Abstract
The KRAB‐ZNF (Krüppel‐associated box domain zinc finger) gene family is composed of a large number of highly homologous genes, gene isoforms, and pseudogenes. The proteins encoded by these genes, whose expression is often tissue‐specific, act as epigenetic suppressors contributing to the addition of repressive chromatin marks and DNA methylation. Due to its high complexity, the KRAB‐ZNF family has not been studied in sufficient detail, and the involvement of its members in carcinogenesis remains mostly unexplored. In this study, we aimed to provide a comprehensive description of cancer‐associated KRAB‐ZNFs using publicly available The Cancer Genome Atlas pan‐cancer datasets. We analyzed 6727 tumor and normal tissue samples from 16 cancer types. Here, we showed that a small but distinctive cluster of 16 KRAB‐ZNFs is commonly upregulated across multiple cancer cohorts in comparison to normal samples. We confirmed these observations in the independent panels of lung and breast cancer cell lines and tissues. This upregulation was also observed for most of the KRAB‐ZNF splicing variants, whose expression is simultaneously upregulated in tumors compared to normal tissues. Finally, by analyzing the clinicopathological data for breast and lung cancers, we demonstrated that the expression of cancer‐associated KRAB‐ZNFs correlates with patient survival, tumor histology, and molecular subtyping. Altogether, our study allowed the identification and characterization of KRAB‐ZNF factors that may have an essential function in cancer biology and thus potential to become novel oncologic biomarkers and treatment targets.
Keywords: breast cancer, cancer, epigenetics, KRAB‐ZNF, lung cancer, TCGA
Abbreviations
- ACC
adrenocortical carcinoma
- BLCA
bladder urothelial carcinoma
- BRCA
breast invasive carcinoma
- CESC
cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
cholangiocarcinoma
- COAD
colon adenocarcinoma
- DLBC
lymphoid neoplasm diffuse large B‐cell lymphoma
- ESCA
esophageal carcinoma
- GBM
glioblastoma multiforme
- HNSC
head and neck squamous cell carcinoma
- KAP1
KRAB‐associated protein‐1
- KICH
kidney chromophobe
- KIPAN
pan‐kidney cohort
- KIRC
kidney renal clear cell carcinoma
- KIRP
kidney renal papillary cell carcinoma
- KRAB‐ZNF
Krüppel‐associated box domain zinc finger
- LGG
brain lower grade glioma
- LIHC
liver hepatocellular carcinoma
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- OV
ovarian serous cystadenocarcinoma
- PAAD
pancreatic adenocarcinoma
- PCPG
pheochromocytoma and paraganglioma
- PRAD
prostate adenocarcinoma
- READ
rectum adenocarcinoma
- SARC
sarcoma
- STAD
stomach adenocarcinoma
- STES
stomach and esophageal carcinoma
- TCGA
The Cancer Genome Atlas
- TGCT
testicular germ cell tumors
- THCA
thyroid carcinoma
- THYM
thymoma
- TRIM28
Tripartite‐motif containing 28
- UCEC
uterine corpus endometrial carcinoma
- UCS
uterine carcinosarcoma
- UVM
uveal melanoma
1. Introduction
Carcinogenesis is a complex process in which normal cells acquire pathologic properties, such as sustained proliferation, disrupted apoptosis, dysregulation of metabolism, and escape from the immune system control (Hanahan and Weinberg, 2011). The process is driven mainly by the aberrant functioning of mutated genes. However, epigenetic changes, such as DNA methylation and histone modifications, play a pivotal role in the initiation and progression of cancer. Overall, the DNA methylation level in cancer cells is decreased, which can contribute to the activation of oncogenes. Simultaneously, tumor suppressor genes (TSGs) can be inactivated by promoter hypermethylation (Llinas‐Arias and Esteller, 2017). Alterations to the DNA methylation profile frequently correlate with changes affecting chromatin state. CpG island hypermethylation in cancer cells is associated with a decrease in histone active marks: histone H3 and H4 acetylation, H3K4 trimethylation, and gain of repressive marks: H3K9me3 and H3K27me3 (Llinas‐Arias and Esteller, 2017). Moreover, the expression of many chromatin modifiers is often deregulated in the tumor (Dawson and Kouzarides, 2012). Yet, the exact mechanisms involved in the regulation of the epigenetic landscape in cancer cells remain poorly characterized.
Krüppel‐associated box domain zinc finger proteins (KRAB‐ZNFs) are the most abundant family of epigenetic repressors found only in tetrapod vertebrates (Lupo et al., 2013; Ma et al., 2014). The human genome harbors over 800 KRAB‐ZNF transcripts encoding functional proteins, splicing variants, and pseudogenes (Corsinotti et al., 2013; Huntley et al., 2006) that may be involved in development, metabolism, proliferation, and carcinogenesis (Ecco et al., 2017; Lupo et al., 2013). Upon binding to DNA, KRAB‐ZNFs trigger transcriptional repression through interaction with KAP1 (KRAB‐associated protein‐1), also known as TRIM28 (Tripartite‐motif containing 28). TRIM28 acts as a scaffold for a multimolecular entity comprising the heterochromatin protein 1 (HP1), the H3K9me3‐specific histone methyltransferase SETDB1, and the histone deacetylase‐containing complex NuRD. Together, these proteins silence transcription by triggering the formation of heterochromatin (Czerwinska et al., 2017; Ecco et al., 2017; Groner et al., 2010). Moreover, we and others have shown that at least in a stem cell context, KRAB‐ZNFs may mediate methylation of the DNA sequences adjacent to their binding motifs (Oleksiewicz et al., 2017; Quenneville et al., 2011; Wiznerowicz et al., 2007).
A growing number of reports indicate that certain members of the KRAB‐ZNF family are involved in various aspects of carcinogenesis. For example, ZNF545 negatively controls cellular proliferation, and its expression is reduced in multiple malignancies due to promoter hypermethylation (Xiang et al., 2017; Xiao et al., 2014). A tumor suppressor role was also described for ZBRK1 (Chen et al., 2015; Lin et al., 2010) and ZNF307 (Liang et al., 2017). Another member of the KRAB‐ZNF family, ZNF224, was demonstrated to exert both tumor suppressor and oncogenic functions depending on the cellular context. Specifically, in a chronic myelogenous leukemia cell line, ZNF224 was shown to augment the signaling of Wilms’ Tumor 1 (WT1), which is a known TSG that controls the expression of genes involved in differentiation, apoptosis, and cell cycle progression (Florio et al., 2010). In contrast, in bladder cancer and chronic lymphocytic leukemia, ZNF224 serves as an oncogene that mediates enhanced cell growth and resistance to apoptosis and chemotherapy (Busiello et al., 2017; Cho et al., 2016; Harada et al., 2010). Oncogenic properties were also reported for ZKSCAN3. This KRAB‐ZNF is upregulated in bladder cancer, while its knockdown induces apoptosis and reduces the viability of cancer cells in in vitro and in vivo experiments (Kawahara et al., 2016). Interestingly, most reports demonstrate that KRAB‐ZNFs act as TSGs and only a few studies focus on their tumor promotion potential. It is highly likely, however, that more KRAB‐ZNF factors are involved in carcinogenesis. Thus, there is an urgent need to search for such genes and to systematically characterize their functions in a cancer setting.
In the current study, we aimed to identify and characterize KRAB‐ZNF factors specific to cancer by exploring the KRAB‐ZNF expression signature in multiple cancer types. To this end, we used the transcriptomic datasets for various patient cohorts deposited in The Cancer Genome Atlas (TCGA) repositories. Our analysis allowed for identification of a small but distinct cluster of cancer‐associated KRAB‐ZNFs, whose expression becomes commonly upregulated in many tumors. Furthermore, we showed that their expression correlates with various clinicopathological features, including patient survival, as well as histological and molecular subtypes. Such properties point toward a putative role of these KRAB‐ZNFs in tumor biology. If this is the case, KRAB‐ZNFs may become novel targets of anticancer therapies or biomarkers for clinical patient management. Nevertheless, such a hypothesis requires further experimental testing. Altogether, these observations provide grounds for future studies exploring KRAB‐ZNF functions related to carcinogenesis.
2. Materials and methods
2.1. TCGA datasets
RNA‐seq and clinicopathological data (data release: 2015‐11‐01) were downloaded from: http://gdac.broadinstitute.org/runs/stddata__2015_11_01/data/. A snapshot of the data from this release is available in the RTCGA package on Bioconductor 10.18129/B9.bioc.RTCGA website (https://www.bioconductor.org/packages/release/bioc/html/RTCGA.html). Counts for RNA‐Seq reads for each gene isoform were downloaded from the TCGA database (https://cancergenome.nih.gov/abouttcga/aboutdata/datalevelstypes). Gene isoforms listed on the official TCGA releases were used. The patient cohorts used were as follows: bladder urothelial carcinoma [BLCA, Normal (N): 19, Tumor (T): 408], breast invasive carcinoma (BRCA, N: 112, T: 1093), cholangiocarcinoma (CHOL, N: 9, T: 36), esophageal carcinoma (ESCA, N: 11, T: 184), head and neck squamous cell carcinoma (HNSC, N: 44, T: 520), kidney chromophobe (KICH, N: 25, T: 66), kidney renal clear cell carcinoma (KIRC, N: 72, T: 533), kidney renal papillary cell carcinoma (KIRP, N: 32, T: 290), liver hepatocellular carcinoma (LIHC, N: 50, T: 371), lung adenocarcinoma (LUAD, N: 59, T: 515), lung squamous cell carcinoma (LUSC, N: 51, T: 501), prostate adenocarcinoma (PRAD, N: 52, T: 497), stomach and esophageal carcinoma (STES, N: 15, T: 221), thyroid carcinoma (THCA, N: 59, T: 501), and uterine corpus endometrial carcinoma (UCEC, N: 11, T: 370). KRAB‐ZNF isoform expression across various tumor types was also tested in the additional cohorts, including: adrenocortical carcinoma (ACC, n = 79), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC, n = 304), colon adenocarcinoma (COAD, n = 285), COADREAD (n = 379), lymphoid neoplasm diffuse large B‐cell lymphoma (DLBC, n = 15), glioblastoma multiforme (GBM, n = 153), GBMLGG (n = 669), brain lower grade glioma (LGG, n = 516), ovarian serous cystadenocarcinoma (OV, n = 303), pancreatic adenocarcinoma (PAAD, n = 178), pheochromocytoma and paraganglioma (PCPG, n = 179), rectum adenocarcinoma (READ, n = 94), sarcoma (SARC, n = 259), SKCM (n = 103), stomach adenocarcinoma (STAD, n = 37), testicular germ cell tumors (TGCT, n = 150), thymoma (THYM, n = 120), uterine carcinosarcoma (UCS, n = 57), and uveal melanoma (UVM; n = 80). Complete names for each cohort are provided in the Abbreviations section.
2.2. Independent validation set of lung and breast cancer tissue samples
Lung cancer and matched normal tissues were collected from 58 patients operated on between 1995–2005 at the Liverpool Heart and Chest Hospital, UK. All patients provided informed consent, and ethical approval was obtained from the Liverpool Ethics Committee. The patient group represents a typical cohort with an operable lung tumor as decided according to standard treatment protocols. The samples were snap frozen and microdissected to ensure 80% cancer cell content. To test KRAB‐ZNF expression in breast cancer, we used commercially available Breast Cancer cDNA Array II purchased from Origene (Rockville, MD, USA) that also provided the clinical and molecular information of patients from whom samples were collected. The molecular data included ER, PR, and HER2 statuses of patients, assessed through immunohistochemistry. The tumors with weakly positive HER2 staining were also analyzed through the fluorescent in situ hybridization assay to detect HER2 amplification. The detailed characterization of clinicopathological and molecular data for lung and breast cancer cohorts used in this study is presented in Table 1. The expression level of the majority of KRAB‐ZNF factors remained low and sometimes undetected in our qPCR assays, which explains the differing sample numbers in the analyses for different factors.
Table 1.
