 0.66, P < .0001), positive regulation of cytokine biosynthetic process (r = 0.4, P < .0001) and response to cytokines (r = 0.39, P < .0001). In the Rembrandt and CGGA datasets, the DDR scores were also positively correlated with immunosuppression (Fig. S5b, r = 0.32, P < .0001 in Rembrandt, r = 0.36, P < .0001 in CGGA-Agilent and r = 0.37, P < .0001 in CGGA-RNAseq), the diversification of immune receptors (r = 0.9, P < .0001 in Rembrandt, r = 0.86, P < .0001 in CGGA-Agilent and r = 0.89, P < .0001 in CGGA-RNAseq), regulation of the cytokine biosynthetic process (r = 0.35, P < .0001 in Rembrandt) and positive regulation of response to cytokine stimulus (r = 0.34, P < .0001 in CGGA-Agilent and r = 0.49, P < .0001 in CGGA-RNAseq).
3.2. Estimation of immune cell types and cytokines in glioma tissues with DNA damage repair alterations
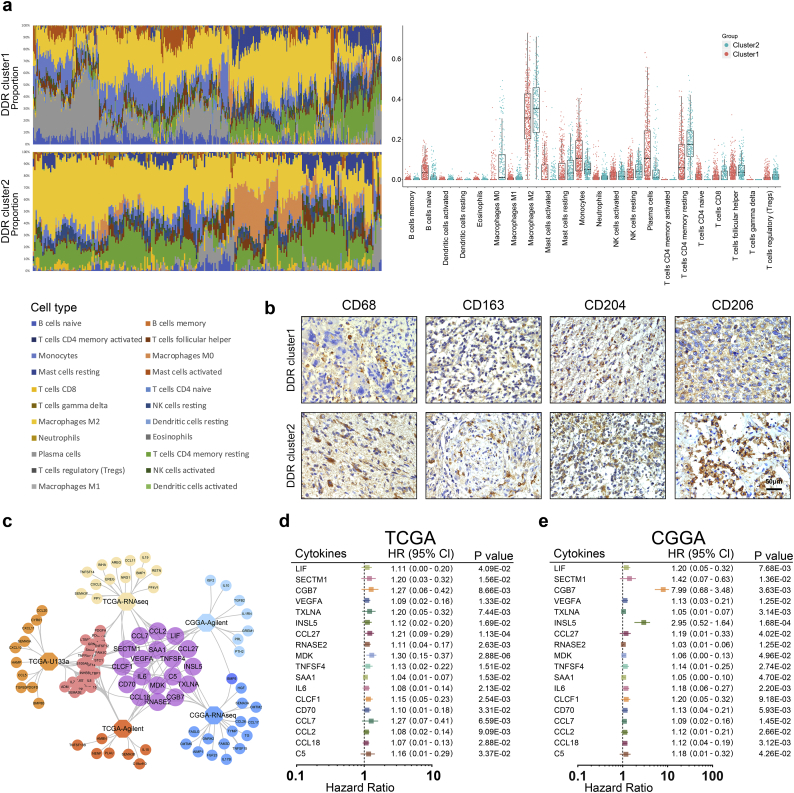
We investigated whether distinct patterns based on the 22 immune cell proportions could be discerned within the DDR clusters by conducting hierarchical clustering of all samples. The relative proportions of 22 immune cell types between the DDR clusters were estimated using CIBERSORT [42] (http://cibersort.stanford.edu/). Normalized gene expression data were used to infer the relative proportions of 22 types of infiltrating immune cells. The proportions of 22 immune cells were depicted with stacked bar charts and their distributions with box plots in a summary of all TCGA samples (Fig. 2a). In the TCGA dataset, M2 macrophages had a larger proportion in DDR cluster2 than in DDR cluster1, and the average score of cluster2 was higher than cluster1 (P = 1.89E-03). This indicated that tumor-associated M2 macrophages were more frequent in the DDR cluster2 samples compared with the DDR cluster1 samples. This result was also validated with analyses of the Rembrandt and CGGA datasets (Fig. S6a-c, P = 1.75E−05 in Rembrandt, P = 4.16E−04 in CGGA-Agilent, P = 1.42E−01 in CGGA-RNAseq).
Fig. 2.
Gliomas with DDR alterations exhibit distinct infiltrating immune cells phenotypes and cytokines expressions.
(a) CIBERSORT scores were estimated with normalized gene expression data to infer the relative proportions of twenty-two types of immune cells. The proportions of twenty-two immune cells were depicted in a summary of all TCGA samples by stacked bar charts and their distributions were represented in box plots. (b) IHC assays were performed to detect the M2 microglia markers CD68, CD163, CD204 and CD206 in clinical glioma tissues (20×). (c) Overlap of cytokines associated with poor outcomes in the TCGA and CGGA datasets. (d-e) Cox regression analyses of DDR-related cytokines in GME for the patients with GBM in the TCGA and CGGA datasets.
To further validate the results in clinical tissue samples, we used IHC to detect the phenotypes of microglia, which are tissue-resident macrophages in the CNS [43,44]. The expression of γH2AX was used to represent the DDR alterations for each patient [3] (Fig. S6d, P = .0111). CD68 (P = .0356), CD163 (P = .0299), CD204 (P = .0394), and CD206 (P = .0341), characterized as M2 macrophage markers [[45], [46], [47]], had higher expression levels in DDR cluster2 than in DDR cluster1 samples (Fig. 2b, Fig. S6e).
