Abstract
Mycobacteria use type VII secretion systems to secrete proteins across their highly hydrophobic diderm cell envelope. Pathogenic mycobacteria, such as Mycobacterium tuberculosis and Mycobacterium marinum, have up to five of these systems, named ESX-1 to ESX-5. Most of these systems contain a set of five conserved membrane components, of which the four Ecc proteins form the core membrane-embedded secretion complex. The fifth conserved membrane protein, mycosin protease (MycP), is not part of the core complex but is essential for secretion, as it stabilizes this membrane complex. Here we investigated which MycP domains are required for this stabilization by producing hybrid constructs between MycP1 and MycP5 in M. marinum and analyzed their effect on ESX-1 and ESX-5 secretion. We found that both the protease and transmembrane domain are required for the ESX system-specific function of mycosins. In addition, we observed that the transmembrane domain strongly affects MycP protein levels. We also show that the extended loops 1 and 2 in the protease domain are probably primarily involved in MycP stability, whereas loop 3 and the MycP5-specific loop 5 are dispensable. The atypical propeptide, or N-terminal extension, is required only for MycP stability. Finally, we show that the protease domain of MycPP1, encoded by the esx-P1 locus on the pRAW plasmid, is functionally redundant to the protease domain of MycP5. These results provide the first insight into the regions of mycosins involved in interaction with and stabilization of their respective ESX complexes.
Keywords: mycobacteria, protein secretion, serine protease, protein stability, protein complex, Mycobacterium marinum, T7SS, type VII secretion system
Introduction
Mycobacteria, including Mycobacterium tuberculosis, have a set of specialized secretion systems, called type VII secretion (T7S)2 systems, to secrete proteins across their highly hydrophobic cell envelope (1). This group of bacteria contains up to five genomic clusters coding for these T7S systems, named ESX-1 to ESX-5 (1, 2). Three of these systems, ESX-1, ESX-3, and ESX-5, have been shown to be functional and to play critical roles in virulence and bacterial physiology (3–6). The ESX-1 system is required for phagosomal rupture both in M. tuberculosis and Mycobacterium marinum (3, 7, 8), and is therefore a crucial factor for completing the intraphagocytic stage of these pathogens. The importance of ESX-1 for virulence is further illustrated by the absence of the esx-1 region in the live vaccine strain Mycobacterium bovis BCG (4). The ESX-3 and ESX-5 systems have very different functions, as they have been linked to iron and fatty acid uptake, respectively (5, 6, 9). Because of their role in metabolite and nutrient uptake, both of these systems are essential for in vitro growth in pathogenic species. However, the essentiality of the ESX-5 system can be circumvented in M. marinum by permeabilizing the unique mycobacterial outer membrane, either by mutating genes involved in the biosynthesis of the outer membrane lipid phthiocerol dimycocerosate or by the introduction of MspA, an outer membrane porin from the nonpathogenic Mycobacterium smegmatis (6).
In the last decade, information has been obtained about the composition of the membrane-embedded ESX complexes (10, 11) and the structure and (possible) functioning of several of its components, e.g. of EccB, EccC, and EccD (12, 13). This has recently seen a strong leap forward with the first structure of the core ESX-5 secretion complex of Mycobacterium xenopi, imaged by negative-stain EM (14). From the observed 6-fold symmetry together with stoichiometry measurements, it can be deduced that the ESX-5 complex consists of six copies of each of the four conserved membrane components that make up the complex, i.e. EccB5, EccC5, EccD5, and EccE5 (14). The data on the M. xenopi ESX-5 complex match the composition determined previously for the ESX-1 and ESX-5 systems of M. marinum and the ESX-5 system of M. bovis BCG by protein pulldown experiments (10, 11). Notably, the fifth membrane component, the mycosin protease (MycP), was not detected in significant amounts in any of these purified complexes. Despite this, mycosins are essential for the proper functioning of the ESX-1 and ESX-5 systems, as mycP1 or mycP5 knockouts result in a secretion deficiency by the respective systems in different species (11, 15), and transposon mutagenesis experiments indicate that MycP3 is also essential in M. tuberculosis (16, 17). Furthermore, mycosins are widely conserved in T7S, not only in all mycobacterial ESX systems but also in T7S clusters of other Actinobacteria, suggesting that they have a critical function in T7S (18, 19). Recently, we showed that both MycP1 and MycP5 stabilize their respective ESX complexes in M. marinum, explaining their essentiality for T7S in mycobacteria (11).
Mycosins belong to the family of subtilisin-like serine proteases, and the available crystal structures indeed show a subtilisin-like fold (20, 21). However, there are also some differences from classical subtilisins, most prominent for the so-called propeptide. Although subtilisins contain an N-terminal propeptide that blocks the active site, which is cleaved off upon protein maturation to activate the protease, for mycosins, this N-terminal extension is not processed but remains part of the mature protease domain. Furthermore, this extension is not blocking the substrate binding pocket but is elongated and tightly wrapped around the protease domain (20–22). As the N-terminal region of mycosins does not contain any of the typical characteristics of a propeptide (22), we refer to it as the N-terminal extension (NE) of mycosins in this study. Compared with typical subtilisins, the mycosins have a deeper active-site groove because of the presence of the N-terminal extension in the mature protein and three extended loops that surround the active site (20, 21). This suggests that mycosins have a higher substrate selectivity compared with most subtilisin-like proteases. Finally, mycosins also have a C-terminal transmembrane (TM) domain, connected to the protease domain by an ∼28-amino-acid-long linker region. Because of the presence of a typical signal sequence preceding the N-terminal extension and a positively charged short C-terminal tail, the protease domain is predicted to be localized in the periplasm.
So far, the number of identified substrates of mycosins is extremely limited. Only MycP1 has been shown to cleave the C terminus of EspB, one of the substrates of the ESX-1 system, in M. tuberculosis and M. marinum (11, 15). Surprisingly, the proteolytic activity of MycP1 is not required for the essential function of this protein in secretion (11, 15). Also, an active-site mutant of mycP5 can still mediate ESX-5 secretion in M. marinum (11). This discrepancy suggests that the second essential function of mycosins is not dependent on their proteolytic activity. As already mentioned, mycosins are involved in stabilization of their respective ESX complex without having a role in complex assembly (11). This conclusion is based on the observation that we could detect the ESX-1 or ESX-5 complex in a mycP1 or mycP5 knockout, respectively, when we cross-linked cell envelope fractions prior to detergent extraction of the membrane-embedded ESX complexes (11). Therefore, the ESX complex is apparently formed in the absence of mycosin but disassociates rapidly upon detergent extraction. The proteolytic activity of MycP is not required for this function, as an active site mutation did not affect the stabilizing effect of MycP1 or MycP5 on its respective ESX complex (11). However, the MycP domain responsible for this function has yet to be identified.