Clinical and histopathological parameters available for breast and lung cancer tumor cohorts
| BRCA | No. | % | Lung | No. | % |
|---|---|---|---|---|---|
| Tumor stage | Tumor stage | ||||
| T1 | 14 | 32.6 | T1 | 3 | 5.2 |
| T2 | 20 | 46.5 | T2 | 44 | 75.9 |
| T3 | 3 | 7.0 | T3 | 8 | 13.8 |
| T4 | 2 | 4.7 | T4 | 3 | 5.2 |
| TX | 4 | 9.3 | |||
| Nodal status | Nodal status | ||||
| Negative | 16 | 37.2 | Negative | 27 | 46.6 |
| Positive | 20 | 46.5 | Positive | 31 | 53.4 |
| NX | 7 | 16.3 | |||
| Gender | Gender | ||||
| Male | 0 | 0 | Male | 44 | 75.9 |
| Female | 43 | 100 | Female | 14 | 24.4 |
| ER status | Differentiation | ||||
| Negative | 10 | 23.3 | Good | 19 | 32.8 |
| Positive | 25 | 58.1 | Moderate | 31 | 53.4 |
| Equivocal | 1 | 2.3 | Poor | 7 | 12.1 |
| No data | 7 | 16.3 | |||
| PR status | Histology | ||||
| Negative | 9 | 20.9 | SqCCL | 32 | 55.2 |
| Positive | 25 | 58.1 | AdenoCa | 24 | 41.8 |
| Equivocal | 2 | 4.6 | Anaplastic Ca | 1 | 1.7 |
| No data | 7 | 16.3 | |||
| HER2+ | NSCLC | 1 | 1.7 | ||
| Negative | 28 | 65.1 | Follow‐up | ||
| Positive | 8 | 18.6 | Alive | 4 | 6.9 |
| No data | 7 | 16.3 | Dead | 52 | 89.7 |
| Triple negative breast cancer | 4 | 9.3 | Age, mean age (range) | 67.1 | 48.5–87.5 |
Data collected in The Human Protein Atlas (www.proteinatlas.org) (Uhlen et al., 2015) were used as another validation set. Immunohistochemical staining on tissue microarrays for lung cancer was performed on different numbers of samples for KRAB‐ZNF: ZNF205 (n = 12), ZNF320 (n = 10), ZNF485 (n = 11), ZNF643 (n = 11), ZNF695 (n = 9), ZNF707 (n = 11), and ZNF 789 (n = 12). There were no data available for ZNF273, ZNF525, and ZNF714. For breast cancer, the samples were available for: ZNF205 (n = 11), ZNF320 (n = 10), ZNF485 (n = 11), ZNF643 (n = 11), ZNF695 (n = 12), ZNF707 (n = 10), and ZNF789 (n = 12). There were no data for ZNF273, ZNF525, and ZNF714. Images are available at the following URL: ZNF205 BRCA (https://www.proteinatlas.org/ENSG00000122386-ZNF205/pathology/tissue/breast+cancer#img), LUNG (https://www.proteinatlas.org/ENSG00000122386-ZNF205/pathology/tissue/lung+cancer#img); ZNF320 BRCA (https://www.proteinatlas.org/ENSG00000182986-ZNF320/pathology/tissue/breast+cancer#img), LUNG (https://www.proteinatlas.org/ENSG00000182986-ZNF320/pathology/tissue/lung+cancer#img); ZNF485 BRCA (https://www.proteinatlas.org/ENSG00000198298-ZNF485/pathology/tissue/breast+cancer#img), LUNG (https://www.proteinatlas.org/ENSG00000198298-ZNF485/pathology/tissue/lung+cancer#img); ZNF643 BRCA (https://www.proteinatlas.org/ENSG00000187801-ZFP69B/pathology/tissue/breast+cancer#img), LUNG (https://www.proteinatlas.org/ENSG00000187801-ZFP69B/pathology/tissue/lung+cancer#img); ZNF695 BRCA (https://www.proteinatlas.org/ENSG00000197472-ZNF695/pathology/tissue/breast+cancer#img), LUNG (https://www.proteinatlas.org/ENSG00000197472-ZNF695/pathology/tissue/lung+cancer#img); ZNF707 BRCA (https://www.proteinatlas.org/ENSG00000181135-ZNF707/pathology/tissue/breast+cancer#img), LUNG (https://www.proteinatlas.org/ENSG00000181135-ZNF707/pathology/tissue/lung+cancer#img); ZNF789 BRCA (https://www.proteinatlas.org/ENSG00000198556-ZNF789/pathology/tissue/breast+cancer#img), LUNG https://www.proteinatlas.org/ENSG00000198556-ZNF789/pathology/tissue/lung+cancer#img.
2.3. Cell culture
All cell lines were cultured under standard conditions (37 °C, 5% CO2) in a humidified incubator. For breast cancer, we used nine different cell lines representing distinct molecular subtypes: luminal A (MCF7, T47D), luminal B (BT474), basal (BT20, BT549, HS578T, MDA‐MB231, MDA‐MB468), and HER2 positive (SKBR3). All breast cancer cell lines were cultured in DMEM, High Glucose (Biowest, Nuaillé, France) with 10% fetal bovine serum (FBS; Biowest), 1% penicillin/streptomycin (Gibco, Carlsbad, CA, USA), and 10 ng·mL−1 insulin (Sigma‐Aldrich, Darmstadt, Germany). As a normal control, we used mammary epithelial cell line MCF12A cultured in DMEM/F12 (Gibco) supplemented with 5% FBS (Biowest), 20 ng·mL−1 EGF (Sigma‐Aldrich), 0.5 ng·mL−1 hydrocortisone (Sigma‐Aldrich), 10 ng·mL−1 insulin (Sigma‐Aldrich), and 1% penicillin/streptomycin (Gibco). For lung cancer analysis, we used 2 lung fibroblast cell lines (IMR90 and LUNG14), 7 LUAD cell lines (A549, CALU3, CALU6, H1299, H2073, H358, and SKLU1), five LUSC cell lines (CALU1, HTB59, HTB182, LUDLU1, and SKMES), one small cell lung carcinoma cell line (DMS53), and one large cell lung carcinoma cell line (CORL23). All lung cancer and fibroblast cell lines were cultured in DMEM/F12 (Gibco) with 10% FBS (Biowest) and 1% penicillin/streptomycin (Gibco). As a normal control, we used NHBE (Normal Human Bronchial Epithelium) cells (Lonza CC‐2540) cultured in Bronchial Epithelial Growth Medium BulletKit (Lonza, Basel, Switzerland). All remaining cell lines were purchased from ATCC (Manassas, VA, USA).
2.4. RT‐qPCR expression analysis
For RT‐qPCR expression analysis, RNA was isolated with a Quick‐RNA™ MiniPrep kit (Zymo Research, Irvine, CA, USA) and converted to cDNA with an iScript kit (Bio‐Rad, Hercules, CA, USA) according to the manufacturers’ protocols. Gene expression analysis was performed using the LightCycler 480 SYBR Green Master Mix (Roche, Basel, Switzerland) with 5 μm primer mix on a LightCycler 96 machine (Roche). We selected endogenous control genes based on in‐lab testing and previously published literature: ACTB for BRCA samples (Maltseva et al., 2013) and ESD for lung samples (Gresner et al., 2009). Relative quantification was performed using the comparative Ct method (RQ = 2−ΔΔCt). NHBE and MCF12A cell lines served as calibrators for expression analysis in lung and breast cancer samples, respectively. We used the following primers: ESD: FW 5′ – TGATCAAGGGAAAGATGACCA – 3′, RV 5′ – AACCCTCTTGCAATCGAAAA – 3′; ACTB: FW 5′ – CAGCCATGTACGTTGCTATCCAGG – 3′, RV 5′ – AGGTCCAGACGCAGGATGGCATG – 3′; ZNF205: FW 5′ – CAGAAGAAAAATGGGCTGTCA – 3′, RV 5′ – CGCCTCTCCACTCCTTCTC – 3′; ZNF273: FW 5′ – ACAGCTAAGACGCCAGGACT – 3′, RV 5′ – TGTGAAGTGTCCAGGCATTG – 3′; ZNF320: FW 5′ – CAGAGACGTGATGCTGGAGA – 3′, RV 5′ – TGCCCTGTTGATGACAATGTA – 3′; ZNF485: FW 5′ – GAGTGGAGACACCTGGATGC – 3′, RV 5′ – TTTTGGTTTGGAAGAGAGAAGC – 3′; ZNF525: FW 5′ – AGGGACGTGATGCTGGAG – 3′, RV 5′ – ATTGCCTTGTGCTGTTGATG – 3′; ZNF643: FW 5′ – GTGGGAGGATGTGACTAAGATGT – 3′, RV 5′ – ACTTTCGCCCTGGGTCTC – 3′; ZNF695: FW 5′ – ACAGAAACCTGATCTCCCTTG – 3′, RV 5′ – TTCACGTTCCAGGGCTCTTTC – 3′; ZNF707: FW 5′ – GAGTTCCAGGCAGTGCAGA – 3′, RV 5′ – CGTGGGTTTTCTTGTGAGCC – 3′; ZNF714: FW 5′ – GCCCTGGAATATGAAGATATG – 3′, RV 5′ – TTCTTAACTGTAAATTCTCATGT 3′; ZNF789: FW 5′ – GATGTGATGTTGGAGAACTACAGG – 3′, RV 5′ – CCAGGATCCACTGCTCGT – 3′.
2.5. Statistical analyses
RNA‐seq data downloaded from TCGA were normalized using RSEM (Li and Dewey, 2011), and differential analysis was performed using the DESeq package (Anders and Huber, 2010). To ensure sufficient sample size for the comparative expression analysis between normal and tumor tissues, we chose only those cohorts that contained at least nine normal samples. A heatmap was generated based on the results of the binomial test with a cutoff threshold of a median of absolute log2 fold change > 1/2. A t‐test with Tukey HSD correction was used to perform a comparative analysis of normalized KRAB‐ZNF expression between normal and tumor samples for BRCA, LUAD, and LUSC. T‐test with FDR correction was used to assess the differences between independent groups within clinical and molecular parameters. The statistical significance of changes in KRAB‐ZNF isoforms was calculated using a t‐test with FDR correction with pooled standard deviation. For the variant analysis, protein domains were assessed with the Expasy‐Prosite Database of Protein Domains, Families and Functional Sites Tool (https://prosite.expasy.org). The relationship between patient survival and KRAB‐ZNFs expression was tested with the log‐rank test and plotted with the Kaplan–Meier curves. KRAB‐ZNFs expression was transformed into binary high/low groups based on maximally ranked statistics. For the RT‐qPCR expression analysis, the difference in gene expression between normal and cancer tissues was calculated using the nonparametric Wilcoxon signed‐rank test for paired data (lung cancer) and Mann–Whitney U‐test for unpaired data (BRCA). All analyses described in the current study that utilized KRAB‐ZNF expression in TCGA datasets were deposited into an online application called KRAB‐ZNF Explorer – http://mi2.mini.pw.edu.pl:8080/KRAB_ZNF/ (R. Cylwa, K. Kiełczewski, U. Oleksiewicz, M. Machnik and P. Biecek, unpublished data). KRAB‐ZNF Explorer contains result tables and graphs that allow expression profiling of all KRAB‐ZNF factors in TCGA datasets, including gene and isoform expression in tumor and normal tissues, correlation with clinicopathological parameters, and association with CpG methylation.
3. Results
3.1. A subgroup of KRAB‐ZNF factors is overexpressed in 16 cancer types in TCGA datasets
We first aimed to identify KRAB‐ZNFs that are commonly overexpressed in cancer cells. Since KRAB‐ZNF expression is frequently tissue‐specific, we compared the mRNA level of 381 human KRAB‐ZNFs (Corsinotti et al., 2013) between tumor and normal tissues using RNA‐seq profiling datasets collected in the TCGA project. For the statistical analysis, we removed the data for cancer types with a low number (n < 9) of corresponding normal tissues to ensure sufficient abundance of samples in each set. Thus, the data available for the analysis comprised 6727 tumor and normal tissue samples from 16 cancer types: BLCA, BRCA, CHOL, ESCA, HNSC, KICH, KIPAN, KIRC, KIRP, LIHC, LUAD, LUSC, PRAD, STES, THCA, and UCEC. Our differential expression analysis revealed that out of 381 KRAB‐ZNFs, only a fraction was deregulated in tumors (56 out of 381, 14.7%; with a cutoff = median of absolute log2 fold change > 1/2) (Fig. 1A). Interestingly, the majority of the KRAB‐ZNFs with an altered mRNA level exhibited reduced expression, while only a small but distinct cluster of 16 KRAB‐ZNFs showed upregulation in multiple cancer types (Fig. 1A). The upregulated KRAB‐ZNFs included: ZNF695, ZNF468, ZNF714, ZNF320, ZNF273, ZNF525, ZNF530, ZNF643, ZNF138, ZNF92, ZNF200, ZNF707, ZNF205, ZNF485, ZNF354A, and ZNF789. The highest significant fold change (FC) was observed for ZNF695 in CHOL (54.9, P < 0.001). We also wanted to test the expression status of TRIM28, which encodes the KRAB‐ZNFs co‐factor protein, in the analyzed samples. As expected, TRIM28 showed higher mRNA expression in the majority of analyzed tumors. The TRIM28/KRAB complex triggers epigenetic repression of specific target genes (Oleksiewicz et al., 2017) and thus might be involved in alterations occurring in cancer cells. Among all of the 240 comparisons of tumor versus normal tissues, 148 were statistically significant (61.7%). Within these 148 comparisons, significantly higher expression in normal tissues compared to tumor tissues was observed only in six cases (ZNF138 in KIRC and THCA, ZNF200 in THCA, ZNF273 in PRAD, ZNF485 in THCA, and ZNF789 in KICH).
Figure 1.