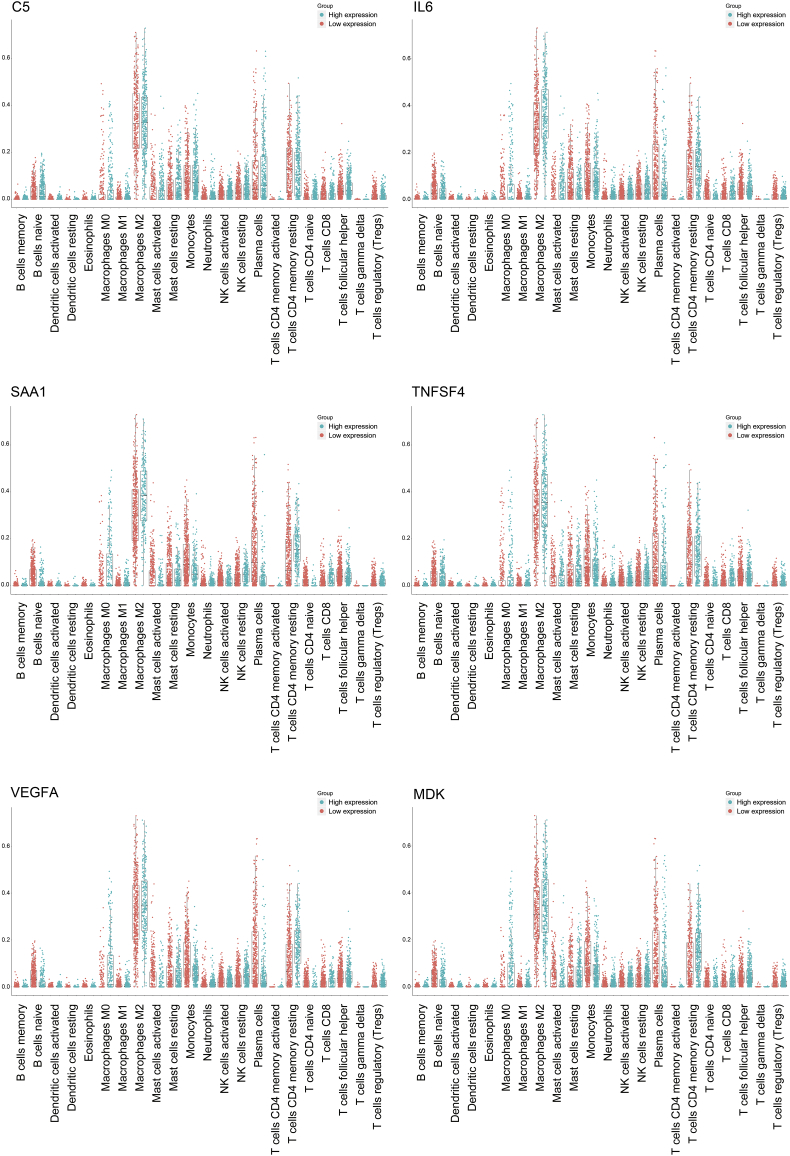
The expression levels of all cytokine genes from the GSEA website was depicted in heatmaps arranged in order of the DDR clusters or the DDR scores (Fig. S7). To further evaluate the correlation between cytokine expression and DDR alterations, we analyzed cytokine genes with either differential expression (FC > 1.2) between the DDR clusters or with positive correlation with the DDR scores (r > 0.2) in the TCGA, Rembrandt and CGGA datasets (Fig. S8). We estimated the survival of patients with GBM across the TCGA, Rembrandt and CGGA datasets. In the TCGA-U133a dataset, 40 of 175 cytokines were significantly correlated with poor prognosis (HR > 1, P < .05). In the TCGA-Agilent dataset, 39 of 201 cytokines were significantly correlated with poor prognosis (HR > 1, P < .05). In the TCGA RNA-seq dataset, 33 of 217 cytokines were significantly correlated with poor prognosis (HR > 1, P < .05). In the CGGA datasets, patients with high expression of cytokines had poor prognosis according to the low expression groups (20 of 213 genes in CGGA-Agilent, 25 of 175 genes in CGGA-RNAseq). Further, we compared the cytokines associated with poor prognosis in the TCGA and CGGA datasets to identify 18 DDR-related cytokines in GME (Fig. 2c). The Cox regression analysis and Kaplan-Meier curves demonstrated that high expression of these DDR-related cytokines in GME indicated an unfavorable prognosis for patients with GBM in the TCGA and CGGA datasets (Fig. 2d-e, Fig. S9a–b).
3.3. DDR-related cytokines derived from glioma cells are synergistic with DDR alterations in vitro and in vivo
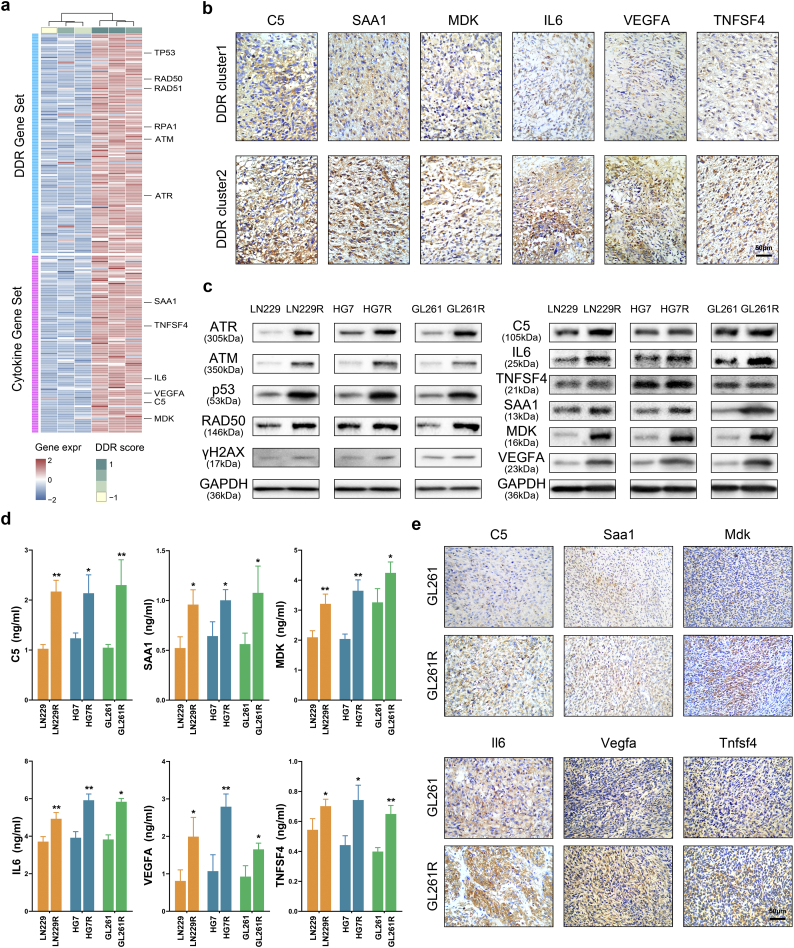
We had established TMZ resistant glioma cells, LN229R, HG7R and GL261R. Compared with LN229, HG7 and GL261 cells (parental cells), TMZ resistant glioma cells had a higher level of the TMZ half maximal inhibitory concentration (IC50) (Fig. S10a, P = 6.29E-05 LN229 vs. LN229R, P = 2.54E-03 HG7 vs. HG7R, P = 9.31E-03 GL261 vs. GL261R). We used gene expression profiling in TMZ-resistant glioma cells and observed that TMZ-resistant glioma cells showed higher DDR scores compared with parental cells (Fig. 3a, P = 4.19E-02). In TCGA dataset, the DDR scores in recurrent tumors are higher than those in new diagnosed tumors (Fig. S10b, P = 1.82E-03). DNA damage repair molecules, such as p53, ATR, ATM and RAD50, were also validated to have higher expression in TMZ-resistant glioma cells than in parental cells, demonstrating that the DDR pathways are altered in TMZ-resistant cells. In addition, six of eighteen DDR-related cytokines (C5, SAA1, TNFSF4, IL6, MDK and VEGFA) in GME were positively correlated with the DDR scores in glioma cells (Fig. 3a, Table S5). In the TCGA and CGGA datasets, higher expression of these six cytokines also correlated with increased malignancy in gliomas (Fig. S11a-c, Table S6). The IHC assay demonstrated that patients in cluster2 (with higher expression levels of γH2AX, as described above) had increased protein expression of DDR-related cytokines derived from glioma cells (Fig. 3b, Fig. S11d). Western blot analyses showed that the higher expression of DDR-related genes was synergistic with higher expression of DDR-related cytokines in glioma cells (Fig. 3c). ELISA assays showed that DDR-related cytokine expression was increased in LN229R, HG7R and GL261R supernatants (Fig. 3d). With xenograft gliomas constructed with the GL261 and GL261R glioma cells, we detected that the expression of six DDR-related cytokines in gliomas formed by the GL261R line was higher than in gliomas formed by the GL261 line (Fig. 3e, Fig. S11e).