Here we set out to identify the domains of MycP1 and MycP5 that are involved in the essential role of mycosins in T7S in M. marinum by producing specific truncated versions and hybrid proteins in which individual domains of MycP1 and MycP5 were exchanged. Our results show that the N-terminal extension is required for structural integrity, whereas both the protease and TM domains of MycP1 and MycP5 are system-specific and essential for the functionality of their respective ESX complexes. By further dissecting the protease domain, we found that the primary function of the extended loops is not related to the stabilization of the membrane complex. Finally, we show that the protease domain of MycPP1 of the plasmid-encoded ESX-P1 system can complement the function of the protease domain of MycP5. This study provides important functional insights into the different domains of the mycosin proteases.
Results
The N-terminal extension and the TM domain are essential for mycosin functioning
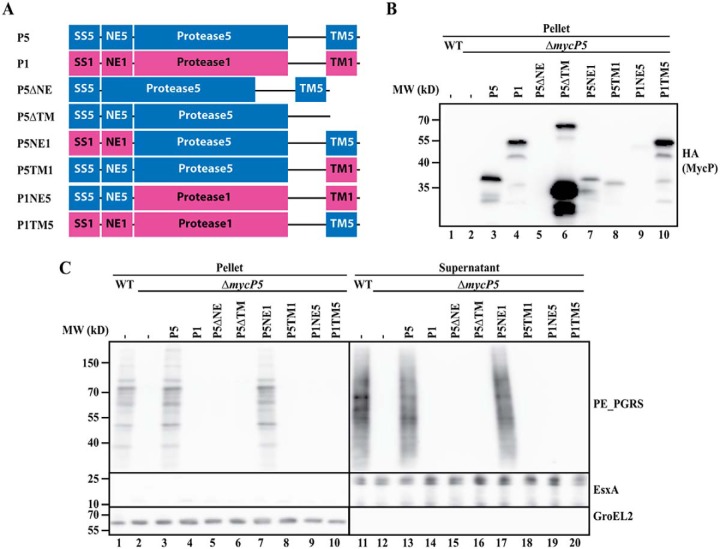
To determine which domains are involved in the essential function of mycosins in T7S, we produced mycP1 and mycP5 truncations in which either the NE or the TM domain was removed (Figs. 1A and 2A) and analyzed their functionality in the previously described mycP1 and of mycP5 knockout strains of M. marinum (11). To be able to dissect the functionality of the different MycP5 constructs in ESX-5 secretion, the essentiality of ESX-5 for in vitro growth was alleviated by introduction of the outer membrane porin MspA (6, 23). As reported previously (11, 15), knockout of both mycP1 and mycP5 resulted in complete secretion deficiency by their respective ESX systems. We showed this deficiency by the absence of the ESX-5 substrate group of PE_polymorphic GC-rich repetitive sequences (PGRS) proteins in the pellet and supernatant for the mycP5 knockout (Fig. 1C, lanes 2 and 12) and the absence of the ESX-1 substrates EsxA and EspB in the supernatant fraction of the mycP1 knockout (Fig. 2C, lane 12). The secretion by the mutants can be complemented by exogenous expression of their respective WT copies bearing a C-terminal HA tag for detection (Figs. 1C, lane 13, P5, and 2C, lane 13, P1). As expected, secretion could not be restored by exogenous expression of MycP1 in the mycP5 knockout strain and vice versa (Figs. 1C, lane 14, and 2C, lane 14). An additional observation was that, although MycP5 is larger than MycP1, the HA signal for MycP5 showed a lower molecular weight compared with MycP1 (Fig. 1B, lanes 3 and 4, P1 and P5). Based on the size of the detected fragment (∼37 kDa), we concluded that MycP5 is processed somewhere in its protease domain, which we will discuss further below.
Figure 1.
A, schematic overview of (hybrid or truncated) mycP5 (blue) and mycP1 (purple) constructs used to complement a mycP5 knockout in M. marinum. The constructs contain the following parts: SS, signal sequence; NE, N-terminal extension; Protease, protease domain (including the linker region between the protease and TM domain); TM, transmembrane domain (including the short C-terminal tail). The numbers indicate the origin of the fragment, i.e. 1 for MycP1 and 5 for MycP5. The exact residues of the swapped domains are indicated in Fig. S4. In addition, all proteins have a C-terminal HA tag. B, immunoblot detection of the HA-tagged constructs expressed in the mycP5 knockout. MW, molecular weight. C, immunoblot analysis of cell pellets and supernatants of WT M. marinum and a mycP5 knockout, complemented with the constructs depicted in A. Proteins were visualized with anti-PE_PGRS (ESX-5 substrates), anti-EsxA (ESX-1 substrate, supernatant loading control), and anti-GroEL2 (cytosolic loading control).
Figure 2.
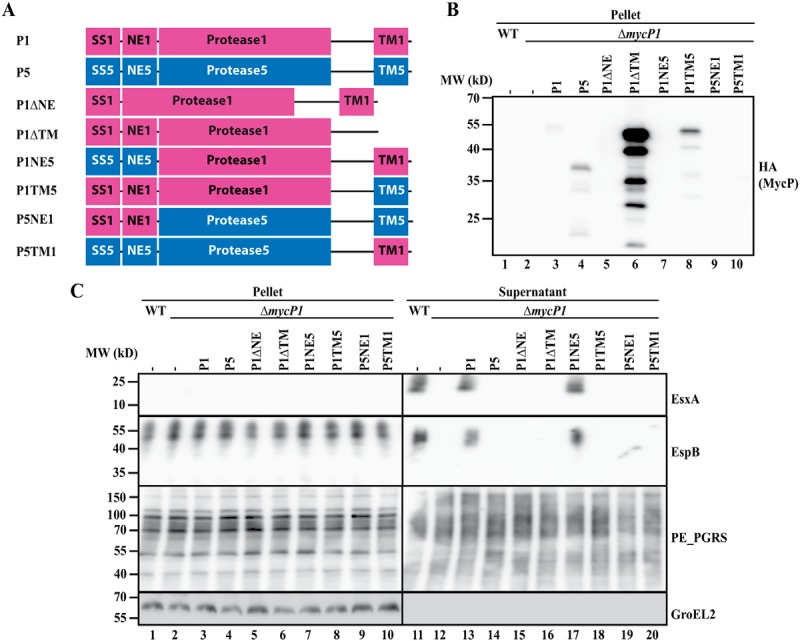
A, schematic overview of (hybrid or truncated) mycP1 (purple) and mycP5 (blue) constructs used to complement a mycP1 knockout in M. marinum. All constructs contain a C-terminal HA tag. See Fig. 1A for an explanation of the abbreviated domains. B, immunoblot detection of the HA-tagged constructs, expressed in the mycP1 knockout. MW, molecular weight. C, immunoblot analysis of cell pellets and supernatants of WT M. marinum and a mycP5 knockout, complemented with the constructs depicted in A. Proteins were visualized with anti-EsxA and anti-EspB (ESX-1 substrates), anti-PE_PGRS (ESX-5 substrates, supernatant loading control), and anti-GroEL2 (cytosolic loading control).