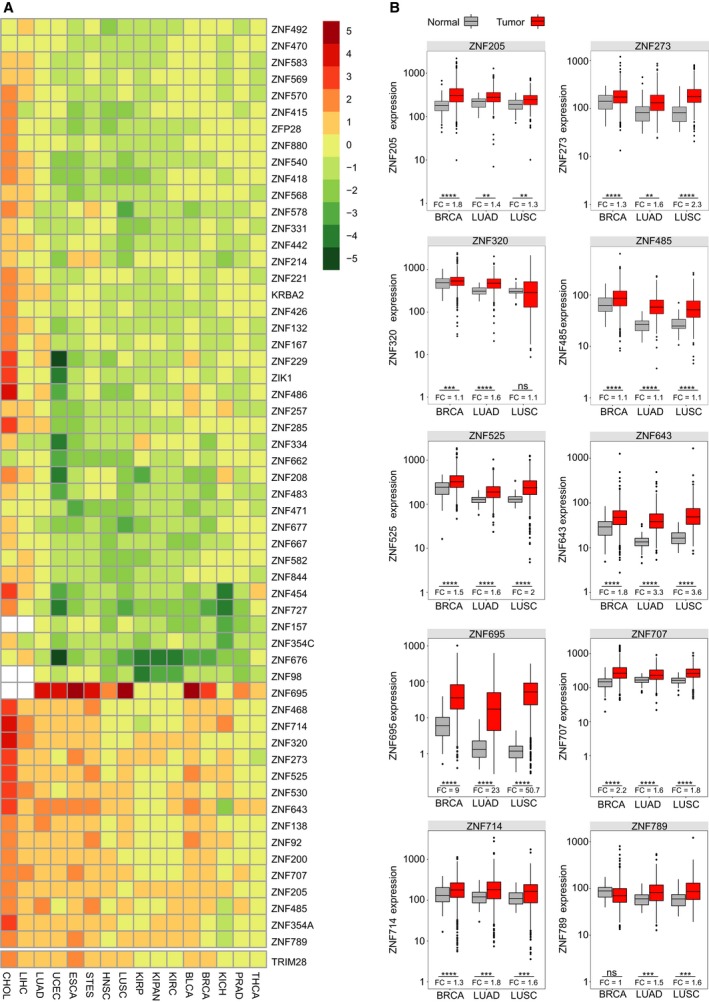
The expression level of selected KRAB‐ZNFs in TCGA samples. (A) Heatmap and supervised clustering of KRAB‐ZNFs with changed expression in 16 tumor cohorts compared to normal tissues. RSEM data were normalized with DESeq. Results were generated based on the binomial test with a cutoff threshold of a median of absolute log2 fold change > 1/2. (B) Boxplots representing selected cancer‐associated KRAB‐ZNF expression in normal and tumor tissue samples of breast and lung cancers. BRCA (N: 112, T: 1093), LUAD (N: 59, T: 515), LUSC (N: 51, T: 501). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 as assessed by t‐test with Tukey HSD correction.
To investigate cancer‐associated KRAB‐ZNFs in more detail, we further narrowed down the analysis to the two most common cancer types: breast cancer (BRCA) and nonsmall cell lung cancer (adenocarcinoma: LUAD and squamous cell carcinoma: LUSC) (Siegel et al., 2017). We chose 10 KRAB‐ZNFs (ZNF205, ZNF273, ZNF320, ZNF485, ZNF525, ZNF643, ZNF695, ZNF707, ZNF714, and ZNF789) and looked closer at their expression profiles in the selected cancer types. First, we compared their expression level in normal and tumor tissues. We found that all of the cancer‐associated KRAB‐ZNFs were significantly upregulated in tumors compared to normal tissues in the TCGA RNA‐seq data (Fig. 1B). The highest difference in the expression was observed in the case of ZNF695 (LUAD: FC = 23.0, P < 0.0001; LUSC: FC = 50.7, P < 0.0001; BRCA: FC = 9.0, P < 0.0001; t‐test with Tukey HSD correction). The smallest significant difference was noticed for ZNF205 in lung cancer (LUAD: FC = 1.4, P < 0.01; LUSC: FC = 1.3, P < 0.01; t‐test with Tukey HSD correction) and ZNF320 in BRCA (FC = 1.1, P < 0.001; t‐test with Tukey HSD correction).
3.2. The overexpression of selected KRAB‐ZNFs is confirmed in the independent validation sets of lung and breast cancer cell lines and tissues
To validate the expression results obtained from TCGA datasets, we analyzed 10 selected KRAB‐ZNFs in the panels of lung and breast cancer cell lines (see point 2.3) and tissues (see point 2.2, Table 1). Specifically, we examined the expression of chosen KRAB‐ZNFs in 16 different lung cancer cell lines and nine breast cancer cell lines representing different tumor subtypes. All of the selected cancer‐associated KRAB‐ZNFs were upregulated in the majority of lung cancer cell lines compared to the normal human bronchial epithelium cell line (NHBE) (Fig. 2A). As for breast cancer, we found that the expression level of most of the examined KRAB‐ZNFs was elevated at least twofold in almost every cell line in comparison with the normal mammary epithelial cell line, MCF12A (Fig. 2B). The only exemption was ZNF789 that showed upregulation in one cell line, MDA‐MD‐M468.
Figure 2.
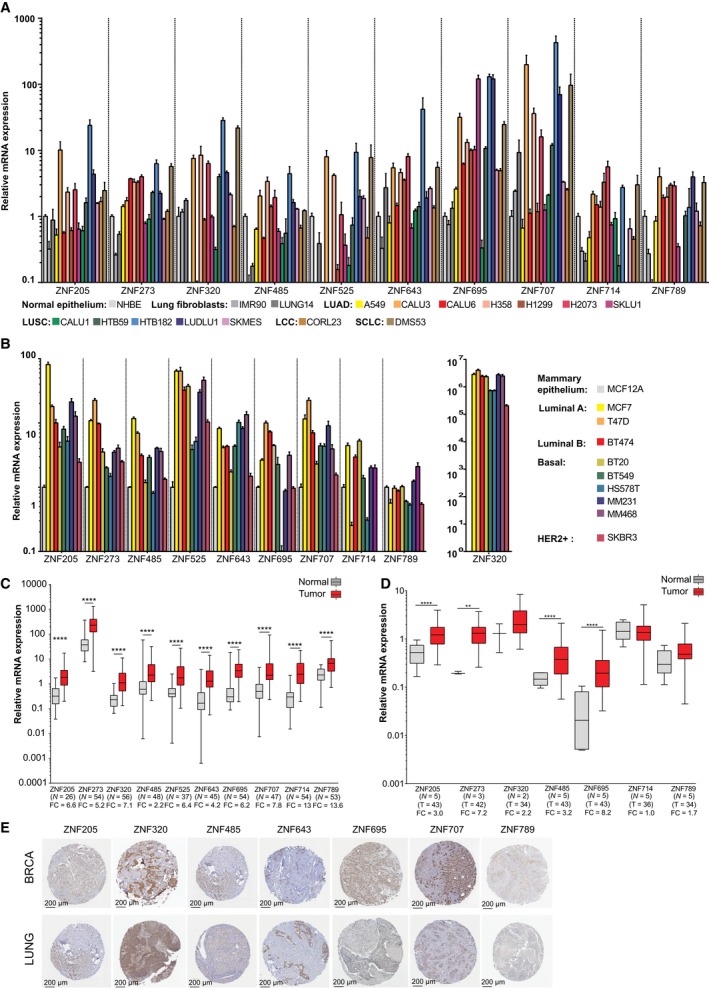
The expression level of selected KRAB‐ZNFs in independent sets of breast and lung cell lines and tissues. (A) RT‐qPCR expression of selected KRAB‐ZNFs in lung cancer cell lines compared to normal human bronchial epithelium. (B) RT‐qPCR expression of selected KRAB‐ZNFs in breast cancer compared to normal mammary epithelium. (A,B) Bars indicate mean expression for three technical replicates ± SD. (C) RT‐qPCR expression of selected KRAB‐ZNFs in lung cancer tissue samples compared to paired normal tissue. P‐values were calculated by Wilcoxon test. (D) RT‐qPCR expression of selected KRAB‐ZNFs in breast cancer tissue compared to normal tissue samples. P‐values were calculated using the Mann–Whitney U‐test. (E) Immunohistochemistry staining for selected KRAB‐ZNFs in lung and breast tissue microarray cancer samples from The Human Protein Atlas. (C,D) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Number of samples in each group and FC values are provided below the graphs.
In the next step, we evaluated KRAB‐ZNFs expression in lung and breast cancer tissue panels (Table 1). We compared the expression between 58 matched pairs of lung cancer and adjacent normal tissues and five normal breast and 43 cancer unmatched tissues. It is noteworthy that for some of the samples, KRAB‐ZNF expression fell below the detection level, and thus, the number of samples used for analysis varied between factors (Fig. 2C,D). Single‐gene qPCR assays confirmed that the expression level of 10 selected KRAB‐ZNFs was at least two times more abundant in lung tumor samples than in matched normal tissues (Fig. 2C). The biggest difference was observed in the case of ZNF714 expression (FC = 13.1, P < 0.0001 as assessed by Wilcoxon test) and the lowest for ZNF485 (FC = 2.2, P < 0.0001). In the breast cancer tissue panel (normal = 5, tumor = 43), we found that four KRAB‐ZNFs (ZNF205, ZNF273, ZNF485, ZNF695) were significantly upregulated in tumor tissue (Fig. 2D). The highest difference was observed for ZNF695 (FC = 8.2, P < 0.001 as assessed by Mann–Whitney U‐test) and the smallest for ZNF205 (FC = 3.0, P < 0.01). We also observed higher expression of ZNF320 and ZNF789 in tumors, but these data failed to reach statistical significance. Mean expression of ZNF714 was similar in normal and tumor samples, whereas ZNF525, ZNF643, and ZNF707 fell below the detection level.
These results were further confirmed on a protein level. We investigated cancer‐associated KRAB‐ZNFs in The Human Protein Atlas (www.proteinatlas.org) (Uhlen et al., 2015) that contains antibody‐based images of selected cancer types. In lung cancer tissue samples, KRAB‐ZNF staining was positive for at least four analyzed tumor samples (ZNF205 – 4/12, ZNF320 – 10/10, ZNF485 – 11/11, ZNF643 – 10/11, ZNF695 – 8/9, ZNF 707 – 10/11) and at least one sample from each set presented nuclear localization of a given factor (Fig. 2E). There were no data published for ZNF273, ZNF525, and ZNF714; and ZNF789 as staining was not detected in any of the analyzed samples. For breast cancer, the number of KRAB‐ZNF‐positive samples was slightly different: (ZNF205 – 6/11, ZNF320 – 10/10, ZNF485 – 11/11, ZNF643 – 11/11, ZNF695 – 12/12, ZNF 707 – 10/10, and ZNF789 – 2/12). No data were available for ZNF273, ZNF525, and ZNF714 (Fig. 2E).
3.3. Most of the cancer‐associated KRAB‐ZNF isoforms become simultaneously upregulated in tumors compared to normal samples
The complexity of the KRAB‐ZNF factors family is linked not only to the large number of its members but also to the high variety of splicing isoforms. As aberrant splicing is a frequent event in carcinogenesis, we wanted to explore the isoform signature for cancer‐associated KRAB‐ZNFs in TCGA datasets. First, we took into consideration the possibility of variant switch between normal and cancer tissue. Out of 600 comparisons between normal (n ≥ 9 per cohort) and cancer tissues (40 isoforms for 10 genes in 15 cohorts), 490 (81.7%) appeared statistically significant in our analysis (P < 0.05 as assessed by t‐test with FDR correction). We divided these variants into six groups based on their status of differential expression. As expected, we found that a majority of splicing variants (84.5%) were overexpressed in cancer tissues compared to their normal counterparts (Fig. 3A). Out of 490 significant isoforms, 21 variants (4.3%) showed expression only in cancer tissues, 220 variants (44.9%) were strongly overexpressed in cancer compared to normal (≥ 2‐fold overexpression, with the highest level reaching 652‐fold change), and 173 variants (35.3%) showed mild overexpression (FC < 2 and ≥ 1.2). It is of note that 21 isoforms that fell below the detection threshold in normal samples included mainly truncated and nonsense variants of ZNF695. The remaining 15.5% isoforms displayed either relatively stable expression (FC < 1.2 and ≥ 0.8 observed in 49 out of 490 variants, 10.0%), mild downregulation (FC < 0.8 and ≥ 0.5 in 21 variants, 4.3%), or strong downregulation (FC from 0.17 to < 0.5 in six variants, 1.2%).
Figure 3.