Fig. 3.
DDR-related cytokines derived from glioma cells are synergistic with DDR alterations in vitro and in vivo.
(a) Gene expression profiling revealed the associations between DDR-related cytokines and the DDR scores or DDR genes. (b) IHC assays were used to detect the expression level of six cytokines, C5, SAA1, MDK, IL6, VEGFA and TNFSF4 in glioma samples within the different DDR clusters (20×). (c) Western blot validation of DDR-associated protein and cytokine expression levels in LN229, LN229R, HG7, HG7R, GL261 and GL261R cells. (d) ELISA assay revealed differential expression of six cytokines in the medium of LN229, LN229R, HG7, HG7R, GL261 and GL261R cells. (e) Cytokine expression in xenograft gliomas formed by GL261 and GL261R cells was detected by IHC assays (20×).
The occurrence of the 1p/19q codeletion decreased as the cytokine scores increased, and was more frequent in cytokine cluster1 than in the cytokine cluster2, estimated by the same method as DDR clusters and DDR scores. However, the Chr7 amplification accompanied by Chr10 loss was positively correlated with higher cytokine scores, and occurred more frequently in cytokine cluster2 than in cytokine cluster1 (Fig. S12, Table S7, Table S8). We also examined the differential proportions of mutations in IDH1 (P = 2.33E-73), TP53 (P = 8.77E-07), ATRX (P = 6.92E-22), EGFR (P = 2.66E-36) and PTEN (P = 3.63E-25) in gliomas in the two clusters (Fig. S13). The cytokine scores demonstrated that high cytokine scores were associated with gliomas characterized by the Mesenchymal subtype, IDH wild-type status and GBM (Fig. S14a-c, Table S9).
3.4. Midkine expression transcriptionally mediated by p53 is upregulated in TMZ resistant glioma cells
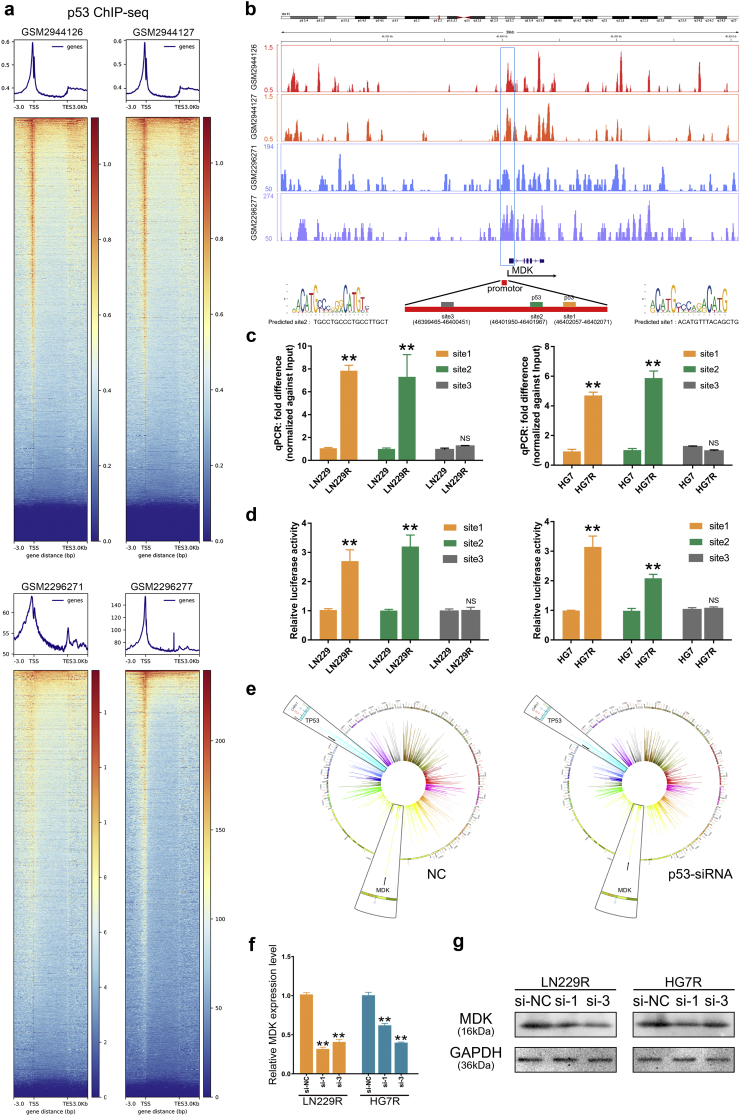
To elucidate the mechanisms through which DDR pathways regulate cytokine expression, we used ChIP-seq data (GSM2944126, GSM2944127, GSM2296271 and GSM2296277) of p53, the key transcriptional factor in DDR pathways. The heatmaps showed all p53 binding sites across the whole genomes of p53 wild-type cells (Fig. 4a). We used the JASPAR database (http://jaspar.genereg.net/) to predict two p53 binding sites (Chr11 46,402,057–46,402,071, Chr11 46,401,950–46,401,967) in the promotor region of the MDK gene (Fig. 4b). Site3 (Chr11 46,399,465–46,400,451) was used as a negative control. The ChIP-qPCR assay validated that p53 protein bound to the predicted site1 and site2 in MDK, and this binding was much higher in LN229R, HG7R and GL261R cells compared with LN229, HG7 and GL261 cells. There were no significant differences in binding at site3 (Fig. 4c). We cloned the predicted sites (site1 and site2) and the negative control (site3) into luciferase reporter plasmids. Cells transfected with promoter fragment (site1 or site2) showed clear induction of luciferase activity in TMZ resistant cells, however cells transfected with negative control fragment had no effect on promoter activation in TMZ resistant cells (Fig. 4d). In the GSE60813 dataset, TP53-knockdown cells had less midkine expression than control cells, further indicating that p53 can transcriptionally regulate the expression of midkine in glioma cells (Fig. 4e). The small interfering RNAs were used to knockdown the p53 expression and observed that they could downregulate MDK expression at mRNA and protein level (Fig. 4f-g, Fig. S14d-e). With transfection of siRNAs (si-1 and si-3) into LN229R, the mRNA expression level of MDK was downregulated relative to si-NC group (P = 3.49E-06 in si-1, P = 1.14E-05 in si-3). The results were similar in HG7R (P = 1.16E-04 in si-1, P = 1.05E-05 in si-3). These results demonstrated that p53 had a transcriptional activation on MDK.
Fig. 4.
P53 transcriptionally upregulates the midkine cytokine in glioma cells.
(a) The heatmap of ChIP-seq data using an antibody against p53 protein. The ChIP-seq data were obtained from the GSM2944126, GSM2944127, GSM2296271 and GSM2296277 datasets. (b) Snapshots of the UCSC genome browser showing the p53 ChIP-seq data at the locus of the MDK gene. Site1 and site2 were predicted by the JASPAR database, and site3 was used as a negative control. (c-d) The ChIP-qPCR assay and the luciferase reporter assay showed that p53 binding in the promotor of the MDK gene was higher in LN229R and HG7R cells compared with the LN229 and HG7 cells. Error bars indicate mean ± SD. ** P < .01; Student's t-test. (e) Circos plots showed the RNA expression levels of TP53 and MDK in p53-NC and p53-siRNA cells. (f-g) Relative mRNA expression level and protein expression level of MDK. In TP53 knockdown groups, the MDK mRNA expression level and protein expression level was lower than those in si-NC group.