Next we determined the effect of deleting the N-terminal extension of MycP5 (P5ΔNE) and MycP1 (P1ΔNE). We found that, for both MycP5 and MycP1, this deletion resulted in complete loss of protein secretion by their respective ESX systems (Figs. 1C, lane 15, and 2C, lane 15). In addition, although we could readily detect HA-tagged WT MycP5 and MycP1 in bacterial pellet fractions, we were unable to detect these truncated proteins via the same C-terminal HA tag in the same bacterial fraction (Figs. 1B, lane 5, and 2B, lane 5), suggesting that the N-terminal extension of mycosins is required for protein stability.
Although deletion of the NE reduced the stability of mycosins, removing the C-terminal TM domain resulted in a strong increase in mycosin levels compared with their WT versions. Although this effect was observed for both MycP5 and MycP1, especially in the case of MycP1ΔTM, the increase in HA signal was of such intensity that the other hybrid constructs were not or barely detectable without overexposing the MycP1ΔTM signal (Figs. 1B, lane 6, P5ΔTM, and 2B, lane 6, P1ΔTM). Therefore, we included an additional immunoblot lacking the truncated MycP1 construct to allow more accurate comparison with WT MycP1 or MycP5 (Fig. S1). Interestingly, for the MycP5 truncate, besides the C-terminal portion of ∼37 kDa, an ∼65 kDa band could be detected, which probably represented the unprocessed full-length protein. However, although present at high levels, the mycosin constructs lacking the TM domain were unable to complement ESX secretion of their respective knockouts (Figs. 1C, lane 16, and 2C, lane 16). Furthermore, expressing the highly stable truncated MycP5ΔTM did not result in a dominant-negative effect on ESX-5–mediated secretion in WT bacteria (Fig. S2). These results show that both the N-terminal extension and the C-terminal TM domain are required for the essential function of mycosins within their respective ESX systems.
Both the protease and TM domain of MycP1 and MycP5 are ESX system–specific
To distinguish between a general role in protein stability (for the N-terminal extension) or subcellular localization (for the TM domain) and a system-specific function of the separate domains, we created hybrid mycP1 and mycP5 constructs in which individual domains were exchanged between the two mycosins, again with a C-terminal HA tag for detection (Figs. 1A and 2A). As deletion of the N-terminal extension resulted in a strong decrease in mycosin levels, we first wanted to confirm whether this domain is only involved in structural stability and does not play a system-specific role. Indeed, although we did observe a decrease in mycosin levels when we exchanged this domain between MycP5 and MycP1 (Figs. 1, A and B, lane 7, P5NE1 and 2, A and B, lane 7, P1NE5, and Fig. S1, lane 5), both constructs were still able to complement ESX-5 or ESX-1 secretion of the mycP5 and mycP1 knockouts, respectively (Figs. 1C, lane 17, and 2C, lane 17). With a similar approach, we exchanged the C-terminal TM domain of the mycosins to investigate whether they have a system-specific role. Although we observed decreased expression of MycP5 with the TM domain of MycP1 (Fig. 1B, lane 8, P5TM1), the opposite MycP1 construct showed substantial expression levels (Fig. 2B, lane 8, P1TM5, and Fig. S1, lane 6). Both constructs were unable to complement secretion by mycP5 or mycP1 knockout (Figs. 1C, lane 18, and 2C, lane 18), strongly suggesting that the TM domain of mycosins does not only function as a membrane anchor but that it has a specific function for its ESX system. Interestingly, we also tested whether all constructs were able to alleviate the essentiality of mycP5, i.e. by mediating growth in the absence of the outer membrane porin MspA (Table S2). All constructs that could complement protein secretion also supported growth of the mycP5 mutant in the absence of MspA. Although P5TM1 was unable to restore ESX-5 secretion in the mycP5 knockout, this construct did allow this mutant strain to grow, albeit with a strongly reduced growth rate. This suggests that this hybrid construct is able to complement the mycP5 mutant to some extent.
Finally, we determined whether the protease domain is also involved in the essential function of mycosins by introducing the hybrid constructs in the opposite mycP knockout, e.g. P1NE5 and P1TM5 in the mycP5 knockout and vice versa (Figs. 1A and 2A). These constructs were also unable to complement the mycosin knockouts and were thus unable to restore secretion of the analyzed ESX systems (Figs. 1C, lanes 19 and 20, and 2C, lanes 19 and 20). As we already showed that exchanging the N-terminal extension has no effect on the functionality of MycP1 or MycP5, the inability of P1TM5 and P5TM1 to complement the mycP5 and mycP1 knockout, respectively, must be contributed to the exchanged protease domain. In conclusion, although the N-terminal extension has a system-specific function in protein stability, both the protease and C-terminal TM domain of mycosins are required and specific for essential function within their respective ESX systems.