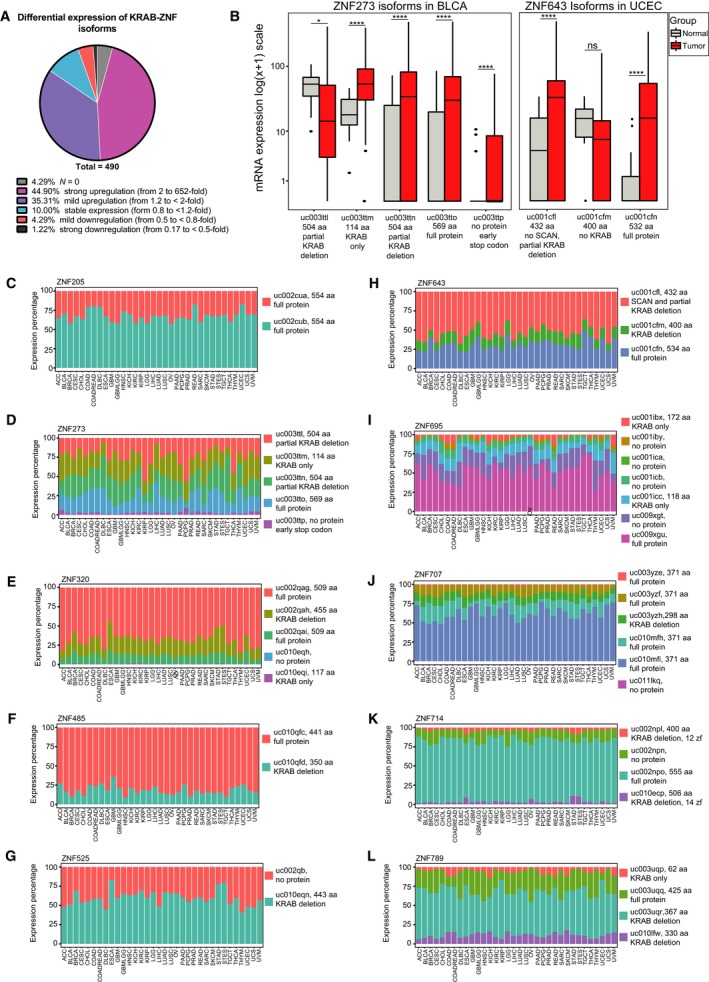
Expression of KRAB‐ZNF splicing isoform in TCGA datasets. (A) Pie chart depicting the percentage of isoforms that appeared statistically significant (n = 490) in our differential expression analysis comparing normal and tumor tissues. The isoforms fell into six categories: with null expression in normal samples, strongly and mildly upregulated in tumor samples, with stable expression, mildly and strongly downregulated in tumor samples. (B) The expression of only two pairs of isoforms is switched between normal and tumor samples. The boxplots represent the isoforms of ZNF273 in BLCA (N: 19, T: 408) and ZNF643 in UCEC (N: 11, T: 370). Statistical significance was calculated based on t‐test with FDR correction: *P < 0.05, ****P < 0.0001. (C–L) Plots demonstrate percentages of total expression of each of the listed splicing variants of 10 selected cancer‐associated KRAB‐ZNFs across 34 TCGA cohorts.
In general, we observed that the expression of the splicing isoforms followed a similar pattern in cancer and normal samples. Thus, to address the question regarding the potential variant switch between cancer and normal tissues, we focused on the expression profiles of the isoforms downregulated in cancer tissues (n = 27). These isoforms fell into three categories. The first category represented 14 isoforms that belong to five KRAB‐ZNFs with higher expression in healthy tissues, out of which only ZNF273 and ZNF789 showed significant downregulation in PRAD and KICH, respectively. The second category comprised 11 isoforms, whose expression was not predominant in the given tissue (i.e., these isoforms have a low or medium expression level), which indicates a low impact on cell function. Finally, the third category harbored two isoforms, whose expression was switched to other isoforms (Fig. 3B). This included ZNF273 in BLCA and ZNF643 in UCEC. ZNF273, the isoform with a 5′ partial deletion of the KRAB domain, was switched to three other isoforms, which could be translated to a full‐length protein, a variant with a C‐terminal partial deletion of the KRAB domain, and a protein devoid of the zinc finger domain. ZNF643, the shortest variant, lacking the SCAN and KRAB domains, was switched to two longer isoforms translated into a 534 aa full‐length protein and a 432 aa protein with a C‐terminal deletion of the SCAN domain and partial deletion of the KRAB domain. Thus, these observations indicate that variant switch is a sporadic event in the case of the analyzed KRAB‐ZNFs.
Next, we compared the expression pattern of the splicing isoforms across 34 cohorts (apart from the cohorts used for the comparative analysis, we used additional datasets, including: ACC, CESC, COAD, COADREAD, DLBC, GBM, GBMLGG, LGG, OV, PAAD, PCPG, READ, SARC, SKCM, STAD, TGCT, THYM, UCS, and UVM). Although the exact expression values differed between various cancer types, the relative expression profile remained uniform across multiple cancers (Fig. 3C–L). However, there were some exemptions to this rule. Four out of five ZNF273 variants (Fig. 3D) occurred in mixed configuration across all cohorts, but all of them showed a relatively high and equal expression level. The isoform expression pattern differed as well in the case of ZNF525 (in DLBC, LIHC, THYM, UCEC, and UCS, Fig. 3G), and ZNF643 (in GBMLLG, LGG, and TGCT, Fig. 3H). There was also some variability in the configuration of isoforms in the case of ZNF695 (Fig. 3I) and ZNF707 (Fig. 3J). Nevertheless, the most abundant isoform did not change. Similarly, in the case of ZNF714 (Fig. 3K) and ZNF789 (Fig. 3L), the variability occurred only among the two isoforms with the lowest expression. The most prevalent isoform expressed in the majority of tumors was the isoform encoding the longest protein. This is not the case for ZNF525 (Fig. 3G) that was expressed in two isoforms: one lacking the KRAB domain and the other that is not translated into protein. Also, in the case of ZNF643 (Fig. 3H), the isoforms with the highest expression level encoded a protein without the SCAN domain and with partial deletion of the KRAB domain, whereas the most abundant ZNF789 variant contained no KRAB domain (Fig. 3L). It is of note, however, that in these two last instances, mRNA coding the full‐length protein were also expressed at a relatively high level.
3.4. Expression of cancer‐associated KRAB‐ZNFs correlates with histology, molecular subtypes, and patient survival in lung tumors
Next, we wanted to explore the clinical significance of KRAB‐ZNF expression in tumor tissues using TCGA datasets and the independent validation set of lung cancer tissues (Table 1). To this end, we compared the expression of cancer‐associated KRAB‐ZNFs with various clinicopathological features, including histology, TNM classification, gender, survival, and smoking history (Table 2). We first utilized the data available for the lung cancer tissue panel (Table 1). We found that ZNF273, ZNF320, and ZNF643 showed higher expression (~ 2‐fold) in LUAD than LUSC (ZNF273: FC = 1.85, P = 0.018; ZNF320: FC = 1.9, P = 0.042; ZNF643: FC = 1.84, P = 0.044, as assessed by t‐test with Tukey HSD correction, Fig. 4A). No other associations were found between KRAB‐ZNF expression and tumor size, differentiation, lymph node status, patient gender, or survival (Table 2).
Table 2.
KRAB‐ZNF expression correlates with various clinicopathological parameters in lung cancer
| Gene | Subtype | Subtype value | Mean ± SEM, Patients’ No. | P adjusted (t‐test) |
|---|---|---|---|---|
| ZNF695 | LUSC histological subtypes | Basaloid | 107.1 ± 17.1, n = 15 | |
| Papillary | 34 ± 10, n = 6 | vs Basaloid P = 0.045 | ||
| Squamous NOS | 62.5 ± 2.5, n = 467 | vs Basaloid P = 0.016 | ||
| ZNF789 | Basaloid | 151.1 ± 22.1, n = 15 | ||
| Squamous NOS | 99.6 ± 2.73, n = 467 | vs Basaloid P = 0.009 | ||
| ZNF525 | LUAD expression subtypes | Squamoid | 220.7 ± 10.9, n = 78 | |
| Bronchioid | 176,4 ± 6.4, n = 89 | vs Squamoid P = 0.002 | ||
| Magnoid | 184.5 ± 11.8, n = 63 | vs Squamoid P = 0.03 | ||
| ZNF643 | Squamoid | 45.3 ± 2.6, n = 78 | vs Magnoid P = 0.005 | |
| Bronchioid | 36.9 ± 2, n = 89 | vs Magnoid P < 10−4 | ||
| Magnoid | 62 ± 6.1, n = 63 | |||
| ZNF695 | Squamoid | 40.5 ± 8.8, n = 78 | vs Bronchioid P = 0.001 | |
| Bronchioid | 11.11 ± 1.3, n = 89 | |||
| Magnoid | 35.5 ± 5.31, n = 63 | vs Bronchioid P = 0.01 | ||
| ZNF707 | Squamoid | 203.5 ± 9.9, n = 78 | P = 0.046 | |
| Magnoid | 249.5 ± 19.7, n = 63 | |||
| ZNF714 | Squamoid | 243.1 ± 20.7, n = 78 | P = 0.03 | |
| Magnoid | 177.8 ± 17.5, n = 63 | |||
| ZNF273 | LUSC molecular subtype | Basal | 193.6 ± 16.8, n = 42 | vs Primitive P = 0.02 |
| Classical | 244.8 ± 18.8, n = 65 | vs Secretory P = 0.006 | ||
| Primitive | 289.9 ± 34.9, n = 27 | |||
| Secretory | 158 ± 11.7, n = 43 | vs Primitive P = 0.0005 | ||
| ZNF320 | Basal | 357.6 ± 45.6, n = 42 | ||
| Classical | 233.1 ± 27.1, n = 65 | vs Primitive P < 10−4 | ||
| Primitive | 527.3 ± 81.4, n = 27 | |||
| Secretory | 388.2 ± 33.4, n = 43 | vs Classical P = 0.025 | ||
| ZNF695 | Basal | 41 ± 5.2, n = 42 | vs Primitive P < 10−4 | |
| Classical | 77.6 ± 6.5, n = 65 | vs Primitive P = 0.009 | ||
| Primitive | 116.5 ± 16.2, n = 27 | |||
| Secretory | 54.9 ± 6.8, n = 43 | vs Primitive P < 10−4 | ||
| ZNF707 | Basal | 232.5 ± 15.5, n = 42 | vs Primitive P = 0.007 | |
| Classical | 186.1 ± 8.6, n = 65 | vs Primitive P < 10−4 | ||
| Primitive | 309.3 ± 29.5, n = 27 | |||
| Secretory | 220.8 ± 11.8, n = 43 | vs Primitive P = 0.001 | ||
| ZNF789 | Basal | 74.2 ± 8.4, n = 42 | vs Primitive P = 0.01 | |
| Classical | 74.4 ± 4, n = 65 | vs Primitive P = 0.007 | ||
| Primitive | 104.2 ± 8.2, n = 27 | |||
| Secretory | 69.6 ± 4.6, n = 43 | vs Primitive P = 0.003 | ||
| ZNF643 | LUSC gender | Female | 77.7 ± 13.2, n = 130 | P = 0.02 |
| Male | 58.3 ± 1.8, n = 370 | |||
| ZNF714 | Female | 211.84 ± 17.6, n = 130 | P = 0.049 | |
| Male | 179.6 ± 7.47, n = 370 | |||
| ZNF695 | LUAD gender | Female | 29.5 ± 2.3, n = 277 | P = 0.011 |
| Male | 41.1 ± 4.1, n = 239 | |||
| ZNF643 | LUAD smoking history | Lifelong Nonsmoker | 41.6 ± 4.3, n = 76 | vs Current smoker P = 0.04 |
| Current smoker | 57.4 ± 4.8, n = 119 | |||
| Current reformed smoker for > 15 years | 40.6 ± 1.8, n = 135 | vs Current smoker P = 0.004 | ||
| ZNF695 | Lifelong Nonsmoker | 23.7 ± 3.9, n = 76 | vs Current smoker P = 0.002 | |
| Current smoker | 51.9 ± 4.9, n = 119 | |||
| Current reformed smoker for > 15 years | 18.3 ± 2.4, n = 135 | vs Current smoker P < 10−4 | ||
| Current reformed smoker for < 15 years | 42.9 ± 5.2, n = 168 | vs Current reformed smoker for > 15 year P = 0.0004 |
Figure 4.