3.5. Midkine derived from glioma cells promotes the M2 polarization of microglia in glioma
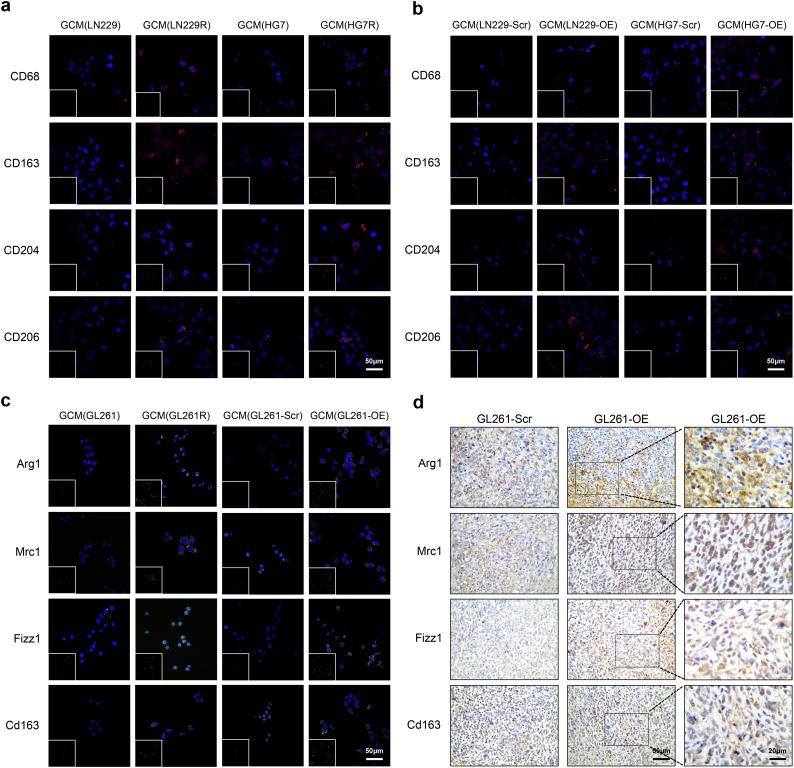
The CIBERSORT analysis detected that the glioma tissues with higher expression of DDR-related cytokines were derived from glioma cells with the M2 macrophage phenotype in the TCGA (Fig. 5, Table S10) and CGGA datasets (Fig. S15, Fig. S16, Tables S11, S12). To determine whether DDR-related cytokines were essential for the microglia polarization phenotype in GME, we performed IF to detect the expression of M2 markers (CD68, CD163, CD204 and CD206 in HMC3 cells and Arg1, Mrc1, Fizz1 [48] and Cd163 [49] in BV-2 cells) (Fig. 6). HMC3 cells cultured with the glioma-conditioned medium (GCM) from LN229R and HG7R cell lines had higher expression of CD68, CD163, CD204 and CD206 than those cultured with GCM from the LN229 and HG7 cell lines. This suggests that the DDR-related cytokines in GME promote human M2 microglia polarization in vitro (Fig. 6a). LN229 (LN229-OE) and HG7 (HG7-OE) glioma cells were transfected to over-express midkine (Fig. S17a-b). The expression of CD68, CD163, CD204 and CD206 in HMC3 cells cultured with GCM from LN229-OE or HG7-OE cells were higher than in those cultured in GCM from LN229-Scr or HG7-Scr (Fig. 6b). The expression of Arg1, Mrc1, Fizz1 and Cd163 was significantly increased in BV-2 cells cultured with GCM from GL261R or GL261-OE compared to those with GCM from GL261 or GL261-Scr (Fig. 6c). An IHC assay validated that tumor tissues formed by GL261-OE had higher expression levels of Arg1, Mrc1, Fizz1 and Cd163 than tumor tissues formed by GL261 cells (Fig. 6d, Fig. S17c. P = 1.86E-02 of Arg1, P = 2.56E-02 of Mrc1, P = 3.02E-02 of Fizz1, P = 2.56E-02 of Cd163). These results revealed that the enhanced levels of midkine secreted by glioma cells also promoted the M2 polarization of HMC3 and BV-2.
Fig. 5.
The association between DDR-related cytokines derived from glioma cells and infiltrating immune cells.
Fig. 6.
Photomicrographs of M2 microglia following GCM stimulation of glioma cells in vitro and in vivo.
(a) The human microglia HMC3 cells were treated with GCM from LN229, LN229R, HG7 and HG7R cells. (b) HMC3 were treated with GCM from LN229-Scr, LN229-OE, HG7-Scr and HG7-OE cells. (c) BV-2 cells were treated with GCM from GL261, GL261R, GL261-Scr and GL261-OE cells. CD68, CD163, CD204 and CD206 were stained in red, while Arg1, Mrc1, Fizz1 and Cd163 were stained in green. Nuclei were stained with DAPI (blue). (d) IHC assays were performed to detect Arg1, Mrc1, Fizz1 and Cd163 in xenograft gliomas formed by GL261-Scr and GL261-OE cells. All images were taken microscopically (20× or 40×). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
DDR plays key roles in maintaining human genomic stability. The maintenance of genome stability is a major challenge faced by cells, as they are continually exposed to endogenous and exogenous factors that generate DNA damage [3]. Conversely, DDR alterations are important determinants of tumor risk, progression, and therapeutic response [40]. Alterations of specific DDR pathways that constitutively activate DDR lead to undesirable outcomes after aggressive treatments for glioma patients with an average overall survival advantage of only 14.6 months after diagnosis [50,51]. However, the mechanism through which DDR promotes glioma malignancy remains unknown.
One hundred and forty genes involved in eight canonical DDR pathways [4,52] were used to calculate the DDR alterations for every sample of TCGA, Rembrandt and CGGA datasets by ssGSEA which estimates the degree of enrichment of DDR pathways in individual samples [12,53]. Besides expression profile, the genetic variations, epigenetic modification and post-transcriptional regulation were also involved in the functional regulations of DDR pathways in cancers [12,14,40,54]. The limitation of the current work is that the assessment of DDR alterations was based on the expression profiles alone. With extended studies on the mutations, epigenetic modification and post-transcriptional regulation of DDR genes, more consequences of DDR alterations on biological processes would need further investigation. It's reported that CNV gain and loss of DDR genes positively correlated with up- and down-regulation of their expression, respectively [55]. Epigenetic silencing of critical genes (e.g., FANCF, BRCA1) through methylation of the promoter region could down-regulate their expression levels [[56], [57], [58]]. Lnc-RI prevents the degradation of RAD51 mRNAs via competitively binding with miR-193a-3p and release of its inhibition of RAD51 expression [59]. Aberrant expression of specific miRNAs might contribute to the variable MGMT expression through post-transcriptional regulation [[60], [61], [62]].
