Abstract
Crimean-Congo hemorrhagic fever virus (CCHFV) is a tick-borne Nairovirus that causes severe hemorrhagic fever with a mortality rate of up to 30% in certain outbreaks worldwide. The virus has wide endemic distribution. There is no effective antiviral therapeutic or FDA approved vaccine for this zoonotic viral illness. The multifunctional CCHFV nucleocapsid protein (N protein) plays a crucial role in the establishment of viral infection and is an important structural component of the virion. Here we show that CCHFV N protein has a distant RNA-binding site in the stalk domain that specifically recognizes the vRNA panhandle, formed by the base pairing of complementary nucleotides at the 5′ and 3′ termini of the vRNA genome. Using multiple approaches, including filter-bonding analysis, GFP reporter assay, and biolayer interferometry we observed an N protein-panhandle interaction both in vitro and in vivo. The purified WT CCHFV N protein and the stalk domain also recognize the vRNA panhandle of hazara virus, another Nairovirus in the family Bunyaviridae, demonstrating the genus-specific nature of N protein-panhandle interaction. Another RNA-binding site was identified at the head domain of CCHFV N protein that nonspecifically recognizes the single strand RNA (ssRNA) of viral or nonviral origin. Expression of CCHFV N protein stalk domain active in panhandle binding, dramatically inhibited the hazara virus replication in cell culture, illustrating the role of N protein-panhandle interaction in Nairovirus replication. Our findings reveal the stalk domain of N protein as a potential target in therapeutic interventions to manage CCHFV disease.
Keywords: viral replication, virology, viral protein, RNA-protein interaction, viral polymerase, C-C chemokine receptor type 5 (CCR5), CCHF, nucleocapsid
Introduction
Crimean-Congo hemorrhagic fever virus (CCHFV)2 is a tick-borne Nairovirus in the Bunyaviridae family. Its infection causes severe hemorrhagic fever with a mortality rate of 5 to 30% in more than 30 countries worldwide (1–4). To date, this zonotic viral illness has been reported in the Balkans, Eastern Europe, Central Asia, Turkey, China, Africa, and the Middle East (5–9). Crimean-Congo hemorrhagic fever (CCHF) is a significant public health concern in Turkey due to high fatality rates associated with this infection (10). Global animal trade, environment, and climate changes have played important roles in the spread of this virus to places where it had not been previously detected. The lack of vaccine and effective antivirals have prompted strict precautions through global health care organizations to prevent the spread of this zoonotic viral illness, especially during the seasonal outbreaks.
Infection in humans usually occurs by either tick bites or direct contact with contaminated blood or tissue samples from the infected hosts (7, 11). When transmitted through tick bites, the incubation period may last from 1 to 3 days, compared with the incubation period of 5 to 6 days when transmission occurs through exposure to contaminated tissue or blood samples. Spread through nosocomial transmission has not been reported (6, 7, 12). The clinical symptoms such as, sudden onset of high fever, chills, myalgias, and arthralgias resemble most other viral infections. However, petechiae and ecchymoses may be clinically observed in CCHFV-infected patients several days post-infection. Blood can also be detected in patients sputum and stool, and continuous hemorrhage may lead to hypovolemic shock (13, 14). Prognosis of CCHFV-infected patients depends upon the imbalance of coagulation factors and severity of leukopenia and thrombocytopenia during infection (14–16).
The viral genome is composed of three negative sense RNA segments (S, M, and L), that encode nucleocapsid protein (N protein), glycoprotein precursor, and RNA-dependent RNA polymerase (RdRp), respectively (17). N protein in association with the viral RNA (vRNA) and complementary RNA (cRNA) forms nucleocapsids that serve as templates for transcription and replication of the viral genome. Unlike cRNA, the nucleocapsids derived from vRNA are selectively packaged into new virions during the assembly process. It still remains unclear how N protein selectively recognizes the vRNA and cRNA inside the host cell and how are the vRNA-derived nucleocapsids selectively packaged into new virions. Recently, the X-ray crystallographic structure of CCHFV N protein at 2.3-Å resolution revealed the presence of head and stalk domains in the N protein structure (18–20). The protein structure is more closely related to the nucleocapsid protein of Arenaviruses (21, 22) compared with the nucleocapsid protein of other Bunyaviruses (23–25). We previously reported that bacterially expressed and purified CCHFV N protein has distinct binding modes for double strand RNA (dsRNA) and single strand RNA (ssRNA) (26). The viral RNA panhandle formed by the base pairing of partially complementary nucleotides at the 5′ and 3′ ends of vRNA is specifically recognized by N protein through the dsRNA-binding mode (26). The highly conserved terminal nine nucleotides from both 5′ and 3′ ends of the vRNA genome are required for high affinity binding of the panhandle (26). In comparison, N protein does not discriminate between viral and nonviral RNAs through ssRNA-binding mode (26). N protein has been reported to interact with the N- and C-terminal regions of the RdRp, suggesting a potential role of N protein-RdRp interaction in transcription and replication of the viral genome (27, 28).
In this article we report the location of panhandle-specific dsRNA and nonspecific ssRNA-binding sites in the stalk and head domains of the N protein, respectively. We show that both WT CCHFV N protein and its stalk domain bind with high affinity to the panhandle of hazara virus, another member of the Nairovirus genus. This suggests that N-panhandle interaction is likely genus-specific. Expression of CCHFV N protein stalk domain with intact panhandle-specific dsRNA-binding site inhibited the hazara virus replication in cell culture, demonstrating the role of N-panhandle interaction in the virus replication cycle. These studies demonstrate that N-panhandle interaction might be a novel target for therapeutic intervention of CCHFV infection.
Results
Purification of head and stalk domains of CCHFV N protein
Using four independent binding approaches, we previously demonstrated that N protein specifically binds to the panhandle RNA structure composed of partially complementary 30 nucleotides from both 5′ and 3′ termini of CCHFV S-segment vRNA (Fig. 1B), with the same affinity as full-length vRNA (Kd ∼ 42 nm). The panhandle-N protein interaction occurred through the dsRNA-binding mode. However, N protein nonspecifically recognized the ssRNA through the ssRNA-binding mode. N protein remained in a unique conformational state when both ssRNA and dsRNA substrates were simultaneously bound to the protein. Competitive binding experiments suggested that N protein might have distinct binding pockets for dsRNA and ssRNA substrates.
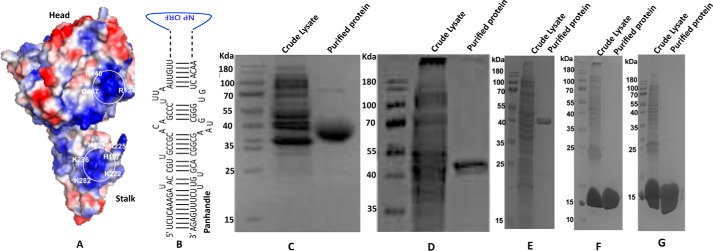
Figure 1.
A, a modeled 3D structure of CCHFV N protein, showing the head and stalk domains. The electrostatic surface potential of N protein was visualized by PyMol. The positively charged surface is colored blue and the negatively charged surface is colored red. The isolated positively charged surfaces constituted by residues Lys-339, Lys-343, Lys-346, Arg-384, Lys-411, His-453, Arg-134, Arg-140, and Gln-467 in the head domain and His-195, His-197, Lys-222, Arg-225, Lys-282, and Lys-286 in the stalk domain likely constitute the distinct RNA-binding pockets. B, a panhandle formed by the partially complementary nucleotides at the 5′ and 3′ termini of the CCHFV S-segment vRNA. C, SDS-PAGE analysis of bacterially expressed and purified head domain of CCHFV N protein. D, head domain mutant 12 in which Arg-339, Lys-343, and Lys-346 were mutated to alanine. E, head domain mutant 34 in which Arg-134 and Arg-140 were converted to alanine. F, stalk domain of CCHFV N protein. G, stalk domain mutant in which His-195 and His-197 were converted to alanine.
Using the X-ray crystal structure of the CCHFV N protein (strain YL04057, PDB code 3U3I), we modeled the 3D structure of CCHFV N protein (strain 10200). Although, the primary sequences of these two strains are slightly different, the modeled structure of strain 10200 is very similar to the X-ray structure of strain YL04057, and shows a distinct stalk domain composed of amino acids from 180 to 300, and a head domain composed of N-terminal 180 amino acids and C-terminal 182 amino acids (Fig. 1A). The electrostatic surface potential, generated by the PyMol viewer software tool, predicted two major RNA-binding regions based on the positively charged grooves at the surface of N protein structure. The isolated positively charged surface constituted by residues Lys-339, Lys-343, Lys-346, Arg-384, Lys-411, His-453, Arg-134, Arg-140, and Gln-467 in the head region, and His-195, His-197, Lys-222, Arg-225, Lys-282, and Lys-286 in the stalk region appear to constitute the two RNA-binding sites in the N protein structure.