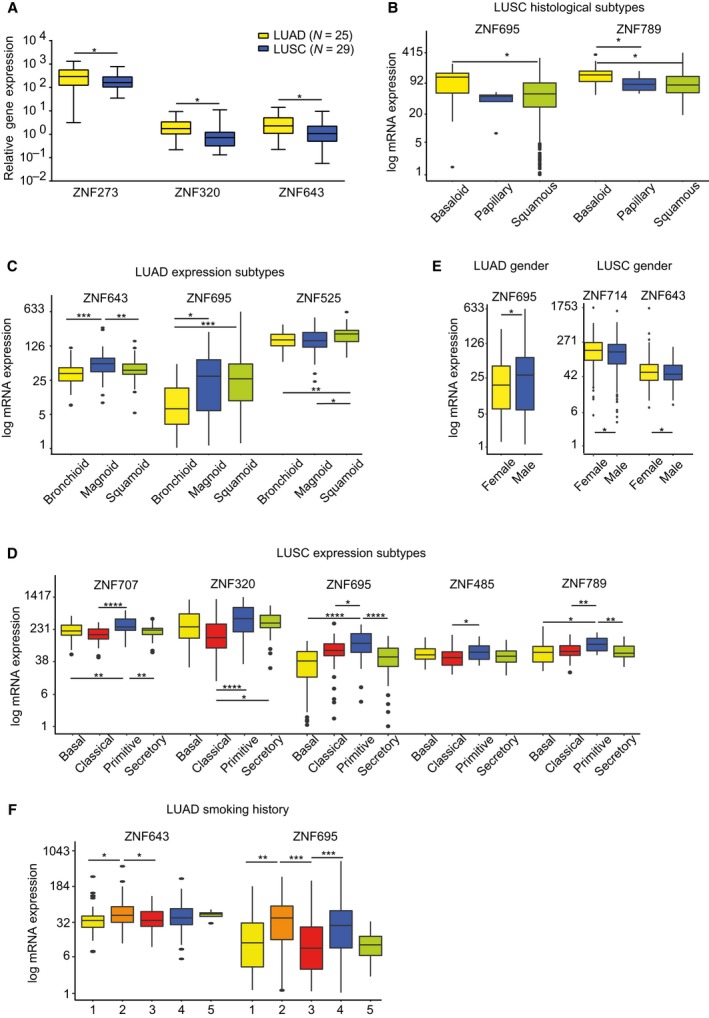
Cancer‐associated KRAB‐ZNFs correlate with various clinical parameters in LUAD and LUSC. (A) Single‐gene qPCR analysis in lung cancer tissue panel shows that ZNF273,ZNF320, and ZNF643 have significantly higher mRNA level in LUAD (n = 25) compared to LUSC (n = 29). (B) In TCGA LUSC histological subtypes, ZNF695 and ZNF789 demonstrate higher expression in the basaloid (n = 15) subtype compared to other subtypes: papillary (n = 6) and/or squamous not otherwise specified (n = 467). (C) Boxplot representation of mRNA expression level of ZNF525,ZNF643, and ZNF695 in TCGA LUAD molecular subtypes: squamoid (n = 78), bronchoid (n = 89), and magnoid (n = 63). (D) Boxplot representation of mRNA expression level of ZNF707,ZNF320,ZNF695,ZNF485, and ZNF789 in TCGA LUSC molecular subtypes: basal (n = 42), classical (n = 65), primitive (n = 27), and secretory (n = 43). (E) The expression of ZNF695 in TCGA LUAD, as well as ZNF714 and ZNF643 in TCGA LUSC correlate with gender. LUAD: female (n = 277), male (n = 239); LUSC: female (n = 130), male (n = 370). (F) ZNF643 and ZNF695 mRNA is more abundant in smokers vs nonsmokers or reformed smokers according to the TCGA dataset. 1: lifelong nonsmoker (n = 76), 2: current smoker (n = 119), 3: current reformed smoker for > 15 years (n = 135), 4: current reformed smoker for ≤ 15 years (n = 168), 5: current reformed smoker, duration not specified (n = 4). (A–F) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 as assessed by t‐test with FDR correction.
We further analyzed the association between KRAB‐ZNF expression patterns and clinical parameters in LUAD and LUSC datasets from the TCGA project. We observed that the majority of significant differences in KRAB‐ZNF expression were related to histology (as noted in the tissue panel), molecular subtypes, and patient survival. In LUSC histological subtypes, we found that ZNF695 and ZNF789 had higher expression in the basaloid subtype than in the papillary and/or squamous subtypes, or in subtypes that were not otherwise specified (Fig. 4B). More associations between KRAB‐ZNF expression and clinical parameters were found in relation to molecular subtypes. We observed that in the LUAD mRNA subtypes, ZNF525 had the highest expression in the squamoid subtype (magnoid and bronchioid), ZNF643 expression was highest in the magnoid subtype, while ZNF695 had the lowest expression in the bronchioid subtype (Fig. 4C). In LUSC, the highest KRAB‐ZNF expression was observed in the primitive subtype, particularly with respect to the classical subtype (Fig. 4D). This relation was true for ZNF320, ZNF485, ZNF695, ZNF707, and ZNF789. In general, no specific trend of association was found between KRAB‐ZNF expression and TNM classification or tumor stage both in LUAD and LUSC. Some associations were observed in the case of gender. In LUAD, ZNF695 expression was higher in males, whereas in LUSC, ZNF643, and ZNF714 expression was higher in females (Fig. 4E). Also, in LUAD, smoking history correlated with expression of ZNF525, ZNF643, and ZNF695 (Fig. 4F). In all cases, the expression level in tumors from current smokers was significantly higher than in reformed smokers or nonsmokers. No such associations were observed in LUSC.
Finally, we looked at the potential correlation of the KRAB‐ZNF expression with patient survival. In our log‐rank analysis, we used maximally selected rank statistics to identify the optimal cutoff point dividing the expression of each of the KRAB‐ZNFs into two groups with high and low mRNA levels. In LUAD patients, longer survival was observed in groups with low expression of ZNF525 (P = 0.006, hazard ratio = 1.83) and ZNF695 (P = 0.006, hazard ratio = 2.4) (Fig. 5A,B) and high expression of ZNF707 (P = 0.034, hazard ratio = 0.66) and ZNF789 (P = 0.002, hazard ratio = 0.5) (Fig. 5C,D). In LUSC patients, high mRNA levels of the majority of KRAB‐ZNF factors correlated with poor prognosis (Fig. 5E–I): ZNF205 (P = 0.009, hazard ratio = 1.6), ZNF320 (P = 0.009, hazard ratio = 1.5), ZNF485 (P = 0.039, hazard ratio = 1.4), ZNF525 (P = 0.017, hazard ratio = 1.6), and ZNF643 (P = 0.031, hazard ratio = 1.9). In contrast, better prognosis was observed in the patients with high expression of ZNF273 (P = 0.005, hazard ratio = 0.6), ZNF695 (P = 0.02, hazard ratio = 0.7), and ZNF789 (P = 0.007, hazard ratio = 0.6) (Fig. 5J–L).
Figure 5.
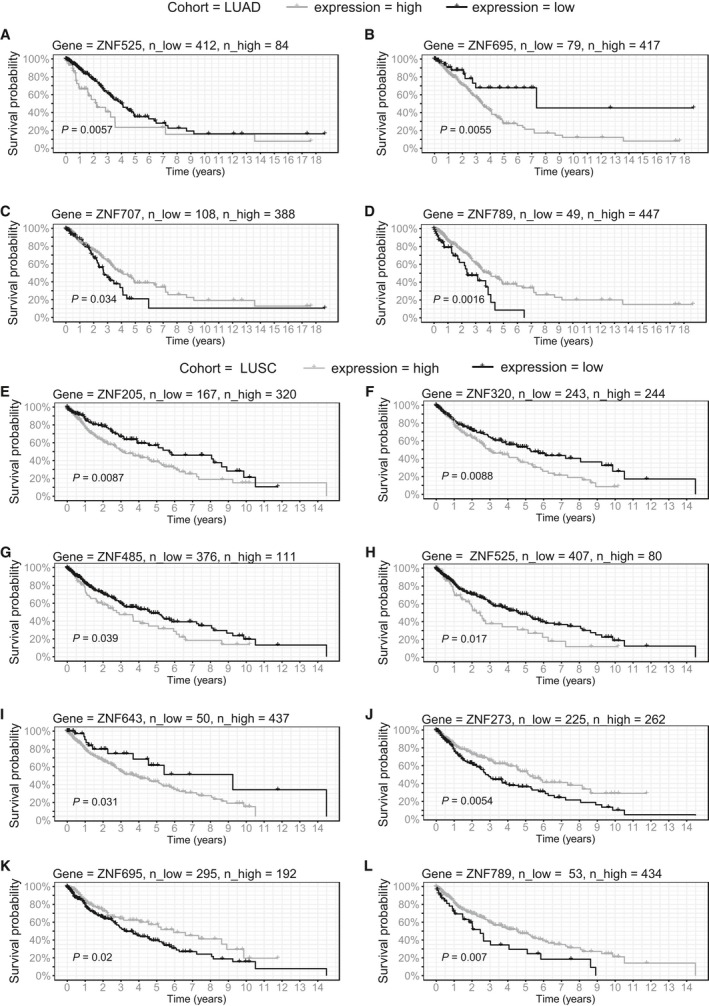
Survival analysis in TCGA LUAD and LUSC (E–L) patients. Kaplan–Meier curves represent ZNF707 (A), ZNF789 (B), ZNF525 (C), and ZNF695 (D) association with patient survival in LUAD and ZNF205 (E), ZNF320 (F), ZNF485 (G), ZNF525 (H), ZNF643 (I), ZNF273 (J), ZNF695 (K), and ZNF789 (L) association with patient survival in LUSC. KRAB‐ZNFs expression was transformed into binary high/low groups based on maximally ranked statistics. P‐values assessed by log‐rank test and number of patients in each group is provided in the figure.
3.5. Expression of cancer‐associated KRAB‐ZNFs correlates with histology, receptor status, molecular subtypes, and patient survival in BRCA
We further examined the clinical significance of KRAB‐ZNF expression in breast cancer. We found multiple interdependencies; however, based on the expression of KRAB‐ZNF in relation to histological and molecular subtypes, we were able to appoint two major categories (Fig. 6). The first one contained KRAB‐ZNFs with the highest expression in basal‐like tumors and included ZNF695, ZNF643, ZNF485, and ZNF273 (Fig. 6A, Table 3). Significantly higher expression in basal‐like tumors in relation to other cancer types (excluding normal‐like) was also observed in the case of ZNF789. As expected, high expression of these factors also correlated with negative staining for estrogen receptor (ER) and progesterone receptor (PR) in the TCGA dataset (Fig. 6B,C, Table 4). However, ZNF273 and ZNF485 correlation with PR status did not reach statistical significance. As far as HER2 staining is concerned (Fig. 6D, Table 4), only ZNF789 expression significantly correlated with HER2‐tumors in the TCGA dataset. Other factors showed either marginal difference or a trend of association with HER2+ tumors (ZNF695). In general, these observations (apart from some observations in the case of ZNF273 and ZNF789) were also confirmed in the independent set of BRCA tissues (Fig. 7). However, the tissue panel results showed mainly a trend that did not reach statistical significance, possibly due to the lower number of samples. Finally, these factors (ZNF273, ZNF485, ZNF695, ZNF643, and ZNF789), together with ZNF714, had significantly increased mRNA levels in the DNA methylation cluster 5 identified for breast cancer (Fig. 6E, Table 5), which also coincides with the basal‐like subtype and the lowest overall DNA methylation level (The Cancer Genome Atlas Network, 2012).
Figure 6.
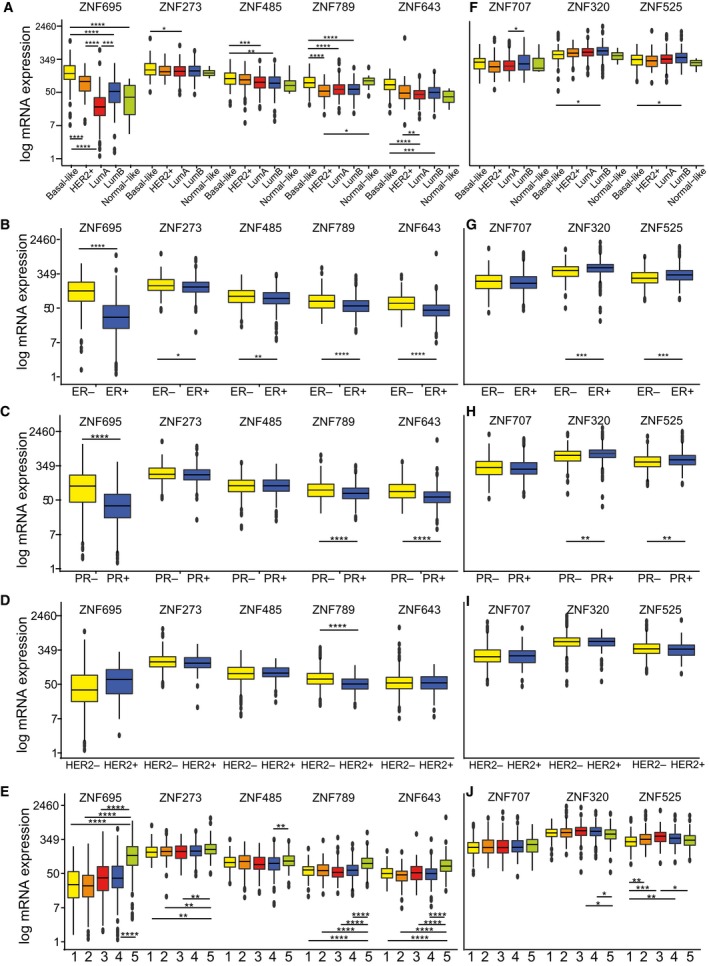
Cancer‐associated KRAB‐ZNF expression correlates with various clinical and molecular parameters in TCGA BRCA patients including: (A, F) histological subgroups: basal‐like (n = 98), HER2+ (n = 58), luminal A (n = 233), luminal B (n = 129), normal‐like (n = 8); (B, G) estrogen receptor (ER) status: ER+ (n = 603), ER− (n = 179); (C, H) progesterone receptor (PR) status: PR+ (n = 526), PR− (n = 253); (D, I) HER2 receptor status: HER2+ (n = 115), HER2− (n = 654); (E, J) methylation clusters: cluster 1: (n = 137), 2: (n = 174), 3: (n = 101), 4: (n = 246), 5: (n = 295). (A–J) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 as assessed by t‐test with FDR correction.
Table 3.