In present study, we observed that low DDR scores indicated lower WHO grades and IDH mutant status, and that high DDR scores showed higher WHO grades and IDH wildtype status. In addition to the clinical features, this study also presents a comprehensive characterization of the molecular landscape of DDR alterations in glioma. Recurrent genomic variations could be involved in the DNA damage repair alterations of glioma. The occurrence of the 1p/19q codeletion, a genomic hallmark of oligodendroglioma, decreased as DDR scores increased. However, Chr7 amplification accompanied by Chr10 loss, a common event in GBM, was positively correlated with higher DDR scores. The somatic mutation spectra based on the DDR clusters revealed that some classical genomic variations were implicated in the DDR alterations. Mutations of IDH1, which reduce NADP+ − dependent IDH activity to cause DNA damage and genome instability in glioma, occurred more frequently in DDR cluster1 (84%) [63]. The chromatin remodeler ATRX, which is one of SWI/SNF-like family proteins, governs genomic stability through the regulation of repetitive sequences [9]. Mutations in ATRX that cause loss of ATRX function were enriched in DDR cluster1. These mutations impair NHEJ activity and result in genetic instability in glioma [64]. TP53 mutations might inhibit the genome caretaker proteins involved in DNA repair and thus evoke genomic instability [34]. EGFR amplification, which was characterized as more frequent in DDR cluster2, activates DNA-dependent protein kinases and stimulates repair of DNA strand breaks [36].
To further investigate the role of DDR in glioma malignancy, we analyzed the common principles of the biological processes associated with DDR and found that DDR alterations exhibit distinct immune phenotypes, cytokine processes and infiltrating immune cell types. The DDR alterations were correlated with immunosuppression, positive regulation of the cytokine biosynthetic process and response to cytokines. DDR alternative tumors appear to avoid host immune-mediated elimination through activation of immunosuppression [65]. The converting immune microenvironment releases a plethora of cytokines and chemokines to communicate with other cells and thereby to orchestrate immune responses [66]. The relative abundance of individual immune cell types differed between DDR alterations [67]. We used the CIBERSORT tool to estimate the abundances of specific immune cell types using gene expression data. The proportion of M2 macrophages is greater in DDR cluster2 than in DDR cluster1, indicating that tumor-associated M2 macrophages were more highly populated in the DDR cluster2 samples compared with the DDR cluster1 samples. We detected M2-phenotypic markers of microglia, the tissue-resident macrophages in the CNS. Microglia can become activated and/or dysregulated in the context of neurodegenerative disease and cancer, and thereby contribute to disease severity [68]. The M2 microglia markers CD68, CD163, CD204, and CD206 were differentially expressed in glioma tissues with DDR alterations.
The DDR has been implicated in activation of the checkpoint induced by the methylating agent TMZ [69]. Critical DNA damage response kinases (e.g., ATM and ATR) are rapidly activated after exposure of cells to TMZ [70]. We observed that the DDR scores of TMZ-resistant glioma cells were higher than that of parental cells. The expression levels of DNA damage response kinases, such as p53, ATR, ATM and RAD50, were higher in TMZ-resistant glioma cells. The effects of TMZ on immunity are variable and result in a decrease of lymphocytes and qualitative dysfunction of T and B cells, contributing to the immunosuppressive environment. TMZ resistance causes the accumulation of immunosuppressive cells which activate immunosuppression through secretion of immunosuppressive cytokines [71]. The DDR-related cytokines (C5, SAA1, TNFSF4, IL6, MDK and VEGFA) were upregulated in TMZ-resistant glioma cells.
C5 is a component of the complement system, which is part of the innate immune system that plays an important role in inflammation, host homeostasis, and host defense against pathogens [72,73]. IL6 amplifies the inflammation response to activate STAT3 and its downstream anti-apoptotic and pro-proliferation genes [74]. TNFSF4 is an important cytokine of the tumor necrosis factor (TNF) ligand family and is associated with susceptibility to systemic sclerosis as well as its clinical and autoantibody subsets [[75], [76], [77]]. SAA1 increases migration and invasion behaviors in glioma cells [78] and binds to several cell surface receptors which are activated during an immune response [79]. Midkine, transactivated by SP1, has biological functions in inducing the glioma cell malignant characteristics [80,81], the induction of macrophage inflammatory proteins [82] and autoimmune responses [83]. VEGFA is a member of the PDGF/VEGF growth factor family [84], which induces proliferation and infiltration of macrophage [85] and contributes to tumor-induced immune suppression [86].
P53, a conventional regulator in DDR alterations, regulates downstream cytokine expression, increases microglia infiltration and converts myeloid cells of the M1-like tumor-suppressing phenotype to the M2-like tumor-promoting phenotype [87]. P53 also reprograms macrophages to a tumor-supportive and anti-inflammatory state [88,89]. We used p53 ChIP-seq data to analyze the p53 binding sites across the whole genomes of p53 wild-type cells and validated two binding sites in the promotor region of the MDK gene. The binding activity was higher in TMZ-resistant glioma cells than in parental cells. Overexpression of midkine stimulates macrophage polarization and contributes to glioma metastases [81,90,91]. After stimulation with GCM of midkine-overexpressing glioma cells, we observed higher expression levels of CD68, CD163, CD204 and CD206 in HMC3 cells and higher expression levels of Arg1, Mrc1, Fizz1 and Cd163 in BV-2 cells. The gliomas formed by GL261-OE have higher protein levels of Arg1, Mrc1, Fizz1 and Cd163, indicating that midkine promotes M2-phenotypic microglia polarization. Our results demonstrated that p53, an important transcription factor in DDR pathways, transactivated MDK to promote communication between glioma cells and microglia via the cytokine process.
In the present work, we observe the clinicopathological characteristics of gliomas with DNA damage repair alterations. Gliomas with DNA damage repair alterations have distinct genomic variation spectrum, and they also exhibit different immune phenotypes, immune cellular types and cytokine processes in gliomas. Overexpression of MDK mediated by p53, the key transcriptional factor in DDR pathways, remodels the GBM immunosuppressive microenvironment by promoting M2 polarization of microglia. Our results suggest a potential role for DDR in regulating the glioma microenvironment and provide promising and realistic targets for glioma immunotherapy.
The following are the supplementary data related to this article.
The clinical and molecular characteristics of samples in the TCGA, Rembrandt and CGGA databases.
qPCR primer sequences.
Q value in line plots calculated by GISTIC 2.0 in the DDR cluster1.
Q value in line plots calculated by GISTIC 2.0 in the DDR cluster2.
The differential expression of DDR related cytokines in GME and DDR related gene.
The correlation between expression of six cytokines and glioma characteristic.
Q value in line plots calculated by GISTIC 2.0 in the cytokine cluster1.
Q value in line plots calculated by GISTIC 2.0 in the cytokine cluster2.
The differential expression of DDR related cytokines in GME and DDR related gene.
The CIBERSORT analysis detected the M2 macrophage phenotype in the TCGA dataset.
The CIBERSORT analysis detected the M2 macrophage phenotype in the CGGA-Agilent dataset.