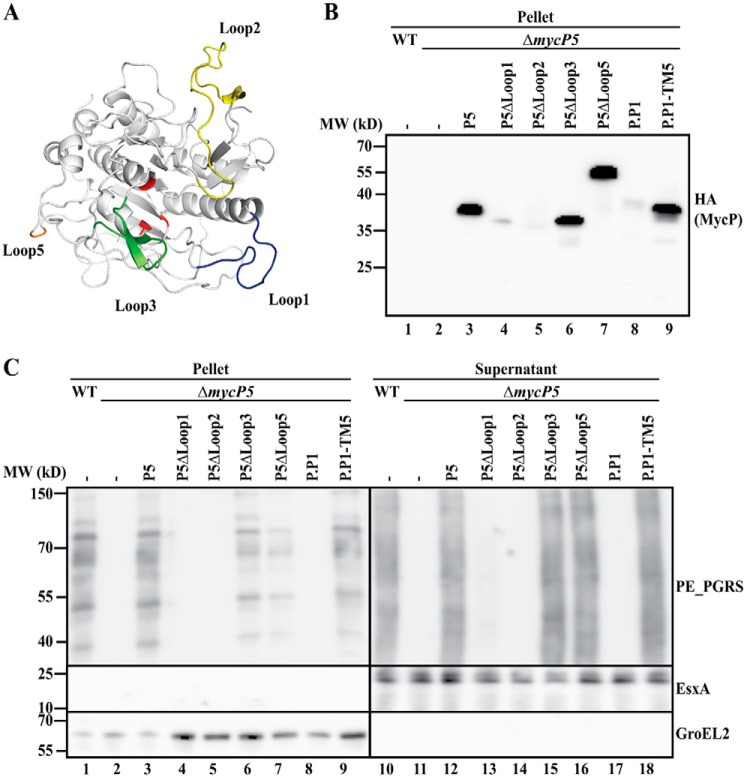
The extended loops of the protease domain are either dispensable or involved in structural stability
Next we set out to identify specific regions within the protease domain that are involved in the essential function of mycosins. A distinctive feature of mycosins compared with classical subtilisins is three extended loops that are present in the protease domain (loops 1–3) (Fig. 3A). In addition to these three conserved loops, MycP5 has an additional large extension to which we will refer as loop 5 in this study. This loop is not present in MycP1. In a structural model of MycP5, produced from the available MycP1 structure from M. smegmatis (20), loop 5 remained unstructured, and we therefore only show the start and end of it in the model shown in Fig. 3A. Because loops 1–3 and loop 5 are unique for mycosins, we hypothesized that they could be involved in the essential function of mycosins. By individually deleting these four loops of MycP5, we investigated their effect on stability and/or functionality. Deleting loop 1 or 2 resulted in reduced stability of MycP5; especially loop 2 deletion showed almost complete loss of MycP5 stability (Fig. 3B). In addition, both constructs resulted in complete loss of ESX-5 secretion (Fig. 3C, lanes 13 and 14). In contrast, deleting loop 3 or loop 5 had little to no effect on MycP5 stability or on ESX-5 secretion (Fig. 3, B, lanes 6 and 7, and C, lanes 15 and 16). Interestingly, deleting loop 5 resulted in a higher detected molecular weight compared with the WT HA-tagged MycP5 (Fig. 3B, lanes 3 and 7), indicating that MycP5 is cleaved somewhere in this loop and that this processing is not required for proper functioning. To investigate whether this processing is performed by MycP5 or by a different component of the ESX-5 system, we introduced both a WT and an active-site mutant variant of HA-tagged MycP5 in M. smegmatis, which lacks the ESX-5 system. However, we found that, in both cases, MycP5 was present as the processed form, showing that the responsible protease is unrelated to the ESX-5 system (Fig. S3C).
Figure 3.
A, model of MycP5, based on the available crystal structure of MycP1. The catalytic triad of the protease is depicted in red, and the three extended loops (1–3) surrounding the catalytic groove are depicted in blue, yellow, and green, respectively. The two residues bordering the extended loop 5 are depicted in orange, whereas loop 5 itself is not shown in this model. B, immunoblot detection of HA-tagged constructs expressed in the mycP5 knockout. P5, WT mycP5; P5ΔLoop1/2/3/5, mycP5 without loop 1, 2, 3, or 5; P.P1, mycPP1; P.P1-TM5, hybrid mycPP1 with the TM region of mycP5. The exact residues of the deleted and swapped regions are indicated in Fig. S4. MW, molecular weight. C, immunoblot analysis of cell pellets and supernatants of WT M. marinum and a mycP5 knockout, complemented with (truncated) mycP5 constructs or (hybrid) mycPP1 with the TM region of mycP5 (see Fig. S4 for the swapped residues). Proteins were visualized with anti-PE_PGRS (ESX-5 substrates), anti-EsxA (ESX-1 substrate, supernatant loading control), and anti-GroEL2 (cytosolic loading control).
Subsequently, we analyzed the role of loop 1 and 2 in protease activity using the ability of purified MycP1 to process its substrate EspB in vitro. Although we were unable to produce stable MycP1 with the deletion of loop 2, deletion of loop 1 resulted in substantial expression of the MycP1 protease domain. This purified protein was also active, as we could not detect a difference in the activity of purified WT MycP1 compared with the loop 1 truncation; in both cases, EspB was successfully processed in vitro (Fig. S3D, lanes 2 and 3, P1 and P1ΔL1). Finally, we exchanged the loop 1 of full-length MycP5 with the loop of MycP1 and analyzed its expression and functionality in the mycP5 knockout strain. This hybrid protein was stable (Fig. S3A, lane 4, P5-L1.P1) and resulted in successful complementation of ESX-5 secretion (Fig. S3B, lane 8). Together, the results show that, although loop 3 and loop 5 are dispensable for MycP5 stability and its essential role in ESX-5 functioning, loop 1 and 2 seem to have a structural role, similar to the N-terminal extension.
The protease domain of MycP5 can be functionally substituted by the protease domain of MycPP1 of the plasmid-encoded ESX-P1 system
As we were unable to obtain further functional insight from analysis of the extended loops in MycP5, and as there are no other distinctive regions or structures within the protease domain, we looked for a different approach. More recently, plasmid-localized esx clusters have been described for mycobacteria, most notably the esx-P1 cluster localized on the pRAW plasmid found in M. marinum E11 (24), involved in bacterial conjugation. Interestingly, the ESX-P1 system is most closely related to the ESX-5 system, e.g. MycPP1 has ∼59.5% identity to MycP5 (Fig. S4 and Table S1), which is substantially higher than the identity of MycP5 to MycP1 (∼46%, Table S1). Because of this high sequence similarity, we set out to investigate to what extent mycPP1 is able to complement the mycP5 knockout of M. marinum M, which does not contain the esx-P1 cluster. Unfortunately, we were able to detect only low protein levels of the HA-tagged MycPP1 (Fig. 3B, lane 8, P.P1), and this construct did not result in successful complementation of the mycP5 knockout (Fig. 3C, lane 17). In addition to this, the HA signal of the MycPP1 protein showed a comparable size as that of MycP5 (Fig. 3B, lane 3), indicating that MycPP1 is also processed, which is in line with the observation that this protein contains a loop 5 extension similar to MycP5 (Fig. S4). As we already determined that the TM domain plays an important role, we exchanged the TM domain of MycPP1 with that of MycP5. Interestingly, this not only resulted in a substantially more stable protein (Fig. 3B, lane 9, P.P1-TM5), but it was also capable of restoring ESX-5 secretion in the mycP5 knockout (Fig. 3C, lane 18). These results show that the protease domain of MycPP1 has functional similarity to that of MycP5 and further strengthens the notion that the TM domain of mycosins has a highly system-specific function.