KRAB‐ZNF expression correlates with breast cancer histology status
| Gene | Subtype value | Mean ± SEM | P‐value (adjusted) |
|---|---|---|---|
| ZNF205 | Basal‐like | 273.3 ± 16 | P = 0.003 |
| Luminal A | 334.2 ± 8.6 | ||
| ZNF273 | Basal‐like | 225.9 ± 14.7 | P = 0.017 |
| Luminal A | 188.1 ± 5.9 | ||
| ZNF320 | Basal‐like | 515.7 ± 25.8 | P = 0.012 |
| Luminal B | 620.9 ± 26.4 | ||
| ZNF485 | Basal‐like | 124.4 ± 6.5 | |
| Luminal A | 97.2 ± 3 | P = 0.0003 vs Basal | |
| Luminal B | 97.4 ± 4.8 | P = 0.002 vs Basal | |
| ZNF525 | Basal‐like | 368.3 ± 16.9 | P = 0.047 |
| Luminal B | 446.6 ± 21.8 | ||
| ZNF643 | Basal‐like | 90.8 ± 4.2 | |
| HER2‐enriched | 78.4 ± 21.2 | P = 0.008 vs Luminal A | |
| Luminal A | 49 ± 1.4 | P < 10−4 vs Basal | |
| Luminal B | 56.9 ± 2.5 | P = 0.0003 vs Basal | |
| ZNF695 | Basal‐like | 197 ± 15.6 | |
| HER2‐enriched | 98 ± 7.8 | P < 10−4 vs Basal | |
| Luminal A | 29.8 ± 3 | P < 10−4 vs Basal | |
| P < 10−4 vs HER2‐enriched | |||
| P = 0.0007 vs Luminal A | |||
| Luminal B | 65.3 ± 4.8 | P < 10−4 vs Basal | |
| Normal‐like | 46.7 ± 13.9 | P < 10−4 vs Basal | |
| ZNF707 | Luminal A | 283.9 ± 10.5 | P = 0.016 |
| Luminal B | 347.3 ± 19.2 | ||
| ZNF714 | Basal‐like | 270.5 ± 18 | |
| HER2‐enriched | 295. 4 ± 26.2 | ||
| Luminal A | 181.7 ± 6.8 | P < 10−4 vs HER2‐enriched | |
| P < 10−4 vs Basal‐like | |||
| P = 0.0002 vs Luminal B | |||
| Luminal B | 251 ± 14.6 | ||
| Normal‐like | 131.5 ± 25.2 | P = 0.028 vs HER2‐enriched | |
| ZNF789 | Basal‐like | 101.7 ± 5.5 | |
| HER2‐enriched | 59.4 ± 4 | P < 10−4 vs Basal | |
| Luminal A | 67 ± 2.5 | P < 10−4 vs Basal | |
| Luminal B | 65.8 ± 2.8 | P < 10−4 vs Basal | |
| Normal‐like | 105.4 ± 19 | P = 0.02 vs HER2‐enriched |
Table 4.
KRAB‐ZNF expression correlates with receptor status in breast cancer patients. ER, estrogen receptor; padj, adjusted P‐value; PR, progesterone receptor. Number of patients in each group: ER+ = 603, ER− = 179, N PR+ = 526, N PR− = 253, HER2+ = 115, HER2− = 653
| Gene | Status | ER status | PR status | HER2 status | |||
|---|---|---|---|---|---|---|---|
| Mean ± SEM | Padj | Mean ± SEM | Padj | Mean ± SEM | Padj | ||
| ZNF205 | − | 263.9 ± 11.9 | 0.015 | 271.6 ± 9.3 | 0.025 | 296.6 ± 5.8 | n.s. |
| + | 302.9 ± 5.6 | 304.8 ± 6.2 | 259.9 ± 10 | ||||
| ZNF273 | − | 215.9 ± 9 | 0.028 | 205.6 ± 6.9 | n.s. | 200.2 ± 4 | n.s. |
| + | 191.4 ± 3.7 | 192.1 ± 4 | 185.5 ± 7.7 | ||||
| ZNF320 | − | 507.4 ± 16.6 | 0.0002 | 528 ± 14 | 0.002 | 583.8 ± 10 | n.s. |
| + | 595.4 ± 9.9 | 297.2 ± 10.7 | 561.3 ± 18.6 | ||||
| ZNF485 | − | 114.8 ± 4.4 | 0.004 | 104.6 ± 3.7 | n.s. | 103 ± 2.1 | n.s. |
| + | 99.1 ± 2 | 101.8 ± 2.1 | 101.9 ± 3.8 | ||||
| ZNF525 | − | 347.8 ± 12.3 | 0.0007 | 366.9 ± 11.1 | 0.01 | 404.8 ± 7.9 | n.s. |
| + | 416. 2 ± 8.7 | 417.6 ± 9.5 | 389.2 ± 20.4 | ||||
| ZNF643 | − | 84.7 ± 3.8 | < 10−4 | 78.1 ± 2.9 | < 10−4 | 63.7 ± 2.4 | n.s. |
| + | 55.9 ± 2.3 | 54.7 ± 2.6 | 57.4 ± 2.6 | ||||
| ZNF695 | − | 169.2 ± 10.3 | < 10−4 | 136 ± 9 | < 10−4 | 72.5 ± 3.9 | n.s. |
| + | 46.8 ± 2.6 | 45.5 ± 2.1 | 77.8 ± 6.2 | ||||
| ZNF707 | − | 299.2 ± 14.3 | n.s. | 296.6 ± 12 | n.s. | 291.1 ± 7.3 | n.s. |
| + | 290.2 ± 7.6 | 290.7 ± 8.1 | 277.7 ± 15.6 | ||||
| ZNF714 | − | 262.1 ± 13.7 | 0.004 | 250.5 ± 10.8 | 0.027 | 225.1 ± 5.9 | n.s. |
| + | 216.4 ± 5.8 | 216 ± 6.2 | 249.5 ± 15.9 | ||||
| ZNF789 | − | 93.8 ± 4.8 | < 10−4 | 86.1 ± 3.8 | < 10−4 | 76.4 ± 1.7 | < 10−4 |
| + | 67.7 ± 1.5 | 67.1 ± 1.5 | 53.7 ± 2.3 | ||||
Figure 7.
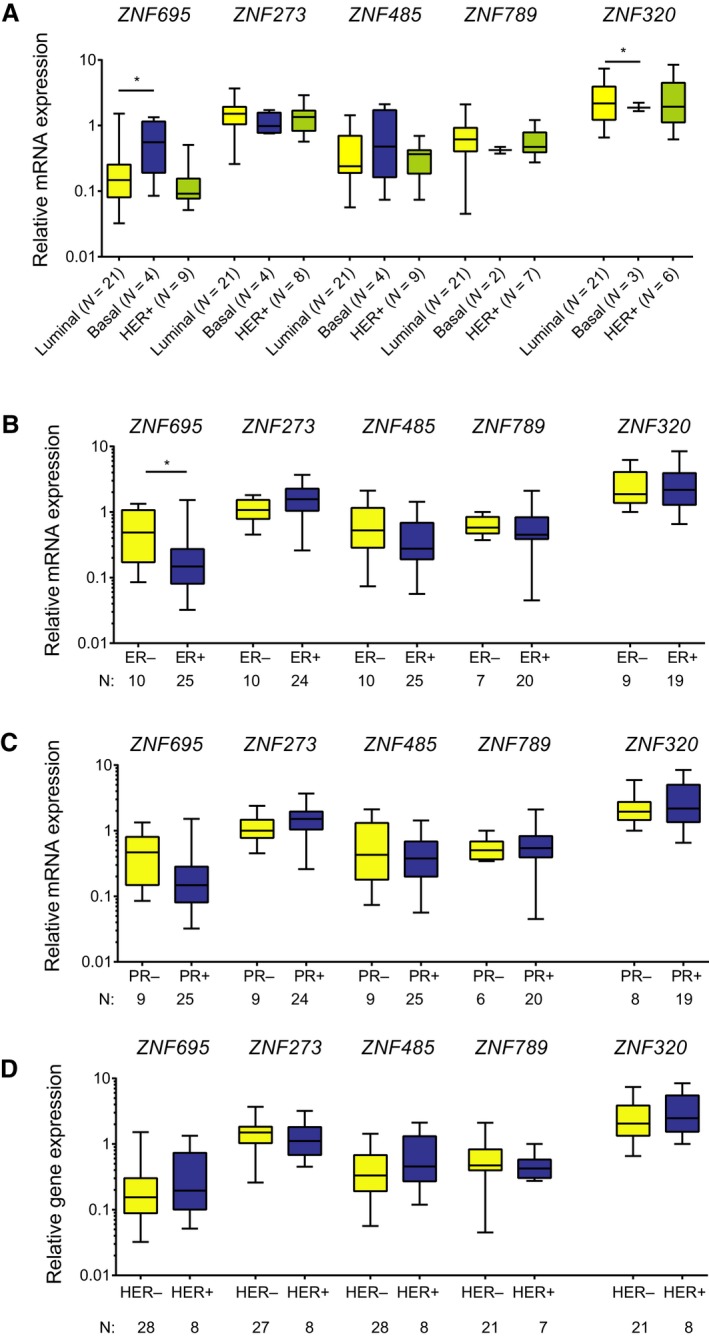
Cancer‐associated KRAB‐ZNF expression correlates with various clinical and molecular parameters in an independent panel of BRCA tissues. This includes histological subgroups (A), estrogen receptor (ER) status (B), progesterone receptor (PR) status (C), and HER2+ receptor status (D). *P < 0.05 as assessed by unpaired t‐test. The number of patients in each group is provided in the figure.
Table 5.
KRAB‐ZNF expression correlated with DNA methylation status in breast cancer. Cluster 1: n = 137, 2: n = 174, 3: n = 101, 4: n = 246, 5: n = 295
| Gene | Methylation cluster | Mean | P‐value (adjusted) |
|---|---|---|---|
| ZNF205 | 1 | 271.3 ± 11.2 | 0.0007 vs 2 |
| 2 | 336.7 ± 11.8 | ||
| 3 | 266.8 ± 12.9 | 0.001 vs 2 | |
| 4 | 293.5 ± 8.8 | 0.02 vs 2 | |
| 5 | 260.2 ± 13.3 | < 10−4 vs 2 | |
| ZNF273 | 1 | 186.8 ± 8.2 | 0.006 vs 5 |
| 2 | 289.2 ± 6.1 | 0.006 vs 5 | |
| 3 | 184.4 ± 8.2 | 0.008 vs 5 | |
| 4 | 200.3 ± 5.7 | ||
| 5 | 226.8 ± 5.7 | ||
| ZNF320 | 1 | 555.5 ± 16.5 | |
| 2 | 586.3 ± 21.2 | ||
| 3 | 626.9 ± 27.6 | 0.016 vs 5 | |
| 4 | 602.9 ± 14.9 | 0.028 vs 5 | |
| 5 | 528.2 ± 19.3 | ||
| ZNF485 | 1 | 12.6 ± 4 | |
| 2 | 102.9 ± 3.7 | ||
| 3 | 98.3 ± 5.6 | ||
| 4 | 97.8 ± 116 | 0.008 vs 5 | |
| 5 | 116 ± 4.6 | ||
| ZNF525 | 1 | 343.6 ± 12.8 | |
| 2 | 422.4 ± 18.2 | 0.005 vs 1 | |
| 3 | 456.2 ± 22.4 | 0.0002 vs 1 | |
| 4 | 414.8 ± 12.3 | 0.008 vs 1 | |
| 5 | 374.2 ± 15.1 | 0.001 vs 3 | |
| ZNF643 | 1 | 55.9 ± 2.2 | < 10−4 vs 5 |
| 2 | 49.5 ± 1.9 | < 10−4 vs 5 | |
| 3 | 62 ± 5.1 | < 10−4 vs 5 | |
| 4 | 57 ± 2.2 | < 10−4 vs 5 | |
| 5 | 95.5 ± 8.9 | ||
| ZNF695 | 1 | 48.3 ± 4.8 | < 10−4 vs 5 |
| 2 | 37 ± 3.1 | < 10−4 vs 5 | |
| 3 | 58.9 ± 7.4 | < 10−4 vs 5 | |
| 4 | 60.3 ± 4.4 | < 10−4 vs 5 | |
| 5 | 180 ± 12.9 | ||
| ZNF714 | 1 | 197 ± 10.6 | 0.0001 vs 5 |
| 2 | 198.8 ± 9.2 | < 10−4 vs 5 | |
| 3 | 247.4 ± 17.3 | ||
| 4 | 227.6 ± 10 | 0.012 vs 5 | |
| 5 | 278.2 ± 14.9 | ||
| ZNF789 | 1 | 64.7 ± 2.4 | < 10−4 vs 5 |
| 2 | 66.7 ± 2.7 | < 10−4 vs 5 | |
| 3 | 64.8 ± 4.8 | < 10−4 vs 5 | |
| 4 | 68.1 ± 2.3 | < 10−4 vs 5 | |
| 5 | 104 ± 4.3 |
The second identified KRAB‐ZNF subgroup included ZNF320, ZNF525, and ZNF707, and its expression was significantly higher in luminal B tumors compared to that in basal‐like and/or luminal A tumors (Fig. 6F, Table 3). Apart from ZNF707, whose expression was not related to the receptor status, mRNA levels of ZNF320 and ZNF525 were significantly higher in ER+ and PR+ tumors (Fig. 6G,H, Table 4). In general, the expression of these KRAB‐ZNFs was not related to HER2 status (Fig. 6I, Table 4). Furthermore, ZNF320 and ZNF525 expression was low in DNA methylation clusters 5 and 1, while the highest expression was noted for cluster 3 (Fig. 6J, Table 5). Cluster 3 correlates with the luminal B subtype and the highest global DNA methylation profile (The Cancer Genome Atlas Network, 2012).