The CIBERSORT analysis detected the M2 macrophage phenotype in the CGGA-RNAseq dataset
Supplementary material
Conflict of interest
The authors had no conflicts of interest.
Author contributions
Xiangqi Meng, Jinquan Cai and Chuanlu Jiang devised the project, the main conceptual ideas and proof outline. Xiangqi Meng, Chunbin Duan and Jinquan Cai worked out most of the technical details and performed the numerical calculations for the suggested experiment. Hengyuan Pang worked out the immunohistochemistry assays, with help from Qun Chen, Bo Han, and Caijun Zha. Dinislam Magafurov verified the numerical results by an independent implementation. Pengfei Wu, Ziwei Li, Shihong Zhao, Ruijia Wang and Lin Lin proposed the experiment in discussions with Chuanlu Jiang and Jinquan Cai. Xiangqi Meng and Jinquan Cai wrote the manuscript.
Contributor Information
Chuanlu Jiang, Email: jcl6688@163.com.
Jinquan Cai, Email: caijinquan666777@126.com.
References
- 1.Yu Z., Jian Z., Shen S.H., Purisima E., Wang E. Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res. 2007;35(1):152–164. doi: 10.1093/nar/gkl1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doucette T., Rao G., Rao A., Shen L., Aldape K., Wei J. Immune heterogeneity of glioblastoma subtypes: extrapolation from the cancer genome atlas. Cancer Immunol Res. 2013;1(2):112–122. doi: 10.1158/2326-6066.CIR-13-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleiner R.E., Verma P., Molloy K.R., Chait B.T., Kapoor T.M. Chemical proteomics reveals a gammaH2AX-53BP1 interaction in the DNA damage response. Nat Chem Biol. 2015;11(10):807–814. doi: 10.1038/nchembio.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dietlein F., Thelen L., Reinhardt H.C. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014;30(8):326–339. doi: 10.1016/j.tig.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y. DNA damage: a trigger of innate immunity but a requirement for adaptive immune homeostasis. Nat Rev Immunol. 2006;6(4):261–270. doi: 10.1038/nri1804. [DOI] [PubMed] [Google Scholar]
- 6.Kytola V., Topaloglu U., Miller L.D., Bitting R.L., Goodman M.M., Agostino D. Mutational landscapes of smoking-related cancers in caucasians and African Americans: precision oncology perspectives at wake forest baptist comprehensive cancer center. Theranostics. 2017;7(11):2914–2923. doi: 10.7150/thno.20355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akgul B., Goktas C. Gene reporter assay to validate microRNA targets in Drosophila S2 cells. Methods Mol Biol. 2014;1107:233–242. doi: 10.1007/978-1-62703-748-8_14. [DOI] [PubMed] [Google Scholar]
- 8.Grun D., Wang Y.L., Langenberger D., Gunsalus K.C., Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol. 2005;1(1):e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han B., Cai J., Gao W., Meng X., Gao F., Wu P. Loss of ATRX suppresses ATM dependent DNA damage repair by modulating H3K9me3 to enhance temozolomide sensitivity in glioma. Cancer Lett. 2018;419:280–290. doi: 10.1016/j.canlet.2018.01.056. [DOI] [PubMed] [Google Scholar]
- 10.Nakahara K., Kim K., Sciulli C., Dowd S.R., Minden J.S., Carthew R.W. Targets of microRNA regulation in the Drosophila oocyte proteome. Proc Natl Acad Sci U S A. 2005;102(34):12023–12028. doi: 10.1073/pnas.0500053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee G.J., Hyun S. Multiple targets of the microRNA miR-8 contribute to immune homeostasis in Drosophila. Dev Comp Immunol. 2014;45(2):245–251. doi: 10.1016/j.dci.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anurag M., Punturi N., Hoog J., Bainbridge M.N., Ellis M.J., Haricharan S. Comprehensive profiling of DNA repair defects in breast cancer identifies a novel class of endocrine therapy resistance drivers. Clin Cancer Res. 2018;24(19):4887–4899. doi: 10.1158/1078-0432.CCR-17-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janabi N., Peudenier S., Heron B., Ng K.H., Tardieu M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci Lett. 1995;195(2):105–108. doi: 10.1016/0304-3940(94)11792-h. [DOI] [PubMed] [Google Scholar]
- 16.Li B., Bedard K., Sorce S., Hinz B., Dubois-Dauphin M., Krause K.H. NOX4 expression in human microglia leads to constitutive generation of reactive oxygen species and to constitutive IL-6 expression. J Innate Immun. 2009;1(6):570–581. doi: 10.1159/000235563. [DOI] [PubMed] [Google Scholar]
- 17.Nakagawa Y., Chiba K. Diversity and plasticity of microglial cells in psychiatric and neurological disorders. Pharmacol Ther. 2015;154:21–35. doi: 10.1016/j.pharmthera.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Jadhav V.S., Krause K.H., Singh S.K. HIV-1 Tat C modulates NOX2 and NOX4 expressions through miR-17 in a human microglial cell line. J Neurochem. 2014;131(6):803–815. doi: 10.1111/jnc.12933. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q., Cai J., Fang C., Yang C., Zhou J., Tan Y. Mesenchymal glioblastoma constitutes a major ceRNA signature in the TGF-beta pathway. Theranostics. 2018;8(17):4733–4749. doi: 10.7150/thno.26550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q., Cai J., Wang Q., Wang Y., Liu M., Yang J. Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/beta-catenin pathway by scaffolding EZH2. Clin Cancer Res. 2018;24(3):684–695. doi: 10.1158/1078-0432.CCR-17-0605. [DOI] [PubMed] [Google Scholar]
- 21.Wei J.W., Cai J.Q., Fang C., Tan Y.L., Huang K., Yang C. Signal peptide peptidase, encoded by HM13, contributes to tumor progression by affecting EGFRvIII secretion profiles in glioblastoma. CNS Neurosci Ther. 2017;23(3):257–265. doi: 10.1111/cns.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang P., Cai J., Yan W., Zhang W., Wang Y., Chen B. Classification based on mutations of TERT promoter and IDH characterizes subtypes in grade II/III gliomas. Neuro-Oncology. 2016;18(8):1099–1108. doi: 10.1093/neuonc/now021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C., Cheng W., Ren X., Wang Z., Liu X., Li G. Tumor purity as an underlying Key factor in glioma. Clin Cancer Res. 2017;23(20):6279–6291. doi: 10.1158/1078-0432.CCR-16-2598. [DOI] [PubMed] [Google Scholar]
- 24.Lambein K., Van Bockstal M., Vandemaele L., Geenen S., Rottiers I., Nuyts A. Distinguishing score 0 from score 1+ in HER2 immunohistochemistry-negative breast cancer. Am J Clin Pathol. 2013;140(4):561–566. doi: 10.1309/AJCP4A7KTAYHZSOE. [DOI] [PubMed] [Google Scholar]
- 25.Sont J.K., De Boer W.I., van Schadewijk W.A., Grunberg K., van Krieken J.H., Hiemstra P.S. Fully automated assessment of inflammatory cell counts and cytokine expression in bronchial tissue. Am J Respir Crit Care Med. 2003;167(11):1496–1503. doi: 10.1164/rccm.2205003. [DOI] [PubMed] [Google Scholar]