The protease and TM domain of MycP are both involved in stabilization of the ESX complex
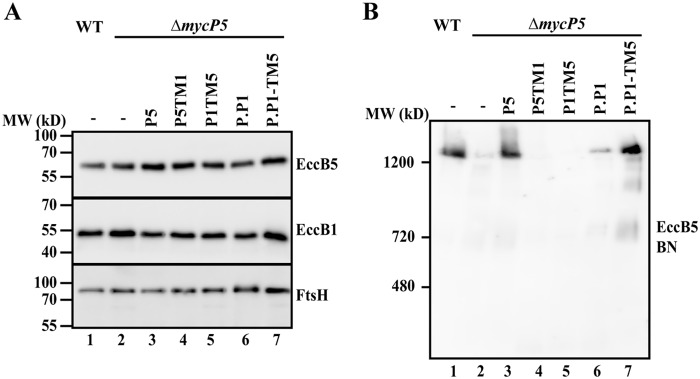
To further confirm that the essential role of the protease and TM domain is linked to the previously determined crucial role of mycosins in the stabilization of their respective ESX complex (11), we investigated the effect of expressing our hybrid constructs in the mycP5 knockout on the ESX-5 membrane complex. First, we established that there were no effects of our introduced constructs on the production or stability of EccB5, one of the components of the ESX-5 membrane complex (Fig. 4A). Next we investigated the stability of the ESX-5 membrane complex. As shown previously (11), the 1.8-MDa ESX-5 complex could be solubilized in a stable form from the WT strain and visualized by blue native PAGE, whereas solubilized complex levels were strongly reduced in the mycP5 knockout (Fig. 4B, lanes 1 and 2). Exchanging either the TM or protease domain of MycP5 with that of MycP1 (P5TM1 and P1TM5) did not restore stability of the ESX-5 complex in the mycP5 knockout (Fig. 4B, lanes 4 and 5), which was expected, as these constructs were also unable to successfully restore secretion (Fig. 1C). In contrast to this, although MycPP1 was unable to complement secretion, we were able to detect a slight increase in ESX-5 complex levels when we introduced MycPP1 (Fig. 4B, lane 6, P.P1). Complementation of the hybrid MycPP1 construct with the TM domain of MycP5 resulted in full restoration of the ESX-5 complex to WT levels (Fig. 4B, lane 7, P.P1-TM5). In conclusion, our results show that the crucial and system-specific role of both the protease and TM domain of mycosins in ESX secretion is consistently linked to the stabilization of their respective ESX complex.
Figure 4.
A, immunoblot analysis of solubilized cell envelope fractions of WT M. marinum and a mycP5 knockout, complemented with WT mycP5 (P5), hybrid constructs of mycP5 with the TM region of mycP1 (P5TM1) or mycP1 with the TM region of mycP5 (P1TM5), WT mycPP1 (P.P1), and a hybrid construct of mycPP1 with the TM region of mycP5 (P.P1-TM5). Proteins were visualized with anti-EccB5 and, as loading controls, anti-EccB1 and anti-FtsH. B, blue native PAGE immunoblot analysis of solubilized cell envelope fractions; the ESX-5 complex was visualized with anti-EccB5.
Discussion
Mycosin is an enigmatic component of the T7S system, as it is a highly conserved and essential component of this system, but its protease activity is dispensable for secretion. Although we were previously able to assign the essential role of mycosins to stabilizing the secretion complex in the mycobacterial cell envelope, the specific mycosin domains involved in this stabilization remain unknown. Here we set out to obtain insight into the role of individual mycosin domains in this process by producing various domain deletions and hybrid mycosin constructs.
It has been shown previously that the N-terminal extension, called the propeptide in subtilisins, is not processed but remains tightly wrapped around the protease domain of mycosins (20, 21). In line with this, our results also strongly suggest that the extension solely has a structural role, as it is able to stabilize protease domains of mycosins from different ESX systems (Figs. 1B, lane 7, P5NE1, and 2B, lane 7, P1NE5). This is further supported by a study by Sun et al. (22), who showed that, although the structural conformation of the protease domain of MycP1 is not altered by deleting the extension, molecular dynamics simulations did show increased flexibility, suggesting decreased stability of the protease domain. This can be explained by the presence of stabilizing interactions between these two domains, i.e. an anti-parallel β-sheet and a disulfide bond. The decreased stability is apparently more severe during in vivo expression in M. marinum (Figs. 1B, lane 5, P5ΔNE, and 2B, lane 5, P1ΔNE) compared with the MycP1 purified from Escherichia coli by Sun et al. (22).
Our results clearly show that both the protease and TM domain are essential and function in a system-specific manner in ESX-mediated secretion and stabilization of the ESX complex (Figs. 1, 2, and 4). However, the importance and role of the TM domain appears to be complicated. Although it is clearly required for secretion and ESX-5 complex stability, the hybrid MycP5 construct with the TM domain of MycP1 (P5TM1) did remove the requirement for MspA in the mycP5 knockout, suggesting that the TM domain of MycP1 is able to take over the role of the equivalent domain of MycP5 to a minor extent. A possible role of the TM domain is to properly localize mycosins to their respective ESX complex by a specific interaction with this membrane complex. Thus, we postulate that a small subpopulation of P5TM1 is still capable of interacting with the ESX-5 complex, possibly only by the protease domain, whereas TM1 only serves as a membrane anchor. This results in a very limited level of functional ESX-5 complexes, showing that this strain is capable of (very limited) growth in the absence of MspA. In addition to this, the expression levels of mycosins seem to be highly influenced by the presence and nature of the TM domain. Although its deletion strongly increased protein levels, exchange of TM domains in the hybrid constructs also affected protein levels; e.g. constructs with the TM domain of MycP5 generally resulted in higher protein levels irrespective of the identity of the protease domain. Especially the increased stability of mycosin in the absence of a TM domain is intriguing, as it suggests that WT mycosin is a relatively unstable protein. The question is then whether a high turnover is somehow related to its function.