The remaining cancer‐associated KRAB‐ZNFs did not show any characteristic features. In the TCGA dataset and tissue panel, ZNF205 expression was relatively equal among the various histological subtypes, although higher expression was noted in the luminal compared to basal‐like tumors (Fig. 8A,B, Table 3). Also, significantly higher expression of ZNF205 was noted in ER+ and PR+ tumors, and a trend of higher expression was observed in HER2‐ tumors (Table 4). The highest expression of ZNF205 was seen in DNA methylation cluster 2 (Table 5). ZNF714 expression (Fig. 8C, Table 4) was highest in HER2‐enriched tumors and lowest in luminal A, and as expected, in the ER−, PR−, and HER2+ tumors in the TCGA dataset. Surprisingly, in the BRCA tissue panel, high ZNF714 expression correlated with ER−, PR+, and HER2− samples (Fig. 8D, Table 4).
Figure 8.
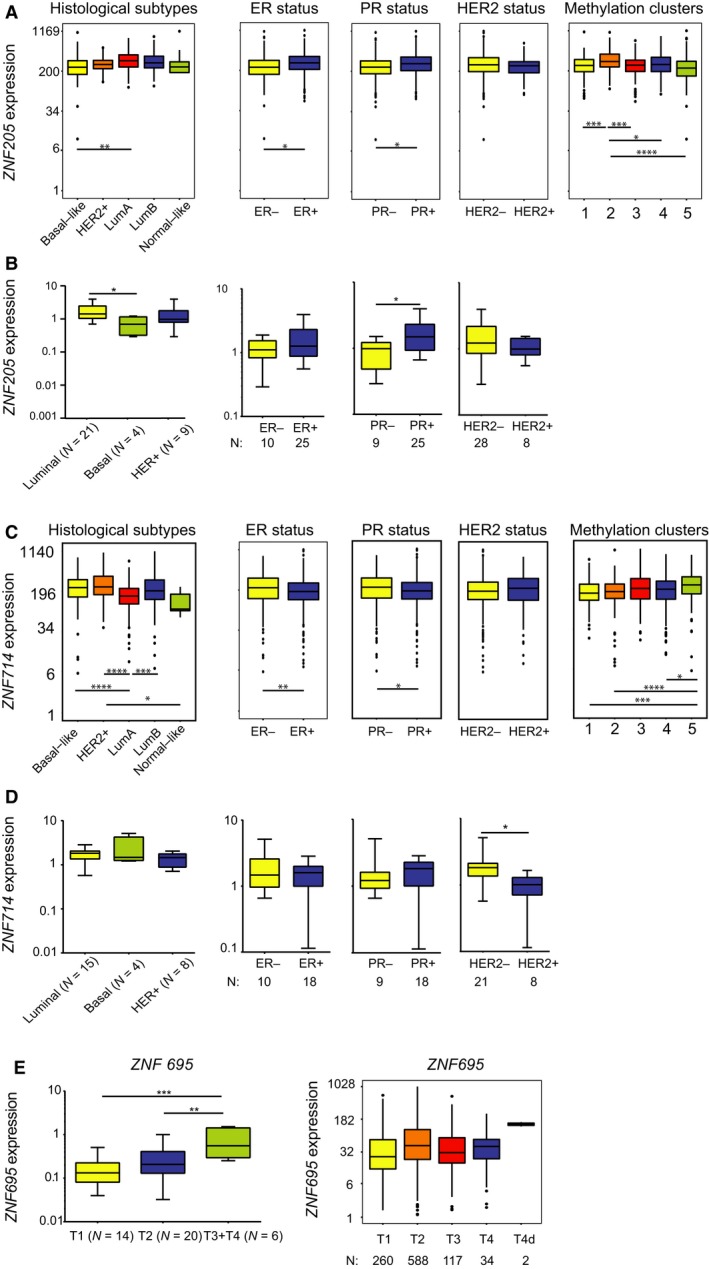
ZNF205 and ZNF714 correlate with various clinical and molecular parameters in BRCA samples. (A) ZNF205 expression positively correlates with luminal A subtype, ER+, and PR+ tumors, while its expression is the highest in methylation cluster 2 in TCGA datasets. (B) ZNF205 expression positively correlates with luminal A subtype and PR+ tumors in an independent set of breast cancer tissues. (C) ZNF714 expression positively correlates with HER2+, ER−, and PR− tumors, while its expression is the highest in methylation cluster 5 in TCGA datasets. (D) ZNF714 expression positively correlates with HER2− tumors in an independent set of breast cancer tissues. (E) ZNF695 expression positively correlates with advanced tumor stage in an independent set of breast cancer tissues, but not in the TCGA dataset. (A, C) Basal‐like (n = 98), HER2‐enriched (n = 58), luminal A (n = 233), luminal B (n = 129), normal‐like (n = 8), ER+ (n = 603), ER− (n = 179), PR+ (n = 526), PR− (n = 253), HER2+ (n = 115), HER2− (n = 654), methylation cluster 1: (n = 137), 2: (n = 174), 3: (n = 101), 4: (n = 246), 5: (n = 295). (B, D, E) Number of samples provided in the figure. (A–E) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 as assessed by t‐test with Tukey HSD correction (A, C, E) or unpaired t‐test (B, D).
As far as other clinicopathological features are concerned, the expression of the majority of KRAB‐ZNFs did not present any distinct trend of association with TNM classification, tumor stage, or patient gender. Only ZNF695 exhibited a positive correlation between expression and tumor size in the BRCA tissue panel, but not in the TCGA dataset (Fig. 8E). Finally, our survival analysis indicated that the expression of KRAB‐ZNFs may act as a risk factor. Patients with high expression of five out of 10 analyzed KRAB‐ZNFs presented significantly shorter overall survival than those with low expression (Fig. 9A–E). This included ZNF273 (P = 0.003, hazard ratio = 1.8), ZNF320 (P < 0.001, hazard ratio = 2.0), ZNF485 (P < 0.0001, hazard ratio = 2.3), ZNF525 (P = 0.003, hazard ratio = 1.8), and ZNF643 (P = 0.002, hazard ratio = 2.4). In contrast, high expression was associated with better prognosis in the case of ZNF205 (P < 0.001, hazard ratio = 0.5), ZNF707 (P = 0.001, hazard ratio = 0.5), and ZNF789 (P = 0.017, hazard ratio = 0.5) (Fig. 9F–H).
Figure 9.
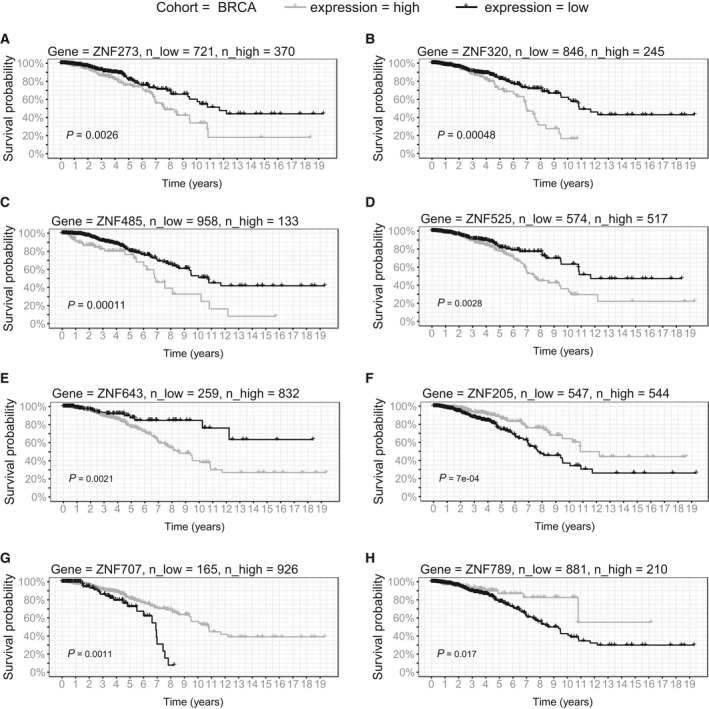
Survival analysis in TCGA BRCA patients. Kaplan–Meier curves represent ZNF273 (A), ZNF320 (B), ZNF485 (C), ZNF525 (D), ZNF643 (E), ZNF205 (F), ZNF707 (G), and ZNF789 (H) association with patient survival in LUSD. KRAB‐ZNFs expression was transformed into binary high/low groups based on maximally ranked statistics. P‐values assessed by log‐rank test and number of patients in each group is provided in the figure.
4. Discussion
The KRAB‐ZNF family is highly complex because it comprises a large number of homologous genes, gene isoforms, and pseudogenes. Such an enormous variety has so far hampered detailed molecular characterization of these proteins. While a growing number of studies reports KRAB‐ZNF involvement in multiple aspects of tumor biology, we aimed to provide a comprehensive portrayal of cancer‐associated KRAB‐ZNFs using publicly available TCGA pan‐cancer datasets. The results obtained from TCGA data were further validated in the independent sets of lung and breast cancer cell lines and tissues. Interestingly, we found many KRAB‐ZNF factors commonly downregulated and only a subset of a few KRAB‐ZNFs upregulated in multiple cancer types. This observation is in agreement with the results reported by Addison et al. (2015). Their study indicated that in the breast cancer TCGA datasets, the majority of KRAB‐ZNFs were downregulated, and only a small fraction was expressed at a higher level. These alterations were attributed to genomic changes involving nonsense mutations, genomic deletions, and amplifications (Addison et al., 2015). Similarly, we observed a small subgroup of KRAB‐ZNFs, overexpressed in multiple cancer types (Fig. 1A). KRAB‐ZNFs are known for their repressive function acting through epigenetic mechanisms, such as deposition of H3K9me3 and DNA methylation (Czerwinska et al., 2017; Ecco et al., 2017; Groner et al., 2010). It is well established that KRAB‐ZNFs bind to the sequences of transposable elements (TEs; Ecco et al., 2017). This leads not only to the restraint of TE expression and transposition (Jacobs et al., 2014), but also to the domestication of their regulatory sequences (e.g., promoters, enhancers) for the advantage of a cell (Ecco et al., 2016; Imbeault et al., 2017; Trono, 2015). It is noteworthy that the genes under the control of KRAB‐ZNF may undergo not only repression, but also activation (Busiello et al., 2017; Frietze et al., 2010; Lupo et al., 2013). At this stage, it is difficult to envisage how the cancer‐associated KRAB‐ZNFs affect tumor behavior, and further functional analyses are necessary to answer this question. From the set of upregulated KRAB‐ZNFs, only few were previously linked to cancer. Nevertheless, their exact molecular functions remain unknown. This includes ZNF695 (Juarez‐Mendez et al., 2013; Li et al., 2015, 2017; Takahashi et al., 2015), ZNF320 (Chernova et al., 2001), ZNF138 (Tommerup and Vissing, 1995), ZNF200 (Peedicayil et al., 2010), ZNF707 (Nesslinger et al., 2007), and ZNF354A (von Eyben, 2004). Interestingly, cancer‐related upregulation of ZNF695, ZNF714, and ZNF138 mirrors the differential expression observed between pluripotent stem cells and differentiated cells (Oleksiewicz et al., 2017). This observation underlines the similarities between cancer and stem cell expression profiles.
We further focused our attention on the differential expression of splicing isoforms. We observed that the majority of variants was detected both in normal and cancer tissues, but as expected, they had a higher level in tumors. The upregulation of the isoforms in malignant tissue seemed to occur simultaneously rather than through a variant switch mechanism. The differential expression of KRAB‐ZNF splicing isoforms in cancer was reported only by Juarez‐Mendez and colleagues (Juarez‐Mendez et al., 2013), who demonstrated a specific increase of ZNF695 variants in ovarian cancer compared to normal cells. Interestingly, we frequently observed increased expression of the variants that are not translated to a protein or are short forms devoid of KRAB or zinc finger domains. It is likely that this phenomenon occurs due to the utilization of similar activation mechanisms that augment the expression of all involved isoforms. However, additional work is required to test this hypothesis. Also, future studies are needed to elucidate the relevance of individual isoforms for the functioning of a tumor cell.
Out of all analyzed cancer‐associated KRAB‐ZNFs, ZNF695 seems to be the most interesting factor. In general, it showed a striking upregulation in many examined tumor types compared to their normal counterparts. Moreover, ZNF695 expression was higher in more aggressive basal‐like and HER2‐enriched breast cancers associated with early relapse and poor clinical outcome (Kennecke et al., 2010; Sorlie et al., 2001, 2003). This observation stays in agreement with the recent study by Li et al. (2015), who also revealed high levels of ZNF695 in basal‐like and HER2‐enriched tumors in three large breast cancer cohorts. In their study, ZNF695 was classified as one of 16 master regulators capable of differentiating luminal and nonluminal BRCA subtypes. ZNF695 was also shown to upregulate cell cycle progression genes (e.g., CDK1, PLK1) cooperatively with other master regulators in nonluminal breast cancers (Li et al., 2015). Also, in lung cancer, ZNF695 might be indirectly associated with proliferation. We observed that in LUAD ZNF695 expression is significantly higher in bronchioid and magnoid mRNA subtypes that demonstrate overrepresentation of growth and proliferation pathways, respectively (Hayes et al., 2006). In LUSC, ZNF695 revealed the highest level in the primitive mRNA subtype with overexpressed proliferation genes (Wilkerson et al., 2010). Furthermore, we have previously shown that ZNF695 was upregulated in pluripotent stem cells compared to more specialized cell types, whereas its knockdown resulted in the loss of self‐renewal properties and differentiation of pluripotent stem cells (Oleksiewicz et al., 2017). Its involvement in the stem cell phenotype was particularly evident in breast cancers, in which ZNF695 expression was the most abundant in basal‐like tumors, known for their high content of stem cells (Honeth et al., 2008; Park et al., 2010). Thus, it is tempting to speculate that ZNF695 may contribute to cancer stem cell identity, at least in basal‐like breast tumors. Nevertheless, further studies are needed to validate such an association.