To determine whether the head and stalk domains harbored the panhandle-specific dsRNA and nonspecific ssRNA-binding sites, we cloned and purified the head and stalk domains of the CCHFV N protein in bacteria (Fig. 1, C and F). To delineate whether isolated positively charged surfaces in the head and stalk domains constitute the RNA-binding sites, we generated head and stalk domain mutants in which positively charged residues in the hypothetical RNA-binding pockets were converted to neutral amino acid alanine. In head domain mutant 12 (Fig. 1D) the three positively charged residues, Arg-339, Lys-343, and Lys-346, were converted to alanine, using site-directed mutagenesis. A similar approach was used to incorporate two point mutations in the head domain mutant 34 (Fig. 1E) replacing Arg-134 and Arg-140 with alanine. In the stalk mutant (Fig. 1G) two positively charged residues His-195 and His-197 were converted to alanine. All the mutants (Fig. 1, C–G) were purified by native procedure on a Ni-NTA column, using the C-terminal His tag. All the mutants were highly expressed in bacteria and their size matched with their respective molecular weights on SDS-PAGE (Fig. 1).
Binding of purified head and stalk domains of CCHFV N protein with the synthetic RNA in vitro
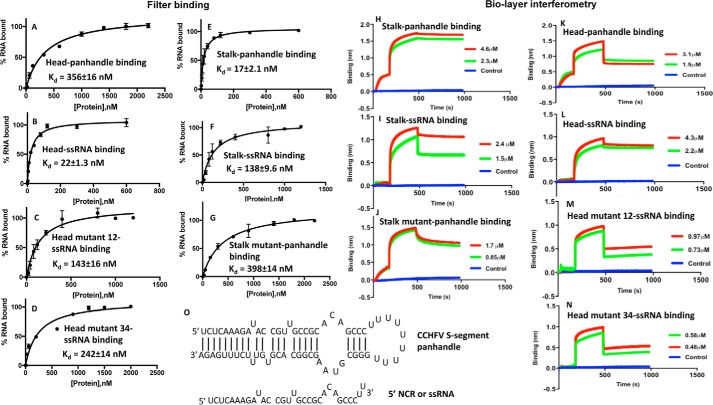
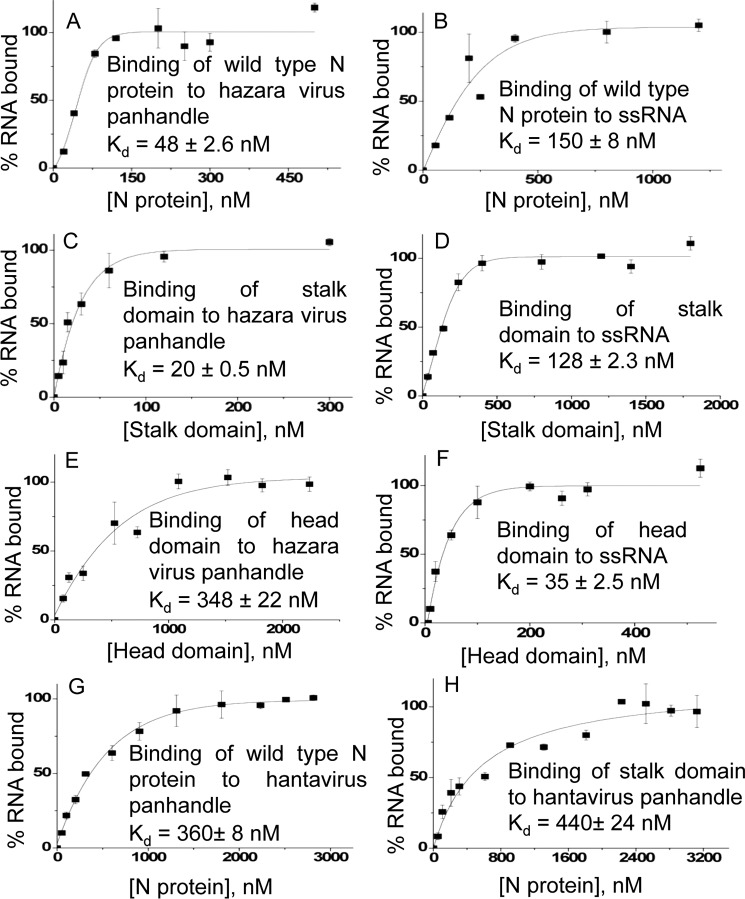
We previously reported that WT CCHFV N protein specifically binds to the panhandle with a Kd value of 42 nm at 80 mm salt concentration (26). The Kd value did not change at higher salt concentrations. In comparison, N protein nonspecifically bound to the single strand 5′ noncoding region (5′ NCR) of CCHFV S-segment RNA or any nonviral ssRNA with a Kd value of 125 nm at 80 mm salt concentration. The Kd value significantly increased with increasing salt concentrations. To identify the panhandle-specific dsRNA and nonspecific ssRNA-binding sites in the WT CCHFV N protein, we synthesized the panhandle composed of 30 nucleotides from both 5′ and 3′ termini of CCHFV S-segment vRNA, separated by a six-residue uracil loop (Fig. 2O) by in vitro T7 transcription reaction, as previously reported (26). The same strategy was used to synthesize the terminal 30 nucleotides of the 5′ NCR, represented as ssRNA in the text (Fig. 2O). Both panhandle and ssRNA were labeled with [32P]CTP during synthesis. We studied the binding of purified head and stalk domains with the synthetic panhandle and ssRNA sequence using two independent experimental approaches. An examination by filter-binding analysis revealed that the stalk domain bound to the panhandle RNA (Kd = 17 ± 2.1 nm) with 8-fold higher affinity compared with the ssRNA (Kd = 138 ± 9.6 nm) (Fig. 2, E and F). Conversely, the binding affinity of the head domain with the ssRNA (kd = 22 ± 1.3 nm) was 16-fold higher compared with the panhandle (Kd = 356 ± 16 nm) (Fig. 2, A and B). These data clearly shows that stalk and head domains preferably bind to panhandle and ssRNA, respectively. The dissociation constants for the binding of stalk and head domains with panhandle and ssRNA, respectively, are consistent with the previously reported dissociation constants for the binding of WT N protein with panhandle and ssRNA (26). Point mutations replacing the two positively charged amino acids His-195 and His-197 to alanine in the positively charged surface grove of the stalk domain (stalk mutant) abrogated the binding with the panhandle RNA (Fig. 2G and Table 1). Similarly, point mutations replacing the positively charged amino acids Arg-339, Lys-343, and Lys-346 (head mutant 12), and Arg-134 and Arg-140 (head mutant 34) with alanine abrogated the binding to the head domain with the ssRNA (Fig. 2, C and D, and Table 1). This suggests that isolated positively charged groves on the stalk and head domains, predicted by the PyMol viewer, likely constitute the panhandle and ssRNA-binding sites, respectively.
Figure 2.
Binding studies performed by filter-binding assay and biolayer interferometry. Binding profiles for the association of stalk domain, stalk mutant domain, head domain, head mutant domain 16, and head mutant domain 34 with CCHFV S-segment vRNA panhandle and ssRNA sequence, generated by filter-binding assay, are shown in A–G. Interaction of each domain with the type of RNA is mentioned inside each panel. Binding results from A–G were also confirmed using biolayer interferometry (BLI). Representative BLI sensograms showing over time association and dissociation of protein with RNA are shown in H–N. The sensograms were generated at two protein concentrations, shown by red and blue in H–N (see “Experimental procedures” for details). Again each panel is internally labeled to show the name of the protein domain and interacting RNA. The dissociation constants (Kd) were calculated as described under “Experimental procedures.” The CCHFV S-segment panhandle and ssRNA used in A–N are shown in O. The panhandle sequence composed of 30 nucleotides from both 5′ and 3′ termini of CCHFV S-segment vRNA, separated by a six-residue uracil loop, was folded by M-fold. It generated the secondary structure, shown in O.
Table 1.
Binding parameters calculated by two independent methods
| Protein | RNA | Biolayer interferometry |
Filter binding |
||
|---|---|---|---|---|---|
| Kd ± S.D. | Ka | Kdis | Kd ± S.D. | ||
| nm | m−1 s−1 | s−1 | nm | ||
| Head | Panhandle | 274 ± 12 | 2.09 ± 104 | 5.737 × 10−3 | 356 ± 9.0 |
| Head | ssRNA | 4.3 ± 0.7 | 7.612 ± 103 | 3.276 × 10−5 | 22 ± 1.3 |
| Head mutant 12 | ssRNA | 115 ± 5.5 | 2.646 ± 104 | 4.448 × 10−3 | 143 ± 16 |
| Head mutant 34 | ssRNA | 218 ± 10 | 8.134 ± 104 | 6.456 × 10−3 | 242 ± 14 |
| Stalk | Panhandle | 4.0 ± 0.5 | 1.62 ± 104 | 6.514 × 10−6 | 17 ± 2.1 |
| Stalk | ssRNA | 90 ± 9.0 | 2.055 ± 104 | 1.858 × 10−3 | 138 ± 9.6 |
| Stalk mutant | Panhandle | 323 ± 11 | 7.452 ± 104 | 2.411 × 10−2 | 398 ± 14 |
To further confirm the results from filter-binding analysis, we monitored the association and dissociation kinetics of synthetic panhandle and ssRNA with head and stalk mutants, using biolayer interferometry on a BLITZ (ForteBio) instrument, as previously reported (26)(see “Experimental procedures” for details). The respective kinetic profiles are shown in Fig. 2, H–N, and the corresponding binding data are shown in Table 1. It is evident from Table 1 that dissociation constants calculated by biolayer interferometry followed the same trend and are in agreement with the dissociation constants calculated by the filter-binding analysis. The high affinity binding of panhandle and ssRNA with the stalk and head domains, respectively (Fig. 2, H and L, and Table 1), further confirms their binding sites in these respective domains. Interaction of head domain mutants with panhandle and stalk mutant with ssRNA was undetectable.