In contrast to the TM domain, the protease domain of MycP5 is absolutely required for ESX-5–mediated secretion and stabilization of the ESX-5 membrane complex, a role that cannot be replaced by the protease domain of MycP1. The essential role of the protease is not related to its protease activity, as we and others showed previously that active-site mutants in MycP1 and MycP5 are fully able to mediate secretion and membrane complex stability of the respective systems (11, 15). We therefore tried to further dissect the specific features of MycP5 that are essential for ESX-5 secretion by deleting its distinctive extended loops in the protease domain. However, none of the loops appear to be involved in the essential and system-specific function. Notably, deletion of loop 2 results in a strong reduction of MycP5 levels (Fig. 3B, lane 5), and as this is also the most strongly conserved loop (Table S1), we expect it to play a structural role in, e.g., establishing a stable protein conformation. This is also supported by the observation that we were unable to produce stable MycP1 lacking loop 2 in E. coli and the presence of a cysteine residue, which forms a disulfide bridge with the core protease structure (20, 21). However, as we were still able to detect low levels of MycP5 lacking loop 2 in M. marinum, we cannot exclude that it is involved in the system-specific functioning of MycP5. In addition to this, although deleting loop 1 also resulted in lower expression levels and an ESX-5 secretion defect, MycP5 with loop 1 of MycP1 is fully functional, showing that this loop is not involved in the system-specific functioning of the protease. Deleting loop 1 in MycP1 did not affect protease activity in vitro, suggesting that this loop is also not directly involved in substrate recognition. These combined results suggest that loop 1 and loop 2 have a general structural role in mycosins. In contrast, loop 3 and loop 5 can be removed in MycP5 without affecting protease integrity and ESX-5 functionality. Interestingly, MycP5 and MycPP1 appear to be processed in the larger loop 5, which is not conserved in all mycosins (e.g. Fig. S4). The two resulting MycP fragments are expected to form a single stable unit because of the structural requirement of the N-terminal extension (Figs. 1B, lane 5, P5ΔNE, and 2B, lane 5, P1ΔNE). The observed cleavage is not an autoproteolytic process; it is not dependent on either proteolytic active MycP5 or on a functional ESX-5 system, as we also observed this processing when we expressed HA-tagged MycP5 and a corresponding HA-tagged active site mutant in M. smegmatis that lacks ESX-5 (Fig. S3C). In addition, for the MycP5 construct with the TM domain deletion, some full-length protein could be observed, showing that the involved protease is not able to fully process this truncate, either because of the high overexpression or because this construct does not localize to the inner membrane. Although we cannot exclude that loop 5 is required for full functionality of the putative protease activity of MycP5, it is remarkable that this loop is only present in a subset of MycP proteins, including MycP2 and mycosins from plasmid-borne ESX systems (25).
The observation that the hybrid construct with the protease domain of MycPP1 (Fig. 3, B, lane 9, P.P1-TM5, and C, lane 18) of the ESX-P1 system and the TM domain of MycP5 also results in successful complementation of the mycP5 knockout is not very surprising, considering the high similarity of the two protease domains (Fig. S4 and Table S1). Interestingly, WT MycPP1 was not able to complement the mycP5 knockout, whereas its presence did result in a small increase in ESX-5 complex levels. ESX-5–mediated secretion was not observed, nor did this complementation allow the bacterium to grow in the absence of MspA. Although this again suggests an important role of the TM domain in the system-specific functioning of MycP, we cannot exclude that the observed different expression levels of the proteases are responsible for the different phenotypes.
Finally, it is interesting to note that almost all tested constructs resulted in an “all-or-nothing” phenotype regarding secretion. Except for the partial complementation of P5TM1, which resulted in a strongly reduced growth rate of the bacterium in the absence of MspA, secretion was either present or absent in the complemented knockout strains of mycP1 and mycP5. This is especially surprising, as we observed a high variability in protein levels of the HA-tagged (hybrid or truncated) mycP1 and mycP5 constructs. Apparently, only small amounts of mycosin are required to allow efficient secretion via the ESX-1 and the ESX-5 membrane complex. Because we have good indications that mycosins have a role in the stability of the membrane complex (11), this could also imply that each bacterium needs only a small number of active secretion machineries to secrete substantial amounts of substrate. Furthermore, we also observed that none of the successful complementations of ΔmycP1 resulted in a difference in the processing of EspB, although this is not surprising, as we did not alter the catalytic site in any of the mycP1 constructs.
Our combined results show that, instead of a highly defined region, a major portion of mycosin, comprising both the protease and the TM domain, is involved in the essential role of this component in ESX secretion, i.e. in the stabilization of the respective ESX complex (for an overview, see Table S2). As the protease domain of MycP1 is not functionally redundant to MycP5, opposed to the protease domain of MyPP1, this provides a starting point for future studies to identify the regions and residues in the mycosin protease domain that are involved in interacting with and stabilizing their respective ESX complex.
Experimental procedures
Bacterial strains and culture conditions
M. marinum M (26) was used for all M. marinum experiments. The mycP1 and mycP5 knockout mutants of this strain have been described previously by Ates et al. (6) and van Winden et al. (11). M. marinum was grown at 30 °C on 7H10 agar with 10% Middlebrook OADC (BD Biosciences) or in Middlebrook 7H9 liquid medium with 10% Middlebrook ADC and with 0.05% Tween 80 at 30 °C and 150 rpm. M. smegmatis MC2155 was used for all M. smegmatis experiments and was grown at 37 °C on 7H10 agar with 10% Middlebrook OADC or in Luria-Bertani (LB) liquid medium supplemented with 0.05% Tween 80 at 150 rpm. E. coli strains DH5α and Rosetta were grown at 37 °C on LB agar or in LB liquid medium at 37 °C and 200 rpm. Culture media were supplemented with the appropriate antibiotics at the following concentrations: kanamycin, 25 μg ml−1; hygromycin, 50 μg ml−1; streptomycin, 35 μg ml−1; ampicillin, 100 μg ml−1.
Cloning
The mycP1 and mycP5 M. marinum genes were amplified from M. marinum M genomic DNA and mycPP1 from M. marinum E11 genomic DNA by PCR with anchored primers (EcoRI and HindIII; Table S3). The active site mutant of mycP5 has been created in a previous study (11). The hybrid combinations of mycP1, mycP5, and mycPP1 point mutations and the HA tag were introduced with nested primers (Table S3). All generated constructs were subsequently digested by EcoRI and HindIII and ligated into similarly digested pMV361 (27). The plasmids used to express MycP1mth and EspBmtb have been described previously by Wagner et al. (21). The mycP1mth construct was modified with anchored (NdeI and XhoI) and nested primers (Table S3) to create mycPmthΔloop1, which was ligated into pET-28a following NdeI and XhoI digestion. All generated plasmids were verified by sequencing of the cloned inserts.