5. Conclusions
In conclusion, we performed a TCGA pan‐cancer expression analysis of the KRAB‐ZNF family of genes and showed that a small subset of its members was commonly upregulated in multiple cancer types. We further validated this observation in independent sets of breast and lung cancer tissues and cell lines. Next, we demonstrated that the expression of the majority of cancer‐associated KRAB‐ZNF splicing isoforms was simultaneously upregulated in tumors as compared to normal adjacent tissues. Furthermore, we found that the expression of KRAB‐ZNFs in breast and lung cancer tissues correlated with histological and molecular subtypes, as well as patient survival. Our observations indicated that these KRAB‐ZNFs may play an oncogenic role during carcinogenesis, which makes them potential biomarkers and/or targets of anticancer therapies. Nevertheless, further studies are needed to elucidate their exact molecular functioning.
Author contributions
UO and MM contributed to the conceptualization of this article. UO, MM, RC, KK, PB, and TL involved in methodology and investigation. UO, MM, RC, KK, and PB performed software, validation, and formal analysis. UO and MM wrote and drafted the original manuscript. UO, MM, RC, KK, PB, TL, and AM wrote, reviewed, and edited the manuscript. UO, MM, KK, RC, and PB involved in visualization. UO, PB, and AM involved in supervision of the article. UO and PB contributed to funding acquisition.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgements
This work was financed by the National Science Centre, Poland, grant No. 2015/17/B/NZ2/03689 to Urszula Oleksiewicz. Przemysław Biecek was financially supported by the Polish National Science Centre grant 2016/21/B/ST6/02176.
References
- Addison JB, Koontz C, Fugett JH, Creighton CJ, Chen D, Farrugia MK, Padon RR, Voronkova MA, McLaughlin SL, Livengood RH et al (2015) KAP1 promotes proliferation and metastatic progression of breast cancer cells. Cancer Res 75, 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S and Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busiello T, Ciano M, Romano S, Sodaro G, Garofalo O, Bruzzese D, Simeone L, Chiurazzi F, Fiammetta Romano M, Costanzo P et al (2017) Role of ZNF224 in cell growth and chemoresistance of chronic lymphocytic leukemia. Hum Mol Genet 26, 344–353. [DOI] [PubMed] [Google Scholar]
- Chen K, Yu G, Gumireddy K, Li A, Yao W, Gao L, Chen S, Hao J, Wang J, Huang Q et al (2015) ZBRK1, a novel tumor suppressor, activates VHL gene transcription through formation of a complex with VHL and p300 in renal cancer. Oncotarget 6, 6959–6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova OB, Hunyadi A, Malaj E, Pan H, Crooks C, Roe B and Cowell JK (2001) A novel member of the WD‐repeat gene family, WDR11, maps to the 10q26 region and is disrupted by a chromosome translocation in human glioblastoma cells. Oncogene 20, 5378–5392. [DOI] [PubMed] [Google Scholar]
- Cho JG, Park S, Lim CH, Kim HS, Song SY, Roh TY, Sung JH, Suh W, Ham SJ, Lim KH et al (2016) ZNF224, Kruppel like zinc finger protein, induces cell growth and apoptosis‐resistance by down‐regulation of p21 and p53 via miR‐663a. Oncotarget 7, 31177–31190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsinotti A, Kapopoulou A, Gubelmann C, Imbeault M, Santoni de Sio FR, Rowe HM, Mouscaz Y, Deplancke B and Trono D (2013) Global and stage specific patterns of Kruppel‐associated‐box zinc finger protein gene expression in murine early embryonic cells. PLoS One 8, e56721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerwinska P, Shah PK, Tomczak K, Klimczak M, Mazurek S, Sozanska B, Biecek P, Korski K, Filas V, Mackiewicz A et al (2017) TRIM28 multi‐domain protein regulates cancer stem cell population in breast tumor development. Oncotarget 8, 863–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA and Kouzarides T (2012) Cancer epigenetics: from mechanism to therapy. Cell 150, 12–27. [DOI] [PubMed] [Google Scholar]
- Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P and Trono D (2016) Transposable elements and their KRAB‐ZFP controllers regulate gene expression in adult tissues. Dev Cell 36, 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G, Imbeault M and Trono D (2017) KRAB zinc finger proteins. Development 144, 2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eyben FE (2004) Chromosomes, genes, and development of testicular germ cell tumors. Cancer Genet Cytogenet 151, 93–138. [DOI] [PubMed] [Google Scholar]
- Florio F, Cesaro E, Montano G, Izzo P, Miles C and Costanzo P (2010) Biochemical and functional interaction between ZNF224 and ZNF255, two members of the Kruppel‐like zinc‐finger protein family and WT1 protein isoforms. Hum Mol Genet 19, 3544–3556. [DOI] [PubMed] [Google Scholar]
- Frietze S, Lan X, Jin VX and Farnham PJ (2010) Genomic targets of the KRAB and SCAN domain‐containing zinc finger protein 263. J Biol Chem 285, 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresner P, Gromadzinska J and Wasowicz W (2009) Reference genes for gene expression studies on non‐small cell lung cancer. Acta Biochim Pol 56, 307–316. [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, Bucher P and Trono D (2010) KRAB‐zinc finger proteins and KAP1 can mediate long‐range transcriptional repression through heterochromatin spreading. PLoS Genet 6, e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D and Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Harada Y, Kanehira M, Fujisawa Y, Takata R, Shuin T, Miki T, Fujioka T, Nakamura Y and Katagiri T (2010) Cell‐permeable peptide DEPDC1‐ZNF224 interferes with transcriptional repression and oncogenicity in bladder cancer cells. Cancer Res 70, 5829–5839. [DOI] [PubMed] [Google Scholar]
- Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A, Socinski MA, Perou C and Meyerson M (2006) Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol 24, 5079–5090. [DOI] [PubMed] [Google Scholar]
- Honeth G, Bendahl PO, Ringner M, Saal LH, Gruvberger‐Saal SK, Lovgren K, Grabau D, Ferno M, Borg A and Hegardt C (2008) The CD44+/CD24‐ phenotype is enriched in basal‐like breast tumors. Breast Cancer Res 10, R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley S, Baggott DM, Hamilton AT, Tran‐Gyamfi M, Yang S, Kim J, Gordon L, Branscomb E and Stubbs L (2006) A comprehensive catalog of human KRAB‐associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res 16, 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeault M, Helleboid PY and Trono D (2017) KRAB zinc‐finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550–554. [DOI] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR and Haussler D (2014) An evolutionary arms race between KRAB zinc‐finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez‐Mendez S, Zentella‐Dehesa A, Villegas‐Ruiz V, Perez‐Gonzalez OA, Salcedo M, Lopez‐Romero R, Roman‐Basaure E, Lazos‐Ochoa M, Montes de Oca‐Fuentes VE, Vazquez‐Ortiz G et al (2013) Splice variants of zinc finger protein 695 mRNA associated to ovarian cancer. J Ovarian Res 6, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara T, Inoue S, Ide H, Kashiwagi E, Ohtake S, Mizushima T, Li P, Li Y, Zheng Y, Uemura H et al (2016) ZKSCAN3 promotes bladder cancer cell proliferation, migration, and invasion. Oncotarget 7, 53599–53610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO and Gelmon K (2010) Metastatic behavior of breast cancer subtypes. J Clin Oncol 28, 3271–3277. [DOI] [PubMed] [Google Scholar]
- Li R, Campos J and Iida J (2015) A gene regulatory program in human breast cancer. Genetics 201, 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B and Dewey CN (2011) RSEM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Kuang L, Zhu B, Chen J, Wang X and Huang X (2017) Identification of prognostic risk factors of acute lymphoblastic leukemia based on mRNA expression profiling. Neoplasma 64, 494–501. [DOI] [PubMed] [Google Scholar]
- Liang Y, Li Q, Chen K, Ni W, Zhan Z, Ye F, Li Y, Fang Y, Zhang F, Chen L et al (2017) Zinc finger protein 307 functions as a tumor suppressor and inhibits cell proliferation by inducing apoptosis in hepatocellular carcinoma. Oncol Rep 38, 2229–2236. [DOI] [PubMed] [Google Scholar]
- Lin LF, Chuang CH, Li CF, Liao CC, Cheng CP, Cheng TL, Shen MR, Tseng JT, Chang WC, Lee WH et al (2010) ZBRK1 acts as a metastatic suppressor by directly regulating MMP9 in cervical cancer. Cancer Res 70, 192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinas‐Arias P, Esteller M (2017) Epigenetic inactivation of tumour suppressor coding and non‐coding genes in human cancer: an update. Open Biology 7, 170152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo A, Cesaro E, Montano G, Zurlo D, Izzo P and Costanzo P (2013) KRAB‐zinc finger proteins: a repressor family displaying multiple biological functions. Curr Genomics 14, 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AN, Wang H, Guo R, Wang YX, Li W, Cui J, Wang G, Hoffman AR and Hu JF (2014) Targeted gene suppression by inducing de novo DNA methylation in the gene promoter. Epigenetics Chromatin 7, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltseva DV, Khaustova NA, Fedotov NN, Matveeva EO, Lebedev AE, Shkurnikov MU, Galatenko VV, Schumacher U and Tonevitsky AG (2013) High‐throughput identification of reference genes for research and clinical RT‐qPCR analysis of breast cancer samples. J Clin Bioinform 3, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesslinger NJ, Sahota RA, Stone B, Johnson K, Chima N, King C, Rasmussen D, Bishop D, Rennie PS, Gleave M et al (2007) Standard treatments induce antigen‐specific immune responses in prostate cancer. Clin Cancer Res 13, 1493–1502. [DOI] [PubMed] [Google Scholar]
- Oleksiewicz U, Gladych M, Raman AT, Heyn H, Mereu E, Chlebanowska P, Andrzejewska A, Sozanska B, Samant N, Fak K et al (2017) TRIM28 and interacting KRAB‐ZNFs control self‐renewal of human pluripotent stem cells through epigenetic repression of pro‐differentiation genes. Stem Cell Reports 9, 2065–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Lee HE, Li H, Shipitsin M, Gelman R and Polyak K (2010) Heterogeneity for stem cell‐related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res 16, 876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peedicayil A, Vierkant RA, Hartmann LC, Fridley BL, Fredericksen ZS, White KL, Elliott EA, Phelan CM, Tsai YY, Berchuck A et al (2010) Risk of ovarian cancer and inherited variants in relapse‐associated genes. PLoS One 5, e8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A et al (2011) In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell 44, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A (2017) Cancer statistics, 2017. CA Cancer J Clin 67, 7–30. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS et al (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98, 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S et al (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 100, 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Yamahsita S, Matsuda Y, Kishino T, Nakajima T, Kushima R, Kato K, Igaki H, Tachimori Y, Osugi H et al (2015) ZNF695 methylation predicts a response of esophageal squamous cell carcinoma to definitive chemoradiotherapy. J Cancer Res Clin Oncol 141, 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumors. Nature 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommerup N and Vissing H (1995) Isolation and fine mapping of 16 novel human zinc finger‐encoding cDNAs identify putative candidate genes for developmental and malignant disorders. Genomics 27, 259–264. [DOI] [PubMed] [Google Scholar]
- Trono D (2015) Transposable elements, polydactyl proteins, and the genesis of human‐specific transcription networks. Cold Spring Harb Symp Quant Biol 80, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A et al (2015) Proteomics. Tissue‐based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, Muldrew K, Miller CR, Randell SH, Socinski MA et al (2010) Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res 16, 4864–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M, Jakobsson J, Szulc J, Liao S, Quazzola A, Beermann F, Aebischer P and Trono D (2007) The Kruppel‐associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J Biol Chem 282, 34535–34541. [DOI] [PubMed] [Google Scholar]
- Xiang S, Xiang T, Xiao Q, Li Y, Shao B and Luo T (2017) Zinc‐finger protein 545 is inactivated due to promoter methylation and functions as a tumor suppressor through the Wnt/beta‐catenin, PI3K/AKT and MAPK/ERK signaling pathways in colorectal cancer. Int J Oncol 51, 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Xiang T, Luo X, Li C, Li Q, Peng W, Li L, Li S, Wang Z, Tang L et al (2014) Zinc‐finger protein 545 inhibits cell proliferation as a tumor suppressor through inducing apoptosis and is disrupted by promoter methylation in breast cancer. PLoS One 9, e110990. [DOI] [PMC free article] [PubMed] [Google Scholar]