- 26.Verhaak R.G., Tamayo P., Yang J.Y., Hubbard D., Zhang H., Creighton C.J. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123(1):517–525. doi: 10.1172/JCI65833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckel-Passow J.E., Lachance D.H., Molinaro A.M., Walsh K.M., Decker P.A., Sicotte H. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoue S., Li W.Y., Tseng A., Beerman I., Elia A.J., Bendall S.C. Mutant IDH1 downregulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell. 2016;30(2):337–348. doi: 10.1016/j.ccell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanel W., Moll U.M. Links between mutant p53 and genomic instability. J Cell Biochem. 2012;113(2):433–439. doi: 10.1002/jcb.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koschmann C., Lowenstein P.R., Castro M.G. ATRX mutations and glioblastoma: impaired DNA damage repair, alternative lengthening of telomeres, and genetic instability. Mol Cell Oncol. 2016;3(3) doi: 10.1080/23723556.2016.1167158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liccardi G., Hartley J.A., Hochhauser D. Importance of EGFR/ERCC1 interaction following radiation-induced DNA damage. Clin Cancer Res. 2014;20(13):3496–3506. doi: 10.1158/1078-0432.CCR-13-2695. [DOI] [PubMed] [Google Scholar]
- 37.Puc J., Keniry M., Li H.S., Pandita T.K., Choudhury A.D., Memeo L. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell. 2005;7(2):193–204. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 38.Guo C., Pirozzi C.J., Lopez G.Y., Yan H. Isocitrate dehydrogenase mutations in gliomas: mechanisms, biomarkers and therapeutic target. Curr Opin Neurol. 2011;24(6):648–652. doi: 10.1097/WCO.0b013e32834cd415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohba S., Mukherjee J., See W.L., Pieper R.O. Mutant IDH1-driven cellular transformation increases RAD51-mediated homologous recombination and temozolomide resistance. Cancer Res. 2014;74(17):4836–4844. doi: 10.1158/0008-5472.CAN-14-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knijnenburg T.A., Wang L., Zimmermann M.T., Chambwe N., Gao G.F., Cherniack A.D. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep. 2018;23(1):239–254. doi: 10.1016/j.celrep.2018.03.076. [e6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin X., Kim L.J.Y., Wu Q., Wallace L.C., Prager B.C., Sanvoranart T. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat Med. 2017;23(11):1352–1361. doi: 10.1038/nm.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prinz M., Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15(5):300–312. doi: 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- 44.Li Q., Barres B.A. Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol. 2018;18(4):225–242. doi: 10.1038/nri.2017.125. [DOI] [PubMed] [Google Scholar]
- 45.Bouhlel M.A., Derudas B., Rigamonti E., Dievart R., Brozek J., Haulon S. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007;6(2):137–143. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 46.Miron V.E., Boyd A., Zhao J.W., Yuen T.J., Ruckh J.M., Shadrach J.L. M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat Neurosci. 2013;16(9):1211–1218. doi: 10.1038/nn.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soldano S., Pizzorni C., Paolino S., Trombetta A.C., Montagna P., Brizzolara R. Alternatively activated (M2) macrophage phenotype is inducible by endothelin-1 in cultured human macrophages. PLoS One. 2016;11(11) doi: 10.1371/journal.pone.0166433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez F.O., Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klopfleisch R. Macrophage reaction against biomaterials in the mouse model – phenotypes, functions and markers. Acta Biomater. 2016;43:3–13. doi: 10.1016/j.actbio.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 50.Squatrito M., Brennan C.W., Helmy K., Huse J.T., Petrini J.H., Holland E.C. Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell. 2010;18(6):619–629. doi: 10.1016/j.ccr.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wainwright D.A., Chang A.L., Dey M., Balyasnikova I.V., Kim C.K., Tobias a. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clin Cancer Res. 2014;20(20):5290–5301. doi: 10.1158/1078-0432.CCR-14-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nickoloff J.A., Jones D., Lee S.H., Williamson E.A., Hromas R. Drugging the cancers addicted to DNA repair. J Natl Cancer Inst. 2017;109(11) doi: 10.1093/jnci/djx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wick W., Weller M., van den Bent M., Sanson M., Weiler M., von Deimling A. MGMT testing-the challenges for biomarker-based glioma treatment. Nat Rev Neurol. 2014;10(7):372–385. doi: 10.1038/nrneurol.2014.100. [DOI] [PubMed] [Google Scholar]
- 55.Chae Y.K., Anker J.F., Carneiro B.A., Chandra S., Kaplan J., Kalyan A. Genomic landscape of DNA repair genes in cancer. Oncotarget. 2016;7(17):23312–23321. doi: 10.18632/oncotarget.8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Esteller M., Silva J.M., Dominguez G., Bonilla F., Matias-Guiu X., Lerma E. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92(7):564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 57.Broustas C.G., Lieberman H.B. DNA damage response genes and the development of cancer metastasis. Radiat Res. 2014;181(2):111–130. doi: 10.1667/RR13515.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lahtz C., Pfeifer G.P. Epigenetic changes of DNA repair genes in cancer. J Mol Cell Biol. 2011;3(1):51–58. doi: 10.1093/jmcb/mjq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen L., Wang Q., Liu R., Chen Z., Zhang X., Zhou P. LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. 2018;46(2):717–729. doi: 10.1093/nar/gkx1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weller M., Stupp R., Reifenberger G., Brandes A.A., van den Bent M.J., Wick W. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010;6(1):39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 61.Kreth S., Limbeck E., Hinske L.C., Schutz S.V., Thon N., Hoefig K. In human glioblastomas transcript elongation by alternative polyadenylation and miRNA targeting is a potent mechanism of MGMT silencing. Acta Neuropathol. 2013;125(5):671–681. doi: 10.1007/s00401-013-1081-1. [DOI] [PubMed] [Google Scholar]
- 62.Zhang W., Zhang J., Hoadley K., Kushwaha D., Ramakrishnan V., Li S. miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression. Neuro-Oncology. 2012;14(6):712–719. doi: 10.1093/neuonc/nos089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dang L., Yen K., Attar E.C. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol. 2016;27(4):599–608. doi: 10.1093/annonc/mdw013. [DOI] [PubMed] [Google Scholar]
- 64.Koschmann C., Calinescu A.A., Nunez F.J., Mackay A., Fazal-Salom J., Thomas D. ATRX loss promotes tumor growth and impairs nonhomologous end joining DNA repair in glioma. Sci Transl Med. 2016;8(328):328ra28. doi: 10.1126/scitranslmed.aac8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mouw K.W., Goldberg M.S., Konstantinopoulos P.A., D'Andrea A.D. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 2017;7(7):675–693. doi: 10.1158/2159-8290.CD-17-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lacy P., Stow J.L. Cytokine release from innate immune cells: association with diverse membrane trafficking pathways. Blood. 2011;118(1):9–18. doi: 10.1182/blood-2010-08-265892. [DOI] [PubMed] [Google Scholar]
- 67.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H. The immune landscape of cancer. Immunity. 2018;48(4):812–30 e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saijo K., Glass C.K. Microglial cell origin and phenotypes in health and disease. Nat Rev Immunol. 2011;11(11):775–787. doi: 10.1038/nri3086. [DOI] [PubMed] [Google Scholar]
- 69.Caporali S., Falcinelli S., Starace G., Russo M.T., Bonmassar E., Jiricny J. DNA damage induced by temozolomide signals to both ATM and ATR: role of the mismatch repair system. Mol Pharmacol. 2004;66(3):478–491. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 70.Annovazzi L., Caldera V., Mellai M., Riganti C., Battaglia L., Chirio D. The DNA damage/repair cascade in glioblastoma cell lines after chemotherapeutic agent treatment. Int J Oncol. 2015;46(6):2299–2308. doi: 10.3892/ijo.2015.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karachi A., Dastmalchi F., Mitchell D., Rahman M. Temozolomide for immunomodulation in the treatment of glioblastoma. Neuro-Oncology. 2018;20(12):1566–1572. doi: 10.1093/neuonc/noy072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ward P.A. The dark side of C5a in sepsis. Nat Rev Immunol. 2004;4(2):133–142. doi: 10.1038/nri1269. [DOI] [PubMed] [Google Scholar]
- 73.Klos A., Wende E., Wareham K.J., Monk P.N. International union of basic and clinical pharmacology. [corrected]. LXXXVII. Complement peptide C5a, C4a, and C3a receptors. Pharmacol Rev. 2013;65(1):500–543. doi: 10.1124/pr.111.005223. [DOI] [PubMed] [Google Scholar]
- 74.Kong L., Zhou Y., Bu H., Lv T., Shi Y., Yang J. Deletion of interleukin-6 in monocytes/macrophages suppresses the initiation of hepatocellular carcinoma in mice. J Exp Clin Cancer Res. 2016;35(1):131. doi: 10.1186/s13046-016-0412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maizels R.M., Yazdanbakhsh M. Immune regulation by helminth parasites: cellular and molecular mechanisms. Nat Rev Immunol. 2003;3(9):733–744. doi: 10.1038/nri1183. [DOI] [PubMed] [Google Scholar]
- 76.Curry W.T., Jr., Gorrepati R., Piesche M., Sasada T., Agarwalla P., Jones P.S. Vaccination with irradiated autologous tumor cells mixed with irradiated GM-K562 Cells stimulates antitumor immunity and T lymphocyte activation in patients with recurrent malignant glioma. Clin Cancer Res. 2016;22(12):2885–2896. doi: 10.1158/1078-0432.CCR-15-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gourh P., Arnett F.C., Tan F.K., Assassi S., Divecha D., Paz G. Association of TNFSF4 (OX40L) polymorphisms with susceptibility to systemic sclerosis. Ann Rheum Dis. 2010;69(3):550–555. doi: 10.1136/ard.2009.116434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Knebel F.H., Albuquerque R.C., Massaro R.R., Maria-Engler S.S., Campa A. Dual effect of serum amyloid a on the invasiveness of glioma cells. Mediat Inflamm. 2013;2013 doi: 10.1155/2013/509089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ganal S.C., MacPherson A.J. An ambulance for retinol. elife. 2014;3 doi: 10.7554/eLife.04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nobata S., Shinozawa T., Sakanishi A. Truncated midkine induces transformation of cultured cells and short latency of tumorigenesis in nude mice. Cancer Lett. 2005;219(1):83–89. doi: 10.1016/j.canlet.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 81.Luo J., Wang X., Xia Z., Yang L., Ding Z., Chen S. Transcriptional factor specificity protein 1 (SP1) promotes the proliferation of glioma cells by up-regulating midkine (MDK) Mol Biol Cell. 2015;26(3):430–439. doi: 10.1091/mbc.E14-10-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sato W., Kadomatsu K., Yuzawa Y., Muramatsu H., Hotta N., Matsuo S. Midkine is involved in neutrophil infiltration into the tubulointerstitium in ischemic renal injury. J Immunol. 2001;167(6):3463–3469. doi: 10.4049/jimmunol.167.6.3463. [DOI] [PubMed] [Google Scholar]
- 83.Kerzerho J., Adotevi O., Castelli F.A., Dosset M., Bernardeau K., Szely N. The angiogenic growth factor and biomarker midkine is a tumor-shared antigen. J Immunol. 2010;185(1):418–423. doi: 10.4049/jimmunol.0901014. [DOI] [PubMed] [Google Scholar]
- 84.Karkkainen M.J., Petrova T.V. Vascular endothelial growth factor receptors in the regulation of angiogenesis and lymphangiogenesis. Oncogene. 2000;19(49):5598–5605. doi: 10.1038/sj.onc.1203855. [DOI] [PubMed] [Google Scholar]
- 85.Sato W., Kosugi T., Zhang L., Roncal C.A., Heinig M., Campbell-Thompson M. The role of VEGF on glomerular macrophage infiltration in advanced diabetic nephropathy. Lab Investig. 2008;88(9):949–961. doi: 10.1038/labinvest.2008.60. [DOI] [PubMed] [Google Scholar]
- 86.Ohm J.E., Gabrilovich D.I., Sempowski G.D., Kisseleva E., Parman K.S., Nadaf S. VEGF inhibits T-cell development and may contribute to tumor-induced immune suppression. Blood. 2003;101(12):4878–4886. doi: 10.1182/blood-2002-07-1956. [DOI] [PubMed] [Google Scholar]
- 87.Vousden K.H., Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2(8):594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 88.Cooks T., Pateras I.S., Jenkins L.M., Patel K.M., Robles A.I., Morris J. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun. 2018;9(1):771. doi: 10.1038/s41467-018-03224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 90.Mashima T., Sato S., Sugimoto Y., Tsuruo T., Seimiya H. Promotion of glioma cell survival by acyl-CoA synthetase 5 under extracellular acidosis conditions. Oncogene. 2009;28(1):9–19. doi: 10.1038/onc.2008.355. [DOI] [PubMed] [Google Scholar]
- 91.Fernandez-Calle R., Vicente-Rodriguez M., Gramage E., de la Torre-Ortiz C., Perez-Garcia C., Ramos M.P. Endogenous pleiotrophin and midkine regulate LPS-induced glial responses. Neurosci Lett. 2018;662:213–218. doi: 10.1016/j.neulet.2017.10.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The clinical and molecular characteristics of samples in the TCGA, Rembrandt and CGGA databases.
qPCR primer sequences.
Q value in line plots calculated by GISTIC 2.0 in the DDR cluster1.
Q value in line plots calculated by GISTIC 2.0 in the DDR cluster2.
The differential expression of DDR related cytokines in GME and DDR related gene.
The correlation between expression of six cytokines and glioma characteristic.
Q value in line plots calculated by GISTIC 2.0 in the cytokine cluster1.
Q value in line plots calculated by GISTIC 2.0 in the cytokine cluster2.
The differential expression of DDR related cytokines in GME and DDR related gene.
The CIBERSORT analysis detected the M2 macrophage phenotype in the TCGA dataset.
The CIBERSORT analysis detected the M2 macrophage phenotype in the CGGA-Agilent dataset.
The CIBERSORT analysis detected the M2 macrophage phenotype in the CGGA-RNAseq dataset
Supplementary material