M. marinum protein secretion and BN-PAGE analysis of ESX-5 complex
For protein secretion analysis, M. marinum strains were grown in 7H9 liquid medium supplemented with 10% Middlebrook ADC and 0.05% Tween 80 until mid-logarithmic phase (A600 of 1–1.4), and then the cells were washed and inoculated in 7H9 liquid medium containing 0.2% dextrose, 0.2% glycerol, and 0.05% Tween 80 at an A600 of 0.5. After 16 h of growth, the cells were pelleted (10 min at 3000 × g), washed with PBS, and resuspended in SDS loading buffer (pellet fraction). Supernatants were passed through an 0.45-μm filter, and the proteins were precipitated with TCA, washed with acetone, and resuspended in SDS loading buffer (supernatant fraction). Alternatively, cell surface proteins were extracted from pelleted cells with 0.5% Genapol X-080 as described previously (11), the cells were repelleted and resuspended in SDS loading buffer (Genapol pellet fraction), and concentrated SDS loading buffer was added to the Genapol X-080–containing supernatant (Genapol supernatant fraction). Proteins from the various fractions were separated on SDS-PAGE gels (with 10% to 16% acrylamide, depending on the size of the analyzed proteins), transferred to a nitrocellulose membrane, and stained with the appropriate antibodies (see below). For both the secretion analysis and the HA expression blots, at least three colonies of each strain were tested. The immunoblots used are an accurate representation of the obtained, consistent results.
For ESX-5 complex analysis, M. marinum was grown to an A600 of 1.2–1.5, pelleted, and resuspended in PBS with 250 mm sucrose. Cells were lysed with a One-Shot Cell disruptor (Constant Systems Ltd.), and unbroken cells were pelleted at 3000 × g. The cell envelope (CE) fraction was separated from the soluble fraction by centrifugation at 200,000 × g for 45 min, and the CE fraction was subsequently resuspended in PBS with 250 mm sucrose. Membrane proteins were extracted from the CE fraction with the mild detergent dodecyl maltoside at a concentration of 0.25%. Solubilized protein (complexes) was either separated under native conditions on a 3–12% NativePage Novex BisTris Protein Gel (Life Technologies) and transferred to a polyvinylidene difluoride membrane or under denaturing conditions on a 12.5% SDS-PAGE gel, which was subsequently transferred to a nitrocellulose membrane. Blots were stained with anti-GroEL2 (Cs44; John Belisle, National Institutes of Health, Bethesda, MD), anti-PE_PGRS (26), anti-EsxA/ESAT-6 (Mab Hyb76-8), anti-HA (HA.11, Covance, Biolegend), anti-EccB5 (12, Colorado State University), anti-EspB (EPFL, Lausanne, Switzerland), anti-EccB1 (11), or anti-FtsH (28) antibodies. Apart from the monoclonal anti-PE_PGRS and anti-EsxA preparations, these are polyclonal rabbit antisera.
Protein expression, purification, and activity assays
His-tagged EspBmtb and MycP1mth were expressed and purified from E. coli Rosetta (DE3) as described previously by van Winden et al. (11). MycP1 activity assays were performed in 20 mm HEPES (pH 7.5), 100 mm at 37 °C for 20 h with 0.15 mg ml−1 MycP1, and 0.25 mg ml−1 EspB. Reactions were stopped by addition of SDS sample buffer and incubation at 94 °C for 5 min. Proteins were separated on a 12.5% SDS-PAGE gel and visualized by Coomassie staining.
Author contributions
V. J. C. v. W., W. B., and E. N. G. H. conceptualization; V. J. C. v. W. and E. N. G. H. formal analysis; V. J. C. v. W., M. P. M. D., and R. U. investigation; V. J. C. v. W. visualization; V. J. C. v. W., M. P. M. D., R. U., W. B., and E. N. G. H. methodology; V. J. C. v. W. and E. N. G. H. writing-original draft; V. J. C. v. W., M. P. M. D., R. U., W. B., and E. N. G. H. writing-review and editing; W. B. and E. N. G. H. supervision; E. N. G. H. project administration.
Supplementary Material
This work was funded by VIDI Grant 864.12.006 (to E. N. G. H.) and ALW Open Grant ALWOP.319 (to M. P. M. D.), both from the Netherlands Organization of Scientific Research (NWO), and by the CCA from Amsterdam University Medical Center (to V. J. C. v. W. and W. B.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4 and Tables S1–S3.
- T7S
- type VII secretion
- NE
- N-terminal extension
- TM
- transmembrane
- PGRS
- polymorphic GC-rich repetitive sequence
- HA
- hemagglutinin
- LB
- Luria-Bertani
- CE
- cell envelope.