Examination of N protein-panhandle binding using an in vivo reporter assay
Because N protein specifically binds to the vRNA panhandle; we asked whether this interaction takes place inside cells during viral infection. We developed a GFP reporter assay to further confirm the interaction between N protein and vRNA panhandle inside the cells. Briefly, the panGFP plasmid was constructed for the expression of GFP reporter mRNA having 5′ and 3′ NCR sequences of CCHFV S-segment vRNA upstream of the start codon and downstream of the stop codon, respectively (Fig. 3A). The expression is driven from a T7 promoter and the 3′ end of the GFP reporter mRNA is defined by the engineered hepatitis delta virus (HDV) ribozyme sequence. The mRNA likely forms a panhandle structure due to complementarity of the nucleotides between the 5′ and 3′ termini (Fig. 3A). Unlike eukaryotic mRNA, the mRNA expressed from panGFP plasmid does not have a 5′ cap and 3′ poly(A) tail. The BSRT7.5 cells stably expressing T7 RNA polymerase (gift from Dr. Conzelman, Germany) were cotransfected with panGFP reporter plasmid along with another plasmid expressing WT CCHFV N protein or head domain, head domain mutant 12, stalk domain, stalk domain mutant, N protein mut1, or N protein mut2. The N protein mut1 and N protein mut2 are derived from WT CCHFV N protein, and have a single point mutation (His-195 to Ala-195) and two point mutations (His-195 and His-197 to Ala) in their stalk domains, respectively. The GFP expression was monitored using either fluorescence microscopy (Fig. 3B) or flow cytometry (Fig. 3D). Interestingly, a strong GFP expression was observed in cells co-expressing the WT CCHFV N protein (Fig. 3, B and D). A weaker GFP signal was observed in cells co-expressing N protein mut1. To ensure that changes in GFP expression were not due to changes in the mRNA levels, we quantified GFP mRNA in cell lysates by real time PCR. It is evident from Fig. 3F that GFP mRNA was equally expressed in cells. An examination of cell lysates by Western blot analysis confirmed the expression of WT N protein, head domain, head domain mutant 34, stalk domain, stalk domain mutant, N protein mut1, and N protein mut2 from the transfected plasmids (Fig. 3C).
Figure 3.
Monitoring the N protein-panhandle binding using an in vivo reporter assay. A, a cartoon showing panGFP plasmid in which GFP (green) flanked by sequences encoding 5′ and 3′ NCR of CCHFV S-segment vRNA (black) was cloned between T7 promoter (blue) and HDV (orange). The mRNA expressed from T7 promoter was folded by M-fold, revealing the formation of panhandle structure by the base pairing of partially complementary nucleotide sequence of the 5′ and 3′ NCR. B, BSRT7.5 cells stably expressing T7 RNA polymerase were seeded in six-well plates and transfected with 2 μg of either empty vector pTriEx1.1 (i), pTriEx-CCHFV-NP plasmid expressing WT CCHFV N protein (ii), pTriExhead plasmid expressing the head domain of the CCHFV N protein (iii), pTriExhead-mutant34 plasmid expressing the head mutant domain 34 (iv), pTriExstalk plasmid expressing the stalk domain of CCHFV N protein (v), pTriExstalk-mutant plasmid expressing the stalk domain mutant (vi), pTriEx CCHFV N protein mut1 plasmid expressing N protein mut1 (vii), or pTriEx CCHFV N protein mut2 plasmid expressing N protein mut2 (viii), as mentioned in the text. Twenty-four hours post-transfection, cells were again transfected with an equal concentration of PanGFP plasmid. Cells were examined under a fluorescence microscope 36 h post second transfection to record the GFP signal (upper eight panels). The experiment was repeated and the cells were stained with DAPI (lower eight panels). The size of the bar in each panel is 100 μm. C, cells from B were lysed and examined by Western blot analysis using anti-His tag antibody to monitor the expression of C-terminally His-tagged fusion proteins from transfected plasmids. D, the cells from B were also examined by FACS. The GFP signal from B (i–viii, upper eight panels) was quantified, normalized related to control (lane 1), and plotted in D. E, the cell lysates from B, containing equal amounts of GFP mRNA, were incubated with Ni-NTA beads. His-tagged proteins bound to washed beads were eluted and total RNA was purified from eluted material. The GFP reporter mRNA in the eluted material was detected using a primer set complementary to the GFP gene. F, total RNA was purified from cell lysates obtained from B. The GFP reporter mRNA was quantified by real time PCR. F shows the equal expression of the reporter GFP mRNA in cells from B.
It is likely that formation of panhandle with a strong secondary structure prevented ribosome engagement and thus inhibited the translation of GFP reporter mRNA in cells. We previously reported that CCHFV N protein does not have the RNA helix unwinding activity (29), and thus the possible unwinding of the panhandle after high affinity binding with N protein to stimulate the translation of GFP reporter mRNA does not seem likely. We also reported previously that CCHFV N protein facilitates mRNA translation possibly by simultaneous binding to the 40S ribosomal subunit and RNA sequence upstream of start codon (29). Based on these previous observations, it is likely that simultaneous binding of N protein with both the 40S ribosomal subunit and the panhandle structure facilitated the ribosome engagement close to the start codon of the GFP reporter mRNA, resulting in the translation of the GFP reporter transcript in cells (Fig. 2B, ii). This is indirectly supported by the observation that incorporation of mutations in the stalk domain of the WT N protein impacted the N protein-mediated translation of the reporter mRNA (Fig. 3B, vii and viii). To confirm that WT N protein and stalk domain remained bound to the panhandle structure in cells, the cell lysates from Fig. 3B were incubated with Ni-NTA beads to pulldown the proteins expressed from transfected plasmids, using the C-terminal His tag. The volume of the lysates was adjusted to ensure an equal amount of GFP mRNA for the respective samples loaded on Ni-NTA beads. Total RNA was purified from the pulldown material and the GFP mRNA was identified by PCR using gene-specific primers. It is evident from Fig. 3E that GFP reporter mRNA co-purified with WT N protein, N protein mut1, and stalk domain, confirming their binding with the panhandle structure. Poor GFP signal due to co-expression of N protein mut1 (Fig. 3B, vii) is likely due to the weaker binding of this mutant with the panhandle. This is indirectly supported by the observation that incorporation of two point mutations in the stalk domain of WT N protein abrogated both panhandle binding (Fig. 3E, viii) and N protein-mediated translation of the GFP reporter mRNA (Fig. 3B, viii).
Disrupting the panhandle structure inhibited the N protein-panhandle interaction in vivo
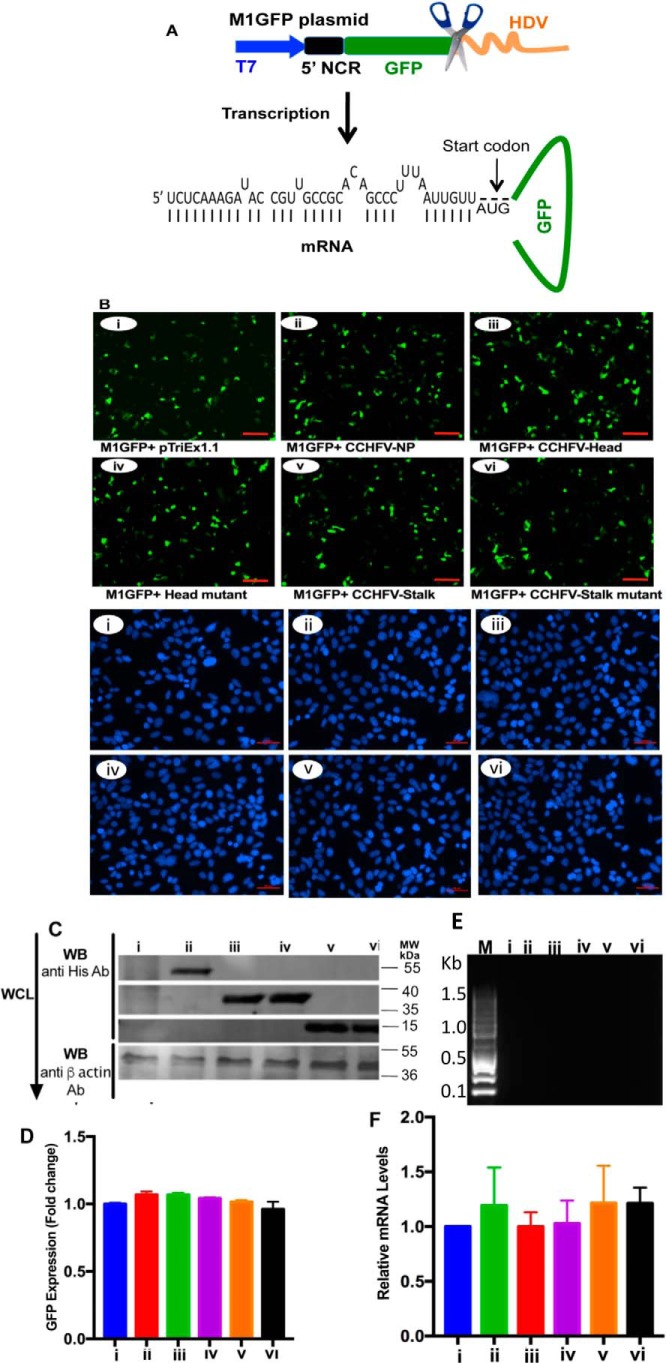
To confirm that panhandle formation inhibited the translation of GFP reporter mRNA (Fig. 3B, i), and N protein-panhandle binding triggered the translation of GFP reporter in cells (Fig. 3B, ii), we generated M1GFP plasmid (Fig. 4A) expressing the GFP reporter mRNA lacking the 3′ NCR sequence of the S-segment mRNA. Lack of this sequence will inhibit the formation of terminal panhandle. The rest of the experiment was performed the same way as described in the legend to Fig. 3, except that panGFP plasmid in Fig. 3 was replaced with the new M1GFP plasmid in this experiment. An examination of the cells by fluorescence microscopy (Fig. 4B) and flow cytometry (Fig. 4D) demonstrated that GFP reporter mRNA lacking the panhandle structure was highly expressed in cells. This confirms the translation inhibition of GFP mRNA by the terminal panhandle in Fig. 3. None of the proteins coexpressed from transfected plasmids (Fig. 4C) affected the translation of GFP mRNA. The proteins expressed from transfected plasmids were pulled down using Ni-NTA beads, as described in the legend to Fig. 3. An examination of the pulled down material by PCR demonstrated that none of the proteins co-purified with the GFP reporter transcript lacking the panhandle structure (Fig. 4E). Quantification of the GFP reporter mRNA in cell lysates by real time PCR revealed that reporter mRNA was equally expressed in cells from the transfected plasmids (Fig. 4F). Taken together, this experiment confirms that formation of the vRNA panhandle inhibited the translation of the GFP reporter transcript in Fig. 3, and its abrogation by the deletion of the 3′ NCR sequence stimulated the translation of the reporter transcript in cells (Fig. 4). Although WT N protein and the head domain nonspecifically bind the 5′ NCR sequence in vitro, they did not efficiently associate with the GFP reporter mRNA in cells (Fig. 4E), likely due to the overwhelmingly available ssRNA-binding sites in the host transcripts.
Figure 4.
Deletion of the 3′ NCR sequence inhibited the N protein-panhandle interaction in vivo. A, a cartoon showing the M1GFP plasmid in which GFP flanked by the 5′ NCR sequence of CCHFV S-segment vRNA was cloned between T7 promoter and HDV sequence. The mRNA expressed from T7 promoter will not form the panhandle structure due to the deletion of 3′ NCR sequence in the mRNA. B, BSRT7.5 cells were transfected with plasmids exactly as described in the legend to Fig. 3B, except the pPanGFP plasmid in Fig. 3B was replaced with M1GFP plasmid in B. Cells were examined by fluorescence microscopy to monitor GFP expression (upper six panels), as described in the legend to Fig. 3B. The experiment was repeated and the cells were stained with DAPI (lower six panels). C, cell lysates from B were tested by Western blot analysis for the expression of proteins from transfected plasmids as described in the legend to Fig. 3C. D, cells from B were examined by FACS analysis and the quantified GFP signal was plotted exactly as described in the legend to Fig. 3C. E, the cell lysates from B, containing equal amounts of GFP mRNA, were loaded on Ni-NTA beads and total RNA was purified from the material eluted from washed beads. GFP mRNA was detected by PCR as described in the legend to Fig. 3E. F, GFP mRNA levels in the cell lysates from B were quantified by real time PCR as described in the legend to Fig. 3F.
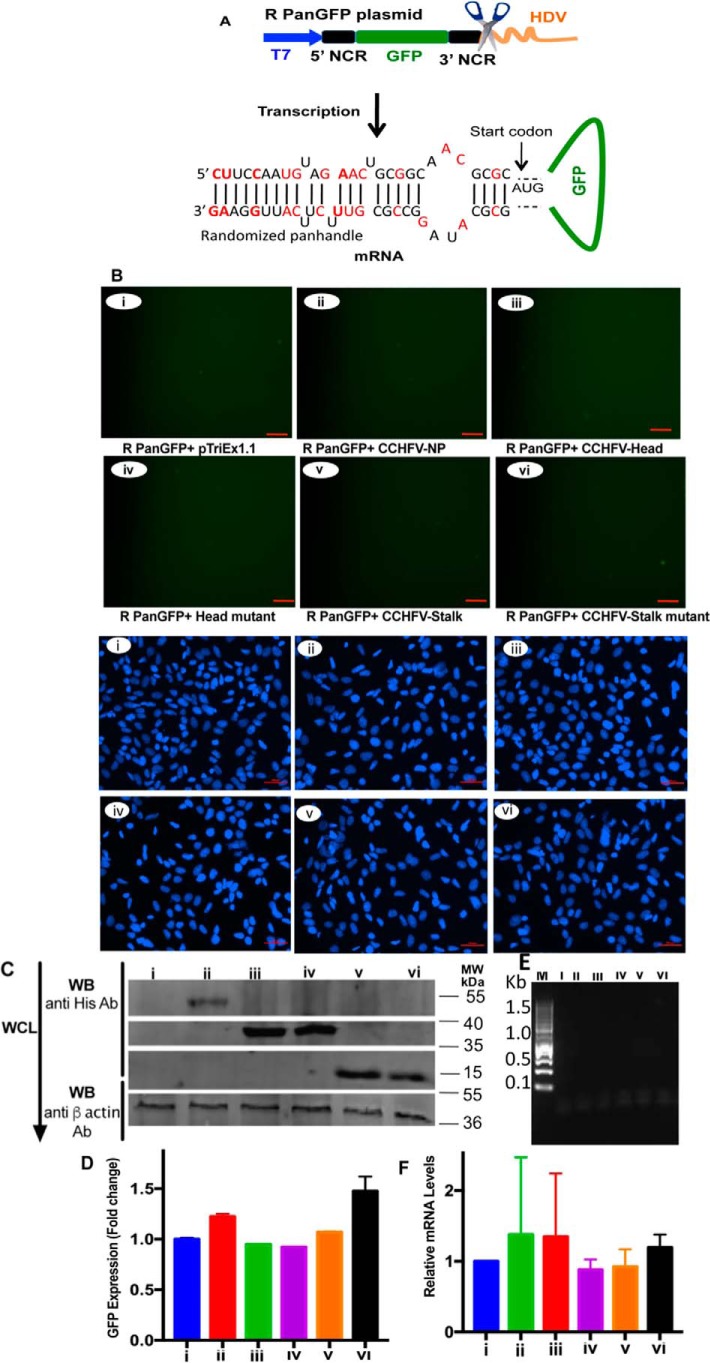
We previously reported that the nine highly conserved nucleotides form both 5′ and 3′ termini of the viral genome are required for specificity and high affinity binding of the panhandle to the N protein (26). Using the same approach as mentioned in Figs. 3 and 4, we generated RpanGFP plasmid (Fig. 5A) expressing the GFP reporter whose 5′ and 3′ termini could anneal and form a mutant panhandle. This mutant panhandle has the same nucleotide composition as WT panhandle but the nucleotide sequence was randomized. It structurally mimics the WT panhandle and does not bind to the N protein with high affinity and specificity, as previously reported (26). The plasmid was cotransfected in cells along with plasmids expressing either WT N protein or head and stalk domain mutants. The experiment was performed exactly as described in the legends to Figs. 3 and 4. It is evident from Fig. 5, B and D, that the reporter transcript failed to undergo translation to generate the GFP signal, although N protein, head and stalk domain mutants were significantly expressed in cells (Fig. 5C). An examination of cell lysates by real time PCR analysis demonstrated that the GFP transcript was equally expressed in cotransfected cells (Fig. 5F). The Ni-NTA pulldown of His-tagged proteins expressed from transfected plasmids demonstrated that GFP reporter mRNA does not co-purify with WT N protein or any of its mutants (Fig. 5E). This experiment further confirms that the strong secondary structure of the mutant panhandle inhibited mRNA translation similar to WT panhandle. However, failure in the recognition of mutant panhandle by the N protein resulted in the failure to overcome the panhandle-induced translation repression, and hence no GFP signal was observed.
Figure 5.
Randomization of 5′ and 3′ NCR sequences inhibited the N protein-panhandle interaction in vivo. A, cartoon showing RpanGFP plasmid expressing a GFP reporter mRNA flanked by randomized sequences of 5′ and 3′ NCR of CCHFV S-segment vRNA. The mutations are shown in red. B, BSRT7.5 cells were transfected with plasmids and examined by fluorescence microscope, except the PanGFP plasmid in Fig. 3B was replaced with RpanGFP plasmid in B. Upper and lower six panels show the GFP expression and DAPI staining, respectively. C, cell lysates from B were examined by Western blot analysis as described in the legend to Fig. 3B. D, cells from B were examined by FACS as described in the legend to Fig. 3D. E, cell lysates from B (upper six panels), containing equal amounts of GFP mRNA, were loaded on Ni-NTA beads and GFP mRNA eluted from washed beads was detected by PCR, as described in the legend to Fig. 3E. F, GFP mRNA in cell lysates from B was quantified by real time PCR exactly as described in the legend to Fig. 3F.
Binding of WT CCHFV N protein, head and stalk domains with other bunyavirus panhandles
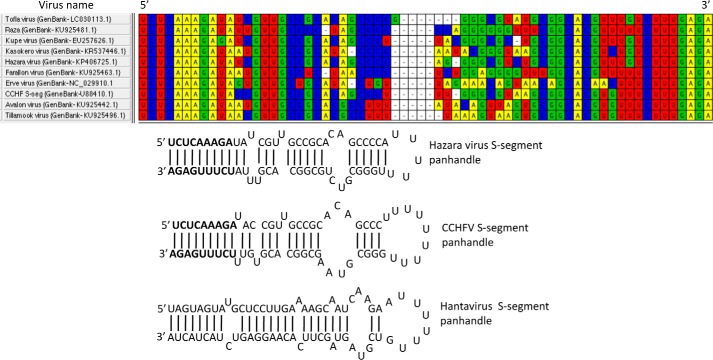
The nucleotide sequence at the 5′ and 3′ termini of the viral genomic RNA in the Nairovirus genus of the Bunyaviridae family is highly conserved (Fig. 6). Because CCHFV N protein requires the terminal nine nucleotides of the panhandle for high affinity binding through the panhandle-specific dsRNA-binding site located in the stalk domain, we asked whether CCHFV N protein binds the panhandles from other members of the Nairovirus genus. The S-segment panhandle and 5′ NCR (ssRNA) sequence (Fig. 6) of the hazara virus, another member of the Nairovirus genus, was synthesized by in vitro T7 transcription reaction, as mentioned under “Experimental procedures.” Binding of WT CCHFV N proteins, head and stalk domains with the synthetic panhandle and ssRNA was examined using a filter-binding approach. It is evident from Fig. 7, A and B, that WT CCHFV N protein bound the hazara virus panhandle with higher affinity compared with the ssRNA. Similar dissociation constants were previously reported for the binding of WT CCHFV N protein with the panhandle and single strand 5′ NCR sequence of the CCHFV S-segment vRNA (26). Unlike the ssRNA, the stalk domain of CCHFV N protein showed high affinity binding with the hazara virus panhandle (compare Fig. 7, C and D). Conversely, unlike the hazara virus panhandle, the head domain of CCHFV N protein showed preferable binding with the ssRNA. Comparing the binding results from Fig. 7, C–F, with Fig. 2 (filter binding), it is clear that stalk domain binds with similar high affinity to the panhandles of both hazara virus and CCHFV. In comparison the head domain bound to the single strand 5′ NCR sequences of both hazara virus and CCHFV.
Figure 6.
Sequence alignments of 5′ and 3′ NCR sequences of Nairovirus S-segment vRNA. Thirty nucleotides from both 5′ and 3′ termini of S-segment vRNA of 10 Nairoviruses were aligned. It is evident that the terminal nucleotides are highly conserved. The 30 nucleotides from both 5′ and 3′ termini of S-segment vRNA, separated by a six-residue uracil loop, of hazara virus and CCHFV were folded by MFold. The folded panhandle structures are shown. The S-segment vRNA panhandle of Andes hantavirus is also shown.
Figure 7.
Binding of CCHFV N protein with other Bunyavirus panhandles. Binding profiles generated by filter-binding analysis for the association of WT CCHFV N protein (A), head domain (C), and stalk domain (E) with the hazara virus S-segment panhandle, composed of 30 nucleotides from both 5′ and 3′ termini of hazara virus S-segment vRNA separated by a six-residue uracil loop. The corresponding binding profiles with a 30-nucleotide long 5′ NCR sequence (ssRNA) of CCHFV S-segment vRNA are shown in B, D, and F, respectively. The binding profiles were generated as described under “Experimental procedures.” The dissociation constants (Kd) calculated from each binding profile are shown in the corresponding panel. Binding profiles for the interaction of WT CCHFV N protein and stalk domain with hantavirus S-segment panhandle are shown in G and H, respectively.
These observations suggest that the panhandle-binding site is likely Nairovirus genus specific. To confirm the Nairovirus genus specificity of the panhandle-binding site, we synthesized the panhandle of hantavirus S-segment RNA, another genus in the Bunyaviridae family. An examination by filter-binding analysis revealed that unlike the Nairovirus panhandles, both WT CCHFV N protein and its stalk domain failed to bind the hantavirus panhandle with high affinity (compare Fig. 7, G and H, with A and C). These results confirm that the panhandle-binding site is specific to the Nairovirus genus.
Inhibition of hazara virus replication by the head domain of CCHFV N protein
We next asked whether N protein-panhandle interaction plays a role in the virus replication cycle. Since both WT CCHFV N protein and its stalk domain bound to both CCHFV and hazara virus panhandles with similar affinity, we tested the role of N protein-panhandle interaction in hazara virus replication in the cell culture. In addition, hazara virus is a commonly used model for CCHFV that has helped to study the CCHFV disease outside the BSL4 facility. Briefly, HEK293T cells were transfected with plasmids expressing WT CCHFV N protein, stalk domain, stalk domain mutant, head domain, head domain mutant 34, N protein mut1, or N protein mut2. Cells were infected 12 h post-transfection with hazara virus and harvested at increasing time intervals post-infection. An examination by Western blot analysis revealed that both WT CCHFV N protein and its mutants were efficiently expressed in cells (Fig. 8B). Total RNA was purified from harvested cells and hazara virus replication was monitored by quantitative estimation of S-segment vRNA by real time PCR analysis. It is evident from Fig. 8A that hazara virus efficiently replicated in cells lacking plasmid transfection. The expression of head domain, N protein mut 1, and N protein mut2 had minor impact upon virus replication. In comparison, the stalk domain harboring the intact panhandle-binding site dramatically inhibited hazara virus replication. The stalk domain mutant deficient in high affinity panhandle binding failed to inhibit hazara virus replication, similar to WT N protein and the head domain mutant. Lack of hazara virus inhibition observed in cells expressing WT CCHFV N protein suggests that CCHFV N protein might substitute the hazara virus N protein during replication. However, the panhandle-specific stalk domain likely competed with the WT hazara virus N protein for panhandle binding, resulting in the inhibition of hazara virus replication.
Figure 8.
Effect of CCHFV N protein expression upon hazara virus replication. HEK293T cells seeded in six-well plates were transfected with empty pTriEx1.1 plasmid, pTriEx-CCHFV-NP plasmid, pTriExhead plasmid, pTriExhead-mutant34 plasmid, pTriExstalk plasmid, pTriExstalk-mutant plasmid, pTriEx CCHFV N protein mut1 plasmid, or pTriEx CCHFV N protein mut2 plasmid, followed by infection with hazara virus 12 h post-transfection at an m.o.i. of 1.0, as described under “Experimental procedures.” Cells were harvested at increasing time points post-infection. Hazara virus replication was monitored by quantitative estimation of S-segment vRNA by real time PCR analysis (A). Western blot analysis was performed to examine the expression of C-terminally His-tagged proteins expressed from transfected plasmids, using anti-His tag antibody (B). HUVEC cells in six-well plates were similarly transfected with empty pTriEx1.1 plasmid, pTriExhead plasmid, or pTriExstalk plasmid, followed by infection with Andes virus at an m.o.i. of 1.0. Andes virus replication was monitored by quantification of S-segment vRNA by real time PCR (C). Expression of stalk and head domains was examined by Western blot analysis (D).
To further test the specificity of N protein-panhandle interaction and its potential role in Nairovirus replication, we tested the impact of stalk and head domains on hantavirus replication, another member in the different genus (hantavirus) within the same Bunyaviridae family. The experiment was repeated exactly as mentioned above except the human umbilical vein endothelial cells (HUVEC) were transfected with plasmids expressing either stalk or head domains of CCHFV N protein, followed by Andes hantavirus infection. An examination by Western blot analysis revealed that both stalk and head domains were efficiently expressed in HUVEC cells (Fig. 8D). Quantitative estimation of hantavurs S-segment RNA, using real time PCR analysis, revealed that neither stalk nor head domain affected the Andes hantavirus replication (Fig. 8C). Taken together, the experiments from Figs. 7 and 8, it is clear that N protein panhandle interaction and its potential roles in virus replication are specific to the Nairovirus genus.
Discussion
The two independent experimental approaches (filter binding and biolayer interferometry) demonstrated that bacterially expressed, the purified stalk domain of CCHFV N protein binds the vRNA panhandle with high affinity (Fig. 2). The binding analysis carried out with the stalk mutant illustrates that the positively charged surface groove on the stalk domain composed of amino acids His-195, His-197, Lys-222, Arg-225, Lys-282, and Lys-286 likely constitutes the panhandle-binding site. Similar in vitro binding analysis carried out with purified head domain and mutants 12 and 34 suggest that a nonspecific ssRNA-binding site is likely composed of amino acids Lys-339, Lys-343, Lys-346, Arg-384, Lys-411, His-453, Arg-134, Arg-140, and Gln-467 in the head domain of CCHFV N protein (Fig. 2). We previously reported that the binding affinity between WT CCHFV N protein and panhandle is not salt sensitive, suggesting that electrostatic interactions might have a little role in the binding event (30). However, in contradiction the point mutations in the stalk domain converting the two positively charged amino acids His-195 and His-197 to alanine impacted the binding. We hypothesize that although both enthalpy and entropy driven components may contribute toward the thermodynamic stability of the N protein-panhandle complex but the entropy component may play a major role. Further experimentation is needed to evaluate the binding energetics to clearly demonstrate the role of electrostatic interaction (enthalpy) in the binding event. However, point mutations in the stalk domain not only remove the positively charged amino acids but also structurally alter the binding pocket thus dramatically impacting the binding with the RNA substrate.
To determine whether specific interaction between CCHFV N protein and vRNA panhandle occurs in cells, a reporter construct (Fig. 3A) expressing GFP mRNA harboring 5′ and 3′ NCR sequences of the CCHFV S-segment vRNA was constructed. The formation of panhandle by the viral NCR sequences inhibited the translation of GFP reporter mRNA in cells (Fig. 3B). Translation suppression by the formation of panhandle was confirmed when the deletion of 3′ NCR sequence triggered GFP reporter expression in cells (Fig. 4B). Interestingly, co-expression of WT CCHFV N protein facilitated the translation of GFP reporter mRNA, confirming the previously reported role of CCHFV N protein in mRNA translation (29). Mutating the panhandle-binding site in the stalk domain impacted the CCHFV N protein-mediated translation of GFP reporter mRNA (Fig. 3). Because the aim of this study was to confirm the interaction between stalk domain and vRNA panhandle, we did not focus on the mechanism by which N protein facilitated the translation of GFP-reported mRNA (Fig. 3). However, this reporter assay will indeed be of potential importance in studying the translation control and identification of potential inhibitors for the CCHFV N protein. The lysates co-expressing the GFP reporter mRNA along with His-tagged WT N protein or stalk domain or head domain were pulled down on Ni-NTA beads. This experiment clearly demonstrated that the GFP reporter mRNA co-purified with WT N protein and stalk domain, confirming the stalk domain-panhandle interaction (Fig. 3, C–F). Single point mutation in the stalk domain (N protein mut1) adversely effected the N protein-panhandle interaction and two point mutations (N protein mut2) dramatically inhibited the binding (Fig. 3E). Interruption in the panhandle formation by the deletion of 3′ NCR sequence (Fig. 4) or expression of GFP reporter mRNA harboring the mutant panhandle (Fig. 5) abrogated the interaction between stalk domain and panhandle, confirmed by the lack of co-purification of the GFP reporter mRNA in the Ni-NTA pulldown assay (Figs. 4 and 5).
We previously reported that N protein requires nine terminal nucleotides from 5′ and 3′ ends of vRNA for high affinity binding. Because these nucleotides are highly conserved in the Nairovirus genus (Fig. 6), it was important to determine whether CCHFV N protein could bind to the panhandles from other viruses in the Nairovirus genus. An examination by filter-binding analysis revealed that CCHFV N protein and its stalk domain can bind to the vRNA panhandle of hazara virus, another member of the Nairovirus genus in the Bunyaviridae family. In contrast, the CCHFV N protein failed to bind the vRNA panhandle of Andes hantavirus, another genus of the Bunyaviridae family. This suggests that panhandle-N protein interaction is genus-specific. Interestingly, the co-expression of WT CCHFV N protein did not influence the hazara virus replication in cell culture, suggesting that CCHFV N protein might replace the hazara virus N protein during virus replication. However, unlike the stalk domain mutant deficient in panhandle binding, the co-expression of stalk domain active in panhandle binding dramatically inhibited the hazara virus replication in cells (Fig. 8A). It is likely that the stalk domain of CCHFV N protein competed with the WT hazara virus N protein for panhandle binding and caused virus inhibition. On the other hand, the stalk domain of CCHFV N protein did not influence the hantavirus replication in cell culture (Fig. 8C), consistent with significantly weak interaction between CCHFV stalk domain and hantavirus panhandle (Fig. 7H). These observations clearly demonstrate that N protein-panhandle interaction is specific to Nairovirus genus, and it plays a role in the Nairovirus replication by an unknown mechanism.
Although not shown in this article, it is likely that distinct panhandle- and ssRNA-binding sites in the stalk and head domains of CCHFV N protein might play a role in the transcription initiation in conjunction of viral RdRp. Nairoviruses initiate transcription by a unique cap snatching mechanism during which a short capped RNA fragment cleaved from the host cell mRNA is used as primer to initiate transcription (31). The panhandle structure formed by the termini of vRNA genome has been proposed to serve as promoter for the Bunyaviridae RdRp (32). In addition, the CCHFV N protein has been reported to interact with the RdRp (27, 28). Because N protein binds to both the RdRp and the panhandle, it is possible that the panhandle-binding site in the stalk domain of N protein might help in the recruitment of RdRp on the promoter during transcription initiation. The ssRNA-binding site in the head domain might play a role in the engagement of capped RNA primer at the transcription initiation site. This is consistent with previous reports suggesting that interaction of CCHFV N protein with short ssRNA induces a conformational change that might expose the encapsidated vRNA genome for transcription (19). Moreover, the primary role of the N protein is to form ribonucleocapsids in association with the vRNA and cRNA. It is possible the specific N protein-panhandle interaction might specifically recognize vRNA and cRNA during the early stages of the encapsidation process. The results presented in this article suggest that the panhandle-binding site in the stalk domain might be a novel target for therapeutic intervention of CCHFV disease.
Experimental procedures
Cells and transfection
HeLa and HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and penicillin-streptomycin (100 μg/ml) in a CO2 incubator. HUVEC were maintained in HUVEC growth medium (Lonza EBM, CC-3121). TurboFect transfection reagent was from Thermo Fisher Scientific. The rabbit anti-His tag antibody was from GenScript. The anti-rabbit secondary antibody was from Fisher Scientific.
Plasmids
The plasmid expressing the CCHFV N protein from strain 10200 was a gift from Stuart T. Nichol (CDC, Atlanta, Georgia). The N protein ORF was PCR amplified and cloned between NdeI and XhoI restriction sites in pET30a backbone to generate the plasmid pET-CCHFNP, as previously reported (29). The regions encoding the head domain of CCHFV N protein (amino acids 1–180 and amino acids 300–482) were amplified and joined by overlapping PCR. The region encoding the stalk domain of CCHFV N protein (amino acids 180–300) was also PCR amplified. The PCR products were gel purified and cloned in pET30a vector between NdeI and XhoI restriction sites to generate pEThead and pETstalk plasmids. The pEThead plasmid was modified by site-directed mutagenesis to convert three positively charged amino acid residues Arg-339, Lys-343, and Lys-346 into alanine. The resulting plasmid was referred as pEThead-mutant12 plasmid. Similarly, pEThead plasmid was modified to generate pEThead-mutant 34 in which Arg-134 and Arg-140 were replaced by alanine. The pETstalk plasmid was also modified to generate pETstalk-mutant plasmid in which His-195 and His-197 were converted to alanine. The WT CCHFV NP, head domain, stalk domain, stalk domain mutant, and head domain mutants 16 and 34 were PCR amplified from their respective pET30a backbones, using appropriate primers, and subcloned in pTriEX1.1 backbone for the expression in mammalian cells. The resulting plasmids were referred as pTriEx CCHFV NP, pTriExhead, pTriExstalk, pTriExstalk-mutant, pTriExhead-mutant 16, and pTriEx head-mutant 34, respectively. The pTriEx CCHFV NP plasmid was modified to generate pTriEx CCHFV N protein mut1 and pTriEx CCHFV N protein mut2 constructs in which His-195 and both His-195 and His-197, respectively, were converted to alanine. The panGFP plasmid was constructed by PCR amplification of GFP ORF using a forward primer flanked with 5′ NCR sequence of CCHFV S-segment vRNA and a reverse primer flanked with 3′ NCR sequence of CCHFV S-segment vRNA. The resulting PCR product was fused with HDV ribozyme sequence downstream of the 3′ NCR sequence, using overlapping PCR. The two restriction sites, BamHI and NotI, were incorporated in the final PCR product through forward and reverse primers, respectively. The final PCR product was cloned in PUC vector between BamHI and NotI restriction sites, downstream of the T7 promoter as shown in Fig. 3. The same strategy was used for construction of the RpanGFP plasmid, except the mutations in the 5′ and 3′ NCR sequences were incorporated through forward and reverse primers, as shown in Fig. 5. Similarly, the M1GFP plasmid was constructed, except the 3′ NCR sequence was not incorporated in the plasmid.
Expression and purification of proteins
The WT CCHFV N protein was purified as previously reported (26, 29). The stalk domain, head domain, and their respective mutants were expressed in Escherichia coli as C-terminally His-tagged fusion proteins, and purified on Ni-NTA beads using a native purification protocol according to the manufacturer's instructions. Briefly, E. coli Rosetta (DE3) cells (Stratagene) were transformed with pEThead plasmid, pEThead-mutant12 plasmid, pEThead-mutant 34 plasmid, pETstalk plasmid, or pETstalk-mutant plasmid. The colonies expressing the protein of interest were grown in 500-ml cultures at 37 °C until the OD at 600 nm reached 0.5. The protein expression was induced by the addition of 0.5 mm isopropyl β-d-1-thiogalactopyranoside to the bacterial culture. The cultures were grown for an additional 20 h at 18 °C. Cells were harvested by centrifugation and re-suspended in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm DTT, 0.5% Triton X-100, 5 mm CHAPS, 0.1 mm phenylmethylsulfonyl fluoride). The re-suspended cells were sonicated on ice and cleared by centrifugation. The cleared cell lysates were loaded onto Ni-NTA columns having a 5-ml bed volume (GOLDBIO), pre-equilibrated with the TBS buffer (50 mm Tris, 150 mm NaCl, pH 7.4). The column was then washed with 50 ml of TBS buffer, followed by additional washing with 100 ml of wash buffer (50 mm Tris, 500 mm NaCl, 0.05% Triton X-100, and 20 mm imidazole). The bound protein was finally eluted with the elution buffer containing 250 mm imidazole. The protein in eluted fractions was quantified and stored at −80 °C.
Filter-binding analysis
The panhandle or 5′ NCR sequence (ssRNA) from the S-segment vRNA of either CCHFV or hazara virus was synthesized by in vitro T7 transcription, as previously reported (26, 29, 33, 34). Briefly, a single strand DNA sequence harboring a 5′ T7 promoter, followed by either panhandle or 5′ NCR sequence was synthesized by IDT. The DNA sequence along with two opposing primers was used as template in a PCR to generate the double strand DNA segment. The DNA segment was gel purified and used as template in an in vitro T7 transcription reaction. The RNA was labeled with [α-32P]CTP during synthesis and purified using Tri-Reagent (Ambion) followed by column purification using an miRNAeasy kit (Invitrogen), as previously reported (34). Interaction of bacterially expressed and purified CCHFV N protein and its mutants with the RNA of interest was studied by a filter-binding assay, as previously reported (35). All binding reactions were carried out in RNA-binding buffer (40 mm Tris-HCl, pH 7.4, 80 mm NaCl, 20 mm KCl, 1 mm DTT) at a constant concentration of RNA with increasing concentrations of the protein. Reaction mixtures were incubated at room temperature for 30–45 min and filtered through nitrocellulose membranes under vacuum. Filters were washed with 10 ml of RNA-binding buffer and dried. The amount of RNA retained on the filter at each input concentration of the protein of interest was measured by quantifying the radioactive signal, using a scintillation counter. The binding profiles for the calculation of dissociation constant (Kd), the concentration of the protein at which 50% RNA was bound to the protein, were generated as previously reported (26, 29, 33, 34). Briefly, each radioactive signal was normalized by the mean saturated signal to calculate the percentage of bound RNA at each input protein concentration. The percentage of bound RNA was plotted versus protein concentration and the data points were fit to a binding profile by GraphPad Prism 7.0 software using nonlinear regression analysis based on a single-site binding model.
Biolayer interferometry
Biolayer interferometry was used to monitor the binding affinities of the head domain, stalk domain, and their mutants with panhandle and ssRNA using the BLItz system (ForteBio Inc.), as previously reported (26, 29, 33). Briefly, biotin-labeled panhandle or 5′ NCR was synthesized and loaded onto high precision streptavidin biosensors (catalogue number 18-5019, Forte Bio Inc.), as previously reported (26, 29, 33). All reactions were carried out at room temperature in RNA-binding buffer. After mounting the RNA, the biosensors were equilibrated in RNA-binding buffer and then dipped in the purified protein solutions of interest at varying concentrations for the measurement of association kinetics. The reaction cycles were as follows: initial baseline for 30 s, loading of biotinylated RNA on streptavidin biosensors for 120 s, baseline for 30 s, association of protein with the RNA for 300 s, followed by dissociation phase of 500 s. The kinetic parameters Ka (association rate constant), Kdis (dissociation rate constant), and the binding affinities (Kd = Kdis/Ka) were calculated with the help of inbuilt data analysis software (BLItZ Pro), as previously reported (26, 29, 33).
Andes and hazara virus propagation
All hantavirus experiments were carried out in a Bio-safety level-3 (BSL3) laboratory following appropriate laboratory guidelines. Hazara virus experiments were carried out in a BSL2 laboratory. Andes virus (provided by NIH) and hazara virus (provided by Dr. Ksiazek from World Reference Center for Emerging viruses and Arbaviruses-WRCEVA, UTMB Galveston) were propagated in vero E6 cells by infecting confluent monolayers of cells with the virus at an m.o.i. of 0.01. Andes virus-infected cells were cultured for 13 days in viral growth medium (DMEM/high glucose from Thermo Scientific, catalogue number SH30243.01, containing 2.5% fetal bovine serum). Hazara virus-infected cells were cultured for 7 days in the same viral growth medium. The viral growth medium containing budded virus particles was harvested, cleared by low speed centrifugation, and stored in 1-ml aliquots at −80 °C in DMEM containing 10% fetal bovine serum. Andes virus was quantified as previously described (36). Hazara virus was quantified by the estimation of S-segment vRNA using real time PCR analysis.
Andes and hazara virus replication
HEK293T cells seeded in 6-well plates were transfected with pTriEx CCHFV NP plasmid, pTriExhead plasmid, pTriExstalk plasmid, pTriExstalk-mutant plasmid, pTriExhead-mutant 34 plasmid, pTriEx CCHFV N protein mut1, or pTriEx CCHFV N protein mut2 plasmid, using Turbofect transfection reagent. Twelve hours post-transfection cells were infected with hazara virus at an m.o.i. of ∼1.0 for 1 h with brief shaking every 15 min. The virus inoculum was removed and replaced with fresh growth medium. Cells from well one were harvested 1 h post-infection and referred as zero time point in Fig. 8. Cells in the remaining wells were allowed to grow and harvested at increasing time points post-infection. Cells were lysed and total RNA was purified from half of the cell lysates using Tri-Reagent (Ambion).
Virus replication was monitored by quantitative estimation of viral S-segment RNA by real time PCR using a relative quantification method and β-actin as internal control, as previously reported (37). The remaining half of the lysate was used in Western blot analysis to examine protein expression from transfected plasmids. Replication of Andes virus was monitored similarly, except HUVEC cells grown in HUVEC growth medium (Lonza EBM, CC-3121) were transfected with either pTriExhead plasmid or pTriExstalk plasmid. Virus replication was monitored as mentioned above and discussed in detail in Ref. 34.
Author contributions
S. J. resources; S. J., A. V., and J. R. data curation; S. J. software; S. J. formal analysis; S. J. investigation; S. J., A. L., S. W., A. T. S., A. D. R., N. A. A., F. C., L. O., and M. A. M. methodology; S. M., A. D. R., and M. A. M. writing-review and editing; A. D. R. and M. A. M. conceptualization.
Acknowledgments
We acknowledge Dr. Dominique Griffon (Associate Dean for Research) and Dr. Steven Henriksen (vice president for Research at Western University) for providing the summer research fellowship to Jacquelyn Ragan, salary support to S. Jeeva, and supporting the purchase of a BLItz system (ForteBio Inc.).
This work was supported by National Institutes of Health Grant 1R15AI126395-01A1 (to M. A. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- CCHFV
- Crimean-Congo hemorrhagic fever virus
- RdRp
- RNA-dependent RNA polymerase
- vRNA
- viral RNA
- cRNA
- complementary
- NCR
- noncoding region
- Ni-NTA
- nickel-nitrilotriacetic acid
- HDV
- hepatitis delta virus
- GFP
- green fluorescent protein
- HUVEC
- human umbilical vein endothelial cells
- DMEM
- Dulbecco's modified Eagle's medium
- DAPI
- 4′,6-diamidino-2-phenylindole
- m.o.i.
- multiplicity of infection.
References
- 1. Elliott R. M., Bouloy M., Calisher C. H., Goldbach R., Moyer J. T., Nichol S. T., Pettersson R., Plyusnin A., and Schmaljohn C. S. (2000) Family Bunyaviridae. in Virus Taxonomy: Seventh Report International Committee for the Taxonomy of Viruses (van Regenmortel M. H. V., Fauquet C. M., Bishop D. H. L., Carstens E. B., Estes M. K., Lemon S., Maniloff J., Mayo M. A., McGeogch D., Pringle C. R., and Wickner R. B., eds) pp. 599–621, Academic Press, San Diego [Google Scholar]
- 2. Hoogstraal H. (1979) The epidermology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe and Africa. J. Med. Entomol. 15, 307–417 10.1093/jmedent/15.4.307 [DOI] [PubMed] [Google Scholar]
- 3. Swanepoel R. (1994) Crimean-Congo hemorrhagic fever. in Infectious diseases of livestock, with reference to South Africa (Thomson G. R., Tustin R. C., and Coetzer J. A. W., eds) pp. 723–729, Oxford University Press, Cape Town, South Africa [Google Scholar]
- 4. Swanepoel R., Shepherd A. J., Leman P. A., Shepherd S. P., McGillivray G. M., Erasmus M. J., Searle L.A., and Gill D. E. (1987) Epidemiologicand clinical features of Crimean-Congo hemorrhagic fever in southern Africa. Am. J. Trop. Med. Hyg. 36, 120–132 10.4269/ajtmh.1987.36.120 [DOI] [PubMed] [Google Scholar]
- 5. Ergönül O. (2006) Crimean-Congo haemorrhagic fever. Lancet Infect. Dis. 6, 203–214 10.1016/S1473-3099(06)70435-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uyar Y., Carhan A., Albayrak N., and Altaş A. B. (2010) Evaluation of PCR and ELISA-IgM results in the laboratory diagnosis of Crimean-Congo haemorrhagic fever cases in 2008 in Turkey. Mikrobiyol. Bul. 44, 57–64 [PubMed] [Google Scholar]
- 7. Whitehouse C. A. (2004) Crimean-Congo hemorrhagic fever. Antiviral Res. 64, 145–160 10.1016/j.antiviral.2004.08.001 [DOI] [PubMed] [Google Scholar]
- 8. Sargianou M., and Papa A. (2013) Epidemiological and behavioral factors associated with Crimean-Congo hemorrhagic fever virus infections in humans. Expert Rev. Anti. Infect. Ther. 11, 897–908 10.1586/14787210.2013.827890 [DOI] [PubMed] [Google Scholar]
- 9. Aksoy D., Barut H., Duygu F., Çevik B., Kurt S., and Sümbül O. (2018) Characteristics of headache and its relationship with disease severity in patients with Crimean-Congo hemorrhagic fever. Agri. Agri. 30, 12–17 [DOI] [PubMed] [Google Scholar]
- 10. Oncü S. (2013) Crimean-Congo hemorrhagic fever: an overview. Virologica Sinica 28, 193–201 10.1007/s12250-013-3327-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mourya D. T., Yadav P. D., Shete A. M., Gurav Y. K., Raut C. G., Jadi R. S., Pawar S. D., Nichol S. T., and Mishra A. C. (2012) Detection, isolation and confirmation of Crimean-Congo hemorrhagic fever virus in human, ticks and animals in Ahmadabad, India, 2010–2011. PLoS Negl. Trop. Dis. 6, e1653 10.1371/journal.pntd.0001653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koksal I., Yilmaz G., Aksoy F., Erensoy S., and Aydin H. (2014) The seroprevalance of Crimean-Congo haemorrhagic fever in people living in the same environment with Crimean-Congo haemorrhagic fever patients in an endemic region in Turkey. Epidemiol. Infect. 142, 239–245 10.1017/S0950268813001155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flick R., and Whitehouse C. A. (2005) Crimean-Congo hemorrhagic fever virus. Curr. Mol. Med. 5, 753–760 10.2174/156652405774962335 [DOI] [PubMed] [Google Scholar]
- 14. Günes T. (2006) [Crimean-Congo Hemorrhagic Fever]. Mikrobiyol. Bul. 40, 279–287 [PubMed] [Google Scholar]
- 15. Ergonul O. (2008) Treatment of Crimean-Congo hemorrhagic fever. Antiviral Res. 78, 125–131 10.1016/j.antiviral.2007.11.002 [DOI] [PubMed] [Google Scholar]
- 16. Labuda M., and Nuttall P. A. (2004) Tick-borne viruses. Parasitology 129, S221–245 10.1017/S0031182004005220 [DOI] [PubMed] [Google Scholar]
- 17. Schmaljohn C. S., and Hooper J. W. (2001) Bunyaviridae: the viruses and their replication. in Fields Virology (Knipe D. M., and Howley P.M., eds) 4th Ed., pp. 1581–1602, Williams & Wilkins, Lippincott, Philadelphia, PA [Google Scholar]
- 18. Carter S. D., Surtees R., Walter C. T., Ariza A., Bergeron É., Nichol S. T., Hiscox J. A., Edwards T. A., and Barr J. N. (2012) Structure, function, and evolution of the Crimean-Congo hemorrhagic fever virus nucleocapsid protein. J. Virol. 86, 10914–10923 10.1128/JVI.01555-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y., Dutta S., Karlberg H., Devignot S., Weber F., Hao Q., Tan Y. J., Mirazimi A., and Kotaka M. (2012) Structure of Crimean-Congo hemorrhagic fever virus nucleoprotein: superhelical homo-oligomers and the role of caspase-3 cleavage. J. Virol. 86, 12294–12303 10.1128/JVI.01627-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo Y., Wang W., Ji W., Deng M., Sun Y., Zhou H., Yang C., Deng F., Wang H., Hu Z., Lou Z., and Rao Z. (2012) Crimean-Congo Hemarrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc. Natl. Acad. Sci. U.S.A. 109, 5046–5051 10.1073/pnas.1200808109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hastie K. M., Kimberlin C. R., Zandonatti M. A., MacRae I. J., and Saphire E. O. (2011) Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc. Natl. Acad. Sci. U.S.A. 108, 2396–2401 10.1073/pnas.1016404108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qi X., Lan S., Wang W., Schelde L. M., Dong H., Wallat G. D., Ly H., Liang Y., and Dong C. (2010) Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468, 779–783 10.1038/nature09605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olal D., Dick A., Woods V. L. Jr, Liu T., Li S., Devignot S., Weber F., Saphire E. O., and Daumke O. (2014) Structural insights into RNA encapsidation and helical assembly of the Toscana virus nucleoprotein. Nucleic Acids Res. 42, 6025–6037 10.1093/nar/gku229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raymond D. D., Piper M. E., Gerrard S. R., and Smith J. L. (2010) Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc. Natl. Acad. Sci. U.S.A. 107, 11769–11774 10.1073/pnas.1001760107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reguera J., Malet H., Weber F., and Cusack S. (2013) Structural basis for encapsidation of genomic RNA by La Crosse Orthobunyavirus nucleoprotein. Proc. Natl. Acad. Sci. U.S.A. 110, 7246–7251 10.1073/pnas.1302298110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jeeva S., Pador S., Voss B., Ganaie S. S., and Mir M. A. (2017) Crimean-Congo hemorrhagic fever virus nucleocapsid protein has dual RNA binding modes. PloS One 12, e0184935 10.1371/journal.pone.0184935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bergeron E., Albariño C. G., Khristova M. L., and Nichol S. T. (2010) Crimean-Congo hemorrhagic fever virus-encoded ovarian tumor protease activity is dispensable for virus RNA polymerase function. J. Virol. 84, 216–226 10.1128/JVI.01859-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Levingston Macleod J. M., Marmor H., García-Sastre A., and Frias-Staheli N. (2015) Mapping of the interaction domains of the Crimean-Congo hemorrhagic fever virus nucleocapsid protein. J. Gen. Virol. 96, 524–537 10.1099/vir.0.071332-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jeeva S., Cheng E., Ganaie S. S., and Mir M. A. (2017) Crimean-Congo hemorrhagic fever virus nucleocapsid protein augments mRNA translation. J. Virol. 91, e00636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ghaemi Z., Guzman I., Gnutt D., Luthey-Schulten Z., and Gruebele M. (2017) Role of electrostatics in protein-RNA binding: the global vs the local energy landscape. J. Phys. Chem. B 121, 8437–8446 10.1021/acs.jpcb.7b04318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jin H., and Elliott R. M. (1993) Non-viral sequences at the 5′ ends of Dugbe nairovirus S mRNAs. J. Gen. Virol. 74, 2293–2297 10.1099/0022-1317-74-10-2293 [DOI] [PubMed] [Google Scholar]
- 32. Flick R., Elgh F., and Pettersson R. F. (2002) Mutational analysis of the Uukuniemi virus (Bunyaviridae family) promoter reveals two elements of functional importance. J. Virol. 76, 10849–10860 10.1128/JVI.76.21.10849-10860.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ganaie S. S., Haque A., Cheng E., Bonny T. S., Salim N. N., and Mir M. A. (2014) Ribosomal protein S19 binding domain provides insights into hantavirus nucleocapsid protein-mediated translation initiation mechanism. Biochem. J. 464, 109–121 10.1042/BJ20140449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Salim N. N., Ganaie S. S., Roy A., Jeeva S., and Mir M. A. (2016) Targeting a novel RNA-protein interaction for therapeutic intervention of hantavirus disease. J. Biol. Chem. 291, 24702–24714 10.1074/jbc.M116.750729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mir M. A., Sheema S., Haseeb A., and Haque A. (2010) Hantavirus nucleocapsid protein has distinct m7G cap- and RNA-binding sites. J. Biol. Chem. 285, 11357–11368 10.1074/jbc.M110.102459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hooper J. W., Larsen T., Custer D. M., and Schmaljohn C. S. (2001) A lethal disease model for hantavirus pulmonary syndrome. Virology 289, 6–14 10.1006/viro.2001.1133 [DOI] [PubMed] [Google Scholar]
- 37. Hussein I. T., Cheng E., Ganaie S. S., Werle M. J., Sheema S., Haque A., and Mir M. A. (2012) Autophagic clearance of Sin Nombre hantavirus glycoprotein Gn promotes virus replication in cells. J. Virol. 86, 7520–7529 10.1128/JVI.07204-11 [DOI] [PMC free article] [PubMed] [Google Scholar]