References
- 1. Abdallah A. M., Gey van Pittius N. C., Champion P. A., Cox J., Luirink J., Vandenbroucke-Grauls C. M., Appelmelk B. J., and Bitter W. (2007) Type VII secretion: mycobacteria show the way. Nat. Rev. Microbiol. 5, 883–891 10.1038/nrmicro1773 [DOI] [PubMed] [Google Scholar]
- 2. Gey Van Pittius N. C., Gamieldien J., Hide W., Brown G. D., Siezen R. J., and Beyers A. D. (2001) The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2, RESEARCH0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simeone R., Bobard A., Lippmann J., Bitter W., Majlessi L., Brosch R., and Enninga J. (2012) Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 8, e1002507 10.1371/journal.ppat.1002507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pym A. S., Brodin P., Brosch R., Huerre M., and Cole S. T. (2002) Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46, 709–717 10.1046/j.1365-2958.2002.03237.x [DOI] [PubMed] [Google Scholar]
- 5. Siegrist M. S., Unnikrishnan M., McConnell M. J., Borowsky M., Cheng T.-Y., Siddiqi N., Fortune S. M., Moody D. B., and Rubin E. J. (2009) Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl. Acad. Sci. 106, 18792–18797 10.1073/pnas.0900589106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ates L. S., Ummels R., Commandeur S., van de Weerd R., Sparrius M., Weerdenburg E., Alber M., Kalscheuer R., Piersma S. R., Abdallah A. M., Abd El Ghany M., Abdel-Haleem A. M., Pain A., Jiménez C. R., Bitter W., and Houben E. N. (2015) Essential role of the ESX-5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genet. 11, e1005190 10.1371/journal.pgen.1005190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van der Wel N., Hava D., Houben D., Fluitsma D., van Zon M., Pierson J., Brenner M., and Peters P. J. (2007) M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129, 1287–1298 10.1016/j.cell.2007.05.059 [DOI] [PubMed] [Google Scholar]
- 8. Houben D., Demangel C., van Ingen J., Perez J., Baldeón L., Abdallah A. M., Caleechurn L., Bottai D., van Zon M., de Punder K., van der Laan T., Kant A., Bossers-de Vries R., Willemsen P., Bitter W., et al. (2012) ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell. Microbiol. 14, 1287–1298 10.1111/j.1462-5822.2012.01799.x [DOI] [PubMed] [Google Scholar]
- 9. Serafini A., Boldrin F., Palù G., and Manganelli R. (2009) Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J. Bacteriol. 191, 6340–6344 10.1128/JB.00756-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Houben E. N., Bestebroer J., Ummels R., Wilson L., Piersma S. R., Jiménez C. R., Ottenhoff T. H., Luirink J., and Bitter W. (2012) Composition of the type VII secretion system membrane complex. Mol. Microbiol. 86, 472–484 10.1111/j.1365-2958.2012.08206.x [DOI] [PubMed] [Google Scholar]
- 11. van Winden V. J. C., Ummels R., Piersma S. R., Jiménez C. R., Korotkov K. V., Bitter W., and Houben E. N. G. (2016) Mycosins are required for the stabilization of the ESX-1 and ESX-5 type VII secretion membrane complexes. mBio 7, pii: e01471–16 10.1128/mBio.01471-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wagner J. M., Chan S., Evans T. J., Kahng S., Kim J., Arbing M. A., Eisenberg D., and Korotkov K. V. (2016) Structures of EccB1 and EccD1 from the core complex of the mycobacterial ESX-1 type VII secretion system. BMC Struct. Biol. 16, 5 10.1186/s12900-016-0056-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rosenberg O. S., Dovala D., Li X., Connolly L., Bendebury A., Finer-Moore J., Holton J., Cheng Y., Stroud R. M., and Cox J. S. (2015) Substrates control multimerization and activation of the multi-domain ATPase motor of type VII secretion. Cell 161, 501–512 10.1016/j.cell.2015.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beckham K. S., Ciccarelli L., Bunduc C. M., Mertens H. D., Ummels R., Lugmayr W., Mayr J., Rettel M., Savitski M. M., Svergun D. I., Bitter W., Wilmanns M., Marlovits T. C., Parret A. H., and Houben E. N. (2017) Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis. Nat. Microbiol. 2, 17047 10.1038/nmicrobiol.2017.47 [DOI] [PubMed] [Google Scholar]
- 15. Ohol Y. M., Goetz D. H., Chan K., Shiloh M. U., Craik C. S., and Cox J. S. (2010) Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe 7, 210–220 10.1016/j.chom.2010.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Griffin J. E., Gawronski J. D., DeJesus M. A., Ioerger T. R., Akerley B. J., and Sassetti C. M. (2011) High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7, e1002251 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sassetti C. M., Boyd D. H., and Rubin E. J. (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 10.1046/j.1365-2958.2003.03425.x [DOI] [PubMed] [Google Scholar]
- 18. Houben E. N., Korotkov K. V., and Bitter W. (2014) Take five: type VII secretion systems of mycobacteria. Biochim. Biophys. Acta 1843, 1707–1716 10.1016/j.bbamcr.2013.11.003 [DOI] [PubMed] [Google Scholar]
- 19. Brown G. D., Dave J. A., Gey van Pittius N. C., Stevens L., Ehlers M. R., and Beyers A. D. (2000) The mycosins of Mycobacterium tuberculosis H37Rv: a family of subtilisin-like serine proteases. Gene 254, 147–155 10.1016/S0378-1119(00)00277-8 [DOI] [PubMed] [Google Scholar]
- 20. Solomonson M., Huesgen P. F., Wasney G. A., Watanabe N., Gruninger R. J., Prehna G., Overall C. M., and Strynadka N. C. (2013) Structure of the mycosin-1 protease from the mycobacterial ESX-1 protein type VII secretion system. J. Biol. Chem. 288, 17782–17790 10.1074/jbc.M113.462036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wagner J. M., Evans T. J., Chen J., Zhu H., Houben E. N., Bitter W., and Korotkov K. V. (2013) Understanding specificity of the mycosin proteases in ESX/type VII secretion by structural and functional analysis. J. Struct. Biol. 184, 115–128 10.1016/j.jsb.2013.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun D., Liu Q., He Y., Wang C., Wu F., Tian C., and Zang J. (2013) The putative propeptide of MycP1 in mycobacterial type VII secretion system does not inhibit protease activity but improves protein stability. Protein Cell 4, 921–931 10.1007/s13238-013-3089-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Faller M., Niederweis M., and Schulz G. E. (2004) The structure of a mycobacterial outer-membrane channel. Science 303, 1189–1192 10.1126/science.1094114 [DOI] [PubMed] [Google Scholar]
- 24. Ummels R., Abdallah A. M., Kuiper V., Aâjoud A., Sparrius M., Naeem R., Spaink H. P., van Soolingen D., Pain A., and Bitter W. (2014) Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. mBio 5, e01744–14 10.1128/mBio.01744-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dumas E., Boritsch E. C., Vandenbogaert M., Rodríguez de la Vega R. C., Thiberge J. M., Caro V., Gaillard J. L., Heym B., Girard-Misguich F., Brosch R., and Sapriel G. (2016) Mycobacterial pan-genome analysis suggests important role of plasmids in the radiation of type VII secretion systems. Genome Biol. Evol. 8, 387–402 10.1093/gbe/evw001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abdallah A. M., Verboom T., Hannes F., Safi M., Strong M., Eisenberg D., Musters R. J., Vandenbroucke-Grauls C. M., Appelmelk B. J., Luirink J., and Bitter W. (2006) A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol. Microbiol. 62, 667–679 10.1111/j.1365-2958.2006.05409.x [DOI] [PubMed] [Google Scholar]
- 27. Stover C. K., de la Cruz V. F., Fuerst T. R., Burlein J. E., Benson L. A., Bennett L. T., Bansal G. P., Young J. F., Lee M. H., Hatfull G. F., Snapper S. B., Barletta R. G., Jacobs W. R., and Bloom B. R. (1991) New use of BCG for recombinant vaccines. Nature 351, 456–460 10.1038/351456a0 [DOI] [PubMed] [Google Scholar]
- 28. van Bloois E., Dekker H. L., Fröderberg L., Houben E. N., Urbanus M. L., de Koster C. G., de Gier J. W., and Luirink J. (2008) Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 582, 1419–1424 10.1016/j.febslet.2008.02.082 